Abstract
The chromosomal mercury resistance determinant of Bacillus cereus RC607 confers resistance to inorganic mercury and to organomercurials. The order of genes in the completed mercury resistance determinant is operator-promoter 1 (O/P1) merR1 merT open reading frame 3 (ORF3) ORF4 merA O/P2 merR2 merB2 merB1. The previously undetermined 1-kb DNA sequence between the merA and merB1 genes includes two significant ORFs, whose predicted protein products are homologous with MerR (the transcriptional regulator) and MerB (the organomercurial lyase enzyme). Two transcriptional start sites (promoters), O/P1 at the beginning of the determinant and O/P2 immediately upstream of the sixth ORF, the newly identified merR2, were mapped by reverse transcriptase (RT) primer extension. A long 6.3-kb mRNA traversing all eight ORFs was shown by RT-PCR. Growth sensitivity measurements in liquid media and cellular mercury volatization assays characterized inducibility and differences in functional activity in B. cereus RC607 and after cloning of the mer determinant into plasmids in Escherichia coli.
Metal resistance systems are well known in many bacterial types. The genes governing these resistances are generally (but not always) found on plasmids and encode resistances to toxic metal(loid) ions including Ag+, AsO2−, AsO43−, Cd2+, Co2+, CrO42−, Cu2+, Hg2+, Ni2+, Pb2+, Sb3+, TeO32−, Tl+, and Zn2+ (e.g., references 26 and 27). While most resistance systems function by energy-dependent efflux of toxic ions, some involve enzymatic transformations. The best-known example of these is mercurial resistance (16, 23, 27) that involves one or two enzymes: mercuric reductase, which converts soluble inorganic Hg2+ to Hg0, which is rapidly eliminated from aerobic microbial cultures as a gas, and organomercurial lyase, which cleaves the Hg-C bond of more toxic methylmercury, phenylmercury, and other organomercurials to less toxic inorganic Hg2+. In addition, all mercurial resistance systems have genes for Hg2+ transport to bring extracellular Hg2+ into the cell, where mercuric reductase is found. The logic for this counterintuitive finding of a transport system to bring a toxic compound into the cell is that extracellular Hg2+ itself is highly toxic and needs to be chaperoned from the initial binding site outside the cell to the intracellular reductase enzyme that depends on the high-energy intracellular cofactor NADPH. A regulatory protein, MerR (28, 30), provides tight control of expression, so that the gene products are made only at times of need. MerR is a positively acting regulatory protein that binds to the transcriptional mRNA start site, and on addition of Hg2+, MerR twists and bends the DNA to a conformation suitable for opening and initiation of mRNA synthesis (1, 30).
Although all mercury resistance systems have these functions (and genes) in common, the organization, and sometimes the occurrence, of genes differs between gram-positive and gram-negative bacteria (e.g., references 23 and 27). With one known exception, all mercury resistance systems of gram-negative bacteria start with a divergently (and therefore separately) transcribed merR gene. For mercury resistance systems of low-G+C gram-positive bacteria (18, 34), the merR gene is the first gene of the major transcript. In both gram-positive and -negative bacteria, the genes determining the Hg2+ transport system are promoter proximal, located upstream of the long merA gene for mercuric reductase. Most mercury resistance systems of gram-negative enteric bacteria lack a merB gene for organomercurial lyase (and are therefore called narrow spectrum since they do not confer resistance to most organomercurials). To date, all mercury resistance systems of gram-positive bacteria are broad spectrum and have the merB gene for organomercurial lyase.
The mercury resistance determinant of Bacillus cereus RC607 is unusual in several aspects, and it is also the most thoroughly studied mer system from a gram-positive bacterium. The Bacillus mer resistance determinant is located on the chromosome and not on a plasmid. The initial studies by Wang et al. (33, 34) reported two sequences with a gap in between. The first gene (initially called open reading frame 1 [ORF1] but now renamed merR1) encodes the positively acting homodimeric MerR protein, which has been studied in depth (12–14). We have identified a second regulatory gene, called merR2, in the newly sequenced gap region and the presence of a second operator-promoter region (O/P2) just upstream of merR2. This is the first occasion when two similarly oriented transcriptional start sites have been identified in a mercurial resistance system. The merA gene is long, with 632 codons and two 5′ motifs for metal-binding domains. The structure of the MerA protein of B. cereus RC607 was solved by X-ray crystallography (25) and is used as the model for all mercuric reductases from gram-positive or -negative bacteria (8). B. cereus MerA is still the only mercury resistance protein with a structural solution from crystallography. The crystal structure was missing the first 160 amino acids, forming the metal-binding motifs (25), leading to the suggestion that they lack a fixed position in the protein crystal. A second merB gene, now called merB2, has been identified (below) in the new sequence in B. cereus RC607. This is the first time two merB genes have been found in a single system in gram-positive bacteria, although two merB genes have been found previously in a Pseudomonas strain (17).
Understanding of the genetic and molecular properties of the mercury resistance determinant of B. cereus RC607 is important because very similar systems have been found in other laboratories with isolates of diverse environmental origins. Nakamura and Silver (20), Bogdanova et al. (3), and Hart et al. (11) found chromosomal determinants of mercury resistance with DNA properties similar to those of this Boston Harbor sediment B. cereus RC607 (19) in bacteria from marine sediments in Japan, soil samples from Russian mining sites, and freshwater river sediments in the United Kingdom, respectively. A system identical to that of B. cereus RC607 has now been identified in anaerobic gram-positive marine bacteria (bacilli and clostridia) in Japan (7a, 16a).
MATERIALS AND METHODS
Growth studies.
Resistance to HgCl2 and phenylmercuric acetate (PMA) of B. cereus RC607 (19), Bacillus subtilis 168, and Escherichia coli JM109, JM109(pUC19) (2), JM109(pYW33), and JM109(pYW40) (plasmids are described in reference 34) was measured in Luria-Bertani (LB) broth (2) containing HgCl2 or PMA. LB broth was inoculated with log-phase cells at a turbidity of 2 Klett units (equivalent to 20 μg [wet weight] of cells per ml), and growth (increase in Klett turbidity units) was measured after 20 h at 37°C.
