Abstract
Protein arginine methylation is involved in many biological processes and can be enhanced in cancer. In mammals, these reactions are catalyzed on multiple substrates by a family of nine protein arginine methyltransferases (PRMTs). However, conditions that may regulate the activity of each enzyme and that may help us understand the physiological role of PRMTs have not been fully established. Previous studies had suggested unexpected effects of temperature and ionic strength on PRMT7 activity. Here we examine in detail the effects of temperature, pH, and ionic strength on recombinant human PRMT1, PRMT5, and PRMT7. We confirmed the unusual temperature dependence of PRMT7, where optimal activity was observed at 15 °C. On the other hand, we found that PRMT1 and PRMT5 are most active near physiological temperatures of 37 °C. However, we showed all three enzymes still have significant activity at 0 °C. Furthermore, we determined that PRMT1 is most active at a pH of about 7.7, while PRMT5 activity is not dependent on pH in the range of 6.5 to 8.5. Significantly, PRMT7 is most active at an alkaline pH of 8.5 but shows little activity at the physiological intracellular pH of about 7.2. We also detected decreased activity at physiological salt conditions for PRMT1, PRMT5, and PRMT7. We demonstrate that the loss of activity is due to the increasing ionic strength. Taken together, these results open the possibility that PRMTs respond in cells undergoing temperature, salt, or pH stress and demonstrate the potential for in vivo regulation of protein arginine methylation.
Keywords: protein methylation, histone methylation, S-adenosylmethionine (SAM), protein arginine N-methyltransferase 5 (PRMT5), posttranslational modification (PTM), intracellular stress, ionic strength, temperature dependence, pH effects
Abbreviations: HsH2B, human histone H2B; PRMT, protein arginine methyltransferase
Protein diversity heavily relies on many enzymes that modify amino acid residues. A major site of protein posttranslational modification is methylation at the guanidino group of arginine residues. These modifications create distinct interactions between proteins and play a significant role in biology including transcription of DNA, activation of enzyme activity, and protein degradation (1, 2, 3, 4). In mammals, nine specific enzymes (protein arginine methyltransferases [PRMTs]) have been characterized and are classified based on the specific reaction they catalyze (5, 6, 7). While all PRMTs can initially catalyze monomethylarginine formation, type I PRMTs (PRMT1, 2, 3, 4, 6, and 8) catalyze asymmetric dimethylarginine formation and type II PRMTs (PRMT 5 and 9) catalyze symmetric dimethylarginine formation, while type III PRMTs (PRMT7) only catalyzes monomethylarginine formation. Determination of the in vitro activity of these purified PRMT enzymes can give important clues to their substrate specificity and their possible regulation (8, 9, 10, 11, 12).
For intracellular cytosolic mammalian enzymes, the assumption is usually made that activity will be at or near optimum levels at the physiological pH of about 7.2 (13), an ionic strength of about 120 mM (14), and a temperature of 37 °C (15). However, we were surprised to find that PRMT7 had little activity at 37 °C and maximum activity was noted at colder temperatures (16, 17). Additionally, little activity was found at the physiological salt concentrations that occur within a cell (16). However, these measurements were generally not taken under initial velocity conditions and we were thus interested in a more detailed picture of PRMT7 catalysis, especially under nonphysiological conditions. We were also interested in comparing the catalysis shown by PRMT7 with that of PRMT1, the major type I enzyme, creating asymmetric dimethylarginine, and PRMT5, the major type II enzyme, creating symmetric dimethylarginine residues.
Here we show that human PRMT1, PRMT5, and PRMT7 activity is optimal under varying conditions. Specifically, PRMT7 activity is optimal at subphysiological temperatures in an alkaline environment containing low salt concentrations. We have also shown that PRMT1 and PRMT5 activities are most optimal closer to physiological temperature and pH, but where reaction mixtures contain little salt.
Results
GST-HsPRMT7 is active at physiological temperatures but has optimal activity at subphysiological temperatures
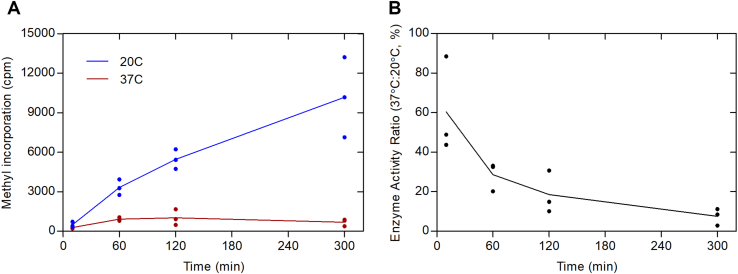
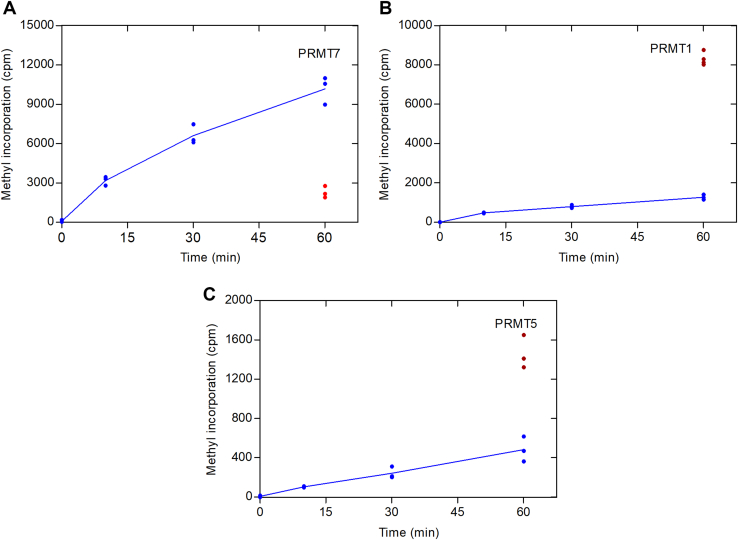
Recent findings have demonstrated that human histone H2B (HsH2B) is an excellent substrate for PRMT7, with methylation sites detected at arginine residues 29, 31, and 33 (10). A synthetic peptide containing residues 23 to 37 (HsH2B (23–37)) was also an excellent substrate (10). Significantly, optimal PRMT7 activity was found at about 20 °C with very little activity at 37 °C (16). Similar effects were found with the bacterially expressed GST human enzyme fusion protein and the insect-expressed untagged mouse enzyme (10). However, because these activities were measured at long incubation times, we wanted to determine the temperature dependence under initial rate conditions. We first determined that GST-HsPRMT7 activity is linear with time up to 1 h at both 20 °C and 37 °C with the HsH2B (23–37) peptide (Fig. 1). At 37 °C, we found only about 25% of the activity as found at 20 °C. We also showed no significant increase of activity when the peptide concentration was raised from 10 μM to 50 μM, indicating that enzyme was saturated with the peptide substrate at the lower concentration. These results now clearly show that PRMT7 activity is not optimal at the physiological temperature.
Figure 1.
