Abstract
Genes involved in allose utilization of Escherichia coli K-12 are organized in at least two operons, alsRBACE and alsI, located next to each other on the chromosome but divergently transcribed. Mutants defective in alsI (allose 6-phosphate isomerase gene) and alsE (allulose 6-phosphate epimerase gene) were Als−. Transcription of the two allose operons, measured as β-galactosidase activity specified by alsI-lacZ+ or alsE-lacZ+ operon fusions, was induced by allose. Ribose also caused derepression of expression of the regulon under conditions in which ribose phosphate catabolism was impaired.
Conversion of the all cis-hexose allose to fructose 6-phosphate in Aerobacter aerogenes requires the activity of three enzymes—allokinase, allose 6-phosphate isomerase, and allulose 6-phosphate epimerase (7). This pathway appears to operate also in Escherichia coli. Thus, five contiguous genes (alsRBACE), expressed as one operon, encode a periplasmic binding protein-mediated transport system (alsB, alsA, and alsC), a putative hexose phosphate epimerase (alsE), and a regulatory protein for allose utilization (alsR). A potential sixth member of the operon (alsK) has been postulated to encode allokinase (9). Ribose utilization requires among other enzymes ribose 5-phosphate isomerase. In E. coli, two ribose phosphate isomerases, A and B, have been identified biochemically (5, 6) and genetically (18, 20). Ribose phosphate isomerase A, encoded by rpiA, is synthesized constitutively, whereas the synthesis of ribose phosphate isomerase B, encoded by rpiB, appears to be increased following growth of cells in ribose-containing medium. A repressor protein, encoded by the rpiR gene, is involved in regulation of rpiB gene expression. Thus, rpiR strains contain elevated activity of ribose phosphate isomerase B (6, 20). The rpiB and rpiR loci are located next to each other at 92.8 min on the linkage map, but are divergently transcribed, with rpiB transcribed clockwise (20).
alsR and rpiR are the same gene (9). In the present work, we show that the rpiB gene product is also involved in allose catabolism, and presumably rpiB encodes allose 6-phosphate isomerase. Hence rpiB will be redesignated alsI. Although nucleotide sequence analysis implies alsK is the distal cistron of an alsRBACEK operon, our results showed that the alsK cistron was neither necessary for allose utilization nor coordinately expressed with the remaining alsRBACE cistrons. Consequently, we have designated alsK as yjcT (15).
Methods.
The E. coli K-12 strains used in this study are listed in Table 1. Growth media (NZY broth or phosphate-buffered AB minimal medium) were described before (8). The carbon sources used were glucose, ribose, xylose, and glycerol at 0.2% each or allose at 0.05 or 0.1%. The growth of cell cultures was monitored in an Eppendorf PCP6121 photometer as optical density at 436 nm. Bacteriophage P1-mediated transduction (13), transformation with plasmid DNA (10), techniques for the growth of bacteriophage λ (16), and lysogenization by recombinant phage (17) have been previously described, as well as methods for the isolation of plasmid DNA (2) and chromosomal DNA (16). Restriction and ligation of DNA were performed as described by the suppliers of restriction endonucleases (Amersham, Promega, and New England Biolabs) and T4 DNA ligase (Amersham). PCR was performed with chromosomal or plasmid DNA as a template by standard procedures with DynaZyme II DNA polymerase (Finzymes, Oy, Finland). For enzyme assays, exponentially growing cells were harvested by centrifugation and disrupted by sonication for 60 s at 0°C and then centrifuged to remove cell debris. The assay of β-galactosidase activity at 30°C (13), or allokinase activity at 37°C (7) and determination of protein content (19) were performed as previously described.
TABLE 1.