Reductase assays.
Whole-cell mercuric reductase assays (e.g., references 21 and 34) for the conversion of Hg2+ to Hg0 was measured with B. cereus RC607 and E. coli JM109(pYW33) as test strains, E. coli JM109(pUC19) as a negative control, and E. coli J53(pGN120) (21) as a positive control. Overnight bacterial cultures were inoculated into fresh LB broth (at 20 μg [wet weight] of cells per ml) and grown at 30°C to a turbidity reading of about 50 to 70 Klett units. An aliquot of the uninduced (UI) cells was harvested by centrifugation and kept on ice. The remaining culture was induced (I) for 1 h by the addition of 1 μM Hg2+. The cell pellets were washed with chilled suspension buffer (50 mM sodium phosphate [pH 7.4], 0.5 mM Na2EDTA) and suspended at the equivalent of 2,000 Klett units. The cell suspension was added to 203Hg2+-containing assay buffer (total volume, 250 μl containing 50 mM sodium phosphate [pH 7.4], 0.5 mM Na2EDTA, 0.2 mM magnesium acetate, 1 mM β-mercaptoethanol, 5 μM HgCl2 [containing 203Hg2+], 0.5 mg of bovine serum albumin fraction V [Sigma Chemical Co., St. Louis, Mo.) per ml, and 250 μg of chloramphenicol per ml) to give a final turbidity value of 200 Klett units. The assay mixture was incubated at 30°C with rapid (200 rpm) shaking, and 25 μl of the assay mixture was periodically removed to 3 ml of water-miscible scintillation fluid. The remaining radioactivity in the samples was counted by a Packard Tri-Carb 1900CA liquid scintillation counter.
DNA sequencing.
To obtain the DNA sequence between the two Bacillus mer determinant sequences of Wang et al. (34), plasmid pYW40 was transformed into E. coli DH5α. Plasmid DNA was isolated and purified by Qiagen (Santa Clarita, Calif.) spin column purification and used for sequencing at the University of Illinois-Urbana DNA Sequencing Facility. The dye terminator dideoxy sequencing reaction protocol of Applied Biosystems was used, and analysis was done with an Applied Biosystems 373A automated DNA sequencer. Primers (boxed in Fig. 1B) were synthesized that started (i) 696 nucleotides (nt) after the stop codon of merA (5′CGCTAGTATCAAGGAAACGG3′; forward) and (ii) 110 nt upstream from the start of merB1 (5′CATAGCTTGTCTGATTTTTGA3′) and in the opposite orientation (reverse). From the first sequence data, a second set of primers was designed and used: 5′AGCTAAGCTGCCTAAAGAATC3′, starting 635 nt down from the start of the first forward primer, and 5′AACTGCCTGCCCATCACGAAT3′, starting 552 nt upstream from the start of the first reverse primer. The sequences were compiled and provided 1,200 nt of double-stranded data including 1,114 previously undetermined positions. Single-stranded sequences from the second set of primers extended in both directions more than 100 nt beyond the initial primers into the previously determined sequences of Wang et al. (34).
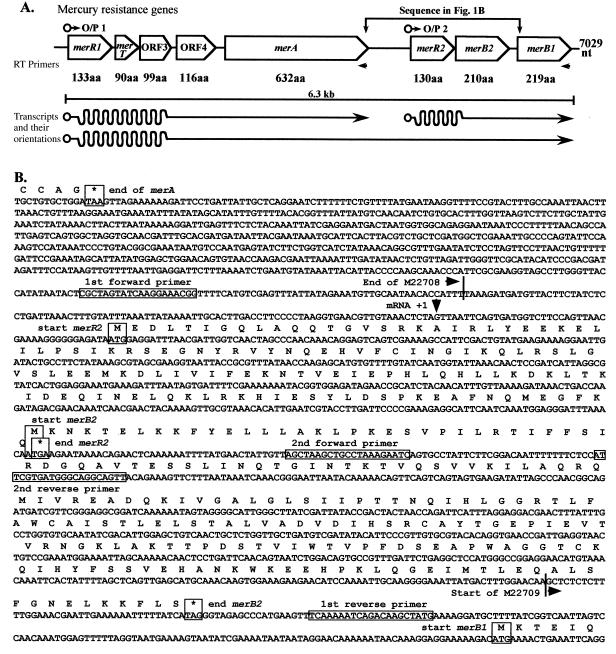
FIG. 1.
(A) Diagram of the chromosomal mercury resistance determinant of B. cereus RC607 showing the two determined O/P regions, the names of the genes, and the sizes of the gene products in amino acids (aa). The locations of the two oligonucleotide primers used for RT-PCR analysis and the three mRNA transcripts are shown. (B) DNA sequence of the region between merA and merB1 (as shown in panel A), with 100 nt per line, showing the newly completed region between the end of the sequence with GenBank accession no. M22708 and the start of the sequence with accession no. M22709. The final amino acid translation and termination codon of merA, the amino acid translation sequences of the new MerR2 and MerB2 protein products, and the first six amino acids of MerB1 are shown. The +1 first mRNA position for O/P2 is marked, and the positions of the four oligomer primers used for sequencing are boxed.
Transcript analysis.
RNA was isolated from uninduced cells or cells exposed to HgCl2 or PMA for 2 h at 37°C during growth in LB broth by using the RNeasy total RNA preparation kit (Qiagen Inc., Santa Clarita, Calif.). E. coli JM109(pYW33) (UI or I by growth with 5, 10, or 25 μM HgCl2 or 5 or 10 μM PMA) and B. cereus RC607 (UI or I with 10 μM HgCl2) were included. The RNA was treated with DNase (RNase free; Life Technologies, Gaithersburg, Md.). For reverse transcriptase PCR (RT-PCR) (10), 1 μg of RNA was used for cDNA synthesis with Superscript II RT in accordance with the manufacturer’s (Life Technologies) protocol. PCR was performed with PlatiTaq polymerase (Life Technologies), and amplification products were visualized in agarose gels after ethidium bromide staining and recorded by a Nucleotech gel documentation system with GelExpert 97 version 2.0.
