Abstract
Dal82p binds to the UISALL sites of allophanate-induced genes of the allantoin-degradative pathway and functions synergistically with the GATA family Gln3p and Gat1p transcriptional activators that are responsible for nitrogen catabolite repression-sensitive gene expression. CAR2, which encodes the arginine-degradative enzyme ornithine transaminase, is not nitrogen catabolite repression sensitive, but its expression can be modestly induced by the allantoin pathway inducer. The dominant activators of CAR2 transcription have been thought to be the ArgR and Mcm1 factors, which mediate arginine-dependent induction. These observations prompted us to investigate the structure of the CAR2 promoter with the objectives of determining whether other transcription factors were required for CAR2 expression and, if so, of ascertaining their relative contributions to CAR2’s expression and control. We show that Rap1p binds upstream of CAR2 and plays a central role in its induced expression irrespective of whether the inducer is arginine or the allantoin pathway inducer analogue oxalurate (OXLU). Our data also explain the early report that ornithine transaminase production is induced when cells are grown with urea. OXLU induction derives from the Dal82p binding site, which is immediately downstream of the Rap1p site, and Dal82p functions synergistically with Rap1p. This synergism is unlike all other known instances of Dal82p synergism, namely, that with the GATA family transcription activators Gln3p and Gat1p, which occurs only in the presence of an inducer. The observations reported suggest that CAR2 gene expression results from strong constitutive transcriptional activation mediated by Rap1p and Dal82p being balanced by the down regulation of an equally strong transcriptional repressor, Ume6p. This balance is then tipped in the direction of expression by the presence of the inducer. The formal structure of the CAR2 promoter and its operation closely follow the model proposed for CAR1.
All nitrogen catabolite repression (NCR)-sensitive genes in Saccharomyces cerevisiae have GATA sequences in their promoters, and their expression is dependent upon one or both of the GATA family transcriptional activators Gln3p and/or Gat1p (also called Nil1p) (for a brief review of the GATA factor field, see the introductions of references 7 and 52). Gln3p has been shown to bind to the GATA sequences, and both Gln3p and Gat1p have the ability to activate transcription of a reporter gene in a lexAp-tethering assay system (7, 52). Expression of the genes encoding components of the arginine- and allantoin-degradative pathways depends upon Gln3p and Gat1p working synergistically with pathway-specific transcription factors (47, 61). Arginine and allantoin pathway gene expression is also subject to induction in response to the presence of small signal metabolites (8, 9, 23, 37–40, 59, 60). In the case of the arginine pathway genes (CAR genes), the inducer is arginine (59) and the transcription factors involved are ArgRIp, ArgRIIp, ArgRIIIp, and Mcm1p. For the allantoin pathway genes (DAL and DUR genes), the inducer is allophanate or its nonmetabolized analogue, oxalurate (OXLU), and the associated transcription factors are Dal82p and Dal81p (8, 9).
The CAR1 promoter has been extensively studied both biochemically and genetically (2–4, 11, 12, 14, 18, 23, 25, 59). It consists of up to 14 cis-acting sites, which bind nine trans-acting factors, Rap1p, Abf1p, Gln3p, Gat1p, ArgRIp, ArgRIIp, ArgRIIIp, Mcm1p, and Ume6p (3, 10, 13, 19, 27–29, 31, 34–36, 42–44, 47, 51, 58). Proteins that bind to the Rap1 and Abf1 sites and a GC-rich sequence are necessary to achieve highly induced transcription levels of CAR1 (27–29, 47). The action of these strong, constitutively acting transcription factors is antagonized by the equally strong negative action of Ume6p (also called Car80p) bound to the URS1 site (1, 3, 42, 47–51). The metabolite-responsive promoter elements are much weaker than those just cited (47) and have been suggested to tip the balance either toward expression when the inducer is present and a repressing nitrogen source is absent or toward quiescence when arginine is absent and/or a repressive nitrogen source such as glutamine is present (47, 50, 51). Three cis-acting sites, which are associated with the ArgRI, ArgRII, ArgRIII, and Mcm1 proteins, mediate arginine-dependent induction (10, 11, 13, 14, 18, 19, 34–36). NCR-sensitive CAR1 expression, on the other hand, is mediated by two GATA sequences associated with Gln3p and Gat1p (47).
Although arginase and ornithine transaminase (encoded by CAR1 and CAR2) catalyze contiguous enzyme reactions in the arginine-degradative pathway of S. cerevisiae, production of the two enzymes is regulated somewhat differently (37–40, 59, 60). While arginine induces production of both arginase and ornithine transaminase, only arginase production is NCR sensitive (14, 37–40, 59, 60). Production of ornithine transaminase, on the other hand, is induced by allophanate, the last intermediate of the allantoin-degradative pathway, while CAR1 expression is not (23). Unlike that of CAR1, the CAR2 promoter has not been characterized.
The allantoin pathway gene promoters appear to be simpler than those in the arginine pathway (8, 9). For example, the DAL7 promoter contains three types of sites, each of which is associated with a different type of regulation. Multiple GATA sequences mediate NCR-sensitive gene expression that depends upon Gln3p and Gat1p (7–9, 51). DAL7 expression is down regulated by the action of Dal80p, which antagonizes transcriptional activation, mediated by Gln3p and Gat1p (9). Finally, inducer-dependent (allophanate or its nonmetabolized analogue OXLU) expression depends upon the UISALL site; Dal82p binds to this site and Dal81p functions in association with it (8, 9, 17). A notable similarity exists in the formal organizations of the CAR and DAL promoters; the action of transcriptional activators is antagonized by repressor proteins, and the balance is shifted by the inducer-responsive transcription factor (11a, 47).
In all cases studied so far, UISALL and Dal82p function only in association with the GATA factors Gln3p and Gat1p (9). Further, genetic data have raised the possibility that Gln3p and/or Gat1p may interact directly with Dal82p (57). A UISALL site placed adjacent to a mutated GATA-containing UASNTR site will suppress its mutant phenotype. This suppression depends upon the presence of wild-type Gln3p and Dal82p and correlates with Dal82p binding to UISALL (57). It does not, however, depend upon Dal81p or an inducer. Dal81p does not appear to bind to DAL promoters (4a, 56).
This work has three objectives, namely, to (i) identify the promoter elements of CAR2 responsible for allophanate-induced expression; (ii) determine whether the overall structure and operation of CAR2 is analogous to those features of CAR1, i.e., whether CAR2 contains strong, opposing positive and negative control elements, with the metabolite-responsive elements shifting the balance; and (iii) determine whether Dal82p can function in association with global transcription factors other than Gln3p and/or Gat1p, and if so, which ones. This work does not address elements and proteins associated with arginine-dependent transcriptional activation. This aspect of CAR2 gene regulation has been reported by Messenguy and Dubois and their colleagues.
MATERIALS AND METHODS
Strains and media.
The S. cerevisiae strain used for β-galactosidase assays was TCY1 (MATα lys2 ura3), and the wild-type strain used for electrophoretic mobility shift assays (EMSAs) was W303-1A (MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1). Strains HEY6 and RDY4 are isogenic derivatives of strain TCY1 containing deletions of the DAL81 and DAL82 genes, respectively. Strain YLS91 (29) (kindly provided by David Shore) is a derivative of W303-1A containing two RAP1 genes; one gene is disrupted by LEU2 (which results in a deletion between the two BamHI sites at nucleotides 818 and ca. 2200), and the other has nucleotides 894 to 1584 deleted, which results in a deletion of amino acids 44 to 303. The RAP1 DNA binding domain is located between residues 361 and 596 of the 827-amino-acid protein (24). Therefore, the truncated protein still retains the DNA binding sites (29). The Escherichia coli strain used for cloning was HB101 (supE44 hsdS20 recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1). Yeast cultures used for Northern blot analysis and β-galactosidase assays were grown in medium containing 0.17% yeast nitrogen base without amino acids or ammonium sulfate (YNB medium) and supplemented with 2% glucose and 0.1% arginine (induced condition) or 0.1% proline (noninduced condition). OXLU was added to YNB-proline medium at a final concentration of 61 mg/ml for the CAR2 induction. The medium used for yeast extract preparations was YPD (1% yeast extract, 2% Bacto Peptone, 2% dextrose).
Northern blot analysis.
Yeast cultures used for Northern blot analyses were grown to the mid-log phase (40 to 60 Klett units). Total RNA was isolated by the method of Carlson and Botstein (5). Poly(A)+ RNA was isolated with oligo(dT)-cellulose (Pharmacia Amersham) columns and resolved on 1.4% agarose–formaldehyde gels (49). The resolved RNAs were transferred to GeneScreen Plus nylon 66 (Dupont, NEN Research Products) membranes. The membranes were then hybridized to double-stranded DNA probes made radioactive by the randomly primed labeling method (Boehringer Mannheim). The procedures followed for hybridization and washing were those described in the GeneScreen Plus protocol manual.
PCR.
PCR was carried out with BamHI-digested plasmid pRS427 containing the CAR2 gene in the BamHI site of plasmid pBR322 as the template (48a). PCR primers used in this study were designed to amplify the CAR2 5′ regulatory region shown in each construct. The PCR mixture consisted of 0.5 μg of template DNA, 100 pmol of each primer, 2.5 U of Taq DNA polymerase, 0.25 mM concentrations of each deoxynucleoside triphosphate, 10 mM Tris-Cl (pH 8.3), 50 mM KCl, and 2.5 mM MgCl2. The PCR cycle program for DNA amplification consisted of one cycle of 94°C (5 min) and 30 cycles of 94°C (1 min), 37°C (1 min), and 72°C (3 min) followed by one cycle of 72°C (5 min). The PCR products were purified with Sephadex G25 spun columns and subjected to enzyme digestion.
