Abstract
Two new, small, early bacteriophage T4 genes, repEA and repEB, located within the origin E (oriE) region of T4 DNA replication, affect functioning of this origin. An important and unusual property of the oriE region is that it is transcribed at early and late periods after infection, but in opposite directions (from complementary DNA strands). The early transcripts are mRNAs for RepEA and RepEB proteins, and they can serve as primers for leading-strand DNA synthesis. The late transcripts, which are genuine antisense RNAs for the early transcripts, direct synthesis of virion components. Because the T4 genome contains several origins, and because recombination can bypass a primase requirement for retrograde synthesis, neither defects in a single origin nor primase deficiencies are lethal in T4 (Mosig et al., FEMS Microbiol. Rev. 17:83–98, 1995). Therefore, repEA and repEB were expected and found to be important for T4 DNA replication only when activities of other origins were reduced. To investigate the in vivo roles of the two repE genes, we constructed nonsense mutations in each of them and combined them with the motA mutation sip1 that greatly reduces initiation from other origins. As expected, T4 DNA synthesis and progeny production were severely reduced in the double mutants as compared with the single motA mutant, but early transcription of oriE was reduced neither in the motA nor in the repE mutants. Moreover, residual DNA replication and growth of the double mutants were different at different temperatures, suggesting different functions for repEA and repEB. We surmise that the different structures and protein requirements for functioning of the different origins enhance the flexibility of T4 to adapt to varied growth conditions, and we expect that different origins in other organisms with multiorigin chromosomes might differ in structure and function for similar reasons.
Bacteriophage T4, one of the largest bacteriophages, encodes most, if not all, of the proteins required for replication and recombination of its DNA. It can initiate replication by several different mechanisms that are usually coordinated with other DNA transactions during the developmental program (for recent reviews, see references 35, 44, 46, and 48). Under certain conditions, different mechanisms can partially substitute for each other (48, 54).
In the first replication mode, DNA synthesis is initiated in the infecting T4 chromosomes from one of several defined origins (24, 31, 33–35, 41, 45, 48, 55, 73). Because host RNA polymerase containing ς70 synthesizes primers for leading-strand DNA synthesis (4, 39, 48, 54) in these T4 origins, origin initiation depends to some extent on the physiological state of the host, and different origins are preferred under different physiological conditions (48, 55). Origin initiation occurs only once during the developmental program in most bacteria infected with wild-type T4. Most replication forks are initiated from intermediates of recombination, whose early formation, in turn, is facilitated by origin-dependent replication (14, 39, 44).
Several T4 origins have been characterized by various methods. Three of them, origin A (oriA), oriE, and oriF, are preferred under usual laboratory conditions, but additional origins are active under other conditions (24, 31, 33–35, 41, 45, 48, 55, 67, 73). Initiations from oriA, oriG, and oriF depend largely on transcripts initiated from motA-dependent middle promoters (4, 12, 21, 22, 34, 35, 48, 55). These promoters are recognized by Escherichia coli RNA polymerase holoenzyme, whose ς70 is modified by T4 AsiA protein, and their activities depend on MotA protein bound to a consensus Mot box at position −30, upstream of the −10 TATA box (1, 27, 63, 64, 66). Therefore, the MotA protein has been proposed to be an origin recognition protein of T4 (6, 35).
However, in the same in vivo experiments in which motA mutations drastically reduced initiation of replication from oriA and oriF, some phage progeny was still produced, and DNA replication was predominantly initiated from oriE (references 12 and 48 and experiments described below), suggesting either that other proteins or RNA (or both) facilitate initiation from oriE or that no origin recognition proteins are required. Here we describe two small genes of the oriE region that affect initiation of DNA replication from this origin.
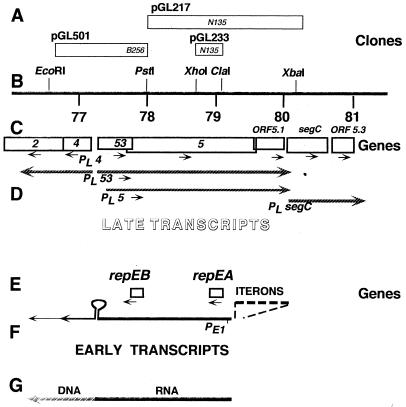
OriE was first described by Halpern et al. (24). The DNA sequence of this region (53) (Fig. 1 and 2) predicted five repeats (iterons) upstream of an early promoter (PE1) facing in the counterclockwise direction, which is opposite from the direction in which the late genes 53 and 5 are transcribed. No other early or middle promoters can be predicted from the DNA sequence of this region, except for a −10 consensus sequence between iterons 2 and 3, which is not associated with a consensus −35 or motA box sequence nor with detectable 5′ transcript ends downstream of it. There are also no predicted factor-independent transcription terminators in this region, but the palindrome shown in Fig. 1 (annotated IR1 in Fig. 2), which contains two divergent late promoters (for the late genes 4, 53, and 5), may facilitate transcription termination by other means. Several 5′ ends of nascent replicating DNA were found downstream of this palindrome (reference 48 and results described below), suggesting that the early transcripts serve as primers for leading-strand DNA synthesis and as mRNAs for two small proteins.
FIG. 1.
A map of the T4 oriE region. (A) Positions of clones used in subcloning and site-directed mutagenesis of repEB and repEA. Italicized names refer to mutations of gene 5 that can be rescued by these clones. (B) Positions of relevant restriction sites in the T4 map (36). (C) Positions of late T4 genes. (D) Positions of late promoters and late transcripts synthesized from this region. (E) Positions of the two early genes described here and the iterons upstream of these genes. (F) Positions of an early promoter and early transcripts. (G) Approximate positions of RNA primer transcripts and of RNA-DNA transition sites, where leading-strand DNA synthesis is primed. Directions of transcription and translation are indicated by arrowheads.
FIG. 2.
DNA sequence of the oriE region and predicted amino acid sequences of the encoded proteins. Proteins encoded in the upper strand are shown above the DNA sequence; those encoded in the lower strand are shown below the DNA sequence. Stop codons are marked with asterisks. Late promoters are underlined and labeled PL with an additional designation of genes that can be expressed from the corresponding transcripts (e.g., PL4). The early promoter (Fig. 1) is labeled PE1. Some inverted repeats are also underlined and labeled IR. IR2 brackets the segment of gene 5 that is homologous to the lysozyme gene e (53). The repeats called iterons upstream of PE1 are underlined in both DNA strands. The GenBank accession no. of this sequence is X15728.
Here we confirm transcription from the early promoter PE1 in wild-type T4 and in a motA mutant. We show that RepEA and RepEB proteins are synthesized from this segment of the genome and that synthesis of RepEB and probably RepEA in T4-infected bacteria is limited to very early times. To investigate the in vivo roles of these proteins, we constructed nonsense mutations in each of the two corresponding genes. Because we anticipated that repEA and repEB are important for T4 DNA replication only when activities of other origins are impaired, we combined repEA1 and repEB1 mutations with the motA mutation sip1 (23, 28) that we had found to be less leaky for motA-dependent transcription than the original motA mutant (42). The sip1 mutation greatly reduces transcript initiation from motA-dependent promoters in oriA and oriF (12, 22), but it allows considerable DNA replication and progeny production, presumably due to initiation from oriE and from recombination intermediates. As expected, in both double mutants DNA replication was much more severely affected than in the single motA mutant. The repEA or repEB mutations did not reduce the abundance of oriE transcripts; however, several oriE priming sites (RNA-DNA transition points) that were present in wild-type and motA mutant DNA were not found in repEB mutant DNA and were altered in the repEA mutant. These and other results indicate that repEB, and probably also repEA, code for oriE-specific DNA replication proteins that perform different functions.
MATERIALS AND METHODS
Bacteria, phages, and plasmids.
The E. coli strains, T4 phages, and plasmids used in this work are listed in Table 1.
TABLE 1.