For identification of transcription start sites by primer extension, 1 μg of RNA was used as the template for cDNA synthesis with Moloney murine leukemia virus RT (Promega Corporation, Madison, Wis.) in accordance with the manufacturer’s protocol. Sequencing ladder reactions were obtained with the Sequenase version 2.0 DNA sequencing kit (Amersham Life Science, Cleveland, Ohio) and the same oligonucleotide primers as used for primer extension. Primer extension and sequencing ladder products were separated on 7% polyacrylamide gels containing 7 M urea and visualized by exposure to X-Omat AR film (Eastman Kodak Company, Rochester, N.Y.).
Nucleotide sequence accession number.
The new sequence and assembled mer determinant of B. cereus RC607 has been assigned GenBank accession no. AF138877.
RESULTS
DNA sequence analysis.
Although this is a report on the transcriptional organization and expression of the mercurial resistance determinant of B. cereus RC607, the presence of a gap in the previous sequence (34) needed to be eliminated. Figure 1A shows the overall completed structure of the chromosomal Bacillus mer determinant with additional data obtained by sequence walking (starting with position 4100 of the sequence with accession no. M22708 and ending with position 216 of the sequence with GenBank accession no. M22709 [Fig. 1B], which is equivalent to positions 4100 to 6205 of the assembled new sequence in the GenBank database). The central 1,114 nt (shown in Fig. 1B) are new data, and the total sequence now consists of 7,029 nt and is available from GenBank under accession no. AF138877. The order of genes in the complete mercury resistance determinant is operator-promoter 1 (O/P1) merR1 merT ORF3 ORF4 merA O/P2 merR2 merB2 merB1 (Fig. 1A). The sequence shown in Fig. 1B starts just before the end of the merA gene (position 4100 of the sequence with GenBank accession no. M22708) and continues to just after the beginning of merB1 (position 210 of the sequence with GenBank accession no. M22709). The final position (nt 4875) of reference 34 (sequence with accession no. M22708 including merA) and the first position (nt 1) of reference 34 (sequence with accession no. M22709 including merB1) are noted in Fig. 1B.
Two significant ORFs were found in the new sequence, as shown in Figure 1. An ORF of 130 codons has a predicted product homologous (29% identical amino acids) to MerR of B. cereus RC607 (34). The corresponding gene is called merR2, and the previous gene is now called merR1. Overlapping the termination codon of merR2 by a single base (ATGA) (Fig. 1B) is the start codon of the second ORF, which is now called merB2, since from sequence homologies it appears to be the gene for an additional organomercurial lyase with (again) 29% of its amino acids identical to those of the previous gene product (now called MerB1). These sequence homologies will be considered further in the Discussion. Wang et al. (34) noted an AT-rich inverted repeat as a strong candidate for termination of transcription after merA. In the region between this proposed transcriptional stop site and the beginning of merR2, an additional transcriptional start site (Fig. 1) was identified by primer extension (see below).
Resistance to inorganic mercury and PMA.
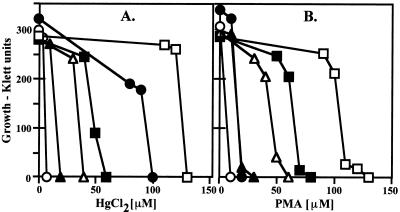
The mercury resistance determinant confers resistance to both Hg2+ and PMA (Fig. 2), in comparison to a control sensitive strain, B. subtilis 168. When it was cloned into plasmid pUC19 in E. coli (pYW33; reference 34), a higher level of resistance was obtained than with B. cereus RC607 (Fig. 2). This difference may reflect differing expression of gene products or different resistance levels of the host cells. Unexpectedly, transformation of the pUC19 control vector into E. coli resulted in a slightly higher level of resistance to both Hg2+ and PMA. The deletion form of pYW33, missing the merO/P1 promoter and the first four genes (pYW40), contains the intact merA, merR2, merB2, and merB1 genes and conferred an intermediate level of resistance to both Hg2+ and PMA (Fig. 2).
FIG. 2.
(A) Growth of B. cereus RC607 and E. coli containing cloned fragments in LB broth with added Hg2+ (A) and phenylmercury acetate (B) at 37°C for 20 h. Symbols: ○, B. subtilis 168 (sensitive control); ●, B. cereus RC607 (resistant); ▴ and ▵, E. coli JM109 and JM109(pUC19) (both sensitive); □ and ■, E. coli JM109 with cloned Bacillus mer fragments in plasmids pYW33 (resistant) and pYW40 (missing O/P1 and transport genes).
Volatilization of radioactive mercury.
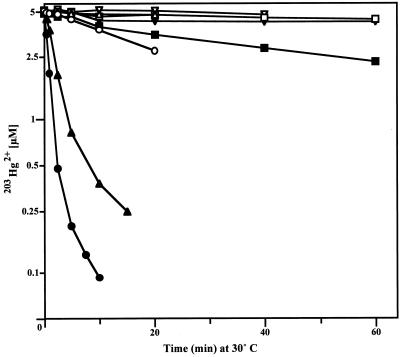
To measure inducibility and overall expression of mercuric reductase, the volatilization of radioactivity from added 5 μM 203Hg2+ was monitored (Fig. 3). B. cereus RC607 showed the most rapid loss of 203Hg2+, and the uninduced Bacillus cells showed approximately 3% of the rate of volatilization of induced cells. With E. coli JM109(pYW33) carrying the cloned Bacillus mer determinant, a much lower rate of volatilization was seen (70 times less than with B. cereus RC607; Fig. 3). This difference is not consistent with the higher resistance level (Fig. 2). How assay conditions such as media, temperature, and timing differences between growth and volatilization experiments explain this is not known. It was not a question of the bacterial species, E. coli versus Bacillus, as E. coli J53(pGN120) cells (21) with a mercury resistance determinant from a gram-negative bacterium volatilized 203Hg2+ rapidly (Fig. 3).
