Abstract
A set of vectors which facilitates the sequential integration of new functions into the Escherichia coli chromosome by homologous recombination has been developed. These vectors are based on plasmids described by Posfai et al. (J. Bacteriol. 179:4426–4428, 1997) which contain conditional replicons (pSC101 or R6K), a choice of three selectable markers (ampicillin, chloramphenicol, or kanamycin), and a single FRT site. The modified vectors contain two FRT sites which bracket a modified multiple cloning region for DNA insertion. After integration, a helper plasmid expressing the flippase (FLP) recombinase allows precise in vivo excision of the replicon and the marker used for selection. Sites are also available for temporary insertion of additional functions which can be subsequently deleted with the replicon. Only the DNA inserted into the multiple cloning sites (passenger genes and homologous fragment for targeting) and a single FRT site (68 bp) remain in the chromosome after excision. The utility of these vectors was demonstrated by integrating Zymomonas mobilis genes encoding the ethanol pathway behind the native chromosomal adhE gene in strains of E. coli K-12 and E. coli B. With these vectors, a single antibiotic selection system can be used repeatedly for the successive improvement of E. coli strains with precise deletion of extraneous genes used during construction.
Plasmid vectors are versatile tools which facilitate the isolation, expression, and analysis of genes (2). Useful characteristics include the facile production of identical DNA for subsequent in vitro and in vivo manipulation, the presence of multiple cloning sites (MCS), selectable markers which allow rapid screening for new or improved traits, and the ease with which they can be established as multiple cellular copies to alter gene expression in recombinant hosts. However, the physiological burdens imposed by multiple copies of plasmid genes, potential for internal rearrangements, and segregational instability are disadvantages for many biotechnological applications (24).
Antibiotic resistance genes are frequently used for plasmid maintenance. Alternative selectable markers based on metabolic deficiencies of the host (7) pose further complications for improvement cycles in production strains. For applications such as deliberate field release, development of organisms for use in food products, and development of biocatalysts for bulk chemicals, special requirements for plasmid maintenance are undesirable.
Many of the problems associated with plasmids can be eliminated by the chromosomal integration of desired traits. Integration tools based on modified transposons (8, 9, 22, 28) and conditional plasmid replicons (10, 15, 17, 20) have been developed. With these tools, integration can be random or precisely directed by DNA fragments homologous to the host genome. However, complications still remain with most integration systems, such as the persistence of selectable markers, transposons, or replicons. For strains in which multiple alterations or continuing improvements are desired, the accumulation of markers and delivery systems can be troublesome. Selectable events may be limited by the availability of functional markers. Integrated DNA (replicons, transposon genes, and selectable marker genes) can serve as a site for homologous recombination events which interfere with targeting or randomness during subsequent constructions. Also, the persistence of replicons and transposons increases the potential for gene transfer to other organisms in the environment.
Replicons and transposons can be eliminated by transformation with purified DNA fragments which lack replication functions (11, 23). Nonantibiotic markers are available but are often less efficient than antibiotics (8, 9, 12). In a few cases, loss of functions, such as tetracycline sensitivity (1), and the absence of a sucrose-sacB system (15, 27) can be selected directly. However, loss of function due to a mutation is typically not a precise event and can result from unstable point mutations, partial deletion of the resistance gene, or extended deletions which impair the host.
Recombinase-based integration systems offer the opportunity to effect precise DNA deletions in vivo (3, 25) and in vitro (6, 11, 13, 32). A novel integration system was initially developed by Szybalski (31) for chromosome walking which uses the flippase (FLP) recombinase and FRT site (68-bp recognition sequence) from the yeast 2μm plasmid (30). Recently, Posfai et al. (26) developed a new set of vectors and helper plasmids which are designed to facilitate this process. Using homologous DNA as a guide, insertion of an FRT site, antibiotic marker, and ultrarare restriction enzyme site can be targeted to any point on the Escherichia coli chromosome. Following two sequential insertions, large fragments of DNA bracketed by the FRT sites can be excised in vivo as a replicating plasmid (30 to 150 kbp) with the aid of a helper plasmid. After excision with this system, one antibiotic marker and one conditional replicon still remain in the chromosome.
