Abstract
The transcriptional organization of the erythromycin biosynthetic gene (ery) cluster of Saccharopolyspora erythraea has been examined by a variety of methods, including S1 nuclease protection assays, Northern blotting, Western blotting, and bioconversion analysis of erythromycin intermediates. The analysis was facilitated by the construction of novel mutants containing a S. erythraea transcriptional terminator within the eryAI, eryAIII, eryBIII, eryBIV, eryBV, eryBVI, eryCIV, and eryCVI genes and additionally by an eryAI −10 promoter mutant. All mutant strains demonstrated polar effects on the transcription of downstream ery biosynthetic genes. Our results demonstrate that the ery gene cluster contains four major polycistronic transcriptional units, the largest one extending approximately 35 kb from eryAI to eryG. Two overlapping polycistronic transcripts extending from eryBIV to eryBVII were identified. In addition, seven ery cluster promoter transcription start sites, one each beginning at eryAI, eryBI, eryBIII, eryBVI, and eryK and two beginning at eryBIV, were determined.
Saccharopolyspora erythraea, a mycelium-forming actinomycete, is the major producer of the clinically important macrolide antibiotic erythromycin. Extensive genetic studies have provided some insight into the genes involved in erythromycin biosynthesis (9, 17, 38). The erythromycin biosynthetic genes are clustered on the S. erythraea chromosome similarly to other secondary metabolic pathway genes (3, 11, 14, 21, 22, 27). The erythromycin gene cluster contains 20 genes involved in the biosynthesis of erythromycin A. The genes involved in the biosynthesis of the polyketide ring, the biosynthesis and attachment of mycarose to the macrolide ring, and the biosynthesis and attachment of desosamine to the macrolide ring have been designated eryA, eryB, and eryC genes, respectively. Additionally, there are three genes encoding modifying enzymes, designated eryF, eryG, and eryK, as well as ermE, encoding the rRNA methylase conferring erythromycin resistance on the host organism. Finally, two open reading frames (ORFs), eryBI, which is not essential for erythromycin A biosynthesis (13), and orf5, encoding a putative type II thioesterase (15), are also located in the ery gene cluster.
The central portion of the biosynthetic cluster contains the three eryA genes encoding a type I polyketide synthase (4, 8). The left flank (conventional ery cluster orientation [see Fig. 2]) contains two eryB genes (eryBII and eryBIII); three eryC genes (eryCI to eryCIII); two genes encoding erythromycin-modifying enzymes, eryF (a C-6 hydroxylase) and eryG (an O-methyltransferase); ermE; eryBI, encoding a proposed β-glucosidase; and orf5, encoding a putative type II thioesterase. The right flank contains four eryB genes (eryBIV to eryBVII), three eryC genes (eryCIV to eryCVI), and eryK (encoding a C-12 hydroxylase [31]).
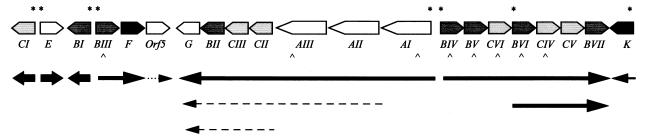
FIG. 2.
Transcriptional map of the 56-kb erythromycin biosynthetic gene cluster illustrating known and predicted transcripts. The thick arrows represent monocistronic transcripts identified by S1 mapping in this study and by Bibb et al. (2). The thin arrows represent polycistronic messages identified in this study, the longest of which extends 35 kb, from eryAI to eryG. The dashed arrows represent putative messages which have not been experimentally verified. The dotted arrow represents a potentially transcribed gene. The asterisks indicate promoter regions. The carets below the genetic map indicate the genes for which mutants containing a transcriptional terminator were constructed in this study.
Previous transcriptional studies by Bibb et al. (2) have shown that ermE and eryCI are transcribed in opposite directions. However, a detailed transcriptional analysis of the entire ery gene cluster has yet to be reported. Reeve and Baumberg recently reported the effects of low levels of phosphate, glucose, and ammonium on ery mRNA expression (25). Here, we present the results of a transcriptional and biochemical analysis of the majority of the erythromycin biosynthetic gene cluster. A series of novel mutants containing either an S. erythraea transcriptional terminator inserted into genes located throughout the ery cluster or an altered eryAI −10 promoter region were constructed and analyzed by either S1 nuclease protection assay, Northern blotting, Western blotting, or bioconversion analysis with erythromycin intermediates. The results indicate that the ery gene cluster contains four major polycistronic transcriptional units, the largest one extending approximately 35 kb, from eryAI to eryG. The transcription start sites for seven ery cluster promoters were also determined.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and plasmids.
The bacterial strains used in this study are described in Table 1. S. erythraea was grown in ABB20 medium (corn flour, 5.7 g/liter; soy flour, 11.5 g/liter; dried brewer’s yeast, 1.5 g/liter; sucrose, 1.0 g/liter; CaCO3, 1.7 g/liter; Edsoy oil, 2.5 ml per 50 ml of medium) from spore preparations maintained on R3M agar plates (16) at 33°C. After 48 h of growth in ABB20 medium, 2.5 ml of cells was transferred to 50 ml of SCM medium (23). S. erythraea CA340, an industrially improved erythromycin-producing strain, was maintained as spore preparations on ABB13 (soytone, 5.0 g/liter; soluble starch, 5.0 g/liter; CaCO3, 3.0 g/liter; MOPS (morpholine propane sulfonic acid), 2.1 g/liter; thiamine-HCl, 0.01 g/liter; FeSO4, 0.012 g/liter) agar plates or as −80°C glycerol stocks and grown under the same conditions as S. erythraea NRRL2338. Escherichia coli was grown either on Luria-Bertani agar plates or in Luria-Bertani broth (29) at 33°C. The antibiotics used for the selection of E. coli plasmids or S. erythraea integrants were ampicillin (100 μg/ml), thiostrepton (20 μg/ml), and hygromycin (80 to 200 μg/ml).
TABLE 1.
