Abstract
Objective
The mechanistic target of rapamycin complex 1 (mTORC1) is a key molecule that links nutrients, hormones, and growth factors to cell growth/function. Our previous studies have shown that mTORC1 is required for β-cell functional maturation and identity maintenance; however, the underlying mechanism is not fully understood. This work aimed to understand the underlying epigenetic mechanisms of mTORC1 in regulating β-cell functional maturation.
Methods
We performed Microarray, MeDIP-seq and ATAC-seq analysis to explore the abnormal epigenetic regulation in 8-week-old immature βRapKO islets. Moreover, DNMT3A was overexpressed in βRapKO islets by lentivirus, and the transcriptome changes and GSIS function were analyzed.
Results
We identified two major epigenetic silencing mechanisms, DNMT3A-dependent DNA methylation and PRC2-dependent H3K27me3 modification, which are responsible for functional immaturity of Raptor-deficient β-cell. Overexpression of DNMT3A partially reversed the immature transcriptome pattern and restored the impaired GSIS in Raptor-deficient β-cells. Moreover, we found that Raptor directly regulated PRC2/EED and H3K27me3 expression levels, as well as a group of immature genes marked with H3K27me3. Combined with ATAC-seq, MeDIP-seq and ChIP-seq, we identified β-cell immature genes with either DNA methylation and/or H3K27me3 modification.
Conclusion
The present study advances our understanding of the nutrient sensor mTORC1, by integrating environmental nutrient supply and epigenetic modification, i.e., DNMT3A-mediated DNA methylation and PRC2-mediated histone methylation in regulating β-cell identity and functional maturation, and therefore may impact the disease risk of type 2 diabetes.
Keywords: β-Cell, Type 2 diabetes, mTORC1, Epigenetic regulation, Dnmt3a, H3K27me3
Abbreviations: mTORC1, the mechanistic target of rapamycin complex 1; T2D, type 2 diabetes; Raptor, regulatory protein associated with mTOR; DNMT3A, DNA methyltransferase 3A; PRC2, polycomb repressor complex 2; H3K27me3, trimethylation of lysine residue 27 on histone 3; EED, embryonic ectoderm development; GSIS, glucose-stimulated insulin secretion; DMR, differential methylation region; DAR, differentially accessible region
Highlights
-
•
Rescued DNMT3A expression in Raptor-deficient islets partially reversed the abnormal induction of immature genes.
-
•
EED/H3K27me3 were impaired in Raptor-ablated β-cell.
-
•
DNA methylation and H3K27me3 are required for mTORC1-dependent epigenetic silencing of immature genes in β-cell.
1. Introduction
Type 2 diabetes is associated with progressive β-cell failure, and involves a gradual loss of β-cell identity, function and survival [1]. Pancreatic β-cell achieves functional maturity within 2 weeks after birth, with a higher threshold for glucose-stimulated insulin secretion (GSIS), as evidenced by inhibition of insulin secretion at basal glucose level and enhanced insulin secretion in response to high glucose [2]. During postnatal maturation, expression of several crucial transcription factors and metabolic genes, i.e., MAFA, NEUROD1, NKX6-1, UCN3 and GCK increases, in order to improve glucose sensing [[3], [4], [5], [6], [7]]. Simultaneously, the expressions of β-cell immature genes [8] and disallowed genes [9] become repressed in these neonatal β-cells as part of the metabolic switch associated with functional maturation [[9], [10], [11]]. It has been suggested that nutrition environment during the fetal and postnatal growth phase, can impact the development and maturation of pancreatic β-cells, and predispose the offspring to diabetes in early or later life [12]. However, how the metabolic signals drive the initial acquisition of glucose responsiveness during the neonatal period of life is still unknown.
mTOR (the mechanistic target of rapamycin) is an evolutionarily conserved serine/threonine kinase, critical for adult pancreatic β-cell metabolism and growth, exists in 1 of 2 complexes defined by regulatory and targeting subunits Raptor (defining mTORC1) or Rictor (mTORC2) [13]. It has been reported that mTORC1 activation during neonatal period and the switch from mTORC1 to AMPK (5′-adenosine monophosphate-activated protein kinase) after weaning, were two critical basis for functional maturation [14]. Raptor is one of the main core components of mTORC1, acts as a substrate recognizing subunit to facilitate the phosphorylation of mTOR [15]. Knockout of Raptor significantly reduced phosphorylation of mTORC1 targets 4E-BP1 and PS6 (Ser240/244) [16]. Using β-cell specific Raptor deficient mice, we and others have found that mTORC1 is required for functional maturation in rodent β-cell [[16], [17], [18]]. In a later study, we implanted insulin pellet to 4-week-old β-cell lineage-tracing βRapKO mice to maintain their glucose at normal range and demonstrated that Raptor directly controls β-cell anabolic metabolism and serves as a checkpoint of β-cell identity and plasticity independent of hyperglycemia [17]. Recently, Helman et al. demonstrated that changes in the nutritional environment sensing by mTORC1 trigger key steps in β-cell maturation after birth [18]; however how this pathway serves as a crosstalk between nutrient sensing and β-cell functional maturation is still not fully understood. Interestingly, our previous data reported that Raptor-deficient immature β-cells had reduced DNMT3A activity, whereas loss of Raptor in Ngn3-progenitor β-cells decreased H3K27 methyltransferase Ezh2 expression and impaired both identity and proliferation potential [19]. The current work builds on our previous work, but takes it further with comprehensive analysis of transcriptional and epigenetic changes in 8-week-old Raptor-deficient immature β-cells, as well established in our previous work [16,17], to explore the role of mTORC1 as a metabolic/epigenetic adaptation to postnatal nutrient environment. In our current study, we identified two major epigenetic silencing mechanisms, DNMT3A-mediated DNA methylation and PRC2-mediated histone methylation, regulated by mTORC1 to suppress a group of immature genes, thus facilitate functional maturation of neonatal β-cell. Manipulating the activities of mTORC1 and remodeling nutrient-epigenetic patterning in pancreatic β-cells might offer new targets to improve β-cell function in diabetes.
2. Methods
2.1. Mice
We used the Cre/loxP system to conditionally delete Raptor in pancreatic β-cells. β-cell specific Raptor knockout mice (βRapKO) were generated by crossing Raptor flox/flox mice (purchased from the Jackson Laboratory, C57BL/6J) with mice expressing Cre recombinase driven by the rat insulin promoter (RIP-Cre, mixed C57BL/6J:129/SvJ), as previously described [16]. βRapKO (Raptorflox/flox RIP-Cre), and their littermates (Raptorflox/flox) were used as WT control. Mice were fed ad libitum and housed in facility on a 12-h light–dark cycle. Genotypes of all mice were determined by PCR analysis of genomic DNA extracted from tails. Male mice were used in all the experiments. All animal experiments were performed in accordance with the principles of Guide for the Care and Use of Experimental Animals of Shanghai Jiao Tong University School of Medicine and approved by the Animal Care Committee of Shanghai Jiao Tong University.
2.2. Immunohistochemistry and morphometric analysis
Pancreata were dissected, fixed and processed as described before [16]. For immunofluorescence, sections of pancreata were stained with primary antibodies listed as following: guinea pig anti-INSULIN (1:200, Abcam ab7842, RRID: AB_306130), rabbit anti-p-S6 (Ser240/244) (1:100, Cell Signaling Technology 5364, RRID: AB_10694233), mouse anti-DNMT3A (1:100, Novus NB120-13888, RRID: AB_789607) and mouse anti-EED (1:1,000, Millipore 09–774, RRID: AB_1587000). Detection was performed using Alexa Fluor 488, 594 and 647 (Jackson ImmunoResearch). Images were acquired using a Zeiss LSM710 confocal microscope and analyzed using ZEN (Zeiss) and ImageJ software.
2.3. Immunoblot analysis
Proteins were extracted from isolated islets of 8-week-old mice, and then quantified by BCA assay. Protein extracts were subjected to electrophoresis on 4–20% polyacrylamide gels (SDS-PAGE) and transferred onto a nitrocellulose membrane. The membrane was incubated with primary antibodies listed as following: mouse anti-DNMT3A (1:1,000, Novus NB120-13888, RRID: AB_789607), mouse anti-TUBULIN (1:10,000, Sigma–Aldrich, T9026, RRID: AB_477593), mouse anti-EED (1:2,000, Millipore 09–774, RRID: AB_1587000), mouse anti-H3K27me3 (1:1,000 Abcam ab6002, RRID: AB_305237) and rabbit anti-H3 (1:3,000, Abcam ab1791, RRID: AB_302613) overnight at 4 °C. TUBULIN were used as an internal control to normalize band intensity.