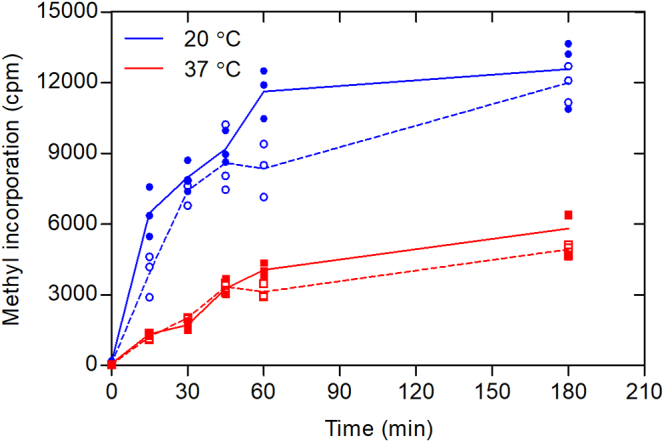
Human PRMT7 is more active at subphysiological temperatures with histone H2B peptide as a substrate. P81 phosphocellulose assay of GST-HsPRMT7 and HsH2B (23–37) was performed at 20 °C and 37 °C as described in the Experimental procedures section. Five micrograms of GST-HsPRMT7 was incubated with peptide and 0.14 μM [3H]AdoMet. Blue lines represent reaction mixtures at 20 °C using 10 μM peptide (closed circles) or 50 μM peptide (open circles). The red line represents reaction mixtures at 37 °C using 10 μM peptide (closed squares) or 50 μM peptide (open squares). HsH2B, histone H2B; PRMT, protein arginine methyltransferase.
Similar results to those shown in Figure 1 with the HsH2B (23–37) peptide substrate were found when we used the GST-GAR protein substrate (Fig. 2). At the 60-min and 120-min timepoints, only about 20 to 30% of the activity at 20 °C was found at 37 °C. Using an alternate assay where the 3H-methylated GST-GAR polypeptides formed in a 2-h assay were fractionated by SDS-PAGE and analyzed by densitometry following fluorimetry, we found 9.2% of the activity at 37 °C as found at 20 °C (data not shown). We also used this latter assay with a recombinant full-length human HsH2B protein and found less than 11% of the activity at 37 °C as found at 20 °C (data not shown).
Figure 2.
Human PRMT7 is more active at subphysiological temperatures with GST-GAR as a substrate.A, P81 phosphocellulose assay of GST-HsPRMT7 activity. GST-HsPRMT7, 6.2 μg GST-GAR, and 0.7 μM [3H]AdoMet was incubated at either 20 °C or 37 °C as described in Experimental procedures. The blue line represents the averages of individual reactions at 20 °C (blue dots). The red line represents individual replicates of reaction mixtures tested at 37 °C (red dots). B, ratios of reactions performed at 37 °C to 20 °C. The black line represents the averages of the individual ratios calculated from A. PRMT, protein arginine methyltransferase.
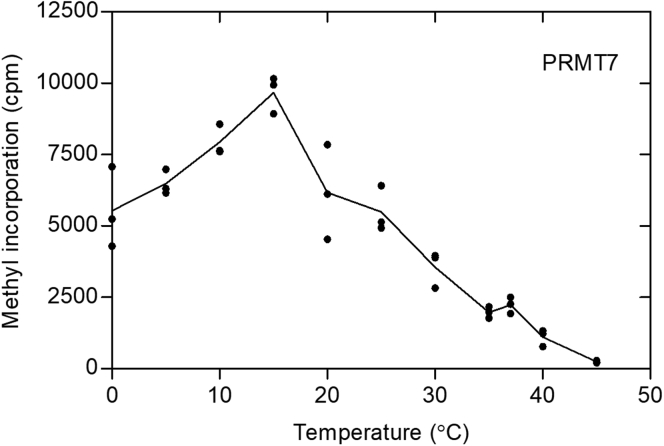
In Figure 3, we measured GST-HsPRMT7 activity at temperature values between 0 °C and 45 °C with the HsH2B (23–37) peptide using the P81 phosphocellulose assay. We found the peak of activity at 15 °C with half activity at 0 °C and at 20 to 25 °C. At the physiological temperature of 37 °C, only about 20% of the activity was found compared to that of 15 °C. These data clearly show that PRMT7 is most active in the cold.
Figure 3.
Human PRMT7 is more active at lower temperatures under steady-state kinetics. P81-based assay of GST-HsPRMT7, 10 μM HsH2B (23–37), and 0.14 μM [3H]AdoMet was carried out as described in Experimental procedures. Reaction mixtures were incubated for 30 min, spotted on P81 phosphocellulose paper, and further assayed as described in Experimental procedures. The data shown are from a single experiment: similar results were seen in three additional replicated experiments. HsH2B, histone H2B; PRMT, protein arginine methyltransferase.
Decreased human PRMT7 activity at 37 °C is not due to enzyme instability
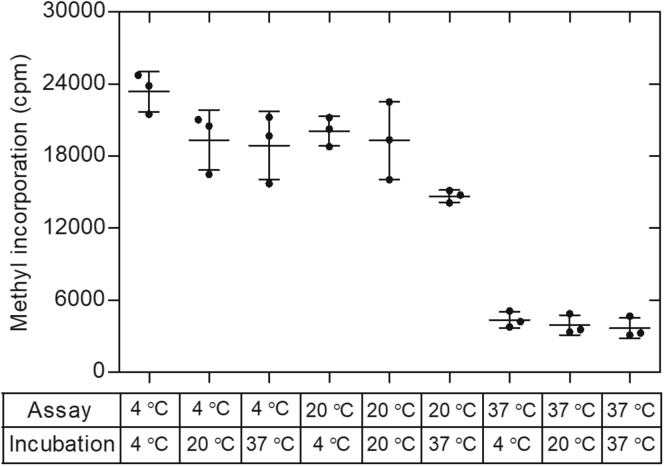
To determine if incubation at 37 °C irreversibly denatured PRMT7, we performed the experiment in Figure 4. Here, we preincubated the enzyme at 4 °C, 20 °C, and 37 °C for 2 h and then assayed activity at each of these temperatures. We found little or no loss of activity when assayed at 4 °C, suggesting that PRMT7 was not denatured at 37 °C. In all cases, similar activities were found when each of the incubated samples was assayed at the indicated temperature. Additionally, previous work had shown that the circular dichroism spectrum of PRMT7 was identical at 4 °C, 17 °C, and 37 °C, suggesting that the enzyme is stable at these temperature ranges (18). Additionally, it was shown that the molecular weight of PRMT7 by sedimentation equilibrium analysis was also identical at these three temperatures (18).
Figure 4.
Human PRMT7 activity is stable from 4 °C to 37 °C. GST-HsPRMT7 was preincubated for 2 h at 4 °C, 20 °C, and 37 °C in the presence of the HsH2B (23–37) substrate and 50 mM K-Hepes, 1 mM DTT at a final pH of 8.5. The enzymatic reaction was then initiated by addition of a final concentration of 0.7 μM [3H]AdoMet and was then incubated for 2 h at the indicated temperature followed by the P81 phosphocellulose assay as described in Experimental procedures. Data are shown for triplicate of samples with standard deviation shown as error bars. HsH2B, histone H2B; PRMT, protein arginine methyltransferase.