Bacterial strains and plasmids
| Plasmid or strain | Description | Reference or source |
|---|---|---|
| E. coli | ||
| pWHM3 | Apr (Thior) E. coli-S. erythraea vector used for making insertions in the S. erythraea chromosome | 34 |
| pAIX-5 | Apr pUC19 containing a 5.5-kb XhoI-XhoI fragment from the eryAI gene to the eryCIV gene | |
| pLitmus | Apr pUC19-based E. coli shuttle vector used for subcloning S. erythraea DNA | New England Biolabs |
| pDPE148A | Apr pLitmus29 containing a 300-bp AvrII fragment containing the S. erythraea rrn terminator | This study |
| pDPE149 | Apr pLitmus29 containing a cloned S. erythraea terminator | This study |
| pDPE27 | Apr pBR322-based E. coli shuttle vector containing the S. erythraea eryCVI, eryBVI, eryCIV, and eryCV genes | This study |
| pDPE45 | Apr pUC18 containing an 2.9-kb HindIII/SstI fragment of S. erythraea DNA extending from an internal SstI site in eryAI to a HindIII site of eryBIV | This study |
| pDPE46 | Apr pDPE45 containing the JVI replicon of Streptomyces phaeochromogenes | 1, this study |
| pARR1 | Apr pDPE45 containing a 300-bp SpeI/XbaI rrn terminator sequence from pDPE148A cloned into the SpeI site | This study |
| pARR2 | Apr (Thior) pWHM3 containing a 4.0-kb HindIII/SstI fragment used to make the eryBIV transcriptional mutant | This study |
| pARR3 | Apr pDPE45 with a 1.59-kb HindIII/NcoI fragment deleted | This study |
| pARR4 | Apr pARR3 containing a 300-bp EcoRI/SstI rrn terminator sequence from pDPE148A | This study |
| pARR5 | Apr pARR4 containing a 1.1-kb SstI/PstI eryAI fragment from pAIX-5 | This study |
| pARR8 | Apr (Thior) pWHM3 containing a 3.3-kb fragment from pARR5 containing the rrn terminator within the eryAI gene | This study |
| pARR9 | Apr; same as pARR5 but containing most of the eryBV gene | This study |
| pARR16 | Apr (Hygr) pJV1-based vector used for constructing eryAI S. erythraea CA340 insertion mutant | This study |
| pARR17 | Apr (Hygr); same as pARR16 but containing an eryAI/rrn terminator cassette | This study |
| pARR47 | Apr pUC18 containing a 200-bp EcoRI/SmaI PCR fragment from the eryAI promoter region; used to introduce half a SmaI site at the eryAI −10 hexamer | This study |
| pARR48 | Apr pARR47 containing an additional 400-bp PCR fragment cloned into the SmaI site; used to create a complete SmaI site at the eryAI −10 hexamer | This study |
| pARR49 | Apr pARR3 containing the mutated eryAI −10 hexamer cloned as a 600-bp BclI/ClaI fragment | This study |
| pARR50 | Apr Thior pWHM3-based vector containing the mutated eryAI −10 hexamer plus additional 1.4-kb eryAI and eryBIV DNA; used to make the eryAI −10 hexamer insertional mutant | This study |
| pKAS132 | Apr pDPE27 containing the rrn terminator cloned into the eryBVI gene | This study |
| pKAS133 | Apr pDPE27 containing the rrn terminator cloned into the eryCIV gene | This study |
| pKAS134 | Apr (Thior) pWHM3 containing a 4.6-kb HindIII/SphI fragment from pKAS132; used for integrating the terminator in the eryBVI gene | This study |
| pKAS135 | Apr (Thior) pWHM3 containing a 4.8-kb HindIII/SphI fragment from pKAS133; used for integrating the terminator in the eryCIV gene | This study |
| pDPE205 | Apr; E. coli shuttle vector containing the rrn terminator cloned into the eryBIV gene | This study |
| pDPE206 | Apr (Thior) pWHM3 containing a 3.0-kb SstI/HindIII fragment from pDPE205; used for integrating the terminator into the eryBIV gene | This study |
| pDPE212 | Apr (Thior) pWHM3 containing a terminator cassette in eryCVI; used for integrating the terminator in eryCVI | This study |
| pDPE218 | Apr (Thior) pWHM3 containing a subcloned rrn terminator within the eryBIII gene; used for integrating the terminator in the eryBIII gene | This study |
| pSAM14-2 | Apr (Hygr), integrative plasmid containing a hygromycin rrn terminator cassette in the eryAIII gene; contains the JV1 replicon | This study |
| S. erythraea | ||
| NRRL2338 | Wild type; erythromycin producer. | |
| eryAI::trrn | Thios NRRL2338 mutant containing an integrated rrn terminator in the eryAI gene obtained from pARR8 | This study |
| eryAIII::trrn | Hygr NRRL2338 mutant containing an integrated rrn terminator in the eryAIII gene obtained from pSAM14-2 | This study |
| eryBV::trrn | Thios NRRL2338 mutant containing an integrated rrn terminator in the eryBV gene obtained from pARR2 | This study |
| eryBVI::trrn | Thios NRRL2338 mutant containing an integrated rrn terminator in the eryBVI gene obtained from pKAS134 | This study |
| eryCIV::trrn | Thios NRRL2338 mutant containing an integrated rrn terminator in the eryCVI gene obtained from pDPE212 | This study |
| eryCVI::trrn | Thios NRRL2338 mutant containing an integrated rrn terminator in the eryCVI gene obtained from pKAS135 | This study |
| ARR50 | ThiosS. erythraea NRRL2338 containing a substituted SmaI site at the eryAI −10 hexamer region by gene replacement | This study |
| CA340 | NRRL2338 derivative; industrially-improved erythromycin producer | |
| eryAI::trrn | HygsS. erythraea CA340 mutant containing an integrated rrn terminator in the eryAI gene obtained from pARR17 | This study |
| eryAIII::trrn | Hygr CA340 mutant containing an integrated rrn terminator in the eryAIII gene obtained from pSAM14-2 | This study |
DNA manipulations.
Restriction digestions, dephosphorylation reactions with calf alkaline phosphatase, and ligation reactions with T4 DNA ligase were performed as directed by the manufacturer. All restriction enzymes and modification enzymes were purchased from New England Biolabs (Beverly, Mass.). S1 nuclease was purchased from Ambion (Austin, Tex.) and Boehringer Mannheim (Indianapolis, Ind.). Chromosomal Southern blotting was performed according to standard procedures (29). All Southern hybridizations were performed at 68°C. Hybridizing fragments were detected by the procedure outlined in the Genius system (Boehringer Mannheim) with the chemiluminescent substrate CDP-Star (Tropix, Bedford, Mass.) as the detection reagent. DNA sequencing reactions were carried out according to the dideoxy chain termination method of Sanger et al. (30) with alkaline-denatured templates (Amersham, Arlington Heights, Ill.) as described by the manufacturer.
Subcloning of an rrn terminator cassette within ery cluster biosynthetic genes.
The eryBIII mutant was constructed by subcloning a 5.1-kb PstI fragment containing a terminator sequence from pDPE149 into the BclI/NcoI sites within the eryBIII gene to generate pDPE218. The S. erythraea strain containing the terminator in eryBIII was designated eryBIII::trrn. The eryBIV mutant was constructed by subcloning a 300-bp BamHI rrn terminator sequence (obtained from pTERM9) into the BclI site located within the eryBIV gene contained on pDPE46 to make plasmid pDPE205. The S. erythraea strain containing the terminator in eryBIV was designated eryBIV::trrn. The eryBV mutant was constructed by first subcloning a 300-bp SpeI/XbaI terminator sequence from pDPE148A into the SpeI site of pDPE45, forming pARR1. A 3.0-kb HindIII/EcoRI fragment from pARR1 was subcloned into HindIII/EcoRI-digested pWHM3 (35), yielding pARR2. The S. erythraea strain containing the terminator in eryBV was designated eryBV::trrn. The eryBVI mutant was constructed by first subcloning a 300-bp BamHI terminator sequence from pTERM12 into the BamHI/BglII site of pDPE27, generating pKAS132. A 4.6-kb HindIII/SphI fragment from pKAS132 was then subcloned into HindIII/SphI-digested pWHM3, forming pKAS134. The S. erythraea strain containing the terminator in eryBVI was designated eryBVI::trrn. To construct the eryCIV mutant, the same 300-bp BamHI terminator sequence from plasmid pTERM12 was ligated into BclI-digested pDPE27, generating pKAS133. A 4.6-kb HindIII/SphI fragment from pKAS133 was then ligated into HindIII/SphI-digested pWHM3 to generate plasmid pKAS135. The S. erythraea strain containing the terminator inserted into the eryCIV gene was designated eryCIV::trrn. The eryCVI mutant was constructed by digesting plasmid pDPE201 with PstI/StuI and inserting the terminator from pTERM9 digested with PstI/StuI to generate pDPE203. Plasmid pEVEH8 was digested with XhoI/MluI, and the 2-kb fragment was added 3′ to the terminator to generate plasmid pDPE204. pDPE204 was then digested with SpeI/NsiI and ligated to pWHM3 digested with XbaI/PstI to generate plasmid pDPE212. The S. erythraea strain containing the terminator inserted into the eryCVI gene was designated eryCVI::trrn. The eryAI mutant was constructed by subcloning a 300-bp SstI/EcoRI fragment from pDPE148A into the SstI/EcoRI site of pDPE45, yielding pARR4. To provide additional eryAI sequence downstream of the terminator, a 1-kb SstI/PstI fragment from pAIX-5 was subcloned into the PstI/EcoRV site of pARR4, yielding pARR5. The SstI fragment end was converted to a blunt end, using T4 DNA polymerase. Finally, a 3.5-kb SspI/PstI fragment from pARR5 was subcloned into similarly digested pWHM3, yielding pARR8. The S. erythraea strain containing the terminator in the eryAI gene was designated eryAI::trrn. The S. erythraea CA340 eryAI mutant was constructed by using the pJV1-based plasmid pMBE-2. pMBE2 was first digested with HindIII and SspI to delete the EcoRI sites. The remaining 5.1 kb of pMBE2 was ligated to a 3.0-kb HindIII/SspI fragment from pARR9, forming pARR16. A 3.0-kb HindIII fragment from pARR9, containing the rrn terminator and an additional 1.0 kb of eryAI DNA, was ligated to HindIII-digested pARR16, forming pARR17. The S. erythraea CA340 strain containing the terminator inserted into the eryAI gene was designated CA340 eryAI::trrn. The eryAIII mutant was constructed by inserting the 5.8-kb XmnI/XbaI fragment from pGM402 into plasmid pCD1, which contains the pJV1 replicon. A 2.0-kb cassette containing the terminator and the hygromycin gene were inserted in the XhoI site located approximately 1.2 kb from the 3′ end of the eryAIII gene, yielding pSAM14-2. The S. erythraea strains containing integrated pSAM14-2 were designated NRRL2338 eryAIII::trrn and CA340 eryAIII::trrn. Protoplast transformation of S. erythraea NRRL2338 was performed according to an adaptation of the procedure originally described by Hopwood et al. (16). pARR17 and pSAM14-2 were transformed into S. erythraea CA340 by electroporation.
