Abstract
Cervical cancer is the fourth largest cancer, affecting women across the globe. Rapid screening is of vital importance for diagnosis and treatment of the disease, especially in developing countries with high risk populations. In this paper, we report a simple, novel and rapid approach for qualitative screening of cervical cancer. A label-free colorimetric technique (“C-ColAur”) involving the in situ formation of gold nanoparticles (Au NPs) in the presence of clinical samples is demonstrated. The as-formed Au NPs, owing to the sample composition produced a characteristic color that can be used for the qualitative detection of malignancy. We demonstrated the proof of principle using clinical samples (cervical fluid) collected from both cancer affected and healthy individuals. The results of the detection technique, “C-ColAur” when compared with those of the existing conventional diagnostic procedures (i.e. Pap smear or biopsy), showed 96.42% sensitivity. With the detection time less than a minute and with no/minimal sample processing requirements, the proposed technique shows great potential for point-of-care as well as clinical screening of cervical cancer.
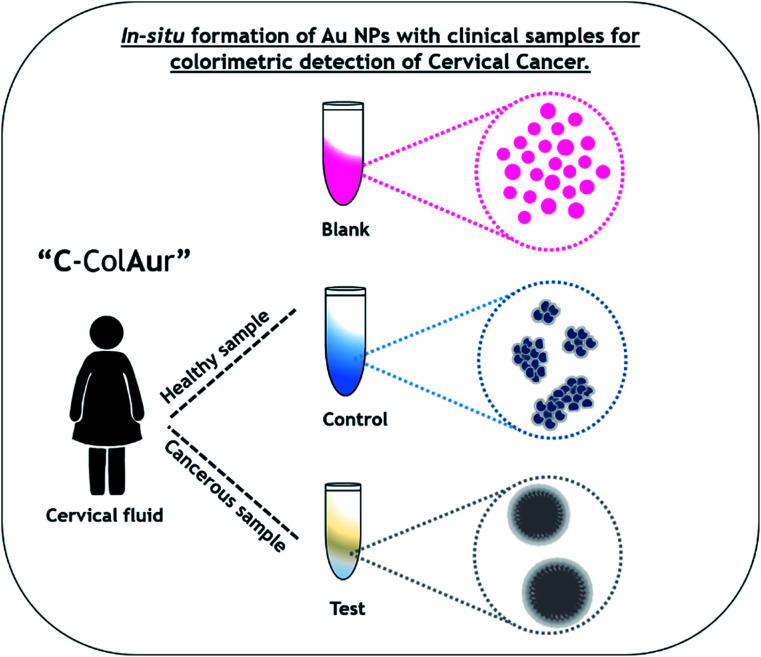
“C-ColAur” technique for colorimetric detection of cervical cancer by in situ formation of gold nanoparticles.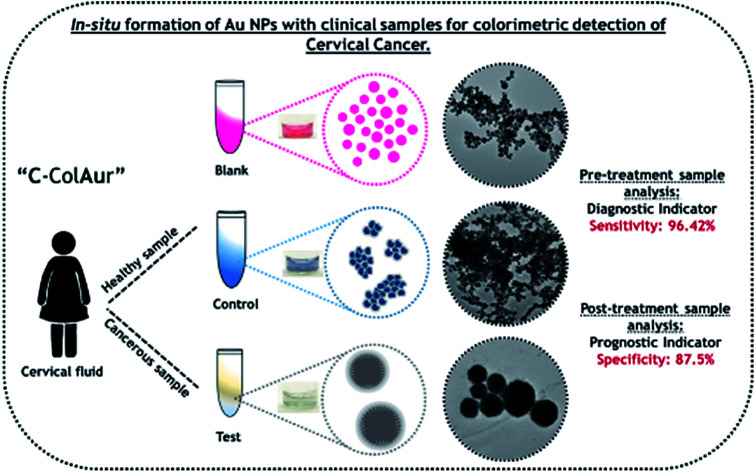
Introduction
Cervical cancer, caused by Human Papilloma Virus (HPV) is the fourth most common cancer and second-largest cause of death in women, worldwide. 569 847 new cases and 311 365 deaths, worldwide were recorded in 2018 (GLOBOCAN 2018) indicating the need for robust screening & treatment strategies. The incidence and mortality are further expected to increase by 36% and 47.5% respectively by 2040.1–5 Cervical cancer has a long pre-clinical phase that spans decades without any symptomatic effects. Organized screening programs have proven to be effective in reducing the mortality of cervical cancer.3,5–7 The success of screening methodologies depend upon increased coverage of the geographical area and higher human participation. The traditional screening/detection techniques of cervical cancer include the papanicolaou (Pap) test, visual inspection with acetic acid (VIA), biopsy and HPV gene testing. The common barriers to these screening procedures are discomfort faced by women during the pelvic examination and need for multiple medical visits.3 Though the currently used screening methods are sensitive, they are expensive and time-consuming, require trained medical personnel, and need sophisticated instrumentation for sample collection, processing and interpretation of results.2,8 The Pap test to identify abnormal cervical cells has been adopted as the standard screening tool in developed nations with sufficient resources. However countries with limited resources, i.e. low and middle-income countries adopted visual inspection or HPV based screening methods, given their lower cost and enormous potential for large scale impact.5,8 Though HPV testing has been considered the most objective and reproducible among all screening tests, it is not cost-effective (especially in developing nations). Hence, VIA has been considered a better and affordable alternate.9–11
The VIA screening test has high sensitivity, providing immediate results, but as mentioned earlier, requires trained medical personnel for pelvic examination. It is also a subjective & qualitative analysis (largely depending on the skill of the medical personnel) and hence suffers from low specificity.12 The currently used screening techniques (cervical smear collection) cause discomfort, pain and embarrassment to women which is one of the prime reasons for under screening in addition to the socio-economic constraints.13,14 Considering the functional and financial limitations of existing screening/diagnostic methods, the development of a rapid point-of-care testing method is very crucial for overcoming the aforementioned limitations. With the emergence of self-sampling for cervical cancer screening, a facile, rapid, and affordable point-of-care screening technique will be extremely beneficial. Among the various sensing mechanisms adapted for screening/diagnosis of diseases, colorimetric assays are more suitable, and hence preferred for rapid testing through visual read-outs.15–17
The advances in nanotechnology led to the development of a variety of biosensors using unique physical and chemical characteristics of nanomaterials. Gold nanoparticles (Au NPs) are one of the most commonly used nanomaterials for various sensing applications, owing to their unique surface plasmon resonance (SPR) based optical properties. Au NPs have received extensive attention, as they can be easily functionalized, and exhibit variations in color corresponding to their size, shape, composition, inter-particle distance and state of aggregation, resulting in rapid and reliable detection.18–22 The typical approach of colorimetric assays involves preparation of a stable colloidal solution/substrate, whose stability gets affected by the addition of the analyte resulting in a color change.