2.4. Islet isolation, cell culture, and treatment
Pancreatic islets were isolated as described before [16]. Briefly, Collagenase P (Roche) solution was injected into the common hepatic bile duct of the mice to perfuse the pancreas. Pancreas were removed and then digested at 37 °C. Islets were handpicked after several purification steps under a microscope. Murine islets isolated from 8-week-old WT and βRapKO mice were initially recovered in 1640 RPMI supplemented with 10% serum, 11.1 mM glucose overnight. For overexpression, lentiviruses that expressed Dnmt3a or GFP were constructed (Genechem). Islets were transfected with LvGFP or LvDnmt3a for 24-h, then culture medium was replaced, and islets were cultured for another 48-h. Islets were collected after 72-h of virus transfection for further molecular studies. For western blotting to evaluate the level of DNMT3A or preparation for RNA sequencing, islets were harvested and stored at −80 °C. For ex vivo GSIS, fresh isolated islets were firstly incubated in Krebs Ringer Bicarbonate HEPES solution containing 2.8 mM glucose for 1-h. Then, a total of 10 islets of similar size were randomly chosen and incubated at 2.8 or 16.7 mM glucose for 1-h. At the end of incubation, the insulin concentrations of the cultured supernatant and insulin content of the islets were calculated (Mouse Insulin ELISA kit, Alpco).
The Mouse insulinoma cells (MIN6 cell line) were purchased from CAMS Cell Culture Center (Beijing, China) and have been authenticated and tested for mycoplasma contamination. MIN6 cells were grown in DMEM medium containing 25.0 mM glucose, 15% FBS, 100 IU/ml penicillin, and 100 ug/ml streptomycin, 10.2 mM l-glutamine, 2.5 mM β-mercaptoethanol at 37 °C in a humidified 5% CO2 atmosphere. Cells were planted on a 6-well plate. When they reached 80% confluence, each well of cells was treated with 25 nmol/L rapamycin (Cell Signaling Technology, Boston, MA, USA) or DMSO for 48-h. At the end of culture, cells were harvested for protein extraction and western blotting.
2.5. RNA sequencing, MeDIP sequencing and microarray
Total RNA was extracted from 8-week-old WT + LvGFP, βRapKO + LvGFP and βRapKO + LvDnmt3a islets using RNeasy micro kit (Qiagen, Germany). Paired-end libraries were synthesized by using the TruSeq® RNA Sample Preparation Kit (Illumina, USA) following TruSeq® RNA Sample Preparation Guide. RNA sequencing was performed on the Illumina HiSeq 2500 (Illumina, USA). The library construction and sequencing were performed at Shanghai Biotechnology Corporation as described before [20]. Genomic DNA of isolated islets was extracted by using DNeasy Blood Tissue Kit (Qiagen, GmBH, Germany). MeDIP was performed using a MagMedIP kit (Diagenode, Denville, NJ, USA) as following the manufacture's instruction. Libraries were prepared and sequenced by Illumina HiSeq 2500 with 50 cycles. Reads were aligned to reference mouse genome (mm10) by Bowtie2 v2.2.5. Peak locations and distributions on the chromosome (Promoter, 5′UTR, CDS, 3′UTR and TTR) were detected by MACS v0.14.2, and differential methylation regions (DMRs) were analyzed by MEDIPS v1.12.0R Bioconductor package. The threshold parameter setting is |fold change| > 1.5 and P value < 0.05. Microarray were performed as previously described [16].
2.6. ATAC sequencing
The ATAC-seq was carried out on 8-week-old WT and βRapKO islets as previously described [21]. Briefly isolated islets were dissolved, qualified and then lysed in lysis buffer (10 mM Tris–HCl (pH 7.4), 10 mM NaCl, 3 mM MgCl2 and NP-40) for 10 min on ice to prepare the nuclei. Immediately after lysis, nuclei were spun at 500g for 5 min to remove the supernatant. Nuclei were then incubated with the Tn5 transposase and tagmentation buffer (Vazyme Biotech, Nanjing, China) at 37 °C for 30 min. After the tagmentation, the stop buffer was added directly into the reaction to end the tagmentation. PCR was performed and aimed products were purified with VAHTSTM DNA Clean Beads (Vazyme, N411) finally. Library concentration was measured using Qubit® DNA Assay Kit in Qubit® 3.0 (Life Technologies, CA, USA) to preliminary quantify. Insert size was assessed using the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA), and after the insert size consistent with expectations, qualified insert size was accurate quantified using qPCR by Step One Plus Real-Time PCR system (ABI, USA). The library preparations were sequenced on an Illumina Hiseq X Ten platform with 150bp paired-end module. Peak annotation was performed using HOMER.
2.7. Statistical analyses
All data were presented as mean ± SEM. Statistical analysis was performed with unpaired Student's t test with a two-tailed distribution for two groups or ANOVA/Bonferroni's correction for multiple groups. P < 0.05 was considered as statistically significant.
2.8. Data and resource availability
Microarray and MeDIP-seq data of this study have been deposited in NCBI GEO (Gene Expression Omnibus) under accession code GSE84404 and GSE95775. RNA-seq data of WT + LvGFP, βRapKO + LvGFP and βRapKO + LvDnmt3a islets have been deposited in GSE130910. The ATAC-seq raw data generated are deposited in the database of the NCBI Sequence Read Archive under the accession code SRP159244. RNA-seq data of purified β-cells from 8-week-old WT, βRapKOGFP and euglycemic βRapKOGFP are available under accession code GSE130792. All data supporting the findings of this study are available with the article. Additional data and resources are available from the corresponding author upon reasonable request.
3. Results
3.1. Loss of mTORC1 decreases DNA methylation and de-represses β-cell immature genes
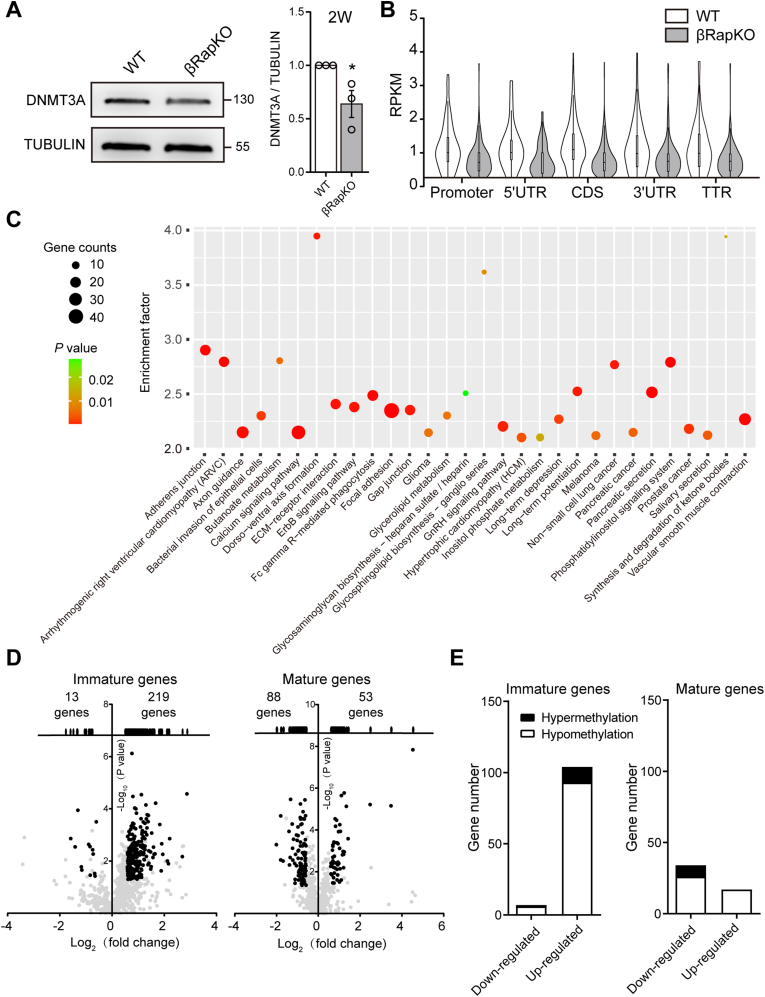
We have previously reported that mTORC1 signaling is required for proper postnatal growth and maturation of β-cell by using β-cell specific Raptor knockout mice (βRapKO), which might occur partly via DNMT3A-mediated de novo methylation [16,20]. We measured DNMT3A expression by quantitative immunofluorescence analysis during the critical postnatal maturation window (P1, P4, P8, P11 and P14) (Supplementary Fig. 1). The dynamic expression pattern of DNMT3A in neonatal β-cells coincided with mTORC1 activities (Supplementary Fig. 2), suggesting that the temporal high induction of DNMT3A and mTORC1 levels during the postnatal period might be crucial for functional maturation of neonatal β-cell. Moreover, we found a significant decrease in DNMT3A protein abundance in islets from 2-week-old Raptor-deficient mice before glycemic level rose [16] (Figure 1A), suggesting a direct effect of mTORC1 on DNMT3A in neonatal β-cell.
