Abstract
The capability of cell-penetrating peptides (CPPs) to enable translocation of cargos across biological barriers shows promising pharmaceutical potential for the transport of drug molecules, as well as nanomaterials, into cells. Herein, we report on the optimization of a CPP, namely sC18, in terms of its translocation efficiency and investigate new CPPs regarding their interaction with silica nanoparticles (NPs). First, alanine scanning of sC18 yielded 16 cationic peptides from which two were selected for further studies. Whereas in the first case, a higher positive net charge and enhanced amphipathicity resulted in significantly higher internalization rates than sC18, the second one demonstrated reduced cellular uptake efficiencies and served as a control. We then attached these CPPs to silica nanoparticles of different sizes (50, 150 and 300 nm) via electrostatic interactions and could demonstrate that the secondary alpha-helical structure of the peptides was preserved. Following this, cellular uptake studies using HeLa cells showed that the tested CPP–NPs were successfully translocated into HeLa cells in a size-dependent manner. Moreover, depending on the CPP used, we realized differences in translocation efficiency, which were similar to what we had observed for the free peptides. All in all, we highlight the high potential of sequential fine-tuning of CPPs and provide novel insights into their interplay with inorganic biologically benign nanoparticles. Given the high cellular permeability of CPPs and their ability to translocate into a wide spectrum of cell types, our studies may stimulate future research of CPPs with inorganic nanocarrier surfaces.
The capability of cell-penetrating peptides (CPPs) to enable translocation of cargos across biological barriers shows promising pharmaceutical potential for the transport of drug molecules, as well as nanomaterials, into cells.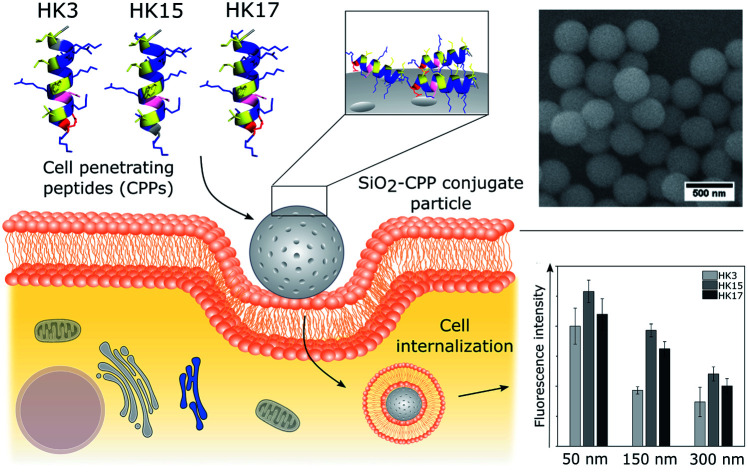
Introduction
Overcoming natural barriers such as cell membranes requires highly efficient internalization mechanisms based on specific biological and physicochemical properties and has become increasingly relevant for targeted delivery of therapeutic substances against a wide range of diseases. Cell-penetrating peptides (CPPs) are short peptide sequences that can translocate across cellular membranes without compromising their integrity and function. Meanwhile, they have been shown to be useful tools for intracellular transport of a broad variety of cargos including drugs, nucleic acids, proteins and other peptides, thereby enhancing their bioavailability.1 CPPs differ considerably in properties like size, charge and amino acid sequence, but they often share a high content of cationic amino acids and/or an amphipathic nature. Although the exact internalization mechanism of CPPs is still highly debated, it is considered to take place via an energy-dependent/endocytotic and/or energy-independent/direct pathway, depending on several factors such as attached cargo, membrane composition, cell type, peptide concentration and sequence.2,3 CPPs have become a widely used tool for the design of multifunctional drug-delivery systems, owing to the possibility of synthetically fine-tuning their sequences as a way to alter their charge, hydrophobicity and solubility.4,5 The combination of CPPs with nanoparticles was shown to result in increased intracellular access of cargoes.6,7 In addition, peptide coatings may provide biological and physical stability for such particles.8 Loading of CPPs onto nanoparticles has been shown to yield highly efficient carriers for anticancer drugs or diagnostic agents,9 to increase the target availability to cell surfaces, and to potentially enhance peptide recognition.10 While most studies on nanoparticle–CPP interactions focused on the use of polymeric, micellar or lipid nanoparticles, relatively few reports are available on the use of inorganic carriers such as gold,11,12 quantum dots10,13,14 or magnetic nanostructures.15 A variety of functionalization strategies including streptavidin–biotin conjugation,13 linkages via polyethylene glycol11 or cysteine-based cross-linking10,12,14,16 were successfully demonstrated to synthesize nanocarriers. However, since covalent linkages can significantly alter the native structure and thus the functionality of the conjugated biomolecule,5 non-covalent approaches via electrostatic interactions represent a valuable, biologically relevant and simple alternative.
With the intent to address the persistent need for efficient delivery vehicles, the present work aims at fine-tuning CPP sequences to increase their internalization efficiency, to attach them to nanoparticle surfaces, and to investigate the biophysical and biological properties of the novel constructs. Silica particles were chosen as inorganic carriers, as they have been employed for multipurpose applications, being prevalent in the field of drug delivery based on their porous structure that is tunable in size and morphology along with their high biocompatibility.17 The CPP we used is sC18, which is derived from the C-terminal domain of the cationic antimicrobial peptide CAP18.18 We recently identified and tested sC18 as a versatile transporter for cytostatic drugs and imaging agents, and additionally demonstrated its potential antimicrobial and anticancer activity.19–21 However, until now, no detailed structure–function relationship study of sC18 has been performed.
Within this work we found out that a simple modification of the peptide sequence, such as the exchange of one amino acid, led to significant changes regarding cellular translocation. Moreover, we demonstrated that besides size-dependent uptake of silica particles, also a peptide-dependent internalization effect was visible for all CPP functionalized particles, which was probably influenced by the structural properties of the attached peptides. Our findings thus contribute to a deeper understanding how fine-tuning CPP-modified inorganic nanoparticles might lead to efficient transport modules.