His-HsPRMT1 and HsPRMT5/MEP50 are most active near physiological temperatures
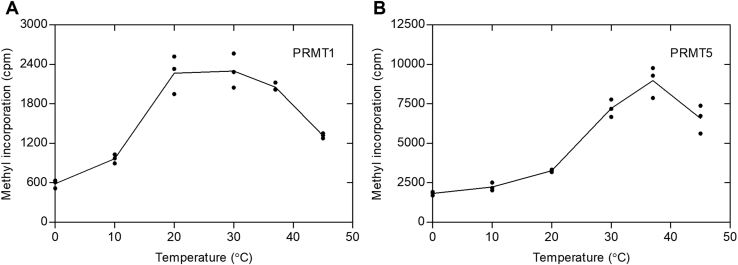
We then wanted to establish the generality of the methyltransferase activity at subphysiological temperatures with the major type I enzyme PRMT1 and the major type II enzyme PRMT5. We thus tested His-HsPRMT1 and HsPRMT5/MEP50 under similar conditions to those used above for GST-HsPRMT7 (Fig. 5). For PRMT1, we found a broad optimal peak between 20 °C and 37 °C with half optimal activities at about 15 °C and 45 °C. For PRMT5, the temperature optimum was found at 37 °C with half optimal activity of 25 °C and over 45 °C. Taken together, these results show that PRMT1 and PRMT5 have temperature optimums near physiological body temperature and show distinct patterns from PRMT7. However, we did detect the activity of both enzymes throughout this temperature range.
Figure 5.
Human PRMT1 and PRMT5/MEP50 are more active near physiological temperatures.In vitro reaction mixtures were incubated at the specified temperatures and assayed using a P81 phosphocellulose assay. Reaction mixtures contained 50 mM K-HEPES, 1 mM DTT and had a final pH of 8.0. Reactions were allowed to incubate for 30 min. A, 80 nM of His-HsPRMT1 was incubated with 10 μM HsH4 (1–21) and 0.14 μM [3H]AdoMet. B, 5.6 nM of HsPRMT5/MEP50 was incubated with 10 μM of HsH4 (1–21), and 0.14 μM [3H]AdoMet. HsH4, histone H4; PRMT, protein arginine methyltransferase.
PRMT1, PRMT5, and PRMT7 are all active at 0 °C
We found that PRMT1, PRMT5, and PRMT7 all displayed activity at 0 °C. To confirm this surprising result, we performed additional time course experiments in assays done entirely on ice (Fig. 6). All of these enzymes were found to have significant activity at 0 °C. GST-HsPRMT7 activity was almost 6-fold higher at 0 °C than at 37 °C. His-HsPRMT1 activity was 6-fold lower in activity at 0 °C than in reaction mixtures that were performed at 37 °C, and HsPRMT5/MEP50 activity was about 3-fold lower at 0 °C than at 37 °C.
Figure 6.
Human PRMT7, PRMT1, and PRMT5/MEP50 are active at 0 °C. P81 phosphocellulose assays of reactions containing GST-HsPRMT7 with HsH2B (23–37), His-HsPRMT1 with HsH4 (1–21), or HsPRMT5/MEP50 with HsH4(1–21) was carried out as described in Experimental procedures. A, Five micrograms of GST-HsPRMT7 was mixed with 10 μM HsH2B (23–37) and 0.14 μM of [3H]AdoMet. Reaction mixtures contained 50 mM K-Hepes, 1 mM DTT, at a final reaction pH of 8.5. B, 40 nM of His-HsPRMT1 was mixed with 10 μM HsH4 (1–21) and 0.14 μM of [3H]AdoMet. Reaction mixtures contained 50 mM K-Hepes, 1 mM DTT, at a final reaction pH of 7.0. C, 5.6 nM of HsPRMT5/MEP50 was mixed with 10 μM HsH4 (1–21) and 0.14 μM [3H]AdoMet. Reaction mixtures contained 50 mM K-Hepes, 1 mM DTT, at a final reaction pH of 7.0. All reaction mixtures were equilibrated to 0 °C for 2 min prior to initiation with [3H]AdoMet. The blue line represents averages of replicates at 0 °C. Red dots represent reactions incubated at 37 °C. HsH2B, histone H2B; HsH4, histone H4; PRMT, protein arginine methyltransferase.
PRMT7 is active at alkaline pH
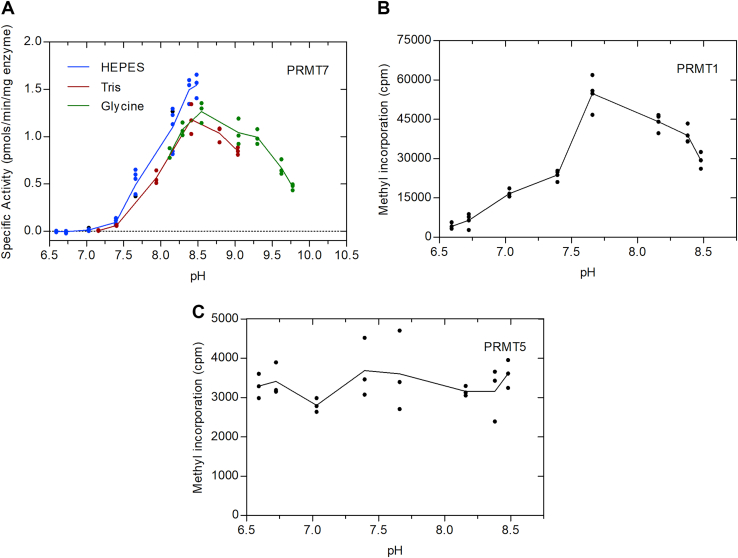
Our finding that PRMT7 is most active at nonphysiological temperatures lead us to examine its activity as a function of pH. As shown in Figure 7A, we were surprised to find little to no activity at the physiological pH of 7.2. This enzyme was found to be more active at alkaline pH values with a pH optimum of 8.4. We performed a control experiment showing that changes in pH did not affect the P81 phosphocellulose assay (Fig. S1). These results suggest that PRMT7 might respond to changes in cellular pH that lead to alkalization.
