Abstract
The use of nanodiamonds for biomedical and consumer applications is growing rapidly. As their use becomes more widespread, so too do concerns around their cytotoxicity. The cytotoxicity of nanodiamonds correlates with their cellular internalisation and circulation time in the body. Both internalisation and circulation time are influenced by the formation of a protein corona on the nanodiamond surface. However, a precise understanding of both how the corona forms and evolves and its influence on cytotoxicity is lacking. Here, we investigated protein corona formation and evolution in response to two classes of nanodiamonds, pristine and aminated, and two types of proteins, bovine serum albumin and fibronectin. Specifically, we found that a corona made of bovine serum albumin (BSA), which represents the most abundant protein in blood plasma, reduced nanodiamond agglomeration. Fibronectin (FN9-10), the second most abundant protein found in the plasma, exhibited a significantly higher nanodiamond binding affinity than BSA, irrespective of the nanodiamond surface charge. Finally, nanodiamonds with a BSA corona displayed less cytotoxicity towards nonphagocytic liver cells. However, regardless of the type of corona (FN9-10 or BSA), both classes of nanodiamonds induced substantial phagocytic cell death. Our results emphasise that a precise understanding of the corona composition is fundamental to determining the fate of nanoparticles in the body.
Protein corona determines fate and cytotoxicity of nanoparticles in biological systems.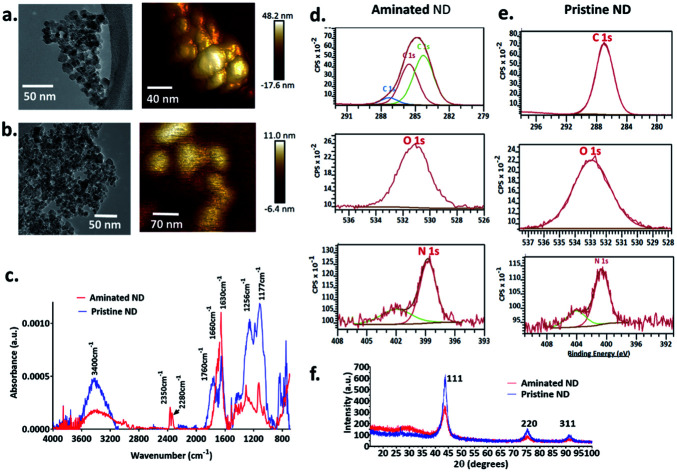
1. Introduction
Nanoparticles have been increasingly used in cosmetics, food and biomedical applications, e.g. drug delivery and medical imaging. The rapid expansion of nanoparticle applications is due to their large surface area to volume ratio and electronic and optical properties.1–6 However, the effect of nanoparticles on human health remains poorly understood, and recent reports on nanoparticle toxicity have raised safety concerns.
Current evidence suggests that the formation of a corona on the nanoparticle surface is the most important parameter that controls nanoparticle toxicity.7–10 The corona arises from the accumulation of adsorbed biomolecules from blood, saliva, synovial fluid or other fluids in the surroundings. Prior studies of corona formation have indicated an association between corona development and the physicochemical parameters of the nanoparticles, such as size,10 shape,11–13 surface charge,8 chemical composition14 and mechanical properties.12,15–17 However, physicochemical properties alone cannot predict corona formation, as the composition of the corona changes dynamically – remodels – when nanoparticles pass through different tissues. The remodelling of the corona is indirectly related to the intrinsic physicochemical characteristics of nanoparticles, and it depends on the exposure time and type of biomolecules present.
Corona remodelling is attributed to the association and dissociation of proteins, as described by Vroman's model.18 Vroman's model, which has been verified in vitro19–22 and in vivo,23 postulates that more abundant proteins with low affinity, such as albumin, bind to the surface of nanoparticles during the early stages of adsorption to form a “soft” corona. During the later stages, the proteins that were initially adsorbed are replaced by less abundant proteins with greater affinity, such as apolipoproteins, fibrinogen and immunoglobulins,23 forming a “hard” corona.24 Notably, the type of serum protein and whether it was denatured upon adsorption influence the reactivity and biodistribution of nanoparticles.25,26 For example, adsorbed opsonins promote the phagocytosis and elimination of nanoparticles from the body, whereas dysopsonins promote longer circulation in the blood and more uniform distribution in tissues.22,27
Understanding the role of corona formation in nanoparticle internalisation can lead to the development of more customized nanoparticles. For example, a corona can reduce the adhesion of nanoparticles to the cell membrane, which has been shown to reduce cytotoxicity for single wall carbon nanotubes and polystyrene nanoparticles.28,29 For drug delivery, however, inhibiting nanoparticle internalisation is an undesired effect, as nanoparticles are typically functionalised with receptor-binding molecules. The formation of a corona effectively coats the molecules and consequently deactivates the ability of nanoparticles to recognise cell receptors.
The molecular composition of a corona can also promote binding of nanoparticles to the cell membrane and consequently enhance the cellular uptake of nanoparticles.27 For example, a corona composed of multiple biomolecules (i.e. proteins like prothrombin, antithrombin III, vitronectin, inter-α-trypsin-inhibitor heavy chain H5, platelet factor 4 and apolipoprotein H) promoted cellular uptake of polystyrene nanoparticles in human cervical carcinoma cells and human mesenchymal stem cells.27 Corona was also shown to enhance cellular uptake of Janus gold–iron oxide and polystyrene nanoparticles in human vascular endothelial and human umbilical vein endothelial cells.30,31
While the rapid formation of a corona on nanoparticles is known to influence their interactions with cells, there is a gap in the fundamental understanding of how a corona forms and how it remodels in response to the microenvironment composition and nanoparticle properties. Comprehensive studies of corona formation are thus essential to analyse the toxicity of nanoparticles with cells and tissues. This new knowledge will also unlock the possibility of designing new classes of nanoparticles with predefined coronas to ensure their safety.
Here we investigated how the corona forms and evolves on nanodiamonds. Nanodiamonds are suggested to be promising carriers for drugs and probes for medical imaging; therefore, it is necessary to establish their safety. Specifically, we investigated:
(1) How the physicochemical characteristics of nanoparticles affect corona formation.
(2) The dynamic process of the association and dissociation of proteins to/from the nanoparticle surface.
(3) How the presence of serum interferes with nanoparticle–cell interaction and toxicity.
(4) How the protein corona modulates nanoparticle–cell interactions and toxicity.
2. Experimental section
2.1. Materials
For all cell-based experiments, we used rat hepatoma cells (Fao) and murine macrophage cells (RAW 264.7). Cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 4500 mg L−1d-glucose, 2 mM l-glutamine (Sigma-Aldrich, Australia), and Penstrep (100 IU mL−1 penicillin and 100 μg mL−1 streptomycin; Gibco, Life Technologies) with and without 10% fetal bovine serum (Sera Laboratories) – referred as a complete medium.
2.2. Physicochemical characterization of nanodiamond particles
The size, shape, morphology, crystal habit and surface area of the nanodiamonds were characterised using atomic force microscopy (AFM), transmission electron microscopy (TEM), nanoparticle tracking analysis (Nanosight), X-ray diffractometry (XRD) and the Brunauer–Emmett–Teller (BET) method. The surface chemical composition was analysed using Fourier transform infrared spectrophotometry (FTIR) and X-ray photoelectron spectroscopy (XPS).
Both aminated nanodiamonds (ANDs) and pristine nanodiamonds (PNDs) were produced using laser-assisted technology with nominal sizes of 5 nm (Ray Techniques Ltd., Israel) and were used as received.
2.3. Expression and purification of fibronectin
Fibronectin (FN9-10) was designed to contain the subunits 9 and 10 (FN9-109 and FN9-1010) – the key cell-binding domains of FN9-10. PCR primers were designed to recognize the cell-binding domains of FN9-10 spanning FN9-109-10: forward ‘FN9-109F’ primer, 5′-GGTACCGGTCTTGATTCCCCAACTGG-3′ (which introduces a KpnI restriction site before the NH2-terminus of FN9-109); reverse ‘FN9-1010R’ primer, 5′-GGTACCTGGTTTGTCAATTTCTGTTCGG-3′ (which incorporates a KpnI restriction site at the COOH-terminus of FN9-1010). PCR was performed in a 30 μL reaction vessel containing 50 mM KCl, 10 mM Tris–HCl (pH 8.3), 1.5 mM MgCl2, 100 μg mL−1 gelatin, 0.2 mM dNTPs, 1.25 units of Ex Taq polymerase (Takara Shuzo Co., Kyoto, Japan), and 50 pmol each of the forward and reverse primers. The thermocycling parameters used in PCR were as follows: denaturation for 1 min at 94 °C; annealing for 1 min at 55 °C; extension for 1 min at 72 °C. After 30 cycles, amplified PCR products were digested with KpnI. After digestion, the PCR product was ligated into the KpnI sites of the pBAD/HisA vector (Invitrogen, Carlsbad, CA), giving rise to a construct (pBAD/HisA-FN9-109-10). Briefly, TOP10 cells were grown overnight in an LB-Amp+ medium at 37 °C for the expression of FN9-109-10. After 6 h, bacteria were pelleted by centrifugation, lysed, and sonicated. A soluble extract was prepared by centrifugation for 20 min at 5000 rpm in a refrigerated centrifuge, and the supernatant was transferred to a fresh tube. The crude protein from the sonicated bacterial supernatant was purified through binding of the His6 tag to the nickel–nitrilotriacetic acid resin column, according to the manufacturer's protocol (Qiagen, Hilden, Germany). The FN9-109-10 was over 90% pure, as determined by Coomassie blue staining of a 12% (v/v) SDS-PAGE gel under reducing conditions.
