Abstract
Human apolipoprotein E (APOE), originally known for its role in lipid metabolism, is polymorphic with three major allele forms, namely, APOEε2, APOEε3, and APOEε4, leading to three different human APOE isoforms. The ε4 allele is a genetic risk factor for Alzheimer’s disease (AD); therefore, the vast majority of APOE research focuses on its role in AD pathology. However, there is increasing evidence for other functions of APOE through the involvement in other biological processes such as transcriptional regulation, mitochondrial metabolism, immune response, and responsiveness to dietary factors. Therefore, the aim of this review is to provide an overview of the potential novel functions of APOE and their characterization. The detection of APOE in various cell organelles points to previously unrecognized roles in mitochondria and others, although it is actually considered a secretory protein. Furthermore, numerous interactions of APOE with other proteins have been detected, providing indications for new metabolic pathways involving APOE. The present review summarizes the current evidence on APOE beyond its original role in lipid metabolism, to change the perspective and encourage novel approaches to future research on APOE and its isoform-dependent role in the cellular metabolism.
Keywords: Mitochondria-associated ER membranes, Protein–protein interactions, Interactome, Proteolytic fragmentation
Introduction
Apolipoprotein E (APOE) is a plasma protein involved in cholesterol metabolism that, as a component of lipoproteins, mediates their cellular uptake by binding to cell surface receptors. In humans, there are three different isoforms resulting from the three allelic variants ε2, ε3, and ε4. APOEε4 carriers show an increased risk of the development of Alzheimer’s disease (AD), which is why a major focus of APOE research is on the relationship between APOE and AD pathology, and most studies focus on the role of APOE in the brain or neuronal cells and astrocytes [1–3]. However, APOE is mainly produced in the liver, and to a lesser extent in astrocytes and microglia. There is much evidence that APOE circulates in two independent pools, since APOE is not able to cross the blood–brain barrier [4], APOE protein levels in plasma are significantly higher than in cerebrospinal fluid (CSF) [5], and plasma and CSF APOE have different posttranslational modifications [6]. Therefore, it must be considered that findings on the role of APOE in the brain may not be directly transferable to APOE in the periphery.
While the ε4 allele shows detrimental effects in the elderly, carrying the ε4 allele actually is beneficial in some aspects, e.g., improved outcome of certain infectious diseases or increased vitamin D levels [7–9]. Therefore, the question arises as to what functions APOE might have in addition to lipid transport and its role in AD pathology. A number of physiological processes in which APOE is involved have already been described, e.g., participation in the immune system [10, 11] or in the response to various nutritional factors [9, 12, 13] reviewed in Egert et al. [14], as well as its binding to a number of genes and thus its function as a transcription factor [15]. Insights into further novel metabolic participations of APOE can be obtained by studying its physical interactions with other proteins. In addition, previously unknown functions may be deduced from the subcellular localization of APOE, since APOE has been detected in various cell organelles although it is primarily a secretory protein [15–17]. Therefore, this review discusses the following topics: The subcellular localization of APOE and proteolytically cleaved APOE fragments, and their resulting roles in mitochondrial metabolism and mitochondria-associated ER membranes (MAMs). The second focus is the interaction of APOE with other proteins, whose function is well understood, and the identification of signalling pathways in which APOE is involved through these interactions.
Protein structure, subcellular trafficking, and cellular processing of APOE
APOE protein structure and isoforms
Expression of the human APOE gene results in a 317-amino-acid-long APOE precursor protein, from which the mature APOE protein with a length of 299 amino acids is formed after cleavage of an N-terminal 18-amino-acid-long signal peptide [18]. The nucleotide exchanges (rs429358 and rs7412) leading to the three main human APOE isoforms result in amino acid exchanges at positions 112 and 158, which are located in the N-terminal domain of the mature protein (Fig. 1a). The APOE4 protein carries arginine residues at both positions, APOE3 has cysteine at position 112 and arginine at position 158, and APOE2 carries cysteine at each of the corresponding positions [19]. These polymorphism of APOE is found exclusively in humans, although APOE is expressed in mammals and other vertebrates [20, 21]. There are some differences in the structural properties of the APOE isoforms that are likely due to the exchange of amino acids, e.g., the domain interaction between the N- and C-termini appears to be affected by the exchange at position 112, such that APOE4 exhibits greater dynamics in structure opening [22, 23]. Furthermore, the stability to chemical denaturation (stability E4 < E3 < E2) and self-association differ between the isoforms [24–26]. These structural differences could have consequences for possible further functions of APOE, since due to the different spatial structures of the isoforms, different regions of the amino acid sequence might be exposed or buried, which could influence, e.g., interactions with other molecules.
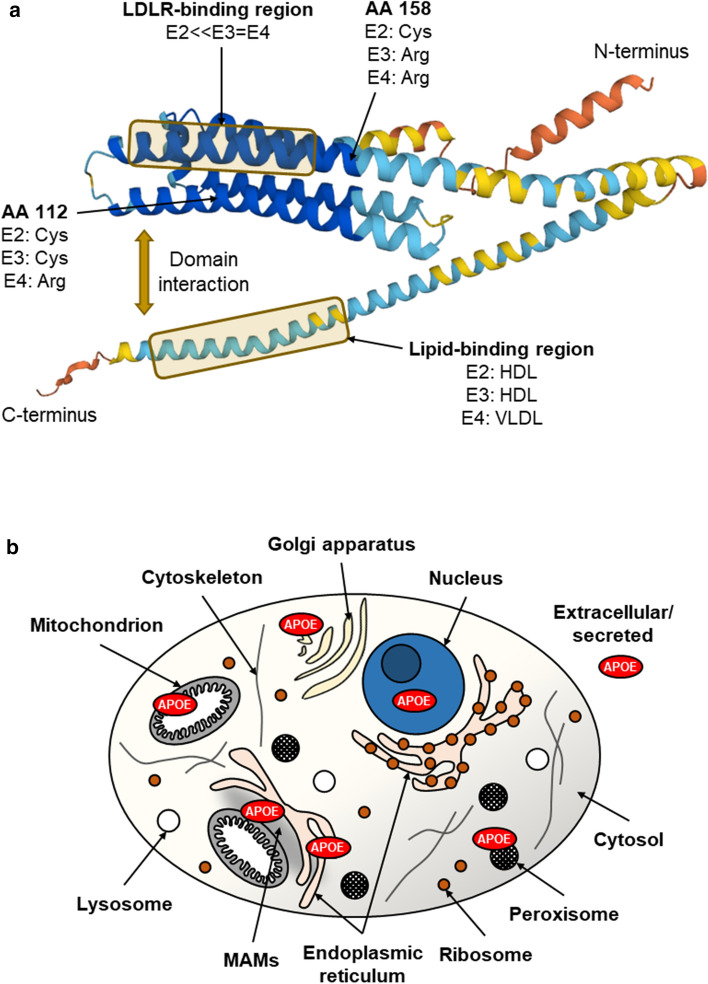
Fig. 1.
Predicted spatial protein structure of human APOE (AlphaFold) and intracellular localisation of APOE. a The APOE isoproteins differ at one and two amino acid (AA) positions, respectively (112 and 158). This affects the structure and thus the function of the protein, e.g., APOE2 shows a significantly reduced affinity for the low-density lipoprotein receptor (LDLR) and the lipid binding preference differs between the isoforms. The different colours of the protein structure indicate the model confidence of the structure prediction (dark blue, very high; light blue, confident; yellow, low; orange, very low). Protein structure image downloaded from https://alphafold.ebi.ac.uk/. HDL high-density lipoprotein, VLDL very low-density lipoprotein. b Schematic illustration of an eukaryotic cell and the individual cell organelles and compartments. APOE is known to be a secreted protein, but has been detected inside the cell in, e.g., mitochondria, peroxisomes, and the nucleus. MAMs mitochondria-associated ER membranes
Because APOEε2 is the least common gene variant, there are only few studies examining this genotype, especially in the context of mitochondrial function which is why the focus of this article is on APOE3 and APOE4.
Subcellular localization of APOE
The APOE protein is a secretory protein but has also been localized inside the cell in many subcellular organelles and compartments (Fig. 1b), such as mitochondria, peroxisomes, and the nucleus [15–17]. Secretory proteins such as APOE contain an amino acid sequence that is recognized by the signal recognition particle and thereby translated directly into the ER. Intracellular proteins are translated from cytosolic ribosomes into the cytosol and contain peptide sequences that are coexpressed and linked to the actual protein sequence at the C- or N-terminus. Examples are the nuclear localization sequence (NLS) or the mitochondrial localization sequence (MLS), which are recognized by proteins on the corresponding membranes that mediate transport into the nucleus or mitochondrion. The APOE protein has been shown to be present in the nucleus and to function as a transcription factor [15, 27], but no NLS has yet been identified in the APOE protein sequence that could explain the nuclear import mechanism of APOE. Similarly, there is much evidence that APOE is present in mitochondria and affects mitochondrial function [16, 17, 28, 29]. However, no MLS has yet been identified in the protein sequence of APOE; consequently, there must be other mechanisms or pathways by which APOE enters the nucleus and passes through mitochondrial membranes into mitochondria. In addition to the localization of APOE in mitochondria, APOE was also detected in MAMs [17, 30]. An overview of the evidence of localization of APOE in these two cell organelles is provided in Table 1. One pathway by which APOE might enter the mitochondria is through interorganelle crosstalk between the endoplasmic reticulum (ER) and mitochondria, as has recently been shown for glycoproteins and misfolded proteins [31]. The secretion of misfolded and structurally altered ER-cargo into mitochondria, may explain the occurrence of APOE fragments in the mitochondrion [32]. Proteolytic cleavage of APOE yields fragments with altered folding characteristics, which can be recognized as misfolded and thus transferred to mitochondria for sequestration. However, this is highly speculative and must be investigated in the future.
Table 1.
