Summary
Genome size (GS) can vary considerably between phylogenetically close species, but the landscape of GS changes in insects remain largely unclear. To better understand the specific evolutionary factors that determine GS in insects, we examined flow cytometry-based published GS data from 1,326 insect species, spanning 700 genera, 155 families, and 21 orders. Model fitting showed that GS generally followed an Ornstein–Uhlenbeck adaptive evolutionary model in Insecta overall. Ancestral reconstruction indicated a likely GS of 1,069 Mb, suggesting that most insect clades appeared to undergo massive genome expansions or contractions. Quantification of genomic components in 56 species from nine families in four insect orders revealed that the proliferation of transposable elements contributed to high variation in GS between close species, such as within Coleoptera. This study sheds lights on the pattern of GS variation in insects and provides a better understanding of insect GS evolution.
Subject areas: Genomics, Phylogenetics, Evolutionary biology
Graphical abstract
Highlights
-
•
The most comprehensive variation pattern of insects genome size (GS) to date
-
•
GS evolution of the Insecta was reflected by the adaptive Ornstein–Uhlenbeck model
-
•
Ancestral state of insect GS was estimated to be ∼1 Gb
-
•
Intrafamily GS variations were driven by recent transpositions and non-coding regions
Genomics; Phylogenetics; Evolutionary biology
Introduction
Genome size (GS) varies greatly in different organisms, with differences of ∼200,000-fold across all eukaryotes (Gregory 2001), ∼12,000-fold across plant species (Leitch et al., 2019) and an ∼7,000-fold difference across all animals (Gregory 2005a). GS is poorly correlated with organismal complexity or number of coding genes (the C- and G-value paradox, Gall 1981; Hahn and Wray 2002), and thus, the biological reasons for this huge variations in GS remain unclear from an evolutionary perspective. Whether the evolutionary causes and consequences of GS variation are adaptive or non-adaptive processes has been widely debated. The non-adaptive theory suggests that variation in GS are introduced through a stochastic and nearly neutral evolutionary process (Petrov 2002; Lynch and Conery 2003), since DNA insertions and deletions mainly occur through random genetic drift (Lynch and Conery 2003; Mohlhenrich and Mueller 2016), with counteracting outcomes between these two processes determining GS (Petrov 2002; Sung et al., 2016; Kapusta et al., 2017). Specifically, DNA can be rapidly added by genome duplication (Vu et al., 2015; Soltis and Soltis 2016; Marburger et al., 2018), tandem repeat amplification (Shah et al., 2020; Majid and Yuan 2021; Stelzer et al., 2021) or transposon transpositions (Kidwell 2002; Naville et al., 2019; Zhang et al., 2020; Lehmann et al., 2021; Oggenfuss et al., 2021). Extraneous and/or slightly deleterious DNA is then gradually deleted by purifying selection (Lynch and Conery 2003). Alternatively, the adaptive hypothesis emphasizes the functional effects resulting from changes in total DNA abundance (Cavalier-Smith 1978, 2005; Gregory and Hebert 1999). Because DNA content directly or indirectly effects the cell volume and specific surface area, this hypothesis argues that GS variation is an adaptive process subject to natural selection for essential cellular processes (e.g., cell division rate or cell metabolism rate) (Cavalier-Smith 1978, 2005; Gregory 2001; Gardner et al., 2020) and fitness-related phenotypes (e.g., body size, developmental rate) (Gregory 2002; Wyngaard et al., 2005).
Insects constitute a major proportion of invertebrates and contain the largest number of reported species, and thus serve as a good system for examining GS evolution. Some intriguing insect traits have been tested for correlations with GS. For example, the reproductive fitness (Arnqvist et al., 2015) of seed beetles and host range of moths in family Noctuidae (Calatayud et al., 2016; Zhang et al., 2019) are both correlated with GS. However, traits such as parasitism by Hymenopterans (Ardila-Garcia et al., 2010), eusociality of Hymenopterans and Blattodeans (Koshikawa et al., 2008; Ardila-Garcia et al., 2010), and flight ability and strategies among dragonflies and damselflies (Odonata) (Ardila-Garcia and Gregory 2009) are all unrelated to GS. Morphological characteristics such as eye width (Liu et al., 2017), antennae length (Liu et al., 2017), egg size (Schmidt-Ott et al., 2009; Church et al., 2019), and body size (1994; Palmer et al., 2003; Tsutsui et al., 2008; Ardila-Garcia and Gregory 2009; Lopes et al., 2009) in adult insects have also been linked to GS, but are not consistently correlated with GS in different clades. However, testing whether these phenotypes have a causal relationship with GS in insects remains difficult since GS is generally dependent on phylogenetic history (Alfsnes et al., 2017).
Several recent studies have conducted small scale comparative phylogenetic investigations to better understand the evolutionary patterns and processes responsible for GS variation in insects. For example, GS in fireflies (family Lampyridae, Coleoptera) (Lower et al., 2017) and flies (subgenus Sophophora, Diptera) (Hjelmen and Johnston 2017) were found to evolve neutrally through a non-adaptive mode. Ancestral reconstruction of GS among crickets (suborder Ensifera, Orthoptera) (Yuan et al., 2021) and butterflies (family Papilionoidea, Lepidoptera) (Liu et al., 2020) suggested that genome expansion or contraction events happened frequently, resulting in a high diversity of GS among these species.
Moreover, genomic analyses in other insects have provided evidence that the abundance of transposable elements (TE) differs widely among species and is positively correlated with GS in insects (Petersen et al., 2019; Wu and Lu 2019). For example, locust (Locusta migratoria, Orthoptera) carries the largest insect genome published to date, ∼6.5 Gb, ∼60% of which is comprised of TEs (Wang et al., 2014). In other large genomes, such as that of the grasshopper Vandiemenella viatica (∼4.5 Gb) (Palacios-Gimenez et al., 2020) or the rice weevil Sitophilus oryzae (770 Mbp) (Parisot et al., 2021), TEs account for an even greater proportion of the genome, exceeding 70%. By contrast, TE represent less than 1% of the genomes of the Antarctic midge (Belgica antarctica, Diptera, 83.9 Mb) (Kelley et al., 2014) and the body louse (Pediculus humanus humanus, Phthiraptera, 108 Mb) (Kirkness et al., 2010). Some studies have reported that the specific accumulation of LINE retrotransposons and DNA transposons leads to GS variation among wood-white butterfly species (genus Leptidea) (Talla et al., 2017) and Asian gypsy moths (genus Lymantria) (Hebert et al., 2019). In addition, the massive horizontal transfer of some TEs, such as the Gypsy, Bel-Pao, and Copia LTR superfamilies, significantly influences GS in mosquitoes (Melo and Wallau 2020).
The majority of these above-mentioned studies were carried out in a small set of species. Because GS may be the outcome of various counteracting factors, it may not be possible to arrive at a single, universal principle that can explain the causes of GS variation through small-scale studies. The recent emergence of numerous insect genome sequencing projects has led to the availability of GS information for more than 1000 insects, either estimated by flow cytometry, K-mer analysis, or genome assembly. To broadly uncover the evolutionary factors driving GS variation in insects, we collected GS data, estimated mainly by flow cytometry, and analyzed differences within and among various insect clades. The results showed that GS varies considerably at different taxonomic levels, even among some close insect species. Comparative phylogenetic analyses showed that different models fit the observed differences in GS in different insect orders, suggesting that different evolutionary history responsible for shaping the GS variation among different insect clades. Analysis of genome content suggested that the expansion of TEs, rather than genome duplication, led the observed variation in GS between phylogenetically close species. Moreover, we found that several specific TEs appear to drive GS variations within different insect clades, suggesting that the type of TE driving changes in GS in each insect group may be the result of lineage-specific evolutionary events.
Results
GS variation in insects
To understand the comprehensive pattern of insect GS variation at broad level, we first collected as much GS data as possible. To this end, we obtained 1,345 records of GS, estimated mainly by flow cytometry, from the Animal Genome Size Database (Gregory 2005a). After removing redundancy, 1,145 insect GS information were kept for further analysis. We also examined previous studies, published before December 2020, to supplement the above insect GS data, which yielded 181 additional records. Ultimately, the GS dataset included 1,326 insect species from 700 genera, 155 families, and 21 orders (Table S1). Among these records, dipteran and coleopteran insects comprised the highest proportion, including 307 and 295 species, respectively, followed by Hymenoptera, Lepidoptera, Odonata and Hemiptera. Among these insect species, we observed a 266-fold variation in GS from the smallest, 68.46 Mb, belonging to the midge Clunio tsushimensis in Diptera, to the largest, 18.23 Gb, in the grasshopper Bryodemella holdereri in Orthoptera. Statistical analyses indicated several significant differences in the maximum, minimum, average, range, and variance of GS across the different insect orders (Table 1 andFigure 1A), among which the Orthoptera GS, 0.91 Gb on average, was significantly larger than any other order (n>10), whereas Order Diptera had the smallest average GS of 381.44 Mb, followed by Hymenoptera with a 434.96 Mb average GS (Wilcoxon rank-sum one-side tests, p< 0.001) (Table S2B). Interestingly, some families carried significantly smaller genomes than those of any other family within the same order, such as Chironomidae in Diptera (average = 127.1 Mb, n = 30), Tenebrionidae in Coleoptera (average = 370.1 Mb, n = 65), Braconidae in Hymenoptera (average = 140.2 Mb, n = 14) and Nymphalidae in Lepidoptera (average = 417.5 Mb, n = 29) (Table S2B).
Table 1.
Statistical summary of 1,326 insect GS from 21 orders
| Min.∗ (Mb) | Max.∗ (Mb) | Mean (Mb) | Mode (Mb) | Range (Max./Min.) | CV.∗(%) | No. of Species | |
|---|---|---|---|---|---|---|---|
| Diptera | 68.46 | 1,858.20 | 381.44 | 207.81 | 27.14 | 84.41 | 307 |
| Coleoptera | 156.48 | 4,909.56 | 771.77 | 351.48 | 31.38 | 75.22 | 295 |
| Hymenoptera | 97.80 | 1,939.70 | 434.96 | 292.40 | 19.83 | 56.98 | 223 |
| Lepidoptera | 214.72 | 1,897.32 | 541.95 | 390.51 | 8.84 | 48.33 | 159 |
| Odonata | 361.86 | 2,308.08 | 1,007.43 | 849.63 | 6.38 | 40.53 | 113 |
| Hemiptera | 176.04 | 6,855.78 | 949.97 | 510.37 | 38.94 | 115.27 | 94 |
| Orthoptera | 1,515.90 | 18,232.00 | 9,052.56 | 9,440.36 | 12.03 | 39.51 | 66 |
| Blattodea | 567.24 | 5,036.70 | 1,810.78 | 1,162.81 | 8.88 | 61.96 | 27 |
| Phasmatodea | 1,330.08 | 7,824.00 | 2,962.44 | 2,338.30 | 5.88 | 57.09 | 12 |
| Archaeognatha | 2,435.22 | 3,804.42 | 3,001.24 | 2,902.40 | 1.56 | 15.62 | 8 |
| Mantodea | 2,855.76 | 4,430.34 | 3,475.81 | 3,087.35 | 1.55 | 18.98 | 5 |
| Thysanoptera | 280.40 | 479.22 | 384.69 | 410.33 | 1.71 | 19.78 | 5 |
| Strepsiptera | 107.58 | 127.14 | 117.36 | – | 1.18 | – | 2 |
| Trichoptera | 258.00 | 455.00 | 356.50 | – | 1.76 | – | 2 |
| Phthiraptera | 107.58 | 107.58 | 107.58 | – | 1.00 | – | 2 |
| Siphonaptera | 459.66 | 459.66 | 459.66 | – | 1.00 | – | 1 |
| Dermaptera | 518.34 | 518.34 | 518.34 | – | 1.00 | – | 1 |
| Embioptera | 2,601.48 | 2,601.48 | 2,601.48 | – | 1.00 | – | 1 |
| Mecoptera | 2,014.68 | 2,014.68 | 2,014.68 | – | 1.00 | – | 1 |
| Zygentoma | 3,022.02 | 3,022.02 | 3,022.02 | – | 1.00 | – | 1 |
| Zoraptera | 1,848.42 | 1,848.42 | 1,848.42 | – | 1.00 | – | 1 |
| Total | 68.46 | 18,232.00 | 1,107.03 | 274.24 | 266.32 | 189.45 | 1,326 |
Note. “∗” Min. is short for minimum; Max. for maximum and CV. for Coefficient of variation. See also Table S1.
