Abstract
PHR1 and PHR2 encode putative glycosylphosphatidylinositol-anchored cell surface proteins of the opportunistic fungal pathogen Candida albicans. These proteins are functionally related, and their expression is modulated in relation to the pH of the ambient environment in vitro and in vivo. Deletion of either gene results in a pH-conditional defect in cell morphology and virulence. Multiple sequence alignments demonstrated a distant relationship between the Phr proteins and β-galactosidases. Based on this alignment, site-directed mutagenesis of the putative active-site residues of Phr1p and Phr2p was conducted and two conserved glutamate residues were shown to be essential for activity. By taking advantage of the pH-conditional expression of the genes, a temporal analysis of cell wall changes was performed following a shift of the mutants from permissive to nonpermissive pH. The mutations did not grossly affect the amount of polysaccharides in the wall but did alter their distribution. The most immediate alteration to occur was a fivefold increase in the rate of cross-linking between β-1,6-glycosylated mannoproteins and chitin. This increase was followed shortly thereafter by a decline in β-1,3-glucan-associated β-1,6-glucans and, within several generations, a fivefold increase in the chitin content of the walls. The increased accumulation of chitin-linked glucans was not due to a block in subsequent processing as determined by pulse-chase analysis. Rather, the results suggest that the glucans are diverted to chitin linkage due to the inability of the mutants to establish cross-links between β-1,6- and β-1,3-glucans. Based on these and previously published results, it is suggested that the Phr proteins process β-1,3-glucans and make available acceptor sites for the attachment of β-1,6-glucans.
PHR1 and PHR2 encode morphological functions of the opportunistic fungal pathogen Candida albicans (27, 41). Their expression varies in relation to the pH of the growth environment, both in vitro and in vivo (7, 27, 41). In vitro expression of PHR1 is detected only when the ambient pH is above 5.5 and increases at more alkaline pHs (41). PHR2 exhibits the inverse pattern, with maximal expression below pH 5 (27). A similar expression pattern during infection is suggested by the virulence pattern of the analogous mutants. Mutants lacking PHR1 are grossly attenuated in an animal model of hematogenous, disseminated candidiasis but retain virulence in a vaginal infection model (7). These models correspond to alkaline and acidic environments, respectively. Conversely, phr2Δ mutants are compromised in their ability to cause vaginal infections but not systemic infections (7). The unique counterbalanced expression pattern of these genes and their in vivo significance have prompted functional studies of the encoded proteins, Phr1p and Phr2p.
The proteins encoded by PHR1 and PHR2 are structurally and functionally related (27). Phr1p and Phr2p belong to a family of related fungal proteins typified by the GAS1 protein of Saccharomyces cerevisiae (30, 45). GAS1 was initially identified as encoding the predominant glycosylphosphatidylinositol-linked cell surface glycoprotein (30, 45). The functional relatedness of Gas1p and Phr1p was established by genetic complementation, and Phr1p is glycosylphosphatidylinositol anchored when it is expressed in yeast (46). The S. cerevisiae genome contains four additional GAS genes (36), and related proteins have been reported for Aspergillus fumigatus (26), Candida maltosa (29), and Schizosaccharomyces pombe (51). The identity between these proteins varies from 32 to 80%.
Aberrant cell morphology is a characteristic phenotype of mutants lacking these genes (27, 29, 34, 41). The cells become enlarged and rounded and have multiple buds (27, 29, 34, 41). The PHR1 and PHR2 mutants are distinctive in that the morphological defects are pH conditional. PHR1 mutants are morphologically abnormal at alkaline pH but normal at acidic pH (41). PHR2 mutants exhibit the inverse relationship between pH and morphology (27). Despite their altered morphology, the mutants remain osmotically stable and maintain normal secretion and cytoskeletal organization (27, 41). Growth is also compromised at the restrictive pH; PHR1 mutants exhibit a greatly reduced growth rate, while PHR2 mutants cease growth (28, 41).
Associated with the abnormal morphology are changes in the polysaccharides of the cell wall. Chitin levels increase 5- to 10-fold in both S. cerevisiae gas1Δ and C. albicans phr1Δ mutants (22, 31, 32, 36). Glucan content is reduced due to a substantial reduction in β-1,6-glucans and a modest reduction in β-1,3-glucans (29, 31, 32, 36). Altered cross-linking of glucans into the cell wall is suggested by an increase in the ratio of alkali-soluble to alkali-insoluble glucan (31, 32). However, this increase is observed in gas1Δ and phr1Δ mutants but not in the analogous epd1Δ mutant of C. maltosa (29). The release of β-1,3-glucans into the culture medium by GAS1 mutants also suggests altered glucan cross-linking (36). Mannoproteins are affected in both amount and localization. Mannan content increases 40%, and the synthesis of a particular mannoprotein, Cwp1p, is increased (22, 37). Approximately 40% of the β-1,6-glucosylated mannoproteins become cross-linked to chitin in the gas1Δ mutant, versus only 4% in normal cells, and these proteins are also released into the culture medium (22, 37). These phenotypes are not all specific to mutation of this family of genes. The increased content of chitin and its increased cross-linkage to β-1,6-glucosylated mannoproteins are also exhibited by FKS1 mutants, which lack β-1,3-glucan synthase, or KNR4 mutants, which are also affected in β-1,3-glucan synthesis (18, 19, 22, 36). Changes in glucan solubility also occur in hkr1Δ cells (50). This has led to the hypothesis that these alterations reflect compensatory changes in cell wall synthesis that are induced to maintain cell wall integrity (19, 31, 36). Thus, none of these phenotypes can be specifically attributed to the loss of Phr1p or its orthologs, and the functional relationship between these proteins and cell wall biosynthesis is unclear.
In this study the structural relationship between Phr1p and family 2 glycosidases (14, 15) was demonstrated, suggesting that Phr1p acts directly upon polysaccharides of the cell wall. By taking advantage of the pH-conditional defects of PHR1 and PHR2 mutants, a temporal analysis of cell wall alterations was performed. Sequential changes in cell wall composition were observed, and these indicated that the decline in β-1,6-glucan content and the increase in chitin content were secondary effects of the mutations. However, the mutations resulted in an immediate four- to fivefold increase in the rate of chitin cross-linking to β-1,6-glucan. Since these glucans are normally cross-linked to β-1,3-glucans and the level of β-1,3-glucans is not substantially altered, the results suggest that Phr1p and Phr2p are critical to the cross-linking of β-1,6- and β-1,3-glucans.
MATERIALS AND METHODS
Strains and growth conditions.
The C. albicans strains used in this study were CAF3 (9); CAS8, a phr1Δ mutant derived from CAF3 (41); CFM2, a phr2Δ mutant (27); and CFM3, a Phr2+ derivative of CFM2 (27). The strains were routinely cultured on YPD agar (44) at 30°C. The medium was adjusted with HCl to pH 4.5 to provide permissive growth conditions for the phr1Δ mutant or with NaOH to pH 7.0 for the phr2Δ mutant. For cell wall analyses, the strains were cultured in Medium 199 containing Earle’s salts and glutamine but lacking sodium bicarbonate (Gibco BRL) and containing 150 mM HEPES. The pH of the medium did not vary more than 0.2 pH unit over the course of the experiments. Since CFM3 and CAS8 are Urd− (9, 41), medium for these strains was supplemented with uridine (25 μg/ml).
Site-specific mutagenesis and mutant strain construction.