Experimental evidence for the localization of APOE in mitochondria and mitochondria-associated ER membranes (MAMs)
| Cellular component | Tissue/cell type | Isolation/labelling | Detection | References |
|---|---|---|---|---|
| Mitochondria | Rat liver | Immunogold labelling of APOE in cryosections | Electron microscopy | [16] |
| Rat liver | Radioactive leucine labelling and Percoll density gradient ultracentrifugation | Anti-VLDL immunoprecipitation, SDS gel electrophoresis, excision of protein bands and radioactivity counting | [17] | |
| APOE4-transfected Neuro2a cells | Not described1 | Western blotting of mitochondrial fractions | [33] | |
| APOE TR mouse hippocampus | Isolation using a commercially available kit 2 and differential centrifugation | Western blotting, LC–MS | [28] | |
| APOE TR mouse primary astrocytes |
Coimmunostaining of Mito-GFP and APOE Isolation using a commercially available kit 3 and differential centrifugation |
Fluorescence microscopy Western blotting of mitochondrial fractions |
[29] | |
| Mitochondria-associated ER membranes | Rat liver | Radioactive leucine labelling and Percoll density gradient ultracentrifugation | Anti-VLDL immunoprecipitation, SDS gel electrophoresis, excision of protein bands and radioactivity counting | [17] |
| Mouse liver and brain | Percoll density gradient ultracentrifugation | LC–MS | [34] | |
| Huh7 cells | Percoll density gradient ultracentrifugation | LC–MS | [35] | |
| Mouse liver | Percoll density gradient ultracentrifugation | LC–MS | [30] | |
| Mouse brain | Percoll density gradient ultracentrifugation | LC–MS | [36] | |
| Mouse liver | Sucrose gradient centrifugation4 | LC–MS | [37] |
APOE has been repeatedly detected in mitochondria and MAMs in rat, mouse, and human cultured cells and tissue using various methodological approaches
TR targeted replacement, LC–MS liquid chromatography–mass spectrometry
1“Mitochondrion-rich fractions” were isolated, but the isolation method was not described
2Benchtop mitochondria isolation kit for rodent tissue (Mitosciences)
3ProteoExtract cytosol/mitochondria fractionation kit (EMD Millipore)
4Analysis of rough ER wrapped around mitochondria, which include MAMs as a subdomain
Proteolytic fragmentation of APOE
APOE fragments are the result of proteolytic cleavage, which is a normal cellular process to degrade proteins that are abnormal or no longer needed and to regulate cellular processes, e.g., activation and inactivation of enzymes by proteolysis. In this process, smaller fragments are formed from a protein, and when fully degraded, the individual amino acids can be recycled. For APOE, fragments of varying length (a few amino acids up to 29 kDa) have been described to be formed by proteolytic cleavage of various proteases, e.g., high-temperature-requirement serine peptidase A1 [38]. Overall, many of the identified APOE fragments are C-terminally truncated fragments found in the brain, where they tend to have detrimental effects and are often associated with AD. Munoz et al. [38] provided a detailed overview of the APOE fragments in different cells and animal models related to AD [38]. These fragments have different physiological functions, some of which have cytotoxic effects [39], decrease Aβ clearance [40], or accumulate in AD brains and cause AD-like neurodegeneration in mice [41].
Independent of AD, there are some fragments related to mitochondria: a 29-kDa fragment (amino acids 1–272) showed increased ER stress, altered composition of MAMs, impaired mitochondrial function, and interaction with mitochondrial proteins [32, 33, 42]. According to Chang et al. [32], APOE4 fragments have a neurotoxic effect in Neuro2a cells, which the authors suggest is due to a mislocalization in mitochondria and associated mitochondrial dysfunction. No conclusions can be drawn about APOE3 fragments, as these have not been studied [32]. A more specific interaction of APOE fragments with mitochondria was demonstrated in Neuro2a cells, where it was shown that a 29-kDa APOE4 fragment (amino acids 1–272) binds directly to mitochondrial respiratory chain proteins and decreases the activity of complexes III and IV to a greater extent than full-length APOE4. APOE3 fragments were not analysed [33]. The 29-kDa fragment lacks a part of the C-terminus involved in domain interaction (amino acids 270–290), which probably results in the spatial structure of this fragment being quite different compared to that of the full-length APOE protein. The altered structure may, for example, expose regions of the amino acid sequence that are normally hidden in full-length APOE, potentially creating binding sites in the APOE fragments for interactions with other molecules that are not present in full-length APOE. In addition to the mitochondrial interaction and localization of APOE fragments, APOE fragments were also shown to be localized in the nucleus, indicating potential activity as a transcription factor, as observed with a 17-kDa APOE fragment [43, 44]. Most studies on APOE fragments conclude that the detected APOE fragments have adverse effects, except for a study in which an extracellular 25-kDa APOE fragment was found to act neurotrophically and to stimulate neurite growth in cultured neurons [38]. Little is known about APOE fragments in other tissues and nonneuronal cells. There is one study on the presence of a 22-kDa APOE fragment in cultured hepatocytes that binds to secreted very low-density lipoprotein (VLDL) [45]. Another category of fragments are small peptides that are only 17–30-amino-acids long and have antimicrobial activity. These peptide sequences originate from the middle of the amino acid sequence of the APOE protein, e.g., they are found in the low-density lipoprotein receptor (LDLR) or heparin binding region [46, 47].
Since an association between APOE fragments and AD is often postulated, one might assume that APOE fragments in CSF could be used as biomarkers for AD. However, no correlation was found between AD or APOE genotype and the concentration of an APOE fragment in CSF or plasma [48]. Overall, the question is whether these APOE fragments, most of which have shown adverse effects, have a function in the brain or whether they are waste products that have not been properly broken down and sometimes happen to have toxic effects. Furthermore, it is unclear whether the same fragments found in the brain are also found in other tissues, such as the liver, and whether they show similar or different effects there.
Based on the growing evidence for the presence of full-length APOE and APOE fragments in mitochondria and MAMs, the potential function of APOE in these two cell organelles will be further addressed in the next two sections.
APOE and its potential role in the mitochondrion
Mitochondrial localization of APOE
The first evidence of mitochondrial APOE dates back to the early 1990s. Immunogold labelling of APOE in rat hepatocytes identified APOE in peroxisomes and the Golgi apparatus but also small amounts of APOE in mitochondria [16]. At the same time, Vance [17] showed that apolipoproteins were enriched in mitochondria, MAMs, and microsome fractions isolated from rat liver, with APOE being the most abundant apolipoprotein. Further evidence for the mitochondrial localization of APOE is provided by an analysis of the mitochondrial hippocampus proteome of APOE3 and APOE4 targeted replacement (TR) mice. Remarkably, in APOE4 mice, APOE protein levels were significantly increased in mitochondria after ischaemic damage, which may indicate stress-dependent mitochondrial translocation [28]. Then, APOE was again shown to accumulate in the mitochondrial fraction of primary astrocytes of APOE TR mice, and colocalization was detected by immunostaining of APOE and mitochondria by confocal microscopy [29]. However, the mechanism or pathway by which APOE enters the mitochondrion remains to be elucidated. The import of molecules into the mitochondrion depends on their size. While small molecules such as calcium and ATP can cross the outer mitochondrial membrane (OMM) through voltage-dependent anion-selective channel 1 (VDAC1), larger molecules such as proteins require the abovementioned MLS to enter the mitochondrial membrane via the translocase of the outer mitochondrial membrane (TOM) complex [49]. In addition to importing small molecules, VDAC1 also plays a role in the protein import of the TOM complex by acting as a coupling factor between the TOM and translocase of the inner mitochondrial membrane (TIM) complex subunits in the mitochondrial intermembrane space [50]. Interestingly, APOE was shown to interact with VDAC1 [51, 52]; however, APOE would still be too large to be imported through VDAC1 channels.
Effects of APOE on mitochondrial function and dynamics
The main function of mitochondria is energy production in the form of ATP via the tricarboxylic acid (TCA) cycle in the mitochondrial matrix and oxidative phosphorylation in the inner mitochondrial membrane (IMM). Altered ATP levels depending on APOE genotype could indicate an influence of APOE on mitochondrial function. This notion was investigated in several studies in APOE-transfected Huh7 cells [53], primary astrocytes isolated from APOE TR mice [29], primary neurons isolated from APOE TR mice [54], and in the brains of APOE TR mice [12], with the result of higher ATP levels in APOE3 cells and mice compared to APOE4. Yin et al. [55] suggested that APOE affects mitochondrial function via the peroxisome proliferator-activated receptor gamma coactivator 1-α (PGC1α) and sirtuin 3 (SIRT3) pathways, as APOE4 mice exhibited reduced PGC1α and SIRT3 expression [55]. However, there are also studies that found no difference between APOE isoforms in total ATP levels but differences in the source of ATP, e.g., higher ATP levels generated by oxidative phosphorylation (OXPHOS) and lower glycolysis-derived ATP in primary endothelial cells from APOE4 mice [56]. Others distinguished between basal and stressed states and found significantly lower ATP levels in stressed APOE4 cells than in APOE3 cells [57].
One reason for the altered ATP levels could be a disruption in the respiratory chain or in the individual complexes of the electron transport chain; therefore, the effect of the APOE genotype on the protein levels of OXPHOS complexes has been studied intensively [53, 57, 58]. The protein levels of complexes I–V were reduced in APOE4 neurons compared to APOE3 neurons [54, 57, 58], while there was no significant difference in astrocytes [57]. In contrast to the decreased protein levels in APOE4 neurons, some subunits of complexes I–V in the entorhinal cortex of APOE3/4 TR mice were increased at the mRNA level compared to APOE3/3 [59]. There are only a few studies examining the effect of APOE on mitochondrial respiration outside the brain, but these studies tend to show no difference between APOE3 and APOE4. In the liver of APOE TR mice and in APOE-transfected Huh7 cells, no isoform-dependent effect on OXPHOS protein levels was found [53], and rat cardiomyocytes treated with recombinant APOE3 and APOE4 showed almost no difference in mitochondrial respiration [52]. The effect of APOE on OXPHOS appears to be dependent not only on the tissue studied (brain versus periphery) but also on the different cell types within the brain. For example, differences in mitochondrial respiration depending on the APOE isoform were observed in the hippocampus but not in the entorhinal cortex [59], and the results are also partially contrasting in neuronal cells and astrocytes [54, 58].
Mitochondria undergo structural changes by fission and fusion and are, therefore, highly dynamic cell organelles. In mitochondrial fusion, two or more organelles become one larger one, induced by specific proteins such as mitofusin 1 and 2 (MFN1/2) and dynamin-like 120 kDa protein (OPA1). In fission events, one mitochondrion is split into two, mediated by proteins including dynamin-related protein 1 (DRP1) and mitochondrial fission 1 protein (FIS1) [60]. APOE appears to have an effect on mitochondrial fusion and fission, although studies show partially conflicting results. In the hippocampus and primary astrocytes of APOE TR mice and human postmortem brain tissue of AD patients, increased mitochondrial fusion was observed with APOE4 [29, 57, 61]. In contrast, in human postmortem brain tissue, both fusion and fission proteins were downregulated in APOEε4 carriers compared to non-APOEε4 carriers (independent of AD status), indicating overall reduced mitochondrial dynamics [62]. APOE-associated with a specific form of low-density lipoprotein (L5-LDL) influences the phosphorylation of DRP1, which leads to an increase in mitochondrial fission in rat heart cells (H9c2 cells), independent of the APOE isoform [52]. An examination of markers of mitochondrial dynamics in Huh7 cells and liver of APOE TR mice also revealed no isoform-dependent differences at the mRNA level (MFN2 and FIS1) [53]. In addition to the possible effects of the APOE isoform, APOE fragments also appear to have an impact on mitochondrial dynamics. Compared with full-length APOE4, a 29-kDa APOE4 fragment showed increased mitochondrial fragmentation and greater fission in the hippocampus of APOE TR mice and neuronal cells. Conclusions on isoform-dependent differences could not be made because APOE3 fragments were not examined [42]. Further evidence of the possible involvement of APOE in mitochondrial metabolism is provided by experiments with APOE knockout mice. Analysis of the liver and hippocampal mitochondrial proteome of APOE knockout mice showed a significant change in the expression of some mitochondrial proteins, such as the subunits of ATP synthase and hexokinase 1 [63, 64].
In summary, there are many indications from different methodological approaches that APOE exhibits a prominent function in the mitochondria (Table 2). However, depending on the tissue and cell type, the APOE isoforms seemingly exhibit different effects on the respiratory chain, ATP levels, and mitochondrial dynamics, which warrants further investigation. This is of particular importance, since APOE is not able to cross the blood–brain and thus, two different APOE pools exist in the human body (brain and periphery), in which the APOE protein shows different concentrations and posttranslational modifications [4–6]. In addition, there is much evidence of proteolytically cleaved APOE fragments in the brain, but almost no studies on fragments in other tissues [38]. This could also lead to tissue-specific APOE isoform-dependent differences. It should be considered that most of the studies on APOE and its mitochondrial involvement were performed in the brain or neuronal cells. However, these results may be not directly transferable to peripheral APOE.
