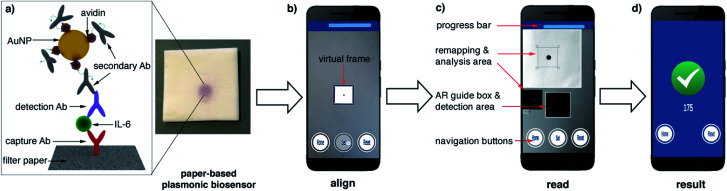
Abstract
Detecting small variations in the levels of IL-6 is crucial for the early diagnosis of sepsis. To be useful in clinical decision-making, this requires detecting IL-6 rapidly in whole blood and with portable readers. Here we introduce immunosensors made of filter paper that use plasmonic nanoprobes to detect IL-6 rapidly in unprocessed blood with an unmodified smartphone. Key aspects of the biosensor fabrication were optimized in order to reduce the assay time without losing sensitivity. This included testing three bioconjugation routes for protein attachment to nanoprobes using gold nanoparticles covered with carboxylate or amine moieties, or polyvinylpyrrolidone (PVP), as starting materials, and using alternating layers of polyelectrolytes to bind the capture antibody to the paper substrate. Smartphone-based signal quantification was achieved with a custom-made app featuring a unique augmented reality guidance system that circumvents the need for smartphone attachments and automates all the steps involved in color quantification. The biosensors were able to detect IL-6 with a limit of detection of 0.1 pg mL−1 and a total assay time within 17 min. They could also detect an increase in IL-6 of only 12.5 pg mL−1 over basal levels in whole blood with 99% confidence. The high sensitivity and rapid turnaround time afforded by the optimized biosensors and the fully automated real-time densitometry app make our biosensors well suited for emergency healthcare situations such as the identification of potential sepsis cases.
Detecting small variations in the levels of IL-6 is crucial for the early diagnosis of sepsis.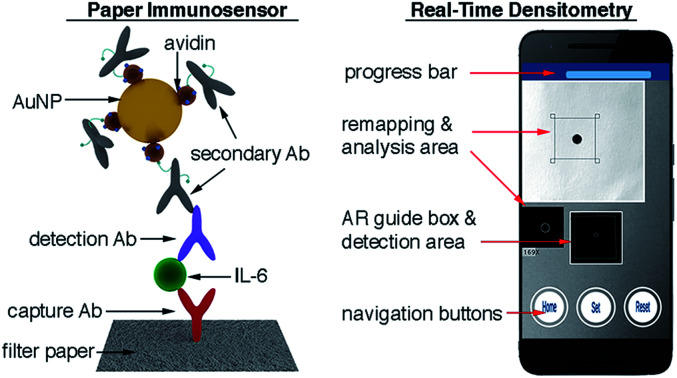
Introduction
Sepsis, an overwhelming response to an infection leading to multiorgan shutdown, can cause death in a few hours if not treated straightaway.1 It is also a major economic burden for healthcare providers, costing US$24 billion per year in the US alone.2 Treating sepsis at a very early stage can improve patient outcomes and reduce associated costs.2 Yet this requires identifying potentially septic patients rapidly, which is difficult when judging only from the presenting symptoms. Fortunately, the serum levels of some proteins such as procalcitonin (PCT), C-reactive protein (CRP) and interleukin 6 (IL-6) fluctuate as sepsis progresses, which can be used to identify at-risk patients.3 Among them, IL-6 levels increase during the first hours of infection when PCT and CRP levels have not changed yet, making it a particularly useful biomarker for improving outcomes with an early diagnosis.4 However, the detection of cytokines such as IL-6, which are found at average concentrations lower than 5.9 pg mL−1 in healthy individuals, requires hour-long ELISA procedures that are too lengthy for early diagnosis schemes.5 Automated immunoassay systems such as Elecsys® IL-6 can drastically reduce the assay time (18 min), but they are bulky, which makes them difficult to implement at some of the places where septic patients could be identified early on, such as the emergency room or an ambulance.
Biosensors are promising candidates for detecting IL-6 in the context of sepsis because they can potentially be deployed in a wide variety of clinical settings.6–8 Indeed, several biosensors have already been proposed whose sensitivity is high enough for detecting clinically relevant concentrations of IL-6 under ideal conditions.9,10 Yet, many of these biosensors still require assay times longer than 30 min, and therefore they are not suitable for aiding clinical decision-making in emergency situations.11,12 Furthermore, the majority of these devices have only been tested in purified serum samples.13–16 Remarkably, a lateral flow test has recently been proposed that can detect IL-6 in unprocessed blood with surface-enhanced Raman spectroscopy (SERS).17 Yet this test requires a microscope for detecting the biomarker at clinically relevant concentrations, which would be cumbersome to implement in bedside diagnosis. A commercial lateral flow immunoassay from Milenia Biotec can detect IL-6 in 20 min, but only at concentrations higher than 50 pg mL−1. In the context of the current state of the art, a biosensor that could detect small variations in the blood levels of IL-6 in less than 20 minutes using portable, user-friendly instrumentation could have a great impact on sepsis care by enabling the identification of at-risk patients rapidly. Such a device could enable monitoring patients in many clinical settings where potential sepsis cases are managed, from primary care centers to ambulances and the emergency department, where it is imperative to identify signs of sepsis rapidly in order to prioritize patients effectively.19
In this manuscript we describe a plasmonic mobile biosensor designed to meet the criteria for rapid IL-6 detection. Our biosensors reduce the assay time because they do not require purification or pretreatment of blood samples, and gold nanoparticles generate intense colorimetric signals with short incubation times. The device setup consists of a paper-based biosensor paired with a smartphone app for colorimetric signal detection. Colorimetric signals are generated by gold nanoprobes modified with avidin and biotinylated antibodies (Fig. 1a). Since IL-6 is found at ultralow concentrations in healthy individuals, and its cut-off values indicating sepsis risk are not much higher (ca. 25 pg mL−1 in newborns and 40–61 pg mL−1 in adults),20 great care was taken to optimize each step of the biosensor fabrication to detect the biomarker rapidly at clinically relevant concentrations in unprocessed blood. This included synthesizing nanoparticles through 3 different routes for obtaining nanoprobes using different bioconjugation strategies as well as testing several approaches for immobilizing biomolecules on paper substrates. The optimized nanoprobes and paper biosensors generated signals in the form of colored spots (Fig. 1a) whose pixel intensity was evaluated in real time with a custom designed companion app (Fig. 1b–d). Smartphone-based densitometry can be challenging for in-field measurements because it requires a specific positioning of the camera with respect to the assay while at the same time compensating for variations in the pixel intensity that did not originate from the biospecific recognition of the analyte.21,22 Our app uses an innovative augmented reality (AR) guidance system to guide the user to the correct positioning of the smartphone, while steps such as white balance calibration, light artifact detection, selection of a region of interest, measurement of pixel intensity, and data validation are automatically performed with real-time image processing and data processing. This means that the user only needs to hover the phone over the assay to obtain reliable densitometry readings within seconds. No cradles, light-tight boxes or special attachments are required to ensure robust measurements. The optimized plasmonic mobile biosensors were able to detect IL-6 with a limit of detection of 0.1 pg mL−1 and a total assay time within 17 min. They were also able to detect IL-6 spiked into unprocessed blood with the same assay time at concentrations well below the cut-off values indicating sepsis. The rapid turnaround time, excellent sensitivity in blood matrices and high portability of the mobile detection scheme make our biosensors well suited for detecting IL-6 in decentralized healthcare schemes such as those involved in the management of potential sepsis cases.
