Abstract
The focus of radiation biodosimetry has changed recently, and a paradigm shift for using molecular technologies of omic platforms in addition to cytogenetic techniques has been observed. In our study, we have used a nonhuman primate model to investigate the impact of a supralethal dose of 12 Gy radiation on alterations in the lung transcriptome. We used 6 healthy and 32 irradiated animal samples to delineate radiation-induced changes. We also used a medical countermeasure, γ-tocotrienol (GT3), to observe any changes. We demonstrate significant radiation-induced changes in the lung transcriptome for total-body irradiation (TBI) and partial-body irradiation (PBI). However, no major influence of GT3 on radiation was noted in either comparison. Several common signaling pathways, including PI3K/AKT, GADD45, and p53, were upregulated in both exposures. TBI activated DNA-damage-related pathways in the lungs, whereas PTEN signaling was activated after PBI. Our study highlights the various transcriptional profiles associated with γ- and X-ray exposures, and the associated pathways include LXR/RXR activation in TBI, whereas pulmonary wound-healing and pulmonary fibrosis signaling was repressed in PBI. Our study provides important insights into the molecular pathways associated with irradiation that can be further investigated for biomarker discovery.
Keywords: MT: bioinformatics, γ-tocotrienol, lung, nonhuman primates, radiation injury, radiation countermeasure, transcriptomics, total-body irradiation, partial-body irradiation
Graphical abstract
The authors demonstrate the effects of ionizing radiation (total-body γ- and partial-body X-ray) exposure on transcriptomic changes in nonhuman primate lung tissue and also investigated the effect of a radiation countermeasure, γ-tocotrienol. Total- and partial-body irradiation identified common and distinct radiation-response-associated molecular pathways without any major influence of γ-tocotrienol.
Introduction
Exposures to ionizing radiation are an undeniable reality of life. Such incidents can bring about harmful health consequences; hence, radiological preparedness is a grave security issue.1,2 Radiation exposures can result in different types of injuries requiring diagnostic and therapeutic interventions. The clinical development of acute radiation syndrome (ARS) depends on the absorbed dose of radiation. Human ARS manifests following exposure to total-body irradiation (TBI) or partial-body irradiation (PBI) at doses >2 Gy delivered at a high dose rate. Some organ systems such as the hematopoietic, gastrointestinal, pulmonary, central nervous, and cutaneous system are more sensitive to irradiation.3 Clinical indications of ARS include the hematopoietic (H-ARS; 2–6 Gy), gastrointestinal (GI-ARS; 6–8 Gy), and neurovascular (NV-ARS; >8 Gy) subsyndromes.4 Individuals exposed to radiation doses resulting in H-ARS or GI-ARS are expected to benefit more from treatment with radiation medical countermeasures (MCMs).
It is important to assess the extent of radiation exposure to provide appropriate and timely medical intervention.5 Physical and biological dosimetry can be used in combination to determine the exposure dose, which provides a tool that accelerates clinical evaluation and response.6 The gold standard for biological dosimetry includes cytogenic analysis of peripheral blood lymphocytes, dicentric chromosome assay, and quantification of metabolites and proteins in the body fluids. However, these methods are labor intensive and unfeasible for population-based screening during a mass causality scenario.7 Hence, accurate and reliable methods suitable for mass screening to differentiate radiation-exposed and unexposed individuals for medical care are warranted. Assays involving genomic, transcriptomic, proteomic, and metabolomic markers have the potential for rapid high-throughput screening of masses and can reliably account for population variability.8, 9, 10 Of these methods, transcriptomic profiling to identify radiation-specific biomarkers is an emerging, high-throughput, reliable, and feasible approach for assessing related damage.11, 12, 13 This advantage of transcriptome profiling over other methods is due to the technological advancements to accurately sequence the transcripts and the availability of bioinformatics algorithms to analyze these profiles. In the recent past, transcriptomic studies have presented insights into various areas of radiation biology and demonstrated great potential for understanding radiation injury, exposure dose determination, and MCM development.11, 12, 13 Several studies from various investigators have profiled transcriptomic changes in various tissues of nonhuman primates (NHPs).11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28 However, the majority of these publications are with baboons, which are no longer used for such studies. Therefore, there is a need for studies performed with rhesus and cynomolgus, which are commonly used NHPs. Though significant progress has been made with transcriptomic studies using radiation injury models of NHPs, additional investigation is required to ensure that signatures are robust.16,18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28
NHP models of radiation injury have several advantages over both small animal models, including rodents, and large animal models, including swine (minipig) and canines.3,29,30 NHPs closely resemble humans and have several advantages with respect to their organ structure, genetic homology sharing with more than 95% DNA sequence similarity, metabolism, response to physiological pathways, cell receptors, suitability for gastrointestinal symptoms (vomiting), long life span, and the ease of sequential sampling for monitoring radiation response and recovery. This model is as close as possible to humans, and is considered the gold standard of all animal models available for drug development and regulatory approval by the United States Food and Drug Administration.31 In brief, the close relationship between NHPs and humans has made NHPs an attractive animal model for preclinical research. γ-Tocotrienol (GT3) is an antioxidant and a component of vitamin E under development as a radioprotector for pre-exposure prophylaxis.32, 33, 34, 35, 36 It has been studied in rodents and NHPs and has demonstrated radioprotective efficacy when administered 24 h before TBI.32,35
In this study, we profiled transcriptomic changes in the lungs of rhesus NHPs exposed to a supralethal dose of radiation inducing severe ARS. We also compared the gene expression changes resulting from TBI and PBI. Furthermore, the changes induced by the promising radiation MCM, GT3, at two time points, day 4 and day 7 post irradiation, were analyzed. The sex-dependent transcriptional changes in response to GT3 and irradiation were also evaluated.
Results
Lung transcriptome profiling for radiation response and drug response
RNA-sequencing (RNA-seq) reads obtained after lung transcriptome sequencing were aligned to the macaque genome using STAR aligner in the two-pass mode. The alignment percentage of uniquely mapped reads ranged from 80 to 89. Filtering based on the expression of the genes having an average expression profile above ten reads was performed. After filtering out lowly expressed genes, 44% of the total genes were included in the downstream analysis. Differential expression of different comparisons was performed using DESeq2. Comparisons for TBI and PBI were performed separately. For each irradiation group, the effect of irradiation (healthy versus irradiated), sex (male versus female), drug treatment (GT3 versus vehicle), and days post exposure (day 7 versus day 4) were profiled using differential expression analysis (Figure 1 and Table 1).
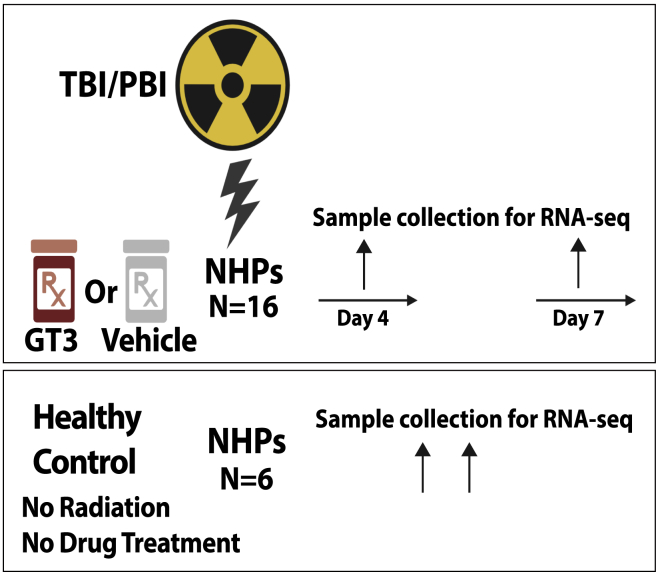
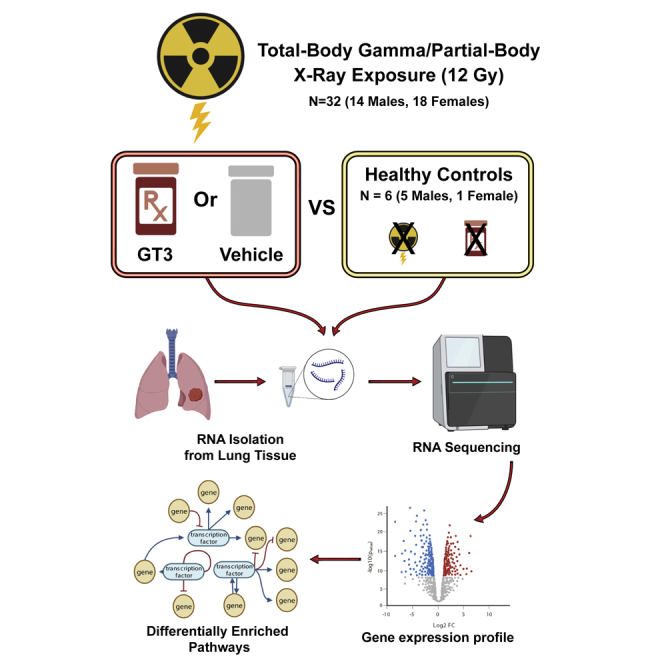
Figure 1.
Experimental design
Nonhuman primates were exposed to either total-body radiation (TBI) or partial-body radiation (PBI). They were compared with healthy controls to identify differentially regulated genes associated with radiation exposure. TBI or PBI exposed NHPs were also treated with γ-tocotrienol (GT3) or vehicle 24 h before radiation. Lung tissue for RNA-seq analysis was collected at days 4 and 7 post exposure.
Table 1.
Different conditions analyzed in this study
| Irradiation | Comparison | Up | Down | Total DE |
|---|---|---|---|---|
| TBI | Healthy vs. TBI-Veh | 419 | 649 | 1,068 |
| Healthy vs. TBI-GT3 | 511 | 753 | 1,264 | |
| TBI-Veh vs. GT3 | 0 | 1 | 1 | |
| TBI-Veh vs. GT3-SD4 | 9 | 14 | 23 | |
| TBI-Veh-SD7 vs. TBI-GT3-SD7 | 4 | 2 | 6 | |
| TBI-M-GT3 vs. TBI-F-GT3 | 26 | 13 | 39 | |
| TBI-M-Veh vs. TBI-F-Veh | 6 | 4 | 10 | |
| PBI | Healthy vs. PBI-Veh | 717 | 867 | 1,584 |
| Healthy vs. PBI-GT3 | 786 | 696 | 1,482 | |
| PBI-M-GT3 vs. PBI-F-GT3 | 14 | 23 | 37 | |
| PBI-M-Veh vs. PBI-F-Veh | 30 | 15 | 45 | |
| PBI-Veh-SD4 vs. PBI-GT3-SD4 | 21 | 124 | 145 | |
| PBI-Veh-SD7_vs. PBI-GT3-SD7 | 2 | 2 | 4 | |
| PBI-Veh vs. PBI-GT3 | 0 | 0 | 0 | |
| TBI/PBI comparison | TBI-M-GT3 vs. PBI-M-GT3 | 51 | 91 | 142 |
| TBI-GT3 vs. PBI-GT3 | 59 | 228 | 287 | |
| TBI-Veh vs. PBI-Veh | 30 | 101 | 131 | |
| TBI-F-GT3 vs._PBI-F-GT3 | 13 | 60 | 73 |
Differentially regulated genes after RNA-seq DESeq2 analysis are shown. M, male; F, female; SD4, day 4; SD7, day 7; DE, differentially expressed genes.