Colorimetric assays using Au NPs are either labelled or label-free. The labelled techniques use Au NPs whose surface is modified/functionalized with a specific label/ligand (DNA, aptamers, peptides, or antibodies). Ligand modified Au NPs are more stable under high ionic strength conditions and undergo controlled aggregations by cross-linking, non-cross-linking, or destabilization strategies.17,20,23 Xia et al. developed a labelled colorimetric method for prostate specific antigen (PSA) detection by inhibition of the formation of Au NPs in serum samples. The method of detection was based on ascorbic acid (AA) induced formation of Au NPs and Cu2+ catalyzed oxidation of AA.24 Wen Ren et al. developed a magnetic probe comprising Fe3O4 core/Au shell nanostructures modified with antibodies for ultrasensitive colorimetric detection of cervical cancer. The magnetic probe successfully detected the protein biomarker, valosin-containing protein (VCP), at a concentration as low as 25 fg mL−1 within 45 min assisted by enzyme-based colorimetric signal amplification.25 Xiao et al. designed and developed a Au/Bi2Se3 hybrid colorimetric biosensor. The catalytic activity of Au/Bi2Se3 was tuned to switch off/on corresponding to the presence of cancer antibody/antigen biomarkers in solution. This colorimetric sensor showed a good generality for the detection of various types of cancer biomarkers, providing an alternative method for cancer diagnosis.16,25 Despite the advantages of improved sensitivity and specificity, labelled techniques are expensive, and time-consuming requiring complex processing of samples and substrates.26
Label-free detection is an affordable alternate to labelled techniques. This technique involves an analyte triggered aggregation (such as electrostatic or hydrogen bonding interactions) of Au NPs.23,27–30 It is cost-effective and requires minimal time for sample processing, and hence more suitable for point-of-care applications. Sina et al. developed a colorimetric technique for detection of the methylscape biomarker using cancer genomes. This method was based on the level of gDNA adsorption on planar and colloidal gold surfaces, respectively resulting in a simple, label-free naked eye platform with an analysis time of less than 10 minutes.31 Mao et al. reported a label-free method for rapid and specific detection of human papilloma virus (HPV). The cysteine (Cys) modified protein (Cys-Sso7d) induced aggregation of Au NPs was highly suppressed in the presence of DNA (HPV 16 & 18 gene fragments) due to the strong binding affinity of Sso7d with dsDNA. The Cys-Sso7d/Au NP probe coupled with PCR showed a specificity & sensitivity of 85.7% and 85.7%, respectively for visual detection of high risk HPV detection.32
The majority of the reported studies on colorimetric analysis for biosensing have deployed Au NPs that were pre-formed. The formation of Au NPs is highly dependent on various factors that trigger or hinder their growth.33 In this paper, we analyse the in situ formation of Au NPs and evaluate their efficacy in detecting cervical cancer using clinical samples (cervical fluids) collected from both healthy and cancer affected subjects. We demonstrate a colorimetric strategy relying on the in situ formation of Au NPs that varied in their size and color depending upon the type of clinical sample leading to screening of cervical cancer. The Au NPs formed in situ were further analysed by spectroscopy and microscopy. The colorimetric detection technique (henceforth to be referred to as “C-ColAur”) was compared with the conventional diagnostic procedures (Pap smear/Biopsy). To the best of our knowledge, this is one of the first reports on the application of in situ formed Au NPs as a colorimetric strategy for screening of cervical cancer.
Experimental section
Materials
Ascorbic acid and trisodium citrate were procured from SRL Chemicals, India. Hydrogen Tetrachloroaurate(iii) (HAuCl4·3H2O), and a phospholipid quantification assay kit (CS0001-1KT) were purchased from Sigma (St. Louis, MO, U.S.A). A BCA protein assay kit was procured from Thermofisher Ltd., India. All the chemicals were used as received without any further purification.
Characterization
The clinical samples were observed under an optical microscope and images were captured (Olympus CKX-53, U.S.A). The absorption spectra were recorded using a UV-Vis spectrophotometer (UV-1800, Shimadzu, Japan). The hydrodynamic diameter, polydispersity index (PDI) and zeta potential were measured by dynamic light scattering (DLS) using a particle size analyzer (Nicomp Nano Z3000 ZLS, U.S.A). The size and morphology of the nanoparticles were characterized by Transmission Electron Microscopy (TEM) (JEOL, JEM 2100, U.S.A). The energy dispersive X-ray analysis (EDS) of Au NPs was performed to understand the elemental composition using a Scanning Electron Microscope (FESEM-JEOL, USA). XRD spectra were obtained using an X-ray Diffractometer (Bruker Discover D8, U.S.A). FT-IR absorption spectra were recorded using a Fourier transform infrared spectrometer (FT-IR, Bruker, U.S.A) in the spectral range of 4000–600 cm−1.