Results and discussion
Structure–function relationship of the CPP sC18
Whereas the primary sequence and the resulting secondary structure of CPPs are essential for their ability to interact with biological membranes, the internalization efficiency depends on many properties such as charge, guanidinium content and amphipathicity, which interplay with each other.22 To gain more insights into this complex combination and to explore novel and more efficient delivery systems, we performed an alanine scan of the recently developed CPP sC18, by substituting each of the 16 amino acids systematically with an alanine residue. All peptides were synthesized by solid phase peptide synthesis (SPPS) using the Fmoc/tBu-strategy. Subsequently, they were labeled at the N-terminal end with 5(6)-carboxyfluorescein (CF), cleaved from the resin, and identified by mass spectrometry (Table 1 and Fig. S1†).
Names, sequences and analytical data of all investigated peptides. CF: 5,6-carboxyfluorescein.
| Code | Sequence | Net charge | MWcalc [Da] | MWexp [Da] |
|---|---|---|---|---|
| HK1 | CF-ALRKRLRKFRNKIKEK-NH2 | +8 | 2442.9 | 2442.4 |
| HK2 | CF-GARKRLRKFRNKIKEK-NH2 | +8 | 2386.8 | 2386.3 |
| HK3 | CF-GLAKRLRKFRNKIKEK-NH2 | +7 | 2343.8 | 2343.4 |
| HK4 | CF-GLRARLRKFRNKIKEK-NH2 | +7 | 2371.8 | 2371.5 |
| HK5 | CF-GLRKALRKFRNKIKEK-NH2 | +7 | 2343.8 | 2343.4 |
| HK6 | CF-GLRKRARKFRNKIKEK-NH2 | +8 | 2386.8 | 2386.3 |
| HK7 | CF-GLRKRLAKFRNKIKEK-NH2 | +7 | 2343.8 | 2343.4 |
| HK8 | CF-GLRKRLRAFRNKIKEK-NH2 | +7 | 2371.8 | 2371,5 |
| HK9 | CF-GLRKRLRKARNKIKEK-NH2 | +8 | 2352.8 | 2352.4 |
| HK10 | CF-GLRKRLRKFANKIKEK-NH2 | +7 | 2343.8 | 2343.3 |
| HK11 | CF-GLRKRLRKFRAKIKEK-NH2 | +8 | 2385.9 | 2385.4 |
| HK12 | CF-GLRKRLRKFRNAIKEK-NH2 | +7 | 2371.8 | 2371.3 |
| HK13 | CF-GLRKRLRKFRNKAKEK-NH2 | +8 | 2368.8 | 2368.3 |
| HK14 | CF-GLRKRLRKFRNKIAEK-NH2 | +7 | 2371.8 | 2371.3 |
| HK15 | CF-GLRKRLRKFRNKIKAK-NH2 | +9 | 2370.9 | 2370.2 |
| HK16 | CF-GLRKRLRKFRNKIKEA-NH2 | +7 | 2371.8 | 2371.5 |
| HK17 | CF-GLRKRLRKFRNKIKEK-NH2 | +8 | 2428.9 | 2428.4 |
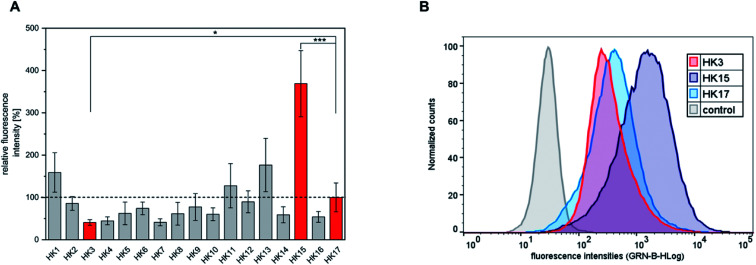
The quantitative intracellular uptake in HeLa cells of the obtained peptides was analyzed by flow cytometry and compared to that of sC18, which was already studied extensively and compared to other CPPs.18 We observed that the replacement of specific amino acids in the sequence of sC18 (HK17) led to significant changes in the cellular internalization efficacy of the peptides (Fig. 1A and S2†). For instance, substitution of positively charged residues such as arginine or lysine with alanine resulted in an expected decrease in the positive net charge of the peptides to +7 (Table 1) and led to decreased internalization efficiencies compared to HK17. This is presumably related to lower electrostatic forces between the positively charged residues and the negatively charged components of the outer cell membrane, which are crucial for membrane binding and translocation. Although the overall mechanism of how CPPs enter cells is not thoroughly elucidated, it is consistently agreed that electrostatic interactions between peptides and outer surface membrane components are key for initiating the following steps in the internalization process.23 Specifically, the most varying uptake rates were obtained by replacing arginine at position three of sC18 by alanine (HK3), which resulted in the lowest cellular uptake. In contrast, substitution of glutamic acid at position 15 with alanine (HK15) led to an increase in charge (+9) and resulted in a 3.7-fold higher internalization of this derivative compared to the native sC18 (Fig. 1B). As already mentioned, amphipathicity of CPPs plays a crucial role in membrane translocation. In fact, it has been speculated that charge distribution alongside the amphipathic helix is one of the major important factors for lipid–peptide interaction.16 A detailed inspection of the formation of amphipathic helices of all peptides generated by using helical wheel projections showed (Fig. S3†) significant differences in charge distribution, which presumably explains the basic differences in translocation efficiency. In general, all peptides demonstrate an amphipathic character containing hydrophobic and hydrophilic residues, although the overall structure is dominated by a more hydrophilic face. For instance, by replacing the negatively charged glutamic acid, which is located within the cationic face of the helix, the only negative charge is removed (HK15) (Fig. S3†). The resulting enhanced amphiphilic structure of the helix probably explains the strong internalization efficiencies, leading to more intense membrane interactions. These results agree with former studies on membrane-active peptides24 and underpin the strong influence of the type and position of each single amino acid on the activity of CPP sequences.