Transcriptomic profile comparisons to understand the effect of TBI
Effect of TBI in lung tissues compared with healthy controls
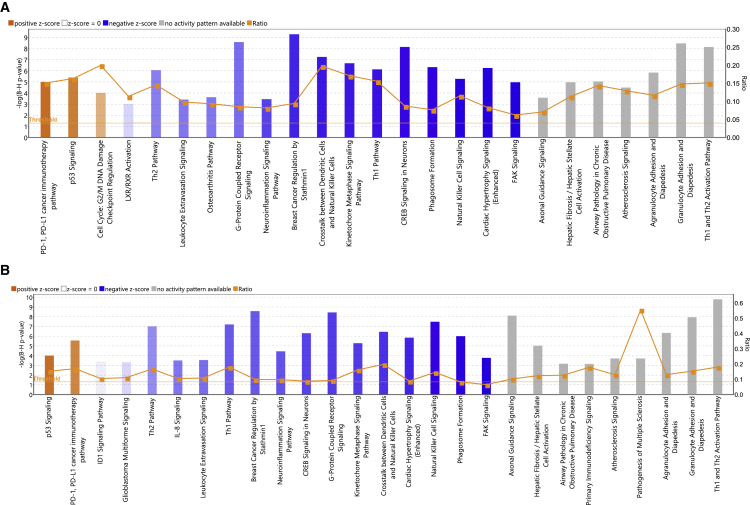
Transcriptomic changes induced in the lung as a result of exposure to 12 Gy 60Co γ-radiation were profiled by comparing the irradiated NHPs treated with vehicle (designated as TBI-Veh) and untreated/unirradiated healthy animals (designated as controls) (Figure 1). A total of 1,068 genes were differentially expressed after irradiation (419 and 649 genes upregulated or downregulated in TBI-Veh, respectively, Table S1). Ingenuity Pathway Analysis (IPA; QIAGEN) was used to analyze these differentially expressed genes, which revealed upregulation of pathways associated with PD-1/PD-L1 cancer immunotherapy, p53 signaling, and cell cycle: G2/M checkpoint regulation (Figure 2A and Table S2). On the other hand, pathways associated with T helper 1/T helper 2 (Th1/Th2), liver X receptor/retinoid X receptor (LXR/RXR) activation, phagosome formation, leukocyte extravasation, natural killer (NK) cell signaling, and neuroinflammation were downregulated after TBI. We observed a strong transcriptional response in the lung after TBI, reflecting the molecular changes associated with irradiation on lung tissues.
Figure 2.
Top enriched canonical pathways identified using IPA in TBI treatment
Both up- and downregulated genes in each comparison are represented here (Benjamini-Hochberg corrected p value ≤0.05). Orange or shades of orange bars indicate the predicted activation state of the canonical pathway, and blue or lighter shades of blue bars indicate a negative Z score and downregulation of the pathway. The activity patterns are not predicted in the pathways with gray bars. The ratio indicates the number of significantly enriched genes compared with the total number of genes associated with that canonical pathway. (A) Canonical pathways enriched in healthy controls (Healthy) versus treatment with vehicle (TBI-Veh). Z-score range is from −4.1 to 2.1. (B) Canonical pathways enriched in Healthy versus pretreatment with GT3 and exposed to radiation (TBI-GT3). Z-score range is from −4.6 to 2.6.
Effect of pretreatment with GT3 before TBI in lung tissues
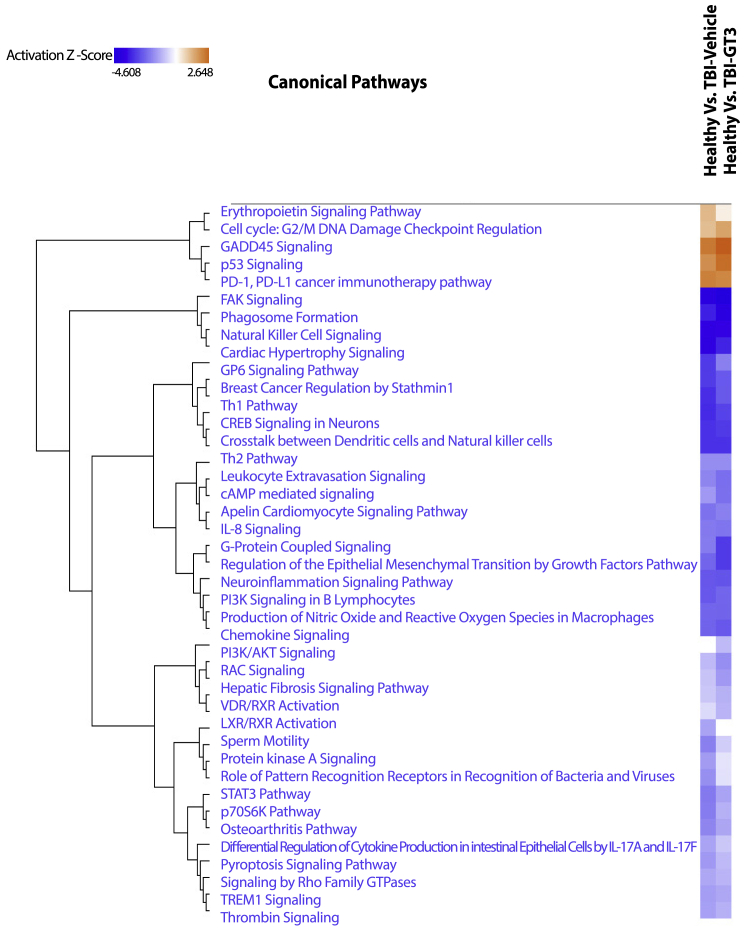
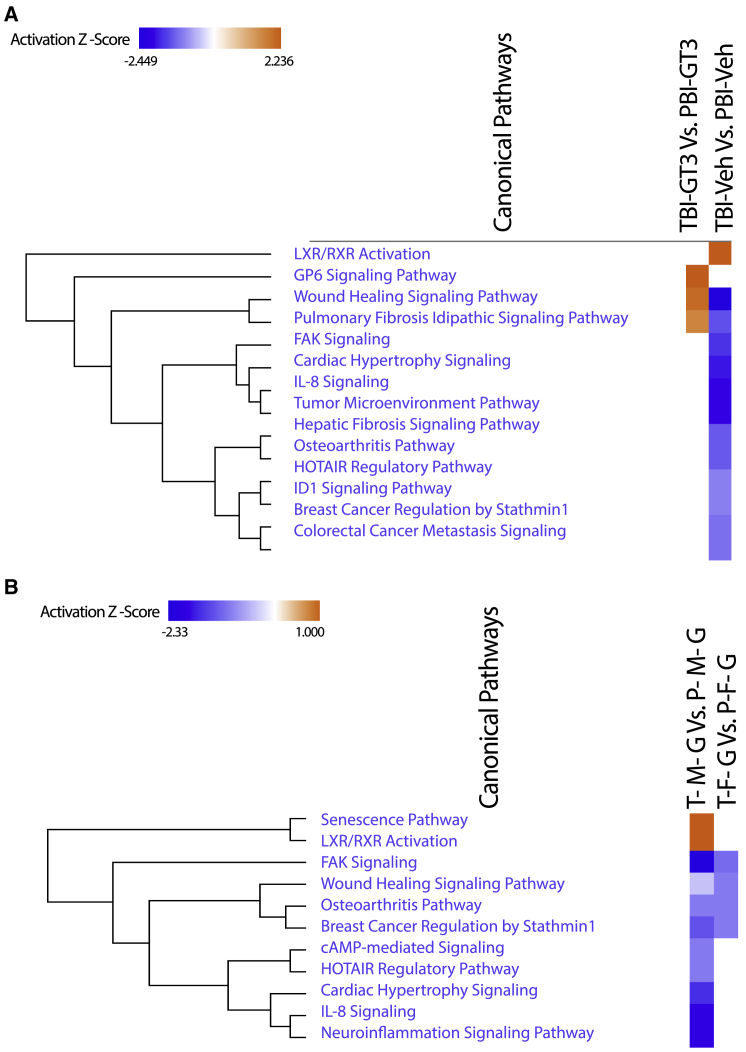
Next, we compared the lung transcriptomic changes associated with NHPs pretreated with GT3 and exposed to radiation (TBI-GT3) to untreated/unirradiated healthy controls. A total of 1,264 genes were differentially expressed after GT3 treatment and TBI (511 and 753 genes upregulated or downregulated in TBI-GT3, respectively, Table S3). IPA analysis revealed several common pathways up- or downregulated, similar to TBI-Veh versus controls (Figure 2B and Table S4). These common pathways included PD-1, PD-L1 cancer immunotherapy pathway, p53 signaling, and focal adhesion kinase (FAK) signaling. We also investigated the differentially expressed genes unique to TBI-GT3 versus controls and TBI-Veh versus controls (Figure S1A). No enriched pathways were identified for genes upregulated only in GT3-treated or vehicle-treated NHPs. Several KEGG pathways, including Th1, Th2, and Th17 cell differentiation, Rap1 signaling pathway, chemokine signaling pathway, Ras signaling pathway, and antigen processing and presentation were enriched among genes downregulated only in TBI-GT3 compared with controls (Figure S1B). Enriched KEGG pathways associated with unique genes downregulated in irradiated (TBI-Veh) included Wnt signaling, chemokine signaling, thyroid hormone synthesis, and insulin secretion (Figure S1C). Using IPA, pathway match analysis identified several common pathways regulated across both comparisons. Several pathways, including p53 signaling, GADD45 signaling, PD-1, PD-L1 cancer immunotherapy pathway, erythropoietin signaling, and cell cycle: G2/M checkpoint regulation, were upregulated in both TBI-GT3 and TBI-Veh. On the other hand, several pathways, including Th1/Th2, NK cell signaling, FAK signaling, and STAT signaling, were downregulated among several others in both comparisons (Figure 3). Since a similar transcriptional response was detected based on common pathways identified across GT3 and healthy controls as well as a vehicle (TBI-GT3 versus controls and TBI-GT3 versus TBI-Veh), GT3 pretreatment was minimally effective in mitigating the effect of TBI.
Figure 3.