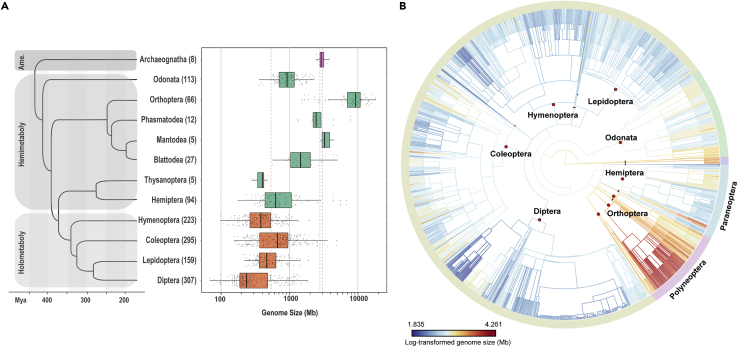
Figure 1.
Patterns of insect GS variation and evolution
(A) GS distribution pattern of 1,326 insect records from 12 orders along the time-calibrated phylogeny cited from Misof et al. (2014). Numbers of GS records in each order are shown in the brackets and orders with <5 species records are not demonstrated here. Dashed lines, of which colors represent the three types of insect metamorphosis (the ametaboly, insects without metamorphosis; the hemimetaboly, insects with incomplete metamorphosis; the holometaboly, insects with complete metamorphosis), indicate the averaged GS of corresponding metamorphosis groups. See also Tables 1, S1 and S2.
(B) Estimated evolution of 1,256 insects GS mapped onto the dendrogram according to the NCBI Taxonomy Tree, with all species names omitted. Colored branches reflect genome size values from small (dark blue) to big (dark red), generated based on the Brownian Motion (BM) process using the function of contMap in R package phytools (Revell 2012). Different insect orders are differentiated by red circles on the branches. The pinkish-purple outermost circle represents the cohort Polyneoptera and the light blue is for the cohort Paraneoptera. See also Table S1 and Figure S1.
The holometabolous insects (clade Holometabola) are thought to possess relatively small genome contents under the selection of the rapid cell division and metabolism rate during the developmental stages with intensive morphological differentiation (Gregory 2002, 2005b). We tested this hypothesis by comparing the difference in GS among the ametabolous orders (including Archaeognatha and Zygentoma), the hemimetabolous orders (including Phthiraptera, Hemiptera, Thysanoptera, Blattodea, Mantodea, Phasmatodea, Embioptera, Orthoptera, Dermaptera, Zoraptera and Odonata), and the holometabolous orders (including Diptera, Mecoptera, Siphonaptera, Lepidoptera, Trichoptera, Coleoptera, Strepsiptera and Hymenoptera). We found that the holometabolous did have the smallest genomes (average GS = 536.7 Mb, n = 990) (Figure 1A), significantly smaller than that of hemimetabolous (p = 1.31E-73, average GS =2,781.4 Mb, n = 327) and ametabolous (p = 0.003, average GS =3,003.5 Mb, n = 9) insects (Figure 1A and Table S2B). Collectively, the similarities and differences in GS showed a great diversity of GS within different insect clades and groups, implying distinct evolutionary histories of insect GS among the studied taxa.
Evolutionary modes and ancestral states of insects GS
To further elucidate the evolutionary history responsible for the observed GS variation pattern, we modeled GS trait evolution within various insect groups and orders. To this end, we first assembled a phylogeny containing 1,256 species out of the 1,326 records in our above GS dataset through NCBI taxonomy database (see method details). Based on this phylogeny, we tested three commonly used models, including Brownian motion (BM), Ornstein-Uhlenbeck (OU) and Early Burst (EB) model, to check which of these evolutionary models could best explain the evolution pattern of 1,256 insects GS using the R package geiger (Harmon et al., 2008). Ignoring the phylogenetic structure (i.e. without phylogenetic signal), the white-noise model (WN), was used as the control. The BM model depicts a stochastic process, thus suggesting that changes in GS were random events. Interpretation of the OU model suggests that changes in GS are the result of selection pressure with a central tendency toward the evolutionary optima. By contrast, the EB model depicts an evolutionary process that varied over time, beginning at a relatively fast rate, then gradually slowing. When all 1,256 insects were considered as a whole, the OU model showed the best fit for log scale representation of GS (Table 2), which also generally showed the best fit for species within the holometabolous orders Coleoptera, Diptera, Hymenoptera, and Lepidoptera (Akaike weights, AW = 0.988, 0.999, 0. 995, 0.996, respectively, Table S3). In contrast, The BM model showed a better fit with the hemimetabolous orders Odonata, Orthoptera and Hemiptera (AW =0.588, 0.599, 0.462, respectively). Taken together, these results showed that GS evolution in Holometabola and Insecta followed the OU model whereas the Hemimetabola was more fit by the BM model, suggesting that adaptive evolution was responsible for GS in insects overall but has less impact on the Hemimetabola.
Table 2.
Summary of best-fit models and estimated ancestral GS in different insect clades
| Taxon | No. of Species analyzed | Best-fit model | Sigma2 | alpha | z0 | Estimated ancestral genome size (Mb) |
|---|---|---|---|---|---|---|
| Insecta | 1,256 | OU | 0.005197 | 0.0178093 | 3.029 | 1,069.05 |
| Odonata | 112 | BM | 0.00115 | 2.972 | 937.56 | |
| Neoptera | 1,135 | OU | 0.005605 | 0.01816975 | 2.931 | 853.10 |
| Polyneoptera | 105 | OU | 0.00389 | 0.0164262 | 3.426 | 2,666.86 |
| Orthoptera | 60 | BM | 0.002146 | 3.678 | 4,764.31 | |
| Paraneoptera | 93 | BM | 0.003993 | 2.709 | 511.68 | |
| Hemiptera | 86 | BM | 0.004062 | 2.951 | 893.31 | |
| Holometabola | 937 | OU | 0.006662 | 0.0399724 | 2.641 | 437.52 |
| Hymenoptera | 216 | OU | 0.005385 | 0.0447022 | 2.52 | 331.13 |
| Coleoptera | 271 | OU | 0.007993 | 0.0413444 | 2.861 | 726.11 |
| Lepidoptera | 154 | OU | 0.004831 | 0.0715872 | 2.695 | 495.45 |
| Diptera | 290 | OU | 0.007891 | 0.0419134 | 2.523 | 333.43 |
Note. All GS data was log10 transformed for model fitting. BM and OU are short for Brownian motion and Ornstein–Uhlenbeck models; Sigma2 represents estimates of rate of size evolution by maximum likelihood (ML); alpha represents the strength of central attraction under the OU models; z0 represents estimated values of the ancestral root state. More parameters in model-fitting are presented in Table S3.
We next traced the ancestral states of GS in the major insect clades or orders by comparing the z0 index from model fitting, which represented the predicted GS value for the root state of the phylogeny. We found that the ancestral GS of insects estimated from 1,256 species was likely around 1,069.05 Mb (Table 2), while the ancestral GS of Neopterans (including Polyneoptera, Paraneoptera and Holometabola) was estimated to be 853.1 Mb. Estimation for Polyneopterans revealed an ancestral GS of 2,666.9 Mb, suggesting this group experienced a dramatic increase from that of their Neopteran ancestors (Figure 1B). Conversely, the GS of Paraneoptera and Holometabola appeared to dramatically decrease from their ancestors to 511.68 and 437.52 Mb, respectively. These findings suggested that GS could evolve from the ancestral node along opposite trends within the infraclass Neoptera. At the Order level, Orthopterans were estimated to possess the largest ancestral GS of 4,764.21 Mb, thus showing extraordinary genomic expansion compared with that of their Polyneopteran ancestors. Coleopterans, which are holometabolous, showed a relatively small increase in estimated ancestral GS (726.11 Mb) compared with that of Holometabola.
In general, the observed GS of extant insects indicated a wide range of genome expansion or contraction compared with estimated ancestral GS. We next examined what proportion of insect species experienced a significant genome expansion or contraction, defined as an increase or decrease of larger than 10%. The results indicated that 16.8% of insects underwent a genome expansion, from that of their progenitors, while 76.5% of species exhibited genome contraction. Notably, GS appeared similar with the ancestral state in only 6.6% of the insect species examined here. Among the different orders, a high number of species within Diptera, Coleoptera and Hemiptera underwent genome contraction, whereas more species in Orthoptera and Hymenoptera experienced a significant genome expansion compared with their respective ancestral GS (Figure S1). These results implied that changes in GS occur frequently in the evolutionary history of insects, leading to high variation in the present state of GS among insects.
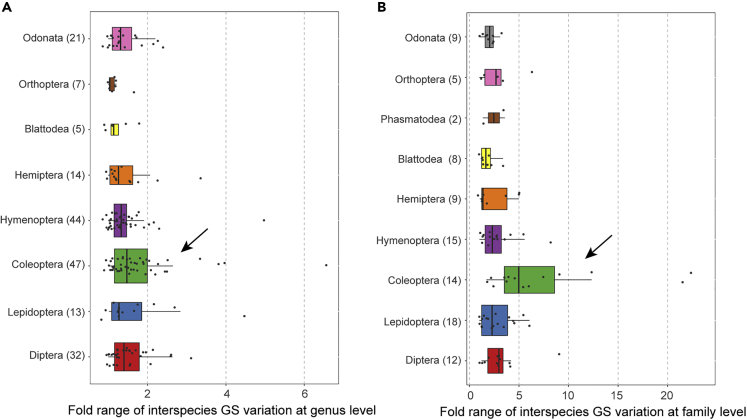
Large interspecies variation in GS within Coleoptera at the genus and family levels
To investigate differences in GS difference among closely related species, we next compared variation in GS between species within genera and within families. The results showed that differences in GS among species of the same genus were generally restricted to less than 2-fold (i.e., 159 out of 187 genera, or 85%, had two or more species with this low variation), suggesting that GS was largely similar within genera. However, we did find genera with large interspecies variation in GS, such as genus Chrysolina in Coleoptera. In particular, we noted that the GS of the leaf beetle Chrysolina americana was 547.7 Mb, while that of Chrysolina carnifex was 3,608.8 Mb, a 6.59-fold range in GS (Figure 2A andTable S4A). However, interspecies comparisons within families showed that GS most commonly (i.e., in 57 of 96 families, or 60%) differed by greater than 2-fold, and by greater than 5-fold in 13 of these families. The largest fold change was observed within Family Curculionidae of Coleoptera (n = 9), which varied from 208 Mb in Dendroctonus ponderosae to 4,910 Mb in Aramigus tessellatus (a 24-fold difference with a coefficient of variation (CV) of 118.9%) (Figure 2B and Table S4B). These findings suggested that Coleopterans apparently showed greater intra genus and intrafamily variation in GS than other clades (Figure 2).
Figure 2.
Fold range (Max./Min.) of interspecies genome size variation at the (A) genus and (B) family level
Numbers in the brackets represent numbers of the investigated taxa (n ≥ 2) in corresponding insect orders. Coleopterans show larger interspecies variation in GS at both genus and family level as indicated by the arrows. See also Table S4.