The glutamate codons encoding residues E169 and E270 of Phr1p were changed to glutamine codons by site-specific mutagenesis. The mutated alleles were generated by inverse-PCR amplification with oligonucleotide primers that incorporated the desired base change. The Q169 allele was produced with the primers RP169 (5′-TTACCAGCAAAAAATCCCAAAAC-3′) and LP169Q (5′-CCAAGTAACTAATAATCGTTCA-3′). RP169 was the inverse complement of nucleotides 481 to 503 of the PHR1 open reading frame. LP169Q was complementary to nucleotides 504 to 525 and incorporated a G-to-C transversion (underlined) at nucleotide 505. The Q270 allele was generated with the primers RP270Q (5′-TTGGGAGAAGAAGGCTGGGAT-3′) and LP270 (5′-TATGGTTGTAATGCTAACCGTCC-3′). RPQ270 was the inverse complement of nucleotides 790 to 810 and incorporated a G-to-C transversion (underlined) at nucleotide position 808. LP270 was complementary to nucleotides 811 to 833 and incorporated a silent A-to-T transversion (underlined) at position 816. As a control, a wild-type allele was generated in the same manner with the primers RPE270 (5′-TTCGGAGAAGAAGGCTGGGAT-3′) and LP270. RPE270 was identical to the wild-type sequence between positions 790 and 810. The silent mutation in RPE270 allowed the PCR-generated allele to be distinguished from the wild-type allele used as the template in the PCR. Plasmid pSMS-54 was used as the template DNA. This plasmid consists of a 2.1-kb EcoRI fragment encoding Phr1p and the 1.3-kb ScaI-XbaI fragment of the C. albicans URA3 gene in pUC18 (41).
The PCR mixture consisted of 1 μg of template DNA, 20 pmol of each primer, 50 nmol of each deoxynucleoside triphosphate, and 2.5 U of TaKaRa LA Taq (Panvera) in 50 μl of 1× LA Taq buffer. The mixture was preincubated 1 min a 94°C and then cycled 10 times with the following incubations: 30 s at 94°C, 30 s at 50°C, and 7.5 min at 68°C, concluding with a 10-min incubation at 72°C. Following amplification, DNA was precipitated from the reaction mixtures and digested with DpnI to fragment the template DNA. The desired product was purified by agarose gel electrophoresis, circularized by treatment with polynucleotide kinase and T4 ligase, and transformed into Escherichia coli XL1-Blue (Strategene). The presence of the desired base changes was verified by nucleotide sequence determination.
To test the effect of the mutations on Phr1p function, the plasmids were integrated into the phr1 locus of strain CAS8 (41). Targeted integration was directed by digestion of the plasmid DNA at the unique SacII site within PHR1. This site lies 5′ of the region deleted from the genomic allele so that recombination results in the newly integrated allele being expressed from the native promoter sequences. Transformed cells were selected as Urd+, and the location and structure of the integrated DNA were determined by Southern blot analysis. Transformation and selection were performed as previously described (27). The transformed strains were examined for pH-dependent morphological defects as described in reference 41.
Quantitation of cell wall carbohydrates.
Strain CAF3 or CAS8 was cultured in YPD medium to stationary phase at 25°C. These cells were inoculated into Medium 199–150 mM HEPES (pH 8.0) at a density of 2.5 × 106 cells/ml and incubated at 25°C for either one culture doubling or six culture doublings. Growth was monitored by measuring the optical density of the culture at 600 nm. For dry weight determinations, quadruplicate cell samples were collected on preweighed nitrocellulose filters (pore size, 0.45 μm) and dried in vacuo prior to being weighed. For carbohydrate analysis cell samples were collected in triplicate or quadruplicate from each culture by centrifugation. The pelleted cells were washed with ice-cold, sterile distilled water and stored at −70°C until use. The cell samples were analyzed directly or after breakage with glass beads to prepare a cell wall fraction. Both methods yielded similar results.
The samples were fractionated as described by Boone et al. (3). They were extracted three times in 0.5 ml of 0.75 M NaOH for 60 min at 75°C. The combined alkali extracts were neutralized with glacial acetic acid and were designated the alkali-soluble fractions. The alkali-insoluble material was washed once with 100 mM Tris (pH 7.5) and once with 10 mM Tris (pH 7.5) and suspended in 1 ml of 10 mM Tris (pH 7.5)–0.01% sodium azide containing 1 mg of the β-1,3-glucanase preparation Zymolyase 100T (ICN Pharmaceuticals). The suspension was incubated 16 h at 37°C with gentle mixing. Insoluble material was removed by centrifugation, and the supernatant solution was designated the Zymolyase-soluble fraction. Control experiments demonstrated that increasing the amount of Zymolyase, extending the incubation period, or treating the insoluble material for a second time failed to release additional carbohydrate. The material remaining after Zymolyase treatment was washed once with sterile distilled water and designated the Zymolyase-insoluble fraction. The hexose contents of these fractions were determined in triplicate by the phenol-sulfuric acid method of Dubois et al. (8), with glucose as a standard. The chitin content was determined as described by Bulawa et al. (4). The resulting values were expressed as micrograms of hexose or hexosamine per milligram (dry weight) of cells. Final values are the averages of results from at least three independent cultures.
Pulse-labeling of glucans.
The control strain, CAF3, and the Phr1− strain, CAS8, were cultured overnight to stationary phase at 25°C in YPD adjusted to pH 4.5. This pH is permissive for growth of CAS8. The overnight culture was used to inoculate 10 ml of Medium 199–150 mM HEPES, adjusted to the nonpermissive pH of 8.0 and preequilibrated to 25°C. The final cell density was 107 cells/ml. Two-milliliter portions of this culture were removed 60, 120, and 240 min postinoculation and mixed with 2 μCi of d-[U-14C]glucose (250 to 360 mCi/mmol; New England Nuclear). The samples were incubated 60 min in the presence of label before being harvested by centrifugation. The labeled cells were washed four times with ice-cold sterile distilled water, mixed with one aliquot of 3H-labeled carrier cells, pelleted by centrifugation, and stored at −70°C until use.
To prepare 3H-labeled carrier cells, a stationary-phase YPD culture of strain CAF3 was used to inoculate 10 ml of Medium 199–150 mM HEPES, pH 8.0, to a final cell density of 107 cells/ml. After 2 h of incubation, 100 μCi of d-[3-3H]glucose was added and incubation was continued for an additional 60 min. The labeled cells were collected by centrifugation and washed three times with ice-cold sterile distilled water. These cells were mixed with unlabeled washed cells from 50 ml of the stationary-phase YPD inoculum and dispensed into 25 equal aliquots.
Strains CFM2 and CFM3 were pulse-labeled in a similar manner except that the YPD medium was adjusted to pH 7.0 and Medium 199 was adjusted to pH 4.5. The cells were labeled with [14C]glucose at a single time point, 240 min postinoculation into Medium 199. [3H]glucose-labeled carrier cells were prepared from the control strain CFM3.
Pulse-chase analysis of glucans.
In the pulse-chase experiments, CAF3 and CAS8 were pulse-labeled as described in the preceding section. Labeling was initiated 4 h postinoculation into Medium 199 and terminated after 60 min. Half of the culture was collected and retained as the prechase sample. The remainder was collected by filtration on a 0.2-μm-pore-size filter and washed with Medium 199–2% glucose–150 mM HEPES adjusted to the permissive pH, 4.5. The washed cells were inoculated into Medium 199–2% glucose–150 mM HEPES, pH 4.5, at the original cell density and incubated 4 h prior to being harvested. The pre- and postchase samples were mixed with [3H]glucose-labeled carrier cells prepared as described in the previous section.
Pulse-chase experiments with the control strain CFM3 and the phr2Δ mutant CFM2 were conducted similarly with several exceptions. The medium was adjusted to pH 7.0 for permissive growth conditions and pH 4.5 for restrictive conditions. The cells were incubated for only 60 min at the restrictive pH prior to being pulse-labeled, and the chase period was 2 h in duration.
Fractionation and analysis of labeled cells.
Alkali-soluble glucans were prepared by the method of Hartland et al. (13). The Zymolyase-soluble fraction was prepared as described by Boone et al. (3) except that the alkali-insoluble material, which contained approximately 1 mg of hexose, was digested with 0.1 mg of Zymolyase 100T. Control experiments demonstrated that digestion was complete under these conditions. After removal of insoluble material by centrifugation, the solubilized material was fractionated by molecular sieve chromatography on a 1.5- by 100-cm column packed with Bio-Gel A 5m (Bio-Rad Laboratories) equilibrated with 0.01% sodium azide. The column was eluted at a flow rate of 0.3 ml/min, and 2-ml fractions were collected. The column was calibrated with linear dextrans.