Comparative IPA analysis identified pathways enriched across healthy versus TBI-Veh and Healthy versus TBI-GT3
The values in the heatmap are the Z scores of each entity in each experiment. Orange indicates a predicted pathway activation and blue a predicted inhibition.
Effect of sex-specific differences on TBI in lung tissues
A comparison of the effect of radiation exposure (TBI-Veh) and GT3 treatment (TBI-GT3) on males and females was also profiled. A total of 39 genes (26 up- and 13 downregulated in TBI-GT3) were differentially expressed in irradiated male NHPs pretreated with GT3 (Table S5). IPA pathway analysis identified signaling pathways including EIF2, regulation of EIF4 and P756K, and vascular endothelial growth factor (VEGF) (Figure S2 and Table S6). Relatively fewer genes were differentially expressed in lungs when male and female NHPs pretreated with the vehicle and exposed to radiation (TBI-Veh) were compared (total differentially expressed genes = 10, six genes upregulated and four genes downregulated in males, Table S7). Our study noted that the lung transcriptional response associated with sex on TBI was very limited, but a slightly higher differential response to GT3 was observed in both sexes.
Effect of days post TBI in lung tissues
A comparison of transcription profiles of TBI NHPs pretreated with GT3 or vehicle was performed to provide insights into the drug’s effectiveness on molecular pathways affected by the drug. However, we identified only a single gene differentially expressed that was upregulated in the GT3-treated animals (Table S8). Transcriptional changes at day 4 and day 7 (TBI-GT3-SD4, TBI-GT3-SD7) compared with vehicle-treated NHPs were slightly different. A total of 23 genes were differentially expressed at day 4 (9 and 14 genes up- and downregulated in vehicle-treated NHPs, respectively, Table S9). On the other hand, these differences diminished on day 7. Only six genes were identified as differentially expressed at day 7 between GT3 and vehicle (Table S10). The results from this comparison indicate the temporal effect of GT3 on TBI.
Transcriptomic profile comparisons to understand the effect of PBI
Effect of PBI in lung tissues compared with healthy controls
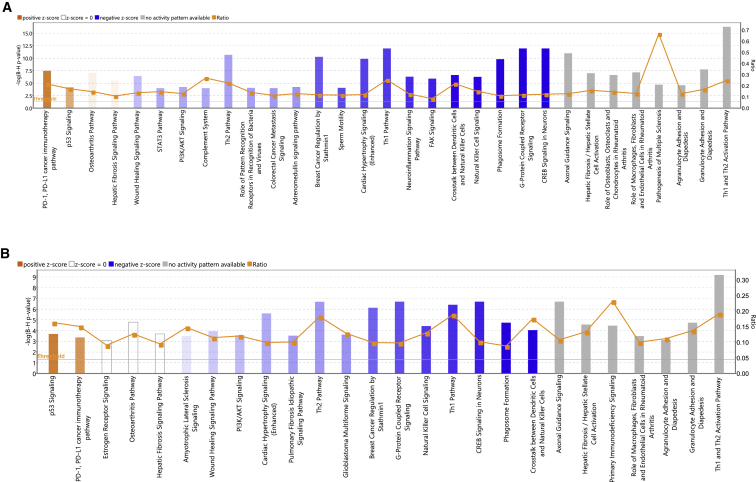
Transcriptomic changes induced in the lung due to PBI to 12 Gy X-rays were profiled by comparing the irradiated NHPs pretreated with vehicle (designated as PBI-Veh) and unirradiated healthy controls (designated as controls). A total of 1,584 genes were differentially expressed after irradiation (717 and 867 genes upregulated or downregulated in PBI-Veh, respectively, Table S11). IPA analysis of these differentially expressed genes revealed activation of pathways associated with PD-1/PD-L1 cancer immunotherapy and p53 signaling after PBI (Figure 4A and Table S12). On the other hand, pathways associated with Th1/Th2, phagosome formation, wound-healing pathway, NK cell signaling, PI3K/AKT signaling, STAT3, FAK signaling, and neuroinflammation were downregulated after PBI. Several of these pathways correlated with the transcriptional changes identified in TBI (72 common pathways including p53 signaling, Th1 and Th2 activation pathway, senescence pathway, wound-healing signaling pathway, and PI3K/AKT signaling). These results show a distinct change in the lung transcriptional profile after PBI, reflecting the direct molecular impact of irradiation on the lungs. This observation was similar to the changes observed after TBI.
Figure 4.
Top enriched canonical pathways identified using IPA in PBI treatment
(A) Canonical pathways enriched in Healthy versus PBI-Veh. Z-score range for this comparison is from −4.2 to 2.1. (B) Canonical pathways enriched in Healthy versus PBI-GT3. Z-score range for this comparison is from −3.4 to 1.9. Both up- and downregulated genes in each comparison are represented here (Benjamini-Hochberg corrected p value ≤0.05). Orange or shades of orange bars indicate the predicted activation state of the canonical pathway, and blue or lighter shades of blue bars indicate a negative Z score and downregulation of the pathway. The activity patterns are not predicted in the pathways with gray bars. The ratio indicates the number of significantly enriched genes compared with the total number of genes associated with that canonical pathway.
Effect of pretreatment with GT3 before PBI in lung tissues
Differential expression analysis of NHPs pretreated with GT3 and exposed to radiation (PBI-GT3) compared with healthy controls identified 1,482 genes (786 and 696 genes with statistically different up- and downregulated gene expression, respectively, Table S13). IPA analysis revealed several common pathways identified in PBI-Veh versus control, for example, activation of pathways associated with PD-1/PD-L1 cancer immunotherapy and p53 signaling after PBI (Figure 4B and Table S14). On the other hand, pathways associated with Th1/Th2, phagosome formation, wound-healing pathway, and PI3K/AKT signaling were downregulated in NHPs pretreated with GT3 and exposed to PBI. A comparable transcriptional response was associated with GT3 and healthy controls and vehicle (PBI-GT3 versus controls and PBI-GT3 versus PBI-Veh), indicating that the GT3 pretreatment was unsuccessful in attenuating the effect of PBI.
Effect of sex-specific differences on PBI in lung tissues
The difference in transcriptional regulation associated with sex-specific differences was also studied and correlated with PBI. We identified the differentially expressed genes in males and females in both comparisons, i.e., NHPs pretreated with GT3 or with vehicle (PBI-M-GT3 versus PBI-F-GT3 and PBI-M-vehicle versus PBI-F-vehicle). A total of 45 genes (30 up- and 15 downregulated) were differentially expressed in male NHPs pretreated with the vehicle and irradiated (Table S15). IPA pathway analysis identified signaling pathways including EIF2 and regulation of EIF4 and P756K to be enriched in males (Figure S3A and Table S16). A total of 37 genes were differentially expressed in lungs when male and female NHPs pretreated with GT3 and exposed to radiation (PBI-GT3) were compared (14 genes upregulated and 23 genes downregulated in males, Table S17). Pathway enrichment analysis identified signaling pathways including EIF2, regulation of EIF4 and P756K, and superoxide radical degradation to be enriched in the males (Figure S3B and Table S18). Our study noted that the lung transcriptional response associated with sex on PBI (both with and without GT3 pretreatment) was limited.
Effect of days post PBI in lung tissues
Transcriptional changes associated with various days post-irradiation were also studied. Lung tissue collected at day 4 (SD4) and day 7 (SD7) post-irradiation were compared. The extent of transcriptional differences was larger at day 4 compared with day 7 (Tables S19 and S20). At day 4 post-irradiation, a comparison of the transcriptional profile of GT3 and vehicle identified 145 differentially regulated genes (21 genes upregulated and 124 genes downregulated in the vehicle). IPA analysis identified glucocorticoid receptor signaling and granulocyte adhesion and diapedesis differentially regulated (Figure S4 and Table S21). Only four genes were differentially expressed on day 7 (two upregulated and two downregulated) in the vehicle compared with GT3. When all samples collected on both days were pooled and compared (PBI-GT3 versus vehicle), no genes were differentially regulated, which indicated that transcriptional changes associated with drug response were time sensitive. The results suggest the temporal effect of GT3 on PBI is comparable with that of TBI.
Comparison of transcriptional changes associated with TBI and PBI
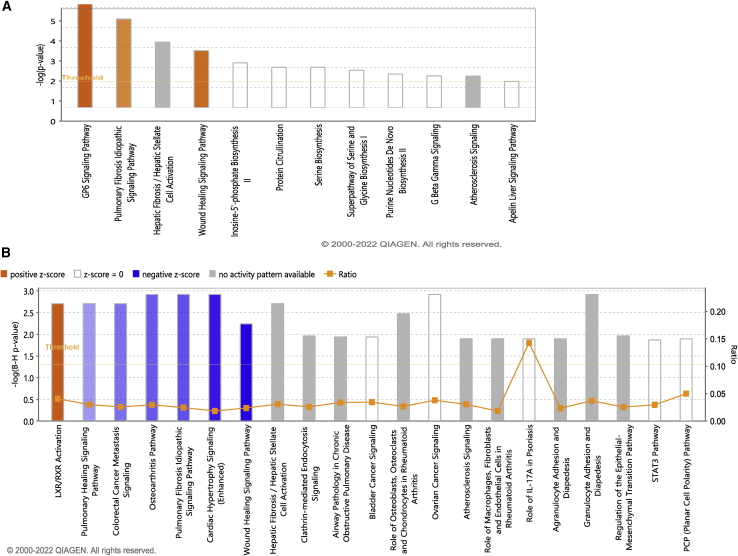
Transcriptional profiles of NHPs with either TBI or PBI were compared. The effect of GT3 in both sexes in two different types of exposure (TBI and PBI) was also examined. The extent of transcriptional differences across TBI and PBI exposure after pretreatment with GT3 (TBI-GT3/PBI-GT3) was larger than with the vehicle treatment (TBI-Veh/PBI-Veh). A total of 287 genes were differentially expressed in the TBI-GT3 versus PBI-GT3 comparison (59 genes upregulated and 228 genes downregulated in TBI, Table S22). IPA analysis identified activation of pathways associated with GP6 signaling, pulmonary fibrosis idiopathic signaling, and wound-healing signaling (Figure 5A and Table S23). On the other hand, 131 genes were differentially expressed in the TBI-Veh versus PBI-Veh comparison (30 genes upregulated and 101 genes downregulated in TBI, Table S24). This comparison highlights the differential lung transcriptional profiles of γ-ray TBI and X-ray PBI. Pathways associated with LXR/RXR activation were activated in TBI, whereas pulmonary wound healing, wound-healing signaling, and pulmonary fibrosis idiopathic signaling was repressed (Figure 5B and Table S25). IPA comparison analysis identified similarities and differences between TBI-GT3 versus PBI-GT3 and TBI-Veh versus PBI-Veh. LXR/RXR activation was higher in vehicle-treated NHPs than in GT3-treated NHPs. On the other hand, the GP6 signaling pathway, wound-healing signaling pathway, and pulmonary fibrosis idiopathic signaling pathway was activated in the GT3-treated NHPs. Several other pathways, including FAK signaling, interleukin-8 (IL-8) signaling, HOTAIR regulatory pathways, and ID-1 signaling pathway were repressed in the vehicle-treated NHPs (Figure 6A).