Bacterial strains
| Strain | Relevant genotypea | Source, reference, or construction |
|---|---|---|
| BW18524 | —b | B. Wanner, Purdue University |
| CC118 | Δ(lac)χ74 recA1 | 11 |
| DPB271 | recD::miniTet | 1 |
| HO340 | —c | 14 |
| HO890 | Δ(rpiA)103::Tetrc | 8 |
| HO1048 | Δ(rpiA-serA)101::KanralsR128c | 20 |
| HO1446 | alsR128c | Ser+ derivative of HO1048 |
| HO1458 | alsI137::Kanrc | 20 |
| HO1686 | Δ(rpiA)103::Tetr/λYYC205[Φ(alsI-lacZ+)139]c | HO890, lysogenization with λYYC205 |
| HO1693 | Δ(rpiA)103::TetralsI137::Kanr/λYYC205[Φ(alsI-lacZ+)139]c | P1(HO1458) × HO1686, Kanr |
| HO1868 | alsI137::Kanr/λYYC205[Φ(alsI-lacZ+)139]c | P1(HO1458) × YYC1060, Kanr |
| HO1973 | alsI137::Kanrb | P1(HO1458) × BW18524, Kanr |
| HO2190 | yjcT8::TnphoA′-2/λYYC205[Φ(alsI-lacZ+)139]c | P1(TP2058) × YYC1060, Kanr |
| HO2376 | Δ(rpiA)103::TetralsE11::TnphoA′-9c | P1(TP2086) × HO890, Kanr |
| P90C | ara Δ(lac-pro) thi | 17 |
| TP1904 | recD::miniTetb | P1(DPB271) × BW18524, Tetr |
| TP2058 | yjcT8::TnphoA′-2b | BW18524, recombination of pTP908 (TP1904) followed by recombinational switching with λTnphoA-132 (Tetr) |
| TP2061 | alsE11::TnphoA′-2b | BW18524, recombination of pTP911 (TP1904) followed by recombinational switching with λTnphoA-132 (Tetr) |
| TP2083 | yjcT8::TnphoA′-9b | TP2058, recombinational switching with λTn5-112 (Kanr) |
| TP2086 | alsE11::TnphoA′-9b | TP2061, recombinational switching with λTn5-112 (Kanr) |
| TP2110 | alsR21::TnphoA′-2b | BW18524, recombination of pTP925 (TP1904) followed by recombinational switching with λTnphoA-132 (Tetr) |
| TP2115 | alsR21::TnphoA′-9b | TP2110, recombinational switching with λTn5-112 (Kanr) |
| YYC1060 | —c/λYYC205[Φ(alsI-lacZ+)139] | HO340, lysogenization with λYYC205 |
| YYC1062 | alsR128/λYYC205[Φ(alsI-lacZ+)139]c | HO1446, lysogenization with λYYC205 |
All of the strains are F−. phoA′ specifies β-galactosidase. Mutations generated by TnphoA-132, TnphoA′-1, or TnphoA′-2 are polar and are transposition proficient. Mutations generated by TnphoA-112 or TnphoA′-9 are nonpolar and are transposition deficient.
Also contains Δ(lac)χ74 ΔphoA532 phnCpΔ(phnCDE)59.
Also contains araC(Am) araD Δ(lac)U169 trp(Am) mal(Am) rpsL relA thi supF.
Isolation and characterization of als and yjcT mutants.
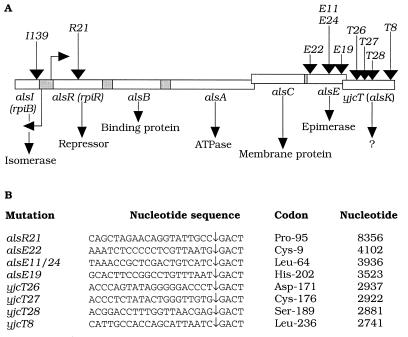
Transposon technology was used to generate one plasmid-harbored alsR::TnphoA′-1 mutation, four alsE::TnphoA′-1 mutations, and four yjcT::TnphoA′-1 mutations (Fig. 1). To avoid effects of a high copy number in an analysis of the regulation of als and yjcT gene expression, allele replacement by homologous recombination was conducted with each of the plasmid-borne als or yjcT mutations. This recombination resulted in the production of strains harboring chromosomally located als or yjcT mutations. The TnphoA′-1 insertions generated polar mutations. For each mutation, a nonpolar version was constructed (Fig. 1). We also constructed an operon fusion allele to the alsI gene [Φ(alsI-lacZ+)139] (Fig. 2). A map of the 10 insertions is shown in Fig. 1A. The nucleotide sequences of the fusion points of the transposon-generated fusions are shown in Fig. 1B. The growth of the als strains, polar as well as nonpolar, on allose was analyzed. The alsR and alsE strains containing polar mutations were Als−, whereas the yjcT strains were Als+. The strains containing nonpolar alsE mutations were also Als−. In contrast, the strains containing nonpolar alsR21 or yjcT8 mutations were Als+. Furthermore, a strain harboring a mutation in the alsI gene (HO1973) was Als−. These results indicated that the alsI and alsE gene products are essential for allose utilization, whereas the repressor, encoded by alsR, is dispensable for allose utilization.