Bacteria, phages, and plasmids used in this study
| Strain, phage, or plasmid | Relevant characteristic or genotype | Reference or source |
|---|---|---|
| E. coli | ||
| B | Wild type, sup0 | Our collection |
| S/6 | Smooth derivative of B; T6 resistant | Our collection |
| CR63 | Wild type, supD | Our collection |
| BL21(DE3) | F−sup0 expresses T7 RNA polymerase | 68 Novagen |
| DH5α | F−endA1 recA1 supE44 thi-1 φ80lacZΔM15 | 60 New England Biolabs |
| Bacteriophages | ||
| T4D | Wild type | Our collection |
| T4D sip1 | motA frameshift mutation | Reference 28 and this work |
| T4D N135 | Gene 5 amber mutation | 53 |
| T4D B256 | Gene 5 amber mutation | 53 |
| repEA1 | Gene repEA amber mutation | This work |
| repEB1 | Gene repEB ochre mutation | This work |
| repEA1-sip1 | repEA motA double mutant | This work |
| repEB1-sip1 | repEA motA double mutant | This work |
| Plasmids | ||
| pET11a | Apr pBR322 origin; expression vector with T7 promoter | Novagen |
| pET11d | Apr pBR322 derivative; expression vector with T7 promoter | Novagen |
| pMal-c2 | Apr ColE1 origin; plasmid to construct fusions with E. coli malE | New England Biolabs |
| pGEM3 | Apr pBR322 derivative | Promega |
| pRV1 | T4 repEA with His tag in pET11d | This work |
| pRV4 | T4 repEB with His tag in pET11d | This work |
| pRV9 | T4 repEB in pET11a | This work |
| pRV10 | T4 repEA in pMal-c2 | This work |
| pRV11 | T4 repEB in pMal-c2 | This work |
| pGL217 | T4 PstI-XbaI fragment, containing C-terminal segment of gene 5, repEA, six iterons and early promoter in pGEM3 | 53 |
| pGL233 | Segment of T4 gene 5 in pGEM3 | 53 |
| pGL501 | T4 fragment containing beginning of gene 5 and repEB in pGEM3 | 53 |
Construction of plasmids.
General procedures followed those previously described (62). The plasmids pRV1 and pRV4, producing six-His-tagged T4 RepEA and RepEB proteins, respectively, were constructed by cloning into pET11d PCR products synthesized from pGL233 or from DNA of wild-type T4 particles, respectively. To facilitate cloning and expression studies, some PCR primers (Table 2) were designed to generate additional restriction and ribosome binding sites.
TABLE 2.
Synthetic oligonucleotides used in this study
| No.a | Sequenceb |
|---|---|
| 1 | GTTATCTAGATAAGGAGGTTTTATTAATGCATCACCATCACCATCATGTGATACTTCTTCATAAG |
| 2 | ATTTAGGTGACACTATA |
| 3 | GTTATCTAGATAAGGAGGTTTTATTAATGCATCACCATCACCATCATGTAATTTTTCAGTTGGA |
| 4 | GTTAGGATCCTTAATTGGTTTGTCGGTG |
| 5 | GTTGGAATTCATGGTGATACTTCTTCAT |
| 6 | GTTGGAATTCATGGTAATTTTTCATTTG |
| 7 | GTTAGGATCCTTAATTGGTTTGTCGGTG |
| 8 | GTTTCATATGGTAATTTTTCAGTTG |
| 9 | GTTAGGATCCTTAATTGGTTTGTCGGTG |
| 10 | TAACCAATGCTGATGGT |
| 11 (316) | GTTCCAGTAGGATGAACTAA |
| 12 (322) | TTATGAAGAAGTATCACC |
| 13 (318) | GAATACCCATAACATCATCACC |
| 14 (323) | CAACTGAAAAATTACCATG |
| 15 (13) | TTTATCTAACCATTCAA |
| 16 (943) | GATTCAGACGATGAAGTAGA |
Numbers in parentheses are our laboratory designations.
The six histidine codons and restriction sites are underlined. The ribosome binding sites and two desired mutations are shown in boldface. All sequences are written in the 5′ to 3′ direction.
Oligo 1 was used as the forward PCR primer to construct pRV1, containing T4 repEA. The reverse primer was the SP6 promoter primer oligo 2, which hybridizes with the vector sequence of pGL233 DNA. The amplified DNA was cleaved with XbaI and BamHI (which cuts in the polylinker of pGL233). The resulting 205-bp fragment, containing T4 repEA, was ligated between the XbaI and BamHI sites of pET11d.
To construct pRV4 containing T4 repEB, the forward PCR primer oligo 3, containing the same six histidine codons and ribosome binding and XbaI restriction sites as primer 1 for pRV1, was used. The reverse PCR primer that hybridized with the end of the gene repEB was oligo 4, which generated the underlined BamHI restriction site. The amplified DNA was cleaved with XbaI and BamHI, and the resulting 184-bp fragment, containing repEB, was ligated between the XbaI and BamHI sites of pET11d.
Plasmid pRV9, from which RepEB without a His tag can be synthesized, was constructed by cloning into pET11a PCR products synthesized from wild-type T4 DNA. The forward primer, oligo 8, generated the underlined NdeI site. The reverse primer, oligo 9, generated the underlined BamHI site (Table 2). The amplified DNA was cleaved with NdeI and BamHI, and the resulting 147-bp fragment, containing repEB, was inserted between the NdeI and BamHI sites of pET11a.
Clones from which RepEA and RepEB fusions with maltose binding protein could be induced were constructed by inserting the corresponding T4 genes into the vector pMal-c2 (New England Biolabs).
All PCR amplifications and mutageneses were performed with Pfu or Turbo Pfu DNA polymerase (Stratagene). All clones constructed from PCR fragments were resequenced to ascertain the absence of gratuitous mutations.
Generating mutations.
To generate the amber mutation repEA1, PCR products were generated from pGL217 (Fig. 1) by using the mutagenic forward primer oligo 11 (the mutated base is printed in bold) and the abutting reverse primer oligo 12. The linear PCR products were circularized with T4 DNA ligase and used to transform E. coli DH5α. The plasmids of several transformants were sequenced, and one, containing only the desired mutation, was used in further studies.
The repEA1 mutation was introduced by homologous recombination into the phage genome. Plasmid-bearing cells were infected with T4 amN135 (gene 5) phage (Fig. 1), and N135+ recombinants were selected from the progeny by plating on sup0 S/6 bacteria. (As described above, the single repEA nonsense mutation does not cause a defective phenotype under these conditions.) One of the N135+ recombinants, found by direct sequencing of several possible candidates to contain the repEA1 mutation, was selected for further studies. This repEA1 mutant was then crossed with the motA mutant sip1. The progeny of this cross was plated on supD CR63 bacteria, which were expected to allow double mutants to grow better than B bacteria. Unexpectedly, the single sip1 mutant proved to be more defective in CR63 than in the sup0 strains B or S/6. Sip1 does not plaque on CR63 at 25°C, and it has variable plaque morphologies and lower efficiencies of plating at intermediate temperatures on CR63 than on E. coli B. At 42°C, sip1 growth could not be tested because CR63 does not grow a lawn. In contrast, on lawns of B or S/6, the single sip1 mutant produces small plaques with turbid halos at 25 and 30°C, and at 34, 37, or 42°C, the plaques are indistinguishable from wild-type plaques. Therefore, as a preliminary selection for double mutants, progeny phage were spot tested on sup0 S/6 hosts at different temperatures (25, 30, 34, 37, and 42°C). The repEA and motA genes of several candidate double mutants, which did not grow on S/6 at 25°C, were resequenced, and one confirmed double mutant was used in subsequent studies.
Using a similar strategy, we generated the repEB1 mutation from plasmid pGL501 by using the mutagenic forward primer oligo 13 and the reverse primer oligo 14. Candidate mutant plasmids were resequenced and crossed with amB256 (gene 5) phage (Fig. 1) by infecting repEB1-pGL501-bearing bacteria with amB256 mutant phage. Several B256+ recombinants were selected, and the presence of the repEB mutation was confirmed by resequencing. The double mutant repEB1 sip1 was constructed by crossing the single repEB1 and sip1 mutants, selecting possible double mutant candidates by spot testing on S/6 at different temperatures, and selecting those that did not grow at 42°C. One double mutant, confirmed by resequencing both repEB and motA genes, was selected for subsequent studies.
Synthesis and purification of His-tagged RepEA and RepEB proteins.
E. coli BL21(DE3), containing plasmids pRV1 or pRV4, was grown at 30°C in 100 ml of Luria broth supplemented with 50 μg of carbenicillin per ml to a density of 1 × 108 to 2 × 108 bacteria per ml. IPTG (isopropyl-β-d-thiogalactopyranoside) was added to 1 mM to induce T7 RNA polymerase. One hour later, the cells were collected by centrifugation at 6,000 × g in a Sorvall GSA rotor for 20 min. The pellet was resuspended in buffer A (20 mM Tris-HCl, pH 7.6, 0.5 M NaCl, 5 mM imidazole, 1 mM Pefabloc). The bacteria were lysed by freeze-thawing at 37°C and subsequent sonication until 99% of the cells were disrupted, as judged by microscopy. Cell debris, containing most of the RepEA and RepEB proteins, was pelleted by centrifugation at 20,000 × g for 30 min. The pellet was resuspended in 10 ml of binding buffer B (6 M guanidinium-HCl [pH 7.6], 0.5 M NaCl, 5 mM imidazole, 0.1% Triton X-100) and was incubated for 1 h at room temperature to dissolve the proteins. Remaining debris was pelleted by centrifuging at 20,000 × g for 30 min. The supernatant was loaded onto a 2-ml nickel-nitrilotriacetic acid column (QIAGEN) that had been previously equilibrated with the same buffer B. Proteins were washed with 10 volumes of buffer B and were subsequently washed again with 10 volumes of buffer C (buffer B made 20 mM in imidazole). The proteins were then renatured on the column by reducing the guanidinium-HCl concentration from 6 to 0.5 M by slowly adding buffer A (1 mM β-mercaptoethanol and 0.1% Triton X-100) and finally 10 volumes of buffer D (20 mM Tris-HCl [pH 7.6], 0.5 M NaCl, 20 mM imidazole, 10 mM MgCl2, 0.1% Triton X-100).