Clinical samples
The clinical samples were collected from the Basavatarakam Indo-American Cancer Hospital & Research Institute, Hyderabad, Telangana, India. The relevant ethical approvals were obtained from the Basavatarakam Indo-American Cancer Hospital & Research Institute, Hyderabad, Telangana, India. Written informed consent was obtained following a protocol approved by the hospital ethics committee (registration number: ECR/7/Inst/AP/2013/RR-16; EC reference number: IECR/2018/125). Study-specific identification codes were assigned, and patient confidentiality was preserved.
Gynecologists collected cervico-vaginal swabs from patients in the lithotomy position using a cytobrush. Respective samples for diagnosis by PAP smear or biopsy were also collected. The contents of the cytobrush were transferred to a storage vial, sealed, coded and transported to the lab where further analysis was performed. Samples were collected from women who visited the hospital for diagnosis/regular check-up (referred to as “pre-treatment samples”). Samples were also collected from women who visited the hospital for follow-up after the therapy, i.e. cancer affected patients who underwent treatment and were called for a follow up (referred to as “post-treatment samples”). A total of 62 samples were collected, of which 14 samples were collected from healthy women, 28 samples were collected from cervical cancer affected women, and 20 samples were collected from women who underwent therapy (radiation/chemotherapy/surgery). All cytology samples collected from patients for diagnosis by Pap smear/biopsy were analysed by the pathology laboratory, Department of Molecular Diagnostics, Basavatarakam Indo-American Cancer Hospital & Research Institute, Hyderabad. Based on the cytology and biopsy outcomes, healthy and cancer affected women were identified, and the results were compared.
Analysis & detection of clinical samples: “C-ColAur” technique
The samples (cervical fluids) were diluted in Milli Q water (1 : 10 dilution) and their absorbance was recorded. The samples were observed under a microscope to understand their composition. The protein and lipid contents in clinical samples were estimated using BCA (bicinchoninic acid) protein assay and phospholipid assay kits respectively following the manufacturer's protocol.
For the colorimetric detection of clinical samples, the following procedure was followed. 100 μL of the clinical sample (1 : 10 dilution) was mixed with 100 μL of 1 mM HAuCl4·3H20, followed by the addition of 200 μL of 2 mM ascorbic acid. The change in the color of the samples after the addition of the reducing agent was photographed. The Au NPs formed without any clinical sample were termed blank (B) NPs, while Au NPs formed with healthy & cancerous cervical fluids were termed control (C) and test (T) Au NPs respectively.
Stability study
The in situ formed blank (B), control (C) & test (T) Au NPs were subjected to salt-based aggregation studies. Saline Sodium Citrate (SSC 5×) buffer was prepared and added to the Au NPs solutions. The corresponding changes in color and absorbance were recorded. Size analysis was performed to understand the change in size with the addition of SSC buffer.31
Sensitivity & specificity of detection
The sensitivity, specificity, negative predictive value (NPV) and positive predictive value (PPV) for the “C-ColAur” technique were calculated using the standard procedures reported in the literature.34,35 Pap smear and biopsy results were considered the gold standard. The results of “C-ColAur” were compared against the gold standard, and the values of sensitivity & specificity were calculated.
Results & discussion
A schematic representation of the detection process using the “C-ColAur” (Cervical cancer detection using Colorimetric sensing with Au NPs) technique is represented in Fig. 1.
Fig. 1. Schematic showing the colorimetric detection of cervical cancer using the “C-ColAur” technique.
The cervico-vaginal samples were collected from the Basavatarakam Indo-American Cancer Hospital & Research Institute, Hyderabad, India. The clinical samples were observed under a light microscope to visualize the morphological differences in the composition of the healthy and cancer affected samples. The healthy sample contained epithelial cells that were discrete, while the cervical cancer affected sample had cancer cell aggregates (Fig. S1A and B†). The cervical fluid samples were diluted (1 : 10 dilution) and analysed for their absorbance. The healthy samples did not show any specific absorption, while cervical cancer affected samples showed absorption at 280 nm corresponding to protein36 (Fig. S1C†).
The proteins and lipids in clinical samples were quantified. The protein and lipid contents in the cancerous samples were observed to be higher, compared to those in the healthy samples (Fig. S1D†).
Blank Au NPs were formed by the ascorbic acid reduction of AuCl4− to Au at ambient temperature. The blank (B) Au NPs, formed in the absence of clinical samples, were very crucial for understanding the variation in the color, size, shape and composition of the control (C) & test (T) Au NPs formed by interactions with healthy and cancerous clinical samples respectively. The cervical fluid samples were individually mixed with HAuCl4, followed by ascorbic acid reduction. This resulted in an immediate color change in the case of healthy samples (bright blue) whereas the cancerous sample showed no specific change in color (Fig. S2†). Biological samples and biomolecular interactions are known to control the dispersion/aggregation of nanoparticles, resulting in their detection/monitoring.20 The rapid change in color was due to the in situ formation of Au NPs. The inset in Fig. 2A shows the Au NPs formed with water (i.e. blank (B)), healthy samples (i.e. control (C)), and cervical cancer affected samples (i.e. test (T)). The color change observed in the clinical samples was specific to the type of the sample (healthy/affected samples – control/test Au NPs). This variation in color could be due to the size/shape/interparticle distance-dependent localized surface plasmon resonance (SPR) of Au NPs.37,38
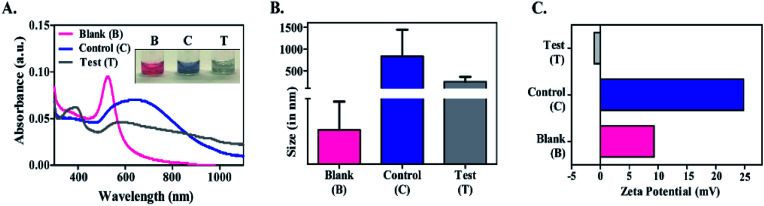
Fig. 2. Clinical sample analysis. (A) Absorbance spectra of blank (B), control (C) & test (T) Au NPs. The inset shows the color of Au NPs. (B) Size analysis and (C) zeta potentials of Au NPs.