FIG. 3.
Volatilization of 203Hg2+ from cells of B. cereus RC607 (○ [UI] or ● [I]), and E. coli J53(pGN120) (▵ [UI] or ▴ [I]), and E. coli JM109(pYW33) (□ [UI] or ■ [I]) and JM109(pUC19) (▿ [UI] or ▾ [I]). Cells were grown in LB broth, I or UI, harvested, and suspended in assay buffer with 5 μM 203HgCl2 at 30°C with aeration and shaking. Samples were removed periodically, and residual radioactivity was counted in scintillation fluid.
Transcription of the mercury resistance determinant.
The transcription from the mercury resistance determinant of B. cereus RC607 was characterized by Northern blot RNA-DNA hybridization, primer extension determination of transcript start sites, and RT-PCR determination of which genes were traversed by a single transcript. Northern blot analysis with RNA from induced Bacillus cells did not show specific transcripts (data not shown). Whether this was due to instability of long transcripts (Fig. 1A) is not clear, but it is not unusual to be unable to isolate intact long transcripts in sufficient amounts for Northern blot analysis (e.g., reference 10).
Primer extension determination of transcriptional start sites.
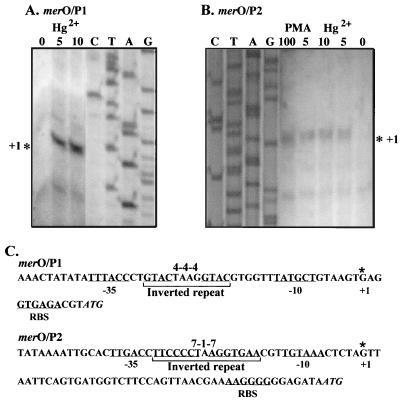
To locate the O/P1 transcriptional start site and to determine whether the second predicted transcriptional start site, O/P2, is used in vivo, primer extension analysis was used (Fig. 4). Sufficient RNA transcript for these experiments was not found in induced cells of B. cereus RC607 (data not shown), so the Bacillus mercury resistance determinant cloned in E. coli was used. RNA isolated from E. coli JM109(pYW33) cells induced with Hg2+ or PMA served as the template for cDNA synthesis using primers situated near the 5′ region of the first gene in each predicted transcript, merR1 (Fig. 4A) or merR2 (Fig. 4B). Specific products mapping to the start sites of both transcripts were obtained (Fig. 4), and the sites are marked in Fig. 1B and 4. O/P1-initiated transcription was induced by addition of Hg2+ (Fig. 4A) or PMA (data not shown). O/P2-initiated transcription was also induced by addition of either Hg2+ or PMA (Fig. 4B). A much longer untranslated mRNA region appears before the presumed ribosomal binding site (RBS) for the first gene after O/P2 than after O/P1 (Fig. 4C). Although 20 nt occur between the predicted −10 and −35 RNA polymerase binding sites of O/P1 (unusually long and associated with the bending and twisting of the DNA region with the 19-nt distance in the better-studied mer O/P in gram-negative bacteria; references 1 and 30), the distance between the predicted RNA polymerase binding sites for O/P2 is 18 nt, closer to the canonical length of 17 ± 1 nt. Both regions between the −10 and −35 sites contain inverted repeats (a perfect 4-4-4 repeat for O/P1 and an imperfect 7-1-7 repeat for O/P2; marked in Fig. 4C). Control reactions showed no corresponding RT transcription product with RNA from an E. coli strain lacking the plasmid-encoded mercury resistance determinant (data not shown). The transcriptional start sites (+1), deduced −10 and −35 RNA polymerase-interacting promoter sequences, proposed RBS, and polypeptide-initiating ATG for both transcripts are indicated in Fig. 4C.
FIG. 4.
Primer extension for start sites of mRNA for mer O/P1 (A) and mer O/P2 (B) with total cellular RNA from E. coli JM109(pYW33) either UI or I with added HgCl2 or PMA (values above the lanes are micromolar concentrations) for 2 h. The transcription start sites determined (marked by asterisks) were mapped against sequencing ladders (lanes C, T, A, and G) using the same oligonucleotide primers. (C) The mer O/P1 and mer O/P2 +1 mRNA initiation nucleotides determined and the predicted −10 and −35 RNA polymerase binding sites are shown. The predicted RBS and start codons for first genes, merR1 and merR2, are marked on the sequence. The perfect 4-4-4 and imperfect 7-1-7 inverted repeats between the −10 and −35 sites are also marked.
RT-PCR transcript analysis.
RT-PCR was used to analyze the bacterial transcripts for the strain RC607 mercury resistance system in both B. cereus and E. coli after cloning into plasmid pYW33. The Bacillus mer determinant was expressed from its own promoter(s) in E. coli, unlike the situation with the previously studied mer operon from plasmid pI258 of another low-G+C gram-positive bacterium, Staphylococcus aureus, which is not expressed in E. coli (6, 18, 29), presumably because of a failure of the MerR protein to interact productively with the heterologous RNA polymerase. Subcloning into E. coli allows comparisons of the mRNA products produced with both gram-positive (homologous) and gram-negative (heterologous) bacterial RNA polymerases, comparable to the volatilization assays shown in Fig. 3.