Plasmid constructions.
Plasmid constructions for the CAR2 5′ deletion analysis were carried out by PCR. PCR products contained CAR2 DNA sequences between various 5′ upstream positions (−634, −530, −470, −420, −380, −350, −302, −251, −233, −205, −177, −165, and −120) and position +3 as well as SalI and BamHI sites at the 5′ and 3′ termini of the products, respectively. The fragments were digested with SalI and BamHI and were cloned into SalI and BamHI sites of plasmid pLG669Z, described in detail by Guarente et al. (20–22).
Plasmid pHP52, used to assay CAR2 upstream activation sequence (UAS)-mediated reporter gene expression, is identical to plasmid pNG15 (27), except that it contains a 2μm yeast replication origin instead of ARS1, and was constructed as follows. (i) The 2.2-kb EcoRI DNA fragment, containing a 2μm replication origin, was cloned into plasmid pNG15 which had been partially digested with EcoRI to delete the TRP1 (also called ARS1)-containing fragment, to yield plasmid pHP51. (ii) The NcoI restriction site downstream of the lacZ gene in plasmid pHP51 was destroyed by NcoI partial digestion and Klenow polymerase reaction to produce plasmid pHP52. (iii) Synthetic oligonucleotides, containing putative CAR2 UAS elements, were cloned into the SalI and EagI sites of the heterologous expression vector pHP52. The core promoter contained in plasmid pNG15 is a modified fragment (+4 to −1100) from the CYC1 gene. The cloning was done with a polylinker that replaced the CYC1 sequence from −228 to −700.
Plasmid pHP200 was constructed to assay the effects of mutating one or more of the cis-acting elements in the context of a full-length CAR2 promoter. PCR primers were designed to amplify CAR2 DNA sequences between positions −126 and +3. SalI and BamHI restriction sites (11 bp apart) were added to the 5′ terminus, and a BglII site was added to the 3′ terminus. The sequence 5′-CCCTTGCCCTTAGCGGCTGACTGGCT-3′ was changed to 5′-CCCTATAGTCGACCGGCTGGGATCCT-3′. The PCR product was digested with SalI and BglII and cloned into plasmid pLG669Z that had been digested with SalI and BamHI; the BamHI-BglII ligation destroyed the BamHI site at the junction and fused the CAR2 sequence from −126 to +3 in frame to lacZ. These steps produced the vector plasmid pHP200. PCR amplification was then used to generate DNA fragments containing wild-type and mutant alleles of the CAR2 5′ regulatory region between positions −302 and −127. These fragments were then cloned into plasmid pHP200 digested with SalI and BamHI, reassembling a full-length CAR2 promoter fused to lacZ. The only substitutions of wild-type sequences were those necessary to introduce the SalI restriction site. This site was situated downstream of the TATA box to avoid any changes in the putative UAS region of the plasmid. β-Galactosidase activities were measured with transformants which contained the set of plasmids constructed in this way. All the DNA fragments derived from synthetic oligonucleotides and PCR products were verified in the recombinant plasmids by sequence analyses (53, 54). Other techniques, including DNA manipulation and DNA gel electrophoresis, were as described in detail by Sambrook et al. (45).
EMSA.
The sequences of synthetic oligonucleotides used as DNA probes and competitors are shown in Fig. 1. Cultures of S. cerevisiae W303-1A and YLS91, used for the preparation of crude extracts, were grown in YPD medium. The protocols for extract preparation and EMSAs were as described by Luche et al. (31) and Kovari et al. (29). Within experimental error, the lanes of the gels (depicted as individual autoradiographs) contained the same amounts of the DNA probes.
FIG. 1.
Oligonucleotides used as radioactive probes and competitors in EMSAs. Sequences homologous to Abf1p, Rap1p, and Dal82p binding sites are indicated with brackets. Mutations in each site, here and in all other figures, are shown as lowercase letters. Numbers in the figure indicate the positions of the bases whose coordinates are given. mt, mutant.
Yeast and bacterial transformation.
Yeast strains were transformed with lithium acetate by the method of Ito et al. (26). E. coli HB101 was transformed by the Tschumper and Carbon modification (55) of the Mandel and Higa method (33).
β-Galactosidase assay.
Assays of β-galactosidase activities were performed in duplicate and also from duplicate or triplicate independent yeast transformants by using the procedures and precautions described in detail earlier (21, 27, 47). Data from repeated experiments generally varied less than 20%, and data from duplicate assays varied less than 5%. The patterns of reporter gene expression observed within each experiment (the results of one experiment are represented in each figure) were invariant upon repetition of the experiment, unless otherwise noted. However, as noted earlier, quantitative comparisons of data are prudent only when all of the results being compared derive from a single experiment (i.e., data from within a figure). Enzyme activities are expressed in units defined by Miller (41) but were based on 10 ml of culture rather than 1 ml.
RESULTS
Induced CAR2 expression.
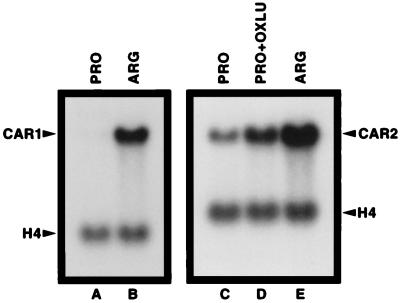
We first established the CAR2 baseline induction pattern using Northern blot analysis. Steady-state levels of CAR1 mRNA (our positive control gene) were much higher in cells provided with arginine as the nitrogen source than with proline, i.e., CAR1 expression was highly inducible. (Fig. 2, lanes A and B). Compared to those of CAR1, CAR2 mRNA levels are less responsive to arginine largely due to the fact that substantial basal-level CAR2 expression occurs when proline is provided as the sole nitrogen source (lanes C and E). CAR2 expression was induced by OXLU but to a much smaller degree than when arginine was used as the inducer (Fig. 2, lanes C and D).
FIG. 2.
Steady-state levels of CAR1 and CAR2 mRNAs in S. cerevisiae grown on various nitrogen sources. Poly(A)+ RNA was prepared from S. cerevisiae Σ1278b cells grown in YNB media containing proline (PRO) or arginine (ARG). OXLU, a gratuitous inducer, was added to YNB-proline medium (PRO+OXLU) at a final concentration of 61 mg/ml for the CAR2 induction. The same designations for growth conditions are used throughout this work. Histone H4 mRNA served as a control for Northern blot loading and transfer efficiencies.
5′ deletion analysis of the CAR2 upstream region.
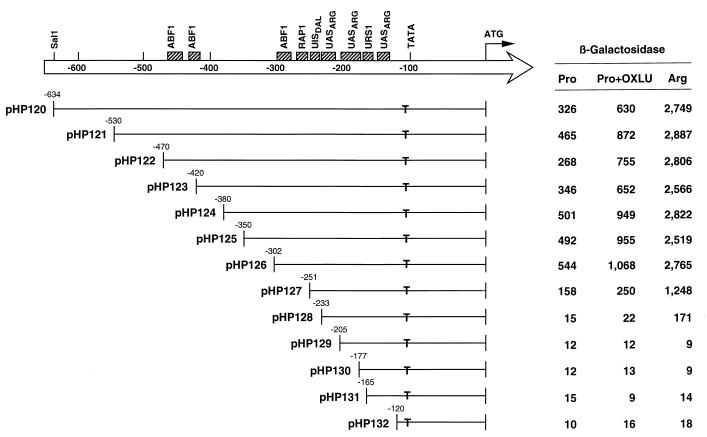
We began dissection of the CAR2 promoter by constructing a CAR2-lacZ fusion plasmid (pHP120) (see Materials and Methods) containing 634 bp of the upstream region. This plasmid supported two- and eightfold-higher levels of β-galactosidase production in response to the presence of OXLU and arginine, respectively (Fig. 3). A series of deletions removing nucleotides −634 to −302 (plasmids pHP120 to pHP126) had little effect on β-galactosidase production, suggesting that this region was not demonstrably required for regulated CAR2 expression (Fig. 3). Deletion of nucleotides −302 to −251 resulted in three-, four-, and twofold decreases in basal, proline- and OXLU-induced, and arginine-induced levels of lacZ expression, respectively (Fig. 3, compare plasmids pHP126 and pHP127). Removal of the next 18 nucleotides to position −233 resulted in further 10-, 11-, and 7-fold decreases in lacZ expression, respectively (Fig. 3, plasmid pHP128). In plasmid pHP128, only low levels of arginine-induced reporter gene expression were observed. Deletion of 28 additional bases to position −205 (plasmid pHP129) resulted in the loss of the remaining arginine-dependent β-galactosidase production. The same result was observed with the three subsequent deletions, the last of which (plasmid pHP132) lacked all sequences 5′ of position −120; the TATA sequence of CAR2 is situated at position −134. These data demonstrate that the region between CAR2 positions −302 and −233 contains elements that are important for high-level expression when arginine is used as the nitrogen source as well as for the more modest level of expression when OXLU is present.
FIG. 3.