The nanoparticles were analysed by UV-visible spectroscopy (Fig. 2A). The blank in situ formed Au NPs showed a characteristic peak at 520 nm, indicating formation of uniform and monodispersed nanoparticles.33,39 The control & test Au NPs showed a broad absorbance ranging between 600–800 nm and 540–640 nm respectively. This red shifted broadening of the SPR could be due to the aggregation-induced coupling of plasmon modes.40–42 Such aggregation of nanoparticles would have been due to the changes in the solvent/sample environment.20,43 In this case, the cervical fluid was the external stimulus that caused Au NPs to form or aggregate differently, leading to a shift in the absorption spectrum, subsequently resulting in the color change. This color change was very much evident in the control than in the test Au NPs, owing to lower intensity (Fig. 2A inset).
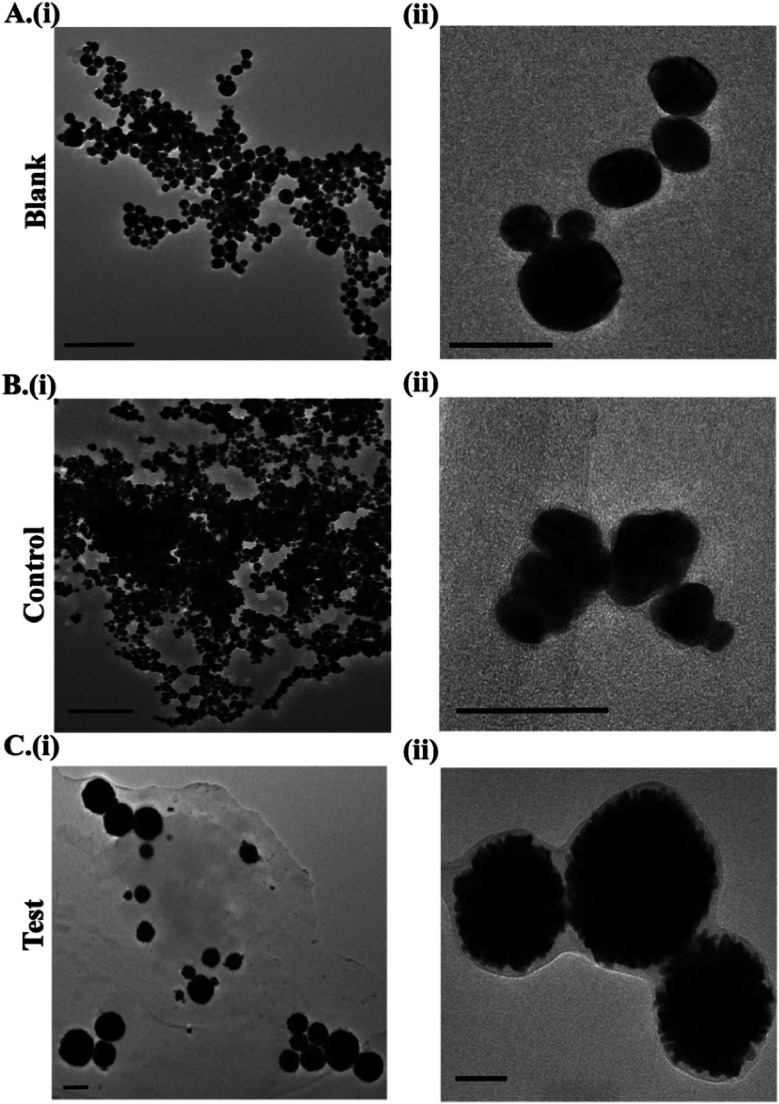
DLS was used to monitor the hydrodynamic size of the in situ formed Au NPs (Fig. 2B). The mean size of the blank Au NPs was found to be 23.4 nm, while that of the control and test Au NPs was 834.8 nm & 252.2 nm, respectively. The zeta potential was recorded as +9.28 mV, +24.83 mV and −1.08 mV for the blank, control and test Au NPs respectively (Fig. 2C). Zeta potential analysis revealed the differences in the surface potential of the Au NPs (ESI Table 1†). The Au NPs were further subjected to TEM analysis to understand the differences in the size/shape (Fig. 3). Blank Au NPs were observed to be spherical and uniformly distributed with an average size of 25–40 nm (Fig. 3A). The control Au NPs were quasi-spherical and smaller in size ranging between 15 and 30 nm (Fig. 3B). They were also found to be aggregated. This aggregation could have resulted in the increased hydrodynamic size as observed in DLS analysis (Fig. 2B). The test Au NPs differed both in size and shape (Fig. 3C) compared to the blank and control Au NPs. The TEM analysis of the test Au NPs showed discrete large sized nanoparticles (250–400 nm). The test Au NPs showed no sign of aggregation, but the particle size was considerably large and the number of particles were low compared to that of the blank & control, which resulted in the reduced absorption intensity as observed in UV-visible analysis (Fig. 2A).
Fig. 3. TEM imaging of (A) blank, (B) control and (C) test Au NPs. *Scale bar in images (i) corresponds to 200 nm and (ii) 50 nm.