Analysis of ery gene cluster mutants by TLC.
All integrants were initially screened by thin-layer chromatography (TLC) as described by Weber et al. (36), for their ability to produce erythromycin A or the predicted intermediate. Bioconversion assays were performed by adding in separate time course experiments the erythromycin intermediates 6-deoxyerythronolide B (6-dEB), erythronolide B (EB), 3-mycarosyl erythronolide B (MEB), erythromycin C, or erythromycin D. These substrates were added at the time of transfer into SCM medium from ABB20 medium. The resulting biotransformation cultures were analyzed for 1 to 5 days as described above. All erythromycin intermediates were added at a final concentration of 25 μg/ml.
RNA isolation.
Ten milliliters of S. erythraea cells was harvested quickly by vacuum filtration onto a Whatman no. 1 filter over ice and washed with 50 ml of cold 10 mM EDTA. The cells were resuspended with 5 ml of extraction buffer (20 mM sodium acetate, 4 M guanidinium isothiocyanate, 1 mM EDTA, pH 8.0), dispersed using a Misonix (Farmingdale, N.Y.) sonicator (50% duty; power setting, 4; 60 pulses; 1-s duration), and vortexed with glass beads. Sodium dodecyl sulfate was added to a final concentration of 2% followed by acidic hot (65°C) phenol (Ambion) extraction. The aqueous phase was extracted with phenol, phenol-chloroform, and chloroform before precipitation. Northern blotting was performed according to established protocols as described by Sambrook et al. (29). Church’s buffer was used in the prehybridization and hybridization reactions (5).
S1 nuclease protection assays.
Single-stranded DNA probes for S1 nuclease protection assays were generated by a modification of the runoff replication procedure described in the manual accompanying the S1 nuclease kit (Ambion). Plasmids containing the appropriate ery cluster gene sequences were uniformly labeled with [32P]dCTP and [32P]dGTP by using sequence-specific primers and Sequenase. In all cases, the sizes of the probes were controlled by linearizing the plasmid with a restriction enzyme that cleaved approximately 200 to 275 bp from the priming site. Single-stranded, uniformly labeled probes were purified from their templates by denaturing polyacrylamide gel electrophoresis (5% acrylamide, 8 M urea–Tris-borate-EDTA). For hybridizations, total S. erythraea RNA was mixed with 104 to 105 Chelenkov counts per min of probe at 50 to 55°C for 18 h. Detection of S1-protected fragments was performed according to the procedure outlined in the manual accompanying the S1 nuclease kit.
Oligonucleotide-directed mutagenesis.
In order to alter the predicted −10 region of eryAI from the native sequence (TATTGT) to an SmaI site (CCCGGG), the following PCR primers were designed: Set A, 5′-ATGAATTCTGCGCGCCCTGGCCCGGGAAGACGAA-3′ and 5′-TCTCCCGGGTCGCCATTGCGTGGTCGTCG-3′, and set B, 5′-TCTCCCGGGTAGGAAGGATCAAGAGGTTGACAT-3′ and 5′-CGGAATTCTGATCAATTGACGGGGAATCA-3′. The PCR product of primer set A was digested with EcoRI and SmaI and subcloned into EcoRI/SmaI-digested pUC18, yielding pARR47. The PCR product of primer set B was digested with SmaI and then subcloned into SmaI-digested pARR47, yielding pARR48. SmaI digestion and sequencing confirmed the generation of an SmaI site at the predicted −10 region of eryAI in the correct orientation. In order to replace the native eryAI −10 region with the altered sequence, pARR48 was digested with BclI and ClaI. This generated 4.0-kb and 400-bp fragments. pARR3, which contains the native eryAI promoter region and an additional 1.6 kb of eryAI and eryBIV DNAs, was digested with BclI and ClaI. The larger fragment was resolved and gel purified from the 400-bp BclI/ClaI fragment and ligated to the mutagenized 400-bp BclI/ClaI fragment, yielding pARR49. Finally, a 2.0-kb EcoRI/HindIII fragment from pARR49, containing the entire eryAI-eryBIV promoter region and an additional 1.6 kb of the eryAI and eryBIV genes, was subcloned into the S. erythraea insertion vector pWHM3 (35), yielding pARR50. pARR50 was protoplast transformed into wild-type S. erythraea as described above.
Other procedures.
Western blotting was performed with rabbit anti-EryG antibodies cross-reacted to soluble cell extracts obtained from various S. erythraea strains. Samples containing 10 μg of protein per lane were loaded onto a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10% acrylamide) gel. The resolved proteins were electrotransferred onto polyvinylidene difluoride membranes (Millipore) at 90 V for 2 h according to standard procedures (29). Cross-reacting proteins were detected with the ECL kit as described by the manufacturer (Amersham). Quantitation of S1-protected fragments was performed with a Storm 860 PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.) equipped with ImageQuant software.
RESULTS
Construction of ery gene cluster mutants containing an inserted rrn terminator.
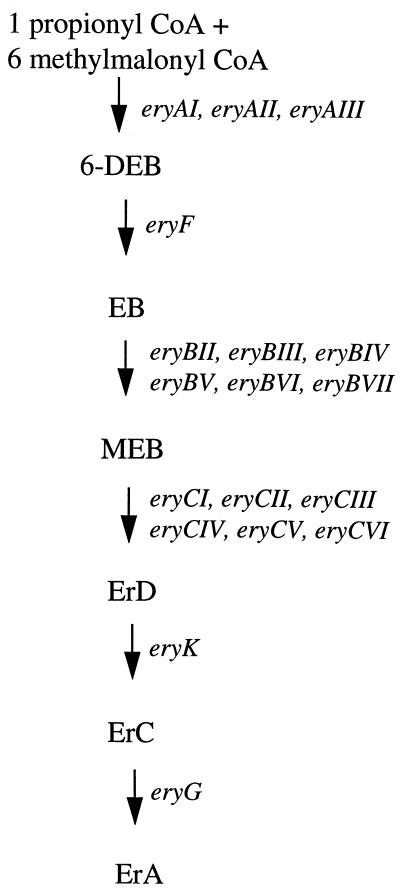
Insertional inactivation of erythromycin biosynthetic cluster genes by targeted mutagenesis was performed by constructing S. erythraea mutants containing an inserted S. erythraea rrn operon terminator at specific sites within the eryAI, eryAIII, eryBIII, eryBIV, eryBV, eryBVI, eryCIV, and eryCVI genes (Table 1). A flow diagram showing the pathway leading to the production of erythromycin A in S. erythraea and the genes involved is given in Fig. 1. Insertion of the rrn terminator allowed the study of the polar effects on downstream ery gene cluster expression by blocking transcription from an upstream promoter(s). The terminator sequence was obtained from the cloned S. erythraea rrnD operon, which has been physically mapped on the chromosome (26). A native S. erythraea terminator was chosen to avoid any differences that might arise among species. The terminator sequence used for insertion mutagenesis was subcloned as a 227-bp fragment, including 22 bp of the 5S portion of the rrn operon. The region was analyzed for secondary structure with the MulFold program (18, 19, 41). Two regions containing secondary structure were identified. The first region, beginning 7 bp downstream from the end of the 5S gene, was predicted to contain a stem structure 17 bp long with a 4-bp loop immediately followed by a thymidine-rich region, characteristic of rho-independent terminators (7). The calculated ΔG of the stem-loop was −30 kcal/mol. A potential second stem-loop structure was identified 30 bp downstream from the first stem-loop structure. It had a predicted 18-bp stem with a 4-bp loop followed by a thymidine-rich region. This stem-loop structure had a predicted ΔG of −24.5 kcal/mol. The engineered S. erythraea DNA containing the terminator was introduced into the chromosome by homologous recombination with vector pWHM3 (35), which replicates poorly in S. erythraea. As an example, the eryBIII mutant was constructed by transforming plasmid pDPE218 into S. erythraea, and the resulting mutant strain containing the terminator sequence in eryBIII was designated eryBIII::trrn. All integrants derived by integration with pWHM3 in S. erythraea NRRL2338 were the result of two separate reciprocal-recombination events which resulted in the eviction of the selectable marker from the chromosome. Integrants derived from plasmid pSAM14-2 required that the hygromycin resistance gene remain in the chromosome. Chromosomal Southern blotting with fragments that overlapped the junctions of the inserted DNA as probes confirmed the correct integration of the terminator sequence in each mutant (data not shown).