Fig. 1. (A) Quantitative cellular uptake of all synthesized peptides obtained in the alanine-scan represented as mean values. HeLa cells were incubated for 30 min with 20 μM peptide solutions. For comparison, the significance between the internalization efficacy of the most outstanding red labeled peptides was determined by students t-test (*p < 0.05; ***p < 0.0005). (B) Flow cytometry fluorescence intensity distributions of HeLa cells treated with HK3, HK15 and HK17.
In the following steps, the focus was kept on the analysis of peptides HK3 and HK15, to compare their behavior with the native peptide HK17 and to understand the influence of the CPP and its functionality in complex delivery systems. As CPPs are known to be able to transport large cargoes such as nanoparticles, this potential was verified for our optimized CPP. For this purpose, these three peptides were attached to silica particles as the model system to investigate whether the structure–function relationship of the CPPs is preserved when coated onto the surface of inorganic nanocarriers.
Interaction of peptides and silica nanoparticles
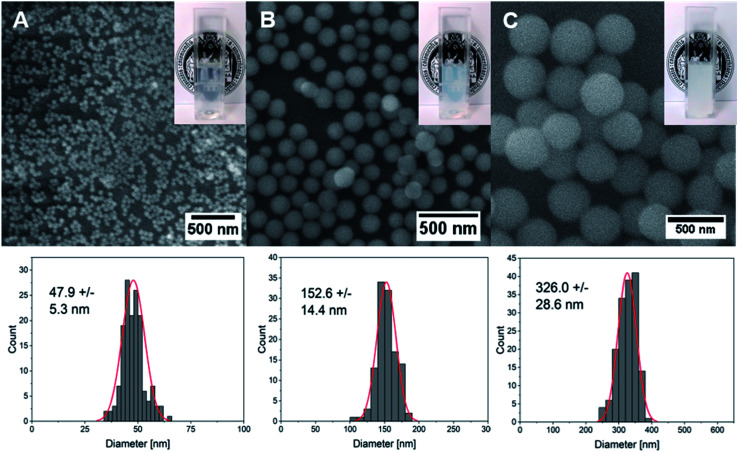
Silica (SiO2) nanoparticles were prepared in three different sizes (average diameter, ca. 50, 150, 300 nm) following the well-known Stöber process25 and subsequently surface-modified with CF-labeled CPPs using a method based on physisorption. The homogeneous size distribution of silica nanoparticles in individual samples was confirmed via scanning electron microscope (SEM) images (Fig. 2A–C) and dynamic light scattering (DLS) measurements (Fig. S4†).
Fig. 2. SEM images and the corresponding size distribution profiles and photographs of aqueous dispersions of silica nanoparticles prepared using the Stöber process with ammonium hydroxide concentrations of (A) 0.22 M, (B) 0.43 M and (C) 0.86 M.
Previous studies have shown that although site-selective binding and quantitative surface coverage might be more controlled and reproducible in terms of covalent attachment of biomolecules to carrier nanoparticles, the native biomolecular structure and function is usually retained when simple electrostatic approaches are employed.26 Moreover, covalent conjugation may change the chemical structure of biomolecules and might thus also alter their functionality. Considering the strong negative charge of the silica particles around −30 mV (Table 2) generated by the presence of deprotonated hydroxyl functionalities on the particle surface, the strongly positively charged peptide was expected to quickly attach to the inorganic vector. Especially, amino acids that contain free amino groups such as lysine and arginine are thought to contribute significantly through Coulomb interactions to form stable peptide–particle conjugates.27 Indeed, the use of dye-labeled peptides allows for the visualization of a clear color change of the particles from white to orange after the attachment (Fig. S5†) which could also be detected as a peak with maximum intensity at around 500 nm using UV-vis spectroscopy (Fig. S4,†Table 2). This peak was even visible after stirring the functionalized particles in a cell culture medium supplemented with 10% FBS. Owing to the presence of strongly negatively charged proteins in serum, surface adsorption of these biomolecules onto CPP-coated NPs may have occurred, and the exchange of CPPs with other proteins to a small extent might have happened. However, the results depicted in Fig. S4† demonstrated clearly that the fluorescence of the CPPs on the particle surface is still detectable after thoroughly stirring the conjugates in serum for two hours. We therefore assume that adsorbed CPPs are not easily exchanged by serum proteins and that they are stable even in their presence. Moreover, a strong change in surface charge from negative to positive values (Table 2) supported the successful biomolecular attachment to silica carriers. Quantitative evaluations of attached peptides performed using a Bradford assay demonstrated an increased peptide concentration per mass of nanoparticles with decreasing particle size which correlates well with the increase in the surface-area-to-volume ratio (Table 2) in smaller particles.