5′ deletion analysis of the CAR2 upstream region. DNA fragments containing the CAR2 regulatory region as well as SalI and BamHI restriction sites at the 5′ and 3′ termini, respectively, were amplified from CAR2 DNA by PCR. The PCR products were digested with SalI and BamHI and cloned into the SalI and BamHI sites of the lacZ expression vector pLG669Z (20–22). The set of plasmids was then used to transform strain TCY1 for analysis. Boxes at the top of the figure represent sequences that are homologous to various transcription factor binding sites. T’s indicate the positions of TATA sequences. Numbers at the left of each insert indicate the 5′ terminus of the remaining CAR2 DNA in the CAR2-lacZ fusion plasmids. Coordinates are indicated relative to the position of the translation start site in this and all subsequent figures.
Demonstration of a putative Abf1p binding to the region from −302 to −233 of CAR2.
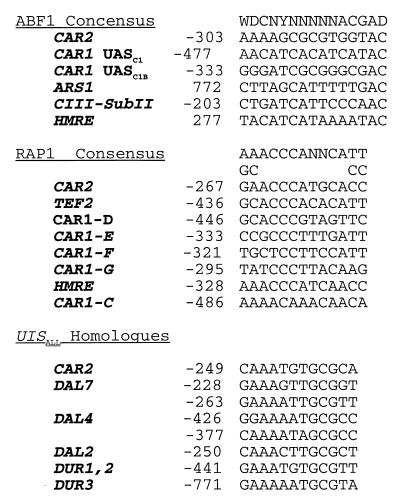
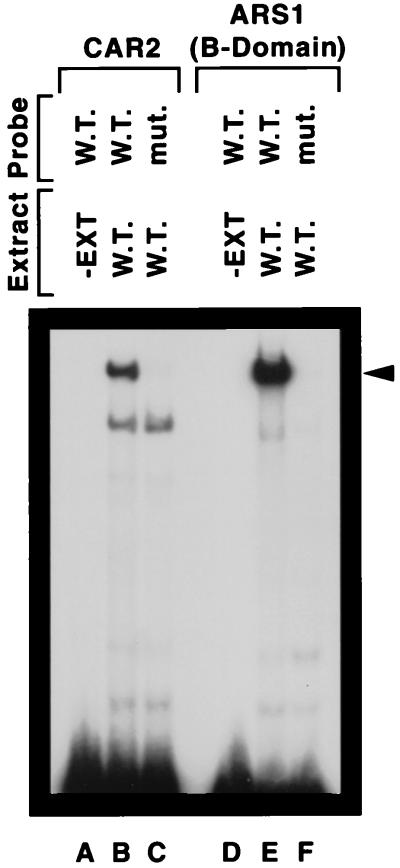
Inspection of the CAR2 upstream region from −302 to −233 revealed sequences homologous to ABF1, RAP1, UISALL and UASARG sites (Fig. 4). Notably absent were GATAA-containing sequences homologous to UASNTR. A priori, the presence of sequences homologous to known transcription factor binding sites is not sufficient reason to conclude that transcription factors bind to them and mediate CAR2 transcription. Therefore, EMSAs were performed to ascertain whether CAR2 promoter fragments containing these homologous sequences were indeed able to bind protein. When radioactively labeled CAR2 fragment HD189 (positions −313 to −271) was incubated with a wild-type yeast crude cell extract (see Materials and Methods), two retarded species were observed (Fig. 5, lane B); no retarded species was observed when extract was omitted from the EMSA reaction mixture (lane A). To assess the binding specificity, we used a mutant allele of fragment HD189 in which sequences homologous to an ABF1 binding site were altered in a way previously shown to destroy Abf1p binding (15, 16, 28). The slower-migrating DNA-protein complex was lost when the mutant promoter fragment was used as the probe (Fig. 5, lane C). These data suggested that only this slower-migrating species was potentially associated with Abf1p binding. The identity of the protein(s) in the more rapidly migrating complex in lanes B and C remains unknown.
FIG. 4.
CAR2 sequences homologous to the binding sites for Abf1p, Rap1p, and Dal82p.
FIG. 5.
Protein binding to ARS1 and CAR2 DNA fragments. DNA fragments labeled with 32P at their 5′ ends and containing the HD189 and ARS1 sequences indicated in Fig. 1 were used as probes. Yeast cell extract (strain W303-1A) was omitted from the reaction mixture resolved in lanes A and D. All reaction mixtures contained a 200-fold excess of sheared calf thymus DNA by sonication as a nonspecific competitor. −EXT, minus extract, W.T., wild type; mut, mutant.
To determine whether the more slowly migrating species possessed a migration rate similar to that of a legitimate Abf1p-DNA complex, we used a well-characterized, similarly sized DNA fragment containing the HMRE ARS1 B domain (15, 16, 28), which was used as an EMSA probe in the analysis of the CAR1 promoter (28) (Fig. 5, lanes D to F). The wild-type HMRE fragment yielded a retarded species with a mobility similar to the one observed with the wild-type CAR2 fragment (lane E); as expected, retardation was not observed with the mutant HMRE fragment (lane F).
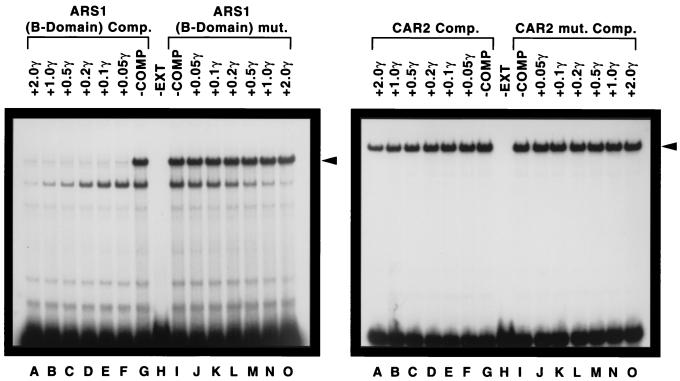
Competition EMSAs further substantiated the specificity of the DNA-protein complex. In the first of these experiments, CAR2 fragment HD189 was used as the radioactive probe and its ability to form a retarded complex was assayed in the presence of increasing amounts of nonradioactive HMRE ARS1 B domain DNA as the competitor (Fig. 6, left panel). The lowest concentration of competitor DNA successfully competed with the CAR2 fragment probe (Fig. 6, left panel, lanes A to H). In contrast, when a mutant allele of the HMRE fragment was used in the assay, no competition was observed (Fig. 6, left panel, lanes H to O). This experiment was then repeated with the probe and competitor reversed, i.e., with the HMRE fragment as the probe and the CAR2 fragment as the competitor. As shown in the right panel of Fig. 6, the CAR2 fragment was only minimally able to compete with the HMRE DNA fragment for protein binding (lanes A to H). This is not a surprising result given the dramatic effectiveness of HMRE as a competitor in the left panel of the figure. Together, the data in Fig. 5 and 6 suggest that a CAR2 fragment containing a sequence from positions −313 to −271 binds to Abf1p. Note that a more rapidly migrating species in panel A (for which the CAR2 DNA probe was used) was equally competed away by increasing amounts of wild-type or mutant HMRE DNA, arguing that this complex does not depend upon sequences homologous to Abf1p binding sites. It is, however, unlikely to be nonspecific binding of protein to DNA because the band was not observed when HMRE DNA was used as the radioactive probe.
FIG. 6.
Competition between CAR2 and ARS1 DNA fragments. An oligonucleotide covering CAR2 positions −313 to −271 (left) or ARS1 positions 748 to 780 and flanked by SalI and EagI restriction sites (right) was labeled at its 5′ end with 32P and used as the probe. Competitor DNA was omitted in the reaction mixtures resolved in lanes G and I. The reaction mixtures in lanes A through F contained decreasing amounts of the unlabeled ARS1 (left) or CAR2 (right) oligonucleotide as a competitor (Comp. and COMP). The reaction mixtures in lanes J through O contained increasing amounts of the unlabeled ars1 mutant (mut.) (left) or car2 mutant (right) oligonucleotides; the amounts that were used are indicated in micrograms (γ). The remaining conditions and procedures used in the experiment were as described in the legend to Fig. 5. Strain W303-1A was the source of the crude cell extract (EXT).
Demonstration of putative Rap1p binding to the region from −302 to −233 of CAR2.
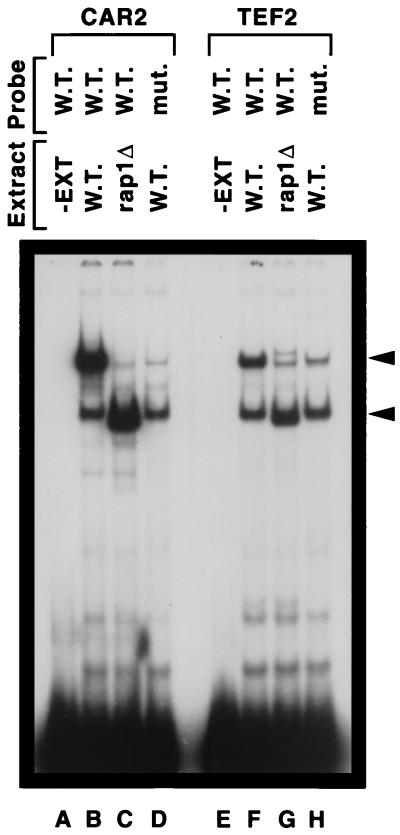
We next investigated a CAR2 sequence contained in DNA fragment HD190 (CAR2 positions −282 to −247) which was homologous to known Rap1p DNA binding sites (Fig. 4). One major and one minor retarded species were observed in the EMSA when extract from the wild-type strain W303-1A was used as the source of protein (Fig. 7, lane B). When the assay was repeated with an extract derived from the rap1 mutant strain YLS91 (carrying a truncated, but binding-competent form of Rap1 [6, 24, 29, 30, 46]), the major retarded species migrated to a new location just below the minor species observed in lane B (Fig. 7, lane C). When a mutant CAR2 DNA probe (fragment HD192, in which the Rap1p site-homologous sequence is mutated) was used in the assay, little if any of the more slowly migrating DNA-protein complex present in lane B was observed (Fig. 7, lane D). The more rapidly migrating species in lane B was also observed in lane D, arguing that it was not associated with the mutated sequence in fragment HD192. The small amount of DNA-protein complex in lanes C and D that possessed the same mobility as the major complex observed in lane B is unlikely to be Rap1p specific because it was also observed in lane C, which did not contain full-length Rap1p. We compared the data shown in Fig. 7, lanes A to D, with that obtained with a similarly sized, well-characterized TEF2 DNA probe previously documented to contain a Rap1p binding site (6, 24, 29, 30, 46). The pattern of data obtained in this control experiment (Fig. 7, lanes E to H) was the same as that observed when the CAR2 fragment was used (lanes A to D). The EMSA using the TEF2 probe also exhibited the nonspecific protein-DNA complexes seen in lanes B to D.