We have modified the plasmids developed by Posfai et al. (26) by adding a second FRT site oriented in the same orientation, bracketing the MCS (Fig. 1). Constructions were performed by standard methods (29). Reagents used in cloning were molecular biology grade and used as directed by the manufacturers. Restriction enzymes, T4 DNA polymerase, and Klenow polymerase were purchased from New England Biolabs (Beverly, Mass.). T4 DNA polymerase was used to produce blunt ends as necessary for cloning. Taq PCR MasterKit was purchased from Qiagen (Santa Clarita, Calif.). PCRs were carried out by using an Eppendorf Mastercycler (Brinkman Instrument Co., Westbury, N.Y.). Primers were obtained from Genosys Biotechnologies (The Woodlands, Tex.). PCR products were ligated into pCR2.1-TOPO (Invitrogen, Carlsbad, Calif.) with topoisomerase. DNA fragments were isolated from gels by using a QIAquick gel extraction kit. DNA fragments were assembled with a Rapid DNA ligation kit (Boehringer Mannheim Corporation, Indianapolis, Ind.). Wizard Plus kits (Promega, Madison, Wis.) were used for plasmid purification. Dideoxy sequencing of plasmids was performed by using fluorescent primers and a LI-COR model 4000L DNA sequencer (Li-Cor, Lincoln, Nebr.) as previously described (16).
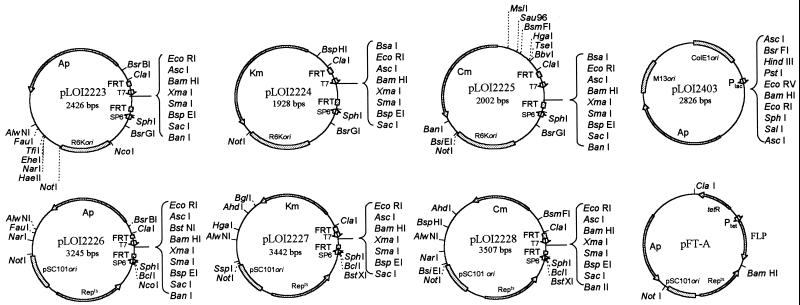
FIG. 1.
Integration vectors and helper plasmids. Plasmids pLOI2223, pLOI2224, and pLOI2225 are integration vectors containing an R6K origin and can be replicated only in a host such as S17-1 which contains λpir. Plasmids pLOI2226, pLOI2227, and pLOI2228 contain a temperature-conditional pSC101 replicon which functions at 30°C but not at 37 to 42°C. Plasmid pLOI2403 contains a high copy replicon with an MCS site bracketed by AscI sites. DNA fragments can be assembled in the pLOI2403 MCS and moved to any integration vector by using AscI. Unique polylinker sites useful for the insertion of passenger DNA and homologous guide fragment are shown on the right side of each plasmid. Additional unique sites are also shown for the insertion of DNA which can be deleted at will after integration with the FLP recombinase. Plasmid pFT-A (ampicillin resistance) was constructed by Posfai et al. (26) and is used as a helper plasmid to express FLP recombinase. A similar plasmid expressing kanamycin resistance (pFT-K) was also constructed by Posfai et al. (26). Both contain a temperature-conditional pSC101 replicon. T7 and SP6 promoters can be used for sequencing. FRT recognition sites are illustrated as rectangles. Selectable markers and replicons are labeled. Complete sequences for pLOI2403 and the six integration vectors (pLOI2223 to pLOI2228) are available from GenBank under the following accession no.: AF172933, AF172934, AF172935, AF172936, AF172937, and AF172938, respectively. Ap, ampicillin; Km, kanamycin; Cm, chloramphenicol; Repts, temperature conditional replication genes. Note that pLOI2403 contains two BsrFI sites.