2.4. Time-dependent remodelling of protein corona composition
The time-resolved remodelling of the protein corona on nanodiamonds was investigated using zeta potential, matrix assisted laser ionization/deionization time of flight spectrometry (MALDI-TOF), micro BCA (bicinchoninic acid) assay and isothermal calorimetry.
2.4.1. Size and charge measurements
The effect of protein corona formation on the size and zeta potential of the nanodiamonds was measured using dynamic light scattering and a zeta sizer (Zetasizer Nano ZS, Malvern Instruments, UK). 100 μg mL−1 of each sample was mixed with 500 μg mL−1 bovine serum albumin (BSA) and FN9-10 protein solution using a vortex mixer for 1 min. Then, all the samples were incubated at 37 °C for 5 min, 30 min, 120 min and 24 h to allow the formation of the protein corona. Size and zeta potential measurements were carried out after each time point.
2.4.2. Quantitative measurements of the composition of the protein corona
To demonstrate that corona formation is a time-resolved process, both aminated and pristine nanodiamonds (500 μg mL−1) were incubated with FN9-10 (250 μg mL−1) and BSA (Sigma Aldrich, USA) (2 mg mL−1). After 5 min, 30 min and 120 min of incubation samples were collected and centrifuged at 15 000g and 4 °C for 15 min, and then washed with deionized water. This process was repeated three times to remove unbound proteins from the surface of nanodiamonds. The pellets obtained were dispersed in 100 μL of deionized water. 0.7 μL of this dispersion was aliquoted and transferred to a MALDI-TOF plate. This was allowed to air dry before adding a sinapic acid matrix (Sigma Aldrich, USA). Measurements were performed using MALDI-TOF (AXIMA Performance, SHIMADZU, Japan) with a 355 nm Nb:YAG laser operating in linear mode with a 120 kV acceleration voltage and a laser repetition rate of 15. For each spectrum, 1000 laser shots were performed. MALDI-TOF enabled the identification of proteins by peptide mass fingerprinting (PMF) and will allow the differentiation of the specific protein bound to the nanodiamond surface.
2.4.3. Assessment of the preferential binding of proteins to the nanodiamond surface
To affirm that proteins exhibit preferential binding to the nanodiamond surface and observe the effect of surface properties on binding affinity, mixtures of both BSA and FN9-10 and both classes of nanodiamonds (pristine and aminated) were used. Time-resolved binding of proteins was investigated using MALDI-TOF. 250 μg mL−1 each of BSA and FN9-10 were vortexed in a 1 : 1 ratio for 5 min. Nanodiamond dispersions (100 μg mL−1) were added to the protein mixture and vortexed for 1 min. Following this, all the samples were incubated at 37 °C for 5 min, 30 min and 120 min to gauge both formation and remodelling of the protein corona. At the end of each time point, samples were collected and analysed using the methodology describe in 2.4.2.
2.4.4. Nano-calorimetric measurements of the corona formation and remodelling
Calorimetric measurements, which measure the binding affinity, stoichiometry and thermodynamic parameters of nanodiamond–protein interactions, were performed using a nanoITC (TA Instruments, Denver, USA) with an effective cell volume of 170 μL. In each experiment, 50 μL of an aqueous FN9-10 solution at 500 μg mL−1 was titrated with 300 μL of an aqueous suspension of pristine and aminated nanodiamonds at 25 μg mL−1 each. An aqueous solution of BSA at 20 mg mL−1 was titrated with a solution containing 5 μg mL−1 each of pristine and aminated nanodiamonds. The experimental temperature was kept constant at 25 °C. The number and injected volume of the titration steps were the same for all measurements (25 × 2 μL). The spacing between injections was set to 300 s.
2.4.5. The measurement of the protein concentration within the corona
The amount of total protein bound to the nanodiamonds was measured using a micro BCA assay as per the manufacturer's protocol (Micro BCA™ Protein Assay Kit, ThermoFisher SCIENTIFIC, USA). Necessary dilution was carried out to maintain the concentration within the linear range of the standard curve using BSA as the standard. The assay sample was prepared by treating aminated and pristine nanodiamonds (500 μg mL−1) with BSA (2 mg mL−1) and FN9-10 (250 μg mL−1). After each time point (5 min, 30 min and 120 min), samples were centrifuged at 15 000g and 4 °C for 15 min, and then washed with deionized water. This process was repeated three times to remove unbound proteins from the surface of nanodiamonds. The pellets obtained were dispersed in 100 μL of deionized water.
2.5. Effect of the corona on nanodiamond uptake, cell viability and immune responses
2.5.1. Formation of the corona on nanodiamonds – preconditioning
Both pristine and aminated nanodiamonds were dispersed in sterile deionized water at 2 mg mL−1. The nanodiamond dispersion was sonicated for 30 min using an ultrasonic probe at 60% amplitude. Fibronectin (FN9-10) and BSA were used as the model proteins for corona studies. FN9-10 and BSA solutions were prepared using deionized water at concentrations of 1 mg mL−1 and 4 mg mL−1, respectively. The nanoparticle dispersion and protein solution were mixed at a ratio of 1 : 1 in an Eppendorf tube using a vortex mixer. This mixture was incubated at 37 °C for 24 h to induce corona formation.
2.5.2. Cellular uptake of nanodiamonds
To visualize the uptake and distribution of nanodiamonds within individual cells, cells were incubated with nanodiamonds and imaged in real time by holotomography using an environmental chamber (3D Cell Explorer, NanoLive, Switzerland).
Fao cells were plated at a density of 5 × 104 cells per mL complete medium in glass bottom dishes (35 mm, Ibidi®, Denmark) functionalized by plasma treatment (Harrick Plasma, USA) for 10 min. Cells were allowed to attach and grow overnight. The medium was replaced with fresh serum-less medium containing nanodiamonds coated with FN9-10 and BSA coronas at concentrations of 10, 25, 50 and 100 μg mL−1. The cells were exposed to nanodiamonds for 48 h. Due to the high quantity of non-transparent nanodiamonds in the cells, images could only be acquired from cells treated with nanodiamonds at a concentration of 10 μg mL−1.
Prior to imaging, cells were washed twice with PBS to remove unbound nanodiamonds. The images were digitally stained using STEVE® software (NanoLive, Lausanne, Switzerland).
2.5.3. Real-time measurements of cell growth and proliferation
The proliferation and morphology of cells (Fao and RAW 264.7 cells) after exposure to nanodiamonds with and without protein coronas (BSA and FN9-10) were determined using live cell imaging (Incucyte™, Essen BioScience, USA). Images were acquired every two hours until 48 h.
2.5.4. Effect of the protein corona on cell viability
The viability of cells treated with nanodiamonds with/without the corona was measured by CCK-8 assay (Dojindo Molecular Technologies Inc., Japan) as per the manufacturer's protocol. Fao cells and RAW 264.7 cells were seeded at a density of 10 000 cells per well on 96-well plates and were allowed to attach overnight. Media was aspirated and replaced with particle-conditioned media containing aminated nanodiamonds or pristine nanodiamonds with and without FN9-10 and BSA coronas. Cells were treated with nanodiamonds at concentrations of 10, 25, 50 and 100 μg mL−1. The effect of nanodiamonds without the corona on cells was investigated under both serum and serum-free conditions, whereas the toxicity of nanodiamonds with the corona was investigated only under serum-free conditions. After 48 h of treatment, cells were washed with PBS and their viability was measured using CCK-8 assay.
2.5.5. Effect of the protein corona on immune response
The effect of the protein corona on the immune response was determined by measuring tumour necrosis factor alpha (TNF-α) release from RAW 264.7 cells. The RAW 264.7 cells were seeded in 48 well plates at a cell density of 1.5 million cells per 500 μL per well. Then, they were incubated (5% CO2, 37 °C) for 2 days until confluency was reached. Once confluent, the supernatant was removed and media with and without serum containing different concentrations of aminated and pristine nanodiamonds (10, 25, 50 and 100 μg mL−1) were added to the cells. Cells were exposed to nanodiamonds overnight and on the following day, 100 ng mL−1 lipopolysaccharide (LPS, LPS-EK, Invitrogen, Carlsbad, USA) was added to the wells to induce inflammation and injury to the cells. The cytokine-containing supernatants, both with LPS treatment and without LPS treatment, were collected after 4 h of incubation. From the collected supernatant, TNF-α release was measured using the enzyme-linked immunosorbent assay (ELISA KIT, Biolegend, CA, USA) as per the manufacturer's protocol.