Figure 5.
Top enriched canonical pathways identified using IPA in GT3 treated TBI and PBI
(A) Canonical pathways enriched in TBI-GT3 versus PBI-GT3. Z-score range for this comparison is from 2.2 to 1.6. (B) Canonical pathways enriched in TBI-Veh versus PBI-Veh. Z-score range for this comparison is from −2.4 to 2.2. Both up- and downregulated genes in each comparison are represented here (Benjamini-Hochberg corrected p value ≤0.05). Orange or shades of orange bars indicate the predicted activation state of the canonical pathway, and blue or lighter shades of blue bars indicate a negative Z score and downregulation of the pathway. The activity patterns are not predicted in the pathways with gray bars. The ratio indicates the number of significantly enriched genes compared with the total number of genes associated with that canonical pathway.
Figure 6.
Comparative IPA analysis identified pathways enriched across (A) TBI-GT3 vs. PBI-GT3 and (B) TBI-Vehicle vs. PBI-Vehicle. The values in the heatmap are the z-scores of each entity in each experiment. Orange indicates a predicted pathway activation, and blue a predicted inhibition.
We also compared the effect of sex-specific differences in TBI or PBI exposure groups. Additional genes were differentially regulated among males compared with females in NHPs pretreated with GT3 and exposed to either TBI or PBI. Among males, 142 genes were differentially regulated (51 genes upregulated and 91 genes downregulated in TBI) in the TBI-M-GT3 versus PBI-M-GT3 comparison (Table S26). Pathway analysis identified only two pathways associated with diapedesis as enriched (Table S27). On the other hand, in females, 73 genes were differentially regulated (13 genes upregulated and 60 genes downregulated in TBI), and pathways associated with wound-healing signaling, FAK signaling, and LXR/RXR activation were repressed in female NHPs pretreated with GT3 and exposed to TBI (Tables S28 and S29). IPA comparison analysis identified a few pathways differentially regulated across male and female NHPs pretreated with GT3 and exposed to either TBI or PBI (Figure 6B). FAK signaling, osteoarthritis pathways, IL-8 signaling, neuroinflammation signaling pathway, cAMP-mediated signaling, HOTAIR regulatory pathway, wound-healing signaling pathways, senescence pathway, and LXR/RXR activation were enriched in both comparisons. FAK signaling pathway, IL-8 signaling, neuroinflammation signaling pathway, cAMP-mediated signaling, and HOTAIR regulatory pathway were repressed to a greater extent in males than females. Similarly, LXR/RXR and senescence pathway activity were higher in males than females.
Gene interactions across total- or partial-body irradiation and various factors analyzed in this study
Gene interactions associated with multiple factors, including sex, days post exposure, and drug treatment, were analyzed separately in NHPs exposed to TBI or PBI. The effect of GT3 treatment in NHPs exposed to TBI identified secretoglobin (SCGB3A2) as differentially regulated. Expression of SCGB3A2 was higher in GT3-treated females than males (Figure S5A). At the same time, vehicle-treated NHPs had opposite expression profiles for this gene. Analysis of GT3’s effect post-irradiation identified ENSMMUG00000061549 (LOC719948) as differentially regulated (Figure S5B). Expression of LOC719948 (PREDICTED: Macaca mulatta olfactory receptor 6K3-like) was higher in GT3-treated NHPs at day 4, whereas vehicle-treated NHPs had higher expression at day 7. Comparison between sex and days post-irradiation identified three genes as differentially regulated: carboxypeptidase b1 (CPB1), serine peptidase inhibitor kazal type 1 (SPINK1), and secretoglobin family 3a member 2 (SCGB3A2). CPB, SCGB3A2, and SPINK1 had higher expression in males on day 4 post irradiation compared with day 7 (Figure S6).
For PBI, the interaction of GT3 treatment with sex among NHPs identified oxysterol binding protein-like 3 (OSBPL3) as differentially expressed. OSBPL3 had higher expression in GT3-treated males than females and lower expression in vehicle-treated males than females (Figure S7). Effects of GT3 treatment at various days post irradiation identified cordon-bleu wh2 repeat protein (COBL), impact rwd domain protein (IMPACT), and uromodulin (UMOD) as differentially regulated (Figure S8). Of these genes, COBL and UMOD had higher expression in GT3-treated NHPs at day 4 than the other groups. On the other hand, IMPACT had higher expression in vehicle-treated NHPs at day 4. Finally, we identified five genes that interacted with sex and days post irradiation. These genes included Itpr interacting domain containing 1 (ITPRID1), Smad family member 1 (SMAD1), and pyruvate carboxylase (PC) (Figure S9).
Discussion
This study compared the lung transcriptional profile of NHPs exposed to a supralethal dose of radiation. Both TBI and PBI study results were analyzed. In addition, we also evaluated the effect of GT3, a promising radioprotector under development, and sex on the effects of irradiation. Transcriptional profiles were remarkably different when healthy NHPs were compared against TBI or PBI NHPs. On the other hand, no major differences were noted between TBI/PBI vehicle- and GT3-treated NHPs. This suggests that GT3 may not be optimally effective as a radioprotector against supralethal doses of radiation. This MCM has demonstrated consistent radioprotection when animals were exposed to lower doses of radiation inducing H-ARS in murine and NHP models.32,34,35 In an earlier study using an NHP model for H-ARS, we reported serum levels of miR-30a-5p, miR-126-5p, and miR-375-3p in GT3-treated and irradiated NHPs (5.8 Gy, 6.5 Gy, and 7.2 Gy, LD50/60, LD50/60, and LD50/60, respectively), which reflected the radioprotective efficacy of GT3; these microRNA levels in GT3-treated animals resembled unirradiated NHP.21 Recently, we have demonstrated that GT3 restores radiation-induced proteomic changes in irradiated murine (11 Gy, LD100/30).37 In the current study we have used a supralethal dose of radiation, which may be responsible for limited transcriptomic changes by GT3.
Gene expression profiles between different conditions exposed to TBI and PBI were analyzed in this study. The differentially regulated pathways identified across all major comparisons are visualized in Figure S10 using IPA Match analysis. Lung transcriptome profile comparison of NHPs exposed to TBI and PBI identified 131 differentially expressed genes. This comparison between TBI-Veh and PBI-Veh reveals the transcriptional differences between γ-ray TBI and X-ray PBI in lungs. Several pathways, including LXR/RXR activation, pulmonary healing signaling pathway, wound-healing signaling pathway, and pulmonary fibrosis idiopathic signaling pathway, were dysregulated after irradiation. It has been demonstrated that lower-energy orthogonal X-ray absorption in tissues is distinct and causes more serious biological damage than higher-energy radionuclide γ-rays.38 Even though our comparisons included TBI and PBI exposure, we were able to identify several pathways that could be associated with the high-energy (γ-ray) or low-energy (X-ray) irradiation. LXR/RXR activation was noted in TBI, whereas pulmonary healing signaling pathway, wound-healing signaling pathway, and pulmonary fibrosis idiopathic signaling pathway was repressed in PBI compared with TBI (Figure 5B). The LXR/RXR activation pathway has been reported to be associated with radiation-induced proteomic changes in the jejunum and blood plasma.39,40 This pathway was activated in TBI NHPs compared with PBI in our analysis. Apolipoprotein E (APOE) was upregulated and IL18RAP, IL1R2, MMP9, and S100A8 were downregulated in TBI. Of these genes, matrix metalloproteinase 9 (MMP9) expression dysregulated by radiation was associated with cell invasiveness and pulmonary metastasis in a subtype of lung carcinoma.41 The pulmonary fibrosis idiopathic signaling pathway was downregulated in TBI, and genes dysregulated in this pathway included BIRC5, COL9A2, FGF9, FZD8, MMP8, MMP9, WNT7A, and WNT9B. Additionally, the development of idiopathic pulmonary fibrosis has been reported as a side effect after thoracic radiotherapy.42, 43, 44 Wnt signaling pathway and its member, wingless-type protein 7a (WNT7A) gene, have been associated with radiosensitivity modulation in lung and mammary gland cancer cells.45,46 Overexpression of WNT7A sensitizes lung cancer cells to radiotherapy, and our analysis indicates that TBI increases the expression of WNT7A in the lungs as a response to ionizing radiation. The wound-healing signaling pathway was upregulated in PBI compared with TBI. Of the genes dysregulated in this pathway, protein expression of vascular endothelial cell growth factor C (VEGFC) has been reported to be induced by exposure to radiation through the activation of the PI3K/Akt/mTOR pathway in lung cancer.47 VEGFC was upregulated in PBI in comparison with TBI in the lung tissue of NHPs, indicating that PBI resulted in increased expression of this gene that could lead to radiation-induced angiogenesis. These pathways have to be further investigated to understand the transcriptional response attributable to variations in radiation energy.
IPA pathway comparison analysis of healthy controls versus TBI-Veh and healthy controls versus TBI-GT3 identified common differentially regulated pathways associated with TBI, along with the response to GT3. Several common pathways were upregulated in both TBI-Veh and TBI-GT3 compared with healthy controls (Figure S10). These pathways included cell cycle: G2/M checkpoint regulation, p53 signaling, GADD45 signaling, PD-1, and PD-L1 cancer immunotherapy. The GADD45 signaling pathway includes GADD45a, which was upregulated in both the TBI samples (TBI-Veh and TBI-GT3) compared with healthy controls. Growth arrest and DNA-damage-inducible gene 45α (GADD45a) has been shown to be transcriptionally activated by several stressors, including UV and ionizing radiation and hyperoxia.48,49 GADD45a is also associated with the maintenance of DNA integrity, apoptosis, DNA methylation excision and repair, and inflammatory lung injury.50, 51, 52 The expression of this gene was also increased in a mouse model of radiation-induced lung injury, and mice deficient in this gene had increased susceptibility to radiation injury.53 Our analysis also indicates that the GADD45a pathway is activated in both vehicle and GT3-treated NHPs, with increased activation levels in GT3-treated NHPs. This might indicate the marginal protective role of GT3 against such a supralethal ionizing radiation exposure.