The Bacillus mer system might synthesize one or two transcripts (Fig. 1A). To identify the number of transcripts and to compare the expression of individual genes, total RNA was isolated from cells grown under induced (I) (growth for 60 min in medium supplemented with Hg2+) and uninduced (UI) conditions. Total RNA was isolated from I and UI cultures of B. cereus and E. coli and used as the template for RT reactions, using primers situated at the 3′ end of merA or merB1 (Fig. 1A). Additional RT oligonucleotide primers corresponding to the 3′ ends of the standardization control genes (for 16S rRNA) from B. cereus and E. coli were included in the same RT reactions as with mercury gene-specific merA and merB1 primers. The products of the RT reactions were templates for amplification of individual genes by PCR.
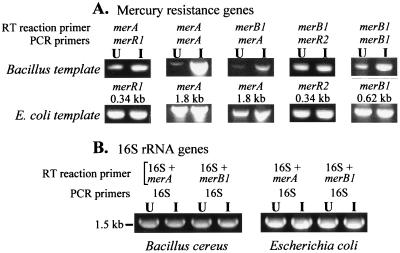
With RNA isolated from B. cereus cells and cDNA synthesized from the merA RT primer, both merR1 and merA were amplified by PCR (Fig. 5A). This shows that merR1 through merA were synthesized as a single transcript. The PCR products were quantitated (Table 1) from the original charge-coupled device camera data shown in Fig. 5. mRNAs for both merR1 and merA were less abundant in the UI cells than in the I cells. However, the induction ratio of apparent transcript abundance was seven times more for merA than for merR1 (Table 1). With total cellular RNA from E. coli JM109(pYW33), merR1 and merA were also PCR amplified by using cDNA from RT with the merA primer. However, the amounts of PCR products amplified for the UI and I E. coli cells were essentially the same (Fig. 5A; Table 1), showing no indication of induction by Hg2+. In control reactions, PCR products were not found when RNA was treated with RNase or when the RT reaction was run without RT (data not shown). PCR amplification products arising from contaminating DNA was ruled out by the absence of products when DNase-treated RNA was used and/or when the RNA sample was directly used as the template for a PCR (results not shown).
FIG. 5.
RT-PCR transcript analysis of the mercury resistance determinant and control 16S rRNA with total cellular RNA from B. cereus RC607 and E. coli JM109(pYW33), UI or I by growth with HgCl2 for 2 h. (A) Mercury resistance genes. RT primers and subsequent PCR primers are indicated for each reaction, as are the sizes of the PCR products detected. (B) Control 16S rRNA gene RT-PCR from the same reactions as with merA or merB1 primers. A quantitative analysis of the ethidium bromide-stained PCR products is shown in Table 1.
TABLE 1.
Quantitation of PCR products from RT-PCRa
| RT primer | PCR primer | Intensity (no. of pixels)
|
Induction ratio (UI:I)
|
||||
|---|---|---|---|---|---|---|---|
|
B. cereus
|
E. coli
|
||||||
| UI | I | UI | I | B. cereus | E. coli | ||
| merA | merR1 | 3,251 | 12,508 | 19,798 | 20,472 | 1:3.9 | 1:1.03 |
| merA | merA | 1,458 | 40,583 | 47,007 | 46,436 | 1:28 | 1:0.99 |
| merB1 | merA | 1,035 | 8,541 | 20,681 | 29,319 | 1:8.3 | 1:1.4 |
| merB1 | merR2 | 6,472 | 8,682 | 12,535 | 16,666 | 1:1.3 | 1:1.3 |
| merB1 | merB1 | 11,028 | 18,579 | 29,252 | 28,902 | 1:1.7 | 1:0.99 |
| 16S + merA | 16S | 25,627 | 24,900 | 38,573 | 42,136 | 1:0.97 | 1:1.09 |
| 16S + merB1 | 16S | 27,423 | 28,416 | 39,608 | 39,150 | 1:1.04 | 1:0.99 |
The ethidium bromide-stained gel was recorded by a Sony charge-coupled device camera attached to a Nucleotech gel documentation system. The RT-PCR amplification products were analyzed by GelExpert 97 version 2.0 software.
Using cDNA from RT with the merB1 primer, PCR products were amplified for three genes, merA, merR2, and merB1 (Fig. 5A). This established that mercury resistance genes merA through merB1 are synthesized as a single transcript and that the transcript does not invariably terminate after merA. Since genes merR1 through merA are cotranscribed and genes merA through merB1 are cotranscribed, all eight genes are considered to be cotranscribed as a single transcript of 6.3 kb. With RNA from B. cereus and the RT product obtained with the merB1 primer, the amounts of PCR products obtained with merA, merR2, and merB1 were greater when I cells were used than when UI cells were used (Fig. 5A; Table 1), although the apparent induction ratios were less than those obtained with the merA gene RT primer. There was no significant indication of inducibility with RT-PCR amplification of merA, merR2, and merB1 in the E. coli background (Table 1). Equivalent amounts of RT-PCR products for the 16S rRNA genes from B. cereus and E. coli were obtained with the I and UI cells (Fig. 5B; Table 1), showing that equivalent amounts of RNA were taken for the reactions.
DISCUSSION
Analysis of DNA sequence.
By combining the two sequences of reference 34 with the new sequence reported here, a continuous total of 7,029 nt (shown in Fig. 1A; available from GenBank under accession no. AF138877) was obtained. The G+C content is not uniform along the sequence. The first 4,875 nt have 39% G+C; the intermediate 2,100 bp in Fig. 1B contain 36% G+C. However, the merB1 gene contains 46% G+C. The hypothesis is that the Bacillus mercury resistance determinant evolved, probably originating with the merR1 through merA genes, with the additional genes merR2, merB2, and merB1 being added subsequently by horizontal transfer from outside this region. The two merR genes and the two merB genes are quite dissimilar in sequence, with only 42 to 43% nt matches upon alignment (analysis not shown), and therefore probably did not arise by gene duplication. It is not clear whether a segment containing the second promoter and the genes merR2 and merB2 was inserted after merA and before merB1 (Fig. 1). The merR2-merB2 region has a rather constant low G+C content, similar to that of the upstream mer region. The distal gene, merB1, alone shows a significantly higher G+C content, indicating a different origin.