2.6. Statistical analysis
Statistical analysis was carried out using GraphPad Prism software. The results were compared using Student's t-test, one-way ANOVA and two-way ANOVA with Tukey's multiple comparison test. Statistical significance was established at p < 0.05.
3. Results and discussion
3.1. Physicochemical characterisation of the nanodiamonds
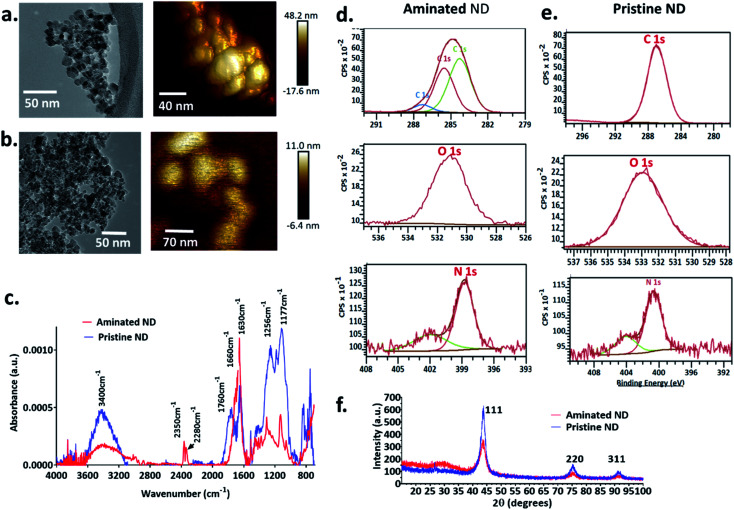
The size of individual nanodiamond particles, both pristine and aminated, was ∼5 nm, but both existed as aggregates. Aggregates of pristine ND aggregates ranged from 22–233 nm and aminated ND aggregates were 10–211 nm (Fig. 1). Pristine NDs were negatively charged with a zeta potential of −8 mV whereas aminated NDs were confirmed to have positive charge with a zeta potential of 20 mV. Details on the physical and chemical properties of NDs are discussed in the ESI file.†
Fig. 1. Physicochemical characterization of nanodiamond particles. TEM and AFM images of (a) aminated nanodiamonds and (b) pristine nanodiamonds. (c) FTIR spectra of nanodiamonds (red spectra: aminated nanodiamonds; blue spectra: pristine nanodiamonds). High-resolution XPS spectra of C, O and N for (d) aminated nanodiamonds and (e) pristine nanodiamonds. (f) XRD spectra for aminated and pristine nanodiamonds.
3.2. Corona formation and colloidal stability of nanodiamonds
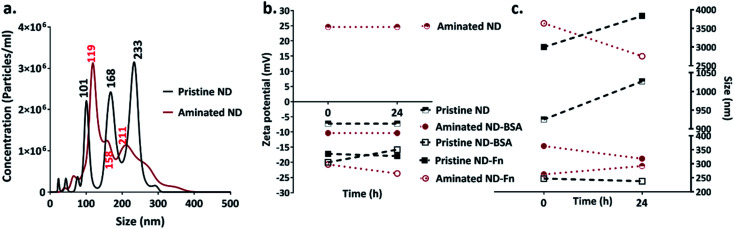
After 5 min of exposure of nanodiamonds (pristine and aminated) to BSA and FN9-10, coronas formed, as shown by the changes in their size and zeta potential (Fig. 2b and c). The exposure of nanodiamonds to BSA prevented agglomeration. The size of pristine nanodiamonds decreased from ∼1000 nm (in water) to 280 nm while the size of aminated nanodiamonds dropped from 3000 nm to 220 nm. Furthermore, both nanodiamond variants dispersed in BSA remained stable until 24 h after exposure, demonstrating the stabilising effects of the BSA corona. Similarly, the zeta potential of the pristine nanodiamonds exposed to BSA dropped from −8 mV to −20 mV and for 24 h it remained at −16 mV (Fig. 2c). A greater change in zeta potential was observed for aminated nanodiamonds after exposure to BSA, as the zeta potential dropped from 20 mV to −11 mV (Fig. 2c).
Fig. 2. Nanoparticle size and charge measurements: (a) size distribution of pristine and aminated nanodiamonds; changes to (b) the zeta potential and (c) size before and after exposure to proteins (BSA and FN9-10).
The drop of particle size and zeta potential of nanodiamonds exposed to BSA may be attributed to colloidal stabilisation by the corona. By inducing steric hindrance and charge stabilisation, colloidal stability affects the ability of nanoparticles to agglomerate, influencing their interactions with cells both in vitro and in vivo.32 Our results showed that BSA exposure can stabilise nanodiamonds in a solution and reduce agglomeration, which confirmed the formation of a corona on the nanodiamond surface.
In contrast, both aminated and pristine nanodiamonds with the FN9-10 corona were not stable; after 24 h, the average size of pristine and aminated nanodiamond agglomerates increased by 800 nm and 500 nm, respectively. FN9-10 exposure also resulted in a negligible drop of the zeta potential from −17 mV to −18 mV for pristine nanodiamonds and from −20 mV to −25 mV for aminated nanodiamonds (Fig. 2b and c), which confirmed that FN9-10 induced changes to the nanodiamond surfaces.
3.3. Time resolved evolution of corona formation on nanodiamonds
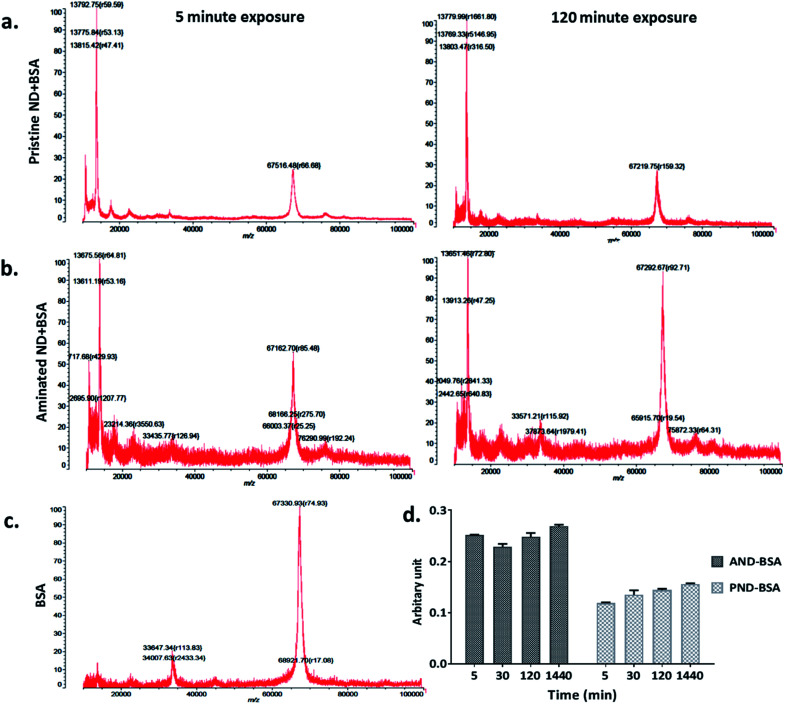
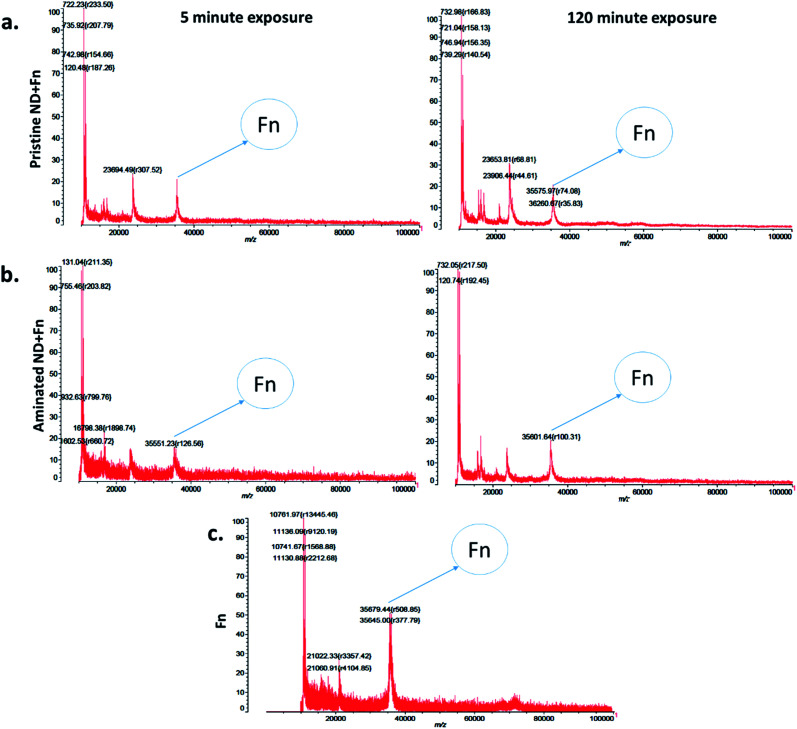
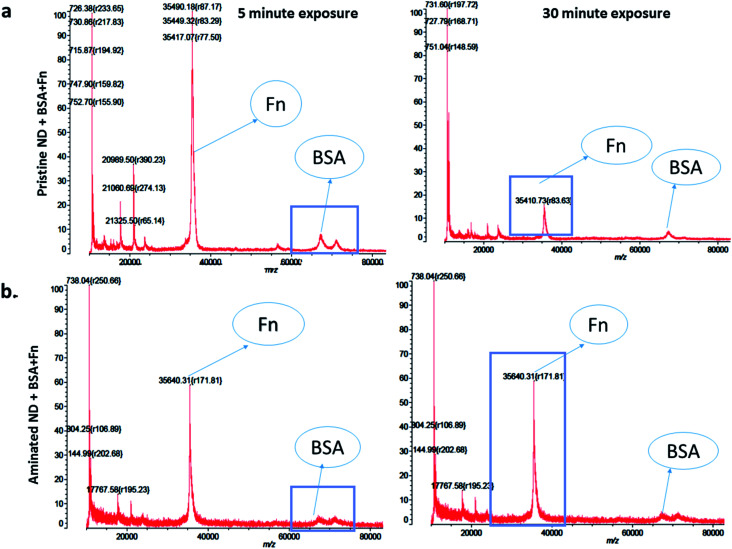
The measurements of corona formation on nanodiamonds using MALDI-TOF showed that BSA and FN9-10 rapidly bind to the surfaces of both types of nanodiamonds (Fig. 3). Relative amounts of each protein on the nanodiamonds increased with time of exposure (Fig. 3a–c). After 5 min of incubation in protein solutions, strong peaks associated with either BSA (m/z of 67 000 kDa; Fig. 3) or FN9-10 (m/z of 35 600 kDa; Fig. 4) were observed for both types of nanodiamonds.