A full set of plasmids (ampicillin, chloramphenicol, and kanamycin resistance plasmids) was made for each conditional replicon (pSC101 and R6K) (Fig. 1). Since both conditional replicons are present at low copy numbers, an additional high copy vector was developed to facilitate constructions by adding an AscI site on either side of the MCS in pLITMUS 38 (New England Biolabs) to produce pLOI2403. AscI recognizes an infrequent sequence and produces a four-base overhang composed only of G and C residues. Using AscI, DNA cloned into the MCS region of pLOI2403 can be transferred from the high copy vector to any of the integration vectors which contain a unique AscI site between the FRT sites. Detailed protocols used in the construction of these plasmids are available upon request.
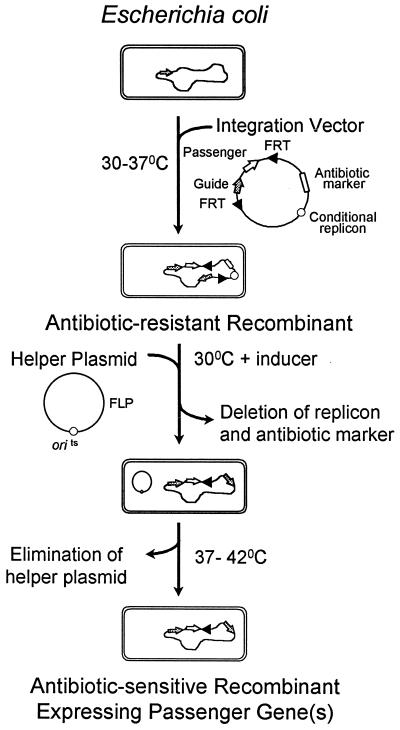
A general procedure for chromosomal integration of DNA with the new vectors with conditional replicons is presented in Fig. 2. The integration of heterologous passenger DNA carrying desired functions can be targeted to any specific chromosomal site by an adjacent fragment of homologous DNA (guide) by using a two-step process (pSC101) or by direct selection at 37 to 42°C (pSC101 or R6K). With a single crossover event, the entire plasmid is incorporated into the chromosome. (If needed, pSC101-based integration vectors can be eliminated by overnight growth and plating at elevated temperatures.) After integration, recombinants are transformed with pFT-A containing the yeast FLP gene under control of the tetracycline promoter and grown under permissive conditions (30°C, pSC101). During growth with chlortetracycline, FLP recombinase is induced and in turn excises the DNA bracketed by concurrently facing FRT sites (selectable marker and replicon) from the chromosome. After growth at 37 to 42°C to eliminate pFT-A, only the passenger gene(s), a single FRT, and the homologous guide fragment should remain in the chromosome.
FIG. 2.
Use of double FRT integration vectors for the insertion of heterologous passenger genes into the chromosome. A guide fragment is ligated into the MCS of an integration vector to target homologous recombination with the chromosome. Since these vectors contain conditional replicons (pSC101 or R6K), integration into the chromosome can be directly selected by antibiotic resistance after transformation or electroporation. A helper plasmid is added to recombinants to express FLP recombinase under the control of the tetracycline promoter. Induction of FLP recombinase with autoclaved chlortetracycline results in precise deletion of DNA encoding the replicon and selectable marker flanked by FRT sites oriented in the same direction. A single FRT site, the passenger DNA, and the guide fragment remain in the chromosome. The helper plasmid with a temperature-conditional replicon can be readily eliminated from cells by growth at 37 to 42°C. Since genes used during construction are excised, the same selection procedure can be repeated in subsequent cycles to provide additional modifications or improvements of the same bacterium.