FIG. 7.
Wild-type and truncated Rap1p binding to CAR2 and TEF2 DNA fragments. DNA fragments 32P labeled at their 5′ ends and containing the HD190 and HD192 sequences indicated in Fig. 1 were used as probes. Yeast cell extract (EXT) was omitted from the reaction mixtures resolved in lanes A and E. A wild-type (strain W303-1A) extract (W.T.) was used in lanes B, D, F, and H. An extract from a rap1 truncation mutant strain (mut.) (YLS91) was used in lanes C and G (rap1Δ).
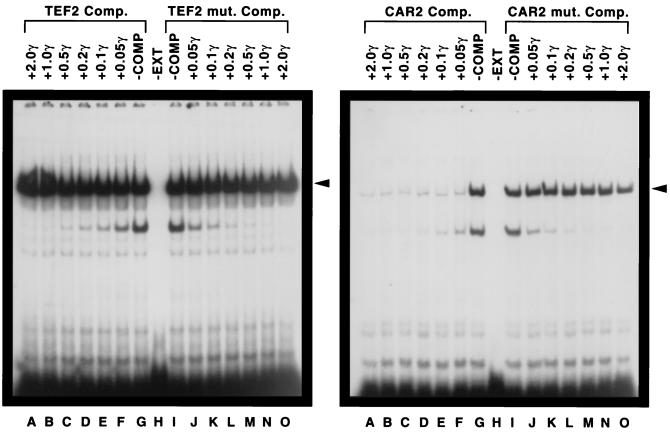
We next assessed the ability of the TEF2 and CAR2 fragments to serve as competitors of one another’s protein binding. At the time this experiment was performed, the Rap1p binding site contained in the TEF2 DNA fragment was the strongest one known (6, 24, 29, 30, 46). As shown in the left panel of Fig. 8, the TEF2 DNA fragment was unable to serve as an effective competitor of the CAR2 fragment for DNA binding (lanes A to H). In the reverse situation, i.e., when the TEF2 fragment was used as the probe and the CAR2 fragment was used as the competitor, strong competition was observed (Fig. 8, right panel). In fact, even at the lowest concentration used, the wild-type CAR2 DNA fragment (HD190) effectively competed with the TEF2 probe (Fig. 8, right panel, lanes A to H). However, when the putative Rap1p binding site contained in the CAR2 fragment was mutated, the fragment was no longer able to serve as the competitor. As with Abf1p, these data support the contention that the CAR2 fragment from positions −282 to −247 contains a putative Rap1p binding site.
FIG. 8.
Competition between CAR2 and TEF2 DNA fragments for protein binding. An oligonucleotide 32P labeled at its 5′ end and covering CAR2 positions −282 to −247 (left) or TEF2 positions −449 to −414 (right) was used as the probe. The reaction mixtures in lanes A through F contained decreasing amounts of the unlabeled TEF2 (left) or CAR2 (right) oligonucleotide as a competitor (Comp. and COMP). The reaction mixtures in lanes J through O contained increasing amounts of the unlabeled TEF2 mutant (mut.) (left) or CAR2 mutant (right) oligonucleotide. The remaining conditions and procedures used in the experiment were as described in the legend to Fig. 6. EXT, extract; γ, microgram amounts.
Demonstration of putative Dal82p binding to the region from −302 to −233 of CAR2.
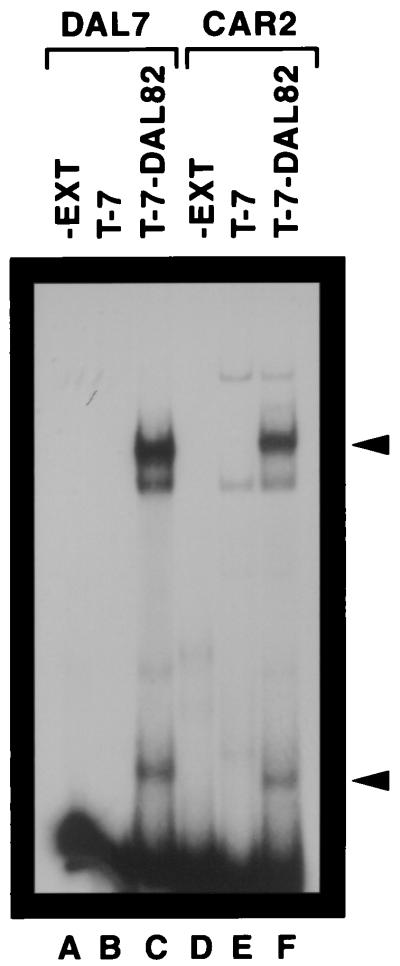
The last CAR2 sequence we investigated was one homologous to UISALL elements that are situated upstream of allantoin pathway genes (Fig. 4). These elements are Dal82p binding sites that mediate induction of DAL and DUR gene expression when allophanate or its analogue, OXLU, is present (9, 17). To determine whether Dal82p bound to the CAR2 UISALL-homologous sequence (−272 to −211), CAR2 fragment RD43 (Fig. 1) was incubated with extracts derived from an E. coli culture transformed with a T-7 expression vector plasmid (pRD4) or one containing a T-7–DAL82 fusion (plasmid pRD41) (Fig. 9, lanes D to F). No signal was observed with extract derived from a transformant containing plasmid pRD4 alone (lane E). In contrast, several DNA-protein complexes were observed with extract derived from cells containing the plasmid bearing the T-7–DAL82 fusion (Fig. 9, lane F). Two of these complexes (indicated by arrows) were specific to the presence of Dal82p. The Dal82p-specific signal with the fastest mobility has been previously shown to derive from a Dal82p degradation product (17). The source of the weak nonspecific signal observed in the lowest-mobility complex is unknown (Fig. 9, lanes E and F). DAL7 DNA fragment RD72X, which contains a well-characterized DAL UISALL element (17), was used in a control experiment to identify the relative mobility of a bona fide UISALL-Dal82p complex (Fig. 9, lanes A to C). In this experiment, the major complexes with the lowest and greatest mobilities were the only ones that were Dal82p specific and migrated to positions similar to those observed in lanes D to F.
FIG. 9.
Protein binding to CAR2 and DAL7 DNA fragments. DNA fragments labeled with 32P at their 5′ ends and containing the JRD72X and RD43 sequences indicated in Fig. 1 were used as probes. An extract (EXT) from IPTG (isopropyl-β-d-thiogalactopyranoside)-induced E. coli cells harboring plasmid pRD4 (T7) or pRD41 (T-7–DAL82) was prepared as described previously (17). EMSA reaction conditions were exactly as described in the work of Dorrington and Cooper (17).
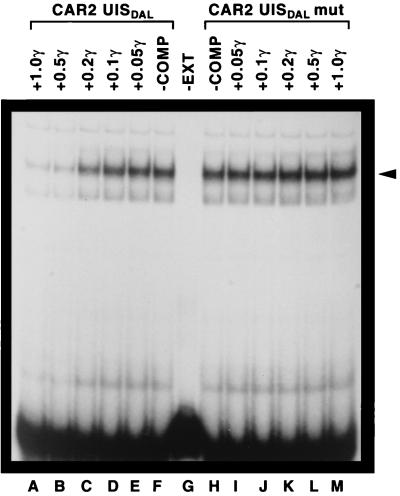
To further demonstrate that the major complex observed in Fig. 9 derived from a bona fide Dal82p-CAR2 UISALL interaction, we performed a competition EMSA using CAR2 wild-type (RD43) and mutant (RD44) fragments as competitors of DAL7 DNA fragment JRD72X (Fig. 10). The wild-type fragment was an effective competitor of JRD72X DNA binding (lanes A to F). Fragment RD44, however, which contained a mutated form of the CAR2 UISALL, was unable to effectively compete with DAL7 for binding to DNA (Fig. 10, lanes H to M). These data led to the conclusion that the CAR2 fragment RD43 contains a sequence capable of binding Dal82p.
FIG. 10.
Competition between CAR2 and DAL7 DNA fragments. An oligonucleotide labeled at its 5′ end with 32P and covering DAL7 positions −241 to −206 (Fig. 1, JRD72X) was used as the probe. The reaction mixtures in lanes A through E contained decreasing amounts of the unlabeled CAR2 wild-type oligonucleotide (Fig. 1, RD43), and lanes I through M contained increasing amounts of the unlabeled CAR2 mutant oligonucleotide (mut) (Fig. 1, RD44). The remaining conditions and procedures used in the experiment were as described in the legend to Fig. 6. COMP, competitor; EXT, extract; γ, microgram quantities.