The p53 pathway was also found to be activated in both vehicle and GT3-treated NHPs, with slightly higher activation in GT3-treated NHPs (Figure S10). p53, known as the “guardian of the genome,” is the master regulator of cellular response to irradiation.54,55 The role of p53 in regulating the cellular response to radiation is cell type dependent and can be different in different cell types.56 In mouse models, p53 loss can lead to resistance to toxicity in the hematopoietic system,57 and on the other hand p53 loss can lead to increased sensitivity in the gastrointestinal system.58,59 We also noted a decrease in the activity of the PI3K/AKT signaling pathway in GT3-treated NHPs compared with vehicle-treated NHPs (Figure 4B). The PI3K/AKT signaling pathway is shown to regulate cell cycle progression and survival.60 A decrease in the PI3K/AKT signaling pathway could increase apoptosis in GT3-treated NHPs. We speculate that radiation-induced p53 activation can lead to an increase in apoptosis in lung tissue.
Animal studies have indicated that the effects of irradiation are manifested in males and females differently, with females being more sensitive to radiation injury.18,61,62 In humans, long-term radiosensitivity is higher in females even when both sexes receive a comparable radiation dose.63,64 In addition, females have a significantly higher risk of dying from radiation-associated cancers.65,66 This study analyzed the transcriptomic differences in males and females when exposed to TBI or PBI and pretreated with GT3. Males had activation of senescence and LXR/RXR pathways when exposed to TBI compared with females (Figure 6B). On the other hand, wound-healing pathways were repressed in females exposed to TBI compared with PBI. The neuroinflammation signaling pathway, IL-8 signaling, HOTAIR regulatory pathway, cAMP-mediated signaling, and FAK signaling were repressed in TBI males compared with PBI. FAK is a cytoplasmic tyrosine kinase associated with integrin signaling, and its inhibition is associated with the induction of senescence and DNA-damage pathway activation in breast and lung cancer cells.67,68 This activation of cellular senescence through inhibition of FAK was also identified in our study. Earlier reports also link inhibition of FAK to sensitization of tumor cell lines to radiation.69
We also identified several genes that interacted with multiple factors studied, including sex at various days post exposure and drug treatment. For example, the expression of SPINK1 in lung tissue exposed to TBI was higher in males than in females at day 4 post-exposure (Figure S6). At day 7 post-exposure, however, females had higher expression of this gene than males. SPINK1 is identified as a plasma marker for radiation resistance in cancer cells, upregulated by radiation-induced hypoxia.70 Reports also suggest that the expression of SPINK1 is modulated by androgen receptor (AR) and repressor element-1 silencing transcription factor, a corepressor of AR,71 indicating that this gene can be differentially regulated in males and females. Among the PBI-treated NHPs, SMAD1 gene expression was higher in males at day 4 post exposure but decreased at day 7 (Figure S9). In females, an opposite trend was noted for this gene. Ionizing radiation produces reactive oxygen species that activate the transforming growth factor β/SMAD pathway, leading to radiation damage.71,72 No reports of sex-specific effects of SMAD1 gene expression linked to radiation response exist, so this interaction needs to be explored further using appropriate experiments.
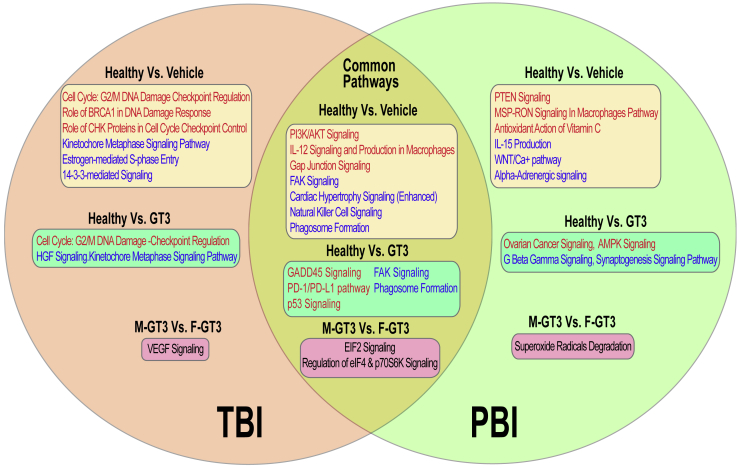
We have summarized major pathways identified in this study in Figure 7. Both common and unique pathways identified among different comparisons in TBI and PBI are represented in this figure. Common pathways upregulated in healthy NHPs compared with vehicle-treated NHPs and exposed to a supralethal dose of radiation included PI3K/AKT signaling, IL-12 signaling and production in macrophages, and gap junction signaling. On the other hand, FAK signaling, NK cell signaling, and phagosome formation pathways were downregulated in both TBI and PBI vehicle-treated NHPs compared with healthy NHPs. Unique upregulated pathways associated with TBI irradiation included cell cycle: G2/M DNA-damage checkpoint regulation, role of BRCA1 in DNA-damage response, and role of CHK proteins in cell cycle checkpoint control. Unique downregulated pathways associated with TBI irradiation included kinetochore metaphase signaling pathway, estrogen-mediated S-phase entry, and 14-3-3-mediated signaling. Interestingly, cell cycle: G2/M DNA-damage checkpoint regulation and kinetochore metaphase signaling pathway were uniquely upregulated or downregulated in TBI, respectively. No common pathways were identified in PBI treatments, showing that NHP response to PBI is variable.
Figure 7.
Common and unique pathways identified in different comparisons across TBI and PBI
Common pathways are represented in the intersection between the two circles. Pathways in red indicate the predicted activation state of the canonical pathway, and blue indicates the downregulation of the pathway. Pathways with no predicted activity patterns are depicted in black.
The lung is an organ that is usually used for studying delayed and late effects of radiation exposure.73, 74, 75 In our study, animals were exposed to a supralethal dose of 12 Gy (either TBI or PBI with 5% bone marrow sparing). This study was designed to investigate early acute effects, and it was not possible to have samples collected for studying the delayed and late effects of radiation exposure, as no animals survived for more than 7 days post-irradiation. Since the NHP study is a rare opportunity, we decided to conduct a transcriptomic study using the lung tissues of these animals. In the future, we plan to conduct such a transcriptomic study with animals exposed to low doses of radiation whereby samples can be collected for delayed and late effects.
Materials and methods
Animals and experimental design
A total of 32 naive rhesus macaques (Macaca mulatta, 14 males and 18 females) were used for this study. They were between 3.5 and 5.5 years of age, weighing 4.35–10.35 kg. Sixteen of these animals were procured from the National Institutes of Health Animal Center (Poolesville, MD, USA), while the remaining 16 were supplied by Covance (Alice, TX, USA). The NHPs were randomly assigned to a TBI or PBI cohort, 16 in each. Each group of 16 animals was further randomly divided; eight received GT3 (37.5 mg/kg, subcutaneously), and the remaining eight received only the vehicle. All animals were maintained in a facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International. Animals were quarantined for 6 weeks prior to the initiation of the experiment. Animal housing, health monitoring, care, and enrichment during the experimental period have been described earlier.35 Animals were fed a primate diet (Teklad T.2050 diet; Harlan Laboratories, Madison, WI, USA) twice daily with at least 6 h between feedings (animals were fed four biscuits each at 7:00 AM and 2:00 PM) and received drinking water ad libitum. All procedures involving animals were approved by the Institutional Animal Care and Use Committee (BIOQUAL, protocol #18-060) and the Department of Defense Animal Care and Use Review Office. This study was carried out in strict accordance with the recommendations made in the Guide for the Care and Use of Laboratory Animals.76
Drug preparation and administration
GT3 and olive oil (vehicle) formulations (50 mg/mL) in 5% Tween-80 in saline were purchased from Callion Pharma (Jonesborough, TN, USA). The quantity of GT3 or vehicle for each NHP was based on individual NHP body weight. The dose of GT3 administered was 37.5 mg/kg.35 Drug and vehicle were administered at the dorsal scapular area (between the shoulder blades) 24 h prior to irradiation. The area surrounding the injection site was shaved at least 48 h before administering the drug so that the site could be easily observed for any adverse skin reactions such as rash/eruption, inflammation, irritation, or abscess formation following GT3 or vehicle administration. Immediately prior to drug injection, the site was wiped with 70% isopropyl rubbing alcohol and allowed to air dry; the drug was administered subcutaneously using a 3-mL disposable luer-lock syringe with a 25-gauge 5/8-inch needle.
Radiation exposure
Total-body irradiation
On the day before irradiation, NHPs were fasted for approximately 12 h. Animals were transported from BIOQUAL to the Armed Forces Radiobiology Research Institute (AFRRI) by BIOQUAL personnel via a climate-controlled transport vehicle. The distance between AFRRI and BIOQUAL was less than 10 miles, and animals were not sedated during the transportation. Upon arrival at AFRRI, the transport crates containing the animals were transferred to a HEPA filtered transport cart which was used to transport the animals through the common hallways and elevators within AFRRI. Animals were removed from the transport cages and placed into a standard housing cage in the radiation staging area. Before sedation for irradiation, the unique tattoo identification numbers were confirmed by study personnel. Animals were then sedated in their cages using the squeeze-back mechanism and were administered a 10–15 mg/kg intramuscular dose of ketamine (100 mg/mL), and up to two NHPs were sedated at a time. Once sedated, the animals were placed in restraint boxes to limit movement and to maintain the proper upright seated position during irradiation. The NHPs’ limbs were secured to the box using ropes that were tied onto a cleat. Animals were then transported to the cobalt facility via elevator, and the attending dosimetrist verified the NHPs’ tattoo identification numbers. A 1.5–5.0 mg/kg intramuscular booster of ketamine was administered to the NHPs to limit movement while being irradiated if needed. For irradiation, two NHPs were placed on the irradiation platform facing away from each other and were exposed to a midline dose of 12 Gy 60Co γ-radiation at a dose rate of 0.6 Gy/min (bilateral, simultaneous exposure) as described earlier.35 To deliver the precise radiation dose, NHP abdominal widths were measured with digital calipers.
Animals were observed throughout the irradiation procedure via in-room cameras. After irradiation, the animals were returned to their cages in the housing area and were monitored for recovery from the procedure. The radiation field in the area of the NHP location was uniform within ±1.5%. The dosimetry for photons was based on the alanine/electron paramagnetic resonance dosimetry system.35,77 This is one of the most precise dosimetry techniques at present, which is used by national standards laboratories for the most critical measurements and calibrations.