TEs and non-coding regions drive GS variation between closely related insect species
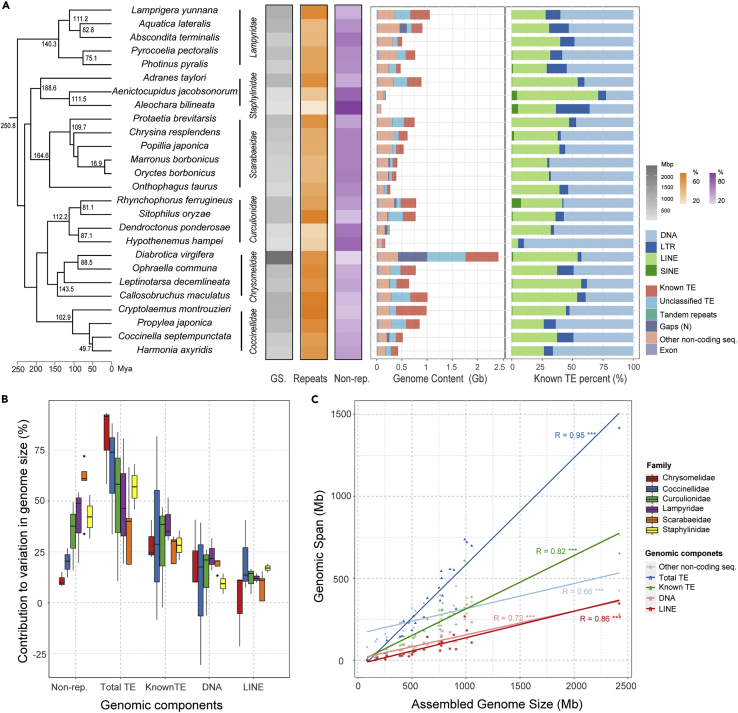
We next sought to identify the main genetic factors contributing to large changes GS in those closely related species. To this end, we quantitatively compared the genome contents of species within the same family level that showed greater than two-fold changes in GS. Since we observed significant variation in Coleopteran GS, we specifically selected 26 beetles from six Coleopteran families for further analysis, as well as 30 other species from three families across three different orders that had high quality genome assemblies (BUSCO value >80%) (Table S5). We found that, among these species, the fold difference in the size of assembled genomes within families ranged from 2.36 in Nymphalidae of Lepidoptera to 10.3 in Staphylinidae of Coleoptera (Table S7A). Since whole genome duplication (WGD) events have been reported to lead to rapid increases in GS for plants and some vertebrates (Ravi and Venkatesh 2008; Hallinan and Lindberg 2011; Soltis and Soltis 2016), we first searched for evidence of possible WGDs in these species. Based on Kernel density curves of Ks age, which should show significant peak(s) if paralogous genes are the product of a WGD event, we ruled out the possibility of WGD as a driver of genome expansion in these species (Figures S2 andS3).
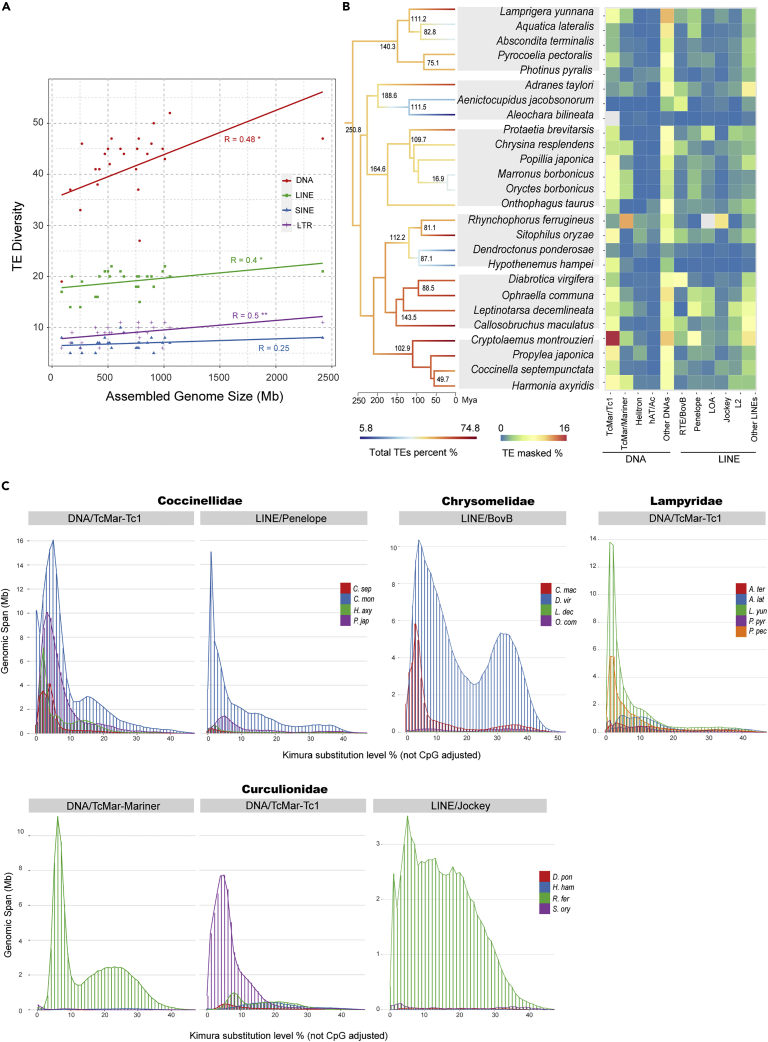
To compare the genome contents of these close species from the same family, we downloaded the genome annotation data for these species from public databases. If annotations were unavailable, we annotated the genomes (Table S5) with an in-house pipeline (see method details). In Coleoptera, genome content analysis showed that the larger genomes contained significantly more repetitive sequences compared to that in smaller genomes within the family (e.g. from 379.1 Mb in Colorado potato beetle Leptinotarsa decemlineata to 1418.8 Mb in the western corn rootworm Diabrotica virgifera, family Chrysomelidae). Furthermore, the repeat sequences mainly consisted of various and highly abundant TEs in these beetle species (e.g., 74.5% TEs in the genome of the mealybug ladybird beetle Cryptolaemus montrouzieri (GS = 988.1 Mb) in family Coccinellidae; 71.4% TE content in the genome of the cowpea weevil Callosobruchus maculatus (GS = 1,007.8 Mb) in family Chrysomelidae; 70.5% TE content in the rice weevil S. oryzae (GS = 770.6 Mb) in family Curculionidae) (Figure 3A and Table S7A).
Figure 3.
Genomic landscapes, contribution to GS and correlation between GS and genomic components of 26 beetles in six families of the order Coleoptera
(A) Genomic landscapes of 26 beetles. Time-calibrated phylogenetic relationship inferred from the BUSCO core gene sets using ML (see method details) is shown on the left, along with the heatmaps of assembled GS (gray), the coverage of repetitive (orange) and non-repetitive (purple) content (%) in each lineage. Genome contents divided into six major genomic components are shown by bar charts on the right, along with the relative abundance of annotated known TEs. See also Tables S5, S6 and S7A.
(B) Contribution of genomic components to the variation in GS among the six individual families of the order Coleoptera, which are calculated compared to the species with the minimum GS in each family. Colors of the boxplots represent the six investigated families. Legends are shown on the far right.
(C) Correlation between assembled GS and contents of the genomic components in 26 beetles. Numbers following the regression lines correspond to Pearson’s correlation coefficient under the phylogenetic independent contrast (PIC) with pvalues (∗∗∗<0.001; 0.001≤∗∗<0.01; 0.01≤∗<0.05). See also Table S8A.
The total content of non-repetitive regions, including exons and other non-coding regions, also varied substantially among these closely related beetles (e.g. from 80 Mb in the rove beetle A. bilineata to 333.3 Mb in A. taylori, family Staphylinidae), although to a lesser extent than repetitive sequences (Figure 3B). However, because exon regions showed comparatively little variance (30 Mb at average level) (Table S6), this observed variability primarily arose from non-coding regions, such as introns and intergenic non-coding DNA. Generally, both TEs and non-coding regions contributed to changes in GS in these beetle species, supported by a significant correlation between total genomic content of TEs and non-coding sequences and GS under phylogenetic correction (r = 0.95, p = 4.53E-13; r = 0.66, p = 3.57E-04, respectively) (Figure 3C and Table S8A). This trend was also evident in correlation analyses of species in family Nymphalidae Order Lepidoptera (Figure S4B and Table S8A). By contrast, the total content of coding sequence of exons was similar across all tested species (20–45 Mb), regardless of differences in total GS (Table S6). In particular, TEs provided the greatest contribution to variation in GS variation and showed a significant positive correlation with GS in both Nymphalidae (r = 0.91, p = 0.0002) and Braconidae (r = 0.97, p = 2.09E-08) corrected by the phylogeny (Figures S4B and S7B and Table S8A).
Interestingly, we found that family Chironomidae of Diptera was an exception to this overall trend in that TEs accounted for only a small fraction of their genomes, ranging from 2.5% in the marine midge Clunio marinus to 8.5% in Chironomus tentans (Figure S9A and Table S7b). Although the total content of TEs was still positively associated with GS in this family (Pearson’s test corrected by phylogeny; r = 0.996, p = 0.0045) (Figure S9B and Table S8A), they contributed little to GS variation between Chironomidae species. Instead, we found that non-coding regions containing non-repetitive sequences ranged from 55.27 Mb in C. marinus to 134.63 Mb in C. tentans in these five midges, accounting for >60% of their genome. Further analysis showed that the total length of non-coding regions was significantly positively correlated with GS, and moreover, provided the largest contribution among different sequence types to intrafamily variation in GS among midge genomes (Pearson’s test corrected by phylogeny; r = 0.98, p = 0.02). Taken together, except in some specific families, the TEs mostly dominated changes in GS among closely related species.
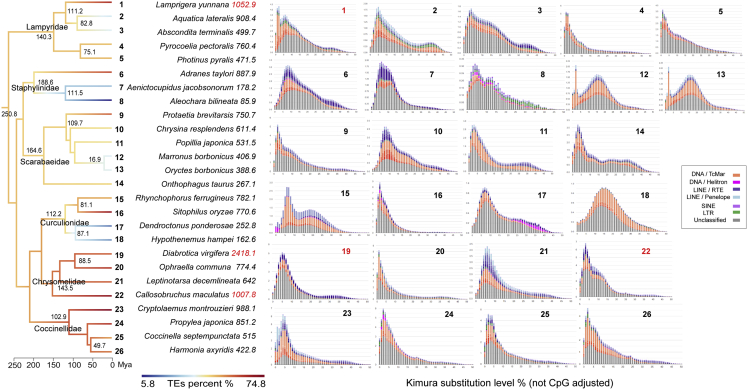
Intrafamilial history of TE expansion
Since GS is strongly affected by the proliferation of TEs in the vast majority of insect families examined here, we next investigated the putative history of TE expansion in various species by estimating Kimura distance between TEs and their consensus sequences to determine whether abrupt events or gradual increase resulted in the current GS. In Coleoptera, the distribution of Kimura distance of five species, such as the red palm weevil Rhynchophorus ferrugineus in Curculionidae, showed a bimodal shape of Kimura distance distribution, with a second peak around 0.2, reflecting the continuous accumulation of TEs (Figure 4). However, eight species such as the coffee berry borer Hypothenemus hampei, showed a bell curve distribution of Kimura distance, with a peak of K2P divergence around 0.15, indicating a relatively ancient burst of TE expansion. Thirteen species, including beetles with GS of more than 1Gb, had an L-shaped distribution with a peak below 0.1, indicating a recent burst of TE transposition. This trend was obvious in common buckeye J. coenia, which had the largest GS we observed in family Nymphalidae of Lepidoptera with an L-shaped distribution with a sharp peak below 0.05 (Figure S5C), and the parasitoid wasp of apple maggot, D. alloeum, in family Braconidae of Hymenoptera (Figure S8). These results suggested that recent TE expansions commonly determine GS in most of the insect species/families examined here.