Fig. 1. Schematic representation of the plasmonic mobile biosensors (a) antibody-decorated gold nanoparticles generate colored spots on paper substrates; (b) the color in the spots is quantified with an app that uses a virtual frame to control the angle and distance between the biosensor and the smartphone; (c) the detection screen appears after aligning the frame in the AR guidance system with the biosensor and pressing the Set button, and the app automatically detects the region of interest (ROI) while at the same time sampling background signals from 4 points around the ROI to avoid interference from uneven lighting.18 (d) When the app has measured 50 valid points, the progress bar stops and the resulting increase in pixel intensity is displayed (see Video S1 in the ESI†). Ab: antibody, IL-6: interleukin 6, AuNP: gold nanoparticle, and AR: augmented reality.
Results and discussion
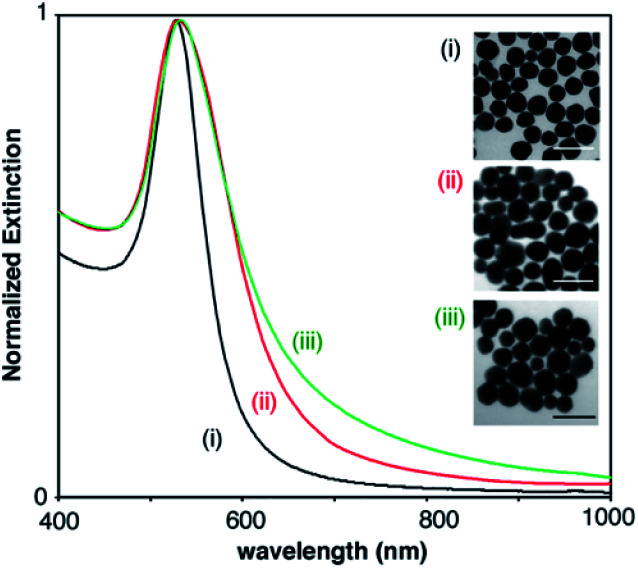
The gold nanoparticles used as starting materials for nanoprobe fabrication were obtained through the reduction of gold ions with either citrate, PAH or PVP molecules. Citrate- and PAH-covered nanoparticles were subsequently modified with SH-PEG-COOH and SH-PEG-NH2 in order to obtain stable carboxylate- and amine-coated nanoparticles, respectively. Fig. 2 shows the extinction spectra and representative TEM images of the 3 types of nanoparticles. Nanoparticle sizes calculated from the TEM images were 45 ± 5 (PVP-coated, (i)), 49 ± 12 nm (carboxylate-coated, (ii)) and 46 ± 10 nm (amine-coated, (iii)). PVP-coated nanoparticles show a narrower size distribution, as evidenced from TEM analysis and the narrower width of the extinction spectra in Fig. 2, because they were obtained with a seeded growth method. The LSPR of PVP-coated nanoparticles, which depends on their size as well as on the coating around them, is 528 nm, whereas nanoparticles coated with carboxylate and amine groups show LSPR centered around 530 and 533 nm, respectively.
Fig. 2. Extinction spectra and representative TEM images of the (i) PVP-coated, (ii) carboxylate-coated, and (iii) amine-coated nanoparticles used as starting materials for developing colorimetric nanoprobes. Scale bars: 100 nm.