This assemblage of the Bacillus mercury resistance determinant does not appear to have been a recent event. At least 95% of the mercury resistance determinants analyzed from Minamata Bay, Japan, marine Bacillus isolates have the same sizes, and apparently the same arrangement, of genes (20) as shown in this report for Massachusetts B. cereus isolate RC607 (19, 33). This similarity includes the 2.1 kb between the end of merA and the beginning of merB1 (Fig. 1B), which shows precisely the same size within the limitations of agarose gel analysis of PCR products (20).
The MerR and MerB protein families.
The MerR2 amino acid sequence is only 29% identical to that of MerR1. In fact, MerR2 is similarly related (not more so) to MerR sequences from gram-negative bacteria and to less-studied MerR paralogs that appear to function in the regulation of other cation-related genes (analysis not shown). The best characterized of these is ZntR (4), which is an E. coli chromosomally encoded MerR paralog that regulates cellular efflux of Zn2+. Thus, MerR2 appears to be a rather remote member of the larger family of proteins homologous to MerR. In contrast, MerR1 is 58% identical to MerR of S. aureus plasmid pI258. MerR2 is still less similar (about 20% of the amino acid are identical) to MerD sequences for the secondary transcriptional regulator from gram-negative bacteria, which are themselves weakly related to MerR, and MerR2 may be hypothesized to play a similar secondary down-regulatory role (28). The three completely invariant cysteines, Cys79, Cys114, and Cys123 in MerR1, of Hg2+-responding MerRs from both gram-positive and -negative bacteria (14, 28) are absent in MerR2 and are replaced with Ile81, Ser116, and Gly126; there are no alternative nearby cysteine residues. Therefore, MerR2 cannot respond to and bind Hg2+ in a manner similar to that of MerR1 (12, 13, 22).
An alternative role for MerR2 may be as a more general transcriptional regulator. In other systems, there are paralogous proteins, such as SoxR of E. coli, which responds to oxygen stress with an iron-sulfur cluster as a sensor (7, 15), and BmrR from B. subtilis, which functions as a positive transcriptional activator of multidrug resistance (35, 36). BmrR has a MerR-like amino-terminal DNA-binding domain and a dissimilar substrate-binding carboxyl-terminal region.
Helmann et al. (12) altered each of the four cysteine residues in B. cereus RC607 MerR1 to alanines and demonstrated that three of the four, Cys79, Cys114, and Cys123, are required for high-affinity binding of Hg2+ to the dimeric protein and also for transcriptional activation in vitro. The fourth cysteine, Cys12, is not required. By in vivo and in vitro heterodimer formation between mutant proteins affected in different residues, Helmann et al. (12, 13) showed that the Cys79 residue is required on one subunit of the MerR dimer and Cys114 and Cys123 are required on the other subunit. The three essential cysteines of Bacillus MerR are also found at equivalent positions in the MerRs of gram-negative bacteria, for which a similar trithiol dimer bridging for Hg2+ binding and activation has been shown (22, 31). The MerR transcriptional regulator of the gram-positive bacterium Streptomyces lividans (5), alone among MerR proteins, is not an activator but is a repressor and shows sequence homology to the ArsR/CadR/SmtB family of metal-responding transcriptional repressors in bacteria (27).
Purification of the MerR1 and MerR2 proteins and quantitative in vitro analysis of binding to mer O/P1 and O/P2 are needed to distinguish between the properties of MerR1 and MerR2 and to understand the function of MerR2.
All known MerB organomercurial lyase sequences are homologous, although the diversity of sequences is great. For example, B. cereus RC607 MerB1 and MerB2 show only 29% identical amino acids. MerB2 is about equally similar to MerB of S. aureus plasmid pI258 as to Bacillus MerB1. Organomercurial resistance was previously associated with the downstream region of the Bacillus mer determinant (34), before the separate merB1 and merB2 genes were known. A more detailed analysis is now needed to establish the substrate specificities of the two protein products, by separately eliminating merB1 and merB2 and testing against a spectrum of organomercurial compounds. Kiyono et al. (17) undertook a similar analysis with the two organomercurial lyase genes (and proteins) from a soil Pseudomonas strain, which was, in fact, the first mercury resistance strain to be studied in depth, more than 30 years ago. The level of understanding of the organomercurial lyase enzyme (32) does not allow the drawing of conclusions with regard to substrate specificity from the protein sequences.
Transcriptional control.
The MerR1 protein binds specifically to the mer O/P1 region in vitro, as shown by gel mobility shift assays and by protection against digestion by a GTAC-cutting restriction endonuclease (14). Runoff transcription assays (14) demonstrated that addition of the MerR protein repressed low-level activity in vitro and that addition of MerR plus Hg2+ resulted in positively regulated higher expression from mer O/P1. However, the precise position of the +1 nucleotide for mRNA synthesis had not been determined prior to the experiment whose results are shown in Fig. 4A and the existence of a second promoter site (Fig. 4C) had not been anticipated. When E. coli RNA polymerase rather than B. subtilis RNA polymerase (14) was used, no in vitro transcription from mer O/P1 occurred. The experiments whose results are shown in Fig. 3 (showing inducible volatilization of radioactive mercury) and Fig. 5 [RT-PCR analysis of mRNA from E. coli JM109(pYW33) cells] demonstrated that the B. cereus RC607 mer determinant can utilize E. coli RNA polymerase in vivo, although not equivalently to that of B. cereus RC607. Previous heterologous-expression studies of the related mer operon of S. aureus plasmid pI258 showed resistance when mer was cloned into B. subtilis but no phenotype when it was cloned into E. coli (18). Northern blot analysis (29) found that in S. aureus, full-length mer operon transcripts were synthesized although the genes are shorter and fewer than with B. cereus RC607. In RT experiments, Skinner et al. (29) identified the +1 mRNA start position equivalent to that shown here for B. cereus RC607 mer O/P1. Furthermore, S. aureus MerR bound and protected the S. aureus mer O/P DNA in DNase footprinting experiments (6) between the −35 and −10 RNA polymerase recognition sites as proposed here and in reference 14 for the Bacillus system. By sequence alignment of O/P regions from different mer systems, Park et al. (24) found that the repeat sequence GTAC----GTAC between the −10 and −35 elements was conserved and may be required for operator function in both gram-positive and -negative bacteria. In B. cereus RC607, mer O/P1 has the GTAC----GTAC repeat in equivalent positions (underlined in Fig. 4C), probably the site for binding of MerR1. mer O/P2 lacks the conserved GTAC----GTAC configuration but has a 7-1-7 repeat between the −10 and −35 elements (underlined in Fig. 4C). The 7-1-7 repeat sequence of mer O/P2 shares only 3 of the 8 nt to the 4-4-4 repeat of mer O/P1. There is, however, a 7-nt region, CTAAGGT, that is conserved between the repeat elements of mer O/P1 and mer O/P2. It is hypothesized that MerR1, if involved in the regulation of mer O/P2, may recognize the 7-nt conserved region near the center of the operator.