Proteins were eluted with buffer A containing 250 mM imidazole. The fractions containing RepEA or RepEB proteins, respectively, were pooled and dialyzed against buffer F (20 mM HEPES [pH 7.6], 3 mM MgCl2, 25 mM KCl, 1 mM EDTA, 1 mM β-mercaptoethanol, 0.1% Triton X-100). Precipitates were spun down in an Eppendorf centrifuge for 30 min. Proteins were concentrated in Amicon Centricon-3 concentrators. Protein concentration was determined by using the BioRad protein assay kit. The proteins were stored at −70°C in 10% glycerol.
Synthesis of labeled RepEB protein without His tags.
E. coli BL21(DE3), containing plasmid pRV9, was grown in 6 ml of M9 medium with carbenicillin (50 μg/ml) at 30°C to a titer of 2 × 108 cells/ml. Then IPTG was added to 0.3 mM, and the cells were grown for 1 h. Fifteen minutes before harvesting, 30 μCi of 35S-labeled amino acids (TRANLABEL; ICN Pharmaceuticals Inc.) was added to the culture. Two minutes before harvesting, incorporation was quenched by adding 60 μl of a 25% casamino acid solution (Difco). Finally, the cells were collected by centrifugation and resuspended in sample buffer for subsequent sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis or in lysis buffer for subsequent immunoprecipitations.
Primer extensions on T4 RNA.
The 5′ ends of transcripts initiated from PE1 were determined by primer extensions using avian myeloblastosis virus reverse transcriptase and oligo 10 as primer on total RNA isolated from T4-infected bacteria using CsCl gradients as described (40) or by using an RNAWiz kit from Ambion according to manufacturer’s instructions. Oligo 10 is complementary to T4 RNA 64 nucleotides downstream of the PE1 promoter. This oligo was end labeled with [γ-32P]ATP (Amersham) and T4 polynucleotide kinase (Promega) and was used for both primer extension and sequencing reactions (62). The products were analyzed on an 8% polyacrylamide denaturing gel.
DNA sequencing.
Plasmid DNA and T4 DNA were sequenced without prior PCR amplification by using reagents and protocols of the Promega femtomole sequencing kit as described (47).
Protein purification for antiserum production.
His-tagged RepEA and RepEB proteins were purified by nickel chelate affinity chromatography as described above. They underwent SDS–20% PAGE. Gel slices, containing the overexpressed His-tagged RepEA or RepEB proteins, were excised and sent to East Acres Biologicals (Southbridge, Mass.) to generate rabbit polyclonal antibodies against the proteins. Only anti-RepEB antiserum was of sufficient concentration and specificity to justify its use in further studies.
Labeling, immunoprecipitations, and immunoblotting of proteins.
E. coli B cells (10 ml for each experiment) were grown in M9 medium, supplemented with biotin and thiamine, to a concentration of 3 × 108 cells/ml. They were infected with 5 to 8 phage particles per cell.
At 2 to 2.5 min after infection, 250 μCi of 35S-labeled amino acids (ICN Pharmaceuticals, Inc.) was added to the culture. At 6.5 min, 5 ml of the infected cells was harvested by centrifugation. At 8 min after infection, an additional 250 μCi was added to the remaining cells, which were harvested 12.5 or 16.5 min (in different experiments) after infection. Each pellet was resuspended in 0.15 ml of an ice-cold solution of 10 mM Tris-HCl (pH 7.7), 20 mM NaCl, 0.5 mM dithiothreitol, 1 mM Pefabloc (Serva), 0.05% Triton X-100, 0.1% sarcosyl, and 0.16 mM EDTA and was lysed gently by freezing in a dry ice-ethanol mixture and thawing at 37°C for five freeze-thaw cycles. Then the cells were sonicated while being kept on ice until 99% of the cells were lysed. The debris was pelleted at 12,000 × g for 20 min at 4°C.
The supernatant was made 1.25 mM in MgCl2 and was incubated with anti-RepEB antiserum overnight at 4°C. Prior to use, protein A-agarose beads (Calbiochem) were washed twice with NET (50 mM Tris-HCl [pH 7.5], 0.15 M NaCl, 5 mM EDTA) containing 0.5% Nonidet P-40 (BRL) and 1 mM methionine and were washed twice with NET containing 0.05% Nonidet P-40 for 2 min per wash and were pelleted between washes for 1 min at 200 × g in a Sorvall centrifuge.
An equal volume of washed protein A-agarose bead slurry (Calbiochem) was added to the antiserum-containing lysate, and the mixture was rotated for 2 h at 4°C. Then the beads were pelleted and washed three times with radioimmunoprecipitation assay buffer (50 mM Tris-HCl [pH 7.7], 0.15 mM NaCl), 1% SDS, 1% Triton X-100, 1% deoxycholate), once with TSA (0.01 M Tris-HCl, 7.7; 0.14 M NaCl), and once with 0.05 M Tris-HCl, pH 6.8. Finally, the beads were boiled in Phastgel (Pharmacia) sample buffer, and proteins were separated by SDS-PAGE and were detected by autoradiography.
For immunoblotting, proteins were separated by PAGE and blotted to nitrocellulose membranes (Schleicher and Schuell). The membranes were blocked and incubated with anti-RepEB serum as described (25). The blots were developed with the chemiluminescence kit of Amersham according to the manufacturer’s instructions. Luminescent bands were detected with Kodak Biomax film.
Measuring T4 DNA replication.
Total T4 DNA replication was measured by incorporation of [3H]thymidine (purchased from ICN Pharmaceuticals, Inc., or NEN) into DNA as described (47). To detect RNA-DNA transition points (i.e., priming sites for leading-strand DNA synthesis), replicating T4 DNA was isolated from infected cells with a DNAzol kit from MRC (Cincinnati, Ohio), according to the manufacturer’s protocol. This DNA was treated with RNase H (Amersham), and RNA-DNA transition points were determined by repetitive primer extensions as described (47), except that Thermosequenase was used instead of Taq DNA polymerase. Purified RNA gave no extension products with this procedure. Oligo 15 (Table 2) for oriE, and oligo 16 for oriF priming sites as controls, were 5′ end labeled with [γ-32P]ATP (Amersham) using T4 polynucleotide kinase (Promega). The products were separated by PAGE and analyzed by autoradiography or by phosphorimaging.
RESULTS
Transcripts are initiated from the promoter PE1 only early after infection.
The DNA sequence of the oriE region (53) suggested that repEA and repEB genes are cotranscribed early from the same promoter PE1 that directs synthesis of primer transcripts for leading-strand synthesis from oriE, and late virion genes are transcribed in the opposite direction later during infection. To facilitate description of the complex expression and replication patterns, the annotated DNA sequence of the oriE region is shown in Fig. 2.
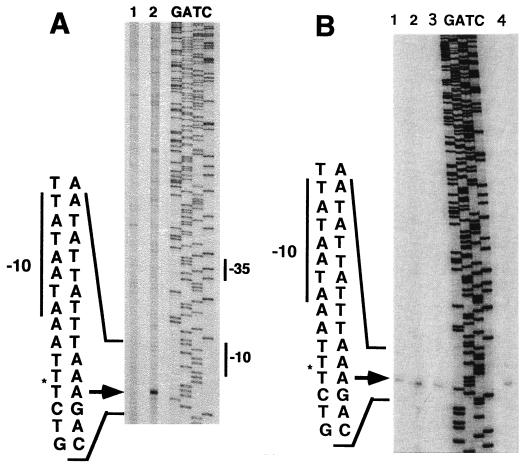
Primer extensions with reverse transcriptase on RNA using several different primers corresponding to this region detected wild-type transcription start sites only at PE1 with primer 10 (Fig. 3A). These transcript 5′ ends were present at nonreduced levels in the motA mutant sip1 (11, 28) and in the repEA and repEB mutants described in the next section (Fig. 3B), indicating that none of these genes is required for transcription from PE1. The reproducible slight overaccumulation of PE1 transcripts in the motA mutant is consistent with earlier evidence that some, but not all, early genes are overexpressed in other motA mutants (7, 23, 42).