To illustrate the utility of these vectors, we have constructed derivatives of E. coli B (strain SE2272, Δfrd) and E. coli K-12 (strain SE2275, Δfrd) in which three heterologous genes were integrated immediately behind adhE in the chromosome. The guide and passenger DNA were cloned into pLOI2403, a high copy plasmid vector. For this construction, the promoterless adhE coding region (guide) was amplified with Genosys ORFmer primers (forward, 5′TTGCTCTTCCATGGCTGTTACTAATGTCGCTGAA3′; reverse, 5′TTGCTCTTCGTTAAGCGGATTTTTTCGCTTTTTTCT3′) and cloned into pCR2.1-TOPO to produce pLOI2408. After EcoRI digestion, the 2.6-kbp adhE region from pLOI2408 was moved into the corresponding site in pLOI2403 to produce pLOI2413. The BamHI site immediately downstream from the 3′ end of the adhE coding region was used to insert a 4.6-kbp BamHI fragment from pLOI510 (23) containing three genes (passenger): a promoterless Zymomonas mobilis pdc without transcriptional terminator and a promoterless Z. mobilis adhB with transcriptional terminator, followed by a complete cat operon with promoter and terminator. In the resulting plasmid (pLOI2230), transcription of the heterologous genes was oriented concurrently with adhE. All constructs containing the Z. mobilis genes were grown in Luria broth (LB) supplemented with glucose (20 g/liter for plates, 50 g/liter for broth).
The 7.2-kbp AscI fragment from pLOI2230 (high copy vector) containing adhE, the artificial operon pdc adhB, and cat was ligated into the low copy integration vector pLOI2224, which contains an R6K replicon (λpir dependent), and transformed into the permissive host S17-1 (8) with selection for kanamycin and chloramphenicol. The resulting clone containing pLOI2231 was used for large-scale plasmid isolation (500 ml) by the alkaline lysis procedure (29).
Approximately 500 ng of pLOI2231 DNA was used for electroporation of SE2272 and SE2275. Both are nonpermissive hosts. Recombinants were readily obtained by selection for either kanamycin (vector) or chloramphenicol (passenger) resistance. Up to 2 h was allowed for expression of the resistance gene prior to spreading on plates for selection. Approximately 1,000 recombinants per 1 μg of DNA (electroporation) were recovered with E. coli K-12 SE2275, a number fivefold higher than that obtained with E. coli B SE2272. Thirty recombinants from each host were screened for the functional expression of alcohol dehydrogenase on indicator plates (5). Based on the rate and intensity of color development, these recombinants expressed higher levels of alcohol dehydrogenase activity than the respective unmodified SE2272 or SE2275 or S17-1 (pLOI2231) harboring promoterless pdc and adhB genes. Unlike the control strains, these recombinants also exhibited a colonial phenotype (large raised colonies on LB containing glucose) that is typical for ethanologenic E. coli (14). Small-scale DNA preparations (seven recombinants per host) were tested for the presence of pLOI2231. None contained plasmids, as tested by gel filtration or based on transformation experiments with S17-1 as the host. These recombinants were presumed to contain chromosomally integrated genes. One clone from each parent, strains FM7 (E. coli B SE2272) and FM19 (E. coli K-12 SE2275), was selected for further study.
Strains FM7 and FM19 were transformed with the helper plasmid (pFT-A) carrying the FLP gene (26) and incubated at 30°C with selection for ampicillin resistance. A mixture of colonies was used to inoculate a broth culture for induction of FLP with autoclaved chlortetracycline (20 μg/ml). After 6 h of incubation at 30°C, the culture was diluted 1:1,000 in LB containing glucose and incubated at 42°C for 16 h to eliminate the helper plasmid. After streaking on solid medium, isolated colonies were screened for the absence of antibiotic markers. Approximately 80% of the colonies were ampicillin and kanamycin sensitive and retained only chloramphenicol resistance and the ethanologenic traits (passenger genes inserted into the MCS). Loss of ampicillin resistance indicated that the helper plasmid had been successfully eliminated while loss of kanamycin resistance confirmed the FLP recombinase-dependent deletion of the vector. These new derivatives of FM7 and FM19 were designated FM18 and FM20, respectively.