Zeta-potential measurements before and after CPP attachment to silica particles, their maximum absorption values, and concentration of attached CPPs on the particle surface as determined by Bradford assay. Absorption maxima for CPP-functionalized particles of 300 nm diameter could not be determined due to the strong scattering of the particles and the low concentration of attached CPPs.
| Sample | ζ-Potential before CPP attachment [mV] | ζ-Potential after CPP attachment [mV] | Lambda max [nm] | Concentration of the CPP [μM] |
|---|---|---|---|---|
| 50-HK3 | −28.3 ± 0.6 | 5.0 ± 0.1 | 498 | 25.28 |
| 50-HK15 | −28.3 ± 0.6 | 12.0 ± 0.8 | 498 | 27.95 |
| 50-HK17 | −28.3 ± 0.6 | 9.1 ± 0.1 | 498 | 28.33 |
| 150-HK3 | −29.3 ± 0.6 | 4.4 ± 0.8 | 501 | 19.33 |
| 150-HK15 | −29.3 ± 0.6 | 22.0 ± 0.4 | 500 | 17.78 |
| 150-HK17 | −29.3 ± 0.6 | 10.9 ± 0.6 | 501 | 22.82 |
| 300-HK3 | −29.9 ± 0.2 | 6.0 ± 0.3 | — | 12.68 |
| 300-HK15 | −29.9 ± 0.2 | 9.8 ± 1.3 | — | 13.13 |
| 300-HK17 | −29.9 ± 0.2 | 7.3 ± 1.0 | — | 10.77 |
SEM and transmission electron microscopy (TEM) studies revealed that the thin CPP layer on the surface of silica particles is almost invisible and does not influence the mean size of the particles (Fig. S5 and S6†).
Cytotoxicity studies and cellular uptake of CPP-functionalized SiO2 particles
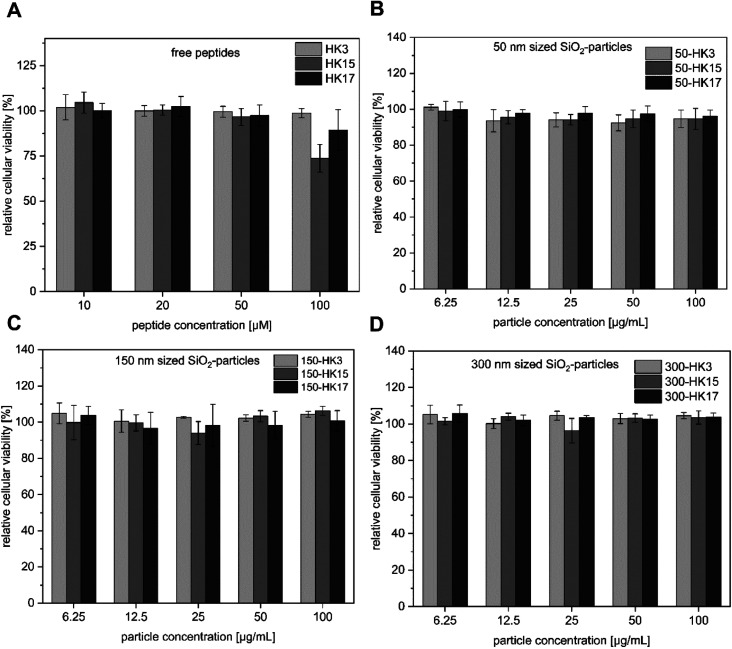
Since high tolerance in a biological environment is an essential prerequisite for delivery vehicles, we first performed cell viability assays including the free peptides and the corresponding CPP-functionalized particles (Fig. 3A–D). Notably, all tested CPP-coated silica particles exhibited no cellular toxicity at concentrations between 6.25 and 100 μg mL−1 (particle concentrations).
Fig. 3. (A–D) Cytotoxicity assay of silica–peptide conjugates (50–300 nm with HK3, HK15 and HK17 surface modification, respectively) and the free peptides.
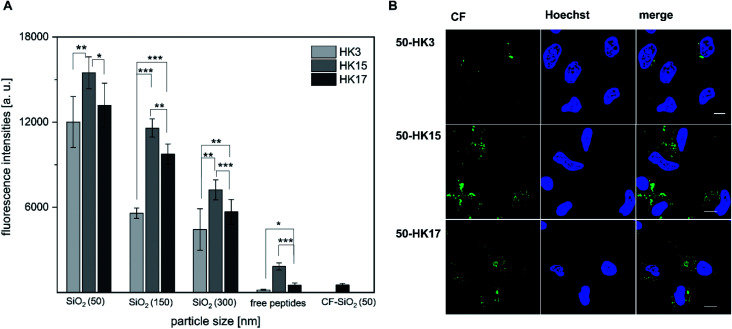
Next, we determined if the functionality of CPPs is preserved when attached to silica nanoparticles. Notably, all CPP–particle constructs were efficiently taken up upon incubation with HeLa cells for only 30 min, an incubation time that is frequently used when working with CPPs (and in particular with sC18).18 Furthermore, the results revealed a size-dependent internalization behavior, as 50 nm particles were preferentially taken up followed by 150 nm and 300 nm particles (Fig. 4A and S7†). This is in accordance with previous studies, in which it was shown that various particle types, including dye functionalized mesoporous silica particles, were most efficiently internalized at mean particle sizes of 50 nm .28–30 However, in addition to the observed size-dependent uptake, a peptide-dependent uptake was also evident. Indeed, similar to the free peptides, HK3-modified particles were internalized to a lower extent compared to HK17, while the fluorescence intensity was highest for the strongly positively charged HK15. In addition, it was recognized that the overall fluorescence intensities were dramatically higher than those for the peptides alone. This is probably due to the rather low concentration of peptides attached to the particles which is challenging to determine and might therefore not concur with the peptide concentration in the free peptide solutions. Furthermore, peptides might benefit from the attachment to the particles by way of improving their overall stability presumably through a shielding effect of the particles (as already observed, see Fig. S4†). Therefore, we were eager to see if the CPP–particle constructs were visible intracellularly when using longer incubation times, e.g. 24 h. We focused on the 50 nm particles, which exhibited the highest internalization rates. Microscopy analysis supported cellular uptake of all tested constructs, albeit intensities were again dependent on the attached CPP. Moreover, we observed fluorescence intensities that might be either the result of cytosolic accumulation or endosomal entrapment. Although the latter might be disadvantageous with respect to future applications, in which endosomal release is important, we recently demonstrated that bioactivity was maintained for other CPP-based conjugates that were also taken up partly by endocytosis.31–33 The highest intracellular intensity levels of the fluorophore were visible in the case of HK15 followed by HK17 and HK3, which confirmed the previously observed trend during flow cytometry measurements of the free peptides (Fig. 1A and B). Additionally, the morphology of HeLa cells was not found to be affected, ruling out any pronounced cytotoxic effects (Fig. 4B).