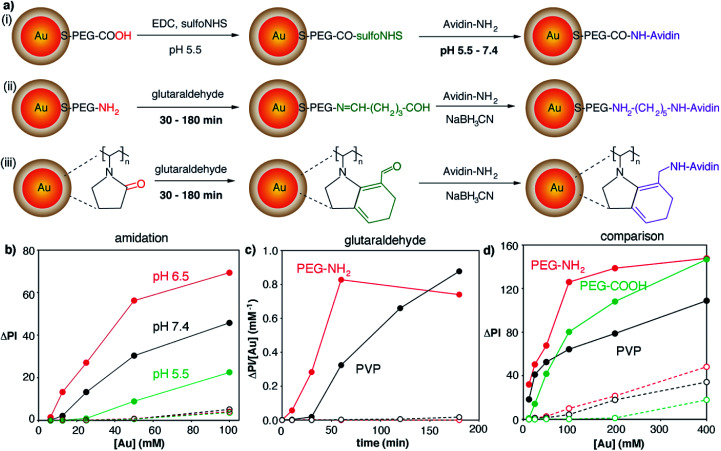
After characterizing the plasmonic nanoparticles we endeavored to optimize all the steps involved in the fabrication of the biosensor, from the covalent attachment of avidin to the nanoprobes to the immobilization of proteins on the paper substrate. Bioconjugation strategies involved amidation with carboxylate-coated nanoparticles or crosslinking with glutaraldehyde using amine- or PVP-coated nanoparticles (Fig. 3a). The amidation reaction involved transforming carboxylate groups into reactive sulfo-NHS esters followed by the addition of the protein. The key parameter to boost the yield of this reaction is the pH of the protein solution, since the reactivity of both the sulfo-NHS ester groups around the nanoparticles and the amines on the protein is pH-dependent.23 Consequently we prepared avidin-modified nanoparticles using protein solutions buffered at pH 5.5, 6.5 or 7.4 in order to discern the best conditions for obtaining nanoprobes. As shown in Fig. 3b, the resulting avidin-decorated nanoprobes were tested with a model biosensor for the detection of biotinylated proteins. As seen in this figure, the highest signals were obtained when the reaction between the sulfo-NHS ester and the protein was performed at pH 6.5. Lower signals for bioconjugation reactions at pH 5.5 and 7.4 are ascribed to lower reaction yields, either because protonated amines have poor reactivity (pH 5.5) or because the sulfo-NHS ester hydrolyzes before the reaction can take place (pH 7.4).23 Control biosensors containing non-biotinylated BSA always yield very low signals, which demonstrates that interactions with the paper substrates modified with biotinylated BSA are specific.
Fig. 3. Bioconjugation reactions for binding avidin to gold nanoparticles; full dots indicate colorimetric signals generated by the biospecific reaction between avidin on the nanoparticles and biotin–BSA on paper substrates; open dots correspond to non-specific interactions with substrates modified with non-biotinylated BSA; (a) schematic representation of the reactions involved in the covalent attachment of avidin to nanoparticles; (b) effect of pH on amidation reactions (i); increase in pixel intensity (ΔPI) generated by avidin-decorated nanoparticles obtained by amidation in which avidin was added at pH 5.5 (green), 6.5 (red) or 7.4 (black); (c) effect of time on bioconjugation reactions using glutaraldehyde crosslinkers; ratio between ΔPI and nanoprobe concentration (ΔPI/[Au]) generated by avidin-decorated nanoparticles obtained after PVP-coated (black, (iii)) or amine-coated (red, (ii)) nanoparticles were reacted with glutaraldehyde for different times; (d) comparison of colorimetric signals (ΔPI) generated by the three types of gold nanoprobes as a function of the concentration ([Au]) of PVP-coated (black), carboxylate-coated (green) and amine-coated (red) gold nanoparticles.
In the case of bioconjugation reactions using glutaraldehyde and amine- or PVP-coated nanoparticles (Fig. 3a), the reaction time with the cross-linker was deemed the most important parameter to be optimized. Since glutaraldehyde is a homobifunctional linker, extended incubation times could lead to nanoparticle agglomeration, whereas short incubation times could result in a low amount of proteins being bound to the nanoprobes. As shown in Fig. 3c (red dots), amine-coated nanoparticles yielded the highest signal per gold concentration when incubated with glutaraldehyde for 1 h, whereas PVP-coated nanoparticles required 3 h to yield the same result (black dots). Control experiments yielded very low signals which demonstrates that the color was mainly generated by biospecific biotin–avidin interactions.
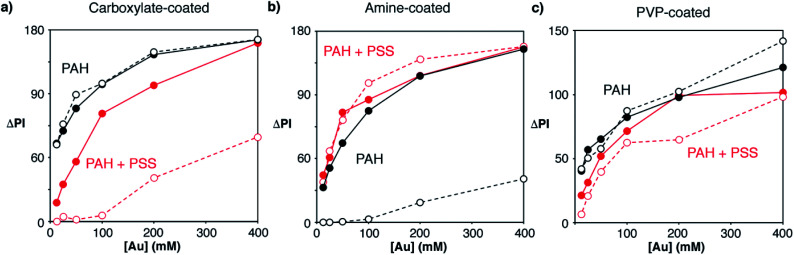
After optimizing the bioconjugation reactions for the 3 proposed types of nanoparticles, the nanoprobes obtained under the best conditions were compared with each other in order to determine which were most appropriate for fabricating our immunosensors. As shown in Fig. 3d, PVP-nanoparticles modified with avidin generated the least intense colorimetric signals (black dots). Amine-nanoparticles (red dots) generated higher signals than carboxylate-counterparts (green dots) when added at the same concentration. However, this increase in the signal originated from non-specific interactions, since higher signals were also observed in control experiments. We then hypothesized that the extent of both specific and non-specific interactions could change depending not only on the type of nanoprobe, but also on the strategy utilized to immobilize proteins on the paper substrate. To test this idea, we compared the signals generated via direct adsorption of biotinylated BSA on paper (Fig. 3d) with those obtained with biosensors fabricated with a modified cellulose matrix (Fig. 4). The paper modifications entailed adding a positively charged polymer PAH, either alone or followed by the addition of negatively charged PSS. These treatments were chosen to evaluate the role of the paper surface charge in both attaching capture biomolecules and establishing non-specific interactions with the nanoparticles, which have different surface charges according to their coatings (zeta potential −2.7, 1.7 and −1.5 mV for carboxylate-, amine-, and PVP-coated nanoparticles modified with avidin, respectively). Experiments with another positively charged polymer (chitosan) were also attempted, but they were discarded because it blocked the pores of the paper, making it difficult to perform washing steps. Similarly, attempts at modifying PAH-modified paper substrates with glutaraldehyde for covalent attachment of proteins were rejected because they resulted in yellow coloration of the filter paper that interfered with colorimetric measurements even after reducing imines with NaCNBH3. Fig. 4 shows the colorimetric signals obtained when biotinylated-BSA was physically adsorbed on PAH-modified paper (black dots) or PAH and PSS (red dots). As shown in Fig. 4a carboxylate-coated nanoparticles modified with avidin generated high non-specific signals when the cellulose matrix was modified with PAH only, but the extent of non-specific interactions decreased upon addition of PSS. This is in agreement with the negative zeta potential value of the nanoparticles, which results in non-specific electrostatic interactions with the matrix covered with positively charged PAH. As seen in Fig. 4b amine-coated nanoparticles show the opposite behavior, that is, high non-specific signals are generated when the matrix is negatively charged through the modification with PSS. PVP-coated nanoparticles yielded non-specific signals in all cases (Fig. 4c), and therefore were discarded from further appraisal. Comparison of the specific signals obtained under the best conditions revealed that biosensors combining carboxylate-coated nanoparticles and unmodified paper substrates, or amine-coated nanoparticles and cellulose modified with PAH, were best suited for colorimetric detection (Fig. S2†). The first design was chosen for further studies to expedite the fabrication process and reduce manufacturing costs, since it does not require modification of the paper substrate.