Partial-body irradiation
NHPs were fasted, transported, and sedated as described for the TBI. For PBI, NHPs were irradiated one at a time using a 4 MV photon beam from an Elekta Infinity clinical linear accelerator (LINAC). Anterior/posterior measurements of the NHPs at the location of the absorbed dose target (“core of the abdomen”) were measured with a digital caliper. Measurements from the NHPs’ crown of the head to knee, hip, and foot were taken. Each NHP was placed in the custom-built positioning device attached to the LINAC couch. The NHPs’ limbs were secured to the platform using restraints that were attached to the device. Animals were exposed to PBI with 5% bone marrow sparing. To achieve 5% bone marrow sparing, the irradiation field excluded the tibia, ankles, and feet of the animal. The field size of approximately 80 cm along its diagonal with a collimator angle of 45° was used to provide a field size that was large enough to fit the crown-to-knee of each NHP inside the field (crown-to-knee lengths of the NHPs varied 54–75 cm). The heart rate and temperature of NHPs were continuously monitored throughout the irradiation procedure (Advisor vital signs monitor; Smiths Medical, Dublin, OH, USA). An absorbed dose of 12 Gy was delivered, with a dose rate of approximately 1.3 Gy/min. To ensure a uniform irradiation field, half of the absorbed dose was delivered with a beam along the anterior-posterior direction (gantry angle 0°), and the other half was delivered along the posterior-anterior direction (gantry angle 180°).
The calculation of the number of monitor units (MUs) required to deliver the requested absorbed dose was based on the dose rate to the abdominal core of the NHP. Each NHP was positioned supine on the platform with its coronal midline at 149 cm from the LINAC target. However, the dose rate to the abdominal core depends on the thickness of tissue through which the beam must travel from the surface to the core of the NHP. To account for the effect of variations in attenuation of the beam due to variations in the anterior-posterior separation of each NHP, the AP separation of each NHP was measured prior to irradiation.
To determine absorbed dose rates to NHPs of various AP separations, the absorbed dose rates to the center of a series of cylindrical water-filled polymethylmethacrylate NHP phantoms were measured using a Farmer ionization chamber (0.6 mL sensitive volume). Phantoms with diameters of 5.08, 6.99, 10.16, and 12.7 cm were used for these dose rate measurements. These phantom diameters covered the range of AP separations of the NHPs that were experimentally irradiated (7–11 cm). The number of MUs required to deliver the requested absorbed dose to the abdominal core was adjusted for the AP separation of each individual NHP. The Farmer ionization chamber used for the dosimetry measurements in this study was calibrated at a National Institute of Standards and Technology-traceable accredited dosimetry calibration laboratory. The calibration was in terms of absorbed dose-to-water. A water-to-soft-tissue correction factor was applied to the dose rates determined by these irradiations. This ionization chamber-based dosimetry system is based on the calculation of the absorbed dose rate that is described in the American Association of Physicists in Medicine Task Group 51 protocol.78 The absorbed dose rates determined from ionization chamber measurements were used to determine a function of dose rate versus phantom diameter (where phantom diameter corresponds to the AP separation of the NHP). This empirical function was then used to determine the dose rate to the abdominal core of each NHP (based on its individual AP separation that was measured prior to irradiation). This function enables calculation of the absorbed dose rate to the cores of NHPs that are intermediate in size between that of the phantoms in which the dosimetry measurements were performed.
Euthanasia
For animals exposed to 12 Gy, either TBI or PBI, three in each treatment group were scheduled to be euthanized for tissue collection on day 4, three on day 7, and two on day 10 post irradiation. However, no animal survived beyond day 7. Moribundity instead of mortality was used to relieve the animal from pain and distress. Euthanasia was carried out as per the American Veterinary Medical Association guidelines when animals reached a point of no return.79
Lung tissue collection
Lung tissue samples from six healthy/unirradiated animals were used as the control. Immediately after collection, all samples were put into a sterile storage tube and placed on dry ice. Samples were stored at −80°C until used. For TBI, six animals were euthanized as scheduled on day 4. The remaining ten animals were euthanized on day 7. Similarly, for PBI, tissue samples from six animals were collected on day 4, and all remaining animals were collected on day 7. The experimental protocol is presented in Figure 1.
RNA isolation
Total RNA was isolated from frozen lung tissue following the manufacturer’s protocol for the RNeasy Lipid Tissue Mini kit (Qiagen, Germantown, MD, USA) and quantified by fluorometry using a Qubit 4 fluorometer (Invitrogen, Carlsbad, CA, USA). The quality of RNA was analyzed on a Bioanalyzer Eukaryote Total RNA Pico Chip (Agilent 2100; Agilent, Santa Clara, CA, USA). The average RNA integrity number (RIN) score across all samples was above the recommended minimum RIN of 7. Total RNA samples were stored at −80°C until use.
Library preparation and sequencing
The library for RNA-seq was prepared with 500 ng of total RNA input using a TrueSeq stranded mRNA Library Prep kit (Illumina, San Diego, CA, USA) with barcoded adapters. Library size distribution was determined using a Bioanalyzer DNA 1000 kit (Agilent 2100), and the library yield and concentration were determined using the KAPA Library Quantification kit for Illumina (Kapa Biosystems, Wilmington, MA, USA). Clustering and sequencing were performed on the NextSeq 500 (Illumina) with paired-end reads of 75 bp in length.16
Data processing and analysis
Sequencing data were demultiplexed, and FASTQ files were generated using bcl2fastq2 software (version 2.20.0, Illumina). Sequencing quality control was performed using the FastQC tool.80 The reads were aligned to the macaque genome, Macaca mulatta Mmul_10.105, using the STAR spliced read aligner81 and the latest Ensembl gene transfer format (GTF) file. The average percent alignment of reads to the reference genome was 85%.
The read count for each gene was obtained using the --quantMode TranscriptomeSAM GeneCounts command from STAR aligner. Principal component analysis was performed to visualize main treatment differences. Differential gene expression analysis was performed using DESeq2.82 A gene was identified as differentially expressed if the false discovery rate (FDR) cutoff using the Benjamini-Hochberg procedure for multiple testing correction was ≤0.05 and the absolute fold change was above 1.5. DESeq2 multifactor design was used to analyze the paired samples in this study, including irradiation, drug treatment, sex, and days post irradiation.
Differentially expressed genes in different comparisons were used to perform pathway analysis using IPA83 and KEGG pathways using ShinyGO v0.741.84 In all the comparisons, KEGG pathway analyses of the up- and downregulated genes were performed separately to reveal the differentially regulated pathways. FDR corrected p ≤ 0.05 was set as the cutoff criterion for the enrichment, and the gene lists were compared against all protein-coding genes in the genome. A comparison of enriched pathways across different conditions was performed using the IPA Match analysis feature. Canonical pathways were identified as significantly enriched from the differentially regulated genes using the right-tailed Fisher’s exact test. Only canonical pathways with FDR corrected p value of ≤0.05 were considered.
Data and materials availability
All data generated or analyzed during this study are included in this published article (and its supplementary information). RNA-seq FASTQ files have been uploaded to the NCBI SRA database with BioProject accession number PRJNA820316.
Acknowledgments
The authors thank the Bioinformatics and Systems Biology Core (BSBC) facility at University of Nebraska Medical Center for providing the computational infrastructure and support. BSBC receives support from Nebraska Research Initiative (NRI) and NIH awards (2P20GM103427, 5P30CA036727, 2U54GM115458). The authors also acknowledge the Holland Computing Center of the University of Nebraska-Lincoln for computational resources, which receives support from the NRI. The opinions or assertions contained herein are the private views of the authors and are not necessarily those of the Uniformed Services University of the Health Sciences or the Department of Defense, USA.
Author contributions
Study design, V.K.S.; performance of the study, V.K.S., S.Y.W., A.D.C., and O.O.F.; transcriptomic data acquisition, J.S.; transcriptomic data analysis, N.N.V. and S.P.; drafting of the manuscript, V.K.S., S.Y.W., A.D.C., N.N.V., S.S., and C.G.; revision of manuscript content, VK..S., N.N.V., and C.G.; funding acquisition, V.K.S. All authors have read and approved the final submitted manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2022.08.006.
Supplemental information
References
- 1.Carter A.B., May M.M., Perry W.J. The day after: action following a nuclear blast in a U.S. city. Wash. Q. 2007;30:19–32. [Google Scholar]
- 2.Benjamin G.C., McGeary M., McCutchen S.R. The National Academies Press; 2009. Assessing Medical Preparedness to Respond to a Terrorist Nuclear Event: Workshop Report. [PubMed] [Google Scholar]
- 3.Williams J.P., Brown S.L., Georges G.E., Hauer-Jensen M., Hill R.P., Huser A.K., Kirsch D.G., MacVittie T.J., Mason K.A., Medhora M.M., et al. Animal models for medical countermeasures to radiation exposure. Radiat. Res. 2010;173:557–578. doi: 10.1667/RR1880.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCann D.G.C. Radiation poisoning: current concepts in the acute radiation syndrome. Am. J. Clin. Med. 2006;3:13–21. [Google Scholar]
- 5.Chaudhry M.A. Biomarkers for human radiation exposure. J. Biomed. Sci. 2008;15:557–563. doi: 10.1007/s11373-008-9253-z. [DOI] [PubMed] [Google Scholar]
- 6.Trompier F., Burbidge C., Bassinet C., Baumann M., Bortolin E., De Angelis C., Eakins J., Della Monaca S., Fattibene P., Quattrini M.C., et al. Overview of physical dosimetry methods for triage application integrated in the new European network RENEB. Int. J. Radiat. Biol. 2017;93:65–74. doi: 10.1080/09553002.2016.1221545. [DOI] [PubMed] [Google Scholar]