The size and morphology of the test Au NPs resembled those of liposome coated gold nanostructures.44–46 The formation of such large sized quasi-spherical Au NPs could be attributed to the presence of cancer cell aggregates (mainly the cell membrane and proteins) in the cervical fluid sample (Fig. S1D†). These cellular constituents could have acted as templates for the formation of such quasi-spherical nanostructures. We hypothesize that the cell membrane (composed of lipids), interacted with HAuCl4 in the presence of ascorbic acid yielding Au NPs of sizes and morphology comparable to those of liposomes (synthesized from lipids) coated with gold.47,48 In high resolution TEM imaging, a layer of coating was observed for both the control & test Au NPs (Fig. 3B and C(ii)). The characteristic SAED patterns and lattice fringes observed in the HRTEM images of the Au NPs confirmed their crystalline structure (Fig. S3–S5†).
The elemental composition of the blank, control and test Au NPs was analysed (Fig. S6†). All the samples showed major weight% of Au followed by carbon (C), oxygen (O) and nitrogen (N). The XRD spectra (Fig. S7†) of the blank, control and test Au NPs exhibited a distinct peak at 2θ ≈ 38° corresponding to the standard Bragg reflection (111) of the face center cubic (fcc) lattice, which is typical of Au NPs. Additionally, a peak at 2θ = 44° was observed for blank Au NPs, indexed as (200) corresponding to the metallic fcc lattice of Au.49–51 The FT-IR analysis of the nanoparticles showed a peak at 1630 cm−1 corresponding to the stretching mode of the C O bond (Fig. S8†), typical to Au NPs formed by ascorbic acid reduction.52
An experiment was performed to understand the influence of clinical samples on pre-formed Au nanoparticles. Au NPs were synthesized by the citrate reduction method.53 The variations in the absorbance and hydrodynamic diameters of these nanoparticles with the addition of clinical samples were evaluated (Fig. S9†). Gold nanoparticles formed using the citrate reduction method (Au NPs) were wine-red colored with an absorption peak at 520 nm. With the addition of clinical samples (Hs: healthy, Cs: cancerous sample), no change in color was observed (inset, Fig. S9A†). The absorption peak at 520 nm also remained unaffected with the addition of the clinical samples (Fig. S9A†), indicating that the addition of clinical samples to pre-formed Au NPs shows no colorimetric detection. A change in the hydrodynamic size and zeta potential of the Au NPs (Fig. S9B and C†) was noted with the addition of clinical samples which can be attributed to the components of clinical samples. This experiment proved that the colorimetric detection technique “C-ColAur” involving the in situ formation of Au NPs was remarkable in differentiating the healthy and cancerous samples with colors specific to the type of sample, which was not observed with pre-formed Au NPs.
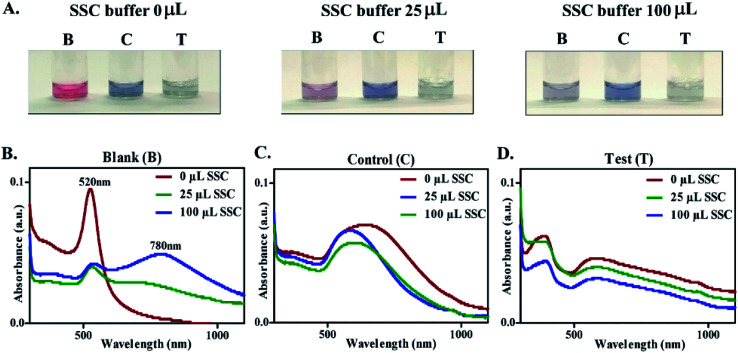
As reported in the literature, the majority of the Au NP based colorimetric techniques use inter-particle cross linking aggregation which occurs by direct interactions, such as antigen–antibody and DNA hybridization.20,28,29,31 In order to understand the biological relevance of the surface coating observed on control & test Au NPs and its role in prevention of salt-induced aggregation, a stability test was performed with SSC buffer. The blank, control and test Au NPs were treated with SSC buffer (0, 25, 100 μL) and were observed for variation in absorbance with subsequent changes in color. Upon the addition of SSC buffer, the blank NPs showed an immediate color change (indicating their aggregation), while no color change was observed for the control and test Au NPs (Fig. 4A). The extent of aggregation was proportional to the shift in the absorption peak.54 For blank Au NPs, aggregation increased progressively, characterized by a gradual decrease of the plasmon peak at 520 nm with the appearance of a new peak between 765 and 810 nm (Fig. 4B). The color change of the resultant solution from red to purple, and further to blue was observed indicating the increasing degree of aggregation (Fig. 4A and S10†).20,55,56 The particle size and zeta potential of Au NPs treated with SSC buffer were also recorded (ESI Table 2†). A significant increase in size with the addition of SSC buffer has been observed for the blank Au NPs. The control & test Au NPs remained unaffected with treatment of SSC buffer depicting their stability (i.e. no aggregation). Unlike the blank Au NPs, there was no significant shift in the wavelength (Fig. 4C and D). The above mentioned findings could be attributed to the interaction of the constituents of clinical samples with the precursors resulting in the formation of Au NPs with a protective coating, making them immune to salt-induced aggregation. This experiment thus confirmed the biological relevance of the surface coating on Au NPs leading to label-free detection of clinical samples.
Fig. 4. Salt induced aggregation of Au NPs. (A) Images showing the change in color of the blank (B), control (C), and test (T) Au NPs upon addition of different concentrations of SSC buffer. The absorbance spectra of (B) blank, (C) control, and (D) test Au NPs treated with SSC buffer.