Figure 1.
De-repression of hypo-methylated immature genes in βRapKO. (A) Western blot showed decreased DNMT3A in islets of 2-week-old βRapKO mice. The results were repeated in three independent experiments with islets obtained from WT (n = 3) and βRapKO (n = 3). (B) The violin plots show methylation distributions of five relevant regions based on genome-wide analysis of DNA extracted from 8-week-old WT and βRapKO islets. (C) Performing pathway analysis of all genes with DMRs obtained from MeDIP-seq [16], the significantly changed pathways are presented. (D) Volcano plots of gene expression of immature genes and mature genes obtained from Microarrays of 8-week-old WT and βRapKO islets. (E) The bar graphs showed the DNA methylation change of these differently expressed immature and mature genes. Black and white bars exhibited hypermethylation and hypomethylation, relatively. Results were presented as mean ± SEM of independent experiment indicated as above. ∗P < 0.05. Differences between groups were assessed by unpaired two-tailed Student's t test.
DNA immunoprecipitation (MeDIP)-seq were performed on islets of 8-week-old βRapKO and WT mice. A total of 22,046 differential methylation regions (DMRs) were identified and annotated to 7,496 differentially methylated genes (DMGs). Overall, we found that methylation values (RPKM) in βRapKO islets were all lower than WT in five relevant regions (Promoter, 5′UTR, CDS, 3′UTR and TTR) (Figure 1B). Pathway analysis of all genes with differential methylation regions (DMRs) identified metabolic pathway (butanoate, glycerolipid, inositol phosphate and ketone bodies), calcium signaling pathway and pancreatic secretion pathway were significantly enriched (Figure 1C).
Recently, Qiu et al. [8] identified 1,024 β-cell mature genes and 692 β-cell immature genes based on fetal and adult mouse β-cell RNA sequencing. Based on this dataset, we found that Raptor preferentially regulated 232 β-cell immature genes and 141 β-cell mature genes (Figure 1D). Interestingly, 94.4% (219 of 232) of β-cell immature genes were up-regulated following Raptor deletion, however, β-cell mature genes showed comparable up- and down-regulatory trends (Figure 1D). We then evaluated the methylation level of these genes and found that the upregulated immature genes that were hypo-methylated in Raptor-deficient β-cells accounts for a large part of maturation-associated genes with altered methylation (Figure 1E). These data suggest that mTORC1-DNA methylation silencing of immature genes is a key mechanism of β-cell functional maturation.
3.2. Rescue DNMT3A activity partially restores functional maturity in βRapKO islets
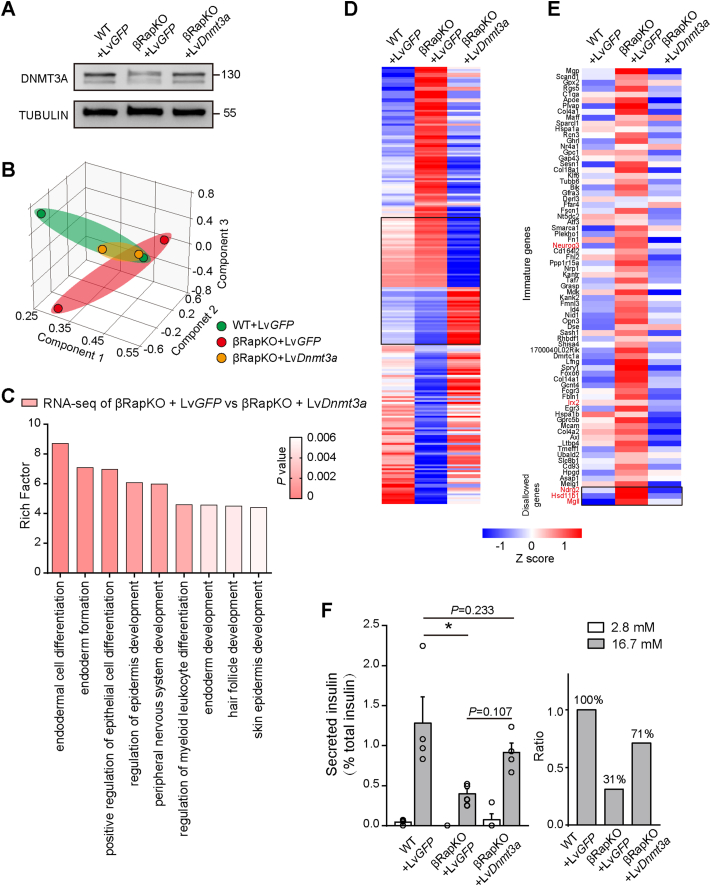
To investigate whether restored DNMT3A activity could rescue immature phenotype of Raptor-deficient β-cells, we isolated 8-week mutant islets and overexpressed DNMT3A using Dnmt3a-encoding lentivirus ex vivo (βRapKO + LvDnmt3a). The expression of DNMT3A was significantly decreased in βRapKO + LvGFP islets, while overexpression of DNMT3A in mutant islets could successfully raise its expression level to similar ranges as in WT + LvGFP islets (Figure 2A). We then performed RNA-seq on WT + LvGFP, βRapKO + LvGFP and βRapKO + LvDnmt3a islets. According to the three-dimensional principal component analysis (PCA) plot of transcriptome profile, the βRapKO + LvGFP differed greatly from WT + LvGFP controls, whereas βRapKO + LvDnmt3a group closely resembled that of the controls (Figure 2B). GO analysis revealed that Dnmt3a-regulated genes were mostly associated with the multiple cell development and differentiation processes (Figure 2C). We further explored the biological processes which are rescued and which ones are not. The data showed that positive regulation of secretion, response to nutrient levels and development growth etc. were significantly enriched between WT + LvGFP and βRapKO + LvGFP, however, did not show significantly different between WT + LvGFP and βRapKO + LvDnmt3a (Supplementary Fig. 3). Response to reactive oxygen species, positive regulation of cell death etc. were both significantly enriched between WT + LvGFP and βRapKO regardless of DNMT3A expression in Raptor-deficiency islets (Supplementary Fig. 3). The heatmap showed that a large number of genes which were dysregulated in Raptor-deficiency islets were rescued after overexpression of DNMT3A (Figure 2D). However, there was a subset of genes that were very similar between WT + LvGFP and βRapKO + LvGFP, but totally dysregulated in the βRapKO + LvDnmt3a (Black square in Figure 2D, enlarged in Supplementary Fig. 4). GO analysis revealed that these genes were associated with the positive regulation of neural precursor cell proliferation (Mdk, Nes, Ngfr) and endodermal cell differentiation (Pdcd4, Chst10).
Figure 2.