Table 2.
Overview of the APOE isoform-dependent effects on mitochondrial and MAM function with the tissue or cell line studied (brain or periphery) indicated
| Readout | Isoform effect | Tissue source | References | ||
|---|---|---|---|---|---|
| Mitochondria | Total ATP level | E3 > E4 | Brain (APOE TR mice) | [12] | |
| Periphery (Huh7 cells1) | [53] | ||||
| Brain (temporal lobes of APOE TR mice) | [55] | ||||
| Brain (primary astrocytes of APOE TR mice) | [29] | ||||
| Brain (primary neurons of APOE TR mice) | [54] | ||||
| E3 < E4 | Brain (entorhinal cortex of APOE TR mice) | [59] | |||
| Brain (primary astrocytes of APOE TR mice) | [54] | ||||
|
Basal: E3 = E4 Stress: E3 > E4 |
Brain (Neuro2a cells2) | [57] | |||
| E3 = E4 | Brain (primary endothelial cells of APOE TR mice) | [56] | |||
| OXPHOS | Protein | E3 > E4 | Brain (primary neurons of NSE-APOE mice) | [58] | |
| Brain (cortical synaptosome of APOE TR mice) | [65] | ||||
| Brain (Neuro2a cells2) | [57] | ||||
| Brain (primary neurons of APOE TR mice) | [54] | ||||
| E3 = E4 | Brain (primary astrocytes of GFAP–APOE mice) | [58] | |||
| Periphery (Huh7 cells1, liver from APOE TR mice) | [53] | ||||
| E3 < E4 | Brain (primary astrocytes of APOE TR mice) | [54] | |||
| mRNA | E3/3 < E3/4 | Brain (entorhinal cortex of APOE TR mice) | [59] | ||
| PGC1α pathway | Protein | E3 > E4 | Brain (temporal lobes of APOE TR mice | [55] | |
| Brain (primary neurons of APOE TR mice) | [54] | ||||
| non-E4 > E4 | Brain (human postmortem brain tissue) | [62] | |||
| mRNA | E3 > E4 | Brain (cortex from APOE TR mice) | [12] | ||
| E3 = E4 | Periphery (Huh7 cells1, liver from APOE TR mice) | [53] | |||
| Mitochondrial respiration (Seahorse) | E3 > E4 | Brain (Neuro2a cells2) | [58] | ||
| Brain (primary neurons of APOE TR mice) | [54] | ||||
| E3 = E4 | Periphery (H2c9 cells, treated with recombinant APOE) | [52] | |||
| Depends on age, brain area and complex | Brain (entorhinal cortex and hippocampus of APOE TR mice) | [59] | |||
| Mitochondrial membrane potential | E3 = E4 | Periphery (Huh7 cells1) | [53] | ||
| E3 > E4 | Brain (primary astrocytes of APOE TR mice) | [29] | |||
| Brain (primary neurons of APOE TR mice) | [54] | ||||
| Mitochondrial translocation/accumulation | E3 < E4 (stress-induced) | Brain (hippocampus of APOE TR mice) | [28] | ||
| E3 < E4 | Brain (primary astrocytes of APOE TR mice) | [29] | |||
| Mitochondria-associated ER membranes | MFN1/2 | Protein | E3 < E4 | Brain (Neuro2a cells2) | [57] |
| Brain (hippocampus of APOE TR mice) | [61] | ||||
| Brain (primary astrocytes of APOE TR mice) | [29] | ||||
| non-E4 > E4 | Brain (human postmortem brain tissue) | [62] | |||
| mRNA | E3 = E4 | Periphery (Huh7 cells1, liver from APOE TR mice) | [53] | ||
| GRP75 | Protein |
GRP75c: E3 < E4 GRP75d: E3 > E4 |
Brain (hippocampus from APOE TR mice) | [66] | |
| E3 = E4 | Brain (Neuro2a cells2) | [58] | |||
| E3 > E4 | Brain (Neuro2a cells2) | [57] | |||
| VDAC1 | Protein | E3 = E4 | Brain (Neuro2a cells2) | [58] | |
| E3 < E4 | Brain (Neuro2a cells2) | [57] | |||
| mRNA | E3 < E4 | Brain (Neuro2a cells2) | [58] | ||
| E3 = E4 | Periphery (Huh7 cells1, liver from APOE TR mice) | [53] | |||
| Phospholipid synthesis | E3 < E4 | Brain (mouse hippocampus cells treated with APOE-containing astrocyte-conditioned media) | [67] | ||
| ER-mitochondria calcium flux | E3 < E4 | Brain (Neuro2a cells2) | [57] | ||
| ER-mitochondria colocalization | E3 < E4 (in trend) | Brain (mouse hippocampus cells treated with APOE-containing astrocyte-conditioned media) | [67] | ||
| VLDL assembly | - | Periphery (liver from CAV1 knockout mice) | [30] | ||
| Periphery (mouse liver) | [37] | ||||
1Transient APOE3- and APOE4-transfected cells
2Stable APOE3- and APOE4-expressing cell lines
Role of APOE in mitochondria-associated ER membranes
Mitochondria interact not only with other mitochondria but also with other organelles, such as the ER. These interactions of the OMM and the ER membrane are described as MAMs and/or mitochondria–ER contacts, so-called “MERCs” (Fig. 2a). These definitions are sometimes used synonymously, although distinctions must be made, since MAMs are the result of subcellular isolation and fractionation of membranes that are associated with mitochondria and ER, and MERCs are the structural and functional organization of interorganelle contacts [68]. For a better understanding, only the term "MAMs" is used in this article.
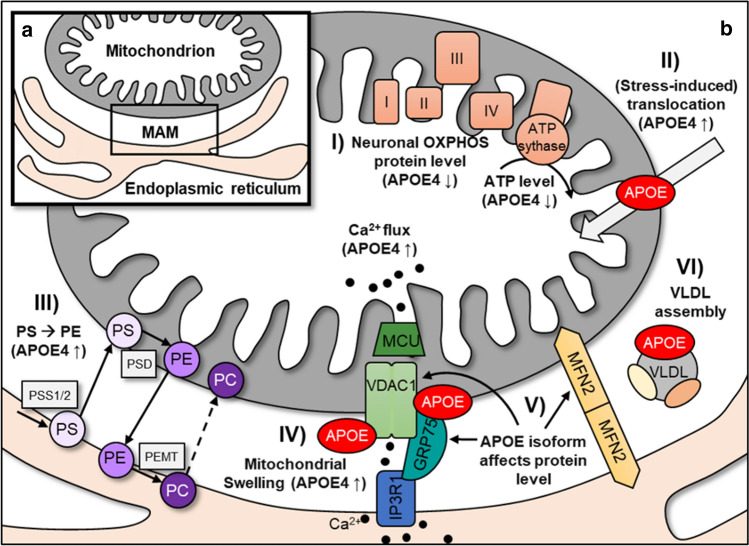
Fig. 2.
Illustration of the assembly of mitochondria and mitochondria–ER contacts and the inherent biochemical pathways with suggested APOE involvement or regulation by the APOE isoform. a Mitochondria-associated ER membrane complex (MAM). b Mitochondrial and MAM pathways in which APOE may be involved. (I) The impact of APOE isoforms on mitochondrial function depends on the cell type and species, but consistently decreased neuronal OXPHOS protein and ATP levels were observed in APOE4. (II) Mitochondrial accumulation and stress-induced translocation are increased in APOE4. (III) The different steps in phospholipid synthesis take place through consecutive exchange of substrates from the ER membrane (synthesis of phosphatidylserine; PS) to the mitochondrion (conversion of PS to phosphatidylethanolamine (PE), which is increased in APOE4) and back. The final methylation step by PEMT is accomplished in the ER membrane, yielding phosphatidylcholine (PC). (IV) Calcium is released from the ER through the IP3R1–GRP75–VDAC1 complex and shuttled to the mitochondrion through the mitochondrial calcium uniporter protein (MCU) into the inner matrix. Increased calcium flux was found in APOE4 Neuro2a cells, and higher mitochondrial swelling was observed in APOE4-treated H9c2 cells, which was caused by the interaction of APOE (derived from the lysosomal degradation of L5-LDL) with VDAC1. The protein–protein interaction of APOE with MAM proteins such as VDAC1 and GRP75 is one explanation for the presence of APOE in MAMs. (V) MFN2 dimers connect the OMM with the ER membrane, acting as MAM tethering proteins. Depending on the tissue and species, APOE affects the expression of MFN1 and MFN2. (VI) MAMs are involved in cholesterol metabolism, and proteomic analyses provide evidence for a possible role of APOE in VLDL assembly in MAMs
Proteome of mitochondria-associated ER membranes and function of mitochondria–ER contacts
The contacts between mitochondria and the ER have been studied for many decades. In 1990, Vance [17] isolated a membrane fraction associated with mitochondria that had many similarities to microsomes but showed a unique and significantly different protein profile compared to mitochondria and ER. This fraction was declared as “fraction X” and later as MAMs [17, 69]. The mammalian MAM protein composition was analysed by mass spectrometry (MS) for the first time by Zhang et al. [70], with an increasing number of proteomic studies analysing MAM fractions in different tissues and cells that have followed [30, 34–36]. From numerous studies in which the MAM proteome has been examined, it has become clear that the MAMs are a dynamic cell structure that is influenced by various parameters, including ageing, diseases such as diabetes, and nutritional factors [36, 71–73]. In addition, the MAM proteome differs by tissue type (liver and brain) [34].
The MAM resident proteins can be classified according to their function. There are tethering proteins that maintain the connection and the distance between the ER and OMM, such as MFN1 and MFN2 [74], which are also important in mitochondrial fusion. Apart from these classical MAM proteins, the assembly of MAMs is very flexible, and proteins can be recruited depending on the state of the cell [75]. Further evidence for the flexibility of MAM assembly is provided by an experimental approach using a synthetic GFP-tagged MAM linker, which demonstrated that contacts between mitochondria and ER can change within minutes and that various stressors, such as starvation and apoptosis, increase the number of contacts [76].
Mitochondria–ER contacts are important in all processes where molecules need to be shuttled between the ER and mitochondria, such as during phospholipid synthesis, since it takes place in both, the OMM and the ER membrane (Fig. 2b) [69, 77]. In addition to lipids, calcium is also transported between the ER (most important cellular calcium storage site) and mitochondria. Calcium is released from the ER through the inositol 1,4,5-trisphosphate receptor 1 (IP3R1) ion channel, crosses the OMM through VDAC1 and accumulates in the mitochondrial matrix by passing through the IMM by way of the mitochondrial calcium uniporter protein (MCU) [78]. The chaperone protein glucose-regulated protein 75 (GRP75) forms a bridge between IP3R1 and VDAC1 [79, 80] and thus, the IP3R1–GRP75–VDAC1 complex has a crucial role in maintaining calcium homeostasis in mitochondria (Fig. 2b).
