Abstract
The afp1 gene, which encodes the antifungal protein AFP1, was cloned from nikkomycin-producing Streptomyces tendae Tü901, using a nikkomycin-negative mutant as a host and screening transformants for antifungal activity against Paecilomyces variotii in agar diffusion assays. The 384-bp afp1 gene has a low G+C content (63%) and a transcription termination structure with a poly(T) region, unusual attributes for Streptomyces genes. AFP1 was purified from culture filtrate of S. tendae carrying the afp1 gene on the multicopy plasmid pIJ699. The purified protein had a molecular mass of 9,862 Da and lacked a 42-residue N-terminal peptide deduced from the nucleotide sequence. AFP1 was stable at extreme pH values and high temperatures and toward commercial proteinases. AFP1 had limited similarity to cellulose-binding domains of microbial plant cell wall hydrolases and bound to crab shell chitin, chitosan, and cell walls of P. variotii but showed no enzyme activity. The biological activity of AFP1, which represents the first chitin-binding protein from bacteria exhibiting antifungal activity, was directed against specific ascomycetes, and synergistic interaction with the chitin synthetase inhibitor nikkomycin inhibited growth of Aspergillus species. Microscopy studies revealed that fluorescein-labeled AFP1 strongly bound to the surface of germinated conidia and to tips of growing hyphae, causing severe alterations in cell morphogenesis that gave rise to large spherical conidia and/or swollen hyphae and to atypical branching.
Proteins with antifungal activity have been isolated from plants, insects, and fungi and characterized. Plant antifungal proteins, including the cysteine-rich small defensins, ribosome-inactivating proteins, lipid transfer proteins, polygalacturonase inhibitor proteins, nonenzymatic chitin-binding proteins, and pathogenesis-related (PR) proteins, appear to comprise defense mechanisms against fungal attack (54). Many of these proteins are rapidly induced upon infection with fungal, bacterial, viral, or viroidal pathogens or by related forms of stress. Among the induced proteins are the PR proteins, which have been classified into families based on amino acid sequence similarities, serological properties, and function (22, 24, 43). Members of the five major families (1 to 5) have been shown to have in vitro antifungal activities. PR proteins of families 2 and 3 (PR-2 and PR-3 proteins) include β-1,3-glucanases and chitinases, respectively, which act synergistically in degrading fungal cell walls and inhibit growth of fungi. Many of the PR-4 proteins have nonenzymatic chitin-binding activity, and PR-5 proteins are related to salt-induced osmotins, which have been shown to permeabilize the fungal plasma membrane and inhibit spore germination and hyphal growth (1, 47, 51). A small histidine-rich antifungal protein and cysteine-rich antifungal proteins have been isolated from insects (8, 20). The latter proteins, which have significant sequence similarity to the 5-kDa plant defensins, inhibit spore germination and cause partial lysis of fungal hyphae. Small, highly basic, and cysteine-rich antifungal proteins not related to plant antifungal proteins have been purified from the extracellular medium of some imperfect ascomycetes (25, 31). These proteins reveal some sequence similarity to phospholipase A2, but their molecular mode of action is not known.
Antifungal proteins are of great biotechnological interest because of their potential use as food and seed preservative agents and for engineering plants for resistance to phytopathogenic fungi (6). Various studies have revealed that transgenic plants overexpressing genes of the PR-1, PR-2, PR-3, and PR-5 families mediate host plant resistance to phytopathogenic fungi, and coexpression of multiple antifungal protein genes in transgenic plants seems to be more effective than expression of single genes (for a review, see reference 54).
Antifungal proteins have not yet been isolated from streptomycetes and other bacteria. Streptomycetes are gram-positive, mycelium-forming soil bacteria that have the capacity to produce a great variety of secreted proteins, including hydrolytic enzymes that degrade organic material in the soil, such as chitin, cellulose, xylan, and starch (33), and enzyme inhibitors (7, 14, 30). Furthermore, streptomycetes have the exceptional ability to produce a broad range of low-molecular-weight antibiotics and other secondary metabolites; many of these compounds have antibacterial and antifungal properties and are used as agents in medicine and agriculture. Streptomyces tendae Tü901 produces antibiotics of various chemical structures, including cyclohexenylglycine, an isoleucine antagonist with antibacterial activity (23); the naphthoquinone compound juglomycin, which has antitumor activity (11); and chlorothricin, a glycosylated macrolide antibiotic that acts as an antagonist of acetyl-coenzyme A in bacteria (9). In addition, the strain synthesizes various nikkomycins, which are peptidyl nucleoside antibiotics. Nikkomycins act as specific inhibitors of chitin synthetases and have high antifungal, insecticidal, and acaricidal activity (for a review, see reference 12).