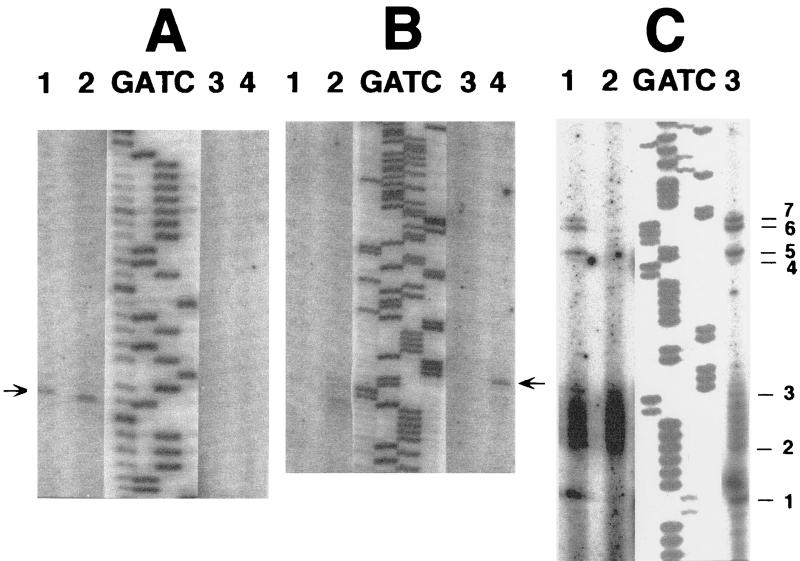
FIG. 3.
Mapping, by primer extensions from primer 10, of the 5′ ends of early transcripts initiated from PE1. (A) Lane 1, RNA isolated from uninfected bacteria; lane 2, RNA isolated 4 min after infection of E. coli B with wild-type T4 at 30°C by using CsCl gradient purification (see Materials and Methods); lanes G, A, T, and C, sequencing reactions using the same primer on DNA of plasmid pGL217. (B) RNA isolated 4 min after infection of E. coli B with an RNAWiz kit (see Materials and Methods). Lane 1, wild-type T4; lane 2, sip1 (motA); lane 3, repEB1; lane 4, repEA1; lanes G, A, T, and C, sequencing reactions using the same primer on DNA of plasmid pGL217.
The PE1-directed transcripts are short-lived. Primer extensions on wild-type RNA samples isolated 6, 8, 10, 12, and 16 min after infection failed to detect these transcripts in samples isolated later than 6 min after infection (data not shown), although early transcripts of other genes (3, 20, 40) were detected in these same preparations.
Synthesis of RepEA and RepEB proteins.
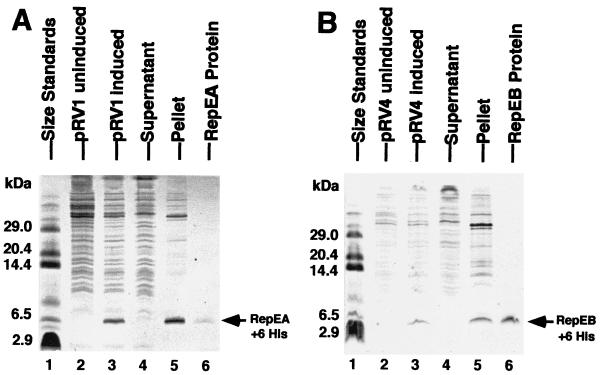
Wild-type repEA and repEB, cloned separately with six-His tags in the T7 promoter expression vector pET11d, and repEB, cloned without His tags in pET11a, produced proteins of the expected sizes in BL21(DE3) after induction of T7 RNA polymerase with IPTG (Fig. 4 and data not shown).
FIG. 4.
Histidine-tagged RepEA (A) and RepEB (B) proteins after induction of cloned repEA and repEB, after centrifugation of the sonicate, and after purification by affinity chromatography as described in Materials and Methods. Proteins were separated by PAGE and were stained with Coomassie blue.
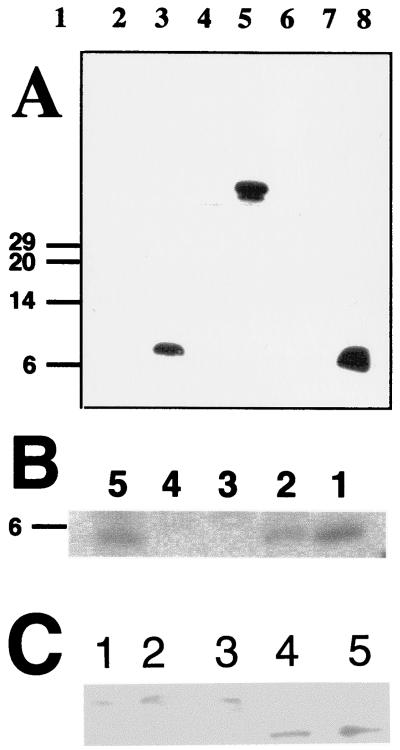
These proteins accumulated in inclusion bodies, were largely insoluble after purification, and resisted many different attempts to permanently solubilize them. Therefore, both proteins were used to produce antibodies. The His-tagged RepEB protein elicited antibodies specific for RepEB protein and fusion proteins containing RepEB, and only those proteins (Fig. 5A). These antibodies were used to monitor the accumulation of untagged RepEB protein in T4-infected bacteria. Figure 5B shows that this protein appeared early after infection, in both wild-type and motA-infected bacteria. Consistent with the transcription analysis (Fig. 3), RepEB protein slightly overaccumulated in the motA mutant. It became undetectable 12.5 and 16.5 min after infection with wild-type T4 (Fig. 5B, lane 3, and data not shown), suggesting that the protein is unstable, like other proteins that control DNA replication origins (32). The single repEB1 mutant and the repEB1 sip1 double mutant produced a truncated protein of the expected size (Fig. 5C).
FIG. 5.
(A) Western blots to test the specificity of anti-RepEB serum used for immunoprecipitations shown in panel B and immunoblots shown in panel C. Total sonicates of unlabeled induced cells bearing plasmids with the fusion genes indicated below were separated by PAGE and processed as described in Materials and Methods. Lane 1, size standards (kDa); lane 2, His-repEA fusion; lane 3, His-repEB fusion; lane 4, mal-repEA fusion, lane 5, mal-repEB fusion; lane 6, mal–β-galactosidase fusion; lane 7, purified His-RepEA protein; lane 8, purified His-RepEB protein. (B) Immunoprecipitations of different 35S-labeled cell lysates with the anti-RepEB antiserum used in panel A. The precipitates were fractionated by PAGE and visualized by autoradiography (see Materials and Methods). Lane 1, RepEB protein induced from pRV4; lane 2, wild-type T4-infected cells harvested 6.5 min after infection; lane 3, wild-type-T4-infected cells harvested 16.5 min after infection; lane 4, uninfected bacteria without plasmids; lane 5, sip1 (motA)-infected bacteria harvested 6.5 min after infection. To compensate for expected differences in repEB expression, extracts 10 times as many T4-infected or uninfected bacteria were precipitated compared with extracts from induced bacteria. (C) Immunoblots of proteins separated by PAGE and probed with the anti-RepEB antiserum used in panel A. Extracts from the following T4 mutants were loaded: lane 1, sip1 (motA); lane 2, repEA1; lane 3, repEA1 sip1; lane 4, repEB1; lane 5, repEB1 sip1.
Synthesis of RepEA protein could not be monitored in T4-infected cells, because antibodies against the His-tagged RepEA protein were not as sufficiently specific as anti-RepEB antibodies to justify their use in similar experiments. However, we surmise that this protein is produced in infected cells, because a mutation in the repEA gene causes an altered phenotype described in the next two sections.
Nonsense mutations in the repEA and repEB genes in combination with a motA mutation affect phage growth.
To test whether the early repEA and repEB genes are important for phage growth, we introduced nonsense mutations into repEA cloned in plasmid pGL217 (53) with the mutagenic oligo 11 and into repEB cloned in plasmid pGL501 with the mutagenic oligo 13 as described in Materials and Methods. These two nonsense mutations were designed so that the amino acid sequence of the base plate lysozyme gp5, encoded in the complementary DNA strand, is not altered. Mutant clones, confirmed by resequencing, were used to introduce the repEA1 and the repEB1 mutations by homologous recombination into the phage genome. To reduce initiation from other origins (see above), each rep mutant was crossed with the motA mutant sip1, and double mutants were selected and confirmed by DNA sequencing as described in Materials and Methods. Our DNA sequencing revealed that sip1 is a frameshift mutation that deletes an A from a run of six A residues in the motA gene (69), changing Lys59 to Asn and causing termination of translation after 20 additional codons.
On lawns of nonsuppressing host bacteria (B or S/6), the single sip1 mutant and the two double mutants had different plaque morphologies and different temperature sensitivities, as described in Materials and Methods. Whereas the repEA1 sip1 double mutant produced only pinpoint plaques at 25 or 30°C, it produced larger plaques at 42°C. In contrast, the repEB1 sip1 double mutant produced no plaques at 42°C and produced pinpoint plaques at 25 or 30°C. At intermediate temperatures (34 and 37°C), plaques of both double mutants were of intermediate sizes. Burst sizes measured after multiple infection (data not shown) confirmed that the different plaque sizes were correlated with differences in latent periods and phage yields. PAGE of 35S-labeled proteins (not shown) revealed no detectable differences in early proteins synthesized by the single sip1 mutant or the repEA sip or repEB sip double mutants. Together with the transcription results described in the preceding section, these results suggested that both repEA and repEB genes are important for motA-independent T4 DNA replication. These inferences were confirmed by the following analyses.
repEA and repEB mutations, in combination with a motA mutation, affect total T4 DNA replication.