Fig. 3. Time resolved measurements of BSA corona formation: (a) MALDI-TOF spectra of pristine nanodiamonds exposed to BSA for 5 min and 120 min. (b) MALDI-TOF spectra of aminated nanodiamonds exposed to BSA for 5 min and 120 min. (c) Spectrum of the BSA standard; (d) micro BCA assay results of aminated and pristine nanodiamonds exposed to BSA.
Fig. 4. Time resolved measurements of fibronectin corona formation: (a) MALDI-TOF spectra of pristine nanodiamonds exposed to FN9-10 for 5 min and 120 min. (b) MALDI-TOF spectra of aminated nanodiamonds exposed to FN9-10 for 5 min and 120 min. (c) Spectra of the FN9-10 standard.
The analysis of peak intensity and its increase with time suggested that BSA has a greater affinity towards aminated nanodiamonds, whereas FN9-10 had similar affinity towards both types. The micro BCA assay showed that the amount of proteins bound to the aminated nanodiamonds was ∼50% higher than that bound to the pristine nanodiamonds (Fig. 3d). After corona formation, the FN9-10-exposed nanoparticles remained relatively stable, as evidenced by the negligible changes in peak intensity (Fig. 4a and b). The better stability of the FN9-10 corona, when compared with the BSA corona, could be attributed to the stronger binding potential of FN9-10 to the surface of nanoparticles.33,34
When nanodiamonds were exposed to the solution of both proteins at the same time, more rapid and stronger binding of FN9-10 was observed (Fig. 5a). This suggests that FN9-10 has higher affinity than BSA towards both types of nanodiamonds. The higher binding affinity of FN9-10 could be explained by differences in the structure. Since BSA and FN9-10 have isoelectric points below 5.5, they both share a charge-induced affinity towards nanodiamonds. However, FN9-10 contains an arginine–glycine–aspartate (RGD) sequence, which offers greater adhesion and higher affinity towards the substrate than albumin.35
Fig. 5. Time-resolved changes in the amount of protein on the nanodiamonds exposed to an equal mixture of BSA and FN9-10 for 5 and 30 min – MALDI-TOF: (a) pristine nanodiamonds and (b) aminated nanodiamonds.
Furthermore, based on the mixed-protein experiment, FN9-10 exhibited a higher binding affinity, as it adsorbed to the nanodiamond more rapidly and at higher quantity than BSA. After 30 min, a corona remodelling event was observed; the amount of both proteins on pristine nanodiamonds decreased (Fig. 5a). In contrast, the amount of BSA and FN9-10 bound to aminated nanodiamonds, despite lower peak intensity, remained constant over time (Fig. 5b). A decrease in the amount of FN9-10 and BSA on pristine nanodiamonds may be associated to Vroman's model,18 due to the relative biochemical and electrical affinity of the BSA and FN9-10 mixture towards pristine NDs and the differences in the surface charge of pristine nanodiamonds.
Taking into consideration the charge of both types of nanodiamonds, our experiments showed more effective binding of proteins to positively charged nanoparticles. Weaker electrostatic interactions between negatively charged pristine nanodiamonds and proteins are likely the reason behind the drop in the protein amount as exposure time increased. This result evidenced that the protein corona remodels with time for nanodiamonds and that the remodelling was associated with the physicochemical properties of nanodiamonds as well as the type of protein.
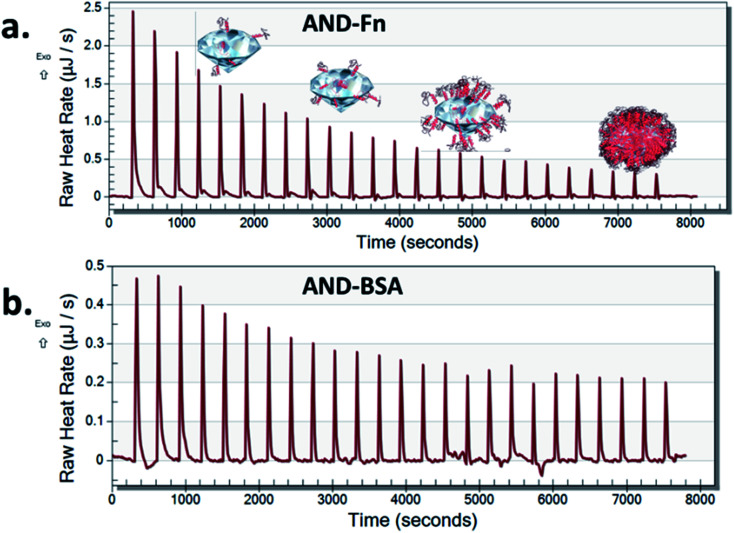
Nano-calorimetric measurements confirmed that FN9-10 had higher binding affinity to both types of nanodiamonds than BSA (Fig. 6 and S2†). Both BSA and FN9-10 adsorbed readily onto the surface of aminated nanodiamonds (Fig. 6) when titrated individually. The heat released at any given time point after titrating FN9-10 with the aminated nanodiamonds was five times higher than that for BSA. The heat released from the first isothermal peak was 2.5 μJ s−1 for FN9-10, whereas for BSA it was 0.46 μJ s−1 (Fig. 6). For both BSA and FN9-10, the heat release decreased to 0.2 μJ s−1 after 2 h of titration. Further titration of BSA and FN9-10 did not induce any changes in the isotherm and the peaks plateaued, suggesting saturation of the protein on the surface. The reason behind the reduction in protein binding and corona growth is not fully understood, but it could be associated with limited protein–protein interactions after the nanodiamond surface was fully covered by proteins.
Fig. 6. Isothermal calorimetry plot of FN9-10 (a) and BSA (b) titrated with aminated nanodiamonds.
Low heat release that remained stable over time was observed when BSA was titrated with pristine nanodiamonds (Fig. S3†). These behaviours could be associated with a lower affinity of BSA to nanodiamond surfaces which promoted sorption and desorption of the protein (release and absorption of heat). This result could be also associated with protein agglomeration in the presence of nanodiamonds, which in turn reduced their ability to bind to individual nanoparticles as the corona.36
Our experiments showed that the composition of the environment that surrounds the nanoparticles is a key factor in corona remodelling. When nanoparticles transition between cells and tissues, the composition of the environment constantly changes. As a consequence of these changes, the corona remodels from transiently bound molecules to long-term binding. As such, nanoparticles carry proteins and other biomolecules from different tissues they pass through.37–39 This finding implies that depending on the route of administration, the corona on the nanoparticles is different.
