Abstract
Most studies on leukemia focus on leukemia cells as isolated objects without considering the bone marrow niche. Pal et al. have recreated the bone marrow niche using induced pluripotent stem cells (iPSCs), identifying CDH2 as a therapeutically druggable leukemia-promoting factor.1
Most studies on leukemia focus on leukemia cells as isolated objects without considering the bone marrow niche. Pal et al. have recreated the bone marrow niche using induced pluripotent stem cells (iPSCs), identifying CDH2 as a therapeutically druggable leukemia-promoting factor.1
Main text
Cancer cells and leukemia blasts, in particular, are capable of shifting their immediate surrounding toward a more permissive microenvironment that promotes oncogenic behavior—enhanced survival and proliferation, diminished differentiation, and, more importantly, development of chemoresistance. This is particularly important considering that the interplay between leukemia and the microenvironment is not taken under consideration in most studies on leukemogenesis. There are also technical obstacles when investigating the crosstalk between the human bone marrow niche and healthy hematopoietic or leukemia cells. Pal et al. overcome these difficulties by using iPSCs.1
The discovery of iPSCs is one of the major breakthroughs of the 20th century, representing huge technological progress and bringing new dimensions to the field of developmental biology.2 From the technological point of view, iPSCs represent the possibility to reprogram almost any human somatic cell into a pluripotent embryonic stem cell-like state, becoming a valuable source of almost every somatic cell. This reprogramming technology is applied in translational research to perform experimental modeling of human diseases and to generate mature somatic cells for therapeutic use. From the perspective of developmental biology, the fact that a simple combination of three to four transcription factors can turn any somatic cell into a pluripotent stem cell shows the tremendous power of transcription factors and surrounding signaling systems in the regulation of cellular behavior, including oncogenesis/leukemogenesis in which a simple failure in this regulation may be fatal. Although highly or partially artificial, iPSCs represent an extremely useful model for translational and cancer research. Pal et al. use the iPSC system to recapitulate the bone marrow niche in vitro in order to investigate the crosstalk between niche components with leukemia blasts. Their ultimate goal was to investigate niche-mediated mechanisms of therapy resistance in leukemia cells in order to identify druggable targets.
In order to recapitulate the bone marrow niche, the authors successfully re-programmed human primary bone marrow mesenchymal cells into iPSCs and subsequently differentiated them into mesenchymal stem cells (iMSC) or vascular niche-like cells (iANG). Performing co-culture assays with leukemia blasts, they found that both mesenchymal cells and vascular niche-like cells support ex vivo proliferation and survival of leukemia cells. iMSC and iANG also inhibited dormancy in leukemia cells contributing to therapy resistance. While iMSC showed therapy protection for both non-cycling and cycling leukemia cells, iANG protected only dormant cells. This may be due to the different physical locations of these two niche cell types in relation to leukemia cells or to the differences in the expression/secretion patterns of homing receptors/adhesion molecules, soluble niche components, growth factors, chemokines, and cytokines.