The monosaccharide compositions of select fractions were determined by high-performance anion-exchange chromatography (HPAEC). The samples were hydrolyzed in 2 M trifluoroacetic acid (TFA) for 2 h at 100°C (11, 13), desalted, and applied to a Dionex CarboPac PA1 column. Isocratic elution with 16 mM NaOH at a flow rate of 1 ml/min was used to separate the sugars. Monosaccharide peaks were detected by pulsed amperometric detection (11), and 14C-labeled sugars were detected by time-resolved liquid scintillation counting.
To quantitate the label incorporated into Zymolyase-insoluble glucans, the material remaining after Zymolyase digestion was washed with distilled water and hydrolyzed in TFA (11, 13). The monosaccharides were fractionated by either thin-layer chromatography (TLC) (13) or HPAEC. For TLC separations, spots were visualized by phosphorimaging and identified by comparison with labeled standards. The spots were excised, eluted with H2O, and quantitated by liquid scintillation counting in a Beckman LS3801 scintillation counter with ScintiSafe 30% cocktail (Fischer Scientific). Internal standards were used to correct for crossover and counting efficiency. The 14C counts were corrected for differences in total incorporation of the cultures to account for differences in cell numbers and growth rate. These corrected values were expressed as either the ratio of 14C to 3H or 14C counts normalized for 3H recovery.
Analysis of Zymolyase-insoluble glucan.
The insoluble material remaining after Zymolyase treatment was further fractionated by digestion with chitinase (23). The pelleted material was washed twice with 50 mM Tris, pH 7.5, and twice with 50 mM potassium phosphate, pH 6.3. The washed material was suspended in 0.1 ml of 50 mM potassium phosphate (pH 6.3)–0.01% sodium azide containing 0.025 U of Serratia marcescens chitinase. Chitinase was either obtained commercially (Sigma) or purified from E. coli expressing the Serratia marcescens chiA gene. The recombinant enzyme was purified as described by Vorgias et al. (49) from E. coli A5178 (43), which was kindly supplied by A. Oppenheim. The digests were incubated at 30°C for 16 h with gentle mixing. Increasing the amount of enzyme or the duration of incubation or repeating the treatment did not increase the yield of released material. The chitinase-solubilized material was fractionated by molecular sieve chromatography on a 1.5- by 100-cm column packed with Bio-Gel P-2 (Bio-Rad Laboratories). The column was equilibrated with 0.1 M acetic acid and eluted at a flow rate of 0.17 ml/min. Two-milliliter fractions were collected. Alternatively, the samples were fractionated on a 1.5- by 10-cm Toyopearl HW-55S column (TosoHaas). The column was equilibrated with 0.1 M acetic acid and eluted at a flow rate of 0.3 ml/min, and 1-ml fractions were collected. The eluted fractions were analyzed for hexose content by the phenol-sulfuric acid assay (8) or for 3H and 14C contents by liquid scintillation counting. The monosaccharide composition of selected fractions was determined by HPAEC following TFA hydrolysis. Smith degradation of chitinase-solubilized material was conducted as described in reference 13).
RESULTS
Relationship between Phr1p and glycosidases.
A basic BLAST search of available databases readily identified a number of Phr1p-related sequences, but no highly significant alignments with proteins of known function were produced (1). It was noted, however, that β-galactosidases from one or more species appeared in seven of the nine search reports when various Phr1p homologs were individually compared with the nonredundant sequences in the GenBank database. The E values ranged from 0.003 to 8.8 with the BLOSUM matrix. This result was surprising given that some of the Phr1p homologs have as little as 33% identity in pairwise comparisons, and this level of identity suggested that there may be some distant relationship between Phr1p and β-galactosidases. To test this possibility, the sequences of Phr1p, Phr2p, and 7 other homologs were compared with the sequences of 12 β-galactosidases of microbial origin. Several highly significant (P = 0) blocks of conserved sequence were identified by the Gibbs sampling strategy (25) as implemented in the program MACAW (42). A subset of these alignments is shown in Fig. 1. The blocks ranged in size from 8 to 34 residues and encompassed the region between amino acids Asn41 and Tyr304 of Phr1p. Global alignment of this region (17) demonstrated that approximately 13% of the residues were conserved in 14 of the 21 compared sequences. Only 4% of the residues were conserved in all 21 sequences, but these included the Phr1p sequences GNE169 and E270Y. Notably, E169 and E270 were aligned with E461 and E537 of E. coli β-galactosidase. E461 and E537 are active-site residues serving as the proton donor and nucleophile, respectively (10). If the alignment was biologically meaningful, then mutation of E169 or E270 was predicted to inactivate Phr1p.
FIG. 1.
Alignment of Phr1p homologs and β-galactosidase sequences. The sequences of Phr1p, Phr2p, and seven related proteins were compared with β-galactosidases from 12 different microorganisms. Only four of the sequences are shown for simplicity. Boxed residues were present in at least 14 of the sequences. Asterisks indicate amino acids that were present in all 21 sequences. The dashes indicate gaps introduced to optimize the alignment. The underlined regions indicate conserved blocks identified by MACAW analysis. Each of the blocks had a P value of 0 and a search space (N) of 1.3460. S. xylosus, Staphylococcus xylosus.
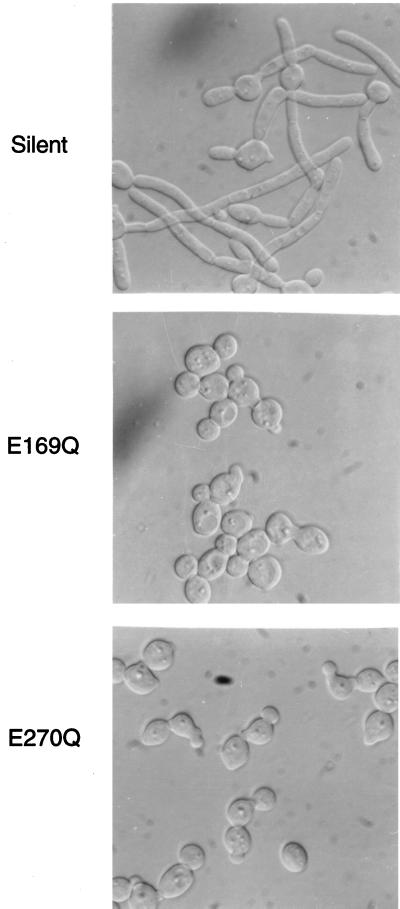
Mutagenesis studies of E. coli lacZ have demonstrated that a glutamine substitution at position E461 or E537 inactivates the protein (2, 5, 6). The analogous missense mutations were introduced into PHR1 at codon 169 or 270, and the mutated genes were tested for their ability to complement the morphological defects of a phr1Δ mutant. As shown in Fig. 2, neither the E169Q nor the E270Q mutant allele was able to complement the phr1Δ mutation. Introduction of a silent mutation into PHR1 yielded a functional protein (Fig. 2). Thus, as predicted from the alignment with β-galactosidases, E169 and E270 are essential for Phr1p activity, which suggests that Phr1p has a distant relationship with these glycosidases. The essential nature of these residues further implies that one or more of the changes in cell wall polysaccharides in phr1Δ mutants may be directly related to the loss of Phr1p activity.
FIG. 2.
Effect of site-specific mutagenesis on the function of PHR1. Mutant alleles of PHR1 were created by site-specific mutagenesis to introduce a silent mutation, an E169Q missense mutation, or an E270Q missense mutation. These mutants were tested for their ability to restore morphological development to a phr1Δ strain. The strains were induced to form hyphae in Medium 199 as previously described (27, 41).
Temporal changes in gross wall structure.
Previous studies demonstrated multiple changes in the polysaccharide composition and organization of the cell walls of PHR1, GAS1, and EPD1 mutants (29, 31, 35, 36). It is unknown which of these differences, if any, are directly related to the mutation. Interpretation of the data is further complicated by the suggestion that some of the observed changes reflect compensatory mechanisms induced to maintain the structural integrity of the wall (19, 31, 36). Since the primary effects of the mutation must occur prior to secondary alterations, temporal analysis can potentially distinguish between these. A temporal analysis of cell wall changes in the phr1Δ mutant was feasible because of its pH-conditional phenotype (41). The cells can be shifted from permissive to restrictive growth conditions, and progressive changes in the cell wall can be measured.