As demonstrated above, the corona remodels dynamically, so it is critical to consider both soft and hard coronas when assessing the impact of nanoparticles on health and the environment. Since the first layer defines how the corona forms and remodels, pre-treating nanoparticles with known functional groups, including proteins, may direct their interactions in specific environments.
3.4. Protein corona determines cytotoxicity of nanodiamonds
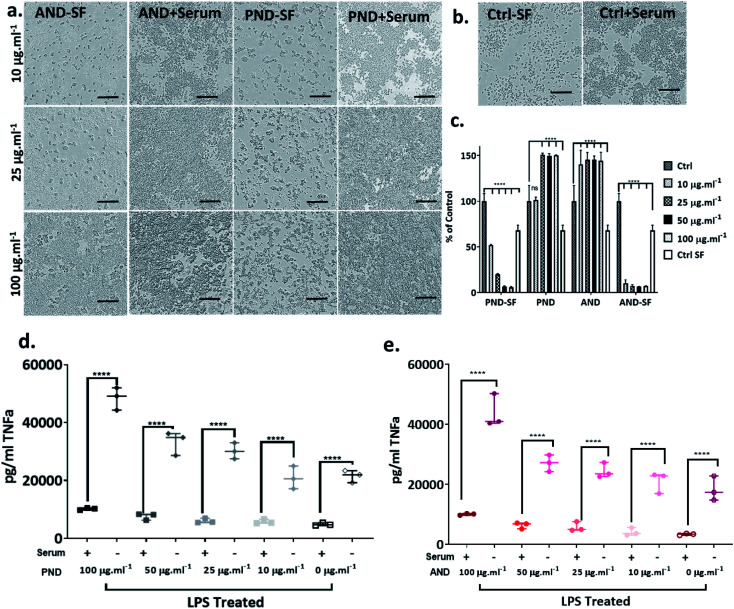
Fao cells cultured in a medium with and without serum were treated with both classes of nanodiamonds ± both coronas. In the absence of serum, the viability of Fao cells treated with pristine and aminated nanodiamonds decreased significantly. The drop of cell viability under serum free conditions was ∼50% for cells treated with 100 μg mL−1 pristine nanodiamonds, while for the same treatment, the drop was only ∼12% in the medium containing serum (Fig. 7a and b). For cells treated with 100 μg mL−1 aminated nanodiamonds, the drop of viability was ∼45% under serum-free conditions and ∼20% in the presence of serum (Fig. 7a and b).
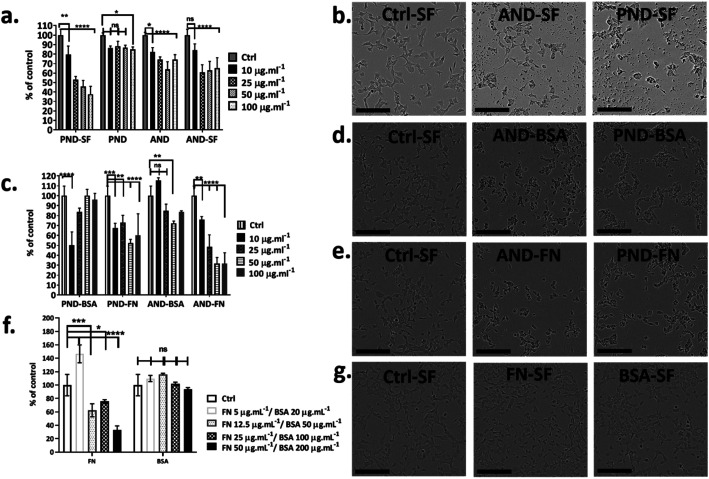
Fig. 7. Effect of the protein corona on nanodiamond cytotoxicity. (a) Metabolic activity of cells exposed to nanodiamonds in the complete medium. (b, d and e) Bright field images of cells before and after exposure to aminated (ANDs) and pristine nanodiamonds (PNDs) with and without coronas (BSA and FN) under serum-free conditions (scale bar 200 μm). (c) Metabolic activity of cells exposed to nanodiamonds with BSA and FN9-10 coronas under serum-free conditions. (f) Metabolic activity of cells exposed to BSA and FN9-10; (*p < 0.05, ****p < 0.001, two-way ANOVA test). (g) Bright field images of cells exposed to BSA and FN.
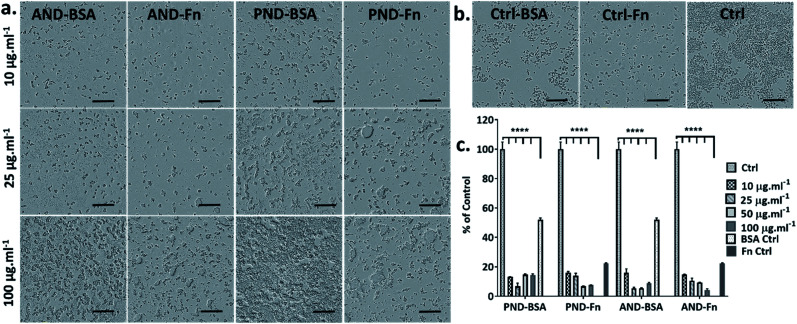
To assess the effect of the protein corona on the toxicity of nanodiamonds, Fao cells were cultured under serum-free conditions and treated with nanodiamonds with BSA and FN-9-10 coronas. When cells were exposed to 10 μg mL−1 pristine nanodiamonds with the BSA corona, viability dropped by ∼50% (Fig. 7c). Interestingly, when cells were treated with these nanodiamonds at higher concentrations (25 μg mL−1), the drop of cell viability was only ∼15%. With a further increase in the nanodiamond concentration (up to 50 and 100 μg mL−1) the viability of cells was comparable to that of control samples. However, for cells treated with aminated nanodiamonds with the BSA corona, the cell viability increased by ∼10% at concentrations of 10 μg mL−1. But it decreased by ∼10–20% for higher concentrations of nanodiamonds (25, 50 and 100 μg mL−1, Fig. 7c).
When Fao cells were treated with pristine nanodiamonds with the FN9-10 corona, a drop in cell viability between 30 and 45% was observed. For example, cell viability dropped by 25% and 45% when cells were treated with nanodiamond concentrations of 25 μg mL−1 and 50 μg mL−1. For cells treated with aminated nanodiamonds with the FN9-10 corona at concentrations of 50–100 μg mL−1, a ∼60% drop in the viability was observed for both concentrations. Overall, a statistically significant drop in cell viability was observed for both types of nanodiamonds with the FN9-10 corona at all concentrations (Fig. 7c) as well as for pristine nanodiamonds with the BSA corona at 10 μg mL−1 and aminated nanodiamonds at 25 μg mL−1 and 50 μg mL−1. The altered cell morphology, reduced cell count and significant drop in cell viability (Fig. 7d and e) showed that nanodiamonds with the FN9-10 corona were cytotoxic and exhibited a higher cytotoxicity than that of nanodiamonds with the BSA corona. We also observed the drop in viability when cells were exposed to fibronectin only at concentrations above 12.5 μg mL−1 (Fig. 7f and g). In contrast, the exposure to low concentrations of BSA (20–50 μg mL−1) led to an increase in cell viability.
Taken together, these results suggested that cytotoxicity of nanodiamonds with the fibronectin corona were independent from the exposure to free fibronectin (unbound fibronectin) in the nanodiamond solution. Cytotoxicity was most likely associated with the enhanced uptake of nanodiamonds with the fibronectin corona and the larger size of agglomerates that induce a different level of injury. Similarly, the increase in viability of cells treated with nanodiamonds with the BSA corona is likely to be associated with more effective delivery of BSA into the cells through nanodiamonds.