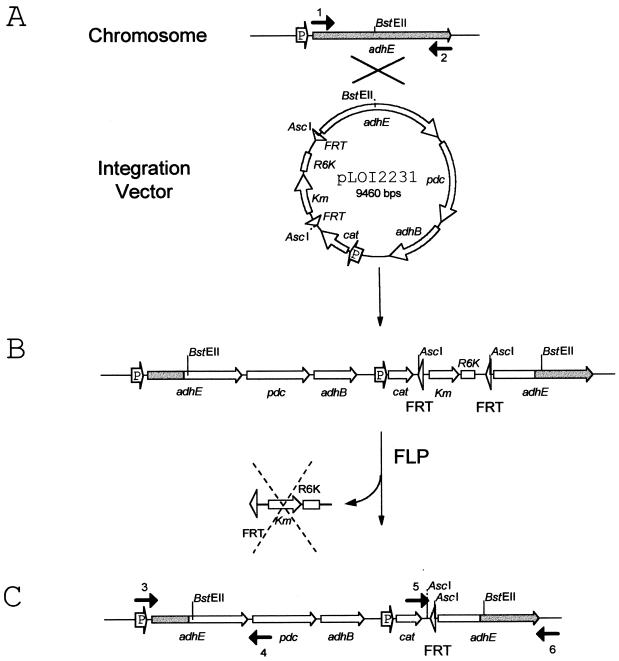
PCR was used to verify the integration events in both FM18 and FM20. Two new sets of primers were designed to amplify the adhE gene, including the unique junctions predicted for pdc (Fig. 3, primers 3 and 4) and cat (Fig. 3, primers 5 and 6) as a result of integration and FLP-mediated deletion. Forward primer 3 hybridizes to the promoter region of adhE, while reverse primer 4 hybridizes to the N-terminal coding region of pdc. Forward primer 5 hybridizes to the C-terminal coding region of the cat gene, and reverse primer 6 hybridizes to the 3′ untranslated portion of the adhE gene. Note that the primers used to clone adhE, forward primer 1 and reverse primer 2, hybridize to the N-terminal and C-terminal coding regions of adhE and are inside of the regions encoded by forward primer 3 and reverse primer 6. All primer sets (SE2272 template, 1 plus 2 and 3 plus 6; FM18 template, 3 plus 4 and 5 plus 6) generated products of the expected sizes (Fig. 4A). Identical results were also obtained with FM20 DNA as the template.
FIG. 3.
Diagram illustrating the insertion of heterologous passenger genes into the chromosomal adhE gene (shaded) and FLP-mediated deletion of the replicon and marker used during construction. Plasmid pLOI2231 is a derivative of pLOI2224 with an AscI cassette containing adhE, pdc, adhB, and cat. Solid arrows above and below the sequences show the location of PCR primers. Promoters for adhE and cat are labeled with a P. No adhE, pdc, or adhB promoters were present in pLOI2231. Selected restriction sites are shown, which are referenced in the text. (A) Alignment of chromosomal adhE and the adhE coding region on pLOI2231. (B) Product of a single crossover event. (C) Product after FLP induction and FLP-mediated excision of the replicon and selectable marker. DNA segment marked by the dashed-line X represents the region excised by FLP recombinase. Arrows with numbers 1 to 6 represent the primers used to amplify the corresponding regions. Sequences for these primers are as follows: primer 1, 5′TTGCTCTTCCATGGCTGTTACTAATGTCGCTGAA3′; primer 2, 5′TTGCTCTTCGTTAAGCGGATTTTTTCGCTTTTTTCT3′; primer 3, 5′GTGAGTGTGAGCGCGGAGT3′; primer 4, 5′TGGCACGAGCATAACCTTC3′; primer 5, 5′CAGTACTGCGATGAGTGGCA3′; and primer 6, 5′GTTGCCAGACAGCGCTACT3′.
FIG. 4.