Recovering the DNMT3A expression in βRapKO partially restored the transcriptome and enhanced GSIS ex vivo. (A) Immunoblot analysis of DNMT3A expression in islets of 8-week-old WT + LvGFP, βRapKO + LvGFP and βRapKO + LvDnmt3a. (B) Principal component analysis (PCA) analysis was based on the transcriptome. Each dot represented an individual. The first three components were plotted in X, Y and Z axis. (C) GO analysis of differentially expressed genes as identified by RNA-seq analysis of 8-week-old βRapKO + LvGFP and βRapKO + LvDnmt3a islets. (D) Heatmap of selected genes in 8-week-old WT + LvGFP, βRapKO + LvGFP and βRapKO + LvDnmt3a. (E) Heatmap of selected immature genes and disallowed genes in islets of WT + LvGFP, βRapKO + LvGFP and βRapKO + LvDnmt3a. (F) Ex vivo GSIS experiments in WT + LvGFP, βRapKO + LvGFP and βRapKO + LvDnmt3a islets were performed. Islets were incubated with Krebs–Ringer bicarbonate buffer for 1-h and stimulated with 2.8 mM and 16.7 mM glucose for 1-h. Secreted insulin was normalized to total insulin in islets. Results were presented as mean ± SEM of independent experiment indicated as above. ∗P < 0.05, Differences between groups were assessed by ANOVA.
Notably, the upregulation of β-cell disallowed genes (Ndrg2, Hsd11b1, Mgll) in βRapKO + LvGFP were reversed following DNMT3A overexpression (Figure 2E). Loss of Raptor induced upregulation of a substantial group of immature genes, including endocrine progenitor marker Neurog3 and α-cell exclusive signature gene Irx2 [22]; whilerestored DNMT3A activity at least partially brought down their expressions to comparable levels as that of WT + LvGFP (Figure 2E). The above data indicates that restored DNMT3A expression could at least partially reverse the transcriptome caused by loss of Raptor, especially the abnormal induction of β-cell immature genes.
Functional maturity of β-cells was recognized by establishment of proper insulin secretion ability in response to glucose [2]. We then performed ex vivo GSIS experiments in 8-week-old WT + LvGFP, βRapKO + LvGFP and βRapKO + LvDnmt3a islets to determine their glucose responsiveness. As expected, loss of Raptor induced impaired GSIS (31.0% of WT + LvGFP, P < 0.05). There was an increased trend in the insulin secretion of βRapKO + LvDnmt3a (about 71.3% of WT + LvGFP, P = 0.233) (Figure 2F). Overexpression of DNMT3A in mutant islets partially restored β-cell function and improved insulin secretion after glucose challenge. Taken together, mTORC1/Dnmt3a activities were required to facilitate glucose responsive insulin secretion in pancreatic β-cells.
3.3. Repressive H3K27me3 dysregulation is another important feature of Raptor-deficient islets
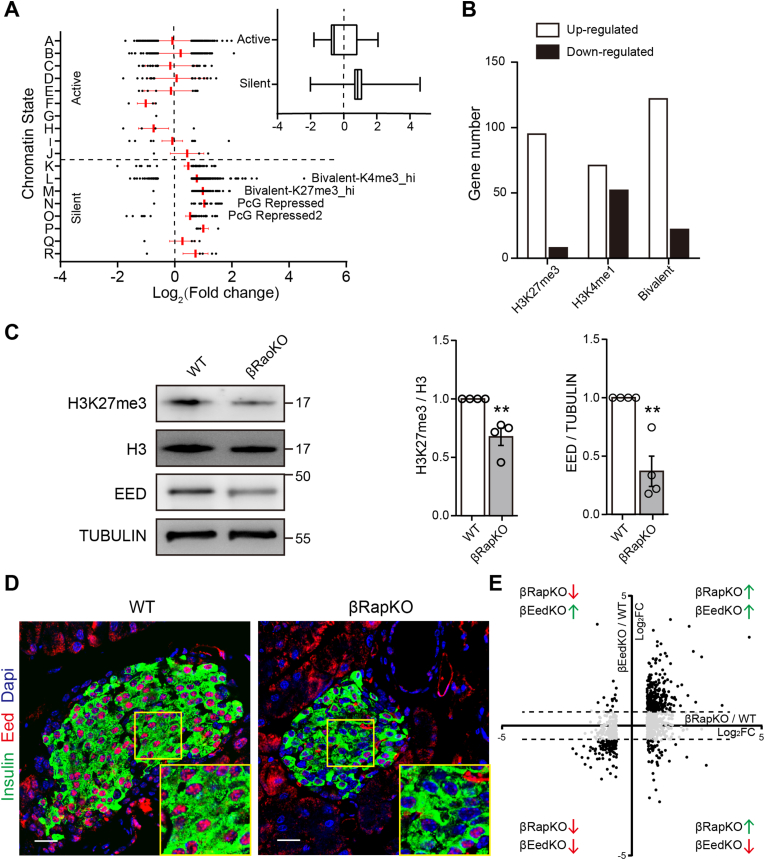
To investigate whether additional possible epigenetic regulation in Raptor-deficient β-cell, we clustered all DEGs in βRapKO into 18-chromatin-state as describe by the resource [23] and grouped state A-J and state K-R genes into two categories “active” and “silent”, according to chromatin-state segmentation [23]. As shown in boxplot (Figure 3A), most DEGs in the “active” category showed minimal variation across the chromatin states. However, the expression of genes with “silent” signatures, i.e., in the L, M, N, O chromatin state were strikingly increased, suggesting that dysregulation occurred in these quiescent chromatin states (Figure 3A). Interestingly, these four chromatin states were all related to H3K27me3: chromatin states L and M were marked with both H3K4me3 and H3K27me3, whereas chromatin states N and O were marked with quiescent intergenic regions (H3K27me2/3). We then cross-analyzed our βRapKO Microarray results with H3K27me3 and H3K4me1 islet ChIP-seq database [24], and found that most of the preferentially upregulated genes induced by the loss of Raptor were those marked with H3K27me3 or bivalently marked (Figure 3B). Contrarily, H3K4me1-marked genes were activated or repressed without remarkable preference (Figure 3B). Thus, we hypothesize that loss of repressive H3K27me3 modification may also dominate in Raptor-deficient β-cells.
Figure 3.
Polycomb dysregulation in βRapKO islets. (A) Variation of differently expressed genes in βRapKO for all chromatin states described as previously study [23]. Then all chromatin states are grouped into two categories active and silent. The boxplots showed the expression change of clustered genes. (B) Gene number of different expressed genes with signatures of H3K27m3, H3K4me1 or bivalent based on available ChIP-seq data [24]. (C) Western blot showed decreased H3K27me3 and EED in islets of 8-week-old βRapKO mice. The results were repeated in four independent experiments with islets obtained from WT (n = 4) and βRapKO (n = 4). H3 and TUBULIN were used as loading controls, respectively. (D) Representative images of immunostaining for insulin (green) and EED (red) in 8-week-old WT and βRapKO (n = 4). (E) The X axis of scatter diagram showed the fold change of all differentially expressed genes in βRapKO. And the Y axis indicated the fold change of gene expression in βEEDKO (GEO: GSE110648 [23]) correspondingly. Results were presented as mean ± SEM of independent experiment indicated as above. ∗∗P < 0.01. Differences between groups were assessed by unpaired two-tailed Student's t test. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To determine whether loss of Raptor caused dysregulation of H3K27me3, we assessed H3K27me3 levels in Raptor-deficient islets by western blot and found a significant decrease in H3K27me3 abundance (decreased 67.5% of WT, Figure 3C). Moreover, the protein abundance of EED, a core subunit of Polycomb repressive complex 2 (PRC2) that methylates H3K27, was also remarkably decreased (Figure 3C). This was also confirmed by double immunofluorescence labeling: EED+ insulin+ cells were decreased in islets from 8-week-old mutants compared to age-matched WT (Figure 3D). Of interest, when we compared the preferentially changed genes in βRapKO and β-cell specific EED knockout islet (βEedKO, GEO: GSE110648 [23]) (Figure 3E), we found that most of the common changed genes were upregulated, including 102 immature genes. These data suggest that mTORC1-dependent EED/H3K27me3 regulation is crucial for proper repression of β-cell immature genes.