Participation of the putative Abf1p, Rap1p, and Dal82p binding sites in transcription.
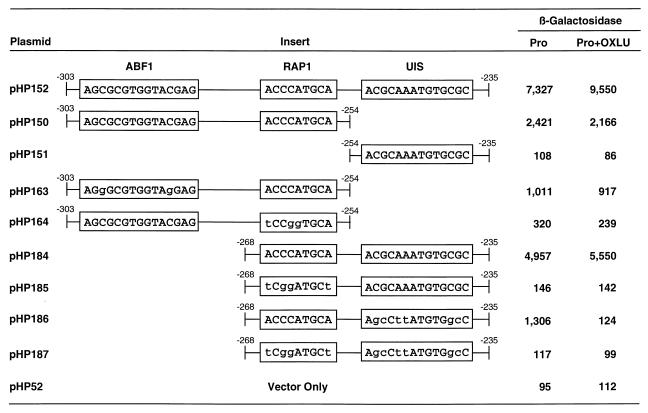
The 70-bp CAR2 fragment (−303 to −235), required for high-level CAR2 expression (Fig. 3) was shown (Fig. 5 to 10) to contain putative Abf1p, Rap1p, and Dal82p protein binding sites. To determine whether each of these putative binding sites participated in transcription, we cloned wild-type and mutant alleles of this fragment into the 2μm-based expression vector pHP52. As shown in Fig. 11, this plasmid supported high-level reporter gene expression but exhibited only a very modest response when OXLU was added to the medium (plasmid pHP152). This modest induction was not surprising because the CAR2 fragment had just been shown to contain putative Abf1 and Rap1 protein binding sites (data in Fig. 5 to 8). If functional Abf1p and Rap1p sites were present, they would participate in activating significant reporter gene expression in the absence of OXLU, which in turn would result in a relatively poor induction response due to the high level of basal expression. To test this possibility, we synthesized a DNA fragment that contained the putative Abf1p and Rap1p binding sequences, cloned it into the expression vector pHP52 (to yield plasmid pHP150), and assayed the reporter gene expression it supported. As anticipated, plasmid pHP150 supported high-level, OXLU-independent reporter gene expression (Fig. 11). This expression, however, was only about one-half to one-fourth the level observed when the putative Dal82p binding site was also present (Fig. 11, compare plasmids pHP152 and pHP150). The slight decrease in activity observed when OXLU was present is a common observation when OXLU is added to cultures; it is due to toxic effects of OXLU on the cell (8, 9). To assess the contribution of the putative Abf1p binding site contained in plasmid pHP150, we mutated the putative Abf1p site in the same way as we had in the EMSA experiments (Fig. 5 and 6). Gene expression supported by the mutant DNA fragment (plasmid pHP163) was about half that supported by its wild-type counterpart (plasmid pHP150) and only about 10% of that observed with plasmid pHP152. This result demonstrates that a mutation that eliminates putative Abf1p binding (Fig. 5 and 6) also decreases reporter gene transcription here and supports the idea that these CAR2 sequences comprise a functional Abf1p binding site.
FIG. 11.
β-Galactosidase production supported by plasmids containing synthetic CAR2 DNA fragments. The synthesized DNA fragments were cloned into the heterologous expression vector pHP52. Plasmids pHP150 and pHP184 contain native CAR2 sequences, whereas their derivatives contain mutated sequences in which the mutations are shown with lowercase letters. The transformation recipient was strain TCY1.
We next mutated the putative Rap1p binding site as we did for the experiments whose results are shown in Fig. 7 and 8 (Fig. 11, plasmid pHP164); this resulted in a seven- to ninefold decrease in lacZ expression compared to that of the wild-type sequence (plasmid pHP150). With the aforementioned data, reporter gene expression supported by plasmid pHP150 can be concluded to be largely due to the sequences homologous to the Abf1p and Rap1p sites. Furthermore, the putative Rap1p binding site appears to drive greater reporter gene expression than the putative Abf1p site.
We next focused on the putative Dal82p binding site situated between CAR2 positions −254 to −235. As before, we synthesized an oligonucleotide containing these positions, cloned it into an expression vector, and assayed its ability to support reporter gene expression. The plasmid containing this oligonucleotide was unable to support β-galactosidase production above the background level (Fig. 11, plasmids pHP151 and pHP52). This was consistent with earlier data showing that the DAL7 UISALL (Dal82p binding site) is unable to support transcription in a heterologous expression vector in the absence of a GATAA-containing UASNTR site (Gln3p and/or Gat1p binding site) (9).
The observations that plasmid pHP150 supports only one-third of the lacZ expression of the parent plasmid pHP152 and that the remainder of plasmid pHP152 (i.e., plasmid pHP151) supports none at all suggest that the putative Dal82p binding site may function synergistically with the putative Rap1p site and/or Abf1p site (Fig. 11). Indeed, the full-length fragment (plasmid pHP152) supported significantly greater transcription than the sum of the levels of transcription of the isolated fragments (plasmids pHP150 and pHP151). To test this suggestion relative to the involvement of the Rap1p site, we synthesized an oligonucleotide containing only the putative Rap1p and Dal82p binding sites (positions −269 to −235) and analyzed its ability to support transcription (Fig. 11, plasmid pHP184). This plasmid supported about two-thirds of the lacZ expression observed with plasmid pHP152 and five- to six-fold-greater expression than with plasmid pHP163. To evaluate the contributions of the individual sites on plasmid pHP184, we first mutated the putative Rap1p site (plasmid pHP185). This mutation decreased β-galactosidase production 30- to 40-fold to a level slightly above background. Mutation of the putative Dal82p binding site of plasmid pHP184 decreased the level of reporter gene expression by about three-fourths. Moreover, the remaining gene expression supported by this plasmid was abolished after addition of OXLU (Fig. 11, plasmid pHP186); this result was surprising and was not reproducible. As expected, mutation of both the putative Rap1p and Dal82p binding sites completely destroyed reporter gene expression (Fig. 11, plasmid pHP187). These data support the contention that Rap1p and Dal82p act synergistically to support reporter gene expression.
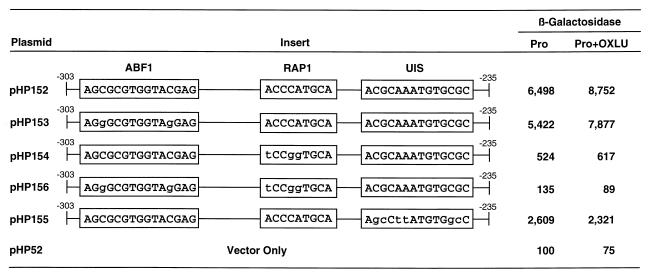
Finally, we analyzed the phenotypes of the above-described set of cis-acting mutations in the context of the entire fragment from −303 to −235 (Fig. 12). Mutation of the putative Abf1p binding site (plasmid pHP153) decreased reporter gene expression by 10 to 20% compared to that of the parent (plasmid pHP152). In contrast, mutation of the putative Rap1p binding site (pHP154) decreased reporter gene expression 12- to 14-fold. In both instances, however, the presence of OXLU resulted in about 20% greater expression. Mutation of both the putative Rap1p and Abf1p sites together eliminated transcription (plasmid pHP156). These data corroborate those shown in Fig. 11.
FIG. 12.
β-Galactosidase production supported by plasmids containing synthetic DNA fragments covering the CAR2 regulatory region between positions −303 and −235, which were cloned into the expression vector pHP52. The transformation recipient and conditions were as described for Fig. 11.
The contribution of a putative Dal82p binding site to overall reporter gene expression supported by plasmid pHP152 appears unimpressive if it is evaluated on the basis of an OXLU response. However, when mutated, the putative Dal82p binding site of plasmid pHP152 exhibits a more dramatic effect. This mutant site supported only 20 to 40% of the reporter gene expression supported by plasmid pHP152 (Fig. 12). Moreover, addition of OXLU in this case did not result in an increase in reporter gene expression but instead resulted in a small decrease. These results suggest that there is a putative Dal82p binding site participating in lacZ expression supported by plasmid pHP152.
Requirement of Dal82p for OXLU-mediated reporter gene expression.