FIG. 1.
Structure of the als operon and location of als-lacZ+ and yjcT-lacZ+ insertions. Mutagenization of the alsRBACE operon and yjcT was performed as follows. Strain CC118 (Δlac) harboring pTP680 (alsR+B+A+C+E+ yjcT+) or pHO390 (yjcT+) was infected with λ::TnphoA′-1 (12). TnphoA′-1 contains a promoterless lacZ+ allele, and a β-galactosidase-producing mutant has acquired an operon fusion. Mutants were selected as kanamycin resistant. Plasmid DNA was isolated and transformed back into strain CC118 and screened for production of β-galactosidase activity by the presence of 5-bromo-4-chloro-3-indolyl galactoside (40 mg liter−1). Insertion of a transposon into the als operon was ascertained by restriction endonuclease analysis. Allele replacement of plasmid-harbored als::TnphoA′-1 or yjcT::TnphoA′-1 mutations and the chromosomal als or yjcT genes was performed by homologous recombination. Restriction endonuclease-linearized plasmid DNA was transformed into an recD strain (TP1904) by selection for kanamycin resistance. Genetic mapping ensured the location of the inserted DNA at 92.8 min on the linkage map. Recombinational switching among transposons was performed as previously described (21). The insertion of TnphoA′-1 generated polar mutations. A recombinational switching, using TnphoA-132 (encoding tetracycline resistance) followed by Tn5-112 (encoding kanamycin resistance), resulted in the isolation of a nonpolar version of each als allele or yjcT8, essentially by removing a transcription terminator located within the right-hand IS50 element of the transposon. The plasmids used were pTP680, which contained a wild-type version of the alsRBACE operon and yjcT in a 7.8-kb DNA fragment of chromosomal origin in pUC19 (22), or pHO390, which contained a PCR-amplified wild-type yjcT allele ligated to the BamHI site of pBR322 (4). The inserted yjcT sequence was confirmed by sequencing. (A) Boxes indicate open reading frames of the alsI and alsRBACE operons and yjcT. Staggered boxes indicate open reading frames with possible overlapping translation (alsA and alsC, and alsE and yjcT). Shaded boxes indicate intercistronic regions. The angled arrows indicate the transcription initiation points before the alsI and alsR cistrons (20). Vertical arrows above the boxes indicate the positions of insertions of als-lacZ+ or yjcT-lacZ+ operon fusions. The alsI-lacZ+ fusion was generated by in vitro techniques (Fig. 2). The presumed gene product of each cistron is indicated below the bar. The plasmids constructed were pTP908 (yjcT8::TnphoA′-1), pTP911 (alsE11::TnphoA′-1), pTP919 (alsE19::TnphoA′-1), pTP922 (alsE22::TnphoA′-1), pTP924 (alsE24::TnphoA′-1), and pTP925 (alsR21::TnphoA′-1), which were isolated from pTP680; and pTP926 (yjcT26::TnphoA′-1), pTP927 (yjcT27::TnphoA′-1), and pTP928 (yjcT28::TnphoA′-1), which were isolated from pHO390. (B) Nucleotide sequence of the points of insertion of TnphoA′-1. Sequencing was performed at the Botanical Institute, University of Copenhagen, in an Applied Biosystems model 377 sequencer by cycle sequencing with dye terminators (ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction kit; Perkin-Elmer) and with the oligodeoxyribonucleotide 5′-GCAGTAATTTCCGAGTCCC-3′ as a primer (Hobolth DNA Syntese, Hillerød, Denmark). A vertical arrow indicates an insertion point. Nucleotides to the left of the arrow originate from the als or yjcT cistrons. Nucleotides to the right of the arrow originate from TnphoA′ sequences. The codon where insertion occurred is indicated together with the nucleotide position of insertion. The latter numbers refer to the nucleotide sequence reported in the database sequence under accession no. AE00482 (3).