Figure 4.
TE divergency landscapes of 26 beetles in Coleoptera
Branches on the time-calibrated phylogeny are colored according to the total abundance of TEs. Species’ Latin names are followed by their GS number, GS >1 Gb are in red. Each species is numbered, and each number corresponds to a small graph on the right showing the TE divergency of species. The yaxis represents genome coverage of various types of TE, and the xaxis represents Kimura substitution level % (TE divergency based on the Kimura distance of TE copies to the corresponding consensus sequences) from 0 to 50. The landscapes are categorized as L-shaped types (the peak value of TE divergency is less than 5), bell-curved types (the peak value of TE divergency is larger than 5) and bimodal-shaped types (two main peaks). See also Figures S10–S15 for zoomed graphs.
Various TEs drive GS changes in different clades of insects
We next investigated differences in the contributions of specific TE types to the observed variation in GS. To this end, we further analyzed the diversity of DNA transposons and retrotransposons in each species, and calculated the abundance of each TE type.
Considering that the number of annotated TE families can serve as an indicator of TE diversity, we tested the relationship between TE diversity and GS. The results showed significant positive correlations between GS and TE diversity of DNA transposons, long interspersed nuclear elements (LINEs), and long terminal repeat retrotransposons (LTRs) across Coleopteran beetles (r = 0.48, p = 0.014; r = 0.4, p = 0.046; r = 0.5, p = 0.009, respectively) (Figure 5A). Similar positive correlations were also observed in other tested clades (Figures S4C and S7C, Table S8B). These findings indicated that the number of TE types detectable in a genome increased with GS. However, when further quantifying these TEs, the major contributions to changes in GS could be attributed to a limited set of specific TEs (Figure 5B and Table S9A). For example, in Coleoptera, DNA/TcMar and three superfamilies of LINE elements such as Penelope, Jockey, and RTE/BovB contributed the most to variation in GS and the landscapes of these TEs expansion history demonstrated considerable transposition activity among different families (Figure 5C).
Figure 5.
Density, genomic abundance and divergency of TEs in 26 beetles
(A) Correlation between TE diversity (the numbers of the annotated TE families) and the assembled GS of 26 beetles in the order Coleoptera. Pearson correlation coefficients are noted near the regression lines with pvalues (∗∗∗<0.001; 0.001≤∗∗<0.01; 0.01≤∗<0.05). See also Table S8B.
(B) Time-calibrated phylogeny of 26 beetles with total TEs coverage mapped on the multi-colored branches. Heatmap on the right represents genome coverage of the top five TEs among all TE types. See also Table S9A.
(C) Kimura substitution level % (TE divergency based on the Kimura distance of TE copies to the corresponding consensus sequences) of several primary TE families which contribute most to variation in GS in four investigated Coleoptera families separately. Coccinellidae: C. sep, Coccinella septempunctata; C. mon, Cryptolaemus montrouzieri; H. axy, Harmonia axyridis; P. jap, Propylea japonica; Chrysomelidae: C. mac, Callosobruchus maculatus; D. vir, Diabrotica virgifera; L. dec, Leptinotarsa decemlineata; O. com, Ophraella communa; Lampyridae: A. ter, Abscondita terminalis; A. lat, Aquatica lateralis; L. yun, Lamprigera yunnana; P. pyr, Photinus pyralis; P. pec, Pyrocoelia pectoralis; Curculionidae: D. pon, Dendroctonus ponderosae; H. ham, Hypothenemus hampei; R. fer, Rhynchophorus ferrugineus; S. ory, Sitophilus oryzae.
In the family of Coccinellidae, DNA/TcMar-Tc1 transposons accounted for more than 160 Mb in C. montrouzieri (16.5% of the genome), ∼130 Mb more than that in Harmonia axyridis. The Kimura distance distributions for both DNA/TcMar-Tc1 transposons and LINE/Penelope elements followed the distribution with a sharp peak around 0–0.1, suggesting a recent burst of transposition by these two types of TE.
In Family Chrysomelidae, the total content of LINE/RTE-BovB retrotransposons reached 204.3 Mb in D. virgifera, accounting for more than 8% of the whole genome, apparently increasing its GS by ∼200 Mb over that of its closest relative Ophraella communa after speciation ∼88.5 Mya. In the Curculionidae family, both DNA/TcMar-Mariner and LINE/Jockey elements accounted for ∼180 Mb in R. ferrugineus (∼23% of genome and 45.4% of total TEs). The blunt peak distribution of Kimura distance for LINE/Jockey, bimodal distribution for LINE/RTE-BovB and DNA/TcMar-Mariner elements all suggested that these transposon types accumulated in a sustained manner over a relatively long evolutionary period in this family.
Among the 11 butterfly species we examined in the Lepidopteran family Nymphalidae, short interspersed nuclear elements (SINEs), especially SINE/tRNA elements accounted for a significant expansion (∼80 Mb) in both GS of Helioconius species H. himera and H. hermathena (Figure S6 and Table S9B) since speciation from their most recent common ancestor 7.7 Mya ago. Kimura distributions of SINE/tRNA TE divergence showed a higher peak below 0.3, indicating further accumulation following the initial transposition burst. In summary, various TEs drive GS changes in different clades of insects. Moreover, either recent bursts or the gradual accumulation of specific types of TEs can explain major changes in GS.
Discussion
The mechanisms driving variation in GS have posed a long-standing question for evolutionary biologists. As one of the most species-rich animal clades, insects represent a good system for studying the mechanism of GS evolution due to wide variability among closely related species. Here, we collected GS data for 1,326 insect species and characterized the changes and contributing factors for 1,256 species within a phylogenetic framework. We found that GS evolution of the holometabolous insects was best-fitted with the OU model, an adaptive process under the “stabilized selection”. In contrast, the hemimetabolous favored the BM model, a neutral evolutionary process. However, since the OU process best explained GS evolution for insects overall, we speculated that selection might still be an important driving force to shape the GS in insects, even in Hemimetabola. Insect genome experienced massive expansion and contraction events under such different evolutionary processes among clades, thus resulting in considerable differences in GS between close species (i.e., within a family) as we observed.
The causes driving these changes in GS have been widely debated and can be generally summarized as the product of either DNA accumulation or DNA elimination. The main mechanisms of DNA accumulation include WGD (Van DePeer et al., 2017), or TE (Maumus et al., 2015; Gilbert et al., 2021) and satellite DNA (Shah et al., 2020; Majid and Yuan 2021; Stelzer et al., 2021) proliferation. WGDs are widely detected in plant species (Soltis et al., 2003; Vu et al., 2015; Soltis and Soltis 2016) and have been reported in animals such as the teleost fish (Ravi and Venkatesh 2008) and mollusks (Hallinan and Lindberg 2011), whereas little evidence has emerged showing WGDs in insects (Li et al., 2018; Roelofs et al., 2020). Here, we also present strong evidence that WGD events did not occur in the insects we examined with large GS, therefore leading us to conclude that it does not likely contribute to variation in GS among close insect species.
Through quantitative characterization of a large set of high-quality insect genomes, our findings reveal that the abundance of various TE types is the major source of differences in GS between close species. That is, variations in GS are predominantly the result of TE accumulation in most insects. Detailed analysis of TE composition showed that DNA transposons or LINE retrotransposons are the main contributors to GS expansion in Coleopteran families. While SINE elements comprise a large proportion of TEs and dominate GS variation in Heliconius butterflies of Family Nymphalidae in Lepidoptera.
Since the transposition of retrotransposons follows a general principle of “copy and paste” (Boeke et al., 1985), genomic expansion driven by increased retrotransposon activity, especially LTR elements, has been widely reported, especially in plants (Sanmiguel and Bennetzen 1998; Piegu et al., 2006; Zedek et al., 2010; Nystedt et al., 2013; Macas et al., 2015; Zhang et al., 2020). Here, we provide additional evidence that LINEs and SINEs can also provide major contributions to changes in GS. In contrast with retrotransposons, most DNA transposons, except for Helitrons and Maverick/Polinton elements (Grabundzija et al., 2016), follow a principle of “cut and paste” transposition (Greenblatt and Brink 1963; Rubin et al., 1982), and thus does not increase their copy numbers or GS despite increased transposition activity. Notably, a relatively unusual abundance of these elements appears responsible for GS enlargement in the dryinid wasp Gonatopus flavifemur (Yang et al., 2021), as well as some beetle genomes examined in this study (Figure 3B). One hypothesis that could explain this effect on GS is cross-species horizontal transposon transfer (HTT) (Schaack et al., 2010; Wallau et al., 2018). Massive HTT events have been reported in insects, with one study estimating that more than 2,000 HTT events occurred over the last 10 Mya (Peccoud et al., 2017), especially among phylogenetically close organisms (Bartolome et al., 2009; El Baidouri et al., 2014). In particular, DNA transposons appear to be more frequently transferred than retrotransposons due to relatively simple structure that is more conducive to persistence in and transfer by the transmission vectors (Gilbert and Feschotte 2018). In the genus of Aedes, TEs that are frequently exchanged through HTT contribute to more than 6% of whole genome and thus drive GS expansion in mosquitoes (Melo and Wallau 2020). More relative explorations are still needed for further understanding the causes for content differences of these active genomic elements among close species.
Beyond that, our results led us to deduce that the evolution of GS is mainly driven by the activity of TE transposition in insects. The activation of TEs is subject to host cues, especially in early stages of gametogenesis and embryo development in which epigenetic reprogramming also enables cell developmental totipotency (Surani and Hajkova 2010; Rowe and Trono 2011). In higher eukaryotes, several surveillance systems have evolved to modulate TE activity. These systems mainly involve epigenetic transcriptional regulation, such as DNA methylation and histone modifications (Karimi et al., 2011; Groh and Schotta 2017) or small RNA-based pathways, such as small interfering RNAs (siRNAs) (Kasschau et al., 2007; Fagegaltier et al., 2009) and PIWI-interacting RNAs (piRNA) (Sienski et al., 2012; Le Thomas et al., 2013; Ernst et al., 2017). In germline cells, piRNA pathways are dedicated to suppressing TE transposition activity and maintaining genomic integrity (Shen et al., 2018; Zhang et al., 2018a). Following vertical transmission of a whole genome from progenitor species, both vertically and horizontally transferred TEs are widely activated in germline cells, copying and inserting themselves across the insect genome. Regulators of the piRNA pathway target potentially deleterious TEs for inactivation, although a proportion of these mobile elements successfully propagate (Wang et al., 2018). Once they escape this initial repression mechanism, the rate of TE amplification is very high (Biemont and Vieira 2006; Naito et al., 2006), and these expanded TEs are then passed to successive generations in reproductive cells.
New TE insertion events also introduce mutations with a range of subsequent effects, including alterations in gene expression, that can affect both genotype and phenotype (Wells and Feschotte 2020; Almojil et al., 2021; Gilbert et al., 2021). Those mutations that lead to positive effects on competitive fitness in the host are retained through positive selection by specific environmental conditions (Wilson 1993; Nagaraju 2001; Daborn et al., 2002). These beneficial mutations become fixed in the expanded genome throughout the population, ultimately leading to new speciation events. A large proportion of the TE insertion mutations contribute relatively neutral effects and are strongly influenced by randomgenetic drift rather than positive selection (Arkhipova 2018). For these mutations, their contribution to genome expansion is eventually regulated by the effective population size of species. In addition, GS is directly correlated with cell metabolic capacity, and bloated or excessive DNA content will strain the cellular energy metabolism normally required for replication and proliferation (Cavalier-Smith 1978, 2005; Gregory and Hebert 1999). Under this cell-level selection, TE sequences under less evolutionary constraint are prone to deletion to reduce GS by replication slippage or genomic rearrangement, such as through illegitimate and unequal homologous recombination (Gray 2000; Devos et al., 2002).