As a first step, the gross composition of cell wall fractions was measured following a shift from permissive to restrictive growth conditions. The phr1Δ mutant strain CAS8 and the parental control strain CAF3 were grown to stationary phase at the permissive pH of 4.5 and incubated an additional period of approximately 8 h. This was done to allow for the presumptive turnover and loss of functionally related Phr2p activity, which was expressed during growth at acidic pH. Thus, there would be no phenotypic lag upon inoculation into fresh medium of restrictive pH. Table 1 shows the results obtained with cells harvested one generation and six generations after inoculation into pH 8.0 medium. After one culture doubling there was no detectable difference between the mutant and parent in the total hexose contents of the alkali-extractable or Zymolyase-solubilized wall fractions. The chitin contents also appeared comparable. There was, however, a notable increase of about 2.5-fold in the amount of Zymolyase-insoluble wall material in the mutant.
TABLE 1.
Hexose contents of cell wall fractions following growth at pH 8.0
| Strain (genotype) | No. of generations | Cell wall fraction
|
Total hexose (avg ± SD) | |||
|---|---|---|---|---|---|---|
| Alkali extracteda | Soluble Zymolyasea | Insoluble Zymolyasea | Chitinb | |||
| CAF3 (PHR1/PHR1) | 1 | 169 ± 9 | 145 ± 51 | 5.8 ± 1.0 | 6.8 ± 1.1 | 337 ± 92 |
| CAS8 (phr1Δ/phr1Δ) | 1 | 172 ± 19 | 130 ± 25 | 15.3 ± 1.8 | 8.1 ± 1.4 | 344 ± 59 |
| CAF3 (PHR1/PHR1) | 6 | 176 ± 17 | 122 ± 5 | 5.4 ± 0.9 | 4.7 ± 0.7 | 303 ± 11 |
| CAS8 (phr1Δ/phr1Δ) | 6 | 157 ± 1 | 100 ± 8 | 28 ± 8 | 25.1 ± 3.4 | 285 ± 17 |
Values are reported as average milligrams of hexose per milligram (dry weight) of cells ± standard deviations of results from at least three experiments.
Values are reported as average milligrams of N-acetylglucosamine per milligram (dry weight) of cells ± standard deviations.
After six generations, additional changes were detected in the mutant, including a slight decline in the hexose content of the Zymolyase-solubilized fraction, a fivefold increase in chitin content, and a further increase in the Zymolyase-insoluble fraction. These delayed results are similar to those previously reported for PHR1, GAS1, and EPD1 mutants. None of these changes were detected when the mutant was grown at pH 4.5 or when a functional copy of PHR1 was introduced (data not shown). Unlike with GAS1 mutants, there was no detectable increase in secretion of polysaccharides into the medium after one generation at the restrictive pH (data not shown) (35, 36). As previously reported for the phr1Δ mutant (32), the total amount of hexoses in the cell wall did not change, suggesting that the mutation has little effect on polysaccharide synthesis per se.
Temporal changes in newly synthesized polysaccharides.
The foregoing results demonstrated that there were time-dependent changes occurring in the cell wall. The delayed appearance of some suggested that they were secondary to a defect(s) associated with the loss of Phr1p. Furthermore, the observation that alterations could be detected after a single generation under restrictive growth conditions suggested that the absence of Phr1p had an immediate effect(s) on cell wall biosynthesis. A limitation of these experiments was that after one generation of growth under restrictive conditions, the cell population consisted of 50% phenotypically mutant cells and 50% phenotypically normal cells, the latter being derived from the initial inoculum. The large fraction of phenotypically normal cells created a high background value against which changes in the mutant cells were measured. This high background obscured subtle changes in newly synthesized polysaccharides. To overcome this limitation, a pulse-labeling scheme was used with the assumption that only newly synthesized wall material would be labeled. Strains CAF3 and CAS8 were grown under nonrestrictive conditions as in the previous experiment. Following inoculation into fresh growth medium at the restrictive pH, the cells were pulse-labeled for 60 min with [14C]glucose. The pulse period was initiated 60, 120, or 240 min postinoculation. The earliest pulse period, 60 min, was chosen based on the observation that budding initiated between 45 and 60 min. The optical density at 600 nm of the culture doubled by approximately 240 min. To control for recovery, the 14C-labeled cells were mixed with a fixed amount of [3H]glucose-labeled CAF3 cells prior to being fractionated.
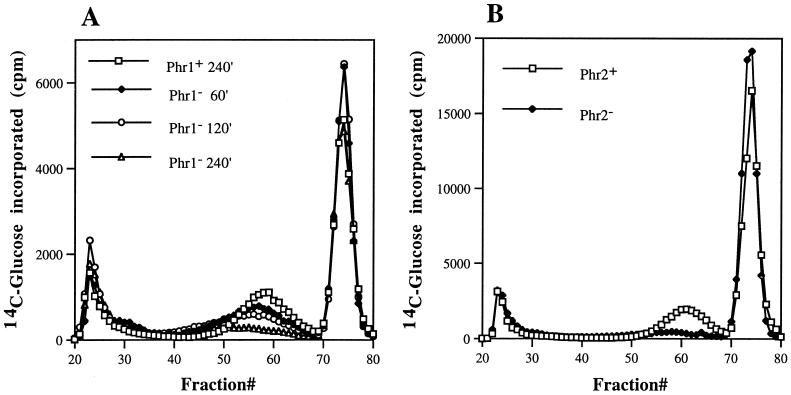
Consistent with the hexose measurements, incorporation of label into alkali-soluble glucans (13) occurred at similar rates in both the mutant and control cells during all three pulse periods (data not shown). However, differences in the levels of Zymolyase-soluble fraction were evident within one generation. Figure 3 shows the elution profile of this fraction when it was separated by molecular sieve chromatography. Three peaks were evident, one of which eluted in the void volume as an intermediate peak of approximately 47 kDa and another of which eluted as a peak of low-molecular-weight material at the end of the column. The last peak contained the products resulting from Zymolyase digestion of β-1,3-glucans. The control strain yielded identical profiles for all three labeling periods. The mutant exhibited only one reproducible change, a progressive decline in the 47-kDa peak. Synthesis of this material by the mutant was comparable to that of the wild type during the initial pulse period but declined by approximately 50% between 120 and 180 min postinoculation. Little or no incorporation occurred between 240 and 300 min. Composition determination by high-performance liquid chromatography demonstrated that >95% of the label in this material was present as glucose. This material was resistant to additional treatment with Zymolyase (results not shown) and apparently represented β-1,6-glucans as defined by Boone et al. (3). Smith degradation destroyed 74 to 78% of this glucan and reduced the remainder to low-molecular-weight products, a result consistent with a predominance of β-1,6-linked residues (data not shown).
FIG. 3.
Elution profile of [14C]glucose-labeled Zymolyase-soluble material fractionated by molecular sieve chromatography. (A) A Phr1+ control strain or a Phr1− null mutant was cultured at the restrictive pH for 60, 120, or 240 min and pulse-labeled for 60 min with [14C]glucose. The Zymolyase-solubilized wall material was prepared and fractionated on a 1.5- by 100-cm column of Bio-Gel A 5m. Since identical profiles were obtained for all three time points of the control strain, only one is shown. (B) Zymolyase-soluble material was prepared from a Phr2+ control strain or a Phr2− mutant cultured 240 min at the restrictive pH and then labeled for 60 min. The material was chromatographed as described for panel A.
The arrested synthesis of this β-1,6-glucan was also evident in unlabeled material. The Zymolyase-soluble fraction from phr1Δ or control cells cultured six generations at pH 8 was chromatographed in an identical manner, and the hexose content of each fraction was determined. The elution profile of the control sample was essentially identical to that of the radiolabeled sample, and the 47-kDa peak was absent from the mutant sample (data not shown). Identical results were obtained with three additional independent PHR1 null mutants. The loss of this glucan would account for the decline in the total hexose content of the Zymolyase-soluble fraction.