Fig. 4. Colorimetric signals (ΔPI) generated by different concentrations of gold nanoprobes ([Au]) modified with avidin when the paper substrate was modified with PAH (black dots) or PAH and PSS (red dots), and biotinylated BSA (full dots) or non-biotinylated BSA (open dots). Nanoprobes modified with avidin were obtained using carboxylate-coated (a), amine-coated (b) or PVP-coated (c) nanoparticles.
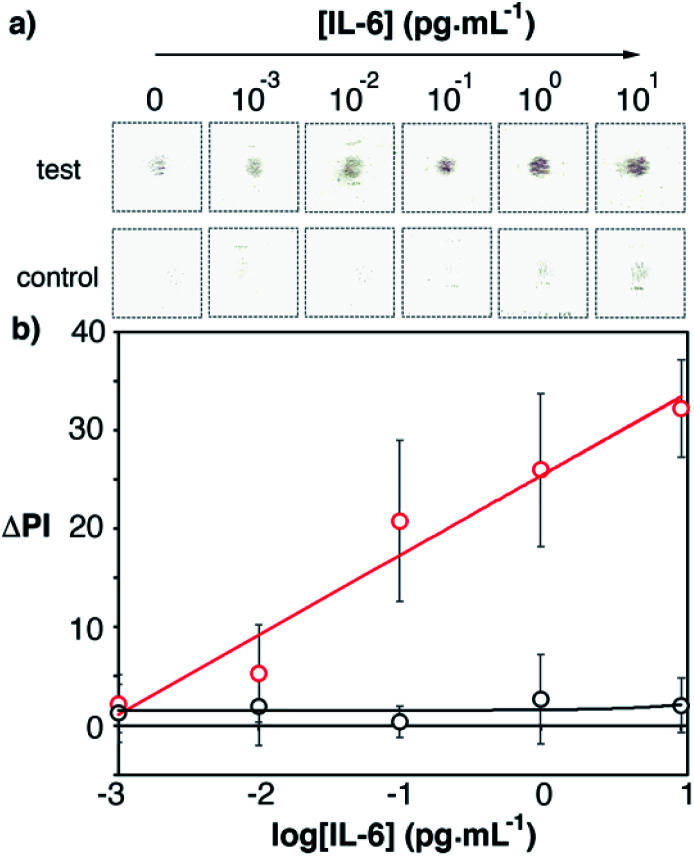
After optimizing the fabrication of the biosensor we sought to determine the best conditions for detecting IL-6. This included optimizing the concentration of capture and detection antibodies, as well as the time required to capture IL-6 (Fig. S3†). Antibody-decorated nanoparticles were obtained by adding biotinylated antibodies to the avidin-modified nanoparticles. From these optimization experiments it was determined that the best conditions for detecting IL-6 were to spot the capture antibody on the paper substrate at a concentration of 10 μg mL−1, and to add the detection antibody at a concentration of 10 μg mL−1 for 5 min (Fig. S3 in the ESI†). The total assay time, including washing steps, was under 17 minutes. Under these conditions, recombinant human IL-6 was detected in buffered solutions in the concentration range between 0.001 and 10 pg mL−1 (Fig. 5, y = 8x + 25, r2 = 0.95). Experiments performed with control biosensors without the capture antibody yielded lower signals, which indicates that the main contribution to the colorimetric output is the biospecific recognition of IL-6 by the antibodies. The limit of detection, expressed as the concentration of the first sample that yields a signal higher than 3 times the standard deviation of the blank, is 0.1 pg mL−1. This limit of detection is much lower than the reported threshold value indicating sepsis, which makes the optimized biosensor a promising technology for the detection of elevated levels of IL-6 in real matrices as detailed below.
Fig. 5. Scanned images (a) and the calibration plot (b) for detecting recombinant IL-6 in PBS with the proposed immunosensors (red dots) and control biosensors without the capture antibody (black dots). [Au] = 400 mM. The total assay time was under 17 min. Error bars are the standard deviation (n = 3).