Fig. 4. (A) Quantitative uptake of CPP functionalized silica particles (50, 150 and 300 nm diameter) into HeLa cells. Uptake was measured after 30 min of incubation using flow cytometry. For comparison, the significance between the internalization efficacies was determined by students t-test (*p < 0.05; **p < 0.005; ***p < 0.0005). (B) Confocal microscopy studies after incubating HeLa cells for 24 h with 50 nm CPP-coated silica nanoparticles. Cell nuclei were stained with Hoechst stain before imaging. Scale bar corresponds to 10 μm.
This concurs with quantitative uptake studies of the coated particles (Fig. 4A) and demonstrates that differences in the peptide functionality which alter the uptake behavior could still be observed after its attachment to inorganic carriers, also when longer incubation times were considered.
CD spectroscopy studies
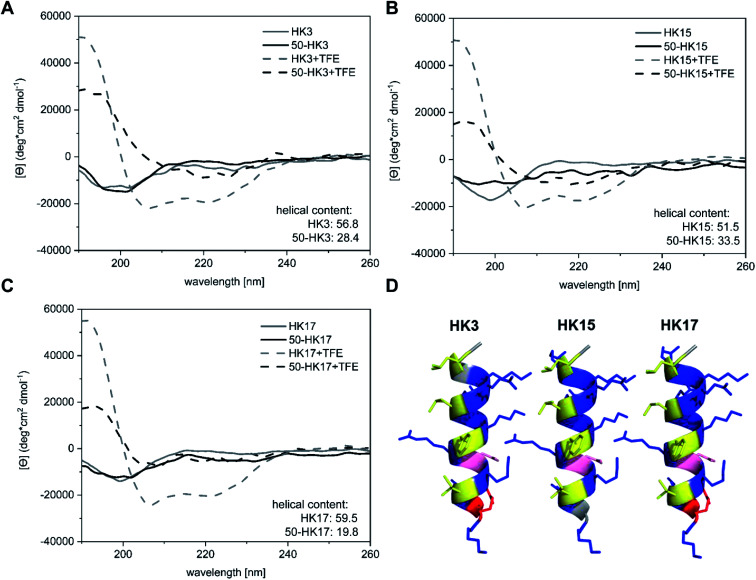
Besides surface charge, the secondary structure is known to play a crucial role in cellular internalization of CPPs. To evaluate if the secondary structure of peptides changed upon electrostatic attachment to particulate carriers, circular dichroism spectroscopy was used. We determined the secondary structure of the free peptides HK3, HK15 and HK17 in comparison to peptide structures on peptide-functionalized particles.
All free peptides formed a random coil structure in phosphate buffer, characterized by a negative minimum around 200 nm (Fig. 5A–C).34 However, upon the addition of the secondary-structure inducing reagent trifluoroethanol (TFE), the formation of an α-helical structure (Fig. 5D) was visible, which is characterized by two minima at 222 nm and 207 nm and a maximum around 190 nm. The tendency of peptides to form an α-helix is correlated to the ellipticity (θ) intensity at 222 nm and is generally expressed as the α-helical content.35 Computational modelling studies have additionally shown that this correlation can also be used to determine potential losses in peptide helicity upon adsorption onto silica particles.36 Evaluation of the peptides used in this study revealed that the highest helicity contents were observed for the free peptide CF-HK17, although only a slight difference is observed compared to CF-HK3 and CF-HK15. Considering the differences in the cell internalization efficiency for these peptides, these results imply that the α-helical conformation itself has a lower impact on their probability to cross cellular barriers and that charge distribution alongside the helix is probably the more important factor.
Fig. 5. CD spectroscopic analyses of (A) CF-HK3, (B) CF-HK15, (C) CF-HK17 and the corresponding functionalized silica particles. The samples were measured in phosphate buffer with and without TFE. (D) Helical structures of the synthesized peptides HK3, HK15 and HK17 were prepared with the PyMOL Molecular Graphics System, Version 2.0 Schrödinger. Hydrophobic residues are marked in yellow, while residues that possess zero hydrophobicity are marked in grey. Hydrophilic uncharged residues are labeled in pink whereas potentially negatively charged residues are labeled in red and potentially positively charged residues are labeled in blue.
When CPP-functionalized nanoparticles were analyzed, again a significant change in structure was visible after the addition of TFE. For 50 nm sized particles (50-HK3, 50-HK15 and 50-HK17), the minima at around 200 nm shifted towards a maximum at 190 nm, while two minima at around 207 nm and 222 nm appeared, indicative of a mainly α-helical structure. This is in accordance with simulation studies that have been performed on the non-covalent adsorption of CPPs (in this case, Antennapedia homeodomain-derived CPPs) onto silica surfaces which revealed conservation of the secondary structure of the peptide on the particle surface.27 Nevertheless, helical contents were significantly lower compared to the free peptides and thus pointed towards a perturbation of the secondary structure upon adsorption of the CPP onto the particles. This observation is in agreement with previously reported simulation studies on peptide silica interactions, where a loss in helicity was detected upon adsorption onto the inorganic surface.36
While a clear change in CD spectroscopy measurements in the presence of TFE was observable for all tested particle–CPP compounds (Fig. S8†), the minima at around 207 nm and 222 nm became less pronounced with increasing particle size. Similarly, a decrease in α-helical content was observed with increasing particle size. In addition to the scattering interferences of CD signals caused by the large size of the particles, previous studies have demonstrated that the carrier size can have a distinct influence on the secondary structure of surface-bound biomolecules. For instance, Vertegel et al. demonstrated that lysozyme adsorption onto silica nanoparticles of three different sizes between 4 and 100 nm led to significant changes in the α-helical content of the protein, whereas a greater loss in alpha helicity was observed with increasing particle size.37 Hence, biomolecules such as CPPs employed in this study exhibit perturbation of their secondary structure upon adsorption onto inorganic carriers of different sizes, presumably related to the change in surface curvature, although the formation of an alpha-helical structure can still be assumed.