To determine if the reduced production of this presumptive β-1,6-glucan was specific to the loss of Phr1p, a phr2Δ mutant was examined. Previous studies had demonstrated that Phr1p and Phr2p are functionally related except that PHR2 functions during growth under acidic conditions and PHR1 acts during growth under alkaline conditions (27). Therefore, the phr2Δ mutant should exhibit a similar loss of β-1,6-glucan when it is cultured at pH 4. A phr2Δ mutant, strain CFM2, was incubated 4 h at the restrictive pH and then pulse-labeled for 1 h. As shown in Fig. 3, incorporation into the Zymolyase-solubilized β-1,6-glucan was barely detectable and dramatically reduced relative to that of the control strain. Thus, mutation of either homolog compromises the production of this β-1,6-glucan fraction.
A second notable difference detected in the total hexose determinations was an increase in the Zymolyase-insoluble fraction of the phr1 mutant. This fraction increased about 2.5-fold within one generation and approximately 5-fold within six generations. Incorporation experiments substantiated the increased synthesis of this glucan fraction. Mutant and control strains were pulse-labeled at the restrictive pH as in the preceding experiments. The wall material remaining after alkali extraction and Zymolyase digestion was hydrolyzed with TFA and fractionated by either TLC or high-performance liquid chromatography. The only labeled monosaccharide detected was glucose. In the control strain CAF3, the amounts of label incorporated were similar at each of the three time points examined (Table 2). Incorporation by the phr1Δ mutant was also constant at each time point but was elevated four- to fivefold above that by the control. The phr2Δ mutant behaved similarly (Table 2). Since the increased synthesis of this glucan fraction was evident within 60 min postinoculation, at the onset of budding and new wall synthesis, it appeared to be a proximate consequence of the absence of Phr1p or Phr2p. It preceded the decline in Zymolyase-solubilized β-1,6-glucan and the increase in chitin content.
TABLE 2.
Incorporation of [14C]glucose into alkali- and Zymolyase-insoluble glucans
| Strain | Phenotype | Time postinoculation (min)a | Avg amt of glucose incorporated (14C/3H) ± SD | Relative incorporationb |
|---|---|---|---|---|
| CAF3 | Phr1+ | 60 | 1.76 ± 0.98 | 1 |
| 120 | 1.51 ± 0.48 | 0.86 | ||
| 240 | 1.58 ± 0.72 | 0.90 | ||
| CAS8 | Phr1− | 60 | 7.77 ± 3.45 | 4.42 |
| 120 | 6.63 ± 3.39 | 3.77 | ||
| 240 | 9.42 ± 3.77 | 5.35 | ||
| CFM3 | Phr2+ | 60 | 1.34 ± 0.10 | 1 |
| 240 | 1.80 ± 0.50 | 1.34 | ||
| CFM2 | Phr2− | 60 | 4.59 ± 0.39 | 3.42 |
| 240 | 6.02 ± 0.68 | 4.49 |
Cultures were incubated for the indicated times prior to addition of [14C]glucose.
Values were calculated relative to the value determined for the 60-min control sample.
Pulse-chase analysis of Zymolyase-insoluble glucan.
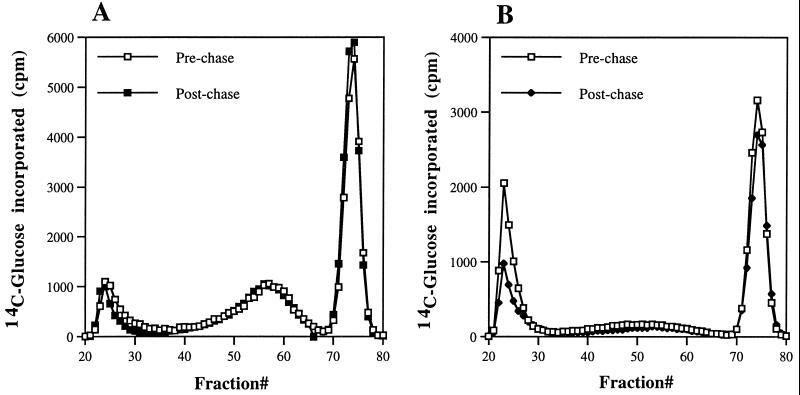
Blocking an intermediate step in a biochemical pathway typically leads to an accumulation of the precursor pools and a decline in product. The increased synthesis of Zymolyase-insoluble glucan and the subsequent decline in Zymolyase-released β-1,6 glucan suggested a possible precursor-product relationship. A pulse-chase experiment was performed to explore this possibility. Cells were incubated with radiolabeled glucose at the nonpermissive pH for 60 min to label the Zymolyase-insoluble glucan and then were shifted to a medium of permissive pH, containing an excess of unlabeled glucose. The amount of label in the glucan was quantitated before and after the shift to permissive growth conditions. A decline in the amount of label following the chase period would indicate that this glucan is an intermediate in cell wall assembly and that Phr1p is required for its processing. The results are tabulated in Table 3. Both the phr1Δ and phr2Δ mutants exhibited the expected increased incorporation into this fraction during the prechase labeling period. However, no diminution in label was evident in either the mutants or the control strains following the chase period. As an additional control, the Zymolyase-solubilized glucan from these samples was fractionated by molecular sieve chromatography to verify that there was no shift of label into the β-1,6-glucan fraction. The pre- and postchase fractionation profiles were indistinguishable (Fig. 4), further supporting the lack of a direct precursor-product relationship. These indistinguishable fractionation profiles suggest that the Zymolyase-insoluble glucan is an end product rather than an intermediate in cell wall assembly and that its accumulation does not reflect a Phr1p-dependent block in its processing.
TABLE 3.
Pulse-chase analysis of alkali- and Zymolyase-insoluble glucans
| Strain | Phenotype | Glucose incorporated (14C/3H)
|
Relative incorporationc
|
||
|---|---|---|---|---|---|
| Prechasea | Postchaseb | Prechase | Postchase | ||
| CAF3 | Phr1+ | 1.95 | 1.97 | 1 | 1.02 |
| CAS8 | Phr1− | 7.29 | 7.18 | 3.75 | 3.69 |
| CFM3 | Phr2+ | 0.76 | 0.98 | 1 | 1.29 |
| CFM2 | Phr2− | 3.97 | 4.90 | 5.23 | 6.45 |
CAF3 and CAS8 were cultured 4 h at the restrictive pH prior to being labeled. CFM3 and CFM2 were cultured 1 h.
CAF3 and CAS8 were cultured 4 h at the permissive pH. CFM3 and CFM2 were cultured 2 h.
Values were calculated relative to the value determined for the control.
FIG. 4.
Pulse-chase analysis of Zymolyase-soluble fraction. The Zymolyase-soluble fraction was prepared from control cells (A) or Phr1− cells (B) and chromatographed on a 1.5- by 100-cm column of Bio-Gel A 5m. The cells were pulse-labeled after 4 h at the restrictive pH, and label was chased for 4 h at the permissive pH.
Characterization of Zymolyase-insoluble glucan.
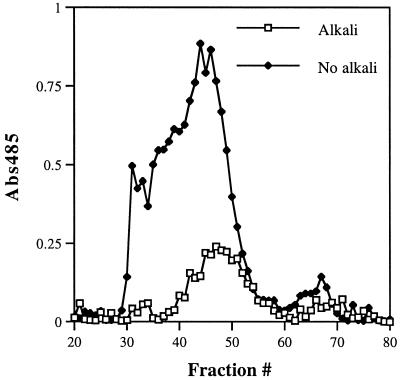
One of the phenotypes associated with a gas1Δ mutation in S. cerevisiae is a large increase in the amount of β-1,6-glucosylated mannoproteins cross-linked to chitin (22, 36). Assuming that the phr1Δ and phr2Δ mutants behaved similarly would account for the increase in Zymolyase-insoluble glucan. To ascertain if this glucan was associated with chitin, the Zymolyase-insoluble material from the mutants was digested with chitinase and separated into soluble and insoluble fractions. The chitinase-soluble material from the phr1Δ mutant had a relative 14C/3H ratio of 6.4 ± 1.7 (average ± standard deviation) compared to that of the CAF3 control sample. In contrast, the relative ratio of the chitinase-insoluble material was 1.5 ± 0.4. Similarly, the relative ratios from the phr2Δ mutant were 4.6 ± 1.1 and 1.4 ± 0.4 for the soluble and insoluble fractions, respectively. Thus, nearly all of the insoluble glucan that accumulates in the mutants was released by chitinase. Fractionation of the chitinase-solubilized glucan by molecular sieve chromatography demonstrated one major peak with a molecular mass 25.2 kDa (Fig. 5). Approximately 80% of this material was destroyed by Smith degradation (data not shown), suggesting that it contained mostly 1,6-linkages. Because the Zymolyase-insoluble glucan is normally a minor component of the cell wall (23, 24) (Table 1), it was not possible to obtain sufficient wild-type material for a comparable analysis.