3.5. Cellular internalisation of nanodiamonds
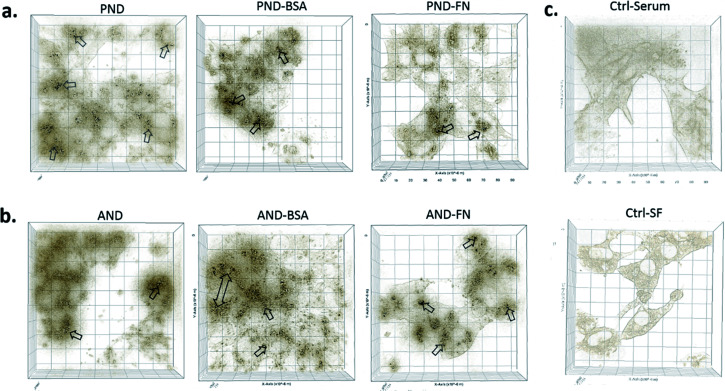
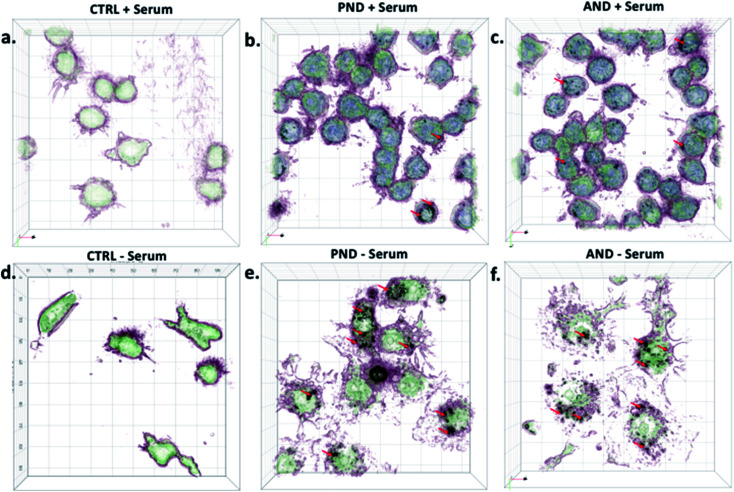
3D imaging of individual cells showed that both types of nanodiamonds with and without the corona coating were internalised by Fao cells (Fig. 8). However, the presence of the corona influenced the amount of nanodiamonds within the cell compartment. Cells exposed to aminated nanodiamonds with FN9-10 showed the highest number of internalised nanoparticles per cell volume, offering a possible explanation for the most significant drop of cell viability in cells treated with these nanodiamonds (Fig. 7c). The cellular internalisation study also showed that the volume of aminated nanodiamonds with the BSA corona per cell volume was higher than that of pristine nanodiamonds with the BSA corona (Fig. 8). These results could also explain the lower drop in viability of cells treated with pristine nanodiamonds with the BSA corona at concentrations above 12.5 μg mL−1 and which could be associated with more effective delivery of BSA into cells at higher nanodiamond concentrations.
Fig. 8. Qualitative analysis of the nanodiamond internalisation by Fao cells. (a) Cells treated with pristine nanodiamonds (PNDs) ± FN9-10 and BSA coronas; (b) cell treated with aminated nanodiamonds (ANDs) ± FN9-10 and BSA coronas; (c) control cells in the presence or absence of serum proteins.
Overall, our studies showed that serum proteins attached to the surface of nanodiamonds may reduce cellular uptake and potentially also their cytotoxicity. Similar results were reported for gold nanoparticles, where reduced cellular uptake and decreased toxicity were observed in phagocytic and non-phagocytic cells exposed to gold nanoparticles with a protein corona.8,10 Another explanation could be that nanodiamonds act as carriers of essential nutrients when they enter the cell. For example, serum proteins such as albumin contain essential nutrients responsible for cell growth and viability. When these proteins are attached to the surface of nanodiamonds as a corona, the internalisation of nanodiamonds delivers them directly into cells.40 In this way, a corona may also reduce or slow down potential negative effects of nanodiamonds on cell viability. However, this phenomenon may not be applicable for all types of proteins and nanoparticles; it assumes that proteins remain in the non-denatured form (functional) and the final effect can also be cell specific.
3.6. Immune response to nanodiamonds
The analysis of the viability of RAW 264.7 macrophage cells cultured in complete media showed that the viability was not compromised by either pristine or aminated nanodiamonds. In fact, at concentrations of nanodiamonds above 25 μg mL−1, the cell viability increased by 40–50% compared with that of control cells (Fig. 9a–c). In contrast, under serum-free conditions, a significant decrease in cell viability was observed (Fig. 9c) for both types of nanodiamonds. A statistically significant drop of cell viability was observed for cells treated with nanodiamonds at concentrations > 25 μg mL−1. The drop of the viability was as high as 90%, which suggests very significant cytotoxicity of nanodiamonds.
Fig. 9. The effect of serum protein on nanodiamond-induced toxicity and proinflammatory response in RAW 264.7 cells: (a) bright field images of cells exposed to aminated and pristine nanodiamonds in the presence and absence of serum proteins (SF-serum free conditions); (b) bright field images of control cells cultured in the presence or absence of serum protein (scale bar 300 μm); (c) metabolic activity of cells exposed to nanodiamonds in the presence and absence of serum proteins; (d and e) proinflammatory response of cells exposed to aminated nanodiamonds in the presence or absence of serum proteins. (****p < 0.001, two-way ANOVA test).
The analysis of inflammatory cytokine production, tissue necrosis factor-alpha (TNFα), showed that nanodiamonds increased the TNFα level under serum-free conditions. Overall, for cells treated with nanodiamonds, the production of TNFα under serum-free conditions was four times higher than in complete media. The production of TNFα was nanodiamond concentration-dependent. For pristine nanodiamonds, TNFα levels were between 20 000 pg mL−1 and 60 000 pg mL−1 and for aminated nanodiamonds, they were between 15 000 pg mL−1 and 45 000 pg mL−1 (Fig. 9d and e).
When RAW 264.7 cells were treated with nanodiamonds overnight and then additionally with lipopolysaccharide (LPS), a pro-inflammatory endotoxin, a statistically significant increase in TNFα production was observed for both types of nanodiamonds (Fig. 9d and e). It is worth noting that the exposure to nanodiamonds alone, without LPS treatment, did not upregulate production of TNFα (Fig. 9d and e). These results suggested that nanodiamonds caused toxicity and made the macrophages susceptible to a secondary toxin, a sign of cell sensitisation.
Qualitative and quantitative analysis of nanodiamond internalisation showed that under serum-free conditions, more nanodiamonds were internalised by the cells (Fig. 10, red arrows). Cells with large quantities of internalised nanodiamonds exhibited a disrupted cell morphology. This result suggests a negative impact of nanodiamonds on cell function and could explain the aforementioned sensitisation to LPS.
Fig. 10. Qualitative analysis of nanodiamond internalisation by RAW 264.7 cells. (a) Control cells cultured in the presence of serum protein; (b) cells treated with pristine nanodiamonds (PNDs) in the presence of serum protein; (c) cells treated with aminated nanodiamonds (ANDs) in the presence of serum protein; (d) control cells cultured in the absence of serum protein; (e) cells treated with pristine nanodiamonds (PNDs) in the absence of serum protein; (f) cells treated with aminated nanodiamonds (ANDs) in the absence of serum protein.
Taken together, our results exemplified that nanodiamonds inhibit immune cell function and sensitise them to secondary toxins. Translating this to human applications, it may mean that nanodiamonds could impair functionality of the immune system and make us more vulnerable to infections or pollution. However, serum proteins, as well as coronas on nanodiamonds, could reduce their cytotoxicity.
When RAW 264.7 cells were cultured under serum-free conditions and treated with both types of nanodiamonds with either the BSA or FN9-10 corona, a drop in cell viability by 80% was observed in all test groups (Fig. 11a–c). Changes to cell morphology, i.e. rounding, and a significant reduction of their number (Fig. 11a) indicated cytotoxicity of nanodiamonds with both types of coronas.
Fig. 11. The effect of the BSA and FN9-10 corona on nanodiamond cytotoxicity in RAW 264.7 cells: (a) bright field images of cells exposed to aminated and pristine nanodiamonds with the BSA and FN9-10 corona; (b) bright field images of RAW 264.7 cells cultured in the presence of BSA, FN9-10 and serum protein (scale bar 300 μm); (c) metabolic activity (CCK-8 assay) of cells exposed to nanodiamonds preconditioned with the BSA and FN9-10 corona. (****p < 0.001, two-way ANOVA test).
Comparative analysis of the viability of Fao and RAW 264.7 cells showed that cytotoxicity of nanodiamonds correlated with the level of nanodiamond internalisation and was dependent on cell type. Fröhlich previously showed that non-phagocytic cells preferentially internalised positively charged nanoparticles, which was reproduced in our experiments.41 We further showed that phagocytic cells internalised both negatively and positively charged nanoparticles effectively.
Our results showed that differences in cytotoxicity of aminated and pristine nanodiamonds are associated primarily with the composition of the corona. The composition of the corona up- or downregulated the internalisation and agglomeration of nanodiamonds, which subsequently impacted the cytotoxicity level. However, the physicochemical properties of nanodiamonds determined how the corona formed and its composition. Therefore, both physicochemical properties (i.e. charge, size, and agglomerate size, which influences sedimentation time) and the composition of the corona are key factors that influence cytotoxicity of nanodiamonds.
To examine the influence of the physicochemical properties on nanodiamonds, we showed that aminated nanodiamonds were cytotoxic to both phagocytic and non-phagocytic cells. We further found that the cytotoxicity of aminated nanodiamonds was higher than that of pristine nanodiamonds, which agrees with the general understanding that cationic nanoparticles are more cytotoxic than anionic ones. In accordance with our findings, it was reported that cationic nanoparticles disrupt plasma-membrane integrity, causing mitochondrial and lysosomal damage, and upregulate autophagosome generation.41 For example, multiple classes of positively charged nanoparticles, including dendrimers, silica, polystyrene, cerium oxide, calcium phosphate–PEI core–shells, silver and gold, have been reported to induce higher cytotoxicity than negatively charged gold nanoparticles.42,43 These observations could explain the higher cytotoxicity of aminated nanodiamonds in our experiments.