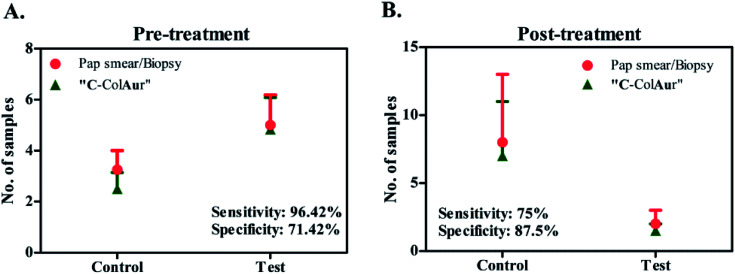
The detection technique “C-ColAur” was validated for its sensitivity and specificity in clinical samples by using PAP smear/biopsy as the gold standard.34,35 A total of 42 samples were collected before treatment. As already mentioned, the control Au NPs (formed with healthy samples) were bright blue, while the test Au NPs (formed with cancerous samples) were found to be colorless. Out of 28 cancerous samples (confirmed by PAP smear/biopsy), the “C-ColAur” technique was able to identify 27 samples correctly (as cancerous). Out of 14 healthy samples, 10 samples were identified correctly (Fig. 5A, ESI Tables 3 and 4†). The “C-ColAur” technique showed a sensitivity of 96.42% and specificity of 71.42% for pre-treatment samples. Such high sensitivity is expected of an ideal screening procedure.57 The test gave a positive predictive value of 87.09% and negative predictive value of 90.9% (ESI Table 5†). 20 samples were collected post treatment to understand the possibility of using “C-ColAur” as a prognostic indicator. The same procedure was followed with post-treatment samples too. “C-ColAur” analysis prediction correlated with the standards (i.e. PAP smear/biopsy) with 17 of the 20 post-treatment samples (Fig. 5B). In the post-treatment sample analysis, “C-ColAur” showed a sensitivity of 75% and specificity of 87.5% (ESI Table 5†). An ideal prognostic indicator is expected to be highly specific,57 thus confirming the ability of “C-ColAur” to be used as a prognostic indicator.
Fig. 5. Sensitivity & specificity of detection. Comparison between conventional (PAP smear/Biopsy) & “C-ColAur” detection procedures for (A) pre-treatment samples and (B) post-treatment samples.
Conclusions
The “C-ColAur” technique is facile & rapid, and requires neither pre-processing of the samples nor any medical expertise or sophisticated equipment. A simple, colorimetric technique with read out time less than a minute was hence demonstrated and validated with clinical samples. The clinical sample analysis showed that the “C-ColAur” technique is comparable to standard diagnostic procedures (PAP smear/biopsy). The “C-ColAur” technique with the pre-treatment samples showed a high sensitivity of ∼96% confirming its efficiency to be used as a screening/diagnostic tool for cervical cancer. In the post-treatment sample analysis, the “C-ColAur” technique showed high specificity (∼87%) indicating its ability to be applied as a prognostic indicator. Further studies with a greater number of clinical samples and validation will help in identifying the specific biomarker responsible for the differences in the formation of Au NPs with the type of sample leading to the development of a robust point-of-care diagnostic/prognostic test for cervico-vaginal diseases.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
The authors thank Padma Shri Prof. Anil K. Gupta for his continuous support, encouragement and belief in the project. The authors acknowledge the Innovation Scholars In-Residence program 2018, hosted by Rashtrapathi Bhavan, New Delhi for the valuable suggestions and discussions with expert committees. The authors acknowledge the support of Dr. T. Subramaneshwara Rao, Medical Director and Chief Oncology Surgeon, Basavatarakam Indo-American Cancer Hospital & Research Institute, Hyderabad, India for necessary permissions to carry out the study. The authors are grateful for the continuous support and efforts of Dr. P. Veena, Dr. P. Sujatha and their respective teams, Dept. of Gynecology, Basavatarakam Indo-American Cancer Hospital & Research Institute, Hyderabad. The authors acknowledge Madhuri Sakaray, research scholar, Centre for Nanoscience & Technology, IST, JNTU Hyderabad for FTIR studies. The authors duly acknowledge DBT-SRISTI-GYTI for the award and grant BIRAC SRISTI PMU-2017/012. The authors also acknowledge MHRD IMPRINT (4291), DST-Inspire (DST/INSPIRE/04/2015/000377), DST-AMT (DST/TDT/AMT/2017/227(G)), and DBT-BIRAC (BIRAC/IKP0866/BIG-14/19), Govt. of India for the research funding. The authors acknowledge the generous financial support by the Vasudha Foundations, Hyderabad, India. Tejaswini Appidi acknowledges the Dept. of Science & Technology, Govt. of India for the DST-Inspire fellowship (IF160291).