3.4. ATAC-seq confirms DNA methylation and H3K27me3 histone methylation are both required for mTORC1-dependent epigenetic silencing
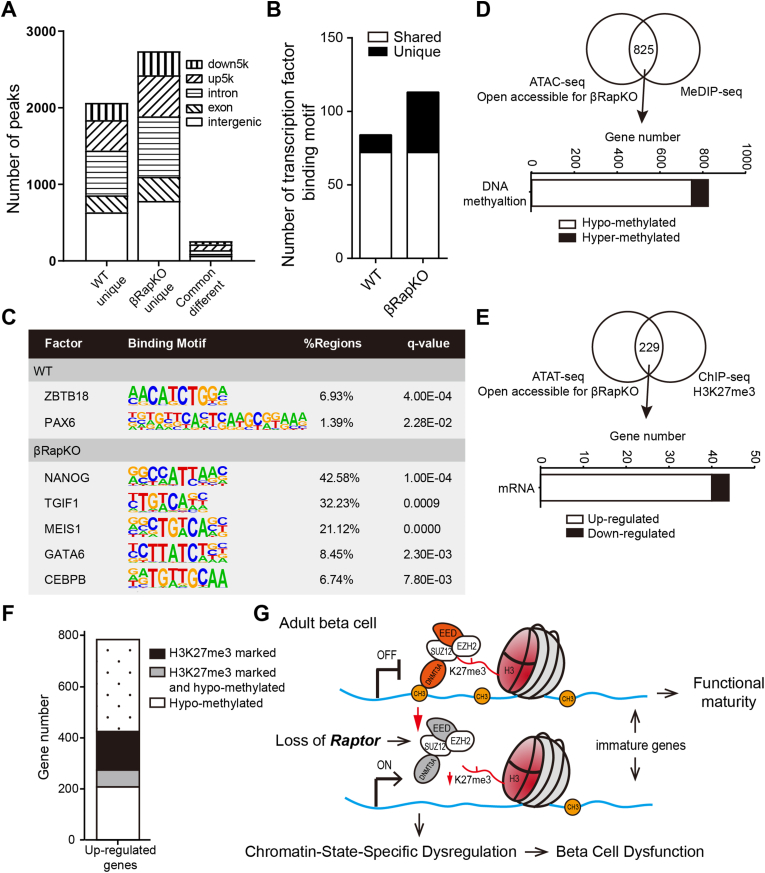
The dynamics of chromatin, which is associated with epigenetic programming of chromatin such as DNA methylation and histone modifications, is critical for β-cell function [10,11,25,26]. Here, we used a sensitive method for integrative epigenetic analysis [27], Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) to globally profile chromatin accessibility in WT and βRapKO islets. Based on ATAC peaks from WT and βRapKO, 2,065 peaks were defined as unique to WT, 2,740 peaks were unique to βRapKO and 260 peaks were common in both (Figure 4A). We used these differentially accessible regions (DARs, including peaks open accessible for WT or βRapKO) obtained from ATAC-seq to predict the regions enriched for TFs. Unique enriched binding sites in βRapKO were around four times that of WT (Figure 4B). Among them, open chromatin regions of βRapKO were enriched for binding sites of TFs, which are known to play important roles in endocrine progenitor cells (NANOG, MEIS1, CEBP/β and GATA6) (Figure 4C), whereas binding site motifs for ZBTB18 and PAX6 were enriched in WT open chromatin regions (Figure 4C).
Figure 4.
Integration of ATAC-seq data with DNA methylation and H3K27me3. (A) MAnorm analysis of ATAC peaks unique open or common different in 8-week-old WT and βRapKO islet. The genomic distribution of each peak was shown with different fill pattern bars. (B) Enriched motifs of ATAC-seq peaks for transcription factor binding sites were identified by HOMER (hypergeometric test). Bar graphs of the number of transcription factor binding motifs in WT and βRapKO. White bars showed the number of transcription factor binding motifs enrich in both WT and βRapKO open chromatin regions, while black bars showed the number of unique ones. (C) Representative transcription factor binding motifs enrich in WT- or βRapKO- specific open chromatin regions. (D) 825 genes open accessible for βRapKO showed differentially DNA methylation level. Bar graph of the number of hypo- (white) and hyper-methylated (black) genes. (E) ATAC-seq analysis of WT and βRapKO showed 229 genes open accessible for βRapKO were marked with H3K27me3. H3K27me3 marked genes were identified by ChIP-seq of adult WT mice [24]. Then observing these gene expression as shown in bar graph, 40 genes showed up-regulated (white), and 4 genes down-regulated (black). (F) Bar graph showed gene number of upregulated genes in 8-week-old βRapKO mice was associated with epigenetic regulations, including DNA methylation and H3K27me3. (G) Schematic diagram of mTORC1-denpendent epigenetic regulation, including two major epigenetic mechanisms, DNA methylation and H3K27me3.
To determine whether altered open accessible chromatin resulted from changes of DNA methylation and/or H3K27me3 histone methylation, we integrated our ATAC-seq with MeDIP-seq and ChIP-seq. All these DARs were annotated to 1,461 genes with WT open chromatin regions and 1,757 genes with βRapKO. Since most immature genes were upregulated with hypomethylation, 1,757 genes with more open accessible for βRapKO were expected to contain genuine targets of epigenetic regulation. Thus, we evaluated the DNA methylation changes of these 1,757 genes and identified 825 genes had DMRs (|fold change| > 1.5, P < 0.05, Figure 4D, Upper panel), among which 90.7% were hypo-methylated (Figure 4D, Lower panel); indicating a predominate way of DNA methylation regulation. Expression of 104 genes in 825 genes with DMRs were significantly changed (|fold change| > 1.5, P < 0.05). Among hypomethylated genes, 65 genes were increased and 29 genes were decreased (Supplementary Fig. 5). Next, to confirm the regulatory role of H3K27me3 in open accessible regions of βRapKO, we integrated data of ATAC-seq, ChIP-seq and Microarray of Raptor-deficient islets. H3K27me3 marked genes were identified by ChIP-seq of adult WT mice [24]. Interestingly, we found 229 genes with open accessible for βRapKO were marked with H3K27me3 (Figure 4E, Upper panel). Among them, 44 genes were preferentially changed at mRNA levels, including 40 up-regulated genes (11 immature genes) (Figure 4E, Lower panel).
In summary, we found more than half of the preferentially upregulated genes in βRapKO islet could be attributed to at least one of the two silencing mechanisms in Raptor-deficient β-cells: 274 genes (34.9%) were hypo-methylated, 217 genes (27.6%) were marked with H3K27me3, and 66 genes (8.4%) had both modification (Figure 4F). As summarized in Figure 4G, chromatin-state-specific epigenetic regulations, including DNA methylation and H3K27me3, are required for mTORC1-dependent epigenetic silencing of immature genes in pancreatic β-cells.
3.5. mTORC1 is epigenetically required for silencing β-cell immature genes
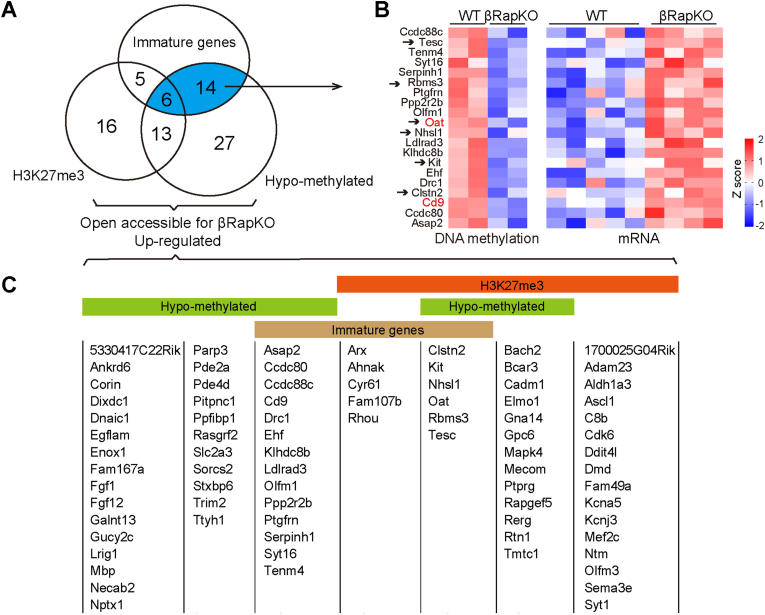
To find out potential candidate genes with chromatin-state-specific dysregulation, we filtered and summarized Raptor-regulated genes with criteria illustrated in Figure 5A. We identified 81 upregulated and open accessible peaks for βRapKO genes that were potentially regulated by DNA methylation and/or H3K27me3 (Figure 5A,C). Among these genes, 25 candidate genes were highly expressed in immature β-cells, including 11 genes with H3K27me3 marked (Figure 5C), 20 with methylation alteration (heatmaps in Figures 5B and 6 with both regulation (Tesc, Rbms3, Oat, Nhsl1, Kit, Clstn2) (Figure 5A,C). Importantly, all these 25 immature genes remained significantly upregulated in purified β-cells from 8-week-old euglycemic βRapKO by implanting an insulin pump, indicating a direct role of Raptor in repression of these immature genes (Supplementary Fig. 6). Indeed, we confirmed a direct induction of immature gene CD9 (Figure 5B,C), a gene functionally related with human β-cell insulin secretion [28] in Min6 cells treated with mTORC1 inhibitor rapamycin (Supplementary Fig. 7). These data further demonstrated that the de-repression of immature genes was directly due to inactivation of mTORC1 signaling.