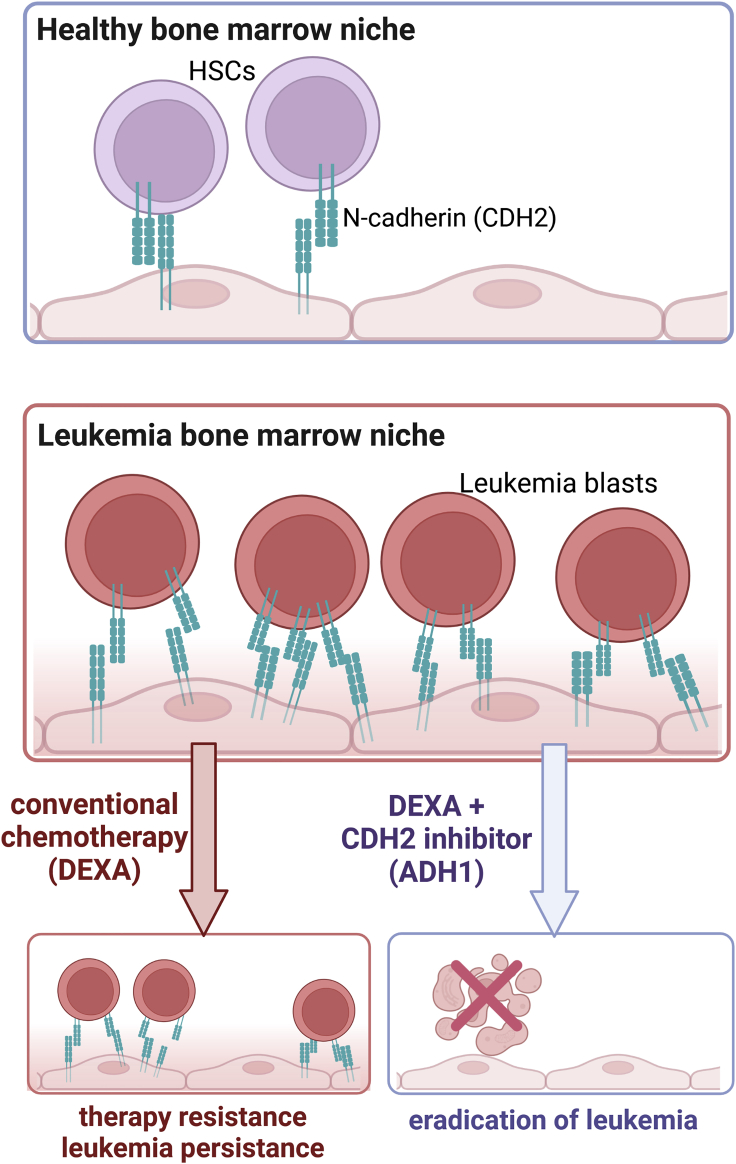
To uncover essential players involved in niche leukemia interactions, the authors compared the gene expression profiles of iPSC-derived niche cells co-cultured with or without patient-derived leukemia blasts, identifying an upregulation of Cadherin 2 (CDH2, N-cadherin) in both niche cells in the presence of leukemia blasts. This finding is especially interesting because CDH2 regulates proliferation, migration, and differentiation of stem cells by inducing β-catenin signaling, phosphorylation of the c-Janus N-terminal kinase (JNK), and activation of small GTPases, such as Rho, Rac, and Cdc42.3, 4, 5, 6 CDH2 also interacts with growth factor receptors, such as VEGFR, EGFR, FGFR, TGFBR, and HGFR, regulating downstream signaling pathways.3 These results suggested that CDH2 upregulation may represent a key initiating event in the emergence of the leukemia-supporting bone marrow niche, and thus the authors hypothesized about the therapeutic potential of targeting CDH2.
They examined the anti-leukemia activity of the CDH2 competitive antagonist ADH-1. ADH-1 or Exherin is a pentapeptide inducing a disruptive effect on tumor vasculature in preclinical models for solid tumors.6 It has been previously evaluated in clinical phase I/II trials in melanoma and other CDH2 positive solid tumors and has been described as a well-tolerated drug with a modest anti-tumor activity, receiving FDA orphan drug status in 2008. Pal et al. showed that, when combined with dexamethasone, ADH-1 was extremely effective in vitro against acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML) cells as well as in vivo in an ALL patient-derived xenograft model using luciferase-tagged leukemic blasts from a very high risk ALL (Figure 1). Of note, no additional toxicity as compared to dexamethasone alone was detected. These findings are of great relevance from different aspects—it demonstrates the importance and applicability of an artificial iPSC system for translational leukemia research; it also provides essential insights into the understanding of the biology of leukemia, particularly the therapy-protective role of the bone marrow environment that is, in turn, re-programmed/re-directed by leukemia cells; and it ultimately identifies CDH2 as an essential adhesion molecule protecting leukemia cells from therapy-induced apoptosis and inhibition of proliferation. The emerging role of CDH2 in hematological malignancies and cancer metastasis, as well as its potential as a therapeutic target, has already been described and discussed by several investigators.7,8 Based on their observations and previous publications, Pal et al. also suggested the potential of CDH2 inhibitor ADH-1 or other drugs with anti-CDH2 activity in combinatorial anti-leukemia therapy. Thus, more comprehensive studies on large cohorts of pediatric and adult leukemia patients should be performed to understand the clinical relevance of CDH2 inhibition for the treatment of leukemia. The investigations on the role of the niche and CDH2, in particular on cancer cell growth and therapy protection using the iPSC system, may also be expanded to other hematological malignancies. It would be interesting to further evaluate the combinatory effects of other factors regulating cell adhesion and tight junctions, such as claudin 2 and caveolin 1, which showed in this study a high upregulation in niche cells upon co-culture with leukemia cells.