Conclusion
The purpose of this work was to study the structure–function relationship of the sC18 CPP family through an alanine scan and to evaluate their interaction with inorganic silica nanoparticles of different sizes. Our results demonstrate that the internalization properties of the peptides rely heavily on each single amino acid and its position within the sequence. A simple exchange of one amino acid with alanine resulted in strong differences in cellular internalization efficiencies. Moreover, after electrostatic attachment of three distinct peptides (HK3, HK15 and HK17) to silica nanoparticles, all peptide–particle compounds were taken up by HeLa cells. However, we observed that the uptake of 50 nm particles coated with CPPs was higher, when compared to the 150 and 300 nm particles, irrespective of their surface coverage. Our findings indicate that the particle size plays a superior role compared to surface functionalization with CPPs in cellular uptake rates. Interestingly, confocal microscopy and flow cytometry studies reflected that differences in cell-penetrating properties were also observed when bound to silica surfaces. Although only electrostatic interactions were used, the formation of the secondary alpha helical structure was less pronounced in conjugates compared to the free biomolecules indicating that chemical surface interactions and attachment of the CPP onto the surface of silica nanoparticles lead to a reduction in the flexibility of the peptide backbone. Moreover, we hypothesize that the peptide net charge, which was highest in the case of HK15 and its particle conjugates, was the key driving force for cellular internalization.
Overall, these results demonstrate the complex interplay between the carrier size as well as surface charge and secondary structure of surface-bound peptides and allow for a deeper understanding in tailoring these conjugates to achieve the highest possible cellular internalization efficiencies. This is of high value for the future design of drug delivery vehicles, for instance, to incorporate drug molecules into the porous carrier structures which would support efficient intracellular access of therapeutics across biological barriers. Furthermore, targeting of specific cells could be achieved by combining CPPs on the surface of the particles with tumor homing peptides.
Experimental procedures
Synthesis of peptide derivatives
All the peptides used in this work were synthesized following the standard Fmoc/tBu-strategy using a Syro I automated multiple solid-phase peptide synthesizer (multiSynTech). The peptides were prepared as C-terminal amides using Rink amide resin, and all peptides were labeled N-terminally with 5(6)-carboxyfluorescein (CF). Labeling of the peptides was performed on resins with fully protected side chains by using 5 eq. of the fluorophore, Oxyma and N,N′-diisopropylcarbodiimide (DIC) (overnight (o/n), RT). Peptides were cleaved from the resin using trifluoroacetic acid (TFA)/triisopropylsilane/water (95 : 2.5 : 2.5, v/v/v) within 3 h at room temperature. Analytical data were obtained by RP-HPLC (Aeris peptide: 2.6 μm XB-C18; LC Column 150 × 3.0 mm (Phenomenex)) using a gradient of 10–60% acetonitrile (ACN) in H2O with 0.1% formic acid for sample separation, followed by electrospray-ionization mass spectrometry (ESI/MS, Thermo Scientific LTQ-XL) measurements. Purification of the peptides was achieved by preparative RP-HPLC (Nucleodur C18ec; 100-5; Macherey-Nagel) using linear gradients of 10–60% B in A (A = 0.1% TFA in water; B = 0.1% TFA in ACN).
Synthesis of silica nanoparticles
Silica particles were prepared using the well-known Stöber process.25 In a typical synthesis, 90.5 mL ethanol and 32.5 mL distilled water were mixed. Then, ammonia (28–30%, Sigma-Aldrich) was added in various concentrations between 0.22 and 0.86 M (1.125 mL (50 nm); 2.25 mL (150 nm); 4.50 mL (300 nm)) depending on the desired particle size. Following this, 7.75 mL tetraethyl orthosilicate (TEOS, Sigma-Aldrich) were added dropwise using a dropping funnel to the solvent mixture which was subsequently stirred for two hours. After that time, the mixture turned milky white and the as-prepared silica nanoparticles were collected as a white precipitate via centrifugation (11 000 rpm, 15 minutes). The as-obtained particles were washed several times with ethanol to remove any residual precursor molecules and dried under ambient conditions.
Preparation of CPP coated silica particles
Silica particles were surface functionalized with different types of CPPs based on a physisorption method. A stable colloidal dispersion of silica particles was prepared by dispersing 2 mg of particles in 1 mL of sterile water (pH was adjusted to 9) using ultrasonication. Then, 80 μL of a 1 mM stock solution of the dye-labeled peptides was added and the dispersion was stirred overnight in the dark. The biofunctionalized particles were separated from the unattached peptides using centrifugation (2 minutes, 15 000 rpm, 15 °C). Finally, the conjugates were washed two times with sterile and filtered water and were stored at 4 °C as 4 mg mL−1 aqueous dispersions until further processing.
Particle characterization
The shape and size distribution of (functionalized) silica particles was investigated using a Nova Nano SEM 430 field-emission scanning electron microscope (SEM) and a ZEISS Leo912 transmission electron microscope (TEM) operated at an accelerating voltage of 120 kV. Zeta potential measurements and dynamic light scattering experiments were performed on aqueous dispersions using a Zetasizer Nano ZS (Malvern Instruments) at a wavelength of 633 nm in folded capillary zeta cells. UV-vis spectroscopy (LAMBDA 950 PerkinElmer) was used to determine the successful functionalization of nanoparticles with dye-labeled CPPs. The peptide concentration on the surface of silica particles was determined with the Roti-Nanoquant dye following the instructions provided with the product.