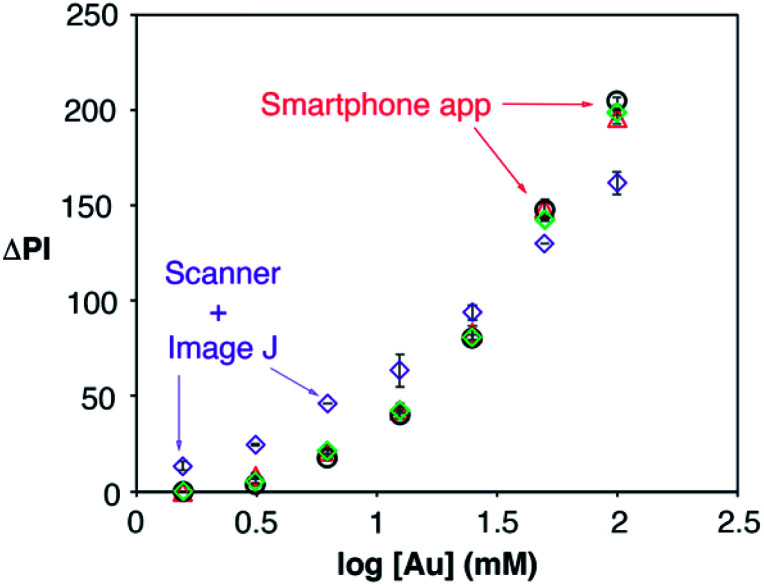
Detecting IL-6 in the context of sepsis at the point of care necessitates the use of unprocessed blood and a portable readout instrument capable of yielding robust results in different locations with varying environmental conditions. The latter was achieved here with a mobile app for performing reliable densitometry at the point of care that only requires hovering a smartphone over the assay in order to quantify colorimetric signals (Video S1†). The app was calibrated using pieces of paper modified with nanoparticle drops at different concentrations, and therefore with different pixel intensities. The calibration was repeated under 3 different room illuminance conditions (200, 570 and 1070 lux) and compared with the same measurements obtained using a desktop scanner and manual image processing with Image J. As shown in Fig. 6, the room illuminance has very little effect on the app performance (coefficient of variation CV 1.7% for [Au] = 12.5 mM). The main difference with the scanner method is that the app yields slightly lower signals when the nanoparticles are spotted at low concentrations. This is attributed to the shorter distance between the assay and the camera in the scanner compared to the smartphone-based approach. These results demonstrate that the proposed app can yield consistent results without using any hardware accessories attached to the smartphone to control imaging conditions, which makes it more economical, portable and hygienic for medical diagnostics.24
Fig. 6. Calibration plot obtained by evaluating the colorimetric signal generated by gold nanoparticles at different concentrations with the densitometry app when the room illuminance was 200 (black dots), 570 (green diamonds) and 1070 lux (red triangles), and with a desktop scanner (purple diamonds); error bars are the standard deviation (n = 3).
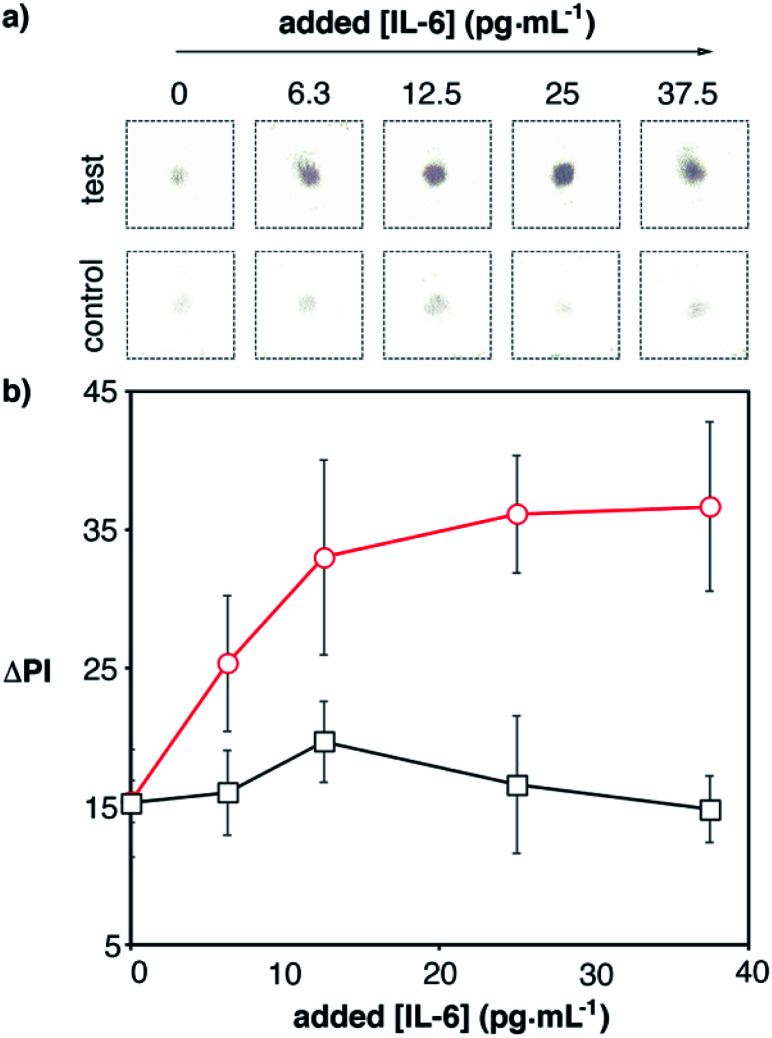
Next we tested whether we could detect elevated levels of IL-6 by combining the app with the optimized paper biosensors. Blood samples from a healthy donor were spiked with recombinant IL-6 and analyzed with our 17 min-long method and real-time densitometry with the mobile app. The characteristic red color of whole blood was efficiently removed with the proposed washing procedure and therefore was not an impediment to perform the assay (Fig. S4 in the ESI†). Since healthy individuals have non-negligible levels of IL-6 (<5.9 pg mL−1), we sought to determine which was the lowest added concentration of IL-6 that could be detected above basal levels with 99% confidence (i.e. above 3 times the standard deviation of the blank (σ), which contains basal levels of IL-6). As can be seen in Fig. 7, control experiments yielded a residual signal due to a small contribution from the color of the sample (Fig. S4†) and non-specific interactions with the whole blood matrix. Experiments with spiked samples yielded higher dose-dependent signals, which demonstrates that the nanoparticles are being anchored to the substrate through specific antibody–antigen interactions. Samples spiked with 12.5 pg mL−1 yielded a signal higher than 3σ. This number is lower than the threshold value indicating sepsis in newborns or adults, which means that our biosensors are suitable for detecting small variations in IL-6 observed in the early stages of sepsis. This, along with the short assay time and robust signal reading afforded by the smartphone app, makes the proposed mobile biosensors ideal for supporting a biomarker-based diagnosis of sepsis in each of the clinical contexts along the chain of care.