Glucose appears to influence mitochondria–ER contacts, as high glucose (25 mM) treatment reduced VDAC1–IP3R1 contacts in cultured hepatocytes by half compared to those of low glucose (5 mM) treatment. Similar effects have been observed in hepatocytes from ob/ob mice (leptin-deficient mice, exhibit obesity and glucose intolerance) [71, 72] and myotubes of obese and diabetic humans, wild-type mice and healthy subjects, respectively [81]. Overall, the protein composition of MAMs and the integrity of mitochondria–ER contacts seem to be important for normal insulin signalling [36, 71]. MAMs may likewise play a significant role in neuronal disease pathology such as AD, since it has been observed that the level of presenilins and γ-secretase activity was increased in MAMs of AD patients, while phospholipid synthesis in fibroblasts was also higher [82, 83]. Interestingly, amyloid beta (Aβ) treatment led to an augmented number of ER-mitochondrial contacts in hippocampal neurons [84].
APOE in mitochondria-associated ER membranes
Vance [17] showed in 1990 that apolipoproteins such as apolipoprotein B (APOB) and apolipoprotein C (APOC), but particularly APOE, were enriched in MAM fractions isolated from rat liver. APOE was also detected in the mitochondrial fraction, but remarkably 15-fold higher in MAM fractions indicating a MAM resident function of APOE [17]. Analyses of the MAM proteome by subcellular isolation of MAM fractions and subsequent liquid chromatography–mass spectrometry (LC–MS) studies in mouse liver, brain, muscle, testes, Huh7 cells, and human testes also indicated the presence of APOE in MAM fractions (Table 1) together with hundreds of other proteins [30, 34–36, 85]. With the exception of Vance [17], previous work has focused on untargeted proteomics to investigate the general composition of MAMs. It is still poorly understood how APOE localizes to MAMs, whether it is permanently present or only during certain conditions, and what physiological function APOE has there. The possible functions of APOE described in the literature and the effects of APOE isoforms on MAMs are shown in Table 2 and Fig. 2b. One possible explanation for how APOE enters the MAM is the physical interaction with the MAM resident proteins GRP75 and VDAC1 [51, 52, 86]. Apart from the protein–protein interaction, the level of different GRP75 isoforms and their phosphorylation have been shown to be regulated in part depending on the APOE isoform. For example, GRP75 isoform c was strongly upregulated in the presence of APOE4, whereas the expression of isoform d was increased with APOE3 [66]. Other authors did not distinguish between GRP75 isoforms and showed downregulation of GRP75 protein expression in APOE4 Neuro2a cells [57] and no effect of APOE on GRP75 protein levels in cultured brain cortical neurons and astrocytes [58]. A further link between APOE and GRP75 became apparent when an APOE4 fragment (amino acids 1–271) was examined, as the fragment caused ER stress and increased GRP75 expression, presumably leading to increased MAM formation and mitochondrial calcium overload [42]. Whether APOE3 fragments have the same effect was not investigated.
Recombinant APOE, which is bound to a specific LDL form, was found to colocalize with VDAC1 after lysosomal degradation of LDL, and leads to increased opening of the mitochondrial permeability transition pore (mPTP) and mitochondrial swelling [52]. This finding could indicate a role of APOE in the regulation of mitochondrial function via the interaction with VDAC1. Multiple proteins have been identified that bind to VDAC1, and some of them regulate VDAC1 function. For example, VDAC1 closure is regulated through the binding of tubulin [87]. It may be conceivable that the binding of APOE to VDAC1 affects the function of the VDAC1–GRP75–IP3R1 complex and, as a result, the shuttling of calcium through MAMs. Increased calcium flux from the ER to the mitochondrion was observed in APOE4 Neuro2a cells compared with APOE3 cells [57], but whether this is the result of a physical interaction of APOE with the VDAC1–GRP75–IP3R1 complex remains to be investigated systematically.
Apart from the physical interaction of APOE with MAM resident proteins, there is evidence for a role of APOE in one of the major functions of mitochondria–ER contacts, the lipid metabolism. Increased conversion of phosphatidylserine (PS) to phosphatidylethanolamine (PE) was observed in human fibroblasts and mouse hippocampus cells treated with APOE4-containing astrocyte-conditioned media compared to APOE3 [67]. However, the protein expression level of the respective enzyme catalysing the conversion from PS to PE was not altered in response to the APOE isoform. The APOE isoform-dependent effect on phospholipid metabolism was not visible in MFN2 knockout cells (decreased ER mitochondrial communication). The authors interpreted this as an indication that the APOE4-dependent effect on phospholipid metabolism is due to increased ER-mitochondrial communication in APOE4-treated cells (defined as MAM activity) [67]. When comparing the hepatic MAM proteome of caveolin 1 (CAV1; main component for caveolae formation) knockout mice and wild-type mice, it was found that APOE protein levels were decreased in the MAM fraction of CAV1 knockout mice. At the same time, increased cholesterol levels were observed in MAM fractions of CAV1 knockout mice [30]. The authors hypothesize that the reduced presence of APOE in MAMs of CAV1 knockout mice disturbs the secretion of VLDL, leading to increased cholesterol levels. There is further evidence of a relationship among cholesterol metabolism, APOE, and the protein composition of MAMs, as shown by an examination of a complex of rough ER wrapped around mitochondria containing MAMs as a subdomain [37]. Transcriptomics and proteomics of these ER-mitochondria complexes in mouse liver revealed an enrichment of proteins involved in VLDL biogenesis, including APOE [37]. These examples demonstrate the possible participation of APOE in phospholipid and cholesterol metabolism in MAMs.
The influence of the APOE isoform on MFN1 and MFN2 expression, which are important representatives of MAM tethering proteins, has been investigated in several studies. In APOE-transfected Neuro2a cells, primary astrocytes from APOE TR mice, and the hippocampus of APOE TR mice, higher MFN1 protein levels were observed in APOE4 [29, 57, 61]. This finding could indicate a possible increased connection between the ER and mitochondria in APOE4 [57]. However, in the human postmortem brain, higher protein levels were found in APOEε3 carriers [62], and in Huh7 cells and APOE TR mouse liver, no differences were found at the mRNA level [53].
Overall, there is some evidence of the participation of APOE in various pathways in MAMs through physical appearance [17], interactions with MAM proteins [52, 86], and influences on MAM functions [30, 42, 67]. These studies provide the first clues about the possible role of APOE in mitochondria–ER contacts, but more systematic research is needed. An overview of the described possible functions of APOE in mitochondria and MAMs is given in Fig. 2b.
Protein–protein interactions
Protein–protein interactions play a crucial role in normal cell function and have been gained scientific interest for decades. The analysis of the human interactome network, for example, has identified up to 100,000 protein–protein interactions [88]. A prominent example of an APOE protein–protein interaction of central importance to its role in lipid metabolism, is the binding to receptors of the LDLR family [89]. A prerequisite for the interaction, however, is the exposure of the specific receptor-binding site in the N-terminus of APOE that is only exposed upon lipid binding [90]. The APOE protein consists of a N- and a C-terminal domain that interact through numerous amino acid residues. There have been a few APOE isoform-dependent differences reported concerning the domain dissociation, folding intermediate formation, and the size of preferentially bound lipid particles [22, 23, 91]. Therefore, it is conceivable that there also exist differences in protein–protein interactions, owing to the differences in binding site exposure, between the APOE isoforms.
APOE protein–protein interactions beyond lipid metabolism
By studying novel protein–protein interactions of APOE, new metabolic pathways involving APOE may be discovered, and potentially novel functions of APOE, apart from its major role in lipid transport and clearance, may be identified. According to PubMed and the BioGRID 4.4 database (Database of Protein, Genetic and Chemical Interactions, https://thebiogrid.org/), approximately 150 proteins interacting with APOE have been identified in human cells and tissues to date. The protein–protein interaction network of an in silico analysis (BioGRID) of the APOE interaction partners discovered thus far is shown in Fig. 3 and illustrates the large number of potential binding partners that may indicate metabolic pathways with previously unknown APOE involvement. Apart from the interactions of APOE with LDLR and other proteins involved in cholesterol metabolism, several other APOE protein–protein interactions have recently been identified. However, the majority of the large number of potential APOE interaction partners that were detected by untargeted so-called “high-throughput” interactome studies, which identify a wide variety of different interactions, have not yet been further investigated for their biological relevance in functional studies or independently confirmed. For example, in 2015, Huttlin et al. [51] published an interactome study in which they identified over 20,000 protein–protein interactions in HEK293T cells by affinity purification and MS, of which 86% of these interactions were previously unknown, including the interactions of APOE with the major histocompatibility complex class II DPα1, matrix metallopeptidase 3, multimerin 1, and VDAC1 [51]. These four proteins were also repeatedly identified as APOE interaction partners in two subsequent interactome studies by Huttlin et al. [92, 93]. Considering only the mitochondrial interactome, APOE was found to be associated with branched-chain alpha-keto acid dehydrogenase subunit α (BCKDHA), subunits of F1-ATP synthase and other respiratory chain complexes (e.g., cytochrome b–c1 complex subunit 1 and 2 (UQCRC1/2), and subunits of NADH dehydrogenase) as well as nonmitochondrial proteins such as cytoplasmic actin, and calnexin, which is located in the ER membrane, among others [94].
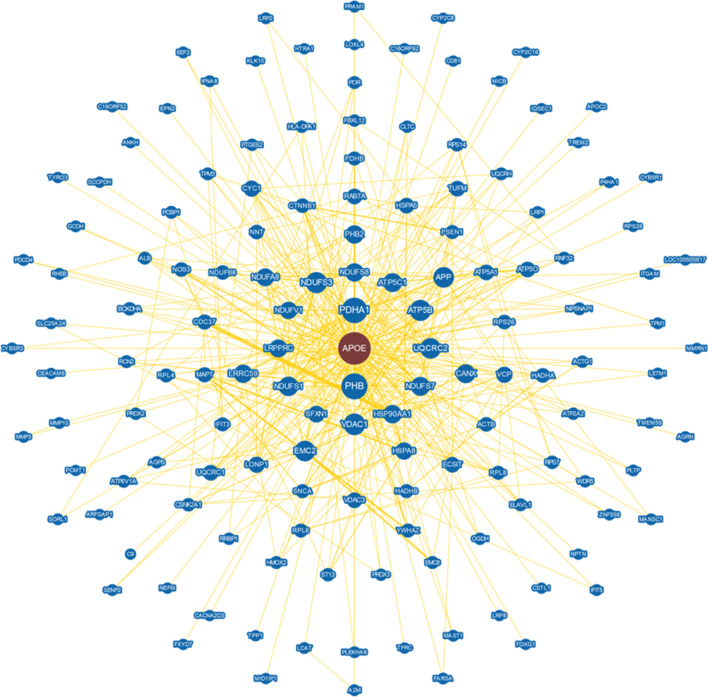
Fig. 3.
Potential APOE protein–protein interactions in human cells and tissue. An in silico analysis performed with BioGRID 4.4 software revealed that there is evidence for 145 potential protein binding partners whose physical interaction with APOE has been demonstrated in at least one study in each case. The larger the blue circle of the corresponding protein is, the stronger the connectivity with APOE, and thicker binding lines represent stronger evidence supporting the association. Image downloaded from https://thebiogrid.org/
Interactions of APOE with nonmitochondrial proteins
Functional studies have already been performed on some APOE protein–protein interactions, partly investigating isoform-dependent differences or examining the physiological significance. These “low-throughput” functional studies are listed in Table 3. Since a markedly large number of mitochondrial proteins were found in the interactome studies and some of them were characterized in more detail, Table 3 is divided into interactions of APOE with nonmitochondrial and mitochondrial proteins. APOE protein–protein interactions with nonmitochondrial proteins include proteins associated with AD pathology, such as the amyloid-β precursor protein (APP) [95, 96]. A network of AD-relevant proteins that interact with each other has been identified by Soler-López et al. [97], which may provide further evidence to elucidate the underlying mechanisms of AD. In addition to AD pathology, APOE has also been shown to bind to synuclein α (SNCA), which accumulates in Parkinson’s disease [98, 99]. However, several other proteins that are not directly related to diseases have been further investigated regarding their interaction with APOE. These include haptoglobin (HPT), which is a haemoglobin binding and transport protein in the plasma and has been shown to bind to APOE via a specific binding site in the APOE amino acid sequence (amino acids 131–150) [100–102].