These collective evolutionary processes could thus explain the variation in GS we detected between close insect species. Whether mutations caused by TE insertions become fixed in the population or TE proliferation is silenced by cellular regulatory mechanisms are all subject to probability. According to this hypothesis, the process of expanding and contracting total GS is always dynamic. In general, further investigations are necessary to better understand the mechanisms by which transposons proliferate or are suppressed and deleted in germline cells, and may shed lights on the evolution of the insect genome and insect genome size.
Limitations of the study
Utilizing published GS data from flow cytometry, we provided a comprehensive analysis of insect GS variation. Nonetheless, compared with millions of insect species recognized by humans so far, the number of insects with GS data are still limited. In addition, information is still absent in some clades such as Neuropterida (snakeflies, dobsonflies and net-winged insects). Selecting more species to fill this gap would deepen the understanding of insect GS evolution. Furthermore, we modeled the evolutionary history of insect GS based on the NCBI taxonomic tree, which is a simplified phylogenetic framework. Producing an ultrametric time-scaled tree covering more species would provide more information for insect GS evolution.
STAR★Methods
Key resources table
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Fei Li (lifei18@zju.edu.cn).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
We examined flow cytometry-based published GS data from 1,326 insect species, spanning 700 genera, 155 families, and 21 orders. The GS dataset (Table S1) was curated from two sources: 1,145 species from the Animal Genome Size Database (http://www.genomesize.com) (Gregory 2005a) and 181 species from different literature sources. The records of GS estimates in our dataset were all measured based on experimental methods, predominantly flow cytometry. As to the 1,345 whole insect GS records from the Animal Genome Size Database (last accessed December 2020), we refined them manually by the following rules: (1) GS of unknown species were removed; (2) GS of the female insects were kept if records of both sexuality are available; (3) GS of the larger insects were kept if the conditions for the measurements of one species records were the same; (4) GS measured by flow cytometry (FCM), Feulgen densitometry (FD), and Feulgen image analysis densitometry (FIA) were prioritized; (5) GS measured from head or brain tissue was preferred (DeSalle et al., 2005; Johnston et al., 2019); and (6) GS was calculated for the arithmetic mean for the range-type records. We then did a large-scale literature review from studies published before December 2020 using web search engines (Scopus, Web of Science, Google Scholar, and Google Search) with the combinations of “genome size”, “C-value”, and “insect” as keywords for the title or abstract. Most of the available GS search results came from insect genome-sequencing papers from the past five years. We extracted the GS data that was measured by flow cytometry rather than sequencing from these results to add to our dataset. We further refined this GS information using the principles mentioned for the filtering of genomes from the Animal Genome Size Database. All scientific names and species taxonomic information in the GS dataset were subsequently cross-referenced to the NCBI Taxonomy database according to taxize package v0.9.99 (Chamberlain and Szocs 2013) on R studio v4.0.4 (Racine 2012).
Method details
Phylogenetic analyses of insect GS
We assembled the phylogenetic relationships of insects based on the NCBI taxonomic tree using a combination of the function class2tree using the R package taxize (v0.9.99) and the information of 1,256 species out of the whole 1,326 records from 21 orders can be searched from insect GS dataset. These 1,256 species with GS data were then mapped onto the cladogram tree for the following analysis of trait evolution analysis. We collapsed unresolved multichotomies in the internal nodes of the tree randomly with the function multi2di of R package phytools v0.7-70 (Revell 2012). All GS values were log-transformed before performing the following analysis.
We fit the evolutionary models for 1256 insect GSs implemented in the fitContinuous function in the geiger package v2.0.7 (Harmon et al., 2008). In addition to the Brownian motion (BM) model (“random walk” of trait evolution), the Ornstein–Uhlenbeck (OU) model (a stable adaptive peak of trait evolution) and the Early Burst (EB) model (declining trait evolution rates through time) were also taken into consideration. Using the stochastic white-noise process (WN) as a blank control, we compared these models by the weighted values of corrected Akaike Information Criterion (AIC) according to the function of aic.w in the phytools package v0.7-70 (Table S3). The GS data of 1,256 insects was superimposed onto the phylogeny for each group using the function contMap in the phytools package v0.7-70.
Genome content analyses among species
Genomic data sources
56 insect species in 9 families from 4 orders were selected to conduct comparative genomic analyses. The genome assemblies and transcriptomes of 34 of these insects, their annotated protein sequence files, and their genome annotation GFF files are publicly available from NCBI (https://www.ncbi.nlm.nih.gov/assembly/), GigaDB (http://www.gigadb.org/), InsectBase 2.0 (http://v2.insect-genome.com/), and BIPAA (https://bipaa.genouest.org/is/). Genome annotation information of the other 22 species was generated in this study (see genome annotation section). All detailed download information is shown in Table S5.
Assessment of the genome assemblies and construction of the phylogenetic framework of the four insect clades
We assessed the quality of genome assemblies of 56 insects by using the BUSCO software v3.0.2b (Simao et al., 2015) (Table S5). Phylogenetic constructions of these four insect clades using ortholog genes from assembled genomes of the 56 investigated species and four outgroup species (Ignelater luminosus, Coleoptera; Venturia canescens, Hymenoptera; Zerene cesonia, Lepidoptera; Drosophila melanogaster, Diptera; Table S5) were identified against the 1,658 BUSCO single-copy reference genes (insect_odb9). The BLAST search e-value was set at a threshold of 1e-10 to filter for high-quality “Complete” ortholog genes in at least 90% of the selected species genomes for the following analyses. MAFFT software v7.310 (Katoh and Standley 2013) was used under the E-INS-i strategy to conduct sequence alignments and trimAl 1.2rev59 software (Capella-Gutierrez et al., 2009) using the “gappout” option to trim ambiguous amino acid sequence loci of 1,385 orthologues in Coleoptera, 868 orthologues in the Braconidae family of Hymenoptera, 1,107 orthologues in the Chironomidae family of Diptera, and 981 orthologues in the Nymphalidae family of Lepidoptera. The trimmed alignments were then concatenated into a supermatrix separately implemented on FASconCAT-G v1.04 (Kuck and Longo 2014). Finally, iqtree v1.6.10 (Nguyen et al., 2015) was used to infer the maximum likelihood (ML) concatenated tree of the super amino acid matrix with the parameters of “-MFP-bb 1000” for the best-fit model and 1000 times ultrafast bootstrapping for the four investigated clades. To further estimate divergence time, phylogenies in these four clades were calibrated in r8s v1.81 (Sanderson 2003) via the penalized-likelihood (PL) and Truncated-Newton (TN) algorithm. In the 26 Coleopterans studied, five nodes were fixed with Curculionidae (155.8 Mya), Chrysomelidae (143.5 Mya), Phytophaga (160–180 Mya), Cucujiformia (199.1–210.8 Mya), and the most recent ancestral (MRCA) node of Staphylinidae and Scarabaeidae (205–215) (Zhang et al., 2018b). In the family of Braconidae in Lepidoptera, two nodes were fixed with the MRCA of C. vestalis and M. cingulum (80–150 Mya) and the MRCA of V. canescens and C. vestalis (120–180 Mya) (Peters et al., 2017). In the family Nymphalidae of Lepidoptera, four nodes were fixed with the MRCA of H. himera and H. hermathena (3.15 Mya) (Kozak et al., 2015), J. coenia and H. hermathena (37–50 Mya), B. anynana and J. coenia (40–60 Mya), and D. plexippus and A. hyperantus (60–80 Mya) (Kawahara et al., 2019). In the family Chironomidae of Diptera, two nodes were fixed with Orthocladinae (160 Mya) and MRCA of B. antarctica and P. steinenii (200–225 Mya) (Cranston et al., 2012).
Repeat sequence identification, quantification, and TE activity analyses
We annotated the repeat sequences of 56 selected species using both de novo and homology-based methods. We used RepeatModeler v2.0.1 (Flynn et al., 2020) to generated species-specific consensus sequence libraries using the flag of “-LTRStruct”. Thisstep integrated the results of three distinct algorithms: RepeatScout v1.0.6 (Price et al., 2005) and RECON v1.08 (Bao and Eddy 2002) for de novo detection and LTR-retriever v2.9.0 (Ou and Jiang 2018) for additional LTR structure detection. We then used RepeatMasker v4.0.7 (http://www.repeatmasker.org) to screen and classify these repetitive sequences against our de novo consensus sequence library and Dfam (v20171107) (Hubley et al., 2016) by the comparison program of RMblast (v2.10.0) (Tarailo-Graovac and Chen, 2009). To calculate the precise quantitative information of the annotated TEs, we utilized “one code to find them all” (Bailly-Bechet et al., 2014), a perl tool that parses the RepeatMasker out-files to avoid multiple-counted TE fragments, thus producing a more accurate statistics for each type of TE.
We estimated the relative activity of TEs by calculating the Kimura substitutional distances between the annotated TE sequences and their consensus sequences through the perl script calcDivergenceFromAlign.pl in RepeatMasker software v4.0.7 under the mode “noCpG”.
Genome annotation, content measuring of genomic components and correlation with GS
We first downloaded available genome annotation information for 34 out of 56 selected species (Table S5). As the rest of 22 species did not have this information available, we downloaded their genome assemblies to conduct our own genome annotation using the following uniform pipelines.
After repeats masking (see Repeats identification parts), protein coding genes were predicted through approaches combining ab-initio gene models and homology and transcriptome evidence-based models. We conducted the ab-initio prediction using the BRAKER2 pipelines v2.1.5 (Bruna et al., 2021) with the mode of “BRAKER with proteins” against the Arthropoda protein database (odb10) obtained from OrthoDB (Zdobnov et al., 2021). This pipeline utilizes GeneMark-ES/ET/EP v4.68 (Bruna et al., 2020) and AUGUSTUS v 3.3.2 (Keller et al., 2011) for gene training. We performed the transcriptome-based prediction on all the genomes of species with available RNA-seq data (Table S5). HISAT2 v2.1.0 (Zhang et al., 2021) and StringTie v2.1.5 (Kovaka et al., 2019) were used to align the transcriptome to the genome and generate a genome-guided transcriptome using the default arguments. TransDecoder v 5.5.0 (https://github.com/TransDecoder/TransDecoder) was then used to identify the intact coding regions in transcriptomes above. We made our homology-based gene predictions using GenomeThreader v1.7.1 (Gremme et al., 2005) against the arthropoda protein database mentioned above. Finally, we used EVidenceModeler (EVM, v1.1.1) (Haas et al., 2008) with default settings to merge all of our gene prediction evidence above into weighted consensus gene structures and PASA v2.4.1 (Haas et al., 2003) to update the EVM output with the gene structure based on the transcript alignments. More detailed parameter settings for our genome annotation pipeline are available at https://github.com/meiyang12/Genome-annotation-pipeline.
We then calculated the genomic coverage of coding and non-coding regions in the non-repetitive sequences for the whole genome of 56 studied species according to the precise genome annotation information that we collected or generated. We cited the method of Francis and Worheide (2017) and used their custom Python script (bitbucket.org/wrf/sequences/gtfstats.py) to calculate the maximum exonic span length coding potential from all CDS regions in the GFF format files. This approach for exonic base calculation will merge all overlapping exon intervals from all the alternative splice sites of annotated genes. The genomic content of other non-coding regions is then counted by the total bases of the downloaded assembly (total GS) minus the repetitive length, the exonic length, and the assembly gaps (N). Considering the impact of phylogenetic similarity, the phylogenetic independent contrasts method (PIC) (Garland et al., 1992) was used to estimate the correlation of the GS between the content of different genomic components by the ape package v5.4-1 (Paradis and Schliep 2019) in R studio v 4.0.4.