Figure 5.
DNMT3A-mediated DNA methylation and PRC2-mediated histone methylation for silencing immature genes in pancreatic β-cell. (A) Venn diagram showed 81 up-regulated genes open accessible for βRapKO with epigenetic dysregulation. Gene expression, DNA methylation, chromatin accessibility was obtained from Microarray, MeDIP-seq and ATAC-seq of WT and βRapKO. H3K27me3 marked genes were identified by ChIP-seq of adult WT mice [24]. (B) Heatmaps of 20 immature genes with hypo-methylated obtained from Microarray and MeDIP-seq, with preferential change by 1.5-fold both at mRNA and DNA methylation levels. Genes pointed by arrows were also marked with H3K27me3. (C) Complete gene list of Figure 5A.
Pancreatic β-cell plays a crucial role in maintaining normal glucose homeostasis. A large-scale rewiring of transcriptional programs have proposed that downregulation of β-cell immature genes is required for transition from β-cell progenitor to mature β-cells [8,29]. Immature genes, such as Mafb [30], Pdgfrα [31] and Prdm16 [32], are highly expressed in β-cell progenitors, promote differentiation or proliferation to expand β-cell mass. However, these declined rapidly in juvenile β-cell after birth. A growing body of evidence suggests that epigenetic regulations, such as DNA methylation [10,33,34], histone modifications (H3K4me3 [35], H3K27me3 [23,25], HDACs [36]) and MicroRNAs [11], are tightly linked to β-cell identity and maturity. Our previous study showed that mTORC1 directly regulates β-cell functional maturation and identity maintenance [13,16,17,19], which requires repression of immature genes and disallowed genes. It remains unclear how mTORC1 regulates immature genes to drive the initial acquisition of glucose responsiveness during β-cell functional maturation.
It has been documented that β-cell specific Dnmt3a knockout mice showed DNA methylation-mediated repression of major developmental metabolic genes, such as HK1 and LDHA, which are required for acquisition of β-cell GSIS function [10]. Here, we have provided evidence that mTORC1 is critical for establishing proper DNA methylation profile in β-cells to facilitate glucose responsiveness. First, we found the expressions of mTORC1/PS6 and DNMT3A to be both induced and peak at P1–P4 in the first maturation wave. Second, loss of Raptor in β-cells resulted in a profound reduction of DNMT3A protein in 2-week βRapKO, independent of hyperglycemia. Third, Raptor ablation induced hypo-methylation and upregulation of a substantial group of genes, including immature genes and disallowed genes. Fourth, ATAC-seq and MeDIP-seq identified that 50% Raptor-induced upregulated genes with more open accessibility were hypo-methylated. Finally, rescued DNMT3A expression in Raptor-deficient islets partially reversed the abnormal induction of immature genes, and restored glucose responsiveness of mutant islets. We also found that disallowed genes (Hsd11b1, Mgll) were de-repressed in βRapKO + LvGFP and reversed after overexpression of DNMT3A. Mgll, a potential coupling factor in insulin secretion [37] and Hsd11b1, a major regulator of tissue-specific glucocorticoid effects [38], were increased in β-cells of human type 2 diabetes [39,40]. Dysregulation of these disallowed genes were reported to be associated with disruption of glucose metabolism and stimulus-secretion coupling [39,40]. Such epigenetic silencing was also observed in hematopoietic stem cell, in which Dnmt3a dominates to repress the stem cell program and silence HSC multipotency genes [41]. Moreover, Sangeeta et al. reported that loss of Dnmt1 hypomethylated and de-repressed Arx, and thus converted β-cell to α-cell [42]. Type 2 diabetic islet has been reported to suffer from decreased methyl donor levels and decreased activity and DNA methyltransferases [43]. Several studies have proposed that establishment of stage-specific DNA methylation shapes the gene expression patterns that guide β-cell differentiation and define the fully functional β-cell phenotype [[44], [45], [46]]. Our findings in diabetic Raptor-deficient immature β-cells and our observations of temporal high induction of DNMT3A and mTORC1 during the first maturation wave reinforced the physiological importance of mTORC1/DNMT3A regulation for β-cell functional maturation. Of course, we should not forget that overexpression of DNMT3A might also lead to caveats: for example, appropriate binding partners needed for the function of DNMT3A may or may not be present at a given stage. Moreover, overexpression of DNMT3A might also lead to a dominant negative effect, a subset of genes was specifically induced which were associated with neural precursor cell proliferation and endodermal cell differentiation.
Polycomb group (PcG) is one of evolutionarily conserved epigenetic regulatory factors and subdivided into two main complexes: Polycomb repressive complex 1 (PRC1) and 2 (PRC2) [47]. The PRC2 established and maintained the H3K27me3 modification which is associated with silencing of genes involved in cell cycle and differentiation [48,49]. In beta-cells, H3K27me3 was primarily observed in genes that promote alternate developmental fates, as well as in several genes whose function was known to be deleterious for mature β-cells [25]. The histone methyltransferase activity of PRC2 is mediated by enhancer of zeste homologue 2 (EZH2), and requires at least two other core components, suppressor of zeste 12 (SUZ12) and embryonic ectoderm development (EED) [48]. We have previously reported that mTORC1/Ezh2 pathway is essential for neonatal β-cell proliferation and identity maintenance in mice [19]. Conditional ablation of Ezh2 in β-cells led to a replication failure and mild diabetes due to decreased H3K27me3 at the Ink4a/Arf locus [50]. We found that loss of Raptor not only decreased Ezh2 expression [19], but also reduced EED protein abundance, both of which contributes to a low level of H3K27me3 in β-cells. Moreover, we found 229 genes with H3K27me3 signature became more accessible for βRapKO and 44 genes were preferentially changed at transcription level. Among them, 40 out of 44 were found upregulated, including 11 immature genes. ALDH1A3 is one of the upregulated genes in Raptor-deficient β-cells with H3K27me3 modification. ALDH1A3 is highly expressed in dedifferentiated cells and serves as a marker for β-cell dedifferentiation [51]. Moreover, Arx, another Raptor/H3K27me3 regulated gene, is an α-cell predominant transcription factor that determines α-cell fate. These results support our previously reported beta to alpha transition and provide another clue to the ways that epigenetic modification may participate in β-cell identity maintenance.
Several studies have shown that human islets from donors with type 2 diabetes had reduced DNA methylation and Polycomb-dependent H3K27m3 levels [23,52,53]. DNA methylation and H3K27me3 could take complementary roles and cooperate in regulating imprinted genes during embryonic development [54]. Viré et al. showed that PRC2 may directly recruit DNMT3A [55]. In the present study, we found Bach2 is one of the genes that are both regulated by DNA methylation and H3K27me3 modification. Excitingly, Bach2 was significantly upregulated in type 2 diabetic β-cells, and β-cell failure can be effectively reversed via BACH2 inhibition [56].
In summary, we have mapped an epigenetic regulatory network in Raptor-deficient immature β-cells and revealed the importance of mTORC1 signaling in maintaining proper DNMT3A-mediated DNA methylation and PRC2-mediated histone methylation for silencing immature genes in pancreatic β-cell. mTORC1/epigenetic induction is especially important during physiological functional maturation after birth which link metabolic signals to immature gene silencing. Some of the mTORC1-dependent epigenetic silenced genes were de-repressed in human type 2 diabetes. These integrated data advance our understanding of the mTORC1, by integrating environmental metabolites and epigenetic modifications, regulates β-cell identity and functional maturation, and impacts disease risk of type 2 diabetes.