Because total T4 DNA replication is initiated in several modes, because there are several origins of replication, and because origin-dependent DNA replication is limited to a single or a few rounds (references 14 and 39 and see above), mutations that affect initiation from a single origin are not expected to alter total DNA synthesis significantly. However, we expected that total T4 DNA replication of the repEA1 or repEB1 mutants would be delayed and reduced when initiation from other origins is reduced. Although most T4 DNA replication is initiated from intermediates of recombination (39), prior origin-dependent DNA replication is needed for generating single-stranded termini that invade homologous regions and initiate subsequent recombination-dependent DNA replication (14, 48).
motA-dependent primer transcripts in oriF (22) and in oriA (12) are respectively reduced to less than 0.2% and less than 2% of the wild-type levels in the motA mutant sip1, and initiation of DNA replication from these origins is reduced accordingly (5, 12) (see Fig. 7C). Total T4 DNA synthesis is also delayed and reduced in motA mutants, largely because many replication and recombination genes, which can be transcribed from both early and middle promoters, are expressed less efficiently in motA than in wild-type T4 (66). It was not known whether reduced expression of replication and recombination genes affects initiation from oriE. However, results described in the next section suggest that the sip1 mutant synthesizes sufficient replication proteins for such initiation.
FIG. 7.
Repetitive primer extensions on nascent T4 DNA (47) of the wild type and sip1, repEA, and repEB mutants. The DNA was isolated as described in Materials and Methods. Products of the repetitive primer extensions were separated by PAGE together with products of sequencing reactions using the same primers and cloned or virion T4 DNA as templates. Because the bands in the sequencing lanes were much darker than in the experimental lanes, lighter exposures of the same gel were scanned and superimposed on the scans of the experimental primer extensions. (A) Repetitive extensions with primer 15 (Table 2) in oriE. Lane 1, wild-type T4; lane 2, sip1; lane 3, repEB1; lane 4, repEA1. The arrowhead corresponds with position 333 in Fig. 2. (B) A segment of a longer run of the same reactions as shown in Fig. 7A. The arrowhead corresponds with position 389 in Fig. 2. (C) Repetitive primer extensions with oligo 16 (Table 2) in oriF on the same nascent DNA samples used for primer extensions with oligo 15 as shown in lanes 1 through 3 in panels A and B. Lane 1, wild-type T4; lane 2, sip1; lane 3, repEB1. The rightmost lane (not numbered) shows the positions of DNA ends 1 through 7 that had been detected in oriF in previous repetitive primer extensions on different nascent DNA preparations (references 47 and 48 and unpublished experiments). Signals at positions 2 and 3 have been consistently weak. The smear between positions 2 and 3 represents PCR products of primer 16 alone. It is also found when no template is added (48). Lanes G, A, T, and C show sequencing reactions using the same primer on DNA of plasmid pGL217 in panels A and B and wild-type T4 virion DNA in panel C.
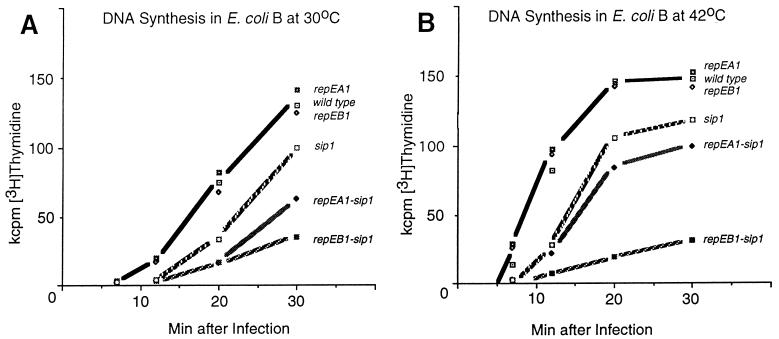
As expected, total DNA synthesis of both repE sip double mutants was significantly more delayed and reduced than synthesis of the single sip mutant (Fig. 6). Like the growth patterns, the residual DNA synthesis patterns of the two double mutants were different at different temperatures. The repEA1 sip1 double mutant was more defective at lower (Fig. 6A) than at higher (Fig. 6B) temperatures. In contrast, the repEB1 sip1 double mutant showed almost no DNA synthesis at 42°C (Fig. 6B). This might indicate that mutant RepEB protein is more defective than mutant RepEA protein, that RepEA protein is dispensable at high temperatures, or that a bypass mechanism can substitute for RepEA but not for RepEB. We prefer the latter explanation, because the phenotype of the repEA1 sip1 mutant is reminiscent of the DNA delay phenotype of primase-deficient and topoisomerase-deficient T4 mutants (44, 55). These mutants grow better at 42°C than at lower temperatures, because at higher temperatures T4 endonuclease VII, which is important for the bypass mechanism, is expressed earlier (54).
FIG. 6.
DNA synthesis measured by incorporation of [3H]thymidine into acid precipitable material (47) after infection of E. coli B with the indicated mutant and wild-type T4 strains at 30 (A) and 42°C (B). The reduced DNA synthesis of the single motA mutant compared with wild-type T4 is expected from the reduced expression of T4 replication and recombination genes in motA mutants (7, 11, 42, 66).
repEB affects primer utilization for leading-strand synthesis at oriE.
To test directly whether the repE mutations affect functioning of oriE, we investigated the use of priming sites for leading-strand DNA synthesis in the single repE and motA mutants by a repetitive primer extension assay on nascent DNA (47). As mentioned earlier, T4 DNA replication uses transcripts to prime origin-dependent leading-strand DNA synthesis (4, 35, 39, 45, 48), and repetitive primer extensions (47) have detected several start sites of leading DNA strands (sites of transitions from RNA to DNA) in oriA, oriF (48), and oriE (mentioned, but not shown, in reference 48). Because primers for this assay are origin specific, single repE mutants can be tested, avoiding possible complications due to delayed synthesis of replication proteins in the motA mutant.
As shown in Fig. 7A, a major leading-strand start site in oriE is detected by extensions from oligo 15 (Table 2) in both the wild type and the motA mutant sip1, but not in (single) repEA1 or repEB1 mutants. Additional start sites that can be detected with a primer further downstream (not shown) are also present in sip1, but are missing in the two rep mutants. Other leading-strand start sites appeared or were enhanced in the single repEA1 mutant, but not in the repEB1 mutant. One example is shown in Fig. 7B. The reasons are not yet understood and are being investigated. It is possible, for example, that these sites reflect altered transcription termination sites.
As controls, we used the same nascent DNA preparations from the wild type and sip1 and repEB1 mutants for extensions with oligo 16 (Table 2), which we had used earlier to detect leading-strand start sites in oriF (47, 48). In the previous experiments we found the seven priming sites marked on the right side of Fig. 7C (among other priming sites). As expected, multiple oriF replication start sites were used in wild-type T4 and in the repEB1 mutant but not in the motA mutant sip1 (Fig. 7C). The absence of these transition points in sip1 DNA is consistent with the drastically reduced transcription from the middle uvsY-oriF promoter in the sip1 mutant (22) and the absence of initiation signals (“comets”) in 2D gels of replicating mutant DNA (4), in which the motA-dependent uvsY promoter (22) was deleted.
Together, these results show that the repEB mutation specifically affects initiation of leading strands from oriE, that the repEA mutation modifies it, and that motA-dependent transcription is not required for initiation of replication at oriE. Presumably, the lower abundance of replication proteins synthesized in the motA mutant sip1 is sufficient to initiate the first round of leading-strand DNA synthesis.
DISCUSSION
The most important conclusions from our results can be summarized as follows: (i) two new, small, early T4 proteins, RepEA and RepEB, encoded in the oriE region, are important for T4 DNA replication; (ii) RepEB is specifically important for the priming of leading-strand DNA synthesis at oriE, whereas RepEA appears to have an auxiliary function, but neither protein is required for transcription from the early promoter; (iii) neither expression of repEA or repEB nor initiation of DNA replication at oriE requires the activator of middle promoters, MotA protein; (iv) the early transcripts encoding RepEA and RepEB, which can also serve as primers for leading-strand DNA synthesis, are short-lived.