PCR fragments containing the adhE gene and junctions. Amplification conditions for primer pairs 1 plus 2 and 3 plus 6 (25 cycles): 45 s at 94°C, 45 s at 60°C, and 60 s at 72°C. Amplification conditions for primer pairs 3 plus 4 and 5 plus 6 (30 cycles): 45 s at 94°C and 60 s at 60°C. DNA was held at 94°C for 3 min prior to the first cycle. Elongation time was increased to 10 min during the final cycle. (A) Full-length PCR products of adhE junctions. Positions and sequences of primers are provided in the legend for Fig. 3. Lane 1, HindIII digest of phage λ DNA (marker sizes, 23.1, 9.4, 6.6, 4.3, 2.3, and 2.0 kbp); lane 2, adhE coding region (2,696 bp) of SE2272 amplified with forward primer 1 and reverse primer 2; lane 3, adhE promoter and 3′ untranslated sequence (2,814 bp) of SE2272 amplified with forward primer 3 and reverse primer 4; lane 4, the adhE and pdc junction (3,108 bp) of FM18 amplified with forward primer 3 and reverse primer 4; lane 5, the cat junction and adhE (3,477 bp) of FM18 amplified with forward primer 5 and reverse primer 6. (B) BstEII digestion of PCR products. A single, central BstEII site was used to cleave PCR products containing adhE into N-terminal and C-terminal fragments. Lane 1 contains a DNA standard (descending, 3.0, 2.0, 1.5, 1.2, and 1.0 kbp). Lanes 2 through 5 contain BstEII-digested PCR products of fragments described for panel A, respectively. Fragment sizes are as follows (N-terminal plus C-terminal fragments): lane 2, 1,226 plus 1,470 bp; lane 3, 1,325 plus 1,489 bp; lane 4, 1,325 plus 1,783 bp; and lane 5, 1,988 plus 1,489 bp.
The adhE gene contains a single central BstEII site which does not occur elsewhere in the PCR products. This site was used to verify the identity of the PCR fragments. As shown in Fig. 4B, all PCR products were cut once to produce fragments containing the N-terminal and C-terminal regions of adhE. Fragments from the adhE coding region alone (primers 1 plus 2) were smaller (N-terminal fragment = 1,226 bp; C-terminal fragment = 1,470 bp) than fragments which included parts of the native adhE promoter (primers 3 plus 4 and 3 plus 6; N-terminal fragment = 1,325 bp) or adhE terminator (primers 3 plus 6 and 5 plus 6; C-terminal fragment = 1,489 bp). The fragment which included part of pdc (primers 3 plus 4) was the largest C-terminal fragment (1,783 bp). The fragment which included part of cat (primers 5 plus 6) was the largest N-terminal fragment (1,988 bp).
The expression of adhE is regulated by a number of factors in E. coli, including cra, adhR, and the abundance of NADH (18, 19, 21). Both message levels and activity are approximately 10-fold higher during anaerobic growth with glucose than during aerobic growth. Since the Z. mobilis genes are integrated behind the adhE coding region to form an operon fusion, expression of pdc should also increase in response to anaerobiosis. Strains FM18 and FM20 were grown in LB containing 50 g of glucose/liter under aerobic and anaerobic conditions. Pyruvate decarboxylase (PDC) activities were determined in heat-treated preparations to eliminate lactate dehydrogenase and other confounding activities (4). Under anaerobic conditions, PDC activities in FM18 and FM20 were 0.254 U per mg of protein and 0.185 U per mg of protein, respectively, levels approximately fourfold higher than the activities observed in cells grown under aerobic conditions. Although these differences in pdc expression were somewhat smaller than the 10-fold reported for native adhE, expression may be modulated by a reduction in NADH, due to concurrent expression of Z. mobilis adhB and availability of acetaldehyde from the PDC-mediated decarboxylation of pyruvate.
The stability of integrated strains was also examined by serial transfers (1,000-fold dilution) in rich medium lacking antibiotics. After 10 sequential transfers, all 100 colonies tested retained both alcohol dehydrogenase expression and chloramphenicol resistance.