Figure 1.
Leukemia cell interaction with bone marrow niche components via N-cadherin causes therapy resistance
Image created with BioRender.com.
Identification of leukemia subtypes and leukemia evolution stage (i.e., leukemia stem cells versus leukemia blasts) in bigger comprehensive studies will shed light on the ultimate role of CDH2 and its therapeutic potential. The difference in the effects of iMSC and iANG on dormant versus proliferating leukemia cells is quite interesting. It is known that N-cadherin-expressing bone marrow stromal progenitor cells maintain reserve hematopoietic stem cells,9 and thus leukemia cells may benefit from these features of MSC for therapy protection. Improving our knowledge of the niche-specific regulation of leukemia cells by comparing the distribution of leukemia cells in the bone marrow depending on the type and subtype of leukemia may provide a better estimation of the therapeutic response to CDH2 inhibitors.
Acknowledgments
Declaration of interests
The authors declare no competing interests.
References
- 1.Pal D., Blair H., Parker J., Hockney S., Beckett M., Singh M., Tirtakusuma R., Nelson R., McNeill H., Angel S., et al. hiPSC-derived bone marrow milieu identifies a clinically actionable driver of niche-mediated treatment resistance in leukaemia. Cell Reports Medicine. 2022;3 doi: 10.1016/j.xcrm.2022.100717. 100724-1–100724–18.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 3.Alimperti S., Andreadis S.T. CDH2 and CDH11 act as regulators of stem cell fate decisions. Stem Cell Res. 2015;14:270–282. doi: 10.1016/j.scr.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blaschuk O.W. Potential therapeutic Applications of N-cadherin antagonists and Agonists. Front. Cell Dev. Biol. 2022;10:866200. doi: 10.3389/fcell.2022.866200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavallaro U., Dejana E. Adhesion molecule signalling: not always a sticky business. Nat. Rev. Mol. Cell Biol. 2011;12:189–197. doi: 10.1038/nrm3068. [DOI] [PubMed] [Google Scholar]
- 6.Yarom N., Stewart D., Malik R., Wells J., Avruch L., J Jonker D. Phase I clinical trial of Exherin (ADH-1) in patients with advanced solid tumors. Curr. Clin. Pharmacol. 2013;8:81–88. doi: 10.2174/1574884711308010011. [DOI] [PubMed] [Google Scholar]
- 7.Kim H.N., Ruan Y., Ogana H., Kim Y.M. Cadherins, Selectins, and Integrins in CAM-DR in leukemia. Front. Oncol. 2020;10:592733. doi: 10.3389/fonc.2020.592733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mrozik K.M., Blaschuk O.W., Cheong C.M., Zannettino A.C.W., Vandyke K. N-cadherin in cancer metastasis, its emerging role in haematological malignancies and potential as a therapeutic target in cancer. BMC Cancer. 2018;18:939. doi: 10.1186/s12885-018-4845-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao M., Tao F., Venkatraman A., Li Z., Smith S.E., Unruh J., Chen S., Ward C., Qian P., Perry J.M., et al. N-Cadherin-Expressing bone and marrow stromal progenitor cells maintain reserve hematopoietic stem cells. Cell Rep. 2019;26:652–669.e6. doi: 10.1016/j.celrep.2018.12.093. [DOI] [PMC free article] [PubMed] [Google Scholar]