Figure 7.
pH dependence of human PRMT7, PRMT1, and PRMT5/MEP50. P81 phosphocellulose assay with either GST-HsPRMT7 and HsH2B (23–37), His-HsPRMT1 and HsH4 (1–21) or HsPRMT5/MEP50 and HsH4 (1–21) was carried out as described in Experimental procedures. A, five micrograms of of GST-HsPRMT7, 10 μM of HsH2B (23–37), and 0.14 μM [3H]AdoMet. The red line represents averages of replicates where reaction mixtures contain 50 mM K-Hepes, 1 mM DTT. The green line represents averages of replicates reaction mixtures containing 50 mM glycine and 1 mM DTT. The blue line represents averages of replicates where reaction mixtures contain 50 mM Tris and 1 mM DTT. Reaction mixtures were incubated at 20 °C for 60 min. B, 40 nM of His-HsPRMT1, 10 μM HsH4 (1–21), and 0.14 μM [3H]AdoMet were mixed with 50 mM K-Hepes and 1 mM DTT and incubated at 37 °C for 30 min. C, 5.6 nM of HsPRMT5/MEP50, 10 μM of HsH4 (1–21) and 0.14 μM [3H]AdoMet were mixed with 50 mM K-Hepes and 1 mM DTT and incubated at 37 °C for 30 min. The black line represents the averages of black dot replicates for His-HsPRMT1 and HsPRMT5/MEP50. In all cases, the final pH was determined from mock mixtures of buffer components, the buffer used in the enzyme preparation, and the solvent for the [3H]AdoMet. HsH2B, histone H2B; HsH4, human histone H4; PRMT, protein arginine methyltransferase.
We also performed pH studies using a substrate of GST-GAR and an SDS-PAGE assay as described in the Experimental procedures (Fig. S2). The results shown here are very similar to those found in Figure 7A with the HsH2B (23–37) peptide and the P81 phosphocellulose assay.
PRMT1 and PRMT5 have distinct pH activity profiles from PRMT7
To compare the activity of the major PRMT1 type I enzyme and the major PRMT5 type II enzyme with PRMT7 as a function of pH, we performed the assays shown in Figure 7, B and C. We found that His-PRMT1 has a pH optimum of about 7.7 with good activity at the physiological pH of 7.2. Additionally, we found that PRMT5/MEP50 is equally active across a wide range of pH values between 6.6 and 8.5. Here, PRMT1 and PRMT5 appeared to differ from PRMT7 in showing activity at the physiological pH.
PRMT7 is inhibited by the addition of salts
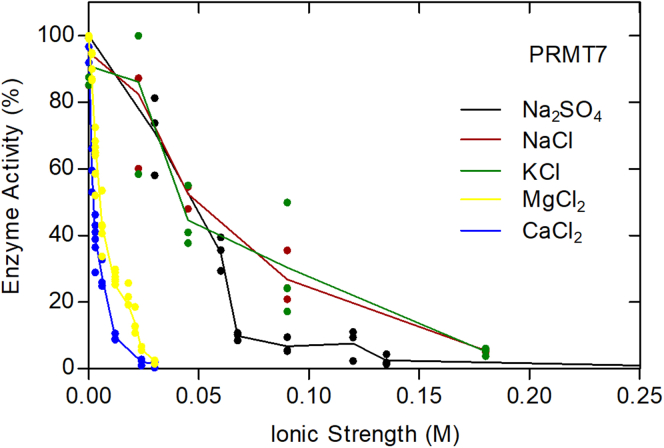
Previous studies have shown that GST-HsPRMT7 activity decreases with increasing NaCl concentrations with little activity seen at physiological salt concentrations, particularly for the HsH2B (23–37) peptide (16). Additionally, a loss of approximately 50% activity was found in 2 mM magnesium and calcium salts (16). Given our results above that PRMT7 has optimal activity outside of the normal physiological range of pH and temperature, we wanted to expand upon these findings. In Figure 8, we show that GST-HsPRMT7 activity decreases not only with NaCl but also with KCl. To ask if this effect is due to ionic strength, we also assayed the enzyme in Na2SO4. We found that the activity is dependent on the ionic strength with half activity at about 50 mM and almost no activity at 150 mM. Since typical intracellular ionic strength values range from about 130 to 270 mM (14), these results suggest that PRMT7 activity is generally repressed under physiological conditions. Control experiments showed that changes in the ionic strength of the incubation mixture did not affect the efficiency of the P81 phosphocellulose assay (Fig. S3). We also performed assays on the effect of ionic strength using GST-GAR as a substrate and the SDS-PAGE assay (Fig. S4). We note that the loss of activity with ionic strength is less with the GST-GAR substrate than with the HsH2B (23–37) peptide, confirming the results of Feng et al. (16).
Figure 8.
Human PRMT7 activity decreases with ionic strength. Five micrograms of GST-HsPRMT7 was mixed with 10 μM HsH2B (23–37) and 0.7 μM [3H]AdoMet. All reaction mixtures contained 50 mM K-Hepes and 1 mM DTT with a final pH of 8.5. Thee black line represents averages when Na2SO4 was added to reaction mixtures. The red line represents averages when NaCl was added to reaction mixtures. The green line represents averages when KCl was added to reaction mixtures. The yellow line represents averages when MgCl2 was added to reaction mixtures. The blue line represents the averages when CaCl2 was added to the reaction mixtures. All reactions were incubated for 10 min at 20 °C. HsH2B, histone H2B; PRMT, protein arginine methyltransferase.
We then examined the effect of calcium and magnesium ions on the activity of PRMT7. Here we found inhibition greater than expected from the ionic strength. With calcium ions, we found half inhibition at approximately 0.7 mM, and with magnesium ions, we found half inhibition at about 1.6 mM (Fig. 8). Since intracellular calcium ion concentrations are submicromolar, we might expect little effect on PRMT activity except perhaps when cells undergo intracellular calcium release. Intracellular free magnesium concentration has been estimated in the cytosol of mammalian cells at about 0.5 mM (19), suggesting little inhibition of PRMT7 with changes in magnesium ion concentration. Taking all of these results together, it appears that PRMT7 may only be active under conditions where intracellular temperature, pH, and ionic conditions are disrupted.
PRMT1 and PRMT5 are also inhibited by the addition of salts
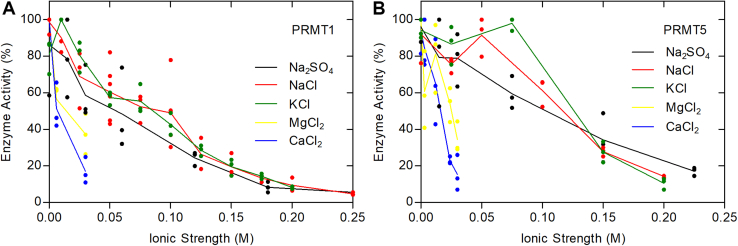
Finally, we sought to determine whether PRMT1 and PRMT5 activity was also affected by ionic strength or calcium and magnesium ions (Fig. 9). For His-HsPRMT1 with the human histone H4 (1–21) peptide, we found half maximal inhibition by ionic strength at about 100 mM and half maximal inhibition by calcium and magnesium ions at about 2 mM. For HsPRMT5/MEP50 under similar conditions, we found half maximal inhibition by ionic strength at about 120 mM, half maximal inhibition of calcium ions at about 5 mM, and half maximal inhibition of magnesium ions at about 8 mM. Interestingly, these enzymes are also inhibited in the physiological ionic strength range, although the inhibition by calcium and magnesium ions is less than that seen with PRMT7. It appears that PRMT1 and PRMT5 would not be significantly inhibited by physiological levels of calcium and magnesium.