Table 3.
Overview of “low-throughput” studies in which targeted APOE protein–protein interactions were investigated and characterized for their biological function
| Interacting proteins | Methods | Tissue/cells | Biological relevance | References |
|---|---|---|---|---|
| Nonmitochondrial | ||||
| A2M | Co-IP WB | Human plasma |
Interaction E2 = E3 > E4 Interaction affected by lipoproteins |
[103] |
| APP |
Co-IP WB Affinity chromatography WB |
COS-1 cells Human plasma |
APP is the precursor of Aβ, which plays an important role in AD pathology due to its neurotoxic properties and accumulation in plaques |
[95] [96] |
| ABCA1 | Co-IP WB | Human skin fibroblasts, human plasma |
Interaction E2 = E3 = E4 APOE might act as a protector for ABCA1 against proteases |
[104] [105] |
| HPT | HPT-coupled beads, loaded with human plasma, pulldown, WB | Human plasma |
Interaction of APOE and HPT via a specific amino acid sequence in APOE (amino acids 131–150) HPT potentially enhances the formation of the APOE–Aβ complex, may be related to AD pathology Inhibition of LCAT activity |
[100,101, 102] |
| NOS1 |
Co-IP WB Coimmunostaining |
HEK293 cells Human hippocampus |
Interaction E2 = E3 = E4 Interaction potentially leads to S-nitrosylation of APOE2 and APOE3, but not APOE4 |
[106] |
|
CDC37 ECSIT PDCD4 ST13 |
Co-IP WB |
SH-SY5Y cells COS-7 cells |
All investigated proteins are related to AD pathology Identification of these protein–protein interactions provides a contribution to the elucidation of possible AD-relevant mechanisms |
[97] |
| SNCA |
Co-IP WB Co-IP ELISA |
Human plasma Human CSF |
SNCA accumulation is a maker for Parkinson’s disease pathology SNCA interacts with APOE and other apolipoproteins Increased APOE level in CSF from Parkinson’s disease patients |
[98] [99] |
| Mitochondrial | ||||
| ATP5A/B | Affinity chromatography WB | Human liver | Potential function in mitochondrial metabolism | [107] |
|
UQCRC1/2 CYC1 |
Co-IP WB | Neuro2a cells |
Interaction E4 fragment > E4 full-length APOE3 was not investigated APOE4 fragment alters mitochondrial activity |
[33] |
| GRP75 | Co-IP WB |
Astrocytes from APOE TR mice Human hippocampus |
Interaction only in E4 cells, not in E3 and E2 Interaction in hippocampus E3 control < E3 AD < E4 AD Different GRP75 isoforms are regulated differently depending on the APOE isoform |
[86] [66] |
| VDAC1 |
Co-IP WB Coimmunostaining |
H9c2 cells |
Interaction E3 = E4 Interaction of APOE (derived from the lysosomal degradation of L5-LDL) with VDAC1 induced mitochondrial swelling (higher in APOE4) |
[52] |
Co-IP co-immunoprecipitation, WB Western blot, ELISA enzyme-linked immunosorbent assay, A2M α2-macroglobulin, APP amyloid precursor protein, ABCA1 ATP-binding cassette transporter, HPT haptoglobin, NOS1 nitric oxide synthase 1, CDC37 cell division cycle 37 HSP90 cochaperone, ECSIT evolutionarily conserved signalling intermediate in the toll pathway, PDCD4 programmed cell death 4, ST13 suppression of tumorigenicity 13 protein, SNCA synuclein α, ATP5A/B ATP synthase subunit α/β, UQCRC1/2 cytochrome b–c1 complex subunit 1/2, CYC1 cytochrome C1, GRP75 glucose-regulated protein 75, VDAC1 voltage-dependent anion-selective channel 1
Interactions of APOE with mitochondrial proteins
Interestingly, among the APOE protein–protein interactions identified thus far are many mitochondrial proteins (Table 3), although APOE is a secretory protein. Investigation of the mitochondrial interactome of human pluripotent embryonal carcinoma stem cells revealed 54 proteins associated with APOE [94]. Some of these interactions have been described before, while others are completely unknown. The interactome study showed the protein–protein interaction of APOE with eight subunits of complex I and with four subunits each of complex III and complex V/F1-ATP synthase of the mitochondrial electron transport chain [94]. The association of APOE with the α- and β-subunits of F1-ATP synthase, UQCRC1/2, and cytochrome C1 (CYC1) has been partly described previously in human liver tissue and APOE-transfected Neuro2a cells [33, 107]. In 1988, APOE was shown to be associated with high affinity for a protein complex in the human liver [107]. The researchers identified the α- and β-subunits of F1-ATP synthase as components of this complex and demonstrated for the first time the interaction of APOE with mitochondrial proteins [107]. Twenty years later, these proteins were again discovered among APOE-associated proteins in APOE4-transfected Neuro2a cells [33]. A C-terminally truncated APOE4 fragment interacted more strongly with the mitochondrial proteins studied than did full-length APOE4. The reason for this could be a more altered spatial structure compared to full-length APOE. Since APOE3 was not studied, no conclusion can be made about potential isoform-dependent differences in terms of the interaction with mitochondrial proteins [33]. The impact of the APOE isoform on mitochondrial metabolism has already been investigated in several studies, especially in the brain and neuronal cells (Table 2), but whether the interaction of APOE with mitochondrial proteins of the electron transport chain is reflected in mitochondrial function, e.g., oxidative phosphorylation or ATP levels, or whether it is causal for disease-associated mitochondrial dysfunction still needs to be explored.
Another interactome study in HEK293T cells emphasized the association of APOE with the mitochondrial channel protein VDAC1, which is localized in the OMM [51]. Based on this information, Chen et al. [52] investigated the effect of the VDAC1–APOE interaction on mitochondrial function in rat cardiomyocytes (H9c2 cells). They treated H9c2 cells with recombinant APOE3 or APOE4 and confirmed the interaction of APOE and VDAC1 by co-immunoprecipitation (co-IP) and immunostaining experiments. They concluded that APOE, derived from the lysosomal degradation of a highly electronegative subtype of LDL (L5), influences the opening and closing of the mPTP, leading to mitochondrial swelling [52]. The interaction with VDAC1 was comparable in APOE3- and APOE4-treated cells, but mitochondrial swelling was greater in APOE4-treated cells.
A large number of proteins have already been identified to interact with VDAC1, including GRP75, which has also been shown to interact with APOE in astrocytes from APOE TR mice and in the human hippocampus [86]. The functional relevance of the GRP75–APOE interaction has not been further examined, but the interaction was observed only in APOE4 astrocytes (not in APOE3 and APOE2) and was different in the human hippocampus depending on the AD or non-AD status and APOE isoforms, with the lowest interaction observed in non-AD APOEε3 carriers, followed by APOEε3 AD, and the highest in APOEε4 AD [86]. Rather than considering the interactions of APOE with GRP75 and VDAC1 separately and assuming that they are dimer protein–protein interactions, these results may also indicate a larger protein complex involving APOE. VDAC1 and GRP75, together with IP3R1, are involved in calcium transport in MAMs (Fig. 2b), which may indicate a role for APOE in this protein complex and thus in mitochondrial calcium homeostasis.
We were able to demonstrate the interaction of APOE with VDAC1 in the liver of APOE TR mice, and its upregulation in response to dietary restriction in APOE3 and APOE4 mice. Another interactor protein identified in this study was BCKDHA, a subunit of an enzyme complex involved in the catabolism of branched-chain amino acids (BCAA). Remarkably, the enzyme activity of this complex was increased in obese APOE4 mice compared to APOE3 mice, but without an effect on plasma BCAA levels in (manuscript in preparation, Rueter et al. 2022).
By identifying and characterizing APOE protein–protein interactions, it becomes clear what multiple potential non-canonical functions of APOE may exist. Many mitochondrial proteins that potentially bind to APOE have been identified in interactome studies, but only a few have been investigated for their biological relevance. Thus, many unanswered questions remain, such as whether protein–protein interactions are permanent or transient and whether they are modifiable by physiological factors, including diet.
Conclusion
In conclusion, novel findings considering its subcellular localization and its unrecognized interaction with other proteins warrants a revision of the traditional view on APOE solely as mediator of lipoprotein-receptor binding. Regarding the large body of evidence existing on the mitochondrial localization, APOE isoform-dependent regulation of mitochondrial functions, and the interaction with mitochondrial proteins, a participation of APOE in the functional network of mitochondria is strongly suggested. However, it remains to be elucidated, by which transfer mechanisms APOE enters the inner mitochondrial matrix and whether this occurs during the process of mitochondria-associated proteostasis. The latter implies that APOE is either proteolytically cleaved or recognized as misfolded, prior to its mitochondrial incorporation. An isoform-dependent mitochondrial accumulation of proteolytic fragments and misfolded intermediates must be considered and may explain the adverse effects on mitochondrial function attributed to APOE4.
The question of whether the association of APOE isoforms with mitochondrial and MAM function has an impact on human health requires further research. Mitochondrial dysfunction in the brain of APOEε4 carriers is discussed as contributing to the development of AD, but it remains unclear whether APOE4 also leads to impaired mitochondrial function in other organs, which would be fatal since mitochondria are essential cell organelles for central processes such as energy metabolism. Because other proteins of lipoprotein metabolism besides APOE have been localized in the MAMs, it is reasonable to assume that APOE plays a role in VLDL synthesis in the hepatic MAMs. Since the lipoprotein binding preference differs between the APOE isoforms and APOE4 has a higher preference for VLDL than APOE3, differences in lipid metabolism in the MAMs may result.
The functional importance of mitochondria–ER contacts has been increasingly recognized and there is growing evidence for a central involvement of APOE. MAM assembly and protein distribution is affected by the cellular energy state and nutritional factors such as high glucose levels, while the APOE genotype has been observed to modulate the responsiveness to dietary energy and micronutrients. Therefore, focussing on APOE isoform-dependent MAM dynamics in response to the availability of cellular energy substrates or dietary micronutrients can be a valuable approach to future research. However, when selecting the target tissue to study the function of APOE in MAMs and mitochondria, it should be noted that the APOE isoproteins might have a different effect in the brain than in the rest of the body, including the liver. In this regard it is also conceivable that the composition and functional dynamics of MAMs differ depending on the type of tissue analysed. Therefore, the direct comparison of APOE isoform-dependent modifications of mitochondrial and MAM function in the brain versus the liver is inevitable.