Author contribution
Q.C.N conceived and designed the study, performed the experiments, analyzed the data, and wrote the manuscript. Q.D.W conceived and designed the study, wrote and reviewed the manuscript. J.J.S, Y.C.W, Y.Q.W and J. W. L. performed the experiments. G.N. and W.Q.W reviewed the manuscript.
Disclosure summary
The authors have nothing to disclose.
Acknowledgements
This study was supported by National Natural Science Foundation of China grants (82070795, 81870527, 82100835) and grants from Shanghai Sailing Program 21YF1426900. We thank Prof D. Accili (Department of Medicine and Naomi Berrie Diabetes Center, Vagelos College of Physicians & Surgeons of Columbia University) and Dr. Yanyun Gu (Department of Endocrine and Metabolic Diseases, Shanghai Institute of Endocrine and Metabolic Diseases, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China) for the helpful discussion and valuable advice. We are grateful to Mrs Ying Huang, Mrs Qin Hang and all members of the Core Facility of Basic Medical Sciences of SJTU for their technical support. We also thank Michelle Lee for her careful editing.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2022.101559.
Contributor Information
Weiqing Wang, Email: wqingw61@163.com.
Qidi Wang, Email: wqd11094@rjh.com.cn.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Weir G.C., Gaglia J., Bonner-Weir S. Inadequate β-cell mass is essential for the pathogenesis of type 2 diabetes. The lancet. Diabetes & endocrinology. 2020;8(3):249–256. doi: 10.1016/S2213-8587(20)30022-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blum B., Hrvatin S.S., Schuetz C., Bonal C., Rezania A., Melton D.A. Functional beta-cell maturation is marked by an increased glucose threshold and by expression of urocortin 3. Nature Biotechnology. 2012;30(3):261–264. doi: 10.1038/nbt.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nishimura W., Takahashi S., Yasuda K. MafA is critical for maintenance of the mature beta cell phenotype in mice. Diabetologia. 2015;58(3):566–574. doi: 10.1007/s00125-014-3464-9. [DOI] [PubMed] [Google Scholar]
- 4.Gu C., Stein G.H., Pan N., Goebbels S., Hornberg H., Nave K.A., et al. Pancreatic beta cells require NeuroD to achieve and maintain functional maturity. Cell Metabolism. 2010;11(4):298–310. doi: 10.1016/j.cmet.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor B.L., Liu F.F., Sander M. Nkx6.1 is essential for maintaining the functional state of pancreatic beta cells. Cell Reports. 2013;4(6):1262–1275. doi: 10.1016/j.celrep.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Meulen T., Donaldson C.J., Caceres E., Hunter A.E., Cowing-Zitron C., Pound L.D., et al. Urocortin3 mediates somatostatin-dependent negative feedback control of insulin secretion. Nat Med. 2015;21(7):769–776. doi: 10.1038/nm.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matschinsky F.M., Wilson D.F. The central role of glucokinase in glucose homeostasis: a perspective 50 Years after demonstrating the presence of the enzyme in islets of langerhans. Frontiers in Physiology. 2019;10:148. doi: 10.3389/fphys.2019.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qiu W.L., Zhang Y.W., Feng Y., Li L.C., Yang L., Xu C.R. Deciphering pancreatic islet beta cell and alpha cell maturation pathways and characteristic features at the single-cell level. Cell Metabolism. 2017;25(5):1194–1205 e1194. doi: 10.1016/j.cmet.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Lemaire K., Granvik M., Schraenen A., Goyvaerts L., Van Lommel L., Gomez-Ruiz A., et al. How stable is repression of disallowed genes in pancreatic islets in response to metabolic stress? PLoS One. 2017;12(8):e0181651. doi: 10.1371/journal.pone.0181651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dhawan S., Tschen S.I., Zeng C., Guo T., Hebrok M., Matveyenko A., et al. DNA methylation directs functional maturation of pancreatic beta cells. Journal of Clinical Investigation. 2015;125(7):2851–2860. doi: 10.1172/JCI79956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez-Sanchez A., Nguyen-Tu M.S., Rutter G.A. DICER inactivation identifies pancreatic beta-cell "disallowed" genes targeted by MicroRNAs. Molecular Endocrinology. 2015;29(7):1067–1079. doi: 10.1210/me.2015-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salinno C., Cota P., Bastidas-Ponce A., Tarquis-Medina M., Lickert H., Bakhti M. Beta-cell maturation and identity in health and disease. International Journal of Molecular Sciences. 2019;20(21) doi: 10.3390/ijms20215417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ni Q., Song J., Wang Y., Sun J., Xie J., Zhang J., et al. Proper mTORC1 activity is required for glucose sensing and early adaptation in human pancreatic beta cells. The Journal of Cinical Endocrinology and Metabolism. 2021;106(2):e562–e572. doi: 10.1210/clinem/dgaa786. [DOI] [PubMed] [Google Scholar]
- 14.Jaafar R., Tran S., Shah A.N., Sun G., Valdearcos M., Marchetti P., et al. mTORC1 to AMPK switching underlies beta-cell metabolic plasticity during maturation and diabetes. Journal of Clinical Investigation. 2019;129(10):4124–4137. doi: 10.1172/JCI127021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nojima H., Tokunaga C., Eguchi S., Oshiro N., Hidayat S., Yoshino K., et al. The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. Journal of Biological Chemistry. 2003;278(18):15461–15464. doi: 10.1074/jbc.C200665200. [DOI] [PubMed] [Google Scholar]
- 16.Ni Q., Gu Y., Xie Y., Yin Q., Zhang H., Nie A., et al. Raptor regulates functional maturation of murine beta cells. Nature Communications. 2017;8:15755. doi: 10.1038/ncomms15755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yin Q., Ni Q., Wang Y., Zhang H., Li W., Nie A., et al. Raptor determines beta-cell identity and plasticity independent of hyperglycemia in mice. Nature Communications. 2020;11(1):2538. doi: 10.1038/s41467-020-15935-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helman A., Cangelosi A.L., Davis J.C., Pham Q., Rothman A., Faust A.L., et al. A nutrient-sensing transition at birth triggers glucose-responsive insulin secretion. Cell Metabolism. 2020;31(5):1004–1016. doi: 10.1016/j.cmet.2020.04.004. e1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y., Sun J., Ni Q., Nie A., Gu Y., Wang S., et al. Dual effect of on neonatal β-cell proliferation and identity maintenance. Diabetes. 2019;68(10):1950–1964. doi: 10.2337/db19-0166. [DOI] [PubMed] [Google Scholar]
- 20.Yin Q., Ni Q., Wang Y., Zhang H., Li W., Nie A., et al. Raptor determines β-cell identity and plasticity independent of hyperglycemia in mice. Nature Communications. 2020;11(1):2538. doi: 10.1038/s41467-020-15935-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui C., Li T., Xie Y., Yang J., Fu C., Qiu Y., et al. Enhancing Acsl4 in absence of mTORC2/Rictor drove beta-cell dedifferentiation via inhibiting FoxO1 and promoting ROS production. Biochimica et Biophysica Acta, Molecular Basis of Disease. 2021;1867(12):166261. doi: 10.1016/j.bbadis.2021.166261. [DOI] [PubMed] [Google Scholar]
- 22.Xin Y., Kim J., Okamoto H., Ni M., Wei Y., Adler C., et al. RNA sequencing of single human islet cells reveals type 2 diabetes genes. Cell Metabolism. 2016;24(4):608–615. doi: 10.1016/j.cmet.2016.08.018. [DOI] [PubMed] [Google Scholar]