Regarding corona composition, nanodiamonds with the fibronectin corona were more cytotoxic (Fig. 7c), and the number of internalised nanodiamonds (including relatively large agglomerates) was greater than that observed for nanodiamonds with the BSA corona (Fig. 8). Since FN9-10 contains the RGD peptide sequence in its structure,44 and RGD can effectively recognise and bind to the integrin receptors on the cell surface, it is possible that the fibronectin corona enhanced the binding of nanodiamonds to the cell membrane. Subsequently, more nanodiamonds could be internalised, inducing cell damage and decreasing cell viability. It has also been reported that the presence of protein aggregates inside cells can contribute to cell damage and disease development.45 Our earlier studies confirmed this when cell-bound nanodiamonds with the fibronectin corona agglomerated following denaturation of the protein.25 These findings could further explain the increased internalisation and cytotoxicity of nanodiamonds with the fibronectin corona.
However, more factors must be considered in determining the cytotoxicity of nanodiamonds. The interplay between nanoparticle properties, environmental conditions, duration of the exposure to nanoparticles and type of cells must be considered. In our case, both pristine and aminated nanodiamonds induced substantial cytotoxicity in both phagocytic and non-phagocytic cells (Fao cells and RAW 264.7 cells) under serum-free conditions (Fig. 7). However, during experiments in the complete medium, cytotoxicity was not observed within 24 h following nanodiamond exposure (Fig. 9c). Importantly, the BSA corona on pristine nanodiamonds reduced internalisation (Fig. 8), while the BSA corona on aminated nanodiamonds increased the viability of Fao cells (Fig. 7c) at a nanodiamond concentration of 10 μg mL−1.
Finally, the observed differences in cytotoxicity of aminated and pristine nanodiamonds with the same type of corona were most likely associated with the differences in the corona structure through differences in the monomer protein structure. We have previously showed that protein denaturation and multimerization may occur when proteins bind to nanoparticles, and these changes to the protein structure depend on the physicochemical properties of nanoparticles.25 Cells have the capacity to recognise protein conformation (functional vs. denatured) and respond accordingly. For example, when BSA binds to the surface of pristine nanodiamonds, the conformation of BSA is retained. Nanodiamonds with the BSA corona can then bind to membrane albumin receptors, which promotes their internalisation, and potentially deliver structurally functional protein to improve cell viability (Fig. 7c).8 In contrast, BSA bound to aminated nanoparticles was partially denatured,8 and we speculated that internalisation was mediated by scavenger receptors and resulted in increased cytotoxicity (Fig. 7c).
4. Conclusions
In the above studies, we demonstrated that the formation of the protein corona on nanodiamonds is a complex and multifactorial process. This process is governed by the interplay between the environmental conditions (i.e. biochemical composition of the surrounding environment and time of exposure) and physicochemical properties of nanodiamonds. Importantly, we showed that the corona remodels with time for nanodiamonds, in accordance with Vroman's model. The remodelling of the corona is an important consideration for biomedical and environmental applications of nanodiamonds because, as we demonstrated here, the composition of the corona is the key factor which determines the stability of nanodiamonds and their interactions with cells.
Overall, our work confirmed that the protein corona modulates internalisation and cytotoxicity of nanodiamonds in non-phagocytic and phagocytic cells. First, we showed that under serum-free conditions, nanodiamonds without the corona caused significant cytotoxicity. However, cytotoxicity was reduced when cells were cultured in complete media. These results highlighted the protective effect of serum proteins. Next, we showed that nanodiamonds with the BSA corona were less cytotoxic to Fao cells than nanodiamonds with the FN9-10 corona. These results correlated with the level of internalisation of nanodiamonds, which qualitatively was greater for aminated nanodiamonds with the fibronectin corona. However, irrespective of the corona type, nanodiamonds caused >80% drop in viability of RAW 264.7 cells (under serum-free conditions). These results emphasised that cytotoxicity is cell type-dependent and that the corona plays a key role in internalisation and cytotoxicity. The demonstrated differences in cytotoxicity are likely to be associated with differences in the nanoparticle internalisation pathway, which is dependent on cell type. Phagocytic cells can internalise significantly higher amounts of nanoparticles, which could result in more evident cytotoxicity.
Furthermore, we showed that nanodiamonds induced proinflammatory responses and immune cells were susceptible to secondary endotoxins. These results suggested that chronic exposure to nanodiamonds can potentially compromise the effectiveness of the immune system.
Finally, we demonstrated that the assessment of risks associated with nanoparticles (nanotoxicity) must consider the presence of the corona. Since the composition and remodelling of the corona determine the fate of nanoparticles in the body, it is necessary to integrate studies of corona formation and remodelling into the assessment framework of nanotoxicity. Corona considerations, while complex and multifactorial, should constitute an essential element of protocols that determine the safety of nanoparticles for regulatory purposes. Notably, our studies highlighted that a precise understanding of corona formation and remodelling is likely to unlock new possibilities to manufacture safer nanoparticles for various applications. For example, nanoparticles with pre-defined coronas can enable the delivery of therapeutics directly to the target in a safe and effective manner – personalized treatments – while reducing potential adverse effects of both nanoparticles and therapeutics.46
Conflicts of interest
The authors declare no competing financial interest.
Supplementary Material
Acknowledgments
W. C. acknowledges the Sydney Nano and SOAR Fellowship for financial support. Authors acknowledge Mr Herman Hau for his support in initial editing of the manuscript. A. G. and K. W. N. acknowledge the Facility for Analysis, Characterization, Testing and Simulation (FACTS), Nanyang Technological University, Singapore, for technical support in electron microscopy analysis.
Electronic supplementary information (ESI) available. See DOI: 10.1039/d0na00231c
References
- Alhaddad A. et al., Nanodiamond as a vector for siRNA delivery to Ewing sarcoma cells. Small. 2011;7:3087–3095. doi: 10.1002/smll.201101193. [DOI] [PubMed] [Google Scholar]
- Baek S. et al., Smart multifunctional drug delivery towards anticancer therapy harmonized in mesoporous nanoparticles. Nanoscale. 2015;7:14191–14216. doi: 10.1039/c5nr02730f. doi: 10.1039/c5nr02730f. [DOI] [PubMed] [Google Scholar]
- Kwon S. et al., Silica-based mesoporous nanoparticles for controlled drug delivery. J. Tissue Eng. 2013;4:2041731413503357. doi: 10.1177/2041731413503357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manus L. M. et al., Gd(iii)-nanodiamond conjugates for MRI contrast enhancement. Nano Lett. 2009;10:484–489. doi: 10.1021/nl903264h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay D. et al., Magnetised thermo responsive lipid vehicles for targeted and controlled lung drug delivery. Pharm. Res. 2012;29:2456–2467. doi: 10.1007/s11095-012-0774-9. [DOI] [PubMed] [Google Scholar]
- Wagstaff A. J. et al., Cisplatin drug delivery using gold-coated iron oxide nanoparticles for enhanced tumour targeting with external magnetic fields. Inorg. Chim. Acta. 2012;393:328–333. doi: 10.1016/j.ica.2012.05.012. [DOI] [Google Scholar]
- Pozzi D. et al., The biomolecular corona of nanoparticles in circulating biological media. Nanoscale. 2015;7:13958–13966. doi: 10.1039/c5nr03701h. doi: 10.1039/c5nr03701h. [DOI] [PubMed] [Google Scholar]
- Fleischer C. C. Payne C. K. Secondary Structure of Corona Proteins Determines the Cell Surface Receptors Used by Nanoparticles. J. Phys. Chem. B. 2014;118:14017–14026. doi: 10.1021/jp502624n. doi: 10.1021/jp502624n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docter D. et al., The nanoparticle biomolecule corona: lessons learned - challenge accepted? Chem. Soc. Rev. 2015;44:6094–6121. doi: 10.1039/c5cs00217f. doi: 10.1039/c5cs00217f. [DOI] [PubMed] [Google Scholar]