Fig. 7. Detection of IL-6 spiked into human blood with plasmonic mobile biosensors; (a) images of colored spots; (b) signal quantification with the smartphone app. Control experiments were performed with biosensors that were not modified with capture antibodies (black squares). The total assay time was under 17 min (Table S3 in the ESI†). Error bars are the standard deviation (n = 3).
Conclusions
In this manuscript we have reported the fabrication and performance of a plasmonic immunosensor for the rapid detection of IL-6 in the context of sepsis. Key aspects of the immunosensor fabrication, including the manufacture of plasmonic nanoprobes decorated with proteins and strategies for attaching capture molecules to the paper substrate, were optimized. We have also introduced a companion app that quantifies the colorimetric signal generated by the plasmonic probe in real time. Our app is unique in that it does not require hardware attachments added to the smartphone in order to stabilize angle and distance parameters; instead it makes use of a software-based AR guidance system. The optimized paper biosensors were able to detect IL-6 under ideal conditions with a low limit of detection of 0.1 pg mL−1 within 17 min. When the biomarker was spiked into serum or blood, the biosensors were able to detect variations in the basal concentration of IL-6 as small as 12.5 pg mL−1 with 99% confidence. The ability to detect small variations in cytokine levels in unprocessed blood using only a paper biosensor and an unmodified smartphone makes the proposed detection scheme ideal to support evidence-based clinical decision-making. In the context of sepsis, it would enable the measurement of biomarkers during triage, when prioritizing high-risk patients is essential for improving sepsis outcomes.19,25 Moreover, the biosensors only require a tiny sample volume (2.5 μL), which makes them suitable for sepsis screening in neonates as well.26 Furthermore, the same strategy could be used to detect other biomarkers27 and pathogens28 by using specific antibodies against them.
Experimental
Materials
Poly(ethylene glycol) 2-mercaptoethyl ether acetic acid (SH-PEG-COOH, Mn 2100), poly(ethylene glycol) 2-mercaptoethyl ether ethylamine (SH-PEG-NH2, Mn 5000), avidin from egg white, 2-(N-morpholino)ethanesulfonic acid (MES), N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC), N-hydroxysulfosuccinimide sodium salt (sulfo-NHS), glutaraldehyde (25% in water), sodium cyanoborohydride (95%), poly(sodium 4-styrenesulfonate) (PSS, 30% in water) and recombinant human interleukin 6 (IL-6) were purchased from Sigma. Poly(allylamine hydrochloride) (PAH) was obtained from Alfa Aesar and bovine serum albumin (BSA, protease free) was purchased from VWR. Phosphate buffered saline (PBS), PBS containing 5 mg mL−1 BSA (PBS–BSA) and PBS containing 0.1% Tween-20 were prepared following standard procedures. Biotinylated proteins were obtained with an EZ-Link biotinylation kit from Thermo Fisher and purified with a PD-desalting column. Monoclonal anti-IL-6 developed in mice (ab11449), polyclonal anti-IL-6 developed in rabbits (ab6672) and anti-rabbit IgG (biotin) preadsorbed developed in donkeys (ab7082) were obtained from abcam.
Nanoprobe fabrication
Detailed protocols for obtaining nanoparticles with an initial coating of citrate, PAH or PVP molecules are available in the ESI.† Citrate-capped nanoparticles were modified with 0.1 mM SH-PEG-COOH overnight to obtain carboxylate-coated nanoparticles, whereas PAH-covered nanoparticles were modified with 0.1 mM SH-PEG-NH2 overnight in order to obtain amine-coated nanoparticles.29 PVP-coated nanoparticles were resuspended in water and used without further purification.
Avidin attachment to carboxylate-coated nanoparticles proceeded as follows. Carboxylate-coated nanoparticles (250 mL) were concentrated to a final volume of 1 mL and washed 5 times with water with the aid of a centrifuge (9000 rpm, 8 min). Then carboxylate moieties were transformed into sulfo-NHS esters via addition of EDC (1 mg) and sulfo-NHS (2 mg) to the nanoparticle suspension in 0.5 M MES buffer of pH 6 (200 μL, [Au] = 100 mM). After 30 min the nanoparticles were pelleted with a centrifuge and the supernatant was substituted with a solution containing avidin (200 μL, 0.1 mg mL−1, overnight). The pH of the avidin solution was varied from 5.5 to 7.4 with phosphate buffer (0.1 M) to test the impact of this parameter on the reaction between the sulfo-NHS esters and amine groups in proteins. Then 400 μL of a blocking solution containing glycine (0.1 M) and BSA (10 mg mL−1) in phosphate buffer (0.1 M, pH 7) was added for at least 30 min followed by washing 5 times with PBST.
Avidin was attached to amine-coated or PVP-coated nanoparticles using glutaraldehyde as a crosslinker with the following procedure.30 First glutaraldehyde (334 μL, 7.5%) in 0.1 M bicarbonate buffer of pH 9 was added to the nanoparticle dispersion (166 μL, [Au] = 100 mM) for different times. Excess reagents were removed with a PD-10 desalting column rather than by centrifugation in order to reduce covalent crosslinking between nanoparticles upon pellet formation. Nanoparticle-rich fractions were identified by their characteristic reddish color. Then avidin (100 μL, 1 mg mL−1) was added to the nanoparticles in the presence of NaCNBH3 (100 μL, 100 mM), which reduces the pH-sensitive imine bonds generated by the reaction between amines and aldehydes and yields stable amine bonds. After overnight incubation unreacted aldehydes were blocked with glutamic acid (0.1 M) in sodium bicarbonate (0.1 M, 100 μL) for 1 h. The resulting avidin-decorated nanoparticles were washed by centrifugation 5 times with PBST.
Nanoparticle suspensions were characterized by transmission electronic microscopy (TEM) with a Hitachi H-600 ABS TEM (carboxylate- and anime-coated nanoparticles) and a JOEL 1011 TEM (PVP-coated nanoparticles) operating at 100 kV. Extinction spectra were taken with a DU 730 Beckman Coulter spectrophotometer. Zeta potential measurements were performed with a Nano Zetasizer (Malvern).