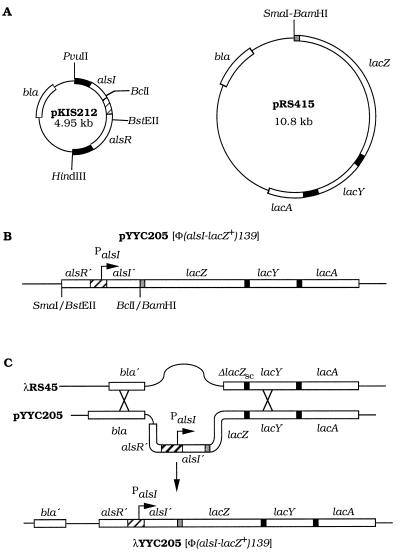
FIG. 2.
Construction of an alsI-lacZ+ gene fusion. Open reading frames are indicated by open double lines, vector sequences are indicated by thin lines, and flanking DNA sequences or intercistronic regions are shown as black or shaded double lines. Relevant restriction endonuclease recognition sites are included. (A) The plasmid pKIS212 contains the alsR and alsI genes (20). The plasmid pRS415 contains a promoterless lac operon, which includes a wild-type lacZ gene with translation initiation sequences. Intercistronic regions, which are not drawn to scale, are shown in black. The shaded region contains part of the trp operon as well as the original W205 fusion (17). (B) Construction of a plasmid-borne gene fusion. DNA of pKIS212 was digested with restriction endonuclease BstEII, followed by incubation with the large fragment of E. coli DNA polymerase I in the presence of the four deoxyribonucleoside triphosphates and digestion with BclI. Plasmid pRS415 was digested with endonucleases SmaI and BamHI. The two DNA species were ligated. Transformation followed by selection for ampicillin resistance in the presence of 5-bromo-4-chloro-3-indolyl galactoside to screen for β-galactosidase synthesis resulted in the isolation of pYYC205. The insert of pYYC205 contained 129 nucleotides of the N-terminal encoding end of the alsR reading frame, the 358 nucleotides of the alsR-alsI intercistronic region (cross-hatched), and 28 nucleotides of the N-terminal encoding end of the alsI reading frame. PalsI indicates the promoter driving transcription of the alsI gene, and an angled arrow indicates the transcription initiation point. alsR′ and alsI′ indicate deletion of the C-terminal encoding ends of the alsR and alsI genes, respectively. The various DNA elements of pYYC205 are not drawn to scale. (C) Isolation of a bacteriophage λ-borne gene fusion by homologous recombination. Bacteriophage λRS45 contains a version of the lacZ cistron that is deleted for the promoter-proximal two-thirds (ΔlacZSC), wild-type versions of the lacY and lacZ cistrons, and a truncated bla gene (bla′). Thus, λRS45 forms white plaques, and lysogens of λRS45 form white colonies in the presence of 5-bromo-4-chloro-3-indolyl galactoside. The lac-bla sequence of λRS45 is homologous to sequences of pYYC205. Consequently homologous recombination (indicated by X) which occurred among plasmid and bacteriophage replicons within the bla sequence and within the lac sequence resulted in the formation of a bacteriophage λ genome carrying the gene fusion. Host strain P90C harboring pYYC205 was infected with λRS45 to allow recombination and to generate a lysate. Strain P90C was infected with this lysate and plated on NZY broth containing 5-bromo-4-chloro-3-indolyl galactoside. Blue plaques, which appeared at a frequency of approximately 4 × 10−3, were restreaked, and one isolate, λYYC205, was kept for further analysis. Insertion of the prophage at the attλ site at 17 min, rather than at alsI at 92.8 min on the linkage map, was confirmed by genetic mapping.