FIG. 1.
Flow diagram indicating the biochemical intermediates and the genes involved in the biosynthesis of erythromycin A. ErD, erythromycin D; ErC, erythromycin C; ErA, erythromycin A; CoA, coenzyme A.
S1 mapping of the transcription start sites for seven ery cluster promoters.
As evidenced by the ery biosynthetic gene cluster map (see below) (Fig. 2), the majority of the promoters identified in this study were predicted to be tandemly arranged and divergently transcribed. Figure 3 shows the results of S1 nuclease mapping of seven transcriptional start sites within four predicted promoter regions. These promoter regions were the 224-bp eryAI-eryBIV intergenic region, the 188-bp eryBI-eryBIII intergenic region, the 83-bp eryCVI-eryBVI intergenic region, and the region immediately upstream of eryK.
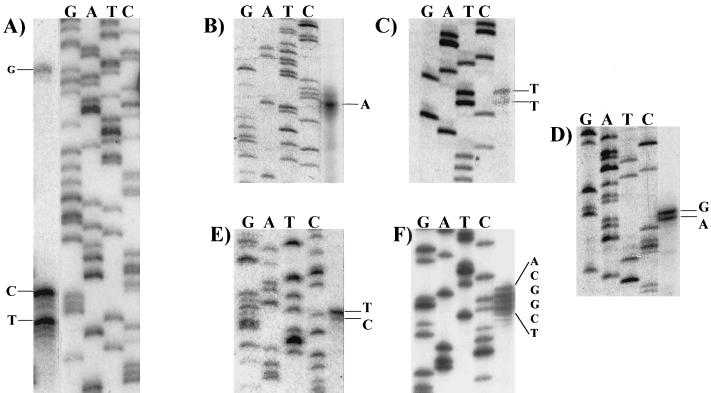
FIG. 3.
S1 nuclease mapping of the 5′ endpoints of ery cluster transcripts. The nucleotide(s) at the side of each panel indicate the likely transcription start site(s). (A) eryBIV; (B) eryAI; (C) eryBI; (D) eryBIII; (E) eryBVI; (F) eryK. The same primer was used to generate the sequence ladder and the 32P-labeled probe for S1 assays. For clarity, similar-intensity images of the S1 and sequence ladder lanes from the same gel were juxtaposed.
To determine the transcription start site for the eryBIV transcript, a 262-bp probe extending from the BclI site in eryBIV to the MluI site located at the beginning of eryAI was used. Three eryBIV protected fragments were identified, beginning 84, 88, and 132 bp upstream of the predicted translation start codon (Fig. 3A). The 84- and 88-bp protected fragments were consistently more abundant than the 132-bp fragment, suggesting that this is the location of the major promoter expressing the eryBIV message (under these experimental conditions). The −35 region of the minor eryBIV promoter is predicted to overlap the −35 region of eryAI. The eryAI transcription start site was identified by using a 314-bp probe starting 76 bp within the eryAI gene. A protected fragment 103 bp in length was observed, beginning 27 bp upstream of the predicted translation start codon (Fig. 3B).
The eryBI transcription start site was identified by using a probe that began 45 bp into eryBI and extended to an NdeI site 35 bp within eryBIII. Two S1-protected fragments were observed, beginning 17 and 18 bp upstream of the predicted translational start for eryBI (Fig. 3C). To determine the eryBIII transcription start site, a 238-bp probe extending from a priming site 70 bp within eryBIII to a BclI site 20 bp upstream of eryBI was used. Two S1-protected fragments were also observed 1 and 2 bp upstream of the predicted GTG translation start codon for eryBIII (Fig. 3D).
The size of the predicted eryBVI-eryCVI intergenic region (83 bp), along with biochemical evidence obtained in this study (see below), suggested that there could be a promoter located upstream of eryBVI (12, 33). We tested this prediction by performing S1 assays with a 345-bp probe that extended from a site within the 5′ end of eryBVI to a StuI site in eryCVI. Two S1-protected fragments were observed (Fig. 3E), beginning 1 and 2 bp upstream of the predicted start codon (12).
The eryK transcription start site was identified by generating a 240-bp probe beginning 65 bp within the eryK gene and extending to a BamHI site within orf21. Several S1-protected fragments beginning 45 to 50 bp upstream of the predicted TTG start site (31) and 9 to 14 bp from the predicted termination codon of orf21 marked the transcription start site for eryK (Fig. 3F). The predicted −35 and −10 promoter regions and mRNA start sites are also shown (see Fig. 8).
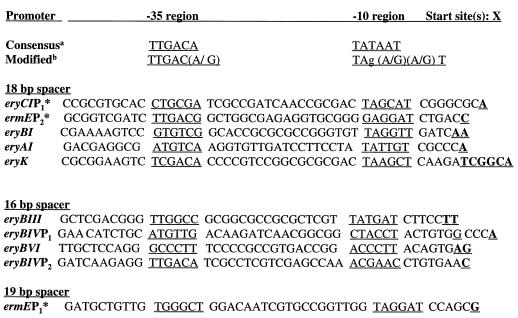
FIG. 8.
Alignment of 10 ery gene cluster promoters. The predicted −10 and −35 regions are underlined. The predicted start sites are boldface and underlined. In five cases, more than one 5′ endpoint was identified in the S1 mapping, and all are underlined. The asterisks mark promoters determined by Bibb et al. (2). a, E. coli consensus for sigma-70 promoters; b, modified E. coli consensus determined for Streptomyces (32).
Characterization of ery gene cluster transcripts by S1 assay.
To test whether the insertion in eryAI::trrn was having a polar effect on transcription of the downstream eryG gene (Fig. 2), S1 assays were performed on total RNAs from NRRL2338 eryAI::trrn and CA340 eryAI::trrn with an eryG probe. A significant reduction of eryG signal in the insertion mutant strains was observed compared to that in the parental background strains (Fig. 4). Quantitation of the hybridizing band in NRRL2338 eryAI::trrn showed an approximately 70% reduction in eryG signal compared to NRRL2338, whereas CA340 eryAI::trrn showed a >90% reduction of eryG signal compared to CA340.
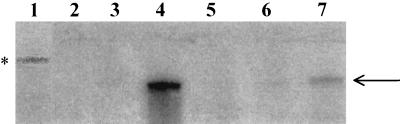
FIG. 4.
Comparison by S1 nuclease protection assay of the mRNA levels of the eryAI transcript in S. erythraea NRRL2338, NRRL2338 eryAI::trrn, CA340, and CA340 eryAI::trrn. Lanes: (1) probe only, untreated; (2) probe only, S1 treated; (3) probe hybridized with 40 μg of Saccharomyces cerevisiae RNA; (4) probe hybridized with 40 μg of S. erythraea CA340 RNA; (5) probe hybridized with 40 μg of S. erythraea CA340 eryAI::trrn RNA; (6) probe hybridized with 40 μg of S. erythraea NRRL2338 eryAI::trrn RNA; (7) probe hybridized with 40 μg of NRRL2338 RNA. A total of 104 cpm of probe was used per S1 nuclease reaction. The asterisk indicates full-length probe. The arrow at the right indicates the S1-protected fragment. For clarity, a lower-intensity exposure of lane 1 was used.