The most thorough analysis of differential synthesis of transcripts over the length of a mer operon was that of Gambill and Summers (9) with the E. coli Tn21 operon, which has only four genes in the major positively activated mer mRNA. Tn21 does not have merB genes. With Northern blot DNA-RNA analysis of abundance, Gambill and Summers (9) concluded that a transcriptional gradient occurred. Distal genes were transcribed more slowly and at lower levels. The quantitative analysis by RT-PCR started in the experiments whose results are shown in Fig. 5 needs to be extended before a quantitative picture of the two promoter sites and relative rates of mRNA synthesis and degradation can be made.
ACKNOWLEDGMENTS
This research was supported by Department of Energy grant ER20056.
We thank Paige Goodlove of the University of Illinois DNA Sequencing Center for care and effort that was truly of a collaborator rather than a support facility. Kunihiko Nakamura (Minamata, Japan) and Elena Bogdanova (Moscow, Russia) contributed to discussions of this work.
REFERENCES
- 1.Ansari A Z, Bradner J E, O’Halloran T V. DNA-bend modulation in a repressor-to-activator switching mechanism. Nature. 1995;374:371–375. doi: 10.1038/374370a0. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology, plus dated supplements. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 3.Bogdanova E S, Bass I A, Minakhin L S, Petrova M A, Mindlin S Z, Volodin A A, Kalyaeva E S, Tiedje J M, Hobman J L, Brown N L, Nikiforov V G. Horizontal spread of mer operons among gram-positive bacteria in natural environments. Microbiology. 1998;144:609–620. doi: 10.1099/00221287-144-3-609. [DOI] [PubMed] [Google Scholar]
- 4.Brocklehurst K R, Hobman J L, Lawley B, Blank L, Marshall S J, Brown N L, Morby A P. ZntR is a Zn(II)-responsive MerR-like transcriptional regulator of zntA in Escherichia coli. Mol Microbiol. 1999;31:893–902. doi: 10.1046/j.1365-2958.1999.01229.x. [DOI] [PubMed] [Google Scholar]
- 5.Brünker P, Rother D, Sedlmeier R, Klein J, Mattes R, Altenbuchner J. Regulation of the operon responsible for broad-spectrum mercury resistance in Streptomyces lividans 1326. Mol Gen Genet. 1996;251:307–315. doi: 10.1007/BF02172521. [DOI] [PubMed] [Google Scholar]
- 6.Chu L, Mukhopadhyay D, Yu H, Kim K-S, Misra T K. Regulation of the Staphylococcus aureus plasmid pI258 mercury resistance operon. J Bacteriol. 1992;174:7044–7047. doi: 10.1128/jb.174.21.7044-7047.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding H, Demple B. Thiol-mediated disassembly and reassembly of [2Fe-2S] clusters in the redox-regulated transcription factor SoxR. Biochemistry. 1998;37:17280–17286. doi: 10.1021/bi980532g. [DOI] [PubMed] [Google Scholar]
- 7a.Endo, G. Personal communication.
- 8.Engst S, Miller S M. Rapid reduction of Hg(II) by mercuric ion reductase does not require the conserved C-terminal cysteine pair using HgBr2 as the substrate. Biochemistry. 1998;37:11496–11507. doi: 10.1021/bi9808161. [DOI] [PubMed] [Google Scholar]
- 9.Gambill B D, Summers A O. Synthesis and degradation of the mRNA of the Tn21 mer operon. J Mol Biol. 1992;225:251–259. doi: 10.1016/0022-2836(92)90919-b. [DOI] [PubMed] [Google Scholar]
- 10.Gupta A. RT-PCR: identification of long multi-gene operons in bacteria. BioTechniques. 1999;27:2–5. doi: 10.2144/99275st04. [DOI] [PubMed] [Google Scholar]
- 11.Hart M C, Elliott G N, Osborn A M, Ritchie D A, Strike P. Diversity amongst Bacillus merA genes amplified from mercury resistant isolates and directly from mercury polluted soil. FEMS Microbiol Ecol. 1998;27:73–84. [Google Scholar]
- 12.Helmann J D, Ballard B T, Walsh C T. The MerR metalloregulatory protein binds mercuric ion as a tricoordinate, metal-bridged dimer. Science. 1990;247:946–948. doi: 10.1126/science.2305262. [DOI] [PubMed] [Google Scholar]
- 13.Helmann J D, Shewchuk L M, Walsh C T. Regulation of gene expression by mercury. Adv Inorg Biochem. 1990;8:33–61. [PubMed] [Google Scholar]
- 14.Helmann J D, Wang Y, Mahler I, Walsh C T. Homologous metalloregulatory proteins from both gram-positive and gram-negative bacteria control transcription of mercury resistance operons. J Bacteriol. 1989;171:222–229. doi: 10.1128/jb.171.1.222-229.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hidalgo E, Leautaud V, Demple B. The redox-regulated SoxR protein acts from a single DNA site as a repressor and an allosteric activator. EMBO J. 1998;17:2629–2636. doi: 10.1093/emboj/17.9.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hobman J L, Brown N L. Bacterial mercury-resistance genes. In: Sigel H, Sigel A, editors. Metal ions in biological systems. Vol. 34. New York, N.Y: Marcel Dekker, Inc.; 1997. pp. 527–568. [PubMed] [Google Scholar]