The addition of allose (0.05%) appeared to potently inhibit the growth with glycerol as the carbon source (0.2%) of strains harboring mutations in alsI (HO1973) or alsE (TP2086), encoding allose 6-phosphate isomerase and allulose 6-phosphate epimerase, respectively. In contrast, the growth of the remaining strains, i.e., those defective in the regulatory protein (TP2115 [alsR]) or YjcT (TP2083) were not inhibited by allose. The lack of growth of the alsE and alsI strains in the presence of allose indicated that a compound, which accumulated in these strains, caused inhibition. It is likely that this compound is allose 6-phosphate in the alsI strain and allose 6-phosphate, allulose 6-phosphate, or both in the alsE strain.
We previously showed that the alsI (rpiB)-encoded enzyme is able to isomerase ribose 5-phosphate and ribulose 5-phosphate. Thus, this enzyme appears to have substrate specificity toward both pentose phosphates and hexose phosphates. A similar situation exists for the Streptococcus mutans galactose 6-phosphate isomerase (encoded by lacAB), which is also able to isomerize ribose 5-phosphate (20).
Regulation of alsI operon expression.
The recombinant Φ(alsI-lacZ+)139 fusion-harboring λ phage was used to lysogenize various E. coli strains. A 58-fold increase in β-galactosidase activity was observed when cells of strain YYC1060 were grown in the presence of allose and compared to the activity of cells grown in the absence of allose (Table 2).
TABLE 2.
Regulation of als regulon expression by allose
| Strain | als allele(s) or plasmida | β-Galactosidase activity (nmol min−1 mg of protein−1) in medium supplemented withb:
|
Activity ratioc | |
|---|---|---|---|---|
| Glycerol | Glycerol + allose | |||
| YYC1060 | Φ(alsI-lacZ+)139 | 438 | 25,300 | 58 |
| TP2115 | alsR21::TnphoA′-9 | 20,300 | 26,900 | 1.3 |
| TP2086 | alsE11::TnphoA′-9 | 201 | 8,630 | 43 |
| TP2083 | yjcT8::TnphoA′-9 | 48.0 | 50.1 | 1.0 |
| BW18524 | als+ | 2.0 | 2.0 | 1.0 |
| HO2190 | Φ(alsI-lacZ+)139 yjcT8 | 451 | 14,800 | 33 |
| TP2083 | yjcT8/pHO390 (yjcT+) | 42.0 | 34.0 | 0.8 |
| TP2083 | yjcT8/pBR322 | 32.0 | 44.0 | 1.4 |
TnphoA′ fusions specify β-galactosidase activity.
Cells were grown without or with allose (0.05%), and β-galactosidase activity was assayed as described in the text.
Ratio of β-galactosidase activity in the presence of allose to β-galactosidase activity in the absence of allose.
The β-galactosidase activity specified by the Φ(alsI-lacZ+)139 fusion contained in host strains, which harbored various genetic lesions of ribose catabolism, and grown on different carbon sources is shown in Table 3. In a wild-type strain (YYC1060), only a modest increase (twofold or less) in alsI gene expression was observed when cells were grown with pentose as a carbon source (xylose, ribose, or both) compared to growth in the presence of glucose. In contrast, alsI gene expression was greatly increased in ribose auxotrophic strains (rpiA or rpiA alsI), when grown on ribose. Thus, with growth in the presence of ribose, the β-galactosidase activity of an rpiA strain (HO1686) increased approximately 25-fold compared to growth in the presence of both ribose and glucose. The increase was less pronounced by growth in the presence of both ribose and xylose: approximately fivefold compared to growth in the presence of both ribose and glucose. Furthermore, alsI gene expression increased 25-fold or more in an rpiA alsI strain (HO1693) grown in the presence of ribose and xylose, compared to growth in the presence of ribose and glucose. A mutation in the alsI gene alone was essentially without effect on alsI gene expression, because the alsI strain HO1868 responded like the wild-type strain YY1060. The β-galactosidase activity in an alsR strain harboring the operon fusion (YYC1062) was increased 20- to 100-fold compared to the activity of the otherwise isogenic alsR+ strain (YYC1060).
TABLE 3.