These results suggested to us the following working hypothesis for initiation of DNA replication from oriE: we propose that one or both of the new repE proteins facilitates loading of helicases to oriE DNA (e.g., to the iteron DNA) and that tracking of these helicases modulates the status of some oriE transcripts (i.e., whether they are terminated or processed and whether they are base paired to DNA, as required for priming, or displaced from the DNA, as required for translation). Experimental tests of this working hypothesis will undoubtedly demand modifications and extensions.
oriE-specific transcripts and proteins are synthesized only briefly after infection.
T4 uses transcripts to prime leading-strand DNA synthesis at origins (4, 35, 39, 44, 48). Formation of primers must include steps by which the nascent RNA is kept base paired or is reinserted into the DNA duplex. Partial digestion of transcripts may occur, and it may or may not be obligatory for priming. Within the framework of our working hypothesis, one or both of the two new RepE proteins synthesized from oriE transcripts facilitates the access of helicases, which in turn facilitates the base pairing of a sister transcript with the DNA template, the priming of DNA synthesis, and perhaps transcription termination, at a distance.
Database searches provided no clue for the functions of RepEA and RepEB proteins. Moreover, RepEA and RepEB proteins, with or without His tags, turned out to be largely insoluble. However, RepEA and RepEB, when fused to maltose-binding protein, are soluble. They bind to single-stranded DNA, preferentially to the iterons marked in Fig. 1 and 2 (70), suggesting that they might facilitate the opening of double-stranded DNA, like origin proteins of other replicons (32, 37). In contrast to origins of other replicons, in T4 DNA the primary targets of such proteins, the iterons, are far away from the major priming sites for leading-strand DNA synthesis (Fig. 1).
The presence of iterons in T4 oriE (Fig. 2) was unexpected at first. Based on bacteria, phage, and plasmid paradigms (15, 17, 32), it is generally assumed that iterons are hallmarks of origins in which primers for leading-strand DNA synthesis are synthesized by primases, which require single-stranded segments for binding to DNA. As mentioned earlier, primers for T4 origin of initiation of leading DNA strands are, however, synthesized by RNA polymerase (4, 39, 48).
Within the framework of our working hypothesis, migration of helicases loaded at the iterons would modulate the status of oriE transcripts (i.e., whether the RNA is base paired to the DNA or to be displaced) far from their target sites. This might depend on competitions between several helicases, (e.g., replicative helicase gp41 [56, 57], the Rho RNA helicase [65, 66], or the UvsW helicase which is, however, a late protein [10]).
Loading of the replicative T4 helicase gp41 by one or both of the RepE proteins can explain another puzzling observation. The gp41 protein, which associates with T4 primase, requires the helper protein gp59 in vitro (56). However, in vivo, gene 59 mutants are proficient in origin DNA replication, and they arrest DNA replication like recombination-deficient T4 mutants (47, 72), suggesting that other proteins load helicases at origins.
The early cessation of transcription through oriE must be a major reason for early cessation of origin-dependent DNA replication (39) at this origin. The small window of time when repEA and repEB are expressed is sufficient to allow a single or few initiations from oriE, but apparent lability of PE1-dependent transcripts and of the RepEB protein, as well as the synthesis of antisense RNA from the late promoters, are all bound to contribute to the apparent early demise of oriE function during phage T4 development.
The early cessation of transcription from PE1 is readily explained by the accumulation of the T4 AsiA protein. This protein sequesters the recognition motif for the −35 region of ς70, and, therefore, inhibits transcription from promoters containing consensus −35 sequences, such as PE1 (13, 64). Most other early T4 promoters have variant −35 sequences (71) and may not be subject to similarly strong asiA-dependent inhibition. The apparent lability of early oriE transcripts (Fig. 3) may be related, in part, to possible base pairing with the late antisense transcripts synthesized from the same region.
We do not know whether the DNA priming sites within oriE (examples are shown in Fig. 7A and B) correspond to transcription termination sites. Unlike oriA (40) and oriF (21, 22, 48), oriE does not contain a classical factor-independent transcription terminator. Transcription termination factor Rho (66) may be required, and the anomalous priming sites of repEA mutant DNA (Fig. 7B) might suggest the possibility that the T4 RepEA protein may also be involved. Distinguishing features of rho-dependent transcription termination sites are largely unknown; apparent 3′ transcript ends near palindromes have been attributed to posttranscriptional decay that stops at RNA hairpins (59, 61).
The untranslated RNA segment between repEA and repEB is unusually large for early T4 transcripts. It is intriguing to note that this untranslated RNA corresponds to a DNA segment encoding the lysozyme segment of the base plate protein gp5 (53). The amino acid sequence of this segment resembles that of the soluble T4 lysozyme gpe, suggesting that a copy of this or a related gene was inserted into an ancestor of the base plate gene 5 during evolution. Such an insertion would have increased the distance between the repEA and repEB genes on the complementary DNA strand (Fig. 2).
repEA and repEB mutants have different phenotypes.
The residual replication of the rep motA double mutants at late times after infection and the residual progeny production may be due to residual initiation of transcription from motA-dependent promoters (12, 22, 35), initiation from other origins (31, 67, 73), and/or, most likely, from recombinational intermediates. Recombinational intermediates can be formed in the absence of prior DNA replication, albeit with a much longer delay than when DNA replication is allowed (8, 14, 44).
However, the different phenotypes of the repEA motA and repEB motA double mutants at different growth temperatures (Fig. 6) require additional comments. One possible scenario to explain why the repEA1 mutant is more leaky at high temperatures than at low temperatures (Fig. 6) postulates a role for RepEA protein, predicted to be hydrophobic, in membrane attachment of T4 DNA. Such membrane association has been observed by many investigators in T4 (18, 19, 29, 30, 41, 43) and is well demonstrated in other organisms (38, 58). This association may be important but not essential (33), since membrane-free in vitro systems can synthesize DNA under optimal conditions with speed similar to that of in vivo systems (2, 32, 56, 57). Another, and possibly related, explanation for the leaky phenotype of the repEA1 mutant at 42°C (Fig. 5B) is that in this mutant T4 primase and/or topoisomerase activities are impaired, and that this deficiency is bypassed by a temperature-dependent recombinational mechanism (50, 52, 54) that requires, among others, T4 endonuclease VII. The T4 topoisomerase is one of several membrane-bound T4 proteins (29, 30).
Different replication origins of T4 have different sequences, different structures, and different requirements for functioning.
The functioning of RepE proteins, the presence of iterons, and the absence of a motA requirement for oriE distinguish this origin from oriA and oriF, suggesting that T4 origins use different initiation mechanisms, which may allow functioning under different conditions. For example, oriE is preferentially used when torsional stress in the replicating T4 DNA is reduced by mutations in T4 topoisomerase and host gyrase or by excessive damage due to 32P decay or oxidation (reference 55 and unpublished observations). oriE is also preferred when T4 infects certain host mutants (nusD) altered in transcription termination factor Rho (67).
These differences, as well as comparisons with T4-related phages (44), support the hypothesis that the T4 genome was assembled during evolution from modular components from several sources (9) and that such processes can generate novel and redundant origins of DNA replication, redundant replication proteins, and complex overlapping transcription patterns (26, 44, 49, 51). The redundancies, in turn, can provide selective advantages under different developmental and environmental conditions because they facilitate coordination of essential viral processes and adjustments of DNA replication and gene expression to different growth conditions (44, 48). Moreover, timed transcription from complementary strands of the same DNA can help to adjust DNA replication and other DNA transactions to optimal growth and progeny production.
Such redundancies are bound to enhance the viability of any organism under different environmental or developmental conditions. The multiple T4 origins may serve as models for the complexities of multiple origins in metazoan chromosomes (16), which may differ in structure and function for reasons similar to those of T4.
ACKNOWLEDGMENTS
This work was supported, in part, by Public Health Service grant GM 13221 to G.M., by Center grant CA 68485 of the National Cancer Institute, by the Vanderbilt-Ingram Cancer Center, by NSF-sponsored Biological Core Facilities, and by the Natural Science Fund of Vanderbilt University.
We thank Anthony Guo and Nancy Colowick for mutating the repEA gene in plasmid GL217 and Jon Weil for the motA mutant sip1.