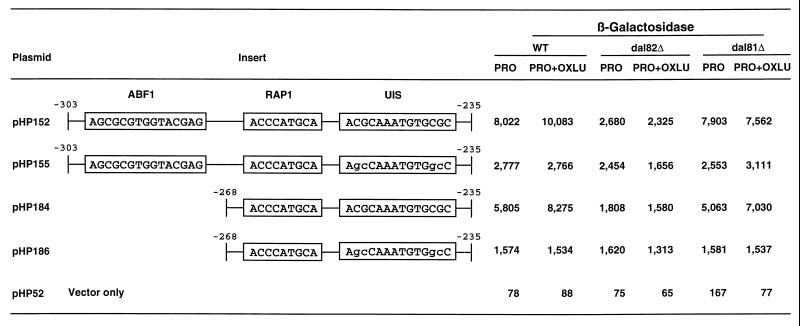
OXLU-induced allantoin pathway gene expression requires participation of the Dal82 and Dal81 proteins (8, 9). Therefore, we determined whether these regulatory proteins were required for reporter gene expression supported by plasmid pHP152. β-Galactosidase production decreased by up to 75% in the dal82 deletion mutant relative to that of the wild type (Fig. 13, plasmid pHP152). Moreover, this decrease was independent of OXLU. However, no decrease in activity was observed in dal81 deletion mutant cells grown without an inducer and decreased only 25% when it was present (Fig. 13, plasmid pHP152). Mutation of the putative Dal82p binding site in plasmid pHP152 resulted in reporter gene expression that was the same as in wild-type, dal82, and dal81 strains (Fig. 13, plasmid pHP155). Incidentally, this was also the level observed with plasmid pHP152 expressed in the dal82 deletion. We repeated the above experiments using vectors containing only the putative Rap1p and Dal82p binding sites (Fig. 13, plasmids pHP184 and pHP186). Results similar to those observed with the entire DNA fragment (plasmid pHP152) were obtained. Again, mutation of the Dal82p binding site or deletion of the DAL82 gene resulted in three- and fivefold decreases in β-galactosidase production in proline medium in the presence and absence of OXLU, respectively. In no case was Dal81p necessary for basal-level activity, indicating that Dal82p but not Dal81p is important for reporter gene expression in the absence of OXLU.
FIG. 13.
β-Galactosidase production in wild-type (TCY1) and dal82 (RDY4) or dal81 (HEY6) deletion mutant strains supported by plasmids containing synthetic DNA fragments covering the CAR2 regulatory regions that were cloned into the expression vector pHP52. WT, wild type.
An important test to determine if the three sites described above in fact function in CAR2 transcription is to evaluate the effects of mutations in these sites in the context of a full-length CAR2 promoter. CAR2, as mentioned earlier, contains sequences homologous to the URS1 site of CAR1, the Ume6p (also called Car80p) repressor binding site (1, 49–51). In our analysis of the CAR1 promoter, we found that the presence of this strong repressor binding site masks some of the effects of mutations in the weaker UAS elements of that promoter (47). The contribution of these weak UASs could be unmasked by mutating the URS1 site. Therefore, we constructed full-length wild-type and mutant UAS alleles in both wild-type and urs1-minus promoter backgrounds (Fig. 13).
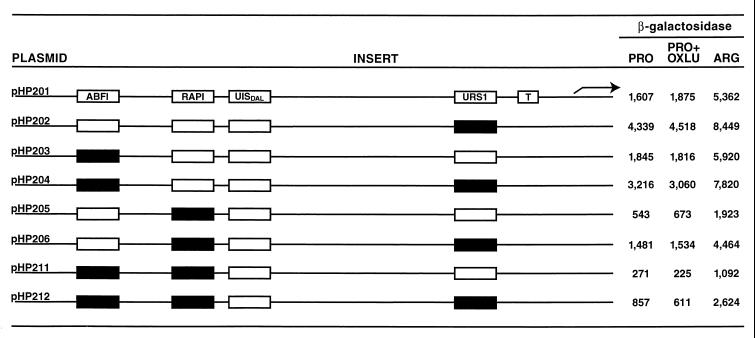
Mutation of the putative Abf1p binding site in the context of a full-length CAR2 promoter did not bring about a demonstrable decrease in β-galactosidase production when the URS1 site was intact (Fig. 14, compare plasmids pHP201 and pHP203). In a urs1 mutant background, however, reporter gene expression decreased about one-third when cultures were grown in proline or proline plus OXLU medium (compare plasmids pHP202 and pHP204). A similar decrease was not observed under conditions of high-level arginine-mediated induction. Mutation of the putative Rap1p binding site, on the other hand, produced about a threefold decrease regardless of the medium used and whether or not the URS1 site was mutated (compare plasmid pHP205 with plasmid pHP206). Mutation of both the putative Abf1p and Rap1p sites resulted in about a 50% decrease relative to the situation when only the Rap1p site was mutated under all conditions tested (Fig. 14, compare pHP205 with pHP211 and pHP206 with pHP212). The double mutant promoter retained only 10 to 20% of the wild-type expression levels regardless of the growth conditions or the presence of a functional URS1 element. This result argues that even though CAR2 is induced by arginine, a large part of its transcription depends upon constitutively functioning Rap1p. A smaller contribution also appears to be made by a second constitutively functioning factor, Abf1p.
FIG. 14.
Genetic analysis of the putative CAR2 Abf1p and Rap1p binding sites in the context of a full-length CAR2 promoter. CAR2 DNA fragments between positions −302 and −132 containing wild-type or mutated putative CAR2 Abf1p and Rap1p binding sites were synthesized chemically or by PCR and cloned into SalI and BamHI sites of plasmid pHP200. Reporter gene expression supported by these plasmids was determined in cells grown in YNB medium containing proline (PRO), arginine (ARG), or proline plus OXLU (PRO+OXLU). Open and closed boxes in the inserts represent wild-type and mutated sites, respectively. T, TATA sequence.
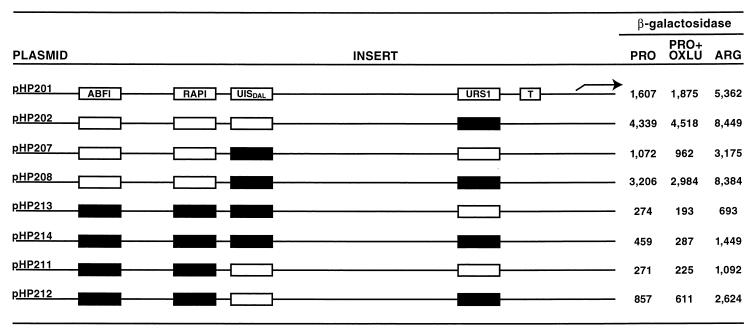
Finally, we assessed the contribution of the putative Dal82p binding site to overall CAR2 expression in a similar manner. Mutation of this site alone reduced β-galactosidase production about 30 to 50% when a wild-type URS1 allele was present (Fig. 15, compare plasmids pHP201 and pHP207). When the analogous experiment was carried out with a urs1 allele, lacZ expression decreased 25 to 35% relative to wild-type expression when proline was provided as the nitrogen source; no difference was observed with arginine as the nitrogen source (Fig. 14, compare plasmid pHP202 with plasmid pHP208).
FIG. 15.
Combinational mutation analysis of the putative CAR2 Abf1p, Rap1p, and Dal82p binding sites in the context of a full-length CAR2 promoter. CAR2 DNA fragments between positions −302 and −132 containing wild-type or mutated putative CAR2 Abf1p, Rap1p, and Dal82p binding sites were synthesized chemically or by PCR and cloned into SalI and BamHI sites of plasmid pHP200. Growth, abbreviations, and other conditions were as described for Fig. 14.
We next assayed the effects of mutating the putative Dal82p binding site with and without the Abf1p and Rap1p sites. The triple mutant possessed only 15 to 30% of the wild-type expression levels, and there was no effect whatever, beyond its slight toxicity, when OXLU was added as the inducer. Further, there was only about one-fourth of the wild-type activity when arginine was provided as the nitrogen source even though the elements necessary for arginine-mediated induction were untouched (Fig. 15, compare plasmid pHP201 with plasmid pHP213). The fold induction with arginine, however, remained the same as in the wild type, indicating the continued functioning of the arginine-dependent transcription factors (the arginine value divided by the proline value). These data demonstrate the level of expression that can be supported by the arginine induction system alone. When the experiment was repeated with a plasmid in which the urs1 site was also mutated, a slightly smaller fraction of wild-type activity was retained (6 to 17%) (Fig. 15, compare plasmid pHP202 with plasmid pHP214). Here, as with plasmid pHP213, there was no OXLU-mediated induction and arginine-induced expression reached only 20% of the wild-type level.
Another way to visualize the contribution of the putative Dal82p binding site is to compare the level of expression observed with mutations in the Abf1p and Rap1p sites with that observed when all three sites (Abf1p, Rap1p, and Dal82p) were mutated. Arginine-induced reporter gene expression in the Abf1p and Rap1p site double mutant was almost twice as great as when the Dal82p site was also mutated in a URS1 allele. There was no difference in the values observed with the double mutant when proline or proline plus OXLU was provided (Fig. 15, compare pHP214 with pHP212 and pHP213 with pHP211). When the experiment was performed with a urs1 allele, β-galactosidase production was up twofold in the double mutant under all conditions compared to that of the triple mutant (Fig. 15, compare plasmids pHP212 and pHP214). Again, however, there was no OXLU-induced expression. The lack of OXLU-induced expression in these experiments is not surprising given the necessity of Rap1p function for the Dal82p site to mediate transcription. These data argue that although the Dal82p site functions synergistically with the Rap1p site with respect to OXLU-induced induction, the Dal82p site also appears to mediate a twofold enhancement of arginine-induced expression.
DISCUSSION
Two criteria must be met to conclude that a promoter contains a given transcription factor binding site: (i) the transcription factor must be shown to bind to the putative site and (ii) the putative site must be shown to be required for or to participate in the gene’s expression. Qualitative EMSAs described in this work fulfilled the first of these criteria by demonstrating that Rap1p, Abf1p, and Dal82p bind to sites in the CAR2 upstream region. Based on crude approximations derived from the competition EMSA data, the Rap1p site in CAR2 is a reasonably strong one; the CAR2 fragment containing the Rap1p site was an effective competitor of the TEF2 fragment containing a strong, well-characterized Rap1p site for protein binding. The Dal82p binding site in CAR2 was also relatively strong in that its ability to serve as a competitor compared favorably with that of an analogous site in a DAL7 fragment, one of the better binding sites found among the allantoin pathway genes. In contrast, Abf1p does not bind to the CAR2 fragment as well as it does to the ARS1 fragment, which contains a well-characterized Abf1p site. The second criterion was fulfilled through genetic analysis of CAR2 promoter fragments cloned into a heterologous expression vector and full-length promoters fused to lacZ. These results demonstrated that sequences needed for Rap1p, Abf1p, and Dal82p binding are also required for maximum levels of CAR2 transcription. Together, the genetic and biochemical results of this work permit us to conclude that the CAR2 promoter contains functional Rap1p, Abf1p, and Dal82p binding sites in addition to the previously identified Ume6p (also called Car80p) and UASARG sites.