- Cheng X. et al., Protein Corona Influences Cellular Uptake of Gold Nanoparticles by Phagocytic and Nonphagocytic Cells in a Size-Dependent Manner. ACS Appl. Mater. Interfaces. 2015;7:20568–20575. doi: 10.1021/acsami.5b04290. doi: 10.1021/acsami.5b04290. [DOI] [PubMed] [Google Scholar]
- Zhang B. et al., Shape dependent cytotoxicity of PLGA-PEG nanoparticles on human cells. Sci. Rep. 2017;7:182–189. doi: 10.1038/s41598-017-07588-9. doi: 10.1038/s41598-017-07588-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai H. et al., Diameter and rigidity of multiwalled carbon nanotubes are critical factors in mesothelial injury and carcinogenesis. Proc. Natl. Acad. Sci. U. S. A. 2011;108:E1330–E1338. doi: 10.1073/pnas.1110013108. doi: 10.1073/pnas.1110013108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B. Zhu M. Li Z. Lung P. S. Chrzanowski W. Kwok C. T. Lu J. Li Q. Cellular fate of deformable needle-shaped PLGA-PEG fibers. Acta Biomater. 2020;112:182–189. doi: 10.1016/j.actbio.2020.05.029. doi: 10.1016/j.actbio.2020.05.029. [DOI] [PubMed] [Google Scholar]
- Jin S. et al., Surface chemistry-mediated penetration and gold nanorod thermotherapy in multicellular tumor spheroids. Nanoscale. 2013;5:143–146. doi: 10.1039/c2nr31877f. doi: 10.1039/c2nr31877f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera Gil P. Oberdörster G. Elder A. Puntes V. Parak W. J. Correlating Physico-Chemical with Toxicological Properties of Nanoparticles: The Present and the Future. ACS Nano. 2010;4:5527–5531. doi: 10.1021/nn1025687. doi: 10.1021/nn1025687. [DOI] [PubMed] [Google Scholar]
- Anselmo A. C. Mitragotri S. Impact of particle elasticity on particle-based drug delivery systems. Adv. Drug Delivery Rev. 2017;108:51–67. doi: 10.1016/j.addr.2016.01.007. [DOI] [PubMed] [Google Scholar]
- Gratton S. E. A. et al., The effect of particle design on cellular internalization pathways. Proc. Natl. Acad. Sci. U. S. A. 2008;105:11613–11618. doi: 10.1073/pnas.0801763105. doi: 10.1073/pnas.0801763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vroman L. Effect of adsorbed proteins on the wettability of hydrophilic and hydrophobic solids. Nature. 1962;196:476–477. doi: 10.1038/196476a0. [DOI] [PubMed] [Google Scholar]
- Tenzer S. et al., Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nat. Nanotechnol. 2013;8:772–781. doi: 10.1038/nnano.2013.181. [DOI] [PubMed] [Google Scholar]
- Casals E. Pfaller T. Duschl A. Oostingh G. J. Puntes V. Time evolution of the nanoparticle protein corona. ACS Nano. 2010;4:3623–3632. doi: 10.1021/nn901372t. [DOI] [PubMed] [Google Scholar]
- Lundqvist M. et al., The evolution of the protein corona around nanoparticles: a test study. ACS Nano. 2011;5:7503–7509. doi: 10.1021/nn202458g. [DOI] [PubMed] [Google Scholar]
- Barrán-Berdón A. L. et al., Time evolution of nanoparticle–protein corona in human plasma: relevance for targeted drug delivery. Langmuir. 2013;29:6485–6494. doi: 10.1021/la401192x. [DOI] [PubMed] [Google Scholar]
- Hadjidemetriou M. Al-Ahmady Z. Kostarelos K. Time-evolution of in vivo protein corona onto blood-circulating PEGylated liposomal doxorubicin (DOXIL) nanoparticles. Nanoscale. 2016;8:6948–6957. doi: 10.1039/c5nr09158f. doi: 10.1039/c5nr09158f. [DOI] [PubMed] [Google Scholar]
- Fong W.-K., et al., in Biological Responses to Nanoscale Particles, Springer, 2019, pp. 101–150 [Google Scholar]
- Khanal D. et al., Biospectroscopy of Nanodiamond-Induced Alterations in Conformation of Intra- and Extracellular Proteins: A Nanoscale IR Study. Anal. Chem. 2016;88:7530–7538. doi: 10.1021/acs.analchem.6b00665. doi: 10.1021/acs.analchem.6b00665. [DOI] [PubMed] [Google Scholar]
- Roach P. Farrar D. Perry C. C. Interpretation of protein adsorption: surface-induced conformational changes. J. Am. Chem. Soc. 2005;127:8168–8173. doi: 10.1021/ja042898o. [DOI] [PubMed] [Google Scholar]
- Ritz S. et al., Protein corona of nanoparticles: distinct proteins regulate the cellular uptake. Biomacromolecules. 2015;16:1311–1321. doi: 10.1021/acs.biomac.5b00108. [DOI] [PubMed] [Google Scholar]
- Lesniak A. et al., Effects of the presence or absence of a protein corona on silica nanoparticle uptake and impact on cells. ACS Nano. 2012;6:5845–5857. doi: 10.1021/nn300223w. [DOI] [PubMed] [Google Scholar]
- Lesniak A. et al., Nanoparticle adhesion to the cell membrane and its effect on nanoparticle uptake efficiency. J. Am. Chem. Soc. 2013;135:1438–1444. doi: 10.1021/ja309812z. [DOI] [PubMed] [Google Scholar]
- Landgraf L. et al., A plasma protein corona enhances the biocompatibility of Au@Fe3O4 Janus particles. Biomaterials. 2015;68:77–88. doi: 10.1016/j.biomaterials.2015.07.049. [DOI] [PubMed] [Google Scholar]
- Ho Y. T. Kamm R. D. Kah J. C. Y. Influence of protein corona and caveolae-mediated endocytosis on nanoparticle uptake and transcytosis. Nanoscale. 2018;10:12386–12397. doi: 10.1039/C8NR02393J. [DOI] [PubMed] [Google Scholar]
- Moore T. L. et al., Nanoparticle colloidal stability in cell culture media and impact on cellular interactions. Chem. Soc. Rev. 2015;44:6287–6305. doi: 10.1039/C4CS00487F. [DOI] [PubMed] [Google Scholar]
- Chrzanowski W. et al., Biointerface: protein enhanced stem cells binding to implant surface. J. Mater. Sci.: Mater. Med. 2012;23:2203–2215. doi: 10.1007/s10856-012-4687-2. [DOI] [PubMed] [Google Scholar]
- Chrzanowski W. et al., Nano-Bio-Chemical Braille for Cells: The Regulation of Stem Cell Responses using Bi-Functional Surfaces. Adv. Funct. Mater. 2015;25:193–205. doi: 10.1002/adfm.201401696. [DOI] [Google Scholar]
- Rahman M., Laurent S., Tawil N., Yahia L. H. and Mahmoudi M., in Protein-nanoparticle interactions, Springer, 2013, pp. 21–44 [Google Scholar]
- Winzen S. et al., Complementary analysis of the hard and soft protein corona: sample preparation critically effects corona composition. Nanoscale. 2015;7:2992–3001. doi: 10.1039/C4NR05982D. [DOI] [PubMed] [Google Scholar]
- Ke P. C. Lin S. Parak W. J. Davis T. P. Caruso F. A Decade of the Protein Corona. ACS Nano. 2017;11:11773–11776. doi: 10.1021/acsnano.7b08008. [DOI] [PubMed] [Google Scholar]
- Hadjidemetriou M. et al., In vivo biomolecule corona around blood-circulating, clinically used and antibody-targeted lipid bilayer nanoscale vesicles. ACS Nano. 2015;9:8142–8156. doi: 10.1021/acsnano.5b03300. [DOI] [PubMed] [Google Scholar]
- Wang H. Ma R. Nienhaus K. Nienhaus G. U. J. S. Formation of a monolayer protein corona around polystyrene nanoparticles and implications for nanoparticle agglomeration. Small. 2019;15:1900974. doi: 10.1002/smll.201900974. [DOI] [PubMed] [Google Scholar]
- Li J. et al., Nanodiamonds as intracellular transporters of chemotherapeutic drug. Biomaterials. 2010;31:8410–8418. doi: 10.1016/j.biomaterials.2010.07.058. [DOI] [PubMed] [Google Scholar]
- Fröhlich E. The role of surface charge in cellular uptake and cytotoxicity of medical nanoparticles. Int. J. Nanomed. 2012;7:5577. doi: 10.2147/IJN.S36111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatoo M. A. et al., Physicochemical properties of nanomaterials: implication in associated toxic manifestations. BioMed Res. Int. 2014;2014:498420. doi: 10.1155/2014/498420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luyts K. Napierska D. Nemery B. Hoet P. H. How physico-chemical characteristics of nanoparticles cause their toxicity: complex and unresolved interrelations. Environ. Sci.: Processes Impacts. 2013;15:23–38. doi: 10.1039/C2EM30237C. [DOI] [PubMed] [Google Scholar]
- Bachman H. Nicosia J. Dysart M. Barker T. H. Utilizing fibronectin integrin-binding specificity to control cellular responses. Advances in Wound Care. 2015;4:501–511. doi: 10.1089/wound.2014.0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamás M. J. Sharma S. K. Ibstedt S. Jacobson T. Christen P. Heavy metals and metalloids as a cause for protein misfolding and aggregation. Biomolecules. 2014;4:252–267. doi: 10.3390/biom4010252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Q. Walkey C. Chan W. C. W. Polyethylene Glycol Backfilling Mitigates the Negative Impact of the Protein Corona on Nanoparticle Cell Targeting. Angew. Chem., Int. Ed. 2014;53:5093–5096. doi: 10.1002/anie.201309464. doi: 10.1002/anie.201309464. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.