Nanoprobe performance
The performance of the avidin-decorated nanoprobes was tested with a model biosensor using paper substrates. The paper substrates were made of Whatman filter paper grade 41 cut into 2.5 × 10 cm pieces and folded like an accordion.31 Biosensors were prepared with 3 different methods. The first method involved spotting biotin–BSA (5 μL, 0.1 mg mL−1 in PBS) on the top paper layer at different concentrations for 5 min (direct adsorption). The second method involved spotting PAH (1% (w/v), 10 μL) and letting it dry (10 min) followed by spotting the protein at different concentrations, whereas the third method involved an additional PSS treatment (1%) after adding the protein followed by washing with water and letting the paper dry (10 min). Next the paper substrates were blocked with PBS–BSA (1 mL). Immediately afterwards avidin-decorated nanoparticles were spotted onto the substrate (2 μL, 4 min) followed by washing with PBST (1 mL, 3 times). Control experiments were performed in the same way but by using non-biotinylated BSA in order to assess the impact on non-specific interactions between the nanoparticles and the paper substrate. The biosensors were left to dry and the colorimetric signals were evaluated with ImageJ after scanning the samples with an MFC-1910F Brother printer-scanner.
Development of an app for real-time densitometry
An Android app was developed on a Unity3D platform (version 2019.1.8) using OpenCV 4.0 APIs with a C# wrapper from Enox Software (OpenCVforUnity version 2.3.3) (see also Table S1 in the ESI†). It was tested on a Huawei POT-LX1 smartphone. The app was transferred to the smartphone via USB connection for debugging and final installation. An 18% standard neutral gray card from a JJC, model number GC-1II was used for camera calibration. Taking the laboratory-based scanner approach as a methodological starting point, we sought to approximate and automate the steps involved in image acquisition and subsequent image processing. A summary comparison of the procedural sequence of steps between these two methods can be found in Table S2.† The source code showing the methods used to automate the image processing steps is available on Github (https://gist.github.com/SMR-83/cdb16aaaf56a3b7c3ba7c02efd1a664b). A statistical analysis was programmed into the app to automatically evaluate the dataset based on a conditional statement in the code which caps the z-score of each data point at 1.5 standard deviations from the mean as the criteria for inclusion in the dataset. A data point with a z-score greater than 1.5 is considered an outlier, and the app is programmed to allow no more than 5 outliers out of 50 measurements for the dataset to be considered valid.
Detection of IL-6 with plasmonic paper-based biosensors
Biosensors for the detection of IL-6 were fabricated as follows. Avidin-nanoparticles (100 μL, [Au] 100 mM) were modified with biotinylated anti-rabbit IgG (10 μL) for 1 h followed by washing 3 times with PBST with the aid of a centrifuge (7000 rpm, 5 min, final volume 25 μL). Capture antibodies (mouse monoclonal anti-IL-6) were immobilized by adding a drop on the paper substrate (2.5 μL, 10 μg mL−1) and letting it dry for at least 10 min. Then the biosensors were blocked with PBS–BSA (1 mL, let dry at room temperature for at least 10 min). To detect IL-6 the paper biosensors were rehydrated with 1 mL of PBS–BSA followed by the addition of IL-6 spiked at different concentrations in PBST, human serum or whole blood (2.5 μL). In order to ensure that the composition of the matrix was not noticeably altered by the spiking procedure, an IL-6 solution with a high concentration of 1 μg mL−1 in PBS was serially diluted more than 104 times with whole serum or blood to yield samples in the concentration range between 50 and 3 pg mL−1. Blood samples were stored in tubes containing EDTA as the anticoagulant. After 5 min the biosensor was washed up to 4 times with PBST (1 mL). Then the detection antibody was added (2.5 μL, 10 μg mL−1) for 5 min and the biosensors were washed once with PBST. Finally, avidin-decorated nanoparticles modified with biotinylated anti-rabbit IgG were added (2 μL, [Au] 400 mM) for 5 min. Then the biosensors were washed 4 times with PBST (1 mL) and the colorimetric signals were evaluated with the mobile densitometry app.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
R. R. acknowledges financial support from FEDER/Ministerio de Ciencia, Innovación y Universidades/Agencia Estatal de Investigación/- Proyecto CTQ2017-82432-R (biosensor development) and Proyecto CTQ2017-92226-EXP (app development). R. R. and S. M. R. acknowledge funding from a Radix fellowship from IdISBa/Impost turisme sostenible/Govern de les Illes Balears. A. A. acknowledges a fellowship for doctoral studies from Roche (“Stop fuga de cerebros” program). R. A. A.-P. and N. P. P. acknowledge financial support from Spanish Ministerio de Economia y Competitividad (CTQ2017-88648R and RYC-2015-19107), the Generalitat de Cataluña (2017SGR883), the Universitat Rovira i Virgili (2018PFR-URV-B2-02), and the Universitat Rovira i Virgili and Banco Santander (2017EXIT-08).