Our results support the conclusion that regulated CAR2 expression depends upon transcriptional activation by constitutively operating global activators (Abf1p and Rap1p) being balanced by a similarly strong repression being exerted by a global repressor, Ume6p (also called Car80p) (51). The balance of positive and negative control is then tipped toward expression when an inducer is present and toward quiescence when it is not. In the case of CAR2, inducers exert their effects through the ArgR proteins in the presence of arginine and Dal81p and Dal82p when OXLU is provided. This formal model of transcriptional regulation is identical to that first proposed for CAR1 (47, 51). It also explains the early observation that ornithine transaminase production is increased with urea as the nitrogen source (23); urea is the precursor of allophanate, the naturally occurring analogue of OXLU (9).
This work also extends our understanding of the Dal81 and Dal82 proteins. Prior to our experiments, functional UISALL sites had been found only upstream of the allantoin pathway genes and were thought to operate exclusively in conjunction with GATA-containing UASNTR elements (9, 17, 57, 61). In this work we demonstrate that UISALL and associated Dal82p can function in association with elements and factors other than the GATA-containing UASNTR elements and their associated Gln3p and/or Gat1p transcriptional activator. This conclusion is supported by the facts that the CAR2 promoter is devoid of UASNTR elements and that the role(s) played by Gln3p and/or Gat1p for the allantoin pathway genes is predominantly provided by Rap1p in the case of CAR2.
The suggestion that Dal82p mediates CAR2 transcription via synergistic association with Rap1p influences the interpretation of earlier experiments concerning Dal82p and Gln3p (57). These experiments showed that placing a functional Dal82p binding site adjacent to a mutant uasNTR site, i.e., one containing a single mutation in the GATA sequence, suppressed the mutant phenotype. Suppression required only a properly situated UISALL element, functional Dal82p, and was not dependent upon Dal81p or an inducer. At the time these suppression data were reported, we suggested that Dal82p bound the UISALL situated adjacent to the mutant uasNTR site, which facilitated or stabilized Gln3p binding to the latter site, potentially through a protein-protein interaction. This scenario remains a viable interpretation of those earlier results. However, the present data demonstrate that Dal82p can also function synergistically with a global transcription factor (Rap1p) having no specific relationship with nitrogen catabolism. Such a result does not particularly support proposing a direct Gln3p-Dal82p interaction to account for the suppression data. An alternative possibility is that interactions mediating Dal82p-dependent stabilization of Gln3p binding may occur more indirectly, perhaps through involvement of components of the SAGA or related core transcription complexes.
Finally, the present work offers some insight into the biochemical characteristics of Dal82p operation. The suppression experiments just discussed are consistent with the idea that UISALL and Dal82p are able to suppress the GATA mutations in the absence of an inducer or Dal81p (57). In correlation with these observations is the repeated demonstration in this work that Dal82p can enhance transcription in association with Rap1p or the transcription factors responsible for arginine-mediated induction, again in the absence of Dal81p or an inducer. In other words, the participation of Dal81p is not essential for all Dal82p functions and neither is an inducer. On the other hand, Dal81p is absolutely required for OXLU-induced gene expression. The separation of Dal81p and Dal82 functions observed here and earlier will significantly influence mechanistic studies aimed at understanding the biochemical functions of these two transcription factors.
ACKNOWLEDGMENTS
We thank the members of the U.T. Yeast Group who read this manuscript and offered suggestions for its improvement. Oligonucleotides used in this study were prepared by the U.T. Molecular Resource Center.
This work was supported by Public Health Service grant GM-35642 from the National Institute of General Medical Sciences and the Academic Research Fund (GE-96-14) of the Ministry of Education, Republic of Korea.
REFERENCES
- 1.Anderson S F, Steber C M, Esposito R E, Coleman J E. UME6, a negative regulator of meiosis in Saccharomyces cerevisiae, contains a C-terminal Zn2Cys6 binuclear cluster that binds the URS1 DNA sequence in a zinc-dependent manner. Protein Sci. 1995;4:1832–1843. doi: 10.1002/pro.5560040918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bechet J, Grenson M, Wiame J-M. Mutations affecting the repressibility of arginine biosynthetic enzymes in Saccharomyces cerevisiae. Eur J Biochem. 1970;12:31–39. doi: 10.1111/j.1432-1033.1970.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 3.Bercy J, Dubois E, Messenguy F. Regulation of arginine metabolism in Saccharomyces cerevisiae: expression of the three ARGR regulatory genes and cellular localization of their products. Gene. 1987;55:277–285. doi: 10.1016/0378-1119(87)90287-3. [DOI] [PubMed] [Google Scholar]
- 4.Bossinger J, Cooper T G. Molecular events associated with the induction of arginase in Saccharomyces cerevisiae. J Bacteriol. 1977;131:163–173. doi: 10.1128/jb.131.1.163-173.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Bricmont P A, Daugherty J R, Cooper T G. The DAL81 gene product is required for induced expression of two differently regulated nitrogen catabolic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:1161–1166. doi: 10.1128/mcb.11.2.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlson M, Botstein D. Two differentially regulated mRNAs with different 5′ ends encode secreted and intracellular forms of yeast invertase. Cell. 1982;28:145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- 6.Chambers A, Stanway C, Tsang J S H, Henry Y, Kingsman A J, Kingsman S M. ARS binding factor 1 binds adjacent to RAP1 at the UASs of the yeast glycolytic genes PGK and PYK1. Nucleic Acids Res. 1990;18:5393–5399. doi: 10.1093/nar/18.18.5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coffman J A, Rai R, Loprete D M, Cunningham T, Svetlov V, Cooper T G. Cross-regulation of four GATA factors that control nitrogen catabolic gene expression in Saccharomyces cerevisiae. J Bacteriol. 1997;179:3416–3229. doi: 10.1128/jb.179.11.3416-3429.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooper T G. Nitrogen metabolism in Saccharomyces cerevisiae. In: Strathern J N, Jones E W, Broach J, editors. The molecular biology of the yeast Saccharomyces: metabolism and gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. pp. 39–99. [Google Scholar]
- 9.Cooper T G. Allantoin degradative system—an integrated transcriptional response to multiple signals. In: Marzluf G, Bambrl R, editors. Mycota III. Berlin, Germany: Springer-Verlag; 1994. pp. 139–169. [Google Scholar]
- 10.Crabeel M, de Rijcke M, Seneca S, Heimberg H, Pfeiffer I, Matisova A. Further definition of the sequence and position requirements of the arginine control element that mediates repression and induction by arginine in Saccharomyces cerevisiae. Yeast. 1995;11:1367–1380. doi: 10.1002/yea.320111405. [DOI] [PubMed] [Google Scholar]
- 11.Cunin R, Dubois E, Vanthienen G, Tinel K, Jacobs A, Crabeel M. Positive and negative regulation of CAR1 expression in Saccharomyces cerevisiae. Mol Gen Genet. 1986;205:170–175. [Google Scholar]
- 11a.Daugherty J, Rai R, Cunningham T S, Cooper T G. Overlapping positive and negative GATA binding sites mediate inducible DAL7 expression in Saccharomyces cerevisiae. J Biol Chem. 1999;274:28026–28034. doi: 10.1074/jbc.274.39.28026. [DOI] [PubMed] [Google Scholar]
- 12.Degols G. Functional analysis of the regulatory region adjacent to the cargB gene of Saccharomyces cerevisiae. Eur J Biochem. 1987;169:193–200. doi: 10.1111/j.1432-1033.1987.tb13597.x. [DOI] [PubMed] [Google Scholar]
- 13.DeRijcke M, Seneca S, Punyammalee B, Glansdorf N, Crabeel M. Characterization of the DNA target site for the yeast ARGR regulatory complex: a sequence able to mediate repression or induction by arginine. Mol Cell Biol. 1992;12:68–81. doi: 10.1128/mcb.12.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deschamps J, Dubois E, Wiame J-M. l-Ornithine transaminase synthesis in Saccharomyces cerevisiae: regulation by inducer exclusion. Mol Gen Genet. 1979;174:225–232. doi: 10.1007/BF00267794. [DOI] [PubMed] [Google Scholar]
- 15.Diffley J F X, Stillman B. Purification of a yeast protein that binds to origins of DNA replication and a transcriptional silencer. Proc Natl Acad Sci USA. 1988;85:2120–2124. doi: 10.1073/pnas.85.7.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diffley J F X, Stillman B. Similarity between the transcriptional silencer binding proteins ABF1 and RAP1. Science. 1989;246:1034–1038. doi: 10.1126/science.2511628. [DOI] [PubMed] [Google Scholar]