To determine whether the terminators in the eryBIV, eryBV, and eryCIV genes were having a polar effect on eryBVII transcription (Fig. 2), S1 assays were performed on total RNAs from eryBIV::trrn, eryBV::trrn, and eryCIV::trrn with a labeled probe that extended entirely within the eryBVII reading frame. Because the entire probe was internal to the eryBVII gene, additional, nonhybridizing DNA (31 bp, composed of a multiple cloning site) was cloned into the plasmid used to make the probe to distinguish between undigested full-length probe and the protected fragment. There was a significant decrease in the amount of eryBVII hybridization signal. The decrease in eryBVII hybridization signal in eryBIV::trrn and eryBV::trrn was similar to the decrease in eryG signal in eryAI::trrn. The results showed a decrease in signal of the hybridizing fragment of roughly 70 and 80% for the eryBIV and eryBV mutants, respectively, compared to NRRL2338 (data not shown). In the case of eryCIV::trrn, no defined protected fragment was observed even after long exposure. This indicated that the terminator insertions in the eryBIV, eryBV, and eryCIV mutant strains had a polar effect on eryBVII expression.
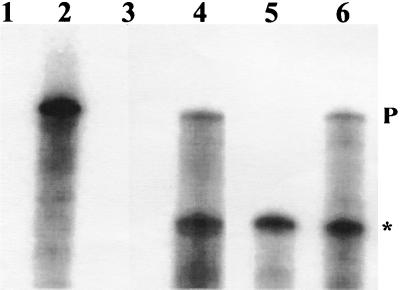
Biochemical evidence (see below) suggested that there could be two promoters within the right flank producing overlapping transcripts. S1 assays were performed on eryBIV::trrn, S. erythraea CA340, and eryAIII::trrn with an eryBVI probe (Fig. 5). eryAIII::trrn was used as the positive control in these experiments. In CA340 (Fig. 5, lane 3) and eryAIII::trrn (lane 6), two protected fragments were observed. The larger protected fragment (330 bp) represents a transcript beginning at the eryBIV promoter, and the smaller protected fragment (180 bp) represents a transcript beginning at the eryBVI promoter, suggesting that the two promoters produce overlapping transcripts. In eryBIV::trrn (lane 5), only the 180-bp protected fragment is observed, suggesting that the inserted terminator in eryBIV is efficiently terminating transcription.
FIG. 5.
The right flank of the ery gene cluster contains two overlapping transcripts from eryBIV to eryBVII. Lane 1, Full-length eryBVI probe treated with S1 nuclease; lane 2, same as lane 1 but not treated with S1 nuclease; lane 3, 40 μg of yeast RNA hybridized with probe; lane 4, 40 μg of S. erythraea CA340 RNA hybridized with probe; lane 5, 40 μg of eryBIV::trrn RNA hybridized with probe; lane 6, 40 μg of CA340 eryAIII::trrn RNA hybridized with probe. All samples were treated with 50 U of S1 nuclease. A total of 3 × 104 Chelenkov counts per min were used per reaction. P, full-length protected fragment; the asterisk marks a shortened S1-protected fragment.
Northern and Western blotting of the eryA mutants reveal a polar effect of the terminator on eryG expression.
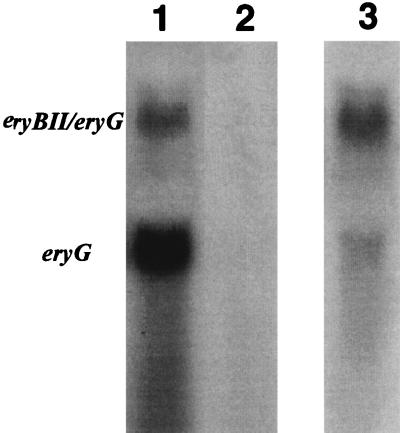
To further analyze ery cluster transcripts in the eryAI-eryG region, Northern blotting was performed on total RNAs extracted from 2-day fermentation cultures of S. erythraea CA340 and CA340 eryAIII::trrn. Hybridization of total RNA from S. erythraea CA340 with an eryG probe revealed two transcripts approximately 2,600 and 1,200 bp in length (Fig. 6). The smaller transcript in NRRL2338 has been described previously by Weber et al. (37). The same Northern blot was probed with the eryBII gene located immediately upstream from eryG. The hybridization pattern was identical to that of the larger transcript from the eryG hybridization (Fig. 6). This indicates that the larger transcript contains eryBII and eryG. In the CA340 eryAIII::trrn mutant, no detectable transcript was observed, even when 15-fold more RNA from the mutant was used in the hybridization.
FIG. 6.
Northern blot analysis of the eryG and eryBII-eryG transcript. Lane 1, 50 μg of CA340 RNA; lane 2, 50 μg of S. erythraea CA340 eryAIII::trrn probed with an eryG probe; lane 3, 50 μg of CA340 RNA probed with an eryBII probe.
Western blot analysis was performed on NRRL2338 and NRRL2338 eryAI::trrn to determine if the O-methyltransferase protein was present in cell extracts of these strains. A cross-reacting band corresponding to EryG protein was observed in NRRL2338 and an E. coli strain overproducing EryG but not in an S. erythraea strain with eryG deleted or in the eryAI::trrn mutant (data not shown). The Northern and Western blot analyses, in conjunction with the S1 studies, provide strong evidence that the eryAI, eryAII, eryAIII, eryBII, eryCII, eryCIII, and eryG genes are primarily cotranscribed from a promoter upstream of eryAI.
Bioconversion analysis of the mutants supplemented with erythromycin intermediates.
To verify that the RNA results correlated with biochemical phenotypes, biotransformation assays were performed on the insertion mutants with erythromycin intermediates. All the mutants were initially screened by TLC assay without supplemented intermediates to confirm the predicted effect of the mutation at the enzymatic level. All of the mutants showed the expected phenotype (Table 2). To test the effectiveness of the terminator to disrupt transcription in the eryA insertion mutants NRRL2338 eryAI::trrn, CA340 eryAI::trrn, and NRRL2338 eryAIII::trrn, the erythromycin intermediates 6-dEB, EB, MEB, and erythromycin C (final concentration, 25 μg/ml) were added to 50-ml shake flask fermentations and assayed for erythromycin A formation over a 4-day period. The results for erythromycin C bioconversion are shown in Fig. 7. The mutants showed a significant reduction in the formation of erythromycin A and the extent of erythromycin A production from all the intermediates compared to that of the wild type, which bioconverted all the intermediates to erythromycin A within 24 h. No dramatic differences in the formation of erythromycin A from the various intermediates were observed. To determine whether the inserted terminator was having an effect other than on termination of transcription, a mutant (S. erythraea ARR50) was isolated in which the predicted eryAI AT-rich −10 hexamer sequence (TATTGT) was replaced with an SmaI site (CCCGGG). No erythromycin A was observed over a 5-day period in 50-ml shake flask fermentations of this strain. Additionally, when 50-ml cultures of ARR50 were supplemented with EB, MEB, erythromycin C, and erythromycin D in biotransformation experiments over a 5-day period, bioconversion to erythromycin A was significantly reduced compared to that of the wild type, as observed similarly in NRRL2338 eryAI::trrn (data not shown). Thus, mutagenesis of eryAI by either insertion of a terminator or alteration of the predicted −10 region resulted in the same phenotype. To determine whether the insertion in the eryB mutant strains eryBIV::trrn, eryBV::trrn, and eryBVI::trrn was having a polar effect on downstream eryC genes, bioconversion assays were performed with supplemented MEB over a 5-day period. The eryBIV::trrn and eryBV::trrn mutants showed a significant reduction in the amount of erythromycin A formed compared to that of S. erythraea NRRL2338. eryBIV::trrn produced erythromycin A after 1 day of fermentation, whereas it required 5 days to detect erythromycin A by the TLC assay with strain eryBV::trrn. Based on the predicted structural (enzymatic) functions of the EryBIV, EryBV, and EryCVI proteins, this shows cotranscription of at least eryBIV, eryBV, and eryCVI. eryBVI::trrn did not produce any erythromycin A, even when the sample loaded for TLC analysis was fivefold concentrated, showing cotranscription (by the same reasoning used for the predicted cotranscription of eryBIV, eryBV, and eryCVI) of at least eryBVI and eryCIV.