Electronic supplementary information (ESI) available. See DOI: 10.1039/d0na00026d
Notes and references
- Wang J. Wu H. Yang Y. Yan R. Zhao Y. Wang Y. Chen A. Shao S. Jiang P. Li Y. Q. Nanoscale. 2018;10:132–141. doi: 10.1039/C7NR06373C. [DOI] [PubMed] [Google Scholar]
- Paoli C. J. Reynolds M. A. Sinha M. Gitlin M. Crouser E. Crit. Care Med. 2018;46:1889–1897. doi: 10.1097/CCM.0000000000003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriquez-Camacho C. Losa J. Biomed Res. Int. 2014;2014:547818. doi: 10.1155/2014/547818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisner M. Clin. Chim. Acta. 2002;323:17–29. doi: 10.1016/S0009-8981(02)00101-8. [DOI] [PubMed] [Google Scholar]
- Meynaar I. A. Droog W. Batstra M. Vreede R. Herbrink P. Crit. Care Res. Pract. 2011;2011:594645. doi: 10.1155/2011/594645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau U. Y. Saxer S. S. Lee J. Bat E. Maynard H. D. ACS Nano. 2016;10:723–729. doi: 10.1021/acsnano.5b05781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong L. Dai H. Zhang S. Lin Y. Anal. Chem. 2016;88:5775–5782. doi: 10.1021/acs.analchem.6b00297. [DOI] [PubMed] [Google Scholar]
- Toma M. Tawa K. ACS Appl. Mater. Interfaces. 2016;8:22032–22038. doi: 10.1021/acsami.6b06917. [DOI] [PubMed] [Google Scholar]
- Russell C. Ward A. C. Vezza V. Hoskisson P. Alcorn D. Steenson D. P. Corrigan D. K. Biosens. Bioelectron. 2019;126:806–814. doi: 10.1016/j.bios.2018.11.053. [DOI] [PubMed] [Google Scholar]
- Hao Z. Pan Y. Shao W. Lin Q. Zhao X. Biosens. Bioelectron. 2019;134:16–23. doi: 10.1016/j.bios.2019.03.053. [DOI] [PubMed] [Google Scholar]
- Yin B. Zheng W. Dong M. Yu W. Chen Y. Joo S. W. Jiang X. Analyst. 2017;142:2954–2960. doi: 10.1039/C7AN00779E. [DOI] [PubMed] [Google Scholar]
- Peng J. Guan J. Yao H. Jin X. Anal. Biochem. 2016;492:63–68. doi: 10.1016/j.ab.2015.09.018. [DOI] [PubMed] [Google Scholar]
- Borse V. Srivastava R. Sens. Actuators, B. 2019;280:24–33. doi: 10.1016/j.snb.2018.10.034. [DOI] [Google Scholar]
- Wei H. Ni S. Cao C. Yang G. Liu G. ACS Sens. 2018;3:1553–1561. doi: 10.1021/acssensors.8b00365. [DOI] [PubMed] [Google Scholar]
- Wu D. Rios-Aguirre D. Chounlakone M. Camacho-Leon S. Voldman J. Biosens. Bioelectron. 2018;117:522–529. doi: 10.1016/j.bios.2018.06.049. [DOI] [PubMed] [Google Scholar]
- Tang C. K. Vaze A. Shen M. Rusling J. F. ACS Sens. 2016;1:1036–1043. doi: 10.1021/acssensors.6b00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. Sun J. Hou Y. Zhang C. Li D. Li H. Yang M. Fan C. Sun B. Biosens. Bioelectron. 2019;141:111432. doi: 10.1016/j.bios.2019.111432. [DOI] [PubMed] [Google Scholar]
- Russell S. M. Alba-Patiño A. Borges M. de la Rica R. Biosens. Bioelectron. 2019;140:111346. doi: 10.1016/j.bios.2019.111346. [DOI] [PubMed] [Google Scholar]
- Reddy Jr B. Hassan U. Seymour C. Angus D. C. Isbell T. S. White K. Weir W. Yeh L. Vincent A. Bashir R. Nat. Biomed. Eng. 2018;2:640–648. doi: 10.1038/s41551-018-0288-9. [DOI] [PubMed] [Google Scholar]
- Hou T. Huang D. Zeng R. Ye Z. Zhang Y. Int. J. Clin. Exp. Med. 2015;8:15238–15245. [PMC free article] [PubMed] [Google Scholar]
- Wang L. J. Chang Y. C. Sun R. Li L. Biosens. Bioelectron. 2017;87:686–692. doi: 10.1016/j.bios.2016.09.021. [DOI] [PubMed] [Google Scholar]
- Vashist S. K. van Oordt T. Schneider E. M. Zengerle R. von Stetten F. Luong J. H. T. Biosens. Bioelectron. 2015;67:248–255. doi: 10.1016/j.bios.2014.08.027. [DOI] [PubMed] [Google Scholar]
- Hermanson G. T., Bioconjugate Techniques, Academic Press, 2nd edn, 2008 [Google Scholar]
- Kong T. You J. B. Zhang B. Nguyen B. Tarlan F. Jarvi K. Sinton D. Lab Chip. 2019;19:1991–1999. doi: 10.1039/C9LC00165D. [DOI] [PubMed] [Google Scholar]
- Stalenhoef J. E. van Nieuwkoop C. Wilson D. C. van der Starre W. E. Delfos N. M. Leyten E. M. S. Koster T. Ablij H. C. van't Wout J. W. van Dissel J. T. J. Infect. 2018;77:18–24. doi: 10.1016/j.jinf.2018.05.007. [DOI] [PubMed] [Google Scholar]
- Molinero-Fernández Á. Moreno-Guzmán M. Arruza L. López M. Á. Escarpa A. ACS Sens. 2019;4:2117–2123. doi: 10.1021/acssensors.9b00890. [DOI] [PubMed] [Google Scholar]
- Russell S. M. Alba-Patiño A. Borges M. de la Rica R. ACS Sens. 2018;3:1712–1718. doi: 10.1021/acssensors.8b00452. [DOI] [PubMed] [Google Scholar]
- Russell S. M. Domenech-Sanchez A. de la Rica R. ACS Sens. 2017;2:848–853. doi: 10.1021/acssensors.7b00259. [DOI] [PubMed] [Google Scholar]
- Tsai D. H. Shelton M. P. DelRio F. W. Elzey S. Guha S. Zachariah M. R. Hackley V. A. Anal. Bioanal. Chem. 2012;404:3015–3023. doi: 10.1007/s00216-012-6418-4. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Lorenzo L. de la Rica R. Álvarez-Puebla R. A. Liz-Marzán L. M. Stevens M. M. Nat. Mater. 2012;11:604–607. doi: 10.1038/nmat3337. [DOI] [PubMed] [Google Scholar]
- Alba-Patiño A. Russell S. M. de la Rica R. Sens. Actuators, B. 2018;273:951–954. doi: 10.1016/j.snb.2018.07.019. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.