Whole genome duplication detection
The potential whole-genome duplication (WGD) events of 56 selected species were detected by wgd tools (Zwaenepoel and Van dePeer 2019) based on the construction of Ks distributions from duplicated paralogous genes in the annotated protein-coding sequences. This package merges the pipelines of paralogues identification according to the all-to-all BLASTP similarity by DIAMOND (v0.9.36.137) (Buchfink et al., 2015), gene family clustering by MCL 14-137 software (Enright et al., 2002) and Ks estimation by the CODEML program on PAML 4.9j software (Yang 2007). We fitted the kernel density estimates (KDEs) to the Ks distribution density using the option of “wgd kde”. All analyses were performed under the default parameters in wgd tools.
Quantification and statistical analysis
All statistical analyses were conducted on the R studio v 4.0.4. GS data in picograms (pg) was first converted into million base pairs (Mb) (1pg = 978 Mb) for analyzing. Nonparametric significant difference analyses by Wilcoxon rank-sum one-side tests were conducted on GS data pairs that did not pass the Shapiro–Wilk test for normality (Table S2).
Acknowledgments
This work was supported by National High Technology Research and Development Program of China (2019YFD1002100, 2021YFD1400100), the Natural Science Foundation of Zhejiang Province (LY22C140005) and the National Science & Technology Fundamental Resources Investigation Program of China (2019FY100400).
Author contributions
F.L designed the research for the whole paper; F.L and K.H coordinated the project; F.L and X.H.Y revised the paper; Y.M annotated the genomes; Y.Y.C conducted data analysis and drafted the paper.
Declaration of interests
The authors declare no competing interests.
Published: September 16, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104873.
Supplemental information
Data and code availability
-
•
This paper analyzes existing, publicly available data, which are included in Supplemental file (Table S5) for details.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact on request.
References
- Alfsnes K., Leinaas H.P., Hessen D.O. Genome size in arthropods; different roles of phylogeny, habitat and life history in insects and crustaceans. Ecol. Evol. 2017;7:5939–5947. doi: 10.1002/ece3.3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almojil D., Bourgeois Y., Falis M., Hariyani I., Wilcox J., Boissinot S. The structural, functional and evolutionary impact of transposable elements in eukaryotes. Genes. 2021;12:918. doi: 10.3390/genes12060918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardila-Garcia A.M., Umphrey G.J., Gregory T.R. An expansion of the genome size dataset for the insect order Hymenoptera, with a first test of parasitism and eusociality as possible constraints. Insect Mol. Biol. 2010;19:337–346. doi: 10.1111/j.1365-2583.2010.00992.x. [DOI] [PubMed] [Google Scholar]
- Ardila-Garcia A.M., Gregory T.R. An exploration of genome size diversity in dragonflies and damselflies (Insecta: Odonata) J. Zool. 2009;278:163–173. [Google Scholar]
- Arkhipova I.R. Neutral Theory, Transposable elements, and eukaryotic genome evolution. Mol. Biol. Evol. 2018;35:1332–1337. doi: 10.1093/molbev/msy083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnqvist G., Sayadi A., Immonen E., Hotzy C., Rankin D., Tuda M., Hjelmen C.E., Johnston J.S. Genome size correlates with reproductive fitness in seed beetles. Proc. Biol. Sci. 2015;282:20151421. doi: 10.1098/rspb.2015.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly-Bechet M., Haudry A., Lerat E. One code to find them all": a perl tool to conveniently parse RepeatMasker output files. Mob. DNA. 2014;5:13–15. [Google Scholar]
- Bao Z., Eddy S.R. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 2002;12:1269–1276. doi: 10.1101/gr.88502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolome C., Bello X., Maside X. Widespread evidence for horizontal transfer of transposable elements across Drosophila genomes. Genome Biol. 2009;10:R22. doi: 10.1186/gb-2009-10-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biemont C., Vieira C. Genetics: junk DNA as an evolutionary force. Nature. 2006;443:521–524. doi: 10.1038/443521a. [DOI] [PubMed] [Google Scholar]
- Boeke J.D., Garfinkel D.J., Styles C.A., Fink G.R. Ty Elements transpose through an rna intermediate. Cell. 1985;40:491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- Bruna T., Hoff K.J., Lomsadze A., Stanke M., Borodovsky M. BRAKER2: automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. NAR Genom. Bioinform. 2021;3:lqaa108. doi: 10.1093/nargab/lqaa108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruna T., Lomsadze A., Borodovsky M. GeneMark-EP+: eukaryotic gene prediction with self-training in the space of genes and proteins. NAR Genom. Bioinform. 2020;2:lqaa026. doi: 10.1093/nargab/lqaa026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B., Xie C., Huson D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- Calatayud P.A., Petit C., Burlet N., Dupas S., Glaser N., Capdevielle-Dulac C., Le Ru B., Jacquin-Joly E., Kaiser-Arnauld L., Harry M., et al. Is genome size of Lepidoptera linked to host plant range? Entomol. Exp. Appl. 2016;159:354–361. [Google Scholar]
- Capella-Gutierrez S., Silla-Martínez J.M., Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Economy, speed and size matter: evolutionary forces driving nuclear genome miniaturization and expansion. Ann. Bot. 2005;95:147–175. doi: 10.1093/aob/mci010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Nuclear volume control by nucleoskeletal DNA, selection for cell volume and cell growth rate, and the solution of the DNA C-value paradox. J. Cell Sci. 1978;34:247–278. doi: 10.1242/jcs.34.1.247. [DOI] [PubMed] [Google Scholar]
- Chamberlain S.A., Szöcs E. taxize: taxonomic search and retrieval in R. F1000Res. 2013;2:191. doi: 10.12688/f1000research.2-191.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church S.H., Donoughe S., de Medeiros B.A.S., Extavour C.G. Insect egg size and shape evolve with ecology but not developmental rate. Nature. 2019;571:58–62. doi: 10.1038/s41586-019-1302-4. [DOI] [PubMed] [Google Scholar]
- Cranston P.S., Hardy N.B., Morse G.E. A dated molecular phylogeny for the Chironomidae (Diptera) Syst. Entomol. 2012;37:172–188. [Google Scholar]
- Daborn P.J., Yen J.L., Bogwitz M.R., Le Goff G., Feil E., Jeffers S., Tijet N., Perry T., Heckel D., Batterham P., et al. A single p450 allele associated with insecticide resistance in Drosophila. Science. 2002;297:2253–2256. doi: 10.1126/science.1074170. [DOI] [PubMed] [Google Scholar]
- Devos K.M., Brown J.K.M., Bennetzen J.L. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 2002;12:1075–1079. doi: 10.1101/gr.132102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSalle R., Gregory T.R., Johnston J.S. Preparation of samples for comparative studies of arthropod chromosomes: visualization, in situ hybridization, and genome size estimation. Methods Enzymol. 2005;395:460–488. doi: 10.1016/S0076-6879(05)95025-8. [DOI] [PubMed] [Google Scholar]
- El Baidouri M., Carpentier M.C., Cooke R., Gao D., Lasserre E., Llauro C., Mirouze M., Picault N., Jackson S.A., Panaud O. Widespread and frequent horizontal transfers of transposable elements in plants. Genome Res. 2014;24:831–838. doi: 10.1101/gr.164400.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright A.J., Van Dongen S., Ouzounis C.A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst C., Odom D.T., Kutter C. The emergence of piRNAs against transposon invasion to preserve mammalian genome integrity. Nat. Commun. 2017;8:1411. doi: 10.1038/s41467-017-01049-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagegaltier D., Bougé A.L., Berry B., Poisot E., Sismeiro O., Coppée J.Y., Théodore L., Voinnet O., Antoniewski C. The endogenous siRNA pathway is involved in heterochromatin formation in Drosophila. Proc. Natl. Acad. Sci. USA. 2009;106:21258–21263. doi: 10.1073/pnas.0809208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn J.M., Hubley R., Goubert C., Rosen J., Clark A.G., Feschotte C., Smit A.F. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl. Acad. Sci. USA. 2020;117:9451–9457. doi: 10.1073/pnas.1921046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis W.R., Wörheide G. Similar ratios of introns to intergenic sequence across animal genomes. Genome Biol. Evol. 2017;9:1582–1598. doi: 10.1093/gbe/evx103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall J.G. Chromosome structure and the C-value paradox. J. Cell Biol. 1981;91:3s–14s. doi: 10.1083/jcb.91.3.3s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner J.D., Laurin M., Organ C.L. The relationship between genome size and metabolic rate in extant vertebrates. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020;375:20190146. doi: 10.1098/rstb.2019.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland T., Harvey P.H., Ives A.R. Procedures for the analysis of comparative data using phylogenetically independent contrasts. Syst. Biol. 1992;41:18–32. [Google Scholar]
- Gilbert C., Feschotte C. Horizontal acquisition of transposable elements and viral sequences: patterns and consequences. Curr. Opin. Genet. Dev. 2018;49:15–24. doi: 10.1016/j.gde.2018.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C., Peccoud J., Cordaux R. Transposable elements and the evolution of insects. Annu. Rev. Entomol. 2021;66:355–372. doi: 10.1146/annurev-ento-070720-074650. [DOI] [PubMed] [Google Scholar]
- Grabundzija I., Messing S.A., Thomas J., Cosby R.L., Bilic I., Miskey C., Gogol-Döring A., Kapitonov V., Diem T., Dalda A., et al. A Helitron transposon reconstructed from bats reveals a novel mechanism of genome shuffling in eukaryotes. Nat. Commun. 2016;7:10716. doi: 10.1038/ncomms10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray Y.H. It takes two transposons to tango: transposable-element-mediated chromosomal rearrangements. Trends Genet. 2000;16:461–468. doi: 10.1016/s0168-9525(00)02104-1. [DOI] [PubMed] [Google Scholar]
- Greenblatt I.M., Alexander Brink R. Transpositions of modulator in maize into divided and undivided Chromosome Segments. Nature. 1963;197:412–413. [Google Scholar]
- Gregory T.R. Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma. Biol. Rev. Camb. Philos. Soc. 2001;76:65–101. doi: 10.1017/s1464793100005595. [DOI] [PubMed] [Google Scholar]
- Gregory T.R. Genome size and developmental complexity. Genetica. 2002;115:131–146. doi: 10.1023/a:1016032400147. [DOI] [PubMed] [Google Scholar]
- Gregory T.R. 2005. Animal Genome Size Database.http://www.genomesize.com [Google Scholar]
- Gregory T.R. The Evolution of the Genome. Academic Press; 2005. Genome size evolution in animals; pp. 3–87. [Google Scholar]
- Gregory T.R., Hebert P.D. The modulation of DNA content: proximate causes and ultimate consequences. Genome Res. 1999;9:317–324. [PubMed] [Google Scholar]
- Gremme G., Brendel V., Sparks M.E., Kurtz S. Engineering a software tool for gene structure prediction in higher organisms. Inf. Softw. Technol. 2005;47:965–978. [Google Scholar]