REFERENCES
- 1.Adelman K, Brody E N, Buckle M. Stimulation of bacteriophage T4 middle transcription by the T4 proteins MotA and AsiA occurs at two distinct steps in the transcription cycle. Proc Natl Acad Sci USA. 1998;95:15247–15252. doi: 10.1073/pnas.95.26.15247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alberts B. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. 1998;92:291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- 3.Barth K A, Powell D, Trupin M, Mosig G. Regulation of two nested proteins from gene 49 (recombination endonuclease VII) and of a lambda RexA-like protein of bacteriophage T4. Genetics. 1988;120:329–343. doi: 10.1093/genetics/120.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belanger K G, Kreuzer K N. Bacteriophage T4 initiates bidirectional DNA replication through a two-step process. Mol Cell. 1998;2:693–701. doi: 10.1016/s1097-2765(00)80167-7. [DOI] [PubMed] [Google Scholar]
- 5.Belanger K G, Mirzayan C, Kreuzer H E, Alberts B M, Kreuzer K. Two-dimensional gel analysis of rolling circle replication in the presence and absence of bacteriophage T4 primase. Nucleic Acids Res. 1996;24:2166–2175. doi: 10.1093/nar/24.11.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson K H, Kreuzer K N. Role of MotA transcription factor in bacteriophage T4 DNA replication. J Mol Biol. 1992;228:88–100. doi: 10.1016/0022-2836(92)90493-4. [DOI] [PubMed] [Google Scholar]
- 7.Brody E, Rabussay D, Hall D H. Regulation of transcription of prereplicative genes. In: Mathews C K, Kutter E M, Mosig G, Berget P B, editors. Bacteriophage T4. Washington, D.C.: American Society for Microbiology; 1983. pp. 174–183. [Google Scholar]
- 8.Broker T R. An electron microscopic analysis of pathways for bacteriophage T4 DNA recombination. J Mol Biol. 1973;81:1–16. doi: 10.1016/0022-2836(73)90243-x. [DOI] [PubMed] [Google Scholar]
- 9.Campbell A C. Phage evolution and speciation. In: Calendar R, editor. The bacteriophages. Vol. 1. New York, N.Y: Plenum Press; 1988. pp. 1–14. [Google Scholar]
- 10.Carles-Kinch K, George J W, Kreuzer K N. Bacteriophage T4 UvsW protein is a helicase involved in recombination, repair and the regulation of DNA replication origins. EMBO J. 1997;16:4142–4151. doi: 10.1093/emboj/16.13.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chace K V, Hall D H. Characterization of new regulatory mutants of bacteriophage T4. II. New class of mutants. J Virol. 1975;15:929–945. doi: 10.1128/jvi.15.4.929-945.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang, A., and G. Mosig. 1999. Unpublished data.
- 13.Colland F, Orsini G, Brody E N, Buc H, Kolb A. The bacteriophage T4 AsiA protein: a molecular switch for sigma 70-dependent promoters. Mol Microbiol. 1998;27:819–829. doi: 10.1046/j.1365-2958.1998.00729.x. [DOI] [PubMed] [Google Scholar]
- 14.Dannenberg R, Mosig G. Early intermediates in bacteriophage T4 DNA replication and recombination. J Virol. 1983;45:813–831. doi: 10.1128/jvi.45.2.813-831.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Del Solar G, Giraldo R, Ruiz-Echevarría M J, Espinosa M, Díaz-Orejas R. Replication and control of circular bacterial plasmids. Microbiol Mol Biol Rev. 1998;62:434–464. doi: 10.1128/mmbr.62.2.434-464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DePamphilis M L. Replication origins in metazoan chromosomes: fact or fiction? Bioessays. 1999;21:5–16. doi: 10.1002/(SICI)1521-1878(199901)21:1<5::AID-BIES2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 17.Eguchi Y, Tomizawa J. Complexes formed by complementary RNA stem-loops—their formations, structures and interaction with ColE1 Rom protein. J Mol Biol. 1991;220:831–842. doi: 10.1016/0022-2836(91)90356-b. [DOI] [PubMed] [Google Scholar]
- 18.Frankel F R. Studies on the nature of replicating DNA in T4-infected Escherichia coli. J Mol Biol. 1966;18:127–143. doi: 10.1016/s0022-2836(66)80081-5. [DOI] [PubMed] [Google Scholar]
- 19.Frankel F R, Majumdar C, Weintraub S, Frankel D. DNA polymerase and the cell membrane after T4 infection. Cold Spring Harbor Symp Quant Biol. 1968;33:495–500. doi: 10.1101/sqb.1968.033.01.057. [DOI] [PubMed] [Google Scholar]
- 20.Frazier M W, Mosig G. The bacteriophage T4 gene mrh whose product inhibits late T4 gene expression in an Escherichia coli rpoH (ς32) mutant. Gene. 1990;88:7–14. doi: 10.1016/0378-1119(90)90053-t. [DOI] [PubMed] [Google Scholar]
- 21.Gruidl M E, Chen T C, Gargano S, Storlazzi A, Cascino A, Mosig G. Two bacteriophage-T4 base plate genes (25 and 26) and the DNA repair gene uvsY belong to spatially and temporally overlapping transcription units. Virology. 1991;184:359–369. doi: 10.1016/0042-6822(91)90852-3. [DOI] [PubMed] [Google Scholar]
- 22.Gruidl M E, Mosig G. Sequence and transcripts of the bacteriophage T4 DNA repair gene uvsY. Genetics. 1986;114:1061–1079. doi: 10.1093/genetics/114.4.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall D H, Snyder R D. Suppressors of mutations in the rII gene of bacteriophage T4 affect promoter utilization. Genetics. 1981;97:1–9. doi: 10.1093/genetics/97.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halpern M E, Mattson T, Kozinski A W. Origins of phage T4 DNA replication as revealed by hybridization to cloned genes. Proc Natl Acad Sci USA. 1979;76:6137–6141. doi: 10.1073/pnas.76.12.6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 26.Hendrix R W, Smith M C, Burns R N, Ford M E, Hatfull G F. Evolutionary relationships among diverse bacteriophages and prophages: all the world’s a phage. Proc Natl Acad Sci USA. 1999;96:2192–2197. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hinton D M, March-Amegadzie R, Gerber J S, Sharma M. Characterization of pre-transcription complexes made at a bacteriophage T4 middle promoter: involvement of the T4 MotA activator and the T4 AsiA protein, a sigma-70 binding protein, in the formation of the open complex. J Mol Biol. 1996;256:235–248. doi: 10.1006/jmbi.1996.0082. [DOI] [PubMed] [Google Scholar]
- 28.Homyk T, Rodriguez A, Weil J. Characterization of T4 mutants that partially suppress the inability of T4 rII to grow in lambda lysogens. Genetics. 1976;83:477–487. [PMC free article] [PubMed] [Google Scholar]
- 29.Huang W M. Membrane-associated proteins of T4-infected Escherichia coli. Virology. 1975;66:508–521. doi: 10.1016/0042-6822(75)90223-8. [DOI] [PubMed] [Google Scholar]
- 30.Huang W M. Inhibition of initiation of bacteriophage T4 DNA replication by perturbation of Escherichia coli host membrane composition. J Virol. 1979;32:917–924. doi: 10.1128/jvi.32.3.917-924.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.King G J, Huang W M. Identification of the origins of T4 DNA replication. Proc Natl Acad Sci USA. 1982;79:7248–7252. doi: 10.1073/pnas.79.23.7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kornberg A, Baker T A. DNA replication. 2nd ed. New York, N.Y: W. H. Freeman; 1992. [Google Scholar]
- 33.Kozinski A W. Origins of T4 DNA replication. In: Mathews C K, Kutter E M, Mosig G, Berget P B, editors. Bacteriophage T4. Washington, D.C.: American Society for Microbiology; 1983. pp. 111–119. [Google Scholar]
- 34.Kreuzer K N, Engman H W, Yap W Y. Tertiary initiation of replication in bacteriophage T4. Deletion of the overlapping uvsY promoter/replication origin from the phage genome. J Biol Chem. 1988;263:11366–11373. [PubMed] [Google Scholar]
- 35.Kreuzer K N, Morrical S W. Initiation of DNA replication. In: Karam J, Drake J W, Kreuzer K N, Mosig G, Hall D H, Eiserling F A, Black L W, Spicer E K, Kutter E, Carlson K, Miller E S, editors. Molecular biology of bacteriophage T4. Washington, D.C.: American Society for Microbiology; 1994. pp. 28–42. [Google Scholar]
- 36.Kutter E, Stidham T, Guttman B, Kutter E, Batts D, Peterson S, Djavakhishvili T, Arisaka F, Mesyanzhinov V, Rüger W, Mosig G. Genomic map of bacteriophage T4. In: Karam J, Drake J W, Kreuzer K N, Mosig G, Hall D H, Eiserling F A, Black L W, Spicer E K, Kutter E, Carlson K, Miller E S, editors. Molecular biology of bacteriophage T4. Washington, D.C.: American Society for Microbiology; 1994. pp. 491–519. [Google Scholar]