Electronic supplementary information (ESI) available: Clinical sample detection process, size analysis, TEM images & data corresponding to clinical samples, and sensitivity & specificity calculations. See DOI: 10.1039/d0na00686f
References
- Mahmoodi P. Fani M. Rezayi M. Avan A. Pasdar Z. Karimi E. Amiri I. S. Ghayour-Mobarhan M. Biofactors. 2019;45:101–117. doi: 10.1002/biof.1465. [DOI] [PubMed] [Google Scholar]
- Zegels G. Van Raemdonck G. A. Tjalma W. A. Van Ostade X. W. Proteome Sci. 2010;8:63. doi: 10.1186/1477-5956-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranberg M. Bech B. H. Blaakær J. Jensen J. S. Svanholm H. Andersen B. BMC Cancer. 2018;18:273. doi: 10.1186/s12885-018-4165-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray F. Ferlay J. Soerjomataram I. Siegel R. L. Torre L. A. Jemal A. Ca-Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- Vu M. Yu J. Awolude O. A. Chuang L. Curr. Probl. Canc. 2018;42:457–465. doi: 10.1016/j.currproblcancer.2018.06.003. [DOI] [PubMed] [Google Scholar]
- Stanczuk G. Baxter G. Currie H. Lawrence J. Cuschieri K. Wilson A. Arbyn M. BMJ Open. 2016;6:e010660. doi: 10.1136/bmjopen-2015-010660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- I. W. G. o. t. E. o. C.-P. Strategies, I. A. f. R. o. Cancer and W. H. Organization, Cervix Cancer Screening, Diamond Pocket Books (P) Ltd., 2005 [Google Scholar]
- Mahmoodi P. Rezayi M. Rasouli E. Avan A. Gholami M. Mobarhan M. G. Karimi E. Alias Y. J. Nanobiotechnol. 2020;18:1–12. doi: 10.1186/s12951-020-0577-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legood R. Gray A. M. Mahé C. Wolstenholme J. Jayant K. Nene B. M. Shastri S. S. Malvi S. G. Muwonge R. Budukh A. M. Int. J. Cancer. 2005;117:981–987. doi: 10.1002/ijc.21220. [DOI] [PubMed] [Google Scholar]
- Sankaranarayanan R. Nene B. M. Shastri S. S. Jayant K. Muwonge R. Budukh A. M. Hingmire S. Malvi S. G. Thorat R. Kothari A. N. Engl. J. Med. 2009;360:1385–1394. doi: 10.1056/NEJMoa0808516. [DOI] [PubMed] [Google Scholar]
- Ardahan M. Temel A. B. Canc. Nurs. 2011;34:158–163. doi: 10.1097/NCC.0b013e3181efe69f. [DOI] [PubMed] [Google Scholar]
- Goel A. Gandhi G. Batra S. Bhambhani S. Zutshi V. Sachdeva P. Int. J. Gynecol. Obstet. 2005;88:25–30. doi: 10.1016/j.ijgo.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Oscarsson M. G. Benzein E. G. Wijma B. E. J. Psychosom. Obstet. Gynecol. 2008;29:23–31. doi: 10.1080/01674820701504619. [DOI] [PubMed] [Google Scholar]
- Dzuba I. G. Díaz E. Y. Allen B. Leonard Y. F. Lazcano Ponce E. C. Shah K. V. Bishai D. Lorincz A. Ferris D. Turnbull B. J. Wom. Health Gend. Base Med. 2002;11:265–275. doi: 10.1089/152460902753668466. [DOI] [PubMed] [Google Scholar]
- Qiu L. Shen Z. Wu Z.-S. Shen G.-L. Yu R. Biosens. Bioelectron. 2015;64:292–299. doi: 10.1016/j.bios.2014.09.008. [DOI] [PubMed] [Google Scholar]
- Xiao L. Zhu A. Xu Q. Chen Y. Xu J. Weng J. ACS Appl. Mater. Interfaces. 2017;9:6931–6940. doi: 10.1021/acsami.6b15750. [DOI] [PubMed] [Google Scholar]
- Ray P. Steckl A. J. ACS Sens. 2019;4:1346–1357. doi: 10.1021/acssensors.9b00301. [DOI] [PubMed] [Google Scholar]
- Chen Y. Zhou S. Li L. Zhu J.-j. Nano Today. 2017;12:98–115. doi: 10.1016/j.nantod.2016.12.013. [DOI] [Google Scholar]
- Saha K. Agasti S. S. Kim C. Li X. Rotello V. M. Chem. Rev. 2012;112:2739–2779. doi: 10.1021/cr2001178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldewachi H. Chalati T. Woodroofe M. Bricklebank N. Sharrack B. Gardiner P. Nanoscale. 2018;10:18–33. doi: 10.1039/C7NR06367A. [DOI] [PubMed] [Google Scholar]
- Huang X. El-Sayed M. A. J. Adv. Res. 2010;1:13–28. doi: 10.1016/j.jare.2010.02.002. [DOI] [Google Scholar]
- Appidi T., Srivastava R. and Rengan A. K., 2019 IEEE 14th Nanotechnology Materials and Devices Conference (NMDC), 2019, pp. 1–4 [Google Scholar]
- Chang C.-C. Chen C.-P. Wu T.-H. Yang C.-H. Lin C.-W. Chen C.-Y. Nanomaterials. 2019;9:861. doi: 10.3390/nano9060861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia N. Deng D. Wang Y. Fang C. Li S.-J. Int. J. Nanomed. 2018;13:2521. doi: 10.2147/IJN.S154046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren W. Mohammed S. I. Wereley S. Irudayaraj J. Anal. Chem. 2019;91:2876–2884. doi: 10.1021/acs.analchem.8b04848. [DOI] [PubMed] [Google Scholar]
- Rhouati A. Catanante G. Nunes G. Hayat A. Marty J.-L. Sensors. 2016;16:2178. doi: 10.3390/s16122178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L. Li N. Chemosensors. 2019;7:53. [Google Scholar]
- Tsai T.-T. Huang C.-Y. Chen C.-A. Shen S.-W. Wang M.-C. Cheng C.-M. Chen C.-F. ACS Sens. 2017;2:1345–1354. doi: 10.1021/acssensors.7b00450. [DOI] [PubMed] [Google Scholar]