Circular dichroism (CD) spectroscopy
CD spectra of the free peptides and the functionalized nanoparticles (20 μM in 10 mM phosphate buffer, pH 7.0, or 10 mM phosphate buffer/2,2,2-trifluoroethanol (TFE) (1 : 1 v/v)) were recorded in triplicate using a Jasco Corp J-715 spectropolarimeter in a range from 190–260 nm. Instrument options were set to 100 mdeg sensitivity, 0.2 nm data pitch, 100 nm min−1 scanning speed and a bandwidth of 1.0 nm. The resulting signal was converted from ellipticity (mdeg) to molar ellipticity [Θ] in deg cm2 dmol−1. In order to ensure that the obtained signals of particle–peptide conjugates were only derived from the peptide itself, reference measurements with the particles only were performed and subtracted from the obtained spectra. The helical content was calculated using the following equation: % α-helix = (([Θ222] − 3000)/(−36 000 − 3000)) × 100.35
Cell culture studies
HeLa (human epithelial cervical cancer; ACC57) cells were cultured as subconfluent monolayers in 10 cm Petri dishes at 37 °C in a humidified atmosphere containing 5% CO2. The cells were maintained in RPMI 1640 medium supplemented with 10% FBS and 4 mM glutamine. When reaching a confluency of ∼80–90%, the cells were split using 0.5 mg mL−1 trypsin-EDTA for cell detachment. All cell experiments were performed at least twice in triplicate. To determine the influence of the peptides and the nanoparticles on the cellular viability, 1.7 × 104 HeLa cells were seeded in 96-well plates and the samples were analyzed as described previously with a resazurin-based viability assay.19
Cellular uptake and internalization studies
To determine the quantitative uptake of the peptides generated in the alanine scan, 3.0 × 104 HeLa cells were seeded in 96-well plates. For the analysis of the functionalized nanoparticles, 1.0 × 105 HeLa cells were seeded in 24-well plates and grown to subconfluency (∼70–80%) overnight. The samples were incubated for 30 min in the appropriate serum-free medium, washed twice with PBS, detached with phenolred-free trypsin and resuspended in phenolred-free medium. Cellular uptake was determined with a Guava easyCyteTM System (Merck) using the GRN-B (525/30) channel, counting 10 000 cells per well. The cellular autofluorescence was subtracted from all measured fluorescence intensities and experiments were performed twice in triplicate.
To gain more insights into the qualitative internalization of the peptides, confocal laser scanning microscopy was done on a Leica TCS SP8 confocal scanning microscope using a 63× immersion oil objective. For this purpose, 3.0 × 104 HeLa cells were seeded in a μ-slide eight-well plate (Ibidi) and grown to subconfluency (∼70–80%). Sample incubation and further processing were performed as already described.21
Conflicts of interest
The authors declare no conflicts of interests.
Supplementary Material
Acknowledgments
The authors would like to acknowledge the infrastructure provided by the Imaging Facility at the Cluster of Excellence – Cellular Stress Responses in Aging-Associated Diseases (CECAD) for confocal microscopy measurements. Moreover, the authors would like to thank Dr Stefan Roitsch for TEM measurements.
Electronic supplementary information (ESI) available: Helical wheel projections of all peptides, HPLC-ESI/MS spectra of the CF-labeled peptides, and CD spectra of 150 nm and 300 nm sized CPP–silica particles in comparison to the free peptides. DLS measurements of silica particles and SEM and TEM measurements after peptide functionalization. Stability tests of CPP conjugates in serum. See DOI: 10.1039/c9na00693a
References
- Zhang D. Wang J. Xu D. J. Controlled Release. 2016;229:130–139. doi: 10.1016/j.jconrel.2016.03.020. [DOI] [PubMed] [Google Scholar]
- Jarver P. Mager I. Langel U. Trends Pharmacol. Sci. 2010;31:528–535. doi: 10.1016/j.tips.2010.07.006. [DOI] [PubMed] [Google Scholar]
- Ramsey J. D. Flynn N. H. Pharmacol. Ther. 2015;154:78–86. doi: 10.1016/j.pharmthera.2015.07.003. [DOI] [PubMed] [Google Scholar]
- Dissanayake S. Denny W. A. Gamage S. Sarojini V. J. Controlled Release. 2017;250:62–76. doi: 10.1016/j.jconrel.2017.02.006. [DOI] [PubMed] [Google Scholar]
- Kebebe D. Liu Y. Wu Y. Vilakhamxay M. Liu Z. Li J. Int. J. Nanomed. 2018;13:1425–1442. doi: 10.2147/IJN.S156616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva S. Almeida A. J. Vale N. Biomolecules. 2019;9(1):pii: E22. doi: 10.3390/biom9010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson E. S. Jiang T. Aguilera T. A. Nguyen Q. T. Ellies L. G. Scadeng M. Tsien R. Y. Proc. Natl. Acad. Sci. U. S. A. 2010;107:4311–4316. doi: 10.1073/pnas.0910283107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spicer C. D. Jumeaux C. Gupta B. Stevens M. M. Chem. Soc. Rev. 2018;47:3574–3620. doi: 10.1039/C7CS00877E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkhani S. M. Valizadeh A. Karami H. Mohammadi S. Sohrabi N. Badrzadeh F. Peptides. 2014;57:78–94. doi: 10.1016/j.peptides.2014.04.015. [DOI] [PubMed] [Google Scholar]