- Groh S., Schotta G. Silencing of endogenous retroviruses by heterochromatin. Cell. Mol. Life Sci. 2017;74:2055–2065. doi: 10.1007/s00018-017-2454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B.J., Delcher A.L., Mount S.M., Wortman J.R., Smith R.K., Hannick L.I., Maiti R., Ronning C.M., Rusch D.B., Town C.D., et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 2003;31:5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B.J., Salzberg S.L., Zhu W., Pertea M., Allen J.E., Orvis J., White O., Buell C.R., Wortman J.R. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008;9:R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M.W., Wray G.A. The g-value paradox. Evol. Dev. 2002;4:73–75. doi: 10.1046/j.1525-142x.2002.01069.x. [DOI] [PubMed] [Google Scholar]
- Hallinan N.M., Lindberg D.R. Comparative analysis of chromosome counts infers three paleopolyploidies in the mollusca. Genome Biol. Evol. 2011;3:1150–1163. doi: 10.1093/gbe/evr087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon L.J., Weir J.T., Brock C.D., Glor R.E., Challenger W. GEIGER: investigating evolutionary radiations. Bioinformatics. 2008;24:129–131. doi: 10.1093/bioinformatics/btm538. [DOI] [PubMed] [Google Scholar]
- Hebert F.O., Freschi L., Blackburn G., Béliveau C., Dewar K., Boyle B., Gundersen-Rindal D.E., Sparks M.E., Cusson M., Hamelin R.C., et al. Expansion of LINEs and species-specific DNA repeats drives genome expansion in Asian Gypsy Moths. Sci. Rep. 2019;9:16413. doi: 10.1038/s41598-019-52840-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjelmen C.E., Johnston J.S. The mode and tempo of genome size evolution in the subgenus Sophophora. PLoS One. 2017;12:e0173505. doi: 10.1371/journal.pone.0173505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubley R., Finn R.D., Clements J., Eddy S.R., Jones T.A., Bao W., Smit A.F.A., Wheeler T.J. The Dfam database of repetitive DNA families. Nucleic Acids Res. 2016;44:D81–D89. doi: 10.1093/nar/gkv1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapusta A., Suh A., Feschotte C. Dynamics of genome size evolution in birds and mammals. Proc. Natl. Acad. Sci. USA. 2017;114:E1460–E1469. doi: 10.1073/pnas.1616702114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston J.S., Bernardini A., Hjelmen C.E. Genome size estimation and quantitative cytogenetics in insects. Methods Mol. Biol. 2019;1858:15–26. doi: 10.1007/978-1-4939-8775-7_2. [DOI] [PubMed] [Google Scholar]
- Karimi M.M., Goyal P., Maksakova I.A., Bilenky M., Leung D., Tang J.X., Shinkai Y., Mager D.L., Jones S., Hirst M., et al. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell. 2011;8:676–687. doi: 10.1016/j.stem.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau K.D., Fahlgren N., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Carrington J.C. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 2007;5:e57. doi: 10.1371/journal.pbio.0050057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara A.Y., Plotkin D., Espeland M., Meusemann K., Toussaint E.F.A., Donath A., Gimnich F., Frandsen P.B., Zwick A., Dos Reis M., et al. Phylogenomics reveals the evolutionary timing and pattern of butterflies and moths. Proc. Natl. Acad. Sci. USA. 2019;116:22657–22663. doi: 10.1073/pnas.1907847116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller O., Kollmar M., Stanke M., Waack S. A novel hybrid gene prediction method employing protein multiple sequence alignments. Bioinformatics. 2011;27:757–763. doi: 10.1093/bioinformatics/btr010. [DOI] [PubMed] [Google Scholar]
- Kelley J.L., Peyton J.T., Fiston-Lavier A.S., Teets N.M., Yee M.C., Johnston J.S., Bustamante C.D., Lee R.E., Denlinger D.L. Compact genome of the Antarctic midge is likely an adaptation to an extreme environment. Nat. Commun. 2014;5:4611. doi: 10.1038/ncomms5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell M.G. Transposable elements and the evolution of genome size in eukaryotes. Genetica. 2002;115:49–63. doi: 10.1023/a:1016072014259. [DOI] [PubMed] [Google Scholar]
- Kirkness E.F., Haas B.J., Sun W., Braig H.R., Perotti M.A., Clark J.M., Lee S.H., Robertson H.M., Kennedy R.C., Elhaik E., et al. Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic lifestyle. Proc. Natl. Acad. Sci. USA. 2010;107:12168–12173. doi: 10.1073/pnas.1003379107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshikawa S., Miyazaki S., Cornette R., Matsumoto T., Miura T. Genome size of termites (Insecta, Dictyoptera, Isoptera) and wood roaches (Insecta, Dictyoptera, Cryptocercidae) Naturwissenschaften. 2008;95:859–867. doi: 10.1007/s00114-008-0395-7. [DOI] [PubMed] [Google Scholar]
- Kovaka S., Zimin A.V., Pertea G.M., Razaghi R., Salzberg S.L., Pertea M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019;20:278. doi: 10.1186/s13059-019-1910-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak K.M., Wahlberg N., Neild A.F.E., Dasmahapatra K.K., Mallet J., Jiggins C.D. Multilocus species trees show the recent adaptive radiation of the mimetic heliconius butterflies. Syst. Biol. 2015;64:505–524. doi: 10.1093/sysbio/syv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuck P., Longo G.C. FASconCAT-G: extensive functions for multiple sequence alignment preparations concerning phylogenetic studies. Front. Zool. 2014;11:81. doi: 10.1186/s12983-014-0081-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Thomas A., Rogers A.K., Webster A., Marinov G.K., Liao S.E., Perkins E.M., Hur J.K., Aravin A.A., Tóth K.F. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 2013;27:390–399. doi: 10.1101/gad.209841.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann R., Kovařík A., Ocalewicz K., Kirtiklis L., Zuccolo A., Tegner J.N., Wanzenböck J., Bernatchez L., Lamatsch D.K., Symonová R. DNA transposon expansion is associated with genome size increase in mudminnows. Genome Biol. Evol. 2021;13:evab228. doi: 10.1093/gbe/evab228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch I.J., Johnston E., Pellicer J., Hidalgo O., Bennett M.D. 2019. Plant DNA C-Values Database.https://cvalues.science.kew.org/ [DOI] [PubMed] [Google Scholar]
- Li Z., Tiley G.P., Galuska S.R., Reardon C.R., Kidder T.I., Rundell R.J., Barker M.S. Multiple large-scale gene and genome duplications during the evolution of hexapods. Proc. Natl. Acad. Sci. USA. 2018;115:4713–4718. doi: 10.1073/pnas.1710791115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Chang Z., Chen L., He J., Dong Z., Yang J., Lu S., Zhao R., Wan W., Ma G., et al. Genome size variation in butterflies (Insecta, Lepidotera, Papilionoidea): a thorough phylogenetic comparison. Syst. Entomol. 2020;45:571–582. [Google Scholar]
- Liu G.C., Dong Z.W., He J.W., Zhao R.P., Wang W., Li X.Y. Genome size of 14 species of fireflies (Insecta, Coleoptera, Lampyridae) Zool. Res. 2017;38:449–458. doi: 10.24272/j.issn.2095-8137.2017.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes D.M., de Carvalho C.R., Clarindo W.R., Praça M.M., Tavares M.G. Genome size estimation of three stingless bee species (Hymenoptera, Meliponinae) by flow cytometry. Apidologie. 2009;40:517–523. [Google Scholar]
- Lower S.S., Johnston J.S., Stanger-Hall K.F., Hjelmen C.E., Hanrahan S.J., Korunes K., Hall D. Genome size in north American fireflies: substantial variation likely driven by neutral processes. Genome Biol. Evol. 2017;9:1499–1512. doi: 10.1093/gbe/evx097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Conery J.S. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- Macas J., Novák P., Pellicer J., Čížková J., Koblížková A., Neumann P., Fuková I., Doležel J., Kelly L.J., Leitch I.J. In depth characterization of repetitive DNA in 23 plant genomes reveals sources of genome size variation in the Legume Tribe Fabeae. PLoS One. 2015;10:e0143424. doi: 10.1371/journal.pone.0143424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majid M., Yuan H. Comparative analysis of transposable elements in Genus Calliptamus grasshoppers revealed that satellite DNA contributes to genome size variation. Insects. 2021;12:837. doi: 10.3390/insects12090837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marburger S., Alexandrou M.A., Taggart J.B., Creer S., Carvalho G., Oliveira C., Taylor M.I. Whole genome duplication and transposable element proliferation drive genome expansion in Corydoradinae catfishes. Proc. Biol. Sci. 2018;285:20172732. doi: 10.1098/rspb.2017.2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maumus F., Fiston-Lavier A.S., Quesneville H. Impact of transposable elements on insect genomes and biology. Curr. Opin. Insect Sci. 2015;7:30–36. doi: 10.1016/j.cois.2015.01.001. [DOI] [PubMed] [Google Scholar]
- Melo E.S.d., Wallau G.L. Mosquito genomes are frequently invaded by transposable elements through horizontal transfer. PLoS Genet. 2020;16:e1008946. doi: 10.1371/journal.pgen.1008946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misof B., Liu S., Meusemann K., Peters R.S., Donath A., Mayer C., Frandsen P.B., Ware J., Flouri T., Beutel R.G., et al. Phylogenomics resolves the timing and pattern of insect evolution. Science. 2014;346:763–767. doi: 10.1126/science.1257570. [DOI] [PubMed] [Google Scholar]
- Mohlhenrich E.R., Mueller R.L. Genetic drift and mutational hazard in the evolution of salamander genomic gigantism. Evolution. 2016;70:2865–2878. doi: 10.1111/evo.13084. [DOI] [PubMed] [Google Scholar]
- Nagaraju J. Identification of a gene associated with Bt resistance in the lepidopteran pest, Heliothis virescens and its implications in Bt transgenic-based pest control. Science. 2001;81:746–747. [Google Scholar]
- Naito K., Cho E., Yang G., Campbell M.A., Yano K., Okumoto Y., Tanisaka T., Wessler S.R. Dramatic amplification of a rice transposable element during recent domestication. Proc. Natl. Acad. Sci. USA. 2006;103:17620–17625. doi: 10.1073/pnas.0605421103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naville M., Henriet S., Warren I., Sumic S., Reeve M., Volff J.N., Chourrout D. Massive changes of genome size driven by expansions of non-autonomous transposable elements. Curr. Biol. 2019;29:1161–1168e6. doi: 10.1016/j.cub.2019.01.080. [DOI] [PubMed] [Google Scholar]
- Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystedt B., Street N.R., Wetterbom A., Zuccolo A., Lin Y.C., Scofield D.G., Vezzi F., Delhomme N., Giacomello S., Alexeyenko A., et al. The Norway spruce genome sequence and conifer genome evolution. Nature. 2013;497:579–584. doi: 10.1038/nature12211. [DOI] [PubMed] [Google Scholar]
- Oggenfuss U., Badet T., Wicker T., Hartmann F.E., Singh N.K., Abraham L., Karisto P., Vonlanthen T., Mundt C., McDonald B.A., et al. A population-level invasion by transposable elements triggers genome expansion in a fungal pathogen. Elife. 2021;10:e69249. doi: 10.7554/eLife.69249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou S., Jiang N. LTR_retriever: a highly accurate and sensitive program for identification of long terminal repeat retrotransposons. Plant Physiol. 2018;176:1410–1422. doi: 10.1104/pp.17.01310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios-Gimenez O.M., Koelman J., Palmada-Flores M., Bradford T.M., Jones K.K., Cooper S.J.B., Kawakami T., Suh A. Comparative analysis of morabine grasshopper genomes reveals highly abundant transposable elements and rapidly proliferating satellite DNA repeats. BMC Biol. 2020;18:199. doi: 10.1186/s12915-020-00925-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer M., Petitpierre E., Pons J. Test of the correlation between body size and DNA content in Pimelia (Coleoptera: Tenebrionidae) from the Canary Islands. Eur. J. Entomol. 2003;100:123–129. [Google Scholar]