Our results with the ethanol pathway genes demonstrate that the new integration vectors can be used to place promoterless genes under the control of a chromosomal promoter. This approach avoids potential problems of lethality or mutation due to unregulated expression in plasmids during construction and integration. These vectors can also be used to replace promoters in chromosomal genes. Additional unique restriction sites are available for the insertion of genes which can be temporarily expressed after integration and subsequently deleted with the replicon and selectable marker. This option, the temporary introduction of new genes, may be useful to test new traits in an isogenic background. Although the vectors described must be propagated in E. coli, they are potentially useful with other organisms. The FLP recombinase is extremely efficient (32) and could be produced intracellularly as a transient expression product after transformation or electroporation of pFT-A.
Nucleotide sequence accession numbers.
Complete sequences for each plasmid have been deposited in GenBank with the following accession no.: AF172933 (pLOI2223), AF172934 (pLOI2224), AF172935 (pLOI2225), AF172936 (pLOI2226), AF172937 (pLOI2227), and AF172938 (pLOI2228).
Acknowledgments
We thank G. Posfai and F. R. Blattner for sharing their plasmid vectors.
This research was supported in part by the Florida Agricultural Experiment Station (publication no. R-06853) and by grants from the U.S. Department of Agriculture, National Research Initiative (98-35504-6177 and 98-35505-6976), and the U.S. Department of Energy, Office of Basic Energy Science (DE-FG02-96ER20222).
REFERENCES
- 1.Bochner B R, Huang H-C, Schieven G L, Ames B N. Positive selection for loss of tetracycline resistance. J Bacteriol. 1980;143:926–933. doi: 10.1128/jb.143.2.926-933.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolivar F, Rodriguez R L, Greene P J, Betlach M C, Heynecker H L, Boyer H W, Crosa J H, Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 3.Cherepanov P P, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 4.Conway T, Osman Y A, Ingram L O. Promoter and nucleotide sequence of the Zymomonas mobilis pyruvate decarboxylase. J Bacteriol. 1987;169:949–954. doi: 10.1128/jb.169.3.949-954.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conway T C, Sewell G W, Osman Y A, Ingram L O. Cloning and sequencing of the alcohol dehydrogenase II from Zymomonas mobilis. J Bacteriol. 1987;169:2591–2597. doi: 10.1128/jb.169.6.2591-2597.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox M M. The FLP protein of the yeast 2-μm plasmid: expression of a eukaryotic recombination system in Escherichia coli. Proc Natl Acad Sci USA. 1983;80:4223–4227. doi: 10.1073/pnas.80.14.4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Degryse E. Stability of host-vector system based on complementation of an essential gene in Escherichia coli. J Biotechnol. 1991;18:29–40. doi: 10.1016/0168-1656(91)90233-l. [DOI] [PubMed] [Google Scholar]
- 8.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposons for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Lorenzo V, Timmis K N. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 10.Hamilton C H, Aldea M, Washburn B K, Babitzke P, Kushner S R. New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hasan N, Koob M, Szybalski W. Escherichia coli genome targeting. I. Cre-lox-mediated in vitro generation of ori-plasmids and their in vivo chromosomal integration and retrieval. Gene. 1994;150:51–56. doi: 10.1016/0378-1119(94)90856-7. [DOI] [PubMed] [Google Scholar]
- 12.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoang T T, Karkhoff-Schweizer R R, Kutchma A J, Schweizer H P. A broad-host-range Flp-FRT recombination system for site-specific excission of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 14.Ingram L O, Conway T, Clark D P, Sewell G W, Preston J F. Genetic engineering of ethanol production in Escherichia coli. Appl Environ Microbiol. 1987;53:2420–2425. doi: 10.1128/aem.53.10.2420-2425.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaniga K, Delor I, Cornelis G R. A wide host suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. J Bacteriol. 1991;109:137–141. doi: 10.1016/0378-1119(91)90599-7. [DOI] [PubMed] [Google Scholar]