Regulation of als regulon expression by pentoses
| Strain | Genotype | β-Galactosidase activity (nmol min−1 mg of protein−1) in minimal medium supplemented witha:
|
||||
|---|---|---|---|---|---|---|
| Glucose | Xylose | Ribose | Ribose + xylose | Ribose + glucose | ||
| YYC1060 | Φ(alsI-lacZ+)139 | 89.6 | 126 | 174 | 132 | 83.8 |
| HO1686 | Φ(alsI-lacZ+)139 rpiA | − | − | 1,510 | 349 | 62.4 |
| HO1693 | Φ(alsI-lacZ+)139 rpiA alsI | − | − | − | 1,590 | 63.8 |
| HO1868 | Φ(alsI-lacZ+)139 alsI | 57.5 | 121 | 132 | 100 | 63.3 |
| YYC1062 | Φ(alsI-lacZ+)139 alsR | 2,480 | 12,100 | 13,000 | 9,300 | 1,910 |
| HO2376 | alsE11::TnphoA′-9 rpiA | − | − | 788 | 221 | 54.0 |
Cells were grown and β-galactosidase activity was assayed as described in the text. −, no growth.
Regulation of alsRBACE operon expression.
Strains harboring a phoA′-1 (lacZ+) gene fusion to the alsR (TP2115) or alsE (TP2086) cistrons were assayed for β-galactosidase activity in extracts of cells grown in the presence or absence of allose (Table 2). In the presence of allose, β-galactosidase activity increased 43-fold compared to the activity in the absence of allose in cells harboring a lacZ fusion to alsE (TP2086). Thus, expression of the alsE cistron appeared to be induced by the presence of allose. Cells harboring an alsR-lacZ+ gene fusion (TP2115) contained a high, constitutive level of β-galactosidase activity. The β-galactosidase activity of a nonfusion strain (BW18524) was negligible. In addition, the expression of the alsRBACE operon was regulated by ribose similarly to that described for the alsI operon. Thus, β-galactosidase activity specified by the alsE11::TnphoA′-9 fusion increased approximately 15-fold in cells grown with ribose or 4-fold in cells grown with ribose and xylose, compared to that in cells grown with ribose and glucose (Table 3).
Lack of involvement of yjcT (alsK) in allose utilization.
The open reading frames of the distal cistron alsE and the following cistron yjcT overlapped by five codons, which may suggest translational coupling of the two cistrons (3). We constructed four independent insertions in yjcT, all of which had similar properties. Most importantly, expression of yjcT apparently was unaffected by allose (Table 2, strain TP2083). Strains with transposon insertions in yjcT were Als+. Furthermore, a yjcT mutation had no effect on expression of the alsRBACE and alsI operons: the introduction of yjcT8 into an alsI139-lacZ+ strain had little effect on the fold of induction of β-galactosidase synthesis (Table 2, strains YYC1060 and HO2190). Supplying yjcT in trans had no effect, as shown by the lack of regulation of the yjcT-lacZ+ fusion strain transformed with pHO390, which contains a wild-type yjcT allele (Table 2, strains TP2083, TP2083/pHO390 and TP2083/pBR322). The allokinase activity in extracts of cells harboring pHO390, (i.e., with yjcT in multicopy) was identical to the activity in extracts of cells harboring pBR322 (0.5 nmol min−1 mg of protein−1). Finally, allokinase activities were similar in cells of wild-type and yjcT strains grown in glycerol (0.3 nmol min−1 mg of protein−1). Allose did not cause induction of allokinase synthesis, and alsR and alsR+ strains contained identical activities of allokinase. These results suggest that the kinase responsible for phosphorylation of allose either has a broad substrate specificity, which may not be subject to induction by allose, or it utilizes a phosphoryl donor different from ATP.
Conclusion.
We have shown that alsI is essential for allose catabolism and that expression of both of the operons, alsI and alsRABCE, is induced by the presence of allose or ribose. In both cases, regulation is dependent on the alsR gene product. Thus, the alsI and alsRBACE operons constitute the als regulon. Apparently the yjcT gene is not a member of the als regulon.