- Paradis E., Schliep K. Ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2019;35:526–528. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- Parisot N., Vargas-Chávez C., Goubert C., Baa-Puyoulet P., Balmand S., Beranger L., Blanc C., Bonnamour A., Boulesteix M., Burlet N., et al. The transposable element-rich genome of the cereal pest Sitophilus oryzae. BMC Biol. 2021;19:241. doi: 10.1186/s12915-021-01158-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccoud J., Loiseau V., Cordaux R., Gilbert C. Massive horizontal transfer of transposable elements in insects. Proc. Natl. Acad. Sci. USA. 2017;114:4721–4726. doi: 10.1073/pnas.1621178114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters R.S., Krogmann L., Mayer C., Donath A., Gunkel S., Meusemann K., Kozlov A., Podsiadlowski L., Petersen M., Lanfear R., et al. Evolutionary history of the Hymenoptera. Curr. Biol. 2017;27:1013–1018. doi: 10.1016/j.cub.2017.01.027. [DOI] [PubMed] [Google Scholar]
- Petersen M., Armisén D., Gibbs R.A., Hering L., Khila A., Mayer G., Richards S., Niehuis O., Misof B. Diversity and evolution of the transposable element repertoire in arthropods with particular reference to insects. BMC Evol. Biol. 2019;19:11–15. doi: 10.1186/s12862-018-1324-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov D.A. Mutational equilibrium model of genome size evolution. Theor. Popul. Biol. 2002;61:531–544. doi: 10.1006/tpbi.2002.1605. [DOI] [PubMed] [Google Scholar]
- Piegu B., Guyot R., Picault N., Roulin A., Saniyal A., Kim H., Collura K., Brar D.S., Jackson S., Wing R.A., et al. Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res. 2006;16:1262–1269. doi: 10.1101/gr.5290206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A.L., Jones N.C., Pevzner P.A. De novo identification of repeat families in large genomes. Bioinformatics. 2005;21:i351–i358. doi: 10.1093/bioinformatics/bti1018. [DOI] [PubMed] [Google Scholar]
- Racine J.S. RStudio: a platform-independent IDE for R and sweave. J. Appl. Econ. 2012;27:167–172. [Google Scholar]
- Ravi V., Venkatesh B. Rapidly evolving fish genomes and teleost diversity. Curr. Opin. Genet. Dev. 2008;18:544–550. doi: 10.1016/j.gde.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Revell L.J. phytools: an R package for phylogenetic comparative biology (and other things) Methods Ecol. Evol. 2012;3:217–223. [Google Scholar]
- Roelofs D., Zwaenepoel A., Sistermans T., Nap J., Kampfraath A.A., Van de Peer Y., Ellers J., Kraaijeveld K. Multi-faceted analysis provides little evidence for recurrent whole-genome duplications during hexapod evolution. BMC Biol. 2020;18:57. doi: 10.1186/s12915-020-00789-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe H.M., Trono D. Dynamic control of endogenous retroviruses during development. Virology. 2011;411:273–287. doi: 10.1016/j.virol.2010.12.007. [DOI] [PubMed] [Google Scholar]
- Rubin G.M., Kidwell M.G., Bingham P.M. The molecular basis of P-M hybrid dysgenesis: the nature of induced mutations. Cell. 1982;29:987–994. doi: 10.1016/0092-8674(82)90462-7. [DOI] [PubMed] [Google Scholar]
- Sanderson M.J. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. [DOI] [PubMed] [Google Scholar]
- Sanmiguel P., Bennetzen J.L. Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotransposons. Ann. Bot. 1998;82:37–44. [Google Scholar]
- Schaack S., Gilbert C., Feschotte C. Promiscuous DNA: horizontal transfer of transposable elements and why it matters for eukaryotic evolution. Trends Ecol. Evol. 2010;25:537–546. doi: 10.1016/j.tree.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Ott U., Rafiqi A.M., Sander K., Johnston J.S. Extremely small genomes in two unrelated dipteran insects with shared early developmental traits. Dev. Genes Evol. 2009;219:207–210. doi: 10.1007/s00427-009-0281-0. [DOI] [PubMed] [Google Scholar]
- Shah A., Hoffman J.I., Schielzeth H. Comparative analysis of genomic repeat content in gomphocerine grasshoppers reveals expansion of satellite DNA and Helitrons in species with unusually large genomes. Genome Biol. Evol. 2020;12:1180–1193. doi: 10.1093/gbe/evaa119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen E.Z., Chen H., Ozturk A.R., Tu S., Shirayama M., Tang W., Ding Y.H., Dai S.Y., Weng Z., Mello C.C. Identification of piRNA binding sites reveals the argonaute regulatory landscape of the C. elegans germline. Cell. 2018;172:937–951.e18. doi: 10.1016/j.cell.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G., Dönertas D., Brennecke J. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151:964–980. doi: 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simao F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Soltis D.E., Soltis P.S., Bennett M.D., Leitch I.J. Evolution of genome size in the angiosperms. Am. J. Bot. 2003;90:1596–1603. doi: 10.3732/ajb.90.11.1596. [DOI] [PubMed] [Google Scholar]
- Soltis P.S., Soltis D.E. Ancient WGD events as drivers of key innovations in angiosperms. Curr. Opin. Plant Biol. 2016;30:159–165. doi: 10.1016/j.pbi.2016.03.015. [DOI] [PubMed] [Google Scholar]
- Stelzer C.P., Blommaert J., Waldvogel A.M., Pichler M., Hecox-Lea B., Mark Welch D.B. Comparative analysis reveals within-population genome size variation in a rotifer is driven by large genomic elements with highly abundant satellite DNA repeat elements. BMC Biol. 2021;19:206. doi: 10.1186/s12915-021-01134-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung W., Ackerman M.S., Dillon M.M., Platt T.G., Fuqua C., Cooper V.S., Lynch M. Evolution of the insertion-deletion mutation rate across the tree of life. G3 (Bethesda) 2016;6:2583–2591. doi: 10.1534/g3.116.030890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani M.A., Hajkova P. Epigenetic reprogramming of mouse germ cells toward totipotency. Cold Spring Harb. Symp. Quant. Biol. 2010;75:211–218. doi: 10.1101/sqb.2010.75.010. [DOI] [PubMed] [Google Scholar]
- Talla V., Suh A., Kalsoom F., Dinca V., Vila R., Friberg M., Wiklund C., Backström N. Rapid increase in genome size as a consequence of transposable element hyperactivity in Wood-White (Leptidea) Butterflies. Genome Biol. Evol. 2017;9:2491–2505. doi: 10.1093/gbe/evx163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarailo-Graovac M., Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinformatics. 2009;25:4. doi: 10.1002/0471250953.bi0410s25. [DOI] [PubMed] [Google Scholar]
- Tsutsui N.D., Suarez A.V., Spagna J.C., Johnston J.S. The evolution of genome size in ants. BMC Evol. Biol. 2008;8:64. doi: 10.1186/1471-2148-8-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van De Peer Y., Mizrachi E., Marchal K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 2017;18:411–424. doi: 10.1038/nrg.2017.26. [DOI] [PubMed] [Google Scholar]
- Vu G.T.H., Schmutzer T., Bull F., Cao H.X., Fuchs J., Tran T.D., Jovtchev G., Pistrick K., Stein N., Pecinka A., et al. Comparative genome analysis reveals divergent genome size evolution in a carnivorous plant genus. Plant Genome. 2015;8:1–14. doi: 10.3835/plantgenome2015.04.0021. [DOI] [PubMed] [Google Scholar]
- Wallau G.L., Vieira C., Loreto É.L.S. Genetic exchange in eukaryotes through horizontal transfer: connected by the mobilome. Mob. DNA. 2018;9:6–16. doi: 10.1186/s13100-018-0112-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Dou K., Moon S., Tan F.J., Zhang Z.Z. Hijacking oogenesis enables massive propagation of LINE and retroviral transposons. Cell. 2018;174:1082–1094.e12. doi: 10.1016/j.cell.2018.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Fang X., Yang P., Jiang X., Jiang F., Zhao D., Li B., Cui F., Wei J., Ma C., et al. The locust genome provides insight into swarm formation and long-distance flight. Nat. Commun. 2014;5:2957. doi: 10.1038/ncomms3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J.N., Feschotte C. A field guide to eukaryotic transposable elements. Annu. Rev. Genet. 2020;54:539–561. doi: 10.1146/annurev-genet-040620-022145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson T.G. Transposable elements as initiators of insecticide resistance. J. Econ. Entomol. 1993;86:645–651. doi: 10.1093/jee/86.3.645. [DOI] [PubMed] [Google Scholar]
- Wu C., Lu J. Diversification of transposable elements in arthropods and its impact on genome evolution. Genes. 2019;10:338. doi: 10.3390/genes10050338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyngaard G.A., Rasch E.M., Manning N.M., Gasser K., Domangue R. The relationship between genome size, development rate, and body size in copepods. Hydrobiologia. 2005;532:123–137. [Google Scholar]
- Yang Y., Ye X., Dang C., Cao Y., Hong R., Sun Y.H., Xiao S., Mei Y., Xu L., Fang Q., et al. Genome of the pincer wasp Gonatopus flavifemur reveals unique venom evolution and a dual adaptation to parasitism and predation. BMC Biol. 2021;19:145. doi: 10.1186/s12915-021-01081-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yuan H., Huang Y., Mao Y., Zhang N., Nie Y., Zhang X., Zhou Y., Mao S. The evolutionary patterns of genome size in Ensifera (Insecta: Orthoptera) Front. Genet. 2021;12:693541. doi: 10.3389/fgene.2021.693541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov E.M., Kuznetsov D., Tegenfeldt F., Manni M., Berkeley M., Kriventseva E.V. OrthoDB in 2020: evolutionary and functional annotations of orthologs. Nucleic Acids Res. 2021;49:D389–D393. doi: 10.1093/nar/gkaa1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zedek F., Smerda J., Smarda P., Bureš P. Correlated evolution of LTR retrotransposons and genome size in the genus eleocharis. BMC Plant Biol. 2010;10:265. doi: 10.1186/1471-2229-10-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Tu S., Stubna M., Wu W.S., Huang W.C., Weng Z., Lee H.C. The piRNA targeting rules and the resistance to piRNA silencing in endogenous genes. Science. 2018;359:587–592. doi: 10.1126/science.aao2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q.J., Li W., Li K., Nan H., Shi C., Zhang Y., Dai Z.Y., Lin Y.L., Yang X.L., Tong Y., et al. The chromosome-level reference genome of tea tree unveils recent bursts of non-autonomous LTR retrotransposons in driving genome size evolution. Mol. Plant. 2020;13:935–938. doi: 10.1016/j.molp.2020.04.009. [DOI] [PubMed] [Google Scholar]
- Zhang S., Gu S., Ni X., Li X. Genome size reversely correlates with host plant range in Helicoverpa species. Front. Physiol. 2019;10:29. doi: 10.3389/fphys.2019.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.Q., Che L.H., Li Y., Dan L., Pang H., Ślipiński A., Zhang P. Evolutionary history of Coleoptera revealed by extensive sampling of genes and species. Nat. Commun. 2018;9:205. doi: 10.1038/s41467-017-02644-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Park C., Bennett C., Thornton M., Kim D. Rapid and accurate alignment of nucleotide conversion sequencing reads with HISAT-3N. Genome Res. 2021;31:1290–1295. doi: 10.1101/gr.275193.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaenepoel A., Van de Peer Y. wgd-simple command line tools for the analysis of ancient whole-genome duplications. Bioinformatics. 2019;35:2153–2155. doi: 10.1093/bioinformatics/bty915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
This paper analyzes existing, publicly available data, which are included in Supplemental file (Table S5) for details.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact on request.