- 16a.Huang C C, Narita M, Yamagata T, Itoh Y, Endo G. Structure analysis of a class II transposon encoding the mercury resistance of the Gram-positive bacterium Bacillus megaterium MB1, a strain isolated from Minamata Bay, Japan. Gene. 1999;234:361–369. doi: 10.1016/s0378-1119(99)00184-5. [DOI] [PubMed] [Google Scholar]
- 17.Kiyono M, Omura T, Fujimori H, Pan-Hou H. Organomercurial resistance determinants in Pseudomonas K-62 are present on two plasmids. Arch Microbiol. 1995;163:242–247. doi: 10.1007/BF00393375. [DOI] [PubMed] [Google Scholar]
- 18.Laddaga R A, Chu L, Misra T K, Silver S. Nucleotide sequence and expression of the mercurial-resistance operon from Staphylococcus aureus plasmid pI258. Proc Natl Acad Sci USA. 1987;84:5106–5110. doi: 10.1073/pnas.84.15.5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahler I, Levinson H S, Wang Y, Halvorson H O. Cadmium- and mercury-resistant Bacillus strains from a salt marsh and from Boston Harbor. Appl Environ Microbiol. 1986;52:1293–1298. doi: 10.1128/aem.52.6.1293-1298.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakamura K, Silver S. Molecular analysis of mercury-resistant Bacillus isolates from sediment of Minamata Bay, Japan. Appl Environ Microbiol. 1994;60:4596–4599. doi: 10.1128/aem.60.12.4596-4599.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nucifora G, Chu L, Silver S, Misra T K. Mercury operon regulation by the merR gene of the organomercurial resistance system of plasmid pDU1358. J Bacteriol. 1989;171:4241–4247. doi: 10.1128/jb.171.8.4241-4247.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Halloran T V. Transition metals in control of gene expression. Science. 1993;261:715–725. doi: 10.1126/science.8342038. [DOI] [PubMed] [Google Scholar]
- 23.Osborn A M, Bruce K D, Strike P, Ritchie D A. Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon. FEMS Microbiol Rev. 1997;19:239–262. doi: 10.1111/j.1574-6976.1997.tb00300.x. [DOI] [PubMed] [Google Scholar]
- 24.Park S J, Wireman J, Summers A O. Genetic analysis of the Tn21 mer operator-promoter. J Bacteriol. 1992;174:2160–2171. doi: 10.1128/jb.174.7.2160-2171.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schiering N, Kabsch W, Moore M J, Distefano M D, Walsh C T, Pai E F. Structure of the detoxification catalyst mercuric ion reductase from Bacillus sp. strain RC607. Nature. 1991;352:168–171. doi: 10.1038/352168a0. [DOI] [PubMed] [Google Scholar]
- 26.Silver S. Genes for all metals—a bacterial view of the periodic table. J Ind Microbiol Biotechnol. 1998;20:1–12. doi: 10.1038/sj.jim.2900483. [DOI] [PubMed] [Google Scholar]
- 27.Silver S, Phung L T. Bacterial heavy metal resistance: new surprises. Annu Rev Microbiol. 1996;50:753–789. doi: 10.1146/annurev.micro.50.1.753. [DOI] [PubMed] [Google Scholar]
- 28.Silver S, Walderhaug M. Gene regulation of plasmid- and chromosome-determined inorganic ion transport in bacteria. Microbiol Rev. 1992;56:195–228. doi: 10.1128/mr.56.1.195-228.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skinner J S, Ribot E, Laddaga R A. Transcriptional analysis of the Staphylococcus aureus plasmid pI258 mercury resistance determinant. J Bacteriol. 1991;173:5234–5238. doi: 10.1128/jb.173.16.5234-5238.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Summers A O. Untwist and shout: a heavy metal-responsive transcriptional regulator. J Bacteriol. 1992;174:3097–3101. doi: 10.1128/jb.174.10.3097-3101.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Utschig L M, Bryson J W, O’Halloran T V. Mercury-199 NMR of the metal receptor site in MerR and its protein-DNA complex. Science. 1995;268:380–385. doi: 10.1126/science.7716541. [DOI] [PubMed] [Google Scholar]
- 32.Walts A E, Walsh C T. Bacterial organomercurial lyase: novel enzymatic protonolysis of organostannanes. J Am Chem Soc. 1988;110:1950–1953. [Google Scholar]
- 33.Wang Y, Mahler I, Levinson H S, Halvorson H O. Cloning and expression in Escherichia coli of chromosomal mercury resistance genes from a Bacillus sp. J Bacteriol. 1987;169:4848–4851. doi: 10.1128/jb.169.10.4848-4851.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Moore M, Levinson H S, Silver S, Walsh C, Mahler I. Nucleotide sequence of a chromosomal mercury resistance determinant from a Bacillus sp. with broad-spectrum mercury-resistance. J Bacteriol. 1989;171:83–92. doi: 10.1128/jb.171.1.83-92.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheleznova E E, Markham P N, Neyfakh A A, Brennan R G. Preliminary structural studies on the multi-ligand-binding domain of the transcription activator, BmrR, from Bacillus subtilis. Protein Sci. 1997;6:2465–2468. doi: 10.1002/pro.5560061122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheleznova E E, Markham P N, Neyfakh A A, Brennan R G. Structural basis of multidrug recognition by BmrR, a transcription activator of a multidrug transporter. Cell. 1999;96:353–362. doi: 10.1016/s0092-8674(00)80548-6. [DOI] [PubMed] [Google Scholar]