FIG. 5.
Effect of alkali extraction on chitinase-solubilized material. A cell wall fraction was prepared from the phr2Δ mutant cultured 4 h at the restrictive pH. Half of the material was extracted with alkali prior to sequential digestion with Zymolyase and chitinase. The other half was not extracted prior to digestion. The chitinase-solubilized material was fractionated on a column of HW-55S, and the hexose in each fraction was measured as described in reference 8.
The foregoing results are consistent with those of the GAS1 mutant except for the absence of mannose. Compositional analysis of the 25.2-kDa chitinase-soluble material detected glucose alone, as was found in the previous analysis of the radiolabeled Zymolyase-insoluble fraction. This difference may reflect procedural differences, since alkali was used to extract mannoproteins prior to Zymolyase digestion. Al alkali extraction step was not used in the S. cerevisiae studies (22, 36). This possibility was tested by preparing the Zymolyase-insoluble, chitinase-soluble fraction from the phr2Δ mutant without prior alkali extraction. The elution profile of the resulting material was substantially different from that with prior alkali extraction and exhibited a number of peaks ranging in size from >250 kDa to approximately 25 kDa (Fig. 5). Compositional analysis of various column fractions demonstrated the presence of glucose and mannose in ratios varying from 28:1 for the highest-molecular-mass material to 2:1 for the 25-kDa peak. Similar results were obtained with the phr1Δ mutant. This suggests that the phr1Δ and phr2Δ mutations, like the gas1Δ mutation, result in an increase in the abundance of chitin-linked glucosylated mannoproteins.
DISCUSSION
In this report, the pH-conditional phenotype of the PHR1 and PHR2 mutants was used to distinguish cell wall changes that occur immediately following the loss of Phr1p or Phr2p. Previous studies examined mutants with a constitutive phenotype or gave no consideration to the duration of cultivation under restrictive conditions (22, 29, 31, 32, 36). The earliest detectable events are, presumably, more directly related to the absence of these proteins. The results demonstrated an immediate increase in the rate of cross-linking of glucans to chitin. This rate increase was followed by a subsequent decline in Zymolyase-releasable, and therefore β-1,3-glucan-linked, β-1,6-glucan and, at a much later time, an increase in chitin content. These changes, but not their temporal order, were noted in previous studies (22, 29, 31, 32, 36). In contrast to previous reports, there was little, if any, change in the content of alkali-insoluble β-1,3-glucan or alkali-soluble glucan. Thus, the ratio of these was unaltered. Release of β-1,3-glucans into the culture medium was not detected in preliminary experiments (unpublished results). The absence of the latter changes during the early stages of growth at the nonpermissive pH suggests that they represent delayed, indirect consequences of the lack of Phr1p or Phr2p.
The earliest detectable event in the mutants was an increased rate of glucan to chitin cross-linking. This glucan contains predominantly β-1,6-linkages, based on its susceptibility to periodate, and is associated with alkali-labile mannans. These features suggest that it corresponds to β-1,6-glucosylated mannoproteins previously demonstrated for C. albicans (20, 21, 40). These proteins are normally linked to β-1,3-glucans of the wall (20, 21, 40). Thus, in phr1Δ and phr2Δ mutants, as in the S. cerevisiae gas1Δ mutant, it appears that β-1,6-glucosylated mannoproteins become cross-linked to chitin and fail to establish their usual linkage to β-1,3-glucan.
There are two possible interpretations of the accumulation of chitin-linked glucans in the mutants. Either the β-1,6-glucosylated mannoproteins are blocked from cross-linking to β-1,3-glucans and diverted to an attachment to chitin or cross-linkage to chitin is a normal intermediate step in their processing and further processing is barred in the absence of Phr1p or Phr2p. The results of the pulse-chase experiments argue against the latter possibility. An incubation period of up to 4 h at the permissive pH failed to chase label from the chitin-linked glucan. This failure does not rule out the existence of spatiotemporal constraints that prevent processing of the glucan at a later time. However, it should be noted that the chitin-linked glucan was not released upon incubation of the Zymolyase-insoluble fractions of the mutants with partially purified Phr1p (unpublished results). Thus, there is no evidence to support the idea that the chitin-linked glucan is a processing intermediate. In the absence of such evidence, it appears that β-1,6-glucosylated mannoproteins are diverted from their normal incorporation path and cross-linked to chitin.
Because the mannoproteins are diverted from their typical linkage to β-1,3-glucan does not imply that cross-linkage to chitin is an abnormal event. In wild-type S. cerevisiae cells 1 to 2% of the wall mannoproteins are attached directly to chitin via their β-1,6-glucan moiety (22). This is not a unique subset of wall proteins since Cwp1p is present in both the β-1,3-glucanase-extractable and -nonextractable, chitin-linked fractions (22, 47). Furthermore, in the PHR1 and PHR2 mutants the increased rate of cross-linking between β-1,6-glucan and chitin was evident immediately upon resumption of cell wall synthesis at the restrictive pH and the rate was constant over the subsequent four hours. These results imply that the cross-linking enzyme(s) is normally present and not induced in response to specific defects in the mutants. Thus, the increased rate of chitin cross-linking seen in the mutants is likely a result of increased substrate availability due to the block in cross-linkage to β-1,3-glucan. From this perspective, the cross-linking of β-1,6-glucosylated mannoproteins to chitin may be seen as a scavenger mechanism to prevent release of those proteins that fail to become properly associated with the β-1,3-glucans.
Other notable outcomes of the temporal analysis of the PHR mutants were revisions in the presumed sequence of events and in the relationship between the increased cross-linkage of glucan to chitin and the increase in the chitin content of the cell wall. Previous studies reported a 5- to 13-fold increase in the chitin contents of gas1Δ and phr1Δ mutants (22, 31, 32). A fivefold increase was documented in the present analysis. However, this increase was detected only after five to six generations under restrictive growth conditions; no increase was detected after one generation. Whereas previous studies had presumed that the increased accumulation of chitin-linked β-1,6-glucosylated mannoproteins was a consequence of the increased amount of chitin (22, 31), precisely the opposite appears to be true. Increased cross-linking to chitin precedes and possibly induces chitin synthesis. The latter suggestion comes from the observation that the chitin binding dyes Calcofluor White and Congo red also stimulate chitin synthesis (38, 39). These dyes bind nascent chitin and prevent cocrystalization into microfibrils (16, 38, 48). In a similar manner, the increased cross-linkage of β-1,6-glucosylated mannoproteins to chitin may interfere with microfibril formation and induce chitin synthesis.
Given that the most immediate consequence of the absence of Phr1p or Phr2p is the diversion of β-1,6-glucosylated mannoproteins from a linkage with β-1,3-glucan to a linkage with chitin, what is Phr1p’s or Phr2p’s role in these processes? Multiple sequence alignments combined with site-specific mutagenesis suggested that Phr1p is related to family 2 glycosidases. Thus, Phr1p and Phr2p are likely to act directly on cell wall polysaccharides. The mutant phenotype would be consistent with their ability either to directly cross-link β-1,6- and β-1,3-glucans or to remodel the glucans in a manner that promotes cross-linkage. Although family 2 glycosidases consists of β-galactosidases and β-glucuronidases, this does not imply that Phr1p and Phr2p act upon either of these substrates. The classification of glycosidases is based upon structure, not substrate specificity (14, 15). Also, significant similarities to family 1 β-glucosidases could also be identified (unpublished results), and limited sequence similarity with plant endoglucanases has been reported (33).