TABLE 2.
S. erythraea mutant strains generated by insertional mutagenesis or alteration of the −10 promoter region
| Straina | Gene Disruptedb | Phenotypec |
|---|---|---|
| eryAI::trrn | eryAI | Null |
| CA340eryAI::trrn | eryAI | Null |
| eryAIII::trrn | eryAIII | Null |
| CA340eryAIII::trrn | eryAIII | Null |
| eryBIII::trrn | eryBIII | 6-dEB |
| eryBIV::trrn | eryBIV | EB |
| eryBV::trrn | eryBV | EB |
| eryBVI::trrn | eryBVI | EB |
| eryCIV::trrn | eryCIV | EB/MEB |
| eryCVI::trrn | eryCVI | EB/MEB |
| ARR50d | eryAI | Null |
All strains are S. erythraea NRRL2338 unless listed as S. erythraea CA340.
Indicates gene containing an inserted 227-bp rrn operon terminator cassette by gene replacement.
Indicates the accumulation of the erythromycin intermediate(s) or the lack of production (null) of any erythromycin intermediate as assayed by TLC.
Contains only an altered eryAI −10 hexamer by gene replacement.
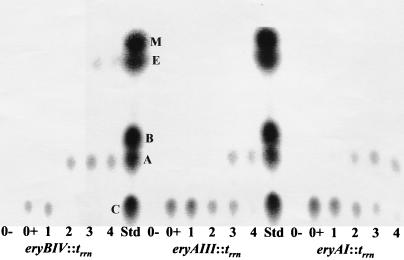
FIG. 7.
TLC assay to test the ability of the eryA transcriptional mutants eryAI::trrn and eryAIII::trrn to bioconvert erythromycin C to erythromycin A. Erythromycin C (25 μg/ml) was added separately to 50-ml SCM cultures containing the eryA mutants and the eryBIV mutant, eryBIV::trrn. One milliliter of the culture broth from 0- to 4-day fermentations was extracted with ethyl acetate and analyzed by TLC. The eryBIV mutant was used as the positive control. Both S. erythraea NRRL2338 (data not shown) and eryBIV::trrn completely bioconverted all the supplemented erythromycin C to erythromycin A in less than 1 day between days 1 and 2 of the fermentation. 0−, no added erythromycin C; 0+, sample taken at the time of supplementation; 1 to 4, day of sampling following addition of erythromycin C; Std., TLC standards (M, 3-mycorosyl erythronolide B; E, erythronolide B; B, erythromycin B; A, erythromycin A; C, erythromycin C).
In eryC insertion mutants constructed on the right flank of the ery cluster, it was expected that the biochemical phenotype would be the same as that observed in the eryB insertion mutants described above (accumulation of only EB), since it was predicted that the insertion would exert a polar effect on downstream eryB genes. In biochemical studies the eryCVI mutant, eryCVI::trrn, accumulated both EB and MEB after 2 days of fermentation, suggesting an effect on the transcription of downstream eryB genes. However, eryCVI::trrn was able to bioconvert all the EB formed to MEB after 4 days. This result is consistent with the S1 assay data suggesting that there is a promoter upstream of eryBVI. The eryCIV mutant, eryCIV::trrn, accumulated equal amounts of EB and MEB. In contrast to eryCVI::trrn, eryCIV::trrn did not bioconvert any of the EB to MEB, even after 4 days of fermentation. This suggests that premature transcription termination of a significant fraction of transcript from both the eryBIV and eryBVI promoters was occurring in eryCIV::trrn (see below).
Previous insertion mutagenesis experiments in the region now known to contain the eryBIII, eryF, and orf5 genes showed that those mutants either were affected in the C-6 hydroxylation step (eryF phenotype) or have an eryH mutation and accumulate EB (EryB phenotype) (34). To determine whether an insertion in eryBIII would have a polar effect on the expression of the downstream eryF gene, TLC assays were performed on eryBIII::trrn over a 5-day period. This strain accumulated only 6-dEB early in the fermentation (24 to 48 h after inoculation), indicating a polar effect on the transcription of eryF.
Thus, although the S1, Northern, and Western assay data suggest an almost complete absence of transcript downstream from the inserted terminators, biochemical evidence suggests either that a low level of read-through of the terminator is occurring or that there are secondary (minor) promoters producing low levels of the downstream transcripts.
DISCUSSION
Transcriptional arrangement of the erythromycin biosynthetic gene cluster.
Figure 2 shows a schematic representation of the transcriptional organization of the 56-kb ery gene cluster based on previous work by Bibb et al. (2) and the results presented in this study. The present work has identified through S1 assay, Northern blot analysis, Western blotting, and biochemical assays that the ery biosynthetic gene cluster is primarily transcribed as four polycistronic messages: eryAI-eryG, eryBIV-eryBVII, eryBVI-eryBVII, and eryBIII-eryF. The largest transcript is approximately 35 kb and contains most of the left flank of the biosynthetic gene cluster. The presence of a very large message on the left flank of the ery cluster was not totally unexpected, since previous studies (8, 28, 33) predicted the possibility of translational coupling of many genes in the eryAI-eryG region. Bioconversion of erythromycin C to erythromycin A was observed in the eryAI transcriptional mutants at a much reduced rate, suggesting that there is a possible minor promoter(s) in the region downstream of the terminator insertions or that read-through of the inserted terminator is occurring. The S1 assay with the 5′ end of eryG as a probe was in agreement with the prediction made from the biochemical data, since greatly reduced levels of eryG message were detected in the eryA mutants. Possible locations for secondary promoters include the region between the insertions in eryAI and eryAIII and the region downstream of the eryAIII insertion.
In order to confirm the transcriptional organization of the eryAI-eryG region suggested from the analysis of the transcriptional terminator mutants, a strain was constructed by oligonucleotide-directed mutagenesis to alter the predicted eryAI −10 hexamer region. The eryAI −10 region in S. erythraea is A+T rich, and therefore it was expected that altering the hexamer to all G+C would dramatically affect expression from that promoter. This strain had a phenotype similar to that of the eryA insertion mutants in TLC and biotransformation assays. No erythromycin A production was observed. This strain also bioconverted erythromycin intermediates in a manner similar to that of the eryAI terminator insertion mutant. This confirms that the predicted −10 region plays an important role in gene expression at the eryAI promoter and that the mutation has a polar effect on downstream eryG transcription.
We have identified two major promoter regions expressing the deoxysugar genes on the right flank of the ery gene cluster. These promoters produce overlapping transcripts, one beginning upstream of eryBIV and extending to eryBVII (about 8.0 kb) and the other beginning at eryBVI and extending to eryBVII (about 4.8 kb). These data provide additional evidence of the effectiveness of the terminator in disrupting transcription. Although the biochemical data indicated that some MEB is bioconverted to erythromycin A in eryBIV::trrn, presumably by read-through of the terminator, no S1-protected fragment corresponding to the large mRNA (8.0 kb) was detected. This suggests that eryBIV message has been severely reduced in eryBIV::trrn but that some transcript is being made, which is undetectable in the S1 assay, to allow a low level of enzyme production and bioconversion.