Figure 9.
Human PRMT1 and PRMT5/MEP50 activities are reduced with increasing ionic strength. P81 phosphocellulose of His-HsPRMT1 or HsPRMT5/MEP50 with HsH4 (1–21) was carried out as described in Experimental procedures. A, 40 nM of His-HsPRMT1, 10 μM HsH4 (1–21), and 0.7 μM [3H]AdoMet were incubated with 50 mM K-Hepes and 1 mM DTT at a final pH of 7.5. Reaction mixtures were incubated at 37 °C for 15 min. B, 5.6 nM of HsPRMT5/MEP50, 10 μM HsH4 (1–21), and 0.7 μM [3H]AdoMet were incubated with 50 mM K-Hepes and 1 mM DTT at a final pH of 7.5. Reaction mixtures were incubated at 37 °C for 30 min. The black line represents averages when Na2SO4 was added to reaction mixtures. The red line represents averages when NaCl was added to reaction mixtures. The green line represents averages when KCl was added to reaction mixtures. The yellow line represents averages when MgCl2 was added to reaction mixtures. The blue line represents the averages when CaCl2 was added to the reaction mixtures. HsH4, human histone H4; PRMT, protein arginine methyltransferase.
Discussion
In this study, we were surprised to find that human PRMT1, PRMT5, and PRMT7 had optimal catalytic activities in vitro under nonphysiological conditions. For example, all three enzymes displayed markedly reduced activity at ionic strengths comparable to the cytosol of mammalian cells. While PRMT1 and PRMT5 were active at a range of pH values, PRMT7 had little activity at the expected intracellular pH of 7.2 and was only fully active at alkaline pH values. Finally, all of the three enzymes displayed a significant catalytic activity at low temperatures, including 0 °C. Significantly, human PRMT7 had markedly reduced activity at the physiological temperature of 37 °C, while PRMT1, PRMT5, and PRMT9 (17) had near-optimal activity at 37 °C. We note that the PRMT7 from the soil nematode worm Caenorhabditis elegans has a distinct temperature profile with optimal activity at about 20 °C near that of its environment in nature (17).
We wanted to know if the optimal activity of PRMTs under these nonphysiological conditions would mean that PRMTs are responding to environmental stresses. We wanted to ask under what conditions would PRMTs be exposed to temperatures lower than 37 °C, under what conditions would PRMTs be exposed to pH values above 7.2, and under what conditions would PRMTs be exposed to ionic strengths lower than what is generally found in mammalian cells.
Humans generally maintain their temperature at 37 °C even when exposed to environmental cold. However, skin cells and cells lining the lungs can be significantly affected (20, 21). We note that mammalian cells have “cold shock” proteins that are induced by a cold shock response (22, 23, 24). One example of cold response is that optimal human sperm production is maintained when the testis is 2 to 7 °C below body temperature (25). It has been suggested that the TRPM8 sensor protein is important for the cold shock response in testes (21). Moderately reduced temperatures have also been found to suppress apoptosis of neural stem cells although it is not clear when such cells would be exposed to lower temperatures (26).
Interestingly, a comparison of genes has associated PRMT7, along with a small group of other genes, to the adaptation of elephantid species to cold environments, particularly of mammoths to the arctic (27).
Many mammalian proteins have been shown to be denatured at subphysiological temperatures (28). However, it is not uncommon for enzymes to adapt to low temperatures, especially those in fish and in bacteria exposed to cold environments (29).
To determine how common is cold activation of enzymes, we searched the literature for publications reporting on the temperature dependence of human or mammalian S-adenosylmethionine methyltransferases. While many of these enzymes show optimal activities around 37 °C as expected, we were interested to find an example of an enzyme that displayed cold activation similar to what we report here for PRMT7. This enzyme is the NS5 RNA methyltransferase from the Dengue virus 4. Optimal activity was found between 22 °C and 30 °C, with less than 20% of this activity found at 37 °C (30). It is presently unclear what the biological advantage is for PRMT7 or the RNA methyltransferase to be cold activated. Additionally, it is unknown why PRMT1, PRMT5, and PRMT7 have significant enzymatic activity at 0 °C.
PRMT7 was shown here to have very little activity at the expected cytosolic pH of about 7.2 in mammalian cells (13, 31). The activity of PRMT7 at pH 7.2 was found to be less than 5% of its optimal activity at pH 8.4. In comparison to pH 7.2, human PRMT1 has about 30% of its optimal activity at pH 7.6, while PRMT5 has a broad optimal range between 6.5 and 8.5. Interestingly, the Dengue virus 4 NS5 RNA methyltransferase also shows enhanced activity at alkaline pH. Its activity at 7.2 is only about 20% of its optimal activity at pH 9.0 (30). A similar situation occurs with the PRDM9 histone H3 lysine 36 methyltransferase where the activity at pH 7.2 is only about 15% of the optimal activity at pH 8.5 (32). All of these results suggest that PRMT7, the NS5 RNA methyltransferase, the histone lysine methyltransferase, and possibly PRMT1 may be regulated by conditions resulting in the alkalization of cells.
PRMT1, PRMT5, and PRMT7 all show inhibition with increasing ionic strength. For PRMT7, this inhibition was much more pronounced with the HsH2B (23–37) peptide than for the GST-GAR substrate. We were intrigued to find that Dengue virus-4 RNA methyltransferase and the PRDM9 histone lysine methyltransferase also showed sensitivity to ionic strength (30, 32). The physiological relevance of such sensitivity is not clear although it has been suggested that cells can respond to changes in ionic strength (14). Normal intracellular ionic strength has been estimated at about 110 to 130 mM in the human embryonic kidney cell line HEK293 reflecting the known intracellular sodium ion and potassium ion levels (14). At this ionic strength, PRMT7 (at least with the HsH2B (23–37) peptide substrate) and the histone lysine methyltransferase PRDM9 have less than 10% activity, while PRMT1, PRMT5, and the Dengue-4 RNA methyltransferase have less than 50% activity.
Finally, PRMT1, PRMT5, and PRMT7 are all inhibited by increasing concentrations of magnesium ions. PRMT7 was found to be inhibited by magnesium ion concentration with half activity found at 1.6 mM, while PRMT1 was half inhibited at 2 mM and PRMT5 was half inhibited at 8 mM. Since the intracellular free magnesium ion concentration has been estimated at about 0.5 mM (19), it appears that the inhibition seen here would not be physiologically relevant. These methyltransferases are also inhibited by calcium ions at levels much greater than the levels found intracellularly.
At present, we do not understand the importance of the subphysiological activities of PRMT1, PRMT5, and PRMT7 and the RNA and protein lysine methyltransferases as described earlier in the study. The similarities in the responses to temperature, pH, and ionic strength are, however, remarkable.
Evidence has been presented that physiological levels of hydrogen peroxide can inhibit human PRMT1 and that this inhibition can be reversed by reducing agents, suggesting a possible regulation of PRMT1 by redox reactions (33). It is clear that PRMT7 activity is dependent upon a reducing environment as well (17, 34). In all of our experiments, we performed incubations in 1 mM DTT (17).