- Kim H. Park M. Hwang J. Kim J. H. Chung D.-R. Lee K.-s. Kang M. ACS Sens. 2019;4:1306–1312. doi: 10.1021/acssensors.9b00175. [DOI] [PubMed] [Google Scholar]
- Aili D. Selegård R. Baltzer L. Enander K. Liedberg B. Small. 2009;5:2445–2452. doi: 10.1002/smll.200900530. [DOI] [PubMed] [Google Scholar]
- Sina A. A. I. Carrascosa L. G. Liang Z. Grewal Y. S. Wardiana A. Shiddiky M. J. Gardiner R. A. Samaratunga H. Gandhi M. K. Scott R. J. Nat. Commun. 2018;9:4915. doi: 10.1038/s41467-018-07214-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J.-Y. Li H.-W. Wei S.-C. Harroun S. G. Lee M.-Y. Lin H.-Y. Chung C.-Y. Hsu C.-H. Chen Y.-R. Lin H.-J. ACS Appl. Mater. Interfaces. 2017;9:44307–44315. doi: 10.1021/acsami.7b13873. [DOI] [PubMed] [Google Scholar]
- Rastegarzadeh S. Abdali S. Talanta. 2013;104:22–26. doi: 10.1016/j.talanta.2012.11.023. [DOI] [PubMed] [Google Scholar]
- Parikh R. Mathai A. Parikh S. Sekhar G. C. Thomas R. Indian J. Ophthalmol. 2008;56:45. doi: 10.4103/0301-4738.37595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevethan R. Front. Public Health. 2017;5:307. doi: 10.3389/fpubh.2017.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad S. Mandal I. Singh S. Paul A. Mandal B. Venkatramani R. Swaminathan R. Chem. Sci. 2017;8:5416–5433. doi: 10.1039/C7SC00880E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Raben K. Chang R. Laube B. Chem. Phys. Lett. 1981;79:465–469. doi: 10.1016/0009-2614(81)85015-4. [DOI] [Google Scholar]
- Thaxton C. S. Georganopoulou D. G. Mirkin C. A. Clin. Chim. Acta. 2006;363:120–126. doi: 10.1016/j.cccn.2005.05.042. [DOI] [PubMed] [Google Scholar]
- Malassis L. Dreyfus R. Murphy R. J. Hough L. A. Donnio B. Murray C. B. RSC Adv. 2016;6:33092–33100. doi: 10.1039/C6RA00194G. [DOI] [Google Scholar]
- Eustis S. El-Sayed M. A. Chem. Soc. Rev. 2006;35:209–217. doi: 10.1039/B514191E. [DOI] [PubMed] [Google Scholar]
- Link S. El-Sayed M. A. J. Phys. Chem. B. 1999;103:4212–4217. doi: 10.1021/jp984796o. [DOI] [Google Scholar]
- Ghosh S. K. Pal T. Chem. Rev. 2007;107:4797–4862. doi: 10.1021/cr0680282. [DOI] [PubMed] [Google Scholar]
- Gunnarsson S. B. Bernfur K. Mikkelsen A. Cedervall T. Nanoscale. 2018;10:4246–4257. doi: 10.1039/C7NR08696B. [DOI] [PubMed] [Google Scholar]
- Rengan A. K. Jagtap M. De A. Banerjee R. Srivastava R. Nanoscale. 2014;6:916–923. doi: 10.1039/C3NR04448C. [DOI] [PubMed] [Google Scholar]
- Rengan A. K. Bukhari A. B. Pradhan A. Malhotra R. Banerjee R. Srivastava R. De A. Nano Lett. 2015;15:842–848. doi: 10.1021/nl5045378. [DOI] [PubMed] [Google Scholar]
- Rengan A. K., Banerjee R. and Srivastava R., 12th IEEE International Conference on Nanotechnology (IEEE-NANO), 2012, pp. 1–4 [Google Scholar]
- Singh S. P. Alvi S. B. Bharadwaj D. Singh A. D. Manda S. V. Srivastava R. Rengan A. K. Int. J. Biol. Macromol. 2018;110:375–382. doi: 10.1016/j.ijbiomac.2017.11.163. [DOI] [PubMed] [Google Scholar]
- Alvi S. B. Appidi T. Deepak B. P. Rajalakshmi P. Minhas G. Singh S. P. Begum A. Bantal V. Srivastava R. Khan N. Biomater. Sci. 2019;7:3866–3875. doi: 10.1039/C9BM00932A. [DOI] [PubMed] [Google Scholar]
- Ren Y. Rao R. Bhusal S. Varshney V. Kedziora G. S. Wheeler R. Kang Y. Roy A. Nepal D. ACS Appl. Nano Mater. 2020;3:8753–8762. doi: 10.1021/acsanm.0c01555. [DOI] [Google Scholar]
- Krishnamurthy S. Esterle A. Sharma N. C. Sahi S. V. Nanoscale Res. Lett. 2014;9:627. doi: 10.1186/1556-276X-9-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelrazek E. M. Abdelghany A. M. Badr S. I. Morsi M. A. J. Mater. Res. Technol. 2018;7:419–431. doi: 10.1016/j.jmrt.2017.06.009. [DOI] [Google Scholar]
- Annur S. Santosa S. J. Aprilita N. H. Orient. J. Chem. 2018;34:2305–2312. [Google Scholar]
- Zhou Y. Pan F.-G. Li Y.-S. Zhang Y.-Y. Zhang J.-H. Lu S.-Y. Ren H.-L. Liu Z.-S. Biosens. Bioelectron. 2009;24:2744–2747. doi: 10.1016/j.bios.2009.01.034. [DOI] [PubMed] [Google Scholar]
- Sato K. Hosokawa K. Maeda M. Nucleic Acids Res. 2005;33:e4. doi: 10.1093/nar/gni007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laromaine A. Koh L. Murugesan M. Ulijn R. V. Stevens M. M. J. Am. Chem. Soc. 2007;129:4156–4157. doi: 10.1021/ja0706504. [DOI] [PubMed] [Google Scholar]
- Reynolds R. A. Mirkin C. A. Letsinger R. L. J. Am. Chem. Soc. 2000;122:3795–3796. doi: 10.1021/ja000133k. [DOI] [Google Scholar]
- McNamara L. A. and Martin S. W., in Principles and Practice of Pediatric Infectious Diseases, Elsevier, 2018, pp. 1–9.e1 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.