Acknowledgments
Barry Wanner, Bob Simons, and Bente Mygind are acknowledged for generously providing plasmids, bacteriophages, and bacterial strains. Charlotte Hansen is acknowledged for running the automated DNA sequencing. Tonny D. Hansen and Anne L. Møller are acknowledged for expert technical assistance. We thank Jan Neuhard for carefully reading the manuscript.
Financial support was obtained from the Danish Center of Microbiology and the Center for Enzyme Research.
REFERENCES
- 1.Biek D P, Cohen S N. Identification and characterization of recD, a gene affecting plasmid maintenance and recombination in Escherichia coli. J Bacteriol. 1986;167:594–603. doi: 10.1128/jb.167.2.594-603.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 4.Bolivar F, Rodriguez R L, Greene P J, Betlach M C, Heynecker H L, Boyer H W, Crosa J H, Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 5.David J, Weismeyer H. Regulation of ribose metabolism in E. coli. II. Evidence for two ribose 5-phosphate isomerase activities. Biochim Biophys Acta. 1970;208:56–67. doi: 10.1016/0304-4165(70)90048-6. [DOI] [PubMed] [Google Scholar]
- 6.Essenberg M K, Cooper R A. Two ribose-5-phosphate isomerases from Escherichia coli K12: partial characterization of the enzymes and consideration of their physiological roles. Eur J Biochem. 1975;55:323–332. doi: 10.1111/j.1432-1033.1975.tb02166.x. [DOI] [PubMed] [Google Scholar]
- 7.Gibbins L N, Simpson F J. The incorporation of D-allose into the glycolytic pathway by Aerobacter aerogenes. Can J Microbiol. 1964;10:829–836. doi: 10.1139/m64-108. [DOI] [PubMed] [Google Scholar]
- 8.Hove-Jensen B, Maigaard M. Escherichia coli rpiA gene encoding ribose phosphate isomerase A. J Bacteriol. 1993;175:5628–5635. doi: 10.1128/jb.175.17.5628-5635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim C, Song S, Park C. The d-allose operon of Escherichia coli K-12. J Bacteriol. 1997;179:7631–7637. doi: 10.1128/jb.179.24.7631-7637.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mandel M, Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970;53:159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- 11.Manoil C, Beckwith J. TnphoA: a transposon probe for protein export signals. Proc Natl Acad Sci USA. 1985;82:8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Metcalf W W, Wanner B L. Mutational analysis of an Escherichia coli fourteen-gene operon for phosphonate degradation, using TnphoA′ elements. J Bacteriol. 1993;175:3430–3442. doi: 10.1128/jb.175.11.3430-3442.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 14.Nilsson D, Hove-Jensen B. Phosphoribosylpyrophosphate synthetase of Bacillus subtilis. Cloning, characterization and chromosomal mapping of the prs gene. Gene. 1987;53:247–255. doi: 10.1016/0378-1119(87)90013-8. [DOI] [PubMed] [Google Scholar]
- 15.Rudd K E. Linkage map of Escherichia coli K-12, edition 10: the physical map. Microbiol Mol Biol Rev. 1998;62:985–1019. doi: 10.1128/mmbr.62.3.985-1019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 17.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 18.Skinner A J, Cooper R A. Genetic studies on ribose-5-phosphate isomerase mutants of Escherichia coli K-12. J Bacteriol. 1974;118:1183–1185. doi: 10.1128/jb.118.3.1183-1185.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith P K, Krohn R I, Hermanson G T, Mallia A K, Gartner F H, Provenzano M D, Fujimoto E K, Goeke N M, Olson B J, Klenk D C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 20.Sørensen K I, Hove-Jensen B. Ribose catabolism of Escherichia coli: characterization of the rpiB gene encoding ribose phosphate isomerase B and of the rpiR gene, which is involved in regulation of rpiB expression. J Bacteriol. 1996;178:1003–1011. doi: 10.1128/jb.178.4.1003-1011.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilmes-Riesenberg M R, Wanner B L. TnphoA and TnphoA′ elements for making and switching fusion for study of transcription, translation, and cell surface localization. J Bacteriol. 1992;174:4558–4575. doi: 10.1128/jb.174.14.4558-4575.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]