It remains to be shown whether or not PRMTs, especially PRMT7, are regulated in vivo by intracellular temperature, pH, or ionic strength. Unfortunately, it has been challenging to measure the individual activities of the nine family members of mammalian PRMTs in cells cultured under normal or low temperature conditions, or under conditions that would affect their intracellular ionic strength or pH. This is particularly a problem for PRMT7, whose activity appears to be generally low in comparison with the major type I PRMT1 enzyme or the major type II PRMT5 enzyme (5, 35). Although evidence has been presented for a number of in vivo sites of PRMT7 methylation (7, 10, 36, 37, 38), it is not clear so far that these are specifically modified by PRMT7 alone and are not also sites for methylation by other PRMTs (39). Additionally, PRMT7 methylation of Arg-17 of a histone H4 peptide has been shown to allosterically activate PRMT5 methylation at the distinct Arg-3 site (9). Such cross talk between PRMTs can complicate the analysis of specific PRMT activity in cells. Finally, it is now clear that reductions in the activity of one PRMT can result in the methylation of its sites by other members of the PRMT family (40, 41). The identification of specific substrates and the generation of specific antibodies raised against their monomethylation site(s) will be important steps in quantitating PRMT7 activity in cells and testing how this activity may respond to changes in their intracellular environment.
Experimental procedures
Preparation of enzymes and protein substrates
Recombinant H. sapiens PRMT7 DNA was cloned into a pGEX-2T plasmid and expressed in BL21 DE3 E. coli. This plasmid encodes the full sequence (residues 1–218) of glutathione S-transferase from Schistosoma japonicum followed by the linker SDLVPRGSST and the full sequence (residues 1–692) of human PRMT7 and was designated as GST-HsPRMT7. This plasmid is available from Addgene.org #34693. Cells were grown in 2L of LB media at 37 °C to an A600 nm of 0.6 to 0.8 in the presence of 100 μg/ml ampicillin. Isopropyl-beta-D-thiogalactoside (CAS # 367–93–1, GoldBio) was then added to a final concentration of 1 mM, and the culture was incubated for an additional 16 to 20 h at 20 °C. Cells were harvested by centrifugation, and 0.5 g of cells was resuspended in 40 ml of 140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4, pH = 7.5, supplemented with an EDTA-free protease inhibitor tablet (Catalog No. PIA32965, Thermo Scientific). The cell suspension was lysed using a Fisher Scientific 550 Sonic Dismembrator with a microtip attachment in four cycles on ice for 1 min, 0.5 s on pulse, and 0.5 s off pulse at 25% amplitude. The lysate was then centrifuged at 27000g for 1 h, and the supernatant was loaded onto a manually packed column (ID 1.5 cm × 15 cm, 4 ml bed volume) containing glutathione Sepharose 4B resin (Catalog No. 17075601, GE Healthcare Life Sciences) equilibrated in the aforementioned buffer at 22 °C. Enzyme was eluted with 10 mM glutathione and 50 mM Tris-HCl, pH 8.0. Purified GST-HsPRMT7 was then dialyzed overnight in 50 mM K-Hepes, 1 mM DTT, pH 8.0, at 4 °C. GST-HsPRMT7 was then mixed with glycerol to a final concentration of 20% and stored at −80 °C. The total GST-HsPRMT7 concentration was determined using a NanoDrop 2000c spectrophotometer by the absorption at 280 nm, assuming that 1 mg/ml would give 1 A.
An N-terminal His-tagged construct of human PRMT1 (designated His-HsPRMT1) was prepared as described previously and purified using a HisTrap Fast Flow (Cytiva 17–5255–01) as per the manufacturer's protocol (42).
Human PRMT5/MEP50 (HsPRMT5/MEP50) FLAG-tag His-tag complex protein was purchased from BioSciences as recombinantly coexpressed and purified proteins in HEK293T cells (0.64 mg/ml, Catalog No. 51045, Lot 170407A; BPS Biosciences), formulated in 40 mM Tris-HCl, pH 8.0, 110 mM NaCl, 2.2 mM KCl, 80 μg/ml FLAG peptide, 20% glycerol, and 3 mM DTT.
A fusion protein of glutathione S-transferase followed by the linker SDLVPRGS and human fibrillarin residues 1 to 145 with substitutions K2E and A145V (GST-GAR) was prepared by expression of plasmid #34697 of Addgene.org. (17). The protein was purified as described for GST-HsPRMT7 above.
Human HsH2B was purchased from NEB, Cat. No. M2505S, as a recombinant protein expressed in E. coli, stored in 300 mM NaCl, 1 mM EDTA, and 20 mM sodium phosphate at pH 7.
Peptides
Synthetic peptides HsH2B (23–37) (Ac-KKDGKKRKRSRKESY, 98.6% purity, MW: 1936.23, Lot No. P17171304) and human histone H4 (1–21) (Ac-SGRGKGGKGLGKGGAKRHRKV, 96.2% purity, MW: 2133.47, Lot No. 89488400001/PE1586) were purchased from Genescript.
In vitro methylation reactions using P81 phosphocellulose
GST-HsPRMT7, His-HsPRMT1, or PRMT5/MEP50 was mixed with a peptide derived from residues 23 to 37 of HsH2B, GST-GAR, or a peptide derived from the acetylated N-terminus of histone H4 (residues 1–21). Reactions were initiated with S-adenosyl-L-[methyl-3H]methionine ([3H]AdoMet) (81.9 Ci/mmol, 7 μM in 9:1 10 mM H2SO4:ethanol; Catalog No. NET155H001MC, PerkinElmer) in a final volume of 30 μl. The reaction mixtures were equilibrated to the desired temperature for 2 min prior to the addition of [3H]AdoMet and incubation in a temperature-regulated water bath. Post incubation, the reaction mixtures were terminated with 0.5 μl of 100% trifluoracetic acid and spun down for 10 s at 10,600g in an Eppendorf microcentrifuge (Model 5415C). All reaction mixtures contained 1 mM DTT and unless otherwise described a final concentration of 50 mM K-HEPES at a final pH of 8.5. To determine the final pH of reaction mixtures, mock mixtures were created containing the appropriate volumes of the enzyme buffer (50 mM K-Hepes, 1 mM DTT, pH 8.0), the [3H]AdoMet solvent (18 mM HCl), and the reaction buffer containing 1 mM DTT and 50 mM K-Hepes, 50 mM glycine, or 50 mM Tris-HCl.
The extent of methylation was determined by a P81 phosphocellulose assay. Twenty-five microliters of reaction mixture was spotted on to a 1 × 1-cm2 of phosphocellulose P81 paper (Catalog No. 05–717-2A, Lab Alley). Papers were dried for 90 min at room temperature and were then washed 3 times with 300 ml of 50 mM sodium bicarbonate, pH = 9.0. The papers were dried again for 90 min in 20-ml scintillation vials (Catalog No. 66021-704, VWR), and 5 ml of scintillation fluid (111177-CS, RPI) was added. Vials were counted in a Beckman LS 6500 Liquid Scintillation Counter for four cycles at 5 min per cycle. The sum of enzyme-only and substrate-only controls was subtracted from individual replicates.