Abbreviations
- A2M
α2-Macroglobulin
- ABCA1
ATP-binding cassette transporter
- AD
Alzheimer’s disease
- APOB
Apolipoprotein B
- APOC
Apolipoprotein C
- APOE
Apolipoprotein E
- APP
Amyloid-β precursor protein
- ATP5A/B
ATP synthase subunit α/β
- BCAA
Branched-chain amino acid
- BCKDHA
Branched-chain alpha-keto acid dehydrogenase subunit α
- CAV1
Caveolin 1
- CDC37
Cell division cycle 37, HSP90 cochaperone
- CSF
Cerebrospinal fluid
- Co-IP
Co-immunoprecipitation
- CYC1
Cytochrome C1
- DRP1
Dynamin-related protein 1
- ECSIT
Evolutionarily conserved signalling intermediate in Toll pathways
- ELISA
Enzyme-linked immunosorbent assay
- ER
Endoplasmic reticulum
- FIS1
Mitochondrial fission 1 protein
- GRP75
Glucose-regulated protein 75
- HPT
Haptoglobin
- IMM
Inner mitochondrial membrane
- IP3R1
Inositol 1,4,5-trisphosphate receptor 1
- LC–MS
Liquid chromatography–mass spectrometry
- LDL
Low-density lipoprotein
- LDLR
Low-density lipoprotein receptor
- MAMs
Mitochondria-associated ER membranes
- MFN1/2
Mitofusin 1/2
- MLS
Mitochondrial localization sequence
- mPTP
Mitochondrial permeability transition pore
- MS
Mass spectrometry
- NLS
Nuclear localization sequence
- NOS1
Nitric oxide synthase 1
- OMM
Outer mitochondrial membrane
- OPA1
Dynamin-like 120 kDa protein
- OXPHOS
Oxidative phosphorylation
- PC
Phosphatidylcholine
- PDCD4
Programmed cell death 4
- PE
Phosphatidylethanolamine
- PEMT
Phosphatidylethanolamine methyltransferase
- PGC1α
Peroxisome proliferator-activated receptor gamma coactivator 1-α
- PS
Phosphatidylserine
- PSD
Phosphatidylserine decarboxylase
- PSS1/2
Phosphatidylserine synthase 1/2
- SIRT3
Sirtuin 3
- SNCA
Synuclein α
- ST13
Suppression of tumorigenicity 13 protein
- TCA
Tricarboxylic acid cycle
- TIM
Translocase of the inner mitochondrial membrane
- TOM
Translocase of the outer mitochondrial membrane
- TR
Targeted replacement
- UQCRC1/2
Cytochrome b–c1 complex subunit 1/2
- VDAC1
Voltage-dependent anion-selective channel 1
- VLDL
Very low-density lipoprotein
- WB
Western blot
Author contributions
JR, PH, and GR wrote and revised the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. This study was supported by the Deutsche Forschungsgemeinschaft (project number 448478889).
Availability of data and material
Not applicable.
Declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261(5123):921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 2.Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, et al. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90(5):1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolters FJ, Yang Q, Biggs ML, Jakobsdottir J, Li S, Evans DS, et al. The impact of APOE genotype on survival: results of 38,537 participants from six population-based cohorts (E2-CHARGE) PLoS ONE. 2019;14(7):e0219668. doi: 10.1371/journal.pone.0219668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu M, Kuhel DG, Shen L, Hui DY, Woods SC. Apolipoprotein E does not cross the blood-cerebrospinal fluid barrier, as revealed by an improved technique for sampling CSF from mice. Am J Physiol Regul Integr Comp Physiol. 2012;303(9):R903–R908. doi: 10.1152/ajpregu.00219.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martínez-Morillo E, Hansson O, Atagi Y, Bu G, Minthon L, Diamandis EP, et al. Total apolipoprotein E levels and specific isoform composition in cerebrospinal fluid and plasma from Alzheimer’s disease patients and controls. Acta Neuropathol. 2014;127(5):633–643. doi: 10.1007/s00401-014-1266-2. [DOI] [PubMed] [Google Scholar]
- 6.Flowers SA, Grant OC, Woods RJ, Rebeck GW. O-glycosylation on cerebrospinal fluid and plasma apolipoprotein E differs in the lipid-binding domain. Glycobiology. 2020;30(2):74–85. doi: 10.1093/glycob/cwz084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oriá RB, Patrick PD, Oriá MOB, Lorntz B, Thompson MR, Azevedo OGR, et al. ApoE polymorphisms and diarrheal outcomes in Brazilian shanty town children. Braz J Med Biol Res. 2010;43(3):249–256. doi: 10.1590/s0100-879x2010007500003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuhlmann I, Minihane AM, Huebbe P, Nebel A, Rimbach G. Apolipoprotein E genotype and hepatitis C, HIV and herpes simplex disease risk: a literature review. Lipids Health Dis. 2010;9:8. doi: 10.1186/1476-511X-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huebbe P, Nebel A, Siegert S, Moehring J, Boesch-Saadatmandi C, Most E, et al. APOE ε4 is associated with higher vitamin D levels in targeted replacement mice and humans. FASEB J. 2011;25(9):3262–3270. doi: 10.1096/fj.11-180935. [DOI] [PubMed] [Google Scholar]
- 10.Jofre-Monseny L, Loboda A, Wagner AE, Huebbe P, Boesch-Saadatmandi C, Jozkowicz A, et al. Effects of apoE genotype on macrophage inflammation and heme oxygenase-1 expression. Biochem Biophys Res Commun. 2007;357(1):319–324. doi: 10.1016/j.bbrc.2007.03.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dose J, Schloesser A, Torres GG, Venkatesh G, Häsler R, Flachsbart F, et al. On a Western diet, APOE ɛ4 is associated with low innate immune sensing, but not APOE ɛ3. J Allergy Clin Immunol. 2018;142(4):1346–1349.e9. doi: 10.1016/j.jaci.2018.05.033. [DOI] [PubMed] [Google Scholar]
- 12.Chin D, Hagl S, Hoehn A, Huebbe P, Pallauf K, Grune T, et al. Adenosine triphosphate concentrations are higher in the brain of APOE3- compared to APOE4-targeted replacement mice and can be modulated by curcumin. Genes Nutr. 2014;9(3):397. doi: 10.1007/s12263-014-0397-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huebbe P, Lange J, Lietz G, Rimbach G. Dietary beta-carotene and lutein metabolism is modulated by the APOE genotype. BioFactors. 2016;42(4):388–396. doi: 10.1002/biof.1284. [DOI] [PubMed] [Google Scholar]
- 14.Egert S, Rimbach G, Huebbe P. ApoE genotype: from geographic distribution to function and responsiveness to dietary factors. Proc Nutr Soc. 2012;71(3):410–424. doi: 10.1017/S0029665112000249. [DOI] [PubMed] [Google Scholar]
- 15.Theendakara V, Peters-Libeu CA, Spilman P, Poksay KS, Bredesen DE, Rao RV. Direct transcriptional effects of apolipoprotein E. J Neurosci. 2016;36(3):685–700. doi: 10.1523/JNEUROSCI.3562-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamilton RL, Wong JS, Guo LS, Krisans S, Havel RJ. Apolipoprotein E localization in rat hepatocytes by immunogold labeling of cryothin sections. J Lipid Res. 1990;31(9):1589–1603. doi: 10.1016/S0022-2275(20)42343-0. [DOI] [PubMed] [Google Scholar]
- 17.Vance JE. Phospholipid synthesis in a membrane fraction associated with mitochondria. J Biol Chem. 1990;265(13):7248–7256. doi: 10.1016/S0021-9258(19)39106-9. [DOI] [PubMed] [Google Scholar]
- 18.Rall SC, Weisgraber KH, Mahley RW. Human apolipoprotein E. The complete amino acid sequence. J Biol Chem. 1982;257(8):4171–4178. doi: 10.1016/S0021-9258(18)34702-1. [DOI] [PubMed] [Google Scholar]
- 19.Weisgraber KH, Rall SC, Jr, Mahley RW. Human E apoprotein heterogeneity. Cysteine-arginine interchanges in the amino acid sequence of the apo-E isoforms. J Biol Chem. 1981;256(17):9077–9083. doi: 10.1016/S0021-9258(19)52510-8. [DOI] [PubMed] [Google Scholar]
- 20.McIntosh AM, Bennett C, Dickson D, Anestis SF, Watts DP, Webster TH, et al. The apolipoprotein E (APOE) gene appears functionally monomorphic in chimpanzees (Pan troglodytes) PLoS ONE. 2012;7(10):e47760. doi: 10.1371/journal.pone.0047760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huebbe P, Rimbach G. Evolution of human apolipoprotein E (APOE) isoforms: gene structure, protein function and interaction with dietary factors. Ageing Res Rev. 2017;37:146–161. doi: 10.1016/j.arr.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 22.Frieden C, Garai K. Structural differences between apoE3 and apoE4 may be useful in developing therapeutic agents for Alzheimer's disease. Proc Natl Acad Sci U S A. 2012;109(23):8913–8918. doi: 10.1073/pnas.1207022109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizuguchi C, Hata M, Dhanasekaran P, Nickel M, Okuhira K, Phillips MC, et al. Fluorescence study of domain structure and lipid interaction of human apolipoproteins E3 and E4. Biochim Biophys Acta. 2014;1841(12):1716–1724. doi: 10.1016/j.bbalip.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morrow JA, Segall ML, Lund-Katz S, Phillips MC, Knapp M, Rupp B, et al. Differences in stability among the human apolipoprotein E isoforms determined by the amino-terminal domain. Biochemistry. 2000;39(38):11657–11666. doi: 10.1021/bi000099m. [DOI] [PubMed] [Google Scholar]
- 25.Clément-Collin V, Barbier A, Dergunov AD, Visvikis A, Siest G, Desmadril M, et al. The structure of human apolipoprotein E2, E3 and E4 in solution. 2. Multidomain organization correlates with the stability of apoE structure. Biophys Chem. 2006;119(2):170–185. doi: 10.1016/j.bpc.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 26.Garai K, Mustafi SM, Baban B, Frieden C. Structural differences between apolipoprotein E3 and E4 as measured by (19)F NMR. Protein Sci. 2010;19(1):66–74. doi: 10.1002/pro.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panin LE, Russkikh GS, Polyakov LM. Detection of apolipoprotein A-I, B, and E immunoreactivity in the nuclei of various rat tissue cells. Biochem Mosc. 2000;65(12):1419–1423. doi: 10.1023/a:1002861008363. [DOI] [PubMed] [Google Scholar]