An important clue is provided by parallel studies of the GEL1 gene of A. fumigatus, which encodes an ortholog of Phr1p. This gene was identified as encoding a novel enzyme that exhibits endo-β-1,3-glucanase, as well as β-1,3-glucanosyltransferase, activity (26). The transferase activity is unique in that it transfers a β-1,3-glucan to the nonreducing end of a second β-1,3-glucan, creating a new β-1,3 linkage between the chains (12). The enzyme did not utilize β-1,6-glucans as substrates (12). Phr1p and Phr2p were shown to have similar catalytic activities (26). Since Phr1p and Phr2p do not have activity toward β-1,6-glucan, it is unlikely that they directly cross-link β-1,6- and β-1,3-glucans.
The foregoing implies that Phr1p and Phr2p act specifically upon the β-1,3-glucans of the cell wall and that their activity is required for the subsequent attachment of β-1,6-glucosylated mannoproteins. Normally, the reducing terminus of the β-1,6-glucan is linked to the nonreducing terminus of the β-1,3-glucan (24). If the role of Phr1p and Phr2p is to generate nonreducing termini in the β-1,3-glucans for the attachment of β-1,6-glucans, this would account for the diversion of β-1,6-glucosylated mannoproteins to a linkage with chitin in the mutants. This hypothesis is made more attractive by the observation that mutations in functionally diverse and unrelated genes such as FSK1 and KNR4 exhibit similar increases in chitin-linked glucans (19, 22, 36). These mutations reduce β-1,3-glucan synthesis (19, 22, 36), which would also reduce the availability of β-1,6-glucan acceptor sites. In the phr1Δ and phr2Δ mutants the amounts of β-1,3-glucans are not altered, suggesting that the role of Phr1p and Phr2p is to create or make available nonreducing termini for the attachment of β-1,6-glucans.
ACKNOWLEDGMENTS
I thank Amadou Fall, Yonghong Zhang, and Abiodun Akintilo for their patience, persistence, and excellent technical assistance in conducting these experiments.
This work was supported by Public Health Service grant GM47727 from the National Institutes of Health and the Burroughs Wellcome Fund scholar award in molecular pathogenic mycology.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bader D E, Ring M, Huber R E. Site-directed mutagenic replacement of Glu-461 with Gln in β-galactosidase (E. coli): evidence that Glu-461 is important for activity. Biochem Biophys Res Commun. 1988;153:301–306. doi: 10.1016/s0006-291x(88)81222-1. [DOI] [PubMed] [Google Scholar]
- 3.Boone C, Sommer S S, Hensel A, Bussey H. Yeast KRE genes provide evidence for a pathway of cell wall β-glucan assembly. J Cell Biol. 1990;110:1833–1843. doi: 10.1083/jcb.110.5.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bulawa C E, Slater M, Cabib E, Au-Young J, Sburlati A, Adair W L J, Robbins P W. The S. cerevisiae structural gene for chitin synthase is not required for chitin synthesis in vivo. Cell. 1986;46:213–225. doi: 10.1016/0092-8674(86)90738-5. [DOI] [PubMed] [Google Scholar]
- 5.Cupples C G, Miller J H. Effects of amino acid substitutions at the active site in Escherichia coli β-galactosidase. Genetics. 1988;120:637–644. doi: 10.1093/genetics/120.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cupples C G, Miller J H, Huber R E. Determination of the roles of Glu-461 in β-galactosidase (Escherichia coli) using site-specific mutagenesis. J Biol Chem. 1990;265:5512–5518. [PubMed] [Google Scholar]
- 7.De Bernardis F, Mühlschlegel F A, Cassone A, Fonzi W A. The pH of the host niche controls gene expression in and virulence of Candida albicans. Infect Immun. 1998;66:3317–3325. doi: 10.1128/iai.66.7.3317-3325.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubois M, Gilles K A, Hamilton J K, Revers P A, Smith F. Colorimetric method for determination of sugars and related substances. Anal Chem. 1956;28:350–356. [Google Scholar]
- 9.Fonzi W A, Irwin M Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gebler J C, Aebersold R, Withers S G. Glu-537, not Glu-461, is the nucleophile in the active site of (lacZ) β-galactosidase from Escherichia coli. J Biol Chem. 1992;267:11126–11130. [PubMed] [Google Scholar]
- 11.Hardy M R. Monosaccharide analysis of glycoconjugates by high-performance anion-exchange chromatography with pulsed amperometric detection. Methods Enzymol. 1989;179:76–82. doi: 10.1016/0076-6879(89)79115-1. [DOI] [PubMed] [Google Scholar]
- 12.Hartland R P, Fontaine T, Debeaupuis J-P, Simenel C, Delepierre M, Latgé J-P. A novel β-(1-3)-glucanosyltransferase from the cell wall of Aspergillus fumigatus. J Biol Chem. 1996;271:26843–26849. doi: 10.1074/jbc.271.43.26843. [DOI] [PubMed] [Google Scholar]
- 13.Hartland R P, Vermeulen C A, Klis F M, Sietsma J H, Wessels J G H. The linkage of (1-3)-β-glucan to chitin during cell wall assembly in Saccharomyces cerevisiae. Yeast. 1994;10:1591–1599. doi: 10.1002/yea.320101208. [DOI] [PubMed] [Google Scholar]
- 14.Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henrissat B. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1993;293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herth W. Calcofluor White and Congo Red inhibit chitin microfibril assembly of Poterioochromonas: evidence for a gap between polymerization and microfibril formation. J Cell Biol. 1980;87:442–450. doi: 10.1083/jcb.87.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hogging D G, Sharp P M. CLUSTAL: a package for performing multiple sequence alignments on a microcomputer. Gene. 1988;173:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 18.Hong Z, Mann P, Brown N H, Tran L E, Shaw K J, Hare R S, DiDomenico B. Cloning and characterization of KNR4, a yeast gene involved in (1,3)-β-glucan synthesis. Mol Cell Biol. 1994;14:1017–1025. doi: 10.1128/mcb.14.2.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong Z, Mann P, Shaw K J, DiDomenico B. Analysis of β-glucans and chitin in a Saccharomyces cerevisiae cell wall mutant using high-performance liquid chromatography. Yeast. 1994;10:1083–1092. doi: 10.1002/yea.320100810. [DOI] [PubMed] [Google Scholar]
- 20.Kapteyn J C, Dijkgraaf G J P, Montijn R C, Klis F M. Glucosylation of cell wall proteins in regenerating spheroplasts of Candida albicans. FEMS Microbiol Lett. 1995;128:271–277. doi: 10.1111/j.1574-6968.1995.tb07535.x. [DOI] [PubMed] [Google Scholar]
- 21.Kapteyn J C, Montijn R C, Dijkgraff G J P, van den Ende H, Klis F M. Covalent association of β-1,3-glucan with β-1,6-glucosylated mannoproteins in cell walls of Candida albicans. J Bacteriol. 1995;177:3788–3792. doi: 10.1128/jb.177.13.3788-3792.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapteyn J C, Ram A F J, Groos E M, Kollar R, Montijn R C, van den Ende H, Llobell A, Klis F M. Altered extent of cross-linking of β1,6-glucosylated mannoproteins to chitin in Saccharomyces cerevisiae mutants with reduced cell wall β1,3-glucan content. J Bacteriol. 1997;179:6279–6284. doi: 10.1128/jb.179.20.6279-6284.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kollár R, Petrakova E, Ashwell G, Robbins P W, Cabib E. Architecture of the yeast cell wall: the linkage between chitin and β(1-3)-glucan. J Biol Chem. 1995;270:1170–1178. doi: 10.1074/jbc.270.3.1170. [DOI] [PubMed] [Google Scholar]
- 24.Kollár R, Reinhold B B, Petráková E, Yeh H J C, Ashwell G, Drgonová J, Kapteyn J C, Klis F M, Cabib E. Architecture of the yeast cell wall, β(1-6)-glucan interconnects mannoprotein, β(1-3)-glucan, and chitin. J Biol Chem. 1997;272:17762–17775. doi: 10.1074/jbc.272.28.17762. [DOI] [PubMed] [Google Scholar]
- 25.Lawrence C E, Altschul S F, Boguski M S, Liu J S, Neuwald A F, Wootton J C. Detecting subtle sequence signals: a Gibbs sampling strategy for multiple alignment. Science. 1993;262:208–214. doi: 10.1126/science.8211139. [DOI] [PubMed] [Google Scholar]
- 26.Mouyna, I., T. Fontaine, M. Vai, M. Monod, W. A. Fonzi, M. Diaquin, L. Popolo, B. Henrissat, R. P. Hartland, and J. P. Latgé. The glucanosyltransferase of Aspergillus fumigatus (Gel1p) responsible for the elongation of cell wall β(1-3)glucan is GPI-anchored and homologous to the Gas family of proteins. Submitted for publication.