- 7.Sullivan J.M., Prasanna P.G.S., Grace M.B., Wathen L.K., Wallace R.L., Koerner J.F., Coleman C.N. Assessment of biodosimetry methods for a mass-casualty radiological incident: medical response and management considerations. Health Phys. 2013;105:540–554. doi: 10.1097/HP.0b013e31829cf221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh V.K., Seed T.M., Cheema A.K. Metabolomics-based predictive biomarkers of radiation injury and countermeasure efficacy: current status and future perspectives. Expert Rev. Mol. Diagn. 2021;21:641–654. doi: 10.1080/14737159.2021.1933448. [DOI] [PubMed] [Google Scholar]
- 9.Singh V.K., Newman V.L., Romaine P.L., Hauer-Jensen M., Pollard H.B. Use of biomarkers for assessing radiation injury and efficacy of countermeasures. Expert Rev. Mol. Diagn. 2016;16:65–81. doi: 10.1586/14737159.2016.1121102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pannkuk E.L., Fornace A.J., Jr., Laiakis E.C. Metabolomic applications in radiation biodosimetry: exploring radiation effects through small molecules. Int. J. Radiat. Biol. 2017;93:1151–1176. doi: 10.1080/09553002.2016.1269218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amundson S.A. Transcriptomics for radiation biodosimetry: progress and challenges. Int. J. Radiat. Biol. 2021:1–9. doi: 10.1080/09553002.2021.1928784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amundson S.A. The transcriptomic revolution and radiation biology. Int. J. Radiat. Biol. 2021:428–438. doi: 10.1080/09553002.2021.1987562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Port M., Hérodin F., Drouet M., Valente M., Majewski M., Ostheim P., Lamkowski A., Schüle S., Forcheron F., Tichy A., et al. Gene expression changes in irradiated baboons: a summary and interpretation of a decade of findings. Radiat. Res. 2021;195:501–521. doi: 10.1667/RADE-20-00217.1. [DOI] [PubMed] [Google Scholar]
- 14.Zheng J., Wang J., Pouliot M., Authier S., Zhou D., Loose D.S., Hauer-Jensen M. Gene expression profiling in non-human primate jejunum, ileum and colon after total-body irradiation: a comparative study of segment-specific molecular and cellular responses. BMC Genomics. 2015;16:984. doi: 10.1186/s12864-015-2168-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghandhi S.A., Turner H.C., Shuryak I., Dugan G.O., Bourland J.D., Olson J.D., Tooze J.A., Morton S.R., Batinic-Haberle I., Cline J.M., Amundson S.A. Whole thorax irradiation of non-human primates induces persistent nuclear damage and gene expression changes in peripheral blood cells. PLoS One. 2018;13:e0191402. doi: 10.1371/journal.pone.0191402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y., Singh J., Varghese R., Zhang Y., Fatanmi O.O., Cheema A.K., Singh V.K. Transcriptome of rhesus macaque (Macaca mulatta) exposed to total-body irradiation. Sci. Rep. 2021;11:6295. doi: 10.1038/s41598-021-85669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andrews R.N., Dugan G.O., Peiffer A.M., Hawkins G.A., Hanbury D.B., Bourland J.D., Hampson R.E., Deadwyler S.A., Clinea J.M. White matter is the predilection site of late-delayed radiation-induced brain injury in non-human primates. Radiat. Res. 2019;191:217–231. doi: 10.1667/RR15263.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ostheim P., Amundson S.A., Badie C., Bazyka D., Evans A.C., Ghandhi S.A., Gomolka M., Lopez Riego M., Rogan P.K., Terbrueggen R., et al. Gene expression for biodosimetry and effect prediction purposes: promises, pitfalls and future directions - key session ConRad 2021. Int. J. Radiat. Biol. 2021:1–12. doi: 10.1080/09553002.2021.1987571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Majewski M., Ostheim P., Gluzman-Poltorak Z., Vainstein V., Basile L., Schüle S., Haimerl M., Stroszczynski C., Port M., Abend M. Gene expression changes in male and female rhesus macaque 60 days after irradiation. PLoS One. 2021;16 doi: 10.1371/journal.pone.0254344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ostheim P., Haupt J., Schüle S., Herodin F., Valente M., Drouet M., Majewski M., Port M., Abend M. Differentiating total- or partial-body irradiation in baboons using mRNA expression patterns: a proof of concept. Radiat. Res. 2020;194:476–484. doi: 10.1667/RADE-20-00121.1. [DOI] [PubMed] [Google Scholar]
- 21.Fendler W., Malachowska B., Meghani K., Konstantinopoulos P.A., Guha C., Singh V.K., Chowdhury D. Evolutionarily conserved serum microRNAs predict radiation-induced fatality in nonhuman primates. Sci. Transl. Med. 2017;9:eaal2408. doi: 10.1126/scitranslmed.aal2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ostheim P., Haupt J., Herodin F., Valente M., Drouet M., Majewski M., Port M., Abend M. miRNA expression patterns differ by total- or partial-body radiation exposure in baboons. Radiat. Res. 2019;192:579–588. doi: 10.1667/RR15450.1. [DOI] [PubMed] [Google Scholar]
- 23.Port M., Hérodin F., Valente M., Drouet M., Ostheim P., Majewski M., Abend M. Persistent mRNA and miRNA expression changes in irradiated baboons. Sci. Rep. 2018;8:15353. doi: 10.1038/s41598-018-33544-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Port M., Herodin F., Valente M., Drouet M., Lamkowski A., Majewski M., Abend M. First generation gene expression signature for early prediction of late occurring hematological acute radiation syndrome in baboons. Radiat. Res. 2016;186:39–54. doi: 10.1667/RR14318.1. [DOI] [PubMed] [Google Scholar]
- 25.Port M., Hérodin F., Valente M., Drouet M., Lamkowski A., Majewski M., Abend M. Gene expression signature for early prediction of late occurring pancytopenia in irradiated baboons. Ann. Hematol. 2017;96:859–870. doi: 10.1007/s00277-017-2952-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Port M., Hérodin F., Valente M., Drouet M., Ullmann R., Majewski M., Abend M. Pre-exposure gene expression in baboons with and without pancytopenia after radiation exposure. Int. J. Mol. Sci. 2017;18:E541. doi: 10.3390/ijms18030541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Port M., Herodin F., Valente M., Drouet M., Ullmann R., Doucha-Senf S., Lamkowski A., Majewski M., Abend M. MicroRNA expression for early prediction of late occurring hematologic acute radiation syndrome in baboons. PLoS One. 2016;11 doi: 10.1371/journal.pone.0165307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Port M., Majewski M., Herodin F., Valente M., Drouet M., Forcheron F., Tichy A., Sirak I., Zavrelova A., Malkova A., et al. Validating baboon ex vivo and in vivo radiation-related gene expression with corresponding human data. Radiat. Res. 2018;189:389–398. doi: 10.1667/RR14958.1. [DOI] [PubMed] [Google Scholar]
- 29.Singh V.K., Newman V.L., Berg A.N., MacVittie T.J. Animal models for acute radiation syndrome drug discovery. Expert Opin. Drug Discov. 2015;10:497–517. doi: 10.1517/17460441.2015.1023290. [DOI] [PubMed] [Google Scholar]
- 30.Williams J.P., Jackson I.L., Shah J.R., Czarniecki C.W., Maidment B.W., DiCarlo A.L. Animal models and medical countermeasures development for radiation-induced lung damage: report from an NIAID Workshop. Radiat. Res. 2012;177:e0025–e0039. doi: 10.1667/rrol04.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh V.K., Olabisi A.O. Nonhuman primates as models for the discovery and development of radiation countermeasures. Expert Opin. Drug Discov. 2017;12:695–709. doi: 10.1080/17460441.2017.1323863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh V.K., Hauer-Jensen M. Gamma-tocotrienol as a promising countermeasure for acute radiation syndrome: current status. Int. J. Mol. Sci. 2016;17:e663. doi: 10.3390/ijms17050663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghosh S.P., Kulkarni S., Hieber K., Toles R., Romanyukha L., Kao T.C., Hauer-Jensen M., Kumar K.S. Gamma-tocotrienol, a tocol antioxidant as a potent radioprotector. Int. J. Radiat. Biol. 2009;85:598–606. doi: 10.1080/09553000902985128. [DOI] [PubMed] [Google Scholar]
- 34.Kulkarni S., Singh P.K., Ghosh S.P., Posarac A., Singh V.K. Granulocyte colony-stimulating factor antibody abrogates radioprotective efficacy of gamma-tocotrienol, a promising radiation countermeasure. Cytokine. 2013;62:278–285. doi: 10.1016/j.cyto.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 35.Singh V.K., Kulkarni S., Fatanmi O.O., Wise S.Y., Newman V.L., Romaine P.L.P., Hendrickson H., Gulani J., Ghosh S.P., Kumar K.S., Hauer-Jensen M. Radioprotective efficacy of gamma-tocotrienol in nonhuman primates. Radiat. Res. 2016;185:285–298. doi: 10.1667/RR14127.1. [DOI] [PubMed] [Google Scholar]
- 36.Singh V.K., Singh P.K., Wise S.Y., Posarac A., Fatanmi O.O. Radioprotective properties of tocopherol succinate against ionizing radiation in mice. J. Radiat. Res. 2013;54:210–220. doi: 10.1093/jrr/rrs088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosen E., Fatanmi O.O., Wise S.Y., Rao V.A., Singh V.K. Gamma-tocotrienol, a radiation countermeasure, reverses proteomic changes in serum following total-body gamma irradiation in mice. Sci. Rep. 2022;12:3387. doi: 10.1038/s41598-022-07266-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bell B., Vercellino J., Brodin N., Velten C., Nanduri L., Tanaka K., Fang Y., Wang Y., Macedo R., English J., et al. Increased relative biological effectiveness of orthovoltage X-rays compared to γ-rays in preclinical irradiation. bioRxiv. 2022 doi: 10.1101/2022.02.18.480594. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Byrum S.D., Burdine M.S., Orr L., Mackintosh S.G., Authier S., Pouliot M., Hauer-Jensen M., Tackett A.J. Time- and radiation-dose dependent changes in the plasma proteome after total body irradiation of non-human primates: implications for biomarker selection. PLoS One. 2017;12:e0174771. doi: 10.1371/journal.pone.0174771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang W., Yu J., Liu T., Tudor G., Defnet A.E., Zalesak S., Kumar P., Booth C., Farese A.M., MacVittie T.J., Kane M.A. Proteomic evaluation of the natural history of the acute radiation syndrome of the gastrointestinal tract in a non-human primate model of partial-body irradiation with minimal bone marrow sparing includes dysregulation of the retinoid pathway. Health Phys. 2020;119:604–620. doi: 10.1097/HP.0000000000001351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chou C.H., Teng C.M., Tzen K.Y., Chang Y.C., Chen J.H., Cheng J.C.H. MMP-9 from sublethally irradiated tumor promotes Lewis lung carcinoma cell invasiveness and pulmonary metastasis. Oncogene. 2012;31:458–468. doi: 10.1038/onc.2011.240. [DOI] [PubMed] [Google Scholar]