- 28.James R, Searcy JL, Le Bihan T, Martin SF, Gliddon CM, Povey J, et al. Proteomic analysis of mitochondria in APOE transgenic mice and in response to an ischemic challenge. J Cereb Blood Flow Metab. 2012;32(1):164–176. doi: 10.1038/jcbfm.2011.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmukler E, Solomon S, Simonovitch S, Goldshmit Y, Wolfson E, Michaelson DM, et al. Altered mitochondrial dynamics and function in APOE4-expressing astrocytes. Cell Death Dis. 2020;11(7):578. doi: 10.1038/s41419-020-02776-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sala-Vila A, Navarro-Lérida I, Sánchez-Alvarez M, Bosch M, Calvo C, López JA, et al. Interplay between hepatic mitochondria-associated membranes, lipid metabolism and caveolin-1 in mice. Sci Rep. 2016;6:27351. doi: 10.1038/srep27351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cortés-Sanchón A, Santhosh-Kumar H, Mantovani M, Osinnii I, Mateos JM, Kaech A, et al. ER-misfolded proteins become sequestered with mitochondria and impair mitochondrial function. Commun Biol. 2021;4(1):1350. doi: 10.1038/s42003-021-02873-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang S, Ran-Ma T, Miranda RD, Balestra ME, Mahley RW, Huang Y. Lipid- and receptor-binding regions of apolipoprotein E4 fragments act in concert to cause mitochondrial dysfunction and neurotoxicity. Proc Natl Acad Sci U S A. 2005;102(51):18694–18699. doi: 10.1073/pnas.0508254102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakamura T, Watanabe A, Fujino T, Hosono T, Michikawa M. Apolipoprotein E4 (1–272) fragment is associated with mitochondrial proteins and affects mitochondrial function in neuronal cells. Mol Neurodegener. 2009;4:35. doi: 10.1186/1750-1326-4-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poston CN, Krishnan SC, Bazemore-Walker CR. In-depth proteomic analysis of mammalian mitochondria-associated membranes (MAM) J Proteomics. 2013;79:219–230. doi: 10.1016/j.jprot.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 35.Horner SM, Wilkins C, Badil S, Iskarpatyoti J, Gale M, Wang T. Proteomic analysis of mitochondrial-associated ER membranes (MAM) during RNA virus infection reveals dynamic changes in protein and organelle trafficking. PLoS ONE. 2015;10(3):e0117963. doi: 10.1371/journal.pone.0117963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma JH, Shen S, Wang JJ, He Z, Poon A, Li J, et al. Comparative proteomic analysis of the mitochondria-associated ER membrane (MAM) in a long-term type 2 diabetic rodent model. Sci Rep. 2017;7(1):2062. doi: 10.1038/s41598-017-02213-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anastasia I, Ilacqua N, Raimondi A, Lemieux P, Ghandehari-Alavijeh R, Faure G, et al. Mitochondria-rough-ER contacts in the liver regulate systemic lipid homeostasis. Cell Rep. 2021;34(11):108873. doi: 10.1016/j.celrep.2021.108873. [DOI] [PubMed] [Google Scholar]
- 38.Muñoz SS, Garner B, Ooi L. Understanding the role of ApoE fragments in Alzheimer's disease. Neurochem Res. 2018 doi: 10.1007/s11064-018-2629-1. [DOI] [PubMed] [Google Scholar]
- 39.Marques MA, Tolar M, Harmony JA, Crutcher KA. A thrombin cleavage fragment of apolipoprotein E exhibits isoform-specific neurotoxicity. NeuroReport. 1996;7(15–17):2529–2532. doi: 10.1097/00001756-199611040-00025. [DOI] [PubMed] [Google Scholar]
- 40.Bien-Ly N, Andrews-Zwilling Y, Xu Q, Bernardo A, Wang C, Huang Y. C-terminal-truncated apolipoprotein (apo) E4 inefficiently clears amyloid-beta (Abeta) and acts in concert with Abeta to elicit neuronal and behavioral deficits in mice. Proc Natl Acad Sci U S A. 2011;108(10):4236–4241. doi: 10.1073/pnas.1018381108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harris FM, Brecht WJ, Xu Q, Tesseur I, Kekonius L, Wyss-Coray T, et al. Carboxyl-terminal-truncated apolipoprotein E4 causes Alzheimer's disease-like neurodegeneration and behavioral deficits in transgenic mice. Proc Natl Acad Sci U S A. 2003;100(19):10966–10971. doi: 10.1073/pnas.1434398100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang T, Hang W, Chen J, Wu Y, Wen B, Xu K, et al. ApoE4 (Δ272–299) induces mitochondrial-associated membrane formation and mitochondrial impairment by enhancing GRP75-modulated mitochondrial calcium overload in neuron. Cell Biosci. 2021;11(1):50. doi: 10.1186/s13578-021-00563-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Love JE, Day RJ, Gause JW, Brown RJ, Pu X, Theis DI, et al. Nuclear uptake of an amino-terminal fragment of apolipoprotein E4 promotes cell death and localizes within microglia of the Alzheimer's disease brain. Int J Physiol Pathophysiol Pharmacol. 2017;9(2):40–57. [PMC free article] [PubMed] [Google Scholar]
- 44.Rohn TT, Moore ZD. Nuclear localization of apolipoprotein E4: a new trick for an old protein. Int J Neurol Neurother. 2017 doi: 10.23937/2378-3001/1410067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Farkas MH, Weisgraber KH, Shepherd VL, Linton MF, Fazio S, Swift LL. The recycling of apolipoprotein E and its amino-terminal 22 kDa fragment: evidence for multiple redundant pathways. J Lipid Res. 2004;45(8):1546–1554. doi: 10.1194/jlr.M400104-JLR200. [DOI] [PubMed] [Google Scholar]
- 46.Azuma M, Kojimab T, Yokoyama I, Tajiri H, Yoshikawa K, Saga S, et al. A synthetic peptide of human apoprotein E with antibacterial activity. Peptides. 2000;21(3):327–330. doi: 10.1016/S0196-9781(00)00165-0. [DOI] [PubMed] [Google Scholar]
- 47.Pane K, Sgambati V, Zanfardino A, Smaldone G, Cafaro V, Angrisano T, et al. A new cryptic cationic antimicrobial peptide from human apolipoprotein E with antibacterial activity and immunomodulatory effects on human cells. FEBS J. 2016;283(11):2115–2131. doi: 10.1111/febs.13725. [DOI] [PubMed] [Google Scholar]
- 48.Gause JW, Day RJ, Caraway CA, Poon WW, Rohn TT. Evaluation of apolipoprotein E fragmentation as a biomarker for Alzheimer's disease. J. Neurol Neurol Disord. 2017 doi: 10.15744/2454-4981.3.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wiedemann N, Pfanner N. Mitochondrial machineries for protein import and assembly. Annu Rev Biochem. 2017;86(1):685–714. doi: 10.1146/annurev-biochem-060815-014352. [DOI] [PubMed] [Google Scholar]
- 50.Ellenrieder L, Dieterle MP, Doan KN, Mårtensson CU, Floerchinger A, Campo ML, et al. Dual role of mitochondrial porin in metabolite transport across the outer membrane and protein transfer to the inner membrane. Mol Cell. 2019;73(5):1056–1065.e7. doi: 10.1016/j.molcel.2018.12.014. [DOI] [PubMed] [Google Scholar]
- 51.Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, et al. The BioPlex network: a systematic exploration of the human interactome. Cell. 2015;162(2):425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen W-Y, Chen Y-F, Chan H-C, Chung C-H, Peng H-Y, Ho Y-C, et al. Role of apolipoprotein E in electronegative low-density lipoprotein-induced mitochondrial dysfunction in cardiomyocytes. Metabolism. 2020;107:154227. doi: 10.1016/j.metabol.2020.154227. [DOI] [PubMed] [Google Scholar]
- 53.Dose J, Nebel A, Piegholdt S, Rimbach G, Huebbe P. Influence of the APOE genotype on hepatic stress response: studies in APOE targeted replacement mice and human liver cells. Free Radic Biol Med. 2016;96:264–272. doi: 10.1016/j.freeradbiomed.2016.04.031. [DOI] [PubMed] [Google Scholar]
- 54.Qi G, Mi Y, Shi X, Gu H, Brinton RD, Yin F. ApoE4 impairs neuron-astrocyte coupling of fatty acid metabolism. Cell Rep. 2021;34(1):108572. doi: 10.1016/j.celrep.2020.108572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yin J, Nielsen M, Carcione T, Li S, Shi J. Apolipoprotein E regulates mitochondrial function through the PGC-1α-sirtuin 3 pathway. Aging (Albany NY) 2019;11(23):11148–11156. doi: 10.18632/aging.102516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marottoli FM, Trevino TN, Geng X, Arbieva Z, Kanabar P, Maienschein-Cline M, et al. Autocrine effects of brain endothelial cell-produced human apolipoprotein E on metabolism and inflammation in vitro. Front Cell Dev Biol. 2021;9:668296. doi: 10.3389/fcell.2021.668296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Orr AL, Kim C, Jimenez-Morales D, Newton BW, Johnson JR, Krogan NJ, et al. Neuronal apolipoprotein E4 expression results in proteome-wide alterations and compromises bioenergetic capacity by disrupting mitochondrial function. J Alzheimers Dis. 2019;68(3):991–1011. doi: 10.3233/JAD-181184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen H-K, Ji Z-S, Dodson SE, Miranda RD, Rosenblum CI, Reynolds IJ, et al. Apolipoprotein E4 domain interaction mediates detrimental effects on mitochondria and is a potential therapeutic target for Alzheimer disease. J Biol Chem. 2011;286(7):5215–5221. doi: 10.1074/jbc.M110.151084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Area-Gomez E, Larrea D, Pera M, Agrawal RR, Guilfoyle DN, Pirhaji L, et al. APOE4 is associated with differential regional vulnerability to bioenergetic deficits in aged APOE mice. Sci Rep. 2020;10(1):4277. doi: 10.1038/s41598-020-61142-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Giacomello M, Pyakurel A, Glytsou C, Scorrano L. The cell biology of mitochondrial membrane dynamics. Nat Rev Mol Cell Biol. 2020;21(4):204–224. doi: 10.1038/s41580-020-0210-7. [DOI] [PubMed] [Google Scholar]
- 61.Simonovitch S, Schmukler E, Masliah E, Pinkas-Kramarski R, Michaelson DM. The effects of APOE4 on mitochondrial dynamics and proteins in vivo. J Alzheimer’s Dis. 2019;70(3):861–875. doi: 10.3233/JAD-190074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yin J, Reiman EM, Beach TG, Serrano GE, Sabbagh MN, Nielsen M, et al. Effect of ApoE isoforms on mitochondria in Alzheimer disease. Neurology. 2020;94(23):e2404–e2411. doi: 10.1212/WNL.0000000000009582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suski M, Olszanecki R, Madej J, Totoń-Żurańska J, Niepsuj A, Jawień J, et al. Proteomic analysis of changes in protein expression in liver mitochondria in apoE knockout mice. J Proteomics. 2011;74(6):887–893. doi: 10.1016/j.jprot.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 64.Stachowicz A, Olszanecki R, Suski M, Głombik K, Basta-Kaim A, Adamek D, et al. Proteomic analysis of mitochondria-enriched fraction isolated from the frontal cortex and hippocampus of apolipoprotein E knockout mice treated with Alda-1, an activator of mitochondrial aldehyde dehydrogenase (ALDH2) Int J Mol Sci. 2017;18(2):435. doi: 10.3390/ijms18020435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shi L, Du X, Zhou H, Tao C, Liu Y, Meng F, et al. Cumulative effects of the ApoE genotype and gender on the synaptic proteome and oxidative stress in the mouse brain. Int J Neuropsychopharmacol. 2014;17(11):1863–1879. doi: 10.1017/S1461145714000601. [DOI] [PubMed] [Google Scholar]
- 66.Osorio C, Sullivan PM, He DN, Mace BE, Ervin JF, Strittmatter WJ, et al. Mortalin is regulated by APOE in hippocampus of AD patients and by human APOE in TR mice. Neurobiol Aging. 2007;28(12):1853–1862. doi: 10.1016/j.neurobiolaging.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 67.Tambini MD, Pera M, Kanter E, Yang H, Guardia-Laguarta C, Holtzman D, et al. ApoE4 upregulates the activity of mitochondria-associated ER membranes. EMBO Rep. 2016;17(1):27–36. doi: 10.15252/embr.201540614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Giacomello M, Pellegrini L. The coming of age of the mitochondria–ER contact: a matter of thickness. Cell Death Differ. 2016;23(9):1417–1427. doi: 10.1038/cdd.2016.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stone SJ, Vance JE. Phosphatidylserine synthase-1 and -2 are localized to mitochondria-associated membranes. J Biol Chem. 2000;275(44):34534–34540. doi: 10.1074/jbc.M002865200. [DOI] [PubMed] [Google Scholar]