- 37.Learn B A, Um S-J, McMacken R. Cryptic single-stranded-DNA binding activities of the phage λ P and Escherichia coli DnaC replication initiation proteins facilitate the transfer of E. coli DnaB helicase onto DNA. Proc Natl Acad Sci USA. 1997;94:1154–1159. doi: 10.1073/pnas.94.4.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lemon K P, Grossman A D. Localization of bacterial DNA polymerase: evidence for a factory model of replication. Science. 1998;282:1516–1519. doi: 10.1126/science.282.5393.1516. [DOI] [PubMed] [Google Scholar]
- 39.Luder A, Mosig G. Two alternative mechanisms for initiation of DNA replication forks in bacteriophage T4: priming by RNA polymerase and by recombination. Proc Natl Acad Sci USA. 1982;79:1101–1105. doi: 10.1073/pnas.79.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Macdonald P M, Mosig G. Regulation of a new bacteriophage T4 gene, 69, that spans an origin of DNA replication. EMBO J. 1984;3:2863–2871. doi: 10.1002/j.1460-2075.1984.tb02221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marsh R C, Breschkin A M, Mosig G. Origin and direction of bacteriophage T4 DNA replication. II. A gradient of marker frequencies in partially replicated T4 DNA as assayed by transformation. J Mol Biol. 1971;60:213–233. doi: 10.1016/0022-2836(71)90289-0. [DOI] [PubMed] [Google Scholar]
- 42.Mattson T, Richardson J, Goodin D. Mutant of bacteriophage T4D affecting expression of many early genes. Nature. 1974;250:48–50. doi: 10.1038/250048a0. [DOI] [PubMed] [Google Scholar]
- 43.Miller R C. Association of replicative T4 deoxyribonucleic acid and bacterial membranes. J Virol. 1972;10:920–924. doi: 10.1128/jvi.10.5.920-924.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mosig G. Recombination and recombination-dependent DNA replication in bacteriophage T4. Annu Rev Genet. 1998;32:379–413. doi: 10.1146/annurev.genet.32.1.379. [DOI] [PubMed] [Google Scholar]
- 45.Mosig G. Relationship of T4 DNA replication and recombination. In: Mathews C K, Kutter E M, Mosig G, Berget P B, editors. Bacteriophage T4. Washington, D.C.: American Society for Microbiology; 1983. pp. 120–130. [Google Scholar]
- 46.Mosig G. T4 bacteriophage and related bacteriophages. In: Webster R G, Granoff A, editors. Encyclopedia of virology. Vol. 3. San Diego, Calif: Academic Press; 1994. pp. 1376–1383. [Google Scholar]
- 47.Mosig G, Colowick N. DNA replication of bacteriophage T4 in vivo. Methods Enzymol. 1995;262:587–604. doi: 10.1016/0076-6879(95)62046-x. [DOI] [PubMed] [Google Scholar]
- 48.Mosig G, Colowick N, Gruidl M E, Chang A, Harvey A J. Multiple initiation mechanisms adapt phage T4 DNA replication to physiological changes during T4’s development. FEMS Microbiol Rev. 1995;17:83–98. doi: 10.1111/j.1574-6976.1995.tb00190.x. [DOI] [PubMed] [Google Scholar]
- 49.Mosig G, Colowick N E, Pietz B C. Several new bacteriophage T4 genes, mapped by sequencing deletion endpoints between genes 56 (dCTPase) and dda (a DNA-dependent ATPase-helicase) modulate transcription. Gene. 1998;223:143–155. doi: 10.1016/s0378-1119(98)00238-8. [DOI] [PubMed] [Google Scholar]
- 50.Mosig, G., J. Gewin, and L. Davenport. Unpublished data.
- 51.Mosig G, Hall D H. Gene expression: a paradigm of integrated circuits. In: Karam J, Drake J W, Kreuzer K N, Mosig G, Hall D H, Eiserling F A, Black L W, Spicer E K, Kutter E, Carlson K, Miller E S, editors. Molecular biology of bacteriophage T4. Washington, D.C.: American Society for Microbiology; 1994. pp. 127–131. [Google Scholar]
- 52.Mosig G, Lin G, Chang A. Interaction of primase and topoisomerase during lagging strand DNA synthesis of phage T4, and a recombinational bypass mechanism. J Cell Biochem. 1992;16(Suppl. B):77. [Google Scholar]
- 53.Mosig G, Lin G W, Franklin J, Fan W H. Functional relationships and structural determinants of two bacteriophage T4 lysozymes: a soluble (gene e) and a baseplate-associated (gene 5) protein. New Biol. 1989;1:171–179. [PubMed] [Google Scholar]
- 54.Mosig G, Luder A, Ernst A, Canan N. Bypass of a primase requirement for bacteriophage T4 DNA replication in vivo by a recombination enzyme, endonuclease VII. New Biol. 1991;3:1195–1205. [PubMed] [Google Scholar]
- 55.Mosig G, Macdonald P, Lin G, Levin M, Seaby R. Gene expression and initiation of DNA replication of bacteriophage T4 in phage and host topoisomerase mutants. In: Cozzarelli N R, editor. Mechanisms of DNA replication and recombination. A. R. New York, N.Y: Liss; 1983. pp. 173–186. [Google Scholar]
- 56.Nossal N G. The bacteriophage T4 replication fork. In: Karam J, Drake J W, Kreuzer K N, Mosig G, Hall D H, Eiserling F A, Black L W, Spicer E K, Kutter E, Carlson K, Miller E S, editors. Molecular biology of bacteriophage T4. Washington, D.C.: American Society for Microbiology; 1994. pp. 43–53. [Google Scholar]
- 57.Nossal N G, Hinton D M, Hobbs L J, Spacciapoli P. Purification of bacteriophage T4 DNA replication proteins. Methods Enzymol. 1995;262:560–584. doi: 10.1016/0076-6879(95)62045-1. [DOI] [PubMed] [Google Scholar]
- 58.Ogden G B, Pratt M J, Schaechter M. The replicative origin of the E. coli chromosome binds to cell membranes only when hemimethylated. Cell. 1988;54:127–135. doi: 10.1016/0092-8674(88)90186-9. [DOI] [PubMed] [Google Scholar]
- 59.Platt T. Rho and RNA: models for recognition and response. Mol Microbiol. 1994;11:983–990. doi: 10.1111/j.1365-2958.1994.tb00376.x. [DOI] [PubMed] [Google Scholar]
- 60.Raleigh E A, Lech K, Brent R. Selected topics from classical bacterial genetics. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: Wiley Interscience; 1989. p. 1.4. [DOI] [PubMed] [Google Scholar]
- 61.Richardson J P, Greenblatt J. Control of RNA chain elongation and termination. In: Neidhardt C F, Curtiss R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella. I. Washington, D.C.: ASM Press; 1996. pp. 822–848. [Google Scholar]
- 62.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 63.Schmidt M C, Kreuzer N K. Purified MotA protein binds the −30 region of a bacteriophage T4 middle promoter and activates transcription in vitro. J Biol Chem. 1992;267:11399–11407. [PubMed] [Google Scholar]
- 64.Severinova E, Severinov K, Darst S A. Inhibition of Escherichia coli RNA polymerase by bacteriophage T4 AsiA. J Mol Biol. 1998;279:9–18. doi: 10.1006/jmbi.1998.1742. [DOI] [PubMed] [Google Scholar]
- 65.Steinmetz E J, Platt T. Evidence supporting a tethered tracking model for helicase activity of Escherichia coli Rho factor. Proc Natl Acad Sci USA. 1994;91:1401–1405. doi: 10.1073/pnas.91.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stitt B, Hinton D. Regulation of middle-mode transcription. In: Karam J, Drake J W, Kreuzer K N, Mosig G, Hall D H, Eiserling F A, Black L W, Spicer E K, Kutter E, Carlson K, Miller E S, editors. Molecular biology of bacteriophage T4. Washington, D.C.: American Society for Microbiology; 1994. pp. 142–160. [Google Scholar]
- 67.Stitt B L, Mosig G. Impaired expression of certain prereplicative bacteriophage T4 genes explains impaired T4 DNA synthesis in Escherichia coli rho (nusD) mutants. J Bacteriol. 1989;171:3872–3880. doi: 10.1128/jb.171.7.3872-3880.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Studier F W, Rosenberg A H, Dunn J J, Dubendorf J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 69.Uzan M, Brody E, Favre R. Nucleotide sequence and control of transcription of the bacteriophage T4 motA regulatory gene. Mol Microbiol. 1990;4:1487–1496. doi: 10.1111/j.1365-2958.1990.tb02059.x. [DOI] [PubMed] [Google Scholar]
- 70.Vaiskunaite, R., and G. Mosig. Unpublished data.
- 71.Wilkens K, Rüger W. Transcription from early promoters. In: Karam J D, Drake J W, Kreuzer K N, Mosig G, Hall D H, Eiserling F A, Black L W, Spicer E K, Kutter E, Carlson K, Miller E S, editors. Molecular biology of bacteriophage T4. Washington, D.C.: American Society for Microbiology; 1994. pp. 132–141. [Google Scholar]
- 72.Wu R, Ma F, Yeh Y-C. Suppression of DNA-arrested synthesis in mutants defective in gene 59 of bacteriophage T4. Virology. 1972;47:147–156. doi: 10.1016/0042-6822(72)90248-6. [DOI] [PubMed] [Google Scholar]
- 73.Yee J K, Marsh R C. Locations of bacteriophage T4 origins of replication. J Virol. 1985;54:271–277. doi: 10.1128/jvi.54.2.271-277.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]