In vitro methylation gel assay
Recombinant GST-HsPRMT7 was incubated with either GST-GAR as described earlier in the study or recombinant human HsH2B expressed in E. coli (Catalog No. M2505S, New England BioLabs) in a total volume of 30 μl. Post incubation, the reaction mixtures were spun down for 10 s at 10,600g. The reactions were then terminated with 5 μl of 5× sample buffer: 250 mM Tris-HCl, 10% SDS, 30% glycerol, 500 mM DTT, and 0.05% bromophenol blue and heated for 1 min in a 95 °C sand bath. The reaction mixtures were allowed to cool to room temperature before loading either onto a 4 to 12% 15-well ExpressPlus PAGE gel (Catalog No. NC1486979, Fisher Scientific Co) or 4 to 20% 10-well Express PAGE gel (Catalog No. NC0613877, Fisher Scientific Co). The gel was allowed to run for 1 h at 140 V, 400 mA using a BioRad PowerPac 300 electrophoresis Power supply. The gels were stained for 1 h in Coomassie stain: 50% methanol and 10% acetic acid, followed by an overnight destain consisting of 15% methanol and 10% acetic acid. The gels were then soaked in water for minimum 3 h prior to enhancing with En3Hance (Catalog No. 6NE9701, PerkinElmer) for 30 min. The gels were then put into water for 1 h post enhancement, wrapped in gel-drying film (Catalog No. PR-V713, Promega), and dried in a Model 583 Gel Dryer, Biorad for 2 h at 80 °C. The dried gel was placed in an autoradiography cassette, and an HyBlot CL autoradiography film (Catalog No. NC9556985, Thermo Scientific) was placed on top of the gel. The film was exposed for 3 to 30 days at −80 °C before development
Data availability
All data described in the manuscript are contained within the manuscript. Additional data are available upon request.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the content of this article.
Acknowledgments
Author contributions
T. L. L. and S. G. C. conceptualization; T. L. L. and S. G. C. methodology; T. L. L. and S. G. C. formal analysis; T. L. L. investigation; T. L. L. resources; T. L. L. and S. G. C. writing - original draft; T. L. L. and S. G. C. writing - review & editing; T. L. L. and S. G. C. visualization; S. G. C. supervision; S. G. C. project administration; S. G. C. funding acquisition.
Funding and additional information
This work was supported by the National Science Foundation grant MCB-1714569 (to S. G. C.) and by funds from the UCLA Academic Senate Faculty Research Program, the Life Extension Foundation, Inc, and the Elizabeth and Thomas Plott Chair in Gerontology of the UCLA Longevity Center (to S. G. C.). T. L was supported by the National Institutes of Health Ruth L. Kirschstein National Research Service Award GM007185.
Edited by Brian Strahl
Supporting information
References
- 1.Jarrold J., Davies C.C. PRMTs and arginine methylation: cancer’s best-kept secret? Trends Mol. Med. 2019;25:993–1009. doi: 10.1016/j.molmed.2019.05.007. [DOI] [PubMed] [Google Scholar]
- 2.Wu Q., Schapira M., Arrowsmith C.H., Barsyte-Lovejoy D. Protein arginine methylation: from enigmatic functions to therapeutic targeting. Nat. Rev. Drug Discov. 2021;20:509–530. doi: 10.1038/s41573-021-00159-8. [DOI] [PubMed] [Google Scholar]
- 3.Xu J., Richard S. Cellular pathways influenced by protein arginine methylation: implications for cancer. Mol. Cell. 2021;81:4357–4368. doi: 10.1016/j.molcel.2021.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Y., Bedford M.T. Protein arginine methyltransferases and cancer. Nat. Rev. Cancer. 2013;13:37–50. doi: 10.1038/nrc3409. [DOI] [PubMed] [Google Scholar]
- 5.Bedford M.T., Clarke S.G. Protein arginine methylation in mammals: who, what, and why. Mol. Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tewary S.K., Zheng Y.G., Ho M.-C. Protein arginine methyltransferases: insights into the enzyme structure and mechanism at the atomic level. Cell. Mol. Life Sci. 2019;76:2917–2932. doi: 10.1007/s00018-019-03145-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halabelian L., Barsyte-Lovejoy D. Structure and function of protein arginine methyltransferase PRMT7. Life (Basel) 2021;11:768. doi: 10.3390/life11080768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frankel A., Brown J.I. Evaluation of kinetic data: what the numbers tell us about PRMTs. Biochim. Biophys. Acta - Proteins Proteomics. 2019;1867:306–316. doi: 10.1016/j.bbapap.2018.10.010. [DOI] [PubMed] [Google Scholar]
- 9.Jain K., Jin C.Y., Clarke S.G. Epigenetic control via allosteric regulation of mammalian protein arginine methyltransferases. Proc. Nat. Acad. Sci. U. S. A. 2017;114:10101–10106. doi: 10.1073/pnas.1706978114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng Y., Maity R., Whitelegge J.P., Hadjikyriacou A., Li Z., Zurita-Lopez C., et al. Mammalian protein arginine methyltransferase 7 (PRMT7) specifically targets RXR sites in lysine- and arginine-rich regions. J. Biol. Chem. 2013;288:37010–37025. doi: 10.1074/jbc.M113.525345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fulton M.D., Brown T., Zheng Y.G. Mechanisms and inhibitors of histone arginine methylation. Chem. Rec. 2018;18:1792–1807. doi: 10.1002/tcr.201800082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li A.S.M., Li F., Eram M.S., Bolotokova A., dela Seña C.C., Vedadi M. Chemical probes for protein arginine methyltransferases. Methods. 2020;175:30–43. doi: 10.1016/j.ymeth.2019.11.017. [DOI] [PubMed] [Google Scholar]
- 13.Casey J.R., Grinstein S., Orlowski J. Sensors and regulators of intracellular pH. Nat. Rev. Mol. Cell Biol. 2010;11:50–61. doi: 10.1038/nrm2820. [DOI] [PubMed] [Google Scholar]
- 14.Liu B., Poolman B., Boersma A.J. Ionic strength sensing in living cells. ACS Chem. Biol. 2017;12:2510–2514. doi: 10.1021/acschembio.7b00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bisswanger H. Enzyme assays. Perspect. Sci. 2014;1:41–55. [Google Scholar]
- 16.Feng Y., Hadjikyriacou A., Clarke S.G. Substrate specificity of human protein arginine methyltransferase 7 (PRMT7): the importance of acidic residues in the double E loop. J. Biol. Chem. 2014;289:32604–32616. doi: 10.1074/jbc.M114.609271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hadjikyriacou A., Clarke S.G. Caenorhabditis elegans PRMT-7 and PRMT-9 are evolutionarily conserved protein arginine methyltransferases with distinct substrate specificities. Biochemistry. 2017;56:2612–2626. doi: 10.1021/acs.biochem.7b00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jain K. University of California; Los Angeles: 2018. Protein Arginine Methyltransferases: Catalytic Mechanisms and Crosstalk in Epigenetics. PhD Thesis. [Google Scholar]