- 70.Zhang A, Williamson CD, Wong DS, Bullough MD, Brown KJ, Hathout Y, et al. Quantitative proteomic analyses of human cytomegalovirus-induced restructuring of endoplasmic reticulum-mitochondrial contacts at late times of infection. Mol Cell Proteomics. 2011 doi: 10.1074/mcp.M111.009936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tubbs E, Theurey P, Vial G, Bendridi N, Bravard A, Chauvin M-A, et al. Mitochondria-associated endoplasmic reticulum membrane (MAM) integrity is required for insulin signaling and is implicated in hepatic insulin resistance. Diabetes. 2014;63(10):3279–3294. doi: 10.2337/db13-1751. [DOI] [PubMed] [Google Scholar]
- 72.Theurey P, Tubbs E, Vial G, Jacquemetton J, Bendridi N, Chauvin M-A, et al. Mitochondria-associated endoplasmic reticulum membranes allow adaptation of mitochondrial metabolism to glucose availability in the liver. J Mol Cell Biol. 2016;8(2):129–143. doi: 10.1093/jmcb/mjw004. [DOI] [PubMed] [Google Scholar]
- 73.Janikiewicz J, Szymański J, Malinska D, Patalas-Krawczyk P, Michalska B, Duszyński J, et al. Mitochondria-associated membranes in aging and senescence: structure, function, and dynamics. Cell Death Dis. 2018;9(3):332. doi: 10.1038/s41419-017-0105-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456(7222):605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 75.Ilacqua N, Sánchez-Álvarez M, Bachmann M, Costiniti V, Del Pozo MA, Giacomello M. Protein localization at mitochondria-ER contact sites in basal and stress conditions. Front Cell Dev Biol. 2017;5:107. doi: 10.3389/fcell.2017.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang Z, Zhao X, Xu J, Shang W, Tong C. A novel fluorescent reporter detects plastic remodeling of mitochondria-ER contact sites. J Cell Sci. 2018;131(1):jcs208686. doi: 10.1242/jcs.208686. [DOI] [PubMed] [Google Scholar]
- 77.Cui Z, Vance JE, Chen MH, Voelker DR, Vance DE. Cloning and expression of a novel phosphatidylethanolamine N-methyltransferase. A specific biochemical and cytological marker for a unique membrane fraction in rat liver. J Biol Chem. 1993;268(22):16655–16663. doi: 10.1016/S0021-9258(19)85468-6. [DOI] [PubMed] [Google Scholar]
- 78.Baughman JM, Perocchi F, Girgis HS, Plovanich M, Belcher-Timme CA, Sancak Y, et al. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature. 2011;476(7360):341–345. doi: 10.1038/nature10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Szabadkai G, Bianchi K, Várnai P, de Stefani D, Wieckowski MR, Cavagna D, et al. Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J Cell Biol. 2006;175(6):901–911. doi: 10.1083/jcb.200608073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.de Stefani D, Bononi A, Romagnoli A, Messina A, de Pinto V, Pinton P, et al. VDAC1 selectively transfers apoptotic Ca2+ signals to mitochondria. Cell Death Differ. 2012;19(2):267–273. doi: 10.1038/cdd.2011.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tubbs E, Chanon S, Robert M, Bendridi N, Bidaux G, Chauvin M-A, et al. Disruption of mitochondria-associated endoplasmic reticulum membrane (MAM) integrity contributes to muscle insulin resistance in mice and humans. Diabetes. 2018;67(4):636–650. doi: 10.2337/db17-0316. [DOI] [PubMed] [Google Scholar]
- 82.Area-Gomez E, de Groof AJC, Boldogh I, Bird TD, Gibson GE, Koehler CM, et al. Presenilins are enriched in endoplasmic reticulum membranes associated with mitochondria. Am J Pathol. 2009;175(5):1810–1816. doi: 10.2353/ajpath.2009.090219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Area-Gomez E, Del Carmen-Lara-Castillo M, Tambini MD, Guardia-Laguarta C, de Groof AJC, Madra M, et al. Upregulated function of mitochondria-associated ER membranes in Alzheimer disease. EMBO J. 2012;31(21):4106–4123. doi: 10.1038/emboj.2012.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hedskog L, Pinho CM, Filadi R, Rönnbäck A, Hertwig L, Wiehager B, et al. Modulation of the endoplasmic reticulum-mitochondria interface in Alzheimer's disease and related models. Proc Natl Acad Sci U S A. 2013;110(19):7916–7921. doi: 10.1073/pnas.1300677110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang X, Wen Y, Dong J, Cao C, Yuan S. Systematic In-depth proteomic analysis of mitochondria-associated endoplasmic reticulum membranes in mouse and human testes. Proteomics. 2018;18(14):e1700478. doi: 10.1002/pmic.201700478. [DOI] [PubMed] [Google Scholar]
- 86.Londono C, Osorio C, Gama V, Alzate O. Mortalin, apoptosis, and neurodegeneration. Biomolecules. 2012;2(1):143–164. doi: 10.3390/biom2010143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rostovtseva TK, Sheldon KL, Hassanzadeh E, Monge C, Saks V, Bezrukov SM, et al. Tubulin binding blocks mitochondrial voltage-dependent anion channel and regulates respiration. Proc Natl Acad Sci U S A. 2008;105(48):18746–18751. doi: 10.1073/pnas.0806303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Venkatesan K, Rual J-F, Vazquez A, Stelzl U, Lemmens I, Hirozane-Kishikawa T, et al. An empirical framework for binary interactome mapping. Nat Methods. 2009;6(1):83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Weisgraber KH, Innerarity TL, Harder KJ, Mahley RW, Milne RW, Marcel YL, et al. The receptor-binding domain of human apolipoprotein E. Monoclonal antibody inhibition of binding. J Biol Chem. 1983;258(20):12348–12354. doi: 10.1016/S0021-9258(17)44181-0. [DOI] [PubMed] [Google Scholar]
- 90.Chen J, Li Q, Wang J. Topology of human apolipoprotein E3 uniquely regulates its diverse biological functions. Proc Natl Acad Sci U S A. 2011;108(36):14813–14818. doi: 10.1073/pnas.1106420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Williams B, II, Convertino M, Das J, Dokholyan NV. ApoE4-specific misfolded intermediate identified by molecular dynamics simulations. PLoS Comput Biol. 2015;11(10):e1004359. doi: 10.1371/journal.pcbi.1004359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huttlin EL, Bruckner RJ, Paulo JA, Cannon JR, Ting L, Baltier K, et al. Architecture of the human interactome defines protein communities and disease networks. Nature. 2017;545(7655):505–509. doi: 10.1038/nature22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Huttlin EL, Bruckner RJ, Navarrete-Perea J, Cannon JR, Baltier K, Gebreab F, et al. Dual proteome-scale networks reveal cell-specific remodeling of the human interactome. Cell. 2021;184(11):3022–3040.e28. doi: 10.1016/j.cell.2021.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Moutaoufik MT, Malty R, Amin S, Zhang Q, Phanse S, Gagarinova A, et al. Rewiring of the human mitochondrial interactome during neuronal reprogramming reveals regulators of the respirasome and neurogenesis. iScience. 2019;19:1114–1132. doi: 10.1016/j.isci.2019.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hass S, Fresser F, Köchl S, Beyreuther K, Utermann G, Baier G. Physical interaction of ApoE with amyloid precursor protein independent of the amyloid Abeta region in vitro. J Biol Chem. 1998;273(22):13892–13897. doi: 10.1016/S0021-9258(19)57838-3. [DOI] [PubMed] [Google Scholar]
- 96.Calero M, Rostagno A, Ghiso J. Search for amyloid-binding proteins by affinity chromatography. Methods Mol Biol. 2012;849:213–223. doi: 10.1007/978-1-61779-551-0_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Soler-López M, Zanzoni A, Lluís R, Stelzl U, Aloy P. Interactome mapping suggests new mechanistic details underlying Alzheimer's disease. Genome Res. 2011;21(3):364–376. doi: 10.1101/gr.114280.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Emamzadeh FN, Allsop D. α-Synuclein interacts with lipoproteins in plasma. J Mol Neurosci. 2017;63(2):165–172. doi: 10.1007/s12031-017-0967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Paslawski W, Zareba-Paslawska J, Zhang X, Hölzl K, Wadensten H, Shariatgorji M, et al. α-synuclein-lipoprotein interactions and elevated ApoE level in cerebrospinal fluid from Parkinson's disease patients. Proc Natl Acad Sci U S A. 2019;116(30):15226–15235. doi: 10.1073/pnas.1821409116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cigliano L, Pugliese CR, Spagnuolo MS, Palumbo R, Abrescia P. Haptoglobin binds the antiatherogenic protein apolipoprotein E - impairment of apolipoprotein E stimulation of both lecithin:cholesterol acyltransferase activity and cholesterol uptake by hepatocytes. FEBS J. 2009;276(21):6158–6171. doi: 10.1111/j.1742-4658.2009.07319.x. [DOI] [PubMed] [Google Scholar]
- 101.Salvatore A, Cigliano L, Carlucci A, Bucci EM, Abrescia P. Haptoglobin binds apolipoprotein E and influences cholesterol esterification in the cerebrospinal fluid. J Neurochem. 2009;110(1):255–263. doi: 10.1111/j.1471-4159.2009.06121.x. [DOI] [PubMed] [Google Scholar]
- 102.Spagnuolo MS, Maresca B, La Marca V, Carrizzo A, Veronesi C, Cupidi C, et al. Haptoglobin interacts with apolipoprotein E and beta-amyloid and influences their crosstalk. ACS Chem Neurosci. 2014;5(9):837–847. doi: 10.1021/cn500099f. [DOI] [PubMed] [Google Scholar]
- 103.Krimbou L, Tremblay M, Davignon J, Cohn JS. Association of apolipoprotein E with alpha2-macroglobulin in human plasma. J Lipid Res. 1998;39(12):2373–2386. doi: 10.1016/S0022-2275(20)33316-2. [DOI] [PubMed] [Google Scholar]
- 104.Krimbou L, Denis M, Haidar B, Carrier M, Marcil M, Genest J. Molecular interactions between apoE and ABCA1: impact on apoE lipidation. J Lipid Res. 2004;45(5):839–848. doi: 10.1194/jlr.M300418-JLR200. [DOI] [PubMed] [Google Scholar]
- 105.Rawat V, Wang S, Sima J, Bar R, Liraz O, Gundimeda U, et al. ApoE4 alters ABCA1 membrane trafficking in astrocytes. J Neurosci. 2019;39(48):9611–9622. doi: 10.1523/JNEUROSCI.1400-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Abrams AJ, Farooq A, Wang G. S-nitrosylation of ApoE in Alzheimer's disease. Biochemistry. 2011;50(17):3405–3407. doi: 10.1021/bi200266v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Beisiegel U, Weber W, Havinga JR, Ihrke G, Hui DY, Wernette-Hammond ME, et al. Apolipoprotein E-binding proteins isolated from dog and human liver. Arteriosclerosis. 1988;8(3):288–297. doi: 10.1161/01.atv.8.3.288. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.