- Carnevale K. J. F. Muroski M. E. Vakil P. N. Foley M. E. Laufersky G. Kenworthy R. Zorio D. A. R. Morgan Jr T. J. Levenson C. W. Strouse G. F. Bioconjug. Chem. 2018;29:3273–3284. doi: 10.1021/acs.bioconjchem.8b00429. [DOI] [PubMed] [Google Scholar]
- Morshed R. A. Muroski M. E. Dai Q. Wegscheid M. L. Auffinger B. Yu D. Han Y. Zhang L. Wu M. Cheng Y. Lesniak M. S. Mol. Pharm. 2016;13:1843–1854. doi: 10.1021/acs.molpharmaceut.6b00004. [DOI] [PubMed] [Google Scholar]
- Boussoufi F. Gallon S. M. N. Chang R. Webster T. J. Int. J. Nanomed. 2018;13:6199–6205. doi: 10.2147/IJN.S168720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B. Liu Q. Zhang Y. Xu L. Fang X. Langmuir. 2008;24:11866–11871. doi: 10.1021/la802048s. [DOI] [PubMed] [Google Scholar]
- Walther C. Meyer K. Rennert R. Neundorf I. Bioconjug. Chem. 2008;19:2346–2356. doi: 10.1021/bc800172q. [DOI] [PubMed] [Google Scholar]
- Dowaidar M. Nasser Abdelhamid H. Hallbrink M. Langel U. Zou X. J. Biomater. Appl. 2018;33:392–401. doi: 10.1177/0885328218796623. [DOI] [PubMed] [Google Scholar]
- Chen L. Zhang Q. Yuan X. Cao Y. Yuan Y. Yin H. Ding X. Zhu Z. Luo S.-Z. Int. J. Biochem. Cell Biol. 2017;83:71–75. doi: 10.1016/j.biocel.2016.12.011. [DOI] [PubMed] [Google Scholar]
- Farjadian F. Roointan A. Mohammadi-Samani S. Hosseini M. Chem. Eng. J. 2019;359:684–705. doi: 10.1016/j.cej.2018.11.156. [DOI] [Google Scholar]
- Neundorf I. Rennert R. Hoyer J. Schramm F. Lobner K. Kitanovic I. Wolfl S. Pharmaceuticals. 2009;2:49–65. doi: 10.3390/ph2020049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronewold A. Horn M. Randelovic I. Tovari J. Munoz Vazquez S. Schomacker K. Neundorf I. ChemMedChem. 2017;12:42–49. doi: 10.1002/cmdc.201600498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt A. Horn M. Schmauck J. P. Brohl A. Giernoth R. Oelkrug C. Schubert A. Neundorf I. Bioconjug. Chem. 2014;25:2166–2174. doi: 10.1021/bc500510c. [DOI] [PubMed] [Google Scholar]
- Klimpel A. Neundorf I. J. Controlled Release. 2018;291:147–156. doi: 10.1016/j.jconrel.2018.10.029. [DOI] [PubMed] [Google Scholar]
- Kalafatovic D. Giralt E. Molecules. 2017;22(11):pii: E1929. doi: 10.3390/molecules22111929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisa M. D. Chassaing G. Swiecicki J.-M. Biochemistry. 2015;54:194–207. doi: 10.1021/bi501392n. [DOI] [PubMed] [Google Scholar]
- Piotrowska U. Sobczak M. Oledzka E. Chem. Biol. Drug Des. 2017;90:1079–1093. doi: 10.1111/cbdd.13031. [DOI] [PubMed] [Google Scholar]
- Stober W. Fink A. Bohn E. J. Colloid Interface Sci. 1968;26:62–69. doi: 10.1016/0021-9797(68)90272-5. [DOI] [Google Scholar]
- Mathur S., Ilyas S., Wortmann L., Kaur J. and Gessner I., in Biocatalysis & Nanotechnology, Pan Stanford Publishing Pte. Ltd., Singapore, 2017 [Google Scholar]
- Grasso G. Deriu M. A. Prat M. Rimondini L. Verne E. Follenzi A. Danani A. J. Phys. Chem. B. 2015;119:8239–8246. doi: 10.1021/jp512782e. [DOI] [PubMed] [Google Scholar]
- Chithrani B. D. Ghazani A. A. Chan W. C. Nano Lett. 2006;6:662–668. doi: 10.1021/nl052396o. [DOI] [PubMed] [Google Scholar]
- Lu F. Wu S. H. Hung Y. Mou C. Y. Small. 2009;5:1408–1413. doi: 10.1002/smll.200900005. [DOI] [PubMed] [Google Scholar]
- Malugin A. Ghandehari H. J. Appl. Toxicol. 2010;30:212–217. doi: 10.1002/jat.1486. [DOI] [PubMed] [Google Scholar]
- Feni L. Parente S. Robert C. Gazzola S. Arosio D. Piarulli U. Neundorf I. Bioconjug. Chem. 2019;30:2011–2022. doi: 10.1021/acs.bioconjchem.9b00292. [DOI] [PubMed] [Google Scholar]
- Lindenblatt D. Horn M. Gotz C. Niefind K. Neundorf I. Pietsch M. ChemMedChem. 2019;14:833–841. doi: 10.1002/cmdc.201800786. [DOI] [PubMed] [Google Scholar]
- Kuhlmann N. Chollet C. Baldus L. Neundorf I. Lammers M. ChemMedChem. 2017;12:1703–1714. doi: 10.1002/cmdc.201700414. [DOI] [PubMed] [Google Scholar]
- Gokce I. Woody R. W. Anderluh G. Lakey J. H. J. Am. Chem. Soc. 2005;127:9700–9701. doi: 10.1021/ja052632x. [DOI] [PubMed] [Google Scholar]
- Raussens V. Ruysschaert J. M. Goormaghtigh E. Anal. Biochem. 2003;319:114–121. doi: 10.1016/S0003-2697(03)00285-9. [DOI] [PubMed] [Google Scholar]
- Meissner R. H. Schneider J. Schiffels P. Colombi Ciacchi L. Langmuir. 2014;30:3487–3494. doi: 10.1021/la500285m. [DOI] [PubMed] [Google Scholar]
- Vertegel A. A. Siegel R. W. Dordick J. S. Langmuir. 2004;20:6800–6807. doi: 10.1021/la0497200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.