- 42.Marks L.B., Bentzen S.M., Deasy J.O., Kong F.M.S., Bradley J.D., Vogelius I.S., El Naqa I., Hubbs J.L., Lebesque J.V., Timmerman R.D., et al. Radiation dose-volume effects in the lung. Int. J. Radiat. Oncol. Biol. Phys. 2010;76:S70–S76. doi: 10.1016/j.ijrobp.2009.06.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsoutsou P.G., Koukourakis M.I. Radiation pneumonitis and fibrosis: mechanisms underlying its pathogenesis and implications for future research. Int. J. Radiat. Oncol. Biol. Phys. 2006;66:1281–1293. doi: 10.1016/j.ijrobp.2006.08.058. [DOI] [PubMed] [Google Scholar]
- 44.Woodward W.A., Chen M.S., Behbod F., Alfaro M.P., Buchholz T.A., Rosen J.M. WNT/beta-catenin mediates radiation resistance of mouse mammary progenitor cells. Proc. Natl. Acad. Sci. USA. 2007;104:618–623. doi: 10.1073/pnas.0606599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu D., Li L., Yan W. Knockdown of TC-1 enhances radiosensitivity of non-small cell lung cancer via the Wnt/beta-catenin pathway. Biol. Open. 2016;5:492–498. doi: 10.1242/bio.017608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen M.S., Woodward W.A., Behbod F., Peddibhotla S., Alfaro M.P., Buchholz T.A., Rosen J.M. Wnt/beta-catenin mediates radiation resistance of Sca1+ progenitors in an immortalized mammary gland cell line. J. Cell Sci. 2007;120:468–477. doi: 10.1242/jcs.03348. [DOI] [PubMed] [Google Scholar]
- 47.Chen Y.H., Pan S.L., Wang J.C., Kuo S.H., Cheng J.C.H., Teng C.M. Radiation-induced VEGF-C expression and endothelial cell proliferation in lung cancer. Strahlenther. Onkol. 2014;190:1154–1162. doi: 10.1007/s00066-014-0708-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Reilly M.A., Staversky R.J., Watkins R.H., Maniscalco W.M., Keng P.C. p53-independent induction of GADD45 and GADD153 in mouse lungs exposed to hyperoxia. Am. J. Physiol. Lung Cell Mol. Physiol. 2000;278:L552–L559. doi: 10.1152/ajplung.2000.278.3.L552. [DOI] [PubMed] [Google Scholar]
- 49.Hollander M.C., Alamo I., Jackman J., Wang M.G., McBride O.W., Fornace A.J., Jr. Analysis of the mammalian gadd45 gene and its response to DNA damage. J. Biol. Chem. 1993;268:24385–24393. [PubMed] [Google Scholar]
- 50.Hollander M.C., Philburn R.T., Patterson A.D., Wyatt M.A., Fornace A.J., Jr. Genomic instability in Gadd45a-/- cells is coupled with S-phase checkpoint defects. Cell Cycle. 2005;4:704–709. doi: 10.4161/cc.4.5.1675. [DOI] [PubMed] [Google Scholar]
- 51.Sheikh M.S., Hollander M.C., Fornance A.J., Jr. Role of Gadd45 in apoptosis. Biochem. Pharmacol. 2000;59:43–45. doi: 10.1016/s0006-2952(99)00291-9. [DOI] [PubMed] [Google Scholar]
- 52.Meyer N.J., Huang Y., Singleton P.A., Sammani S., Moitra J., Evenoski C.L., Husain A.N., Mitra S., Moreno-Vinasco L., Jacobson J.R., et al. GADD45a is a novel candidate gene in inflammatory lung injury via influences on Akt signaling. FASEB J. 2009;23:1325–1337. doi: 10.1096/fj.08-119073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mathew B., Takekoshi D., Sammani S., Epshtein Y., Sharma R., Smith B.D., Mitra S., Desai A.A., Weichselbaum R.R., Garcia J.G.N., Jacobson J.R. Role of GADD45a in murine models of radiation- and bleomycin-induced lung injury. Am. J. Physiol. Lung Cell Mol. Physiol. 2015;309:L1420–L1429. doi: 10.1152/ajplung.00146.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lindsay K.J., Coates P.J., Lorimore S.A., Wright E.G. The genetic basis of tissue responses to ionizing radiation. Br. J. Radiol. 2007;80:S2–S6. doi: 10.1259/bjr/60507340. [DOI] [PubMed] [Google Scholar]
- 55.Gudkov A.V., Komarova E.A. The role of p53 in determining sensitivity to radiotherapy. Nat. Rev. Cancer. 2003;3:117–129. doi: 10.1038/nrc992. [DOI] [PubMed] [Google Scholar]
- 56.Lee C.L., Blum J.M., Kirsch D.G. Role of p53 in regulating tissue response to radiation by mechanisms independent of apoptosis. Transl. Cancer Res. 2013;2:412–421. [PMC free article] [PubMed] [Google Scholar]
- 57.Lotem J., Sachs L. Hematopoietic cells from mice deficient in wild-type p53 are more resistant to induction of apoptosis by some agents. Blood. 1993;82:1092–1096. [PubMed] [Google Scholar]
- 58.Merritt A.J., Potten C.S., Kemp C.J., Hickman J.A., Balmain A., Lane D.P., Hall P.A. The role of p53 in spontaneous and radiation-induced apoptosis in the gastrointestinal tract of normal and p53-deficient mice. Cancer Res. 1994;54:614–617. [PubMed] [Google Scholar]
- 59.Komarova E.A., Kondratov R.V., Wang K., Christov K., Golovkina T.V., Goldblum J.R., Gudkov A.V. Dual effect of p53 on radiation sensitivity in vivo: p53 promotes hematopoietic injury, but protects from gastro-intestinal syndrome in mice. Oncogene. 2004;23:3265–3271. doi: 10.1038/sj.onc.1207494. [DOI] [PubMed] [Google Scholar]
- 60.Brazil D.P., Yang Z.Z., Hemmings B.A. Advances in protein kinase B signalling: AKTion on multiple fronts. Trends Biochem. Sci. 2004;29:233–242. doi: 10.1016/j.tibs.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 61.Billings P.C., Romero-Weaver A.L., Kennedy A.R. Effect of gender on the radiation sensitivity of murine blood cells. Gravit. Space Res. 2014;2:25–31. [PMC free article] [PubMed] [Google Scholar]
- 62.Kirichuck V.F., Ivanov A.N., Antipova O.N., Krenickiy A.P., Mayborodin A.V., Tupikin V.D. Sex-specific differences in changes of disturbed functional activity of platelets in albino rats under the effect of terahertz electromagnetic radiation at nitric oxide frequencies. Bull. Exp. Biol. Med. 2008;145:75–77. doi: 10.1007/s10517-008-0002-0. [DOI] [PubMed] [Google Scholar]
- 63.Narendran N., Luzhna L., Kovalchuk O. Sex difference of radiation response in occupational and accidental exposure. Front. Genet. 2019;10:260. doi: 10.3389/fgene.2019.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pierce D.A., Shimizu Y., Preston D.L., Vaeth M., Mabuchi K. Studies of the mortality of atomic bomb survivors. report 12, part I. Cancer: 1950-1990. 1996. Radiat. Res. 2012;178 doi: 10.1667/rrav06.1. AV61–87. [DOI] [PubMed] [Google Scholar]
- 65.Delongchamp R.R., Mabuchi K., Yoshimoto Y., Preston D.L. Cancer mortality among atomic bomb survivors exposed in utero or as young children, October 1950-May 1992. Radiat. Res. 1997;147:385–395. [PubMed] [Google Scholar]
- 66.Ozasa K., Shimizu Y., Suyama A., Kasagi F., Soda M., Grant E.J., Sakata R., Sugiyama H., Kodama K. Studies of the mortality of atomic bomb survivors, Report 14, 1950-2003: an overview of cancer and noncancer diseases. Radiat. Res. 2012;177:229–243. doi: 10.1667/rr2629.1. [DOI] [PubMed] [Google Scholar]
- 67.Pylayeva Y., Gillen K.M., Gerald W., Beggs H.E., Reichardt L.F., Giancotti F.G. Ras- and PI3K-dependent breast tumorigenesis in mice and humans requires focal adhesion kinase signaling. J. Clin. Invest. 2009;119:252–266. doi: 10.1172/JCI37160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chuang H.H., Wang P.H., Niu S.W., Zhen Y.Y., Huang M.S., Hsiao M., Yang C.J. Inhibition of FAK signaling elicits lamin A/C-associated nuclear deformity and cellular senescence. Front. Oncol. 2019;9:22. doi: 10.3389/fonc.2019.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Skinner H.D., Giri U., Yang L., Woo S.H., Story M.D., Pickering C.R., Byers L.A., Williams M.D., El-Naggar A., Wang J., et al. Proteomic profiling identifies PTK2/FAK as a driver of radioresistance in HPV-negative head and neck cancer. Clin. Cancer Res. 2016;22:4643–4650. doi: 10.1158/1078-0432.CCR-15-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Suwa T., Kobayashi M., Shirai Y., Nam J.M., Tabuchi Y., Takeda N., Akamatsu S., Ogawa O., Mizowaki T., Hammond E.M., Harada H. SPINK1 as a plasma marker for tumor hypoxia and a therapeutic target for radiosensitization. JCI Insight. 2021;6:e148135. doi: 10.1172/jci.insight.148135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tiwari R., Manzar N., Bhatia V., Yadav A., Nengroo M.A., Datta D., Carskadon S., Gupta N., Sigouros M., Khani F., et al. Androgen deprivation upregulates SPINK1 expression and potentiates cellular plasticity in prostate cancer. Nat. Commun. 2020;11:384. doi: 10.1038/s41467-019-14184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kruse J.J.C.M., Floot B.G.J., te Poele J.A.M., Russell N.S., Stewart F.A. Radiation-induced activation of TGF-beta signaling pathways in relation to vascular damage in mouse kidneys. Radiat. Res. 2009;171:188–197. doi: 10.1667/RR1526.1. [DOI] [PubMed] [Google Scholar]
- 73.Williams J.P., Johnston C.J., Finkelstein J.N. Treatment for radiation-induced pulmonary late effects: spoiled for choice or looking in the wrong direction? Curr. Drug Targets. 2010;11:1386–1394. doi: 10.2174/1389450111009011386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hanania A.N., Mainwaring W., Ghebre Y.T., Hanania N.A., Ludwig M. Radiation-induced lung injury: Assessment and management. Chest. 2019;156:150–162. doi: 10.1016/j.chest.2019.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.MacVittie T.J., Farese A.M., Parker G.A., Jackson W., 3rd The time course of radiation-induced lung injury in a nonhuman primate model of partial-body irradiation with minimal bone marrow sparing: clinical and radiographic evidence and the effect of neupogen administration. Health Phys. 2019;116:366–382. doi: 10.1097/HP.0000000000000968. [DOI] [PubMed] [Google Scholar]
- 76.National Research Council of the National Academy of Sciences . National Academies Press; 2011. Guide for the Care and Use of Laboratory Animals. [Google Scholar]
- 77.Nagy V. Accuracy considerations in EPR dosimetry. Appl. Radiat. Isot. 2000;52:1039–1050. doi: 10.1016/s0969-8043(00)00052-x. [DOI] [PubMed] [Google Scholar]
- 78.Almond P.R., Biggs P.J., Coursey B.M., Hanson W.F., Huq M.S., Nath R., Rogers D.W. AAPM's TG-51 protocol for clinical reference dosimetry of high-energy photon and electron beams. Med. Phys. 1999;26:1847–1870. doi: 10.1118/1.598691. [DOI] [PubMed] [Google Scholar]
- 79.Association A.V.M. 2020. AVMA Guidelines for the Euthanasia of Animals: 2020 Edition.https://www.avma.org/sites/default/files/2020-01/2020-Euthanasia-Final-1-17-20.pdf [Google Scholar]
- 80.Babraham Institute . 2019. FastQC: A Quality Control Tool for High Throughput Sequence Data.https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- 81.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Krämer A., Green J., Pollard J., Jr., Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ge S.X., Jung D., Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2020;36:2628–2629. doi: 10.1093/bioinformatics/btz931. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.