- 17.Dorrington R, Cooper T G. The DAL82 protein of Saccharomyces cerevisiae binds to the DAL upstream induction sequence (UIS) Nucleic Acids Res. 1993;21:3777–3784. doi: 10.1093/nar/21.16.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dubois E, Bercy J, Messenguy F. Characterization of two genes, ARGRI and ARGRIII, required for specific regulation of arginine metabolism in yeast. Mol Gen Genet. 1987;207:142–148. doi: 10.1007/BF00331501. [DOI] [PubMed] [Google Scholar]
- 19.Dubois E, Messenguy F. Isolation and characterization of the yeast ARGRII gene involved in regulating both anabolism and catabolism of arginine. Mol Gen Genet. 1985;198:283–289. doi: 10.1007/BF00383008. [DOI] [PubMed] [Google Scholar]
- 20.Guarente L. Yeast promoters and lacZ fusion designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- 21.Guarente L, Mason T. Heme regulatory transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell. 1983;32:1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- 22.Guarente L, Ptashne M. Fusion of Escherichia coli lacZ to the cytrochrome c gene of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1981;78:2199–2230. doi: 10.1073/pnas.78.4.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hennaut C. Ornithine transaminase synthesis in Saccharomyces cerevisiae: induction by allophanate, intermediate and inducer of the urea degradative pathway adds to arginine induction. Curr Genet. 1981;4:69–72. doi: 10.1007/BF00376788. [DOI] [PubMed] [Google Scholar]
- 24.Henry Y A, Chambers A, Tsang J S H, Kingsman A J, Kingsman S M. Characterization of the DNA binding domain of the yeast RAP1 protein. Nucleic Acids Res. 1990;18:2617–2623. doi: 10.1093/nar/18.9.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoet P R, Wiame J M. On the nature of argR mutations in Saccharomyces cerevisiae. Eur J Biochem. 1974;43:87–92. doi: 10.1111/j.1432-1033.1974.tb03388.x. [DOI] [PubMed] [Google Scholar]
- 26.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kovari L, Sumrada R, Kovari I, Cooper T G. Multiple positive and negative cis-acting elements mediate induced arginase (CAR1) gene expression in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:5087–5097. doi: 10.1128/mcb.10.10.5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kovari L Z, Cooper T G. Participation of ABF-1 protein in expression of the Saccharomyces cerevisiae CAR1 gene. J Bacteriol. 1991;173:6332–6338. doi: 10.1128/jb.173.20.6332-6338.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kovari L Z, Kovari I, Cooper T G. Participation of RAP1 protein in expression of the Saccharomyces cerevisiae arginase (CAR1) gene. J Bacteriol. 1993;175:941–951. doi: 10.1128/jb.175.4.941-951.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurtz S, Shore D. RAP1 protein activates and silences transcription of mating-type genes in yeast. Genes Dev. 1991;5:616–628. doi: 10.1101/gad.5.4.616. [DOI] [PubMed] [Google Scholar]
- 31.Luche R M, Sumrada R, Cooper T G. A cis-acting element present in multiple genes serves as a repressor-protein binding site for the yeast CAR1 gene. Mol Cell Biol. 1990;10:3884–3995. doi: 10.1128/mcb.10.8.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lustig A J, Kurtz S, Shore D. Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science. 1990;250:549–553. doi: 10.1126/science.2237406. [DOI] [PubMed] [Google Scholar]
- 33.Mandel M, Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970;53:159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- 34.Messenguy F, Dubois E. Genetic evidence for a role for MCM1 in the regulation of arginine metabolism in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:2586–2592. doi: 10.1128/mcb.13.4.2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Messenguy F, Dubois E, Boonchird C. Determination of the DNA-binding sequences of ARGR proteins to arginine anabolic and catabolic promoters. Mol Cell Biol. 1991;11:2852–2863. doi: 10.1128/mcb.11.5.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Messenguy F, Dubois E, Descamps F. Nucleotide sequence of the ARGRII regulatory gene and amino acid sequence homologies between ARGRII, PPRI and GAL4 regulatory proteins. Eur J Biochem. 1986;57:77–81. doi: 10.1111/j.1432-1033.1986.tb09640.x. [DOI] [PubMed] [Google Scholar]
- 37.Middlehoven W J. The pathway of arginine breakdown in Saccharomyces cerevisiae. Biochim Biophys Acta. 1964;93:650–652. doi: 10.1016/0304-4165(64)90349-6. [DOI] [PubMed] [Google Scholar]
- 38.Middlehoven W J. The derepression of arginase and ornithine transaminase in nitrogen-starved baker’s yeast. Biochim Biophys Acta. 1968;156:440–443. doi: 10.1016/0304-4165(68)90284-5. [DOI] [PubMed] [Google Scholar]
- 39.Middlehoven W J. Enzyme repression in the arginine pathway of Saccharomyces cerevisiae. Antonie Leeuwenhoek J Microbiol Serol. 1969;35:215–226. doi: 10.1007/BF02219132. [DOI] [PubMed] [Google Scholar]
- 40.Middlehoven W J. Induction and repression of arginase and ornithine transaminase in baker’s yeast. Antonie Leeuwenhoek J Microbiol Serol. 1970;36:1–19. doi: 10.1007/BF02069003. [DOI] [PubMed] [Google Scholar]
- 41.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 42.Park H-D, Luche R M, Cooper T G. The yeast UME6 gene product is required for transcriptional repression mediated by the CAR1 URS1 repressor binding site. Nucleic Acids Res. 1992;20:1909–1915. doi: 10.1093/nar/20.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qui H F, Dubois E, Broen P, Messenguy F. Functional analysis of ARGRI and ARGRIII regulatory proteins involved in the regulation of arginine metabolism in Saccharomyces cerevisiae. Mol Gen Genet. 1990;222:192–200. doi: 10.1007/BF00633817. [DOI] [PubMed] [Google Scholar]
- 44.Rai R, Daugherty J R, Cooper T G. UASNTR functioning in combination with other UAS elements underlies exceptional patterns of nitrogen regulation in Saccharomyces cerevisiae. Yeast. 1995;11:247–260. doi: 10.1002/yea.320110307. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 46.Shore D, Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987;51:721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- 47.Smart W C, Coffman J A, Cooper T G. Combinatorial regulation of the Saccharomyces cerevisiae CAR1 (arginase) gene promoter in response to multiple environmental signals. Mol Cell Biol. 1996;16:5876–5887. doi: 10.1128/mcb.16.10.5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strich R, Surosky R T, Steber C, Dubois E, Messenguy F, Esposito R E. UME6 is a key regulator of nitrogen repression and meiotic development. Genes Dev. 1994;8:796–810. doi: 10.1101/gad.8.7.796. [DOI] [PubMed] [Google Scholar]
- 48a.Sumrada, R. Personal communication.
- 49.Sumrada R, Cooper T G. Isolation of the CAR1 gene from Saccharomyces cerevisiae and analysis of its expression. Mol Cell Biol. 1982;2:1514–1523. doi: 10.1128/mcb.2.12.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sumrada R A, Cooper T G. Point mutation generates constitutive expression of an inducible eukaryotic gene. Proc Natl Acad Sci USA. 1985;82:643–647. doi: 10.1073/pnas.82.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sumrada R A, Cooper T G. Ubiquitous expression sequences control activation of inducible arginase gene in yeast. Proc Natl Acad Sci USA. 1987;84:3997–4001. doi: 10.1073/pnas.84.12.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Svetlov V, Cooper T G. The minimal transactivation region of Saccharomyces cerevisiae Gln3p is localized to 13 amino acids. J Bacteriol. 1997;179:7644–7652. doi: 10.1128/jb.179.24.7644-7652.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tabor S, Richardson C C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci USA. 1987;84:4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taylor G, McPherson M, Hames D. PCRII: a practical approach. New York, N.Y: IRL Press; 1994. [Google Scholar]
- 55.Tschumper G, Carbon J. Sequence of a yeast DNA fragment containing a chromosomal replicator and the TRP1 gene. Gene. 1980;10:1157–1166. doi: 10.1016/0378-1119(80)90133-x. [DOI] [PubMed] [Google Scholar]
- 56.Turoscy V, Cooper T G. Pleiotropic control of five eucaryotic genes by multiple regulatory elements. J Bacteriol. 1982;151:1237–1246. doi: 10.1128/jb.151.3.1237-1246.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Vuuren H J J, Daugherty J R, Rai R, Cooper T G. Upstream induction sequence, the cis-acting element required for response to the allantoin pathway inducer and enhancement of operation of the nitrogen-regulated upstream activation sequence in Saccharomyces cerevisiae. J Bacteriol. 1991;173:7186–7195. doi: 10.1128/jb.173.22.7186-7195.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Viljoen M, Kovari L Z, Kovari I A, Park H-D, van Vuuren H J J, Cooper T G. Tripartite structure of the Saccharomyces cerevisiae arginase (CAR1) gene inducer-responsive upstream activation sequence. J Bacteriol. 1992;174:6831–6839. doi: 10.1128/jb.174.21.6831-6839.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Whitney P A, Magasanik B. The induction of arginase in Saccharomyces cerevisiae. J Biol Chem. 1973;248:6197–6202. [PubMed] [Google Scholar]
- 60.Wiame J-M. The regulation of arginine metabolism in Saccharomyces cerevisiae: exclusion mechanism. Curr Top Cell Regul. 1971;4:1–38. [Google Scholar]
- 61.Yoo H-S, Cooper T G. The DAL7 promoter consists of multiple elements that cooperatively mediate regulation of the gene’s expression. Mol Cell Biol. 1989;9:3231–3243. doi: 10.1128/mcb.9.8.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]