- 19.Vormann J. Magnesium: nutrition and metabolism. Mol. Aspects Med. 2003;24:27–37. doi: 10.1016/s0098-2997(02)00089-4. [DOI] [PubMed] [Google Scholar]
- 20.Juan Y., Haiqiao W., Xie W., Huaping H., Zhong H., Xiangdong Z., et al. Cold-inducible RNA-binding protein mediates airway inflammation and mucus hypersecretion through a post-transcriptional regulatory mechanism under cold stress. Intl J. Biochem. Cell Biol. 2016;78:335–348. doi: 10.1016/j.biocel.2016.07.029. [DOI] [PubMed] [Google Scholar]
- 21.Borowiec A.-S., Sion B., Chalmel F., Rolland A.D., Lemonnier L., De Clerck T., et al. Cold/menthol TRPM8 receptors initiate the cold-shock response and protect germ cells from cold-shock–induced oxidation. FASEB J. 2016;30:3155–3170. doi: 10.1096/fj.201600257R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LLeonart M.E. A new generation of proto-oncogenes: Cold-inducible RNA binding proteins. Biochim. Biophys. Acta - Rev. Cancer. 2010;1805:43–52. doi: 10.1016/j.bbcan.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 23.De Leeuw F., Zhang T., Wauquier C., Huez G., Kruys V., Gueydan C. The cold-inducible RNA-binding protein migrates from the nucleus to cytoplasmic stress granules by a methylation-dependent mechanism and acts as a translational repressor. Exp. Cell Res. 2007;313:4130–4144. doi: 10.1016/j.yexcr.2007.09.017. [DOI] [PubMed] [Google Scholar]
- 24.Phadtare S., Alsina J., Inouye M. Cold-shock response and cold-shock proteins. Curr. Opin. Microbiol. 1999;2:175–180. doi: 10.1016/S1369-5274(99)80031-9. [DOI] [PubMed] [Google Scholar]
- 25.Aldahhan R.A., Stanton P.G. Heat stress response of somatic cells in the testis. Mol. Cell Endocrinol. 2021;527 doi: 10.1016/j.mce.2021.111216. [DOI] [PubMed] [Google Scholar]
- 26.Saito K., Fukuda N., Matsumoto T., Iribe Y., Tsunemi A., Kazama T., et al. Moderate low temperature preserves the stemness of neural stem cells and suppresses apoptosis of the cells via activation of the cold-inducible RNA binding protein. Brain Res. 2010;1358:20–29. doi: 10.1016/j.brainres.2010.08.048. [DOI] [PubMed] [Google Scholar]
- 27.Lynch V.J., Bedoya-Reina O.C., Ratan A., Sulak M., Drautz-Moses D.I., Perry G.H., et al. Elephantid genomes reveal the molecular bases of woolly mammoth adaptations to the arctic. Cell Rep. 2015;12:217–228. doi: 10.1016/j.celrep.2015.06.027. [DOI] [PubMed] [Google Scholar]
- 28.Privalov P.L. Cold denaturation of protein. Crit. Rev. Biochem. Mol. Biol. 1990;25:281–306. doi: 10.3109/10409239009090612. [DOI] [PubMed] [Google Scholar]
- 29.Marshall C.J. Cold-adapted enzymes. Trends Biotech. 1997;15:359–364. doi: 10.1016/S0167-7799(97)01086-X. [DOI] [PubMed] [Google Scholar]
- 30.Dong H., Chang D.C., Hua M.H.C., Lim S.P., Chionh Y.H., Hia F., et al. 2’-O methylation of internal adenosine by flavivirus NS5 methyltransferase. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roos A., Baron W.F. Intracellular pH. Physiol. Rev. 1981;61:296–434. doi: 10.1152/physrev.1981.61.2.296. [DOI] [PubMed] [Google Scholar]
- 32.Eram M.S., Bustos S.P., Lima-Fernandes E., Siarheyeva A., Senisterra G., Hajian T., et al. Trimethylation of histone H3 lysine 36 by human methyltransferase PRDM9 protein. J. Biol. Chem. 2014;289:12177–12188. doi: 10.1074/jbc.M113.523183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morales Y., Cáceres T., May K., Hevel J.M. Biochemistry and regulation of the protein arginine methyltransferases (PRMTs) Arch. Biochem. Biophys. 2016;590:138–152. doi: 10.1016/j.abb.2015.11.030. [DOI] [PubMed] [Google Scholar]
- 34.Zurita-Lopez C.I., Sandberg T., Kelly R., Clarke S.G. Human protein arginine methyltransferase 7 (PRMT7) is a type III enzyme forming ω-NG-monomethylated arginine residues. J. Biol. Chem. 2012;287:7859–7870. doi: 10.1074/jbc.M111.336271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blanc R.S., Richard S. Arginine methylation: the coming of age. Mol. Cell. 2017;65:8–24. doi: 10.1016/j.molcel.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 36.Jain K., Clarke S. PRMT7 as a unique member of the protein arginine methyltransferase family: a review. Arch. Biochem. Biophys. 2019;665:36–45. doi: 10.1016/j.abb.2019.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haghandish N., Baldwin R.M., Morettin A., Dawit H.T., Adhikary H., Masson J.Y., et al. PRMT7 methylates eukaryotic translation initiation factor 2α and regulates its role in stress granule formation. Mol. Biol. Cell. 2019;30:778–793. doi: 10.1091/mbc.E18-05-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szewczyk M.M., Ishikawa Y., Organ S., Sakai N., Li F., Halabelian L., et al. Pharmacological inhibition of PRMT7 links arginine monomethylation to the cellular stress response. Nat. Commun. 2020;11:2396. doi: 10.1038/s41467-020-16271-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao W.-W., Xiao R.Q., Peng B.-L., Xu H.-T., Shen H.-F., Huang M.-F., et al. Arginine methylation of HSP70 regulates retinoid acid-mediated RARβ2 gene activation. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E3327–E3336. doi: 10.1073/pnas.1509658112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dhar S., Vemulapalli V., Patananan A.N., Huang G.L., Di Lorenzo A., Richard S., et al. Loss of the major Type I arginine methyltransferase PRMT1 causes substrate scavenging by other PRMTs. Sci. Rep. 2013;3:1311. doi: 10.1038/srep01311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y., Person M.D., Bedford M.T. Pan-methylarginine antibody generation using PEG linked GAR motifs as antigens. Methods. 2022;200:80–86. doi: 10.1016/j.ymeth.2021.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Debler E.W., Jain K., Warmack R.A., Feng Y., Clarke S.G., Blobel G., et al. A glutamate/aspartate switch controls product specificity in a protein arginine methyltransferase. Proc. Nat. Acad. Sci. U. S. A. 2016;113:2068–2073. doi: 10.1073/pnas.1525783113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data described in the manuscript are contained within the manuscript. Additional data are available upon request.