When the initial biochemical assays were performed on the eryC mutants located on the right flank of the ery gene cluster, an EryB phenotype was expected, since the insertion was predicted to have a polar effect on one or more eryB genes. Surprisingly, both EB and MEB accumulated in the culture supernatants in these strains. These data, taken together with the S1 assay results indicating a promoter upstream of eryBVI, suggest several possibilities: (i) that very little EryBVII is necessary for the predicted 3,5 epimerization reaction despite our not being able to visualize an S1-protected fragment; this would suggest that there is a promoter downstream of eryCIV or enough read-through of the terminator is occurring to allow sufficient production of the epimerase; (ii) that the predicted 3,5 epimerization reaction catalyzed by EryBVII is being carried out by another epimerase, as was suggested by Linton et al. and Salah-Bey et al. (20, 28); or (iii) that the MEB observed is unepimerized. Recently, it has been shown that an eryBVII mutant accumulates mainly EB and small amounts of erythromycin A and erythromycin B analogs in which the neutral sugar residue might contain a 3-C-methyl or 3-O-methyl 4-keto-6-deoxyglucose or the 5-epimer of cladinose (13), indicating that the EryBVII epimerization reaction cannot be complemented by another epimerase under the growth conditions used. This suggests that what we observed by TLC analysis in the eryCIV mutant was EB and possibly unepimerized MEB.
The right flank of the ery gene cluster also contains the eryK gene, which is transcribed in the opposite direction from the eryB and eryC genes. eryK appears to be expressed as a monocistronic message, although we have not ruled out the possibility that this message is derived from a larger transcript. Only one eryK transcript was observed in the S1 assay, suggesting that either the eryK transcript is rapidly processed from a larger transcript or it is monocistronic. We consider the former possibility unlikely, since sequence analysis of a 7.0-kb region upstream of eryK by Pereda et al. (24) indicated no obvious involvement of the ORFs in erythromycin biosynthesis. Along with eryK and the previously described ermE and eryCI genes, eryBI is transcribed as a monocistronic message, bringing the total number of ery cluster genes contained in their own transcriptional units to four. Recently, eryBI was shown not to be essential for erythromycin biosynthesis (13).
Previous work performed on the eryBIII-eryF region (previously designated the eryH region) showed that these genes and possibly the undefined orf5, are probably arranged as an operon, since insertion mutants generated in that study were determined to be affected in the C-6 hydroxylation step (eryF mutant) or to have an eryH mutation and accumulate EB (EryB phenotype) (34). Additionally, the TGA codon of eryF (previously designated orf4) and the predicted ATG start of orf5 overlap by 1 bp, potentially making these two genes translationally coupled (15). Results of TLC assays of supernatants from a strain containing a terminator in eryBIII showed an accumulation of 6-dEB. Additionally, the cultures accumulated other unknown products after several days of growth in shake flask fermentations. These strains are able to bioconvert MEB to erythromycin A, indicating that not all genes involved in downstream synthesis were affected in these mutants. The precise role of the orf5 gene has yet to be determined. Haydock et al. (15) have proposed that this gene encodes a type II thioesterase or acyltransferase. The ORF5 gene product is not essential for erythromycin production, since insertions in the reading frame do not eliminate erythromycin biosynthesis (10). However, a significant decrease in macrolide production (6-dEB in this strain) was observed, along with the production of other unknown compounds, in this mutant compared to other engineered strains. Recently, Cundliffe (6) and Xue et al. (40) have reported similar decreases in macrolide production of mutant strains deficient in type II thioesterases. Thus, it appears that eryBIII, eryF, and possibly orf5 are cotranscribed.
Previous work predicted a promoter region upstream from the eryG translational start site, since a cloned fragment from that region gave promoter activity with a luxAB reporter group system. In addition, the putative transcription start site was determined in S. erythraea by S1 mapping (37). We propose in addition to this prediction and as a possible alternative hypothesis, that what was observed previously was primarily the result of RNA processing of the eryAI-eryG transcript and not a major independently transcribed message. The promoter activity observed might have been due to (i) the introduction and expression of S. erythraea DNA in the heterologous host Streptomyces lividans and (ii) the high copy number of pIJ702-based vectors in S. lividans. It is still possible that there is a minor promoter located in the region upstream of eryG and downstream of the insertion site in eryAIII, but it would be significantly weaker than the eryAI promoter, as indicated by the RNA and bioconversion experiments. The basis for the stability of the eryG-containing transcript (observed in the Northern blot analysis) compared to the mature 35-kb transcript is unknown. Figure 8 shows an alignment of all the ery cluster promoter regions showing the transcription start sites and the predicted −10 and −35 regions. Several promoter sequences have similarity to the E. coli consensus −10 and −35 regions as well as to the modified consensus determined for Streptomyces (32). All of the −10 and −35 regions determined in this study contain a predicted spacer region between 16 and 18 bp. In several cases there is a conserved T at position 6 in the −10 region, which has been shown to be common in Streptomyces promoters (39). In several cases, multiple protected fragments were observed at the 5′ transcription endpoint. The transcriptional studies reported here indicate that the majority of ery biosynthetic genes are transcribed as large polycistronic messages from several key regulatory regions. This work should facilitate future studies directed at improving our understanding of the regulation of this industrially important secondary metabolic pathway.
ACKNOWLEDGMENTS
We thank Mark Satter for constructing the plasmids used to generate the eryBVI and eryCIV mutants and Mike Staver for providing the plasmid containing the cloned S. erythraea rrn operon and pAIX-5. We also acknowledge Marty Babcock and Sandra Splinter for providing the anti-EryG antibodies and technical advice with the Western blotting and Steve Kakavas, Diane Stassi, and Kurt Harris for technical assistance in using the PhosphorImager. Thanks also to Janet Westpheling for suggesting site-specific mutagenesis of the −10 hexamer region.
REFERENCES
- 1.Bailey C R, Bruton C J, Butler M J, Chater K F, Harris J E, Hopwood D A. Properties of in vitro recombinant derivatives of pJV1, a multicopy plasmid from Streptomyces phaeochromogenes. J Gen Microbiol. 1986;132:2071–2078. doi: 10.1099/00221287-132-8-2071. [DOI] [PubMed] [Google Scholar]
- 2.Bibb M J, White J, Ward J M, Janssen G R. The mRNA for the 23S rRNA methylase encoded by the ermE gene of Saccharopolyspora erythraea is translated in the absence of a conventional ribosome binding site. Mol Microbiol. 1994;14:533–545. doi: 10.1111/j.1365-2958.1994.tb02187.x. [DOI] [PubMed] [Google Scholar]
- 3.Caballero J L, Martinez E, Malpartida F, Hopwood D A. Organization and functions of the actVA region of the actinorhodin biosynthetic gene cluster of Streptomyces coelicolor. Mol Gen Genet. 1991;230:401–412. doi: 10.1007/BF00280297. [DOI] [PubMed] [Google Scholar]
- 4.Caffrey P, Bevitt D J, Staunton J, Leadlay P F. Identification of DEBS 1, DEBS 2, and DEBS 3, the multienzyme polypeptides of the erythromycin-producing polyketide synthase from Saccharopolyspora erythraea. FEBS Lett. 1992;304:225–228. doi: 10.1016/0014-5793(92)80624-p. [DOI] [PubMed] [Google Scholar]
- 5.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cundliffe, E. Personal communication.
- 7.Deng Z, Kieser T, Hopwood D A. Activity of a Streptomyces transcriptional terminator in Escherichia coli. Nucleic Acids Res. 1987;15:2665–2675. doi: 10.1093/nar/15.6.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donadio S, Staver M J, McAlpine J B, Swanson S J, Katz L. Modular organization of genes required for complex polyketide biosynthesis. Science. 1991;252:675–679. doi: 10.1126/science.2024119. [DOI] [PubMed] [Google Scholar]
- 9.Donadio S, Stassi D, McAlpine J B, Staver M J, Sheldon P J, Jackson M, Swanson S J, Wendt-Pienkowski E, Wang Y-G, Jarvis B, Hutchinson C R, Katz L. Recent developments in the genetics of erythromycin formation. In: Baltz R H, Hegeman G D, Skatrud P L, editors. Industrial microorganisms: basic and applied molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 257–265. [Google Scholar]
- 10.English, R. S. Unpublished results.