- 27.Mühlschlegel F A, Fonzi W A. PHR2 of Candida albicans encodes a functional homolog of the pH-regulated gene PHR1 with an inverted pattern of expression. Mol Cell Biol. 1997;17:5960–5967. doi: 10.1128/mcb.17.10.5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mühlschlegel F A, Fonzi W A, Hoyer L, Payne T, Poulet F M, Clevenger J, Latgé J-P, Calera J, Beauvis A, Paris S, Monod M, Sturtevant J, Ghannoum M, Nozawa Y, Calderone R. Molecular mechanisms of virulence in fungus-host interactions for Aspergillus fumigatus and Candida albicans. Med Mycol. 1998;36(Suppl. 1):238–248. [PubMed] [Google Scholar]
- 29.Nakazawa T, Horiuchi H, Ohata A, Takagi M. Isolation and characterization of EPD1, an essential gene for pseudohyphal growth of a dimorphic yeast, Candida maltosa. J Bacteriol. 1998;180:2079–2086. doi: 10.1128/jb.180.8.2079-2086.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nuoffer C, Jeno P, Conzelmann A, Riezman H. Determinants for glycophospholipid anchoring of the Saccharomyces cerevisiae GAS1 protein to the plasma membrane. Mol Cell Biol. 1991;11:27–37. doi: 10.1128/mcb.11.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Popolo L, Gilardelli D, Bonfante P, Vai M. Increase in chitin as an essential response to defects in assembly of cell wall polymers in the ggp1D mutant of Saccharomyces cerevisiae. J Bacteriol. 1997;179:463–469. doi: 10.1128/jb.179.2.463-469.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Popolo L, Vai M. Defects in assembly of the extracellular matrix are responsible for altered morphogenesis of a Candida albicans phr1 mutant. J Bacteriol. 1998;180:163–166. doi: 10.1128/jb.180.1.163-166.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Popolo L, Vai M. The Gas1 glycoprotein, a putative wall polymer cross-linker. Biochim Biophys Acta. 1999;1426:385–400. doi: 10.1016/s0304-4165(98)00138-x. [DOI] [PubMed] [Google Scholar]
- 34.Popolo L, Vai M, Gatti E, Porello S, Bonfante P, Balestrini R, Alberghina L. Physiological analysis of mutants indicates involvement of the Saccharomyces cerevisiae GPI-anchored protein gp115 in morphogenesis and cell separation. J Bacteriol. 1993;175:1879–1885. doi: 10.1128/jb.175.7.1879-1885.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ram A F J, Brekelmans S S C, Oehlen L J W M, Klis F M. Identification of two cell cycle regulated genes affecting the β1,3-glucan content of cell walls in Saccharomyces cerevisiae. FEBS Lett. 1995;358:165–170. doi: 10.1016/0014-5793(94)01418-z. [DOI] [PubMed] [Google Scholar]
- 36.Ram A F J, Kapteyn J C, Montijn R C, Caro L H P, Douwes J E, Baginsky W, Mazur P, van den Ende H, Klis F M. Loss of the plasma membrane-bound protein Gas1p in Saccharomyces cerevisiae results in the release of β1,3-glucan into the medium and induces a compensation mechanism to ensure cell wall integrity. J Bacteriol. 1998;180:1418–1424. doi: 10.1128/jb.180.6.1418-1424.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ram A F J, Klis F M. Presented at the Morphogenesis and Metabolism in Opportunistic Fungal Pathogens Conference. 1995. , Poitiers, France, 3 to 8 October 1995. [Google Scholar]
- 38.Roncero C, Durán A. Effect of Calcofluor White and Congo red on fungal cell wall morphogenesis: in vivo activation of chitin polymerization. J Bacteriol. 1985;163:1180–1185. doi: 10.1128/jb.163.3.1180-1185.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roncero C, Valdivieso M H, Ribas J C, Durán A. Effect of Calcofluor White on chitin synthases from Saccharomyces cerevisiae. J Bacteriol. 1988;170:1945–1949. doi: 10.1128/jb.170.4.1945-1949.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanjuán R, Zueco J, Stock R, Font de Mora J, Sentandreu R. Identification of glucan-mannoprotein complexes in the cell wall of Candida albicans using a monoclonal antibody that reacts with a (1,6)-β-glucan epitope. Microbiology. 1995;141:1545–1551. doi: 10.1099/13500872-141-7-1545. [DOI] [PubMed] [Google Scholar]
- 41.Saporito-Irwin S M, Birse C E, Sypherd P S, Fonzi W A. PHR1, a regulated gene of Candida albicans, is required for morphogenesis. Mol Cell Biol. 1995;15:601–613. doi: 10.1128/mcb.15.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schuler G D, Altschul S F, Lipman D J. A workbench for multiple alignment construction and analysis. Proteins Struct Funct Genet. 1991;9:180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- 43.Shapira R, Ordentlich A, Chet I, Oppenheim A B. Control of plant diseases by chitinase expressed from cloned DNA in Escherichia coli. Phytopathology. 1989;79:1246–1249. [Google Scholar]
- 44.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratories; 1986. [Google Scholar]
- 45.Vai M, Gatti E, Lacana E, Popolo L, Alberghina L. Isolation and deduced amino acid sequence of the gene encoding gp115, a yeast glycophospholipid-anchored protein containing a serine-rich region. J Biol Chem. 1991;266:12242–12248. [PubMed] [Google Scholar]
- 46.Vai M, Orlandi I, Cavadini P, Alberghina L, Popolo L. Candida albicans homologue of GGP1/GAS1 gene is functional in Saccharomyces cerevisiae and contains the determinants for glycosylphosphatidylinositol attachment. Yeast. 1996;12:361–368. doi: 10.1002/(SICI)1097-0061(19960330)12:4%3C361::AID-YEA920%3E3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 47.Van der Vaart J M, Caro L H P, Chapman J W, Klis F M, Verrips C T. Identification of three mannoproteins in the cell wall of Saccharomyces cerevisiae. J Bacteriol. 1995;177:3104–3110. doi: 10.1128/jb.177.11.3104-3110.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vannini G L, Poli F, Donini A, Pancaldi S. Effects of Congo red on wall synthesis and morphogenesis in Saccharomyces cerevisiae. Plant Sci Lett. 1983;31:9–17. [Google Scholar]
- 49.Vorgias C E, Kingswell A J, Dauter Z, Oppenheim A B. Crystallization of recombinant chitinase from the cloned chiA gene of Serratia marcescens. J Mol Biol. 1992;226:897–898. doi: 10.1016/0022-2836(92)90640-6. [DOI] [PubMed] [Google Scholar]
- 50.Yabe T, Yamada-Okabe T, Kasahara S, Furuichi Y, Nakajima T, Ichishima E, Arisawa M, Yamada-Okabe H. HKR1 encodes a cell surface protein that regulates both cell wall β-glucan synthesis and budding pattern in the yeast Saccharomyces cerevisiae. J Bacteriol. 1996;178:477–483. doi: 10.1128/jb.178.2.477-483.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoshioka S, Kato K, Nakai K, Okayama H, Nojima H. Identification of open reading frames in Schizosaccharomyces pombe cDNAs. DNA Res. 1997;4:363–369. doi: 10.1093/dnares/4.6.363. [DOI] [PubMed] [Google Scholar]