- 23.Lu T.T., Heyne S., Dror E., Casas E., Leonhardt L., Boenke T., et al. The polycomb-dependent epigenome controls beta cell dysfunction, dedifferentiation, and diabetes. Cell Metabolism. 2018;27(6):1294–1308. doi: 10.1016/j.cmet.2018.04.013. e1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Avrahami D., Li C., Zhang J., Schug J., Avrahami R., Rao S., et al. Aging-dependent demethylation of regulatory elements correlates with chromatin state and improved beta cell function. Cell Metabolism. 2015;22(4):619–632. doi: 10.1016/j.cmet.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Arensbergen J., Garcia-Hurtado J., Moran I., Maestro M.A., Xu X., Van de Casteele M., et al. Derepression of Polycomb targets during pancreatic organogenesis allows insulin-producing beta-cells to adopt a neural gene activity program. Genome Research. 2010;20(6):722–732. doi: 10.1101/gr.101709.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie R., Everett L.J., Lim H.W., Patel N.A., Schug J., Kroon E., et al. Dynamic chromatin remodeling mediated by polycomb proteins orchestrates pancreatic differentiation of human embryonic stem cells. Cell Stem Cell. 2013;12(2):224–237. doi: 10.1016/j.stem.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature Methods. 2013;10(12):1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dorrell C., Schug J., Canaday P.S., Russ H.A., Tarlow B.D., Grompe M.T., et al. Human islets contain four distinct subtypes of beta cells. Nature Communications. 2016;7:11756. doi: 10.1038/ncomms11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanavia T., Huang C., Manduchi E., Xu Y., Dadi P.K., Potter L.A., et al. Temporal transcriptome analysis reveals dynamic gene expression patterns driving beta-cell maturation. Frontiers in Cell and Developmental Biology. 2021;9:648791. doi: 10.3389/fcell.2021.648791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Artner I., Hang Y., Mazur M., Yamamoto T., Guo M., Lindner J., et al. MafA and MafB regulate genes critical to beta-cells in a unique temporal manner. Diabetes. 2010;59(10):2530–2539. doi: 10.2337/db10-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen H., Gu X., Liu Y., Wang J., Wirt S.E., Bottino R., et al. PDGF signalling controls age-dependent proliferation in pancreatic beta-cells. Nature. 2011;478(7369):349–355. doi: 10.1038/nature10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugiyama T., Benitez C.M., Ghodasara A., Liu L., McLean G.W., Lee J., et al. Reconstituting pancreas development from purified progenitor cells reveals genes essential for islet differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(31):12691–12696. doi: 10.1073/pnas.1304507110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ouni M., Saussenthaler S., Eichelmann F., Jahnert M., Stadion M., Wittenbecher C., et al. Epigenetic changes in islets of langerhans preceding the onset of diabetes. Diabetes. 2020;69(11):2503–2517. doi: 10.2337/db20-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bacos K., Gillberg L., Volkov P., Olsson A.H., Hansen T., Pedersen O., et al. Blood-based biomarkers of age-associated epigenetic changes in human islets associate with insulin secretion and diabetes. Nature Communications. 2016;7:11089. doi: 10.1038/ncomms11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhandare R., Schug J., Le Lay J., Fox A., Smirnova O., Liu C., et al. Genome-wide analysis of histone modifications in human pancreatic islets. Genome Research. 2010;20(4):428–433. doi: 10.1101/gr.102038.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lundh M., Galbo T., Poulsen S.S., Mandrup-Poulsen T. Histone deacetylase 3 inhibition improves glycaemia and insulin secretion in obese diabetic rats. Diabetes, Obesity and Metabolism. 2015;17(7):703–707. doi: 10.1111/dom.12470. [DOI] [PubMed] [Google Scholar]
- 37.Rutter G.A., Georgiadou E., Martinez-Sanchez A., Pullen T.J. Metabolic and functional specialisations of the pancreatic beta cell: gene disallowance, mitochondrial metabolism and intercellular connectivity. Diabetologia. 2020;63(10):1990–1998. doi: 10.1007/s00125-020-05205-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morgan S.A., McCabe E.L., Gathercole L.L., Hassan-Smith Z.K., Larner D.P., Bujalska I.J., et al. 11beta-HSD1 is the major regulator of the tissue-specific effects of circulating glucocorticoid excess. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(24):E2482–E2491. doi: 10.1073/pnas.1323681111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fadista J., Vikman P., Laakso E.O., Mollet I.G., Esguerra J.L., Taneera J., et al. Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(38):13924–13929. doi: 10.1073/pnas.1402665111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marselli L., Thorne J., Dahiya S., Sgroi D.C., Sharma A., Bonner-Weir S., et al. Gene expression profiles of Beta-cell enriched tissue obtained by laser capture microdissection from subjects with type 2 diabetes. PLoS One. 2010;5(7):e11499. doi: 10.1371/journal.pone.0011499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Challen G.A., Sun D., Jeong M., Luo M., Jelinek J., Berg J.S., et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nature Genetics. 2011;44(1):23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chakravarthy H., Gu X., Enge M., Dai X., Wang Y., Damond N., et al. Converting adult pancreatic islet alpha cells into beta cells by targeting both Dnmt1 and Arx. Cell Metabolism. 2017;25(3):622–634. doi: 10.1016/j.cmet.2017.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lawlor N., Khetan S., Ucar D., Stitzel M.L. Genomics of islet (Dys)function and type 2 diabetes. Trends in Genetics. 2017;33(4):244–255. doi: 10.1016/j.tig.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parveen N., Dhawan S. DNA methylation patterning and the regulation of beta cell homeostasis. Frontiers in Endocrinology. 2021:651258. doi: 10.3389/fendo.2021.651258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J., Wu X., Zhou Y., Lee M., Guo L., Han W., et al. Decoding the dynamic DNA methylation and hydroxymethylation landscapes in endodermal lineage intermediates during pancreatic differentiation of hESC. Nucleic Acids Research. 2018;46(6):2883–2900. doi: 10.1093/nar/gky063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neiman D., Moss J., Hecht M., Magenheim J., Piyanzin S., Shapiro A.M.J., et al. Islet cells share promoter hypomethylation independently of expression, but exhibit cell-type-specific methylation in enhancers. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(51):13525–13530. doi: 10.1073/pnas.1713736114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schuettengruber B., Bourbon H.M., Di Croce L., Cavalli G. Genome regulation by polycomb and trithorax: 70 Years and counting. Cell. 2017;171(1):34–57. doi: 10.1016/j.cell.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 48.Comet I., Riising E.M., Leblanc B., Helin K. Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nature Reviews Cancer. 2016;16(12):803–810. doi: 10.1038/nrc.2016.83. [DOI] [PubMed] [Google Scholar]
- 49.Li Y., Chen X., Lu C. The interplay between DNA and histone methylation: molecular mechanisms and disease implications. EMBO Reports. 2021;22(5):e51803. doi: 10.15252/embr.202051803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen H., Gu X., Su I.H., Bottino R., Contreras J.L., Tarakhovsky A., et al. Polycomb protein Ezh2 regulates pancreatic beta-cell Ink4a/Arf expression and regeneration in diabetes mellitus. Genes & Development. 2009;23(8):975–985. doi: 10.1101/gad.1742509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim-Muller J.Y., Fan J., Kim Y.J., Lee S.A., Ishida E., Blaner W.S., et al. Aldehyde dehydrogenase 1a3 defines a subset of failing pancreatic beta cells in diabetic mice. Nature Communications. 2016;7:12631. doi: 10.1038/ncomms12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Volkmar M., Dedeurwaerder S., Cunha D.A., Ndlovu M.N., Defrance M., Deplus R., et al. DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. EMBO Journal. 2012;31(6):1405–1426. doi: 10.1038/emboj.2011.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dayeh T., Volkov P., Salo S., Hall E., Nilsson E., Olsson A.H., et al. Genome-wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion. PLoS Genetics. 2014;10(3):e1004160. doi: 10.1371/journal.pgen.1004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen Z., Yin Q., Inoue A., Zhang C., Zhang Y. Allelic H3K27me3 to allelic DNA methylation switch maintains noncanonical imprinting in extraembryonic cells. Science Advances. 2019;5(12):eaay7246. doi: 10.1126/sciadv.aay7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vire E., Brenner C., Deplus R., Blanchon L., Fraga M., Didelot C., et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439(7078):871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 56.Son J., Ding H., Farb T.B., Efanov A.M., Sun J., Gore J.L., et al. BACH2 inhibition reverses β cell failure in type 2 diabetes models. Journal of Clinical Investigation. 2021;131(24) doi: 10.1172/JCI153876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.