- 11.Fernandez-Moreno M A, Martinez E, Boto L, Hopwood D A, Malpartida F. Nucleotide sequence and deduced functions of a set of cotranscribed genes of Streptomyces coelicolor A3(2) including the polyketide synthase for the antibiotic actinorhodin. J Biol Chem. 1992;267:19278–19290. [PubMed] [Google Scholar]
- 12.Gaisser S, Bohm G A, Cortes J, Leadlay P F. Analysis of seven genes from the eryAI-eryK region of the erythromycin biosynthetic gene cluster in Saccharopolyspora erythraea. Mol Gen Genet. 1997;256:239–251. doi: 10.1007/s004380050566. [DOI] [PubMed] [Google Scholar]
- 13.Gaisser S, Bohm G A, Doumith M, Raynal M-C, Dhillon N, Cortes J, Leadlay P F. Analysis of eryBI, eryBIII, and eryBVII from the erythromycin biosynthetic gene cluster in Saccharopolyspora erythraea. Mol Gen Genet. 1998;258:78–88. doi: 10.1007/s004380050709. [DOI] [PubMed] [Google Scholar]
- 14.Gandecha A R, Large S L, Cundliffe E. Analysis of four tylosin biosynthetic genes from the tylLM region of the Streptomyces fradiae genome. Gene. 1997;184:197–203. doi: 10.1016/s0378-1119(96)00595-1. [DOI] [PubMed] [Google Scholar]
- 15.Haydock S F, Dowson J A, Dhillon N, Roberts G A, Cortes J, Leadlay P F. Cloning and sequence analysis of genes involved in erythromycin biosynthesis in Saccharopolyspora erythraea: sequence similarities between EryG and a family of S-adenosylmethionine-dependent methyltransferases. Mol Gen Genet. 1991;230:120–128. doi: 10.1007/BF00290659. [DOI] [PubMed] [Google Scholar]
- 16.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, United Kingdom: John Innes Foundation; 1985. [Google Scholar]
- 17.Hopwood D A, Sherman D H. Molecular genetics of polyketides and its comparison to fatty acid biosynthesis. Annu Rev Genet. 1990;24:37–66. doi: 10.1146/annurev.ge.24.120190.000345. [DOI] [PubMed] [Google Scholar]
- 18.Jaeger J A, Turner D H, Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci USA. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaeger J A, Turner D H, Zuker M. Predicting optimal and suboptimal secondary structure for RNA. Methods Enzymol. 1989;183:281–306. doi: 10.1016/0076-6879(90)83019-6. [DOI] [PubMed] [Google Scholar]
- 20.Linton K J, Jarvis B W, Hutchinson C R. Cloning of the genes encoding thymidine diphosphoglucose 4,6-dehydratase and thymidine diphospho-4-keto-6-deoxyglucose 3,5-epimerase from the erythromycin-producing Saccharopolyspora erythraea. Gene. 1995;153:33–40. doi: 10.1016/0378-1119(94)00809-7. [DOI] [PubMed] [Google Scholar]
- 21.MacNeil D J, Occi J L, Gewain K M, MacNeil T, Gibbons P H, Ruby C L, Danis S J. Complex organization of the Streptomyces avermitilis genes encoding the avermectin polyketide synthase. Gene. 1992;115:119–125. doi: 10.1016/0378-1119(92)90549-5. [DOI] [PubMed] [Google Scholar]
- 22.Martin J F, Liras P. Organization and expression of genes involved in the biosynthesis of antibiotics and other secondary metabolites. Annu Rev Microbiol. 1989;43:173–206. doi: 10.1146/annurev.mi.43.100189.001133. [DOI] [PubMed] [Google Scholar]
- 23.Paulus T J, Tuan J S, Luebke V E, Maine G T, DeWitt J P, Katz L. Mutation and cloning of eryG, the structural gene for erythromycin O-methyltransferase from Saccharopolyspora erythraea, and expression of eryG in Escherichia coli. J Bacteriol. 1990;172:2541–2546. doi: 10.1128/jb.172.5.2541-2546.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pereda A, Summers R, Katz L. Nucleotide sequence of the ermE distal flank of the erythromycin biosynthesis cluster in Saccharopolyspora erythraea. Gene. 1997;193:65–71. doi: 10.1016/s0378-1119(97)00086-3. [DOI] [PubMed] [Google Scholar]
- 25.Reeve L M, Baumberg S. Physiological controls of erythromycin production by Saccharopolyspora erythraea are exerted at least in part at the level of transcription. Biotechnol Lett. 1998;20:585–589. [Google Scholar]
- 26.Reeves A R, Post D A, Vanden Boom T J. Physical-genetic map of the erythromycin-producing organism Saccharopolyspora erythraea. Microbiology. 1998;144:2151–2159. doi: 10.1099/00221287-144-8-2151. [DOI] [PubMed] [Google Scholar]
- 27.Ruan X, Stassi D, Lax S A, Katz L. A second type-I PKS gene cluster isolated from Streptomyces hygroscopicus ATCC 29253, a rapamycin-producing strain. Gene. 1997;203:1–9. doi: 10.1016/s0378-1119(97)00450-2. [DOI] [PubMed] [Google Scholar]
- 28.Salah-Bey K, Doumith M, Michel J-M, Haydock S, Cortes J, Leadlay P F, Raynal M-C. Targeted gene inactivation for the elucidation of deoxysugar biosynthesis in the erythromycin producer Saccharopolyspora erythraea. Mol Gen Genet. 1998;257:542–553. doi: 10.1007/s004380050680. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stassi D, Donadio S, Staver M J, Katz L. Identification of a Saccharopolyspora erythraea gene required for the final hydroxylation step in erythromycin biosynthesis. J Bacteriol. 1993;175:182–189. doi: 10.1128/jb.175.1.182-189.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strohl W R. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 1992;20:961–974. doi: 10.1093/nar/20.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Summers R G, Donadio S, Staver M J, Wendt-Pienkowski E, Hutchinson C R, Katz L. Sequencing and mutagenesis of genes from the erythromycin biosynthetic gene cluster of Saccharopolyspora erythraea that are involved in L-mycarose and D-desosamine production. Microbiology. 1997;143:3251–3262. doi: 10.1099/00221287-143-10-3251. [DOI] [PubMed] [Google Scholar]
- 34.Tuan J S, Weber J M, Staver M J, Leung J O, Donadio S, Katz L. Cloning of genes involved in erythromycin biosynthesis from Saccharopolyspora erythraea using a novel actinomycete-Escherichia coli cosmid. Gene. 1990;90:21–29. doi: 10.1016/0378-1119(90)90435-t. [DOI] [PubMed] [Google Scholar]
- 35.Vara J, Lewandowska-Skarbek M, Wang Y-G, Donadio S, Hutchinson C R. Cloning of genes governing the deoxysugar portion of the erythromycin biosynthesis pathway in Saccharopolyspora erythraea (Streptomyces erythreus) J Bacteriol. 1989;171:5872–5881. doi: 10.1128/jb.171.11.5872-5881.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weber J M, Wierman C K, Hutchinson C R. Genetic analysis of erythromycin production in Streptomyces erythreus. J Bacteriol. 1985;164:425–433. doi: 10.1128/jb.164.1.425-433.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weber J M, Schoner B, Losick R. Identification of a gene required for the terminal step in erythromycin A biosynthesis in Saccharopolyspora erythraea (Streptomyces erythreus) Gene. 1989;75:235–241. doi: 10.1016/0378-1119(89)90269-2. [DOI] [PubMed] [Google Scholar]
- 38.Weber J M, Leung J O, Maine G T, Potenz R H B, Paulus T J, DeWitt J P. Organization of a cluster of erythromycin genes in Saccharopolyspora erythraea. J Bacteriol. 1990;172:2372–2383. doi: 10.1128/jb.172.5.2372-2383.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Westpheling J, Brawner M. Two transcribing activities are involved in expression of the Streptomyces galactose operon. J Bacteriol. 1989;171:1355–1361. doi: 10.1128/jb.171.3.1355-1361.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xue Y, Zhao L, Liu H-W, Sherman D H. A gene cluster for macrolide antibiotic biosynthesis in Streptomyces venezuelae: architecture of metabolic diversity. Proc Natl Acad Sci USA. 1998;95:12111–12116. doi: 10.1073/pnas.95.21.12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]