- 16.Lai X, Davis F C, Hespell R B, Ingram L O. Cloning of cellobiose phosphoenolpyruvate-dependent phosphotransferase genes: functional expression in recombinant Escherichia coli and identification of a putative binding region for disaccharides. Appl Environ Microbiol. 1996;63:355–363. doi: 10.1128/aem.63.2.355-363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Borgne S, Palmeros B, Valle F, Bolivar F, Gosset G. pBRINT-Ts: a plasmid family with a temperature-sensitive replicon, designed for chromosomal integration into the lacZ gene of Escherichia coli. Gene. 1998;223:213–219. doi: 10.1016/s0378-1119(98)00168-1. [DOI] [PubMed] [Google Scholar]
- 18.Leonardo M R, Cunningham P R, Clark D P. Anaerobic regulation of the adhE gene, encoding the fermentative alcohol dehydrogenase of Escherichia coli. J Bacteriol. 1993;175:870–878. doi: 10.1128/jb.175.3.870-878.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leonardo M R, Dailly Y, Clark D P. Role of NAD in regulating the adhE gene of Escherichia coli. J Bacteriol. 1996;178:6013–6018. doi: 10.1128/jb.178.20.6013-6018.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Link A J, Phillips D, Church G M. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J Bacteriol. 1997;179:6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mikulskis A, Aristarkhov A, Lin E C C. Regulation of expression of the ethanol dehydrogenase gene (adhE) in Escherichia coli by catabolite repressor activator protein Cra. J Bacteriol. 1997;179:7129–7134. doi: 10.1128/jb.179.22.7129-7134.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy K C. Use of bacteriophage λ recombination functions to promote gene replacement in Escherichia coli. J Bacteriol. 1998;180:2063–2071. doi: 10.1128/jb.180.8.2063-2071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohta K, Beall D S, Shanmugam K T, Ingram L O. Genetic improvement of Escherichia coli for ethanol production: chromosomal integration of Zymomonas mobilis genes encoding pyruvate decarboxylase and alcohol dehydrogenase II. Appl Environ Microbiol. 1991;57:893–900. doi: 10.1128/aem.57.4.893-900.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peredelchuk M Y, Bennet G N. A method for the construction of E. coli strains with multiple DNA insertions in the chromosome. Gene. 1997;187:231–238. doi: 10.1016/s0378-1119(96)00760-3. [DOI] [PubMed] [Google Scholar]
- 25.Posfai G, Koob M, Hradecna Z, Hasan N, Filutowicz M, Szybalski W. In vivo excision and amplification of large segments of the Escherichia coli genome. Nucleic Acids Res. 1994;22:2392–2398. doi: 10.1093/nar/22.12.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Posfai G, Koob M D, Kirkpatrick H A, Blattner F R. Versatile insertion plasmids for targeted genome manipulations in bacteria: isolation, deletion, and rescue of the pathogenicity island LEE of the Escherichia coli O157:H7 genome. J Bacteriol. 1997;179:4426–4428. doi: 10.1128/jb.179.13.4426-4428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reid J L, Collmer A. An nptI-sacR cartridge for constructing directed, unmarked mutations in gram-negative bacteria by marker exchange-eviction mutagenesis. Gene. 1985;57:239–246. doi: 10.1016/0378-1119(87)90127-2. [DOI] [PubMed] [Google Scholar]
- 28.Rode C K, Obreque V H, Bloch C A. New tools for integrated genetic and physical analyses of the Escherichia coli chromosome. Gene. 1995;166:1–9. doi: 10.1016/0378-1119(95)00630-5. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Senecoff J F, Bruckner R C, Cox M M. The FLP recombinase of the yeast 2-μm plasmid: characterization of its recombination site. Proc Natl Acad Sci USA. 1985;82:7270–7274. doi: 10.1073/pnas.82.21.7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szybalski W. From the double-helix to novel approaches to the sequencing of large genomes. Gene. 1993;135:279–290. doi: 10.1016/0378-1119(93)90078-h. [DOI] [PubMed] [Google Scholar]
- 32.Wild J, Hradecna Z, Posfai G, Szybalski W. A broad-host-range in vivo pop-out and amplification system for generating large quantities of 50- to 100-kb genomic fragments for direct DNA sequencing. Gene. 1996;179:181–188. doi: 10.1016/s0378-1119(96)00487-8. [DOI] [PubMed] [Google Scholar]