In an attempt to isolate nikkomycin biosynthesis genes, we transformed a nikkomycin-nonproducing mutant of S. tendae Tü901 with DNA fragments from S. tendae Tü901 cloned into the multicopy plasmid pIJ699 and screened for antifungally active transformants, using Paecilomyces variotii as the test organism. In the course of these experiments, we cloned a gene for a protein that exhibits antifungal activity. Here, we present the characterization of this antifungal protein, AFP1, and its gene and show that AFP1 is a chitin-binding protein that strongly binds to the cell wall of germinated conidia and to hyphal tips of sensitive fungi and interferes with their growth polarity.
MATERIALS AND METHODS
Microorganisms and plasmids.
S. tendae Tü901/8c, which produces nikkomycins I, J, X, and Z, was obtained from H. Zähner, University of Tübingen, Tübingen, Germany. S. tendae Tü901/NP51 is a nonproducing mutant of S. tendae Tü901/8c which does not synthesize antifungally active nikkomycins (4). S. lividans TK23 was obtained from D. A. Hopwood, John Innes Institute, Norwich, England. Plasmids pIJ699 (21) and pIJ486 (49) were provided by T. Kieser and D. A. Hopwood, John Innes Institute, and were used for cloning in Streptomyces. Escherichia coli JM83 {F− araΔ(lac-proAB) rpsL(Strr) [φ80dlacΔ(lacZ)M15]thi} and vectors pUC18/19 (53) were used for subcloning. The plasmids constructed for use in this study are shown in Fig. 1.
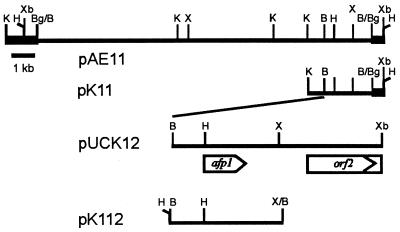
FIG. 1.
Restriction map of the 14.5-kb BamHI insert of pAE11 and subcloned fragments containing the afp1 gene. Bold lines represent segments of the vector pIJ699. The nucleotide sequence of the 2.08-kb BamHI-XbaI insert of pUCK12 was determined. The size and location of the afp1 gene and the 5′ region of orf2 were deduced from the nucleotide sequence and are indicated by an arrow and a box, respectively. Abbreviations for restriction enzymes: B, BamHI; Bg, BglII; H, HindIII; K, KpnI; X, XhoI; Xb, XbaI.
Test organisms for the determination of biological activity of AFP1 were E. coli DSM 498 (strain K-12), Micrococcus luteus DSM 1790, Candida albicans Tü167, Yarrowia lipolytica DSM 70562, Saccharomyces cerevisiae Tü125, Mucor miehei Tü284, Mucor hiemalis Tü179, Aspergillus terreus, Aspergillus fumigatus Tü161, Aspergillus niger Tü504, Cladosporium herbarum DSM 63422, Mycotypha africana DSM 3118, P. variotii Tü137, Penicillium chrysogenum DSM 844, and Trichophyton mentagrophytes Tü510 and were obtained from the culture collection of the Institute of Microbiology I, University of Tübingen (Tü strains) or from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ strains).
Culture conditions.
S. tendae NP51(pK11) and S. lividans(pK112) were grown on solid HA medium (3) containing 30 μg of thiostrepton ml−1 at 27°C. Liquid cultures were incubated on a rotatory shaker at 27°C as described previously (27), and AFP1 production was monitored on nikkomycin production medium SP (27), modified precultivation CRM medium, and HA medium (3). These media were buffered by 50 or 100 mM MES (2-[N-morpholino]ethanesulfonic acid), pH 6.0. Thiostrepton (Sigma) was added at a concentration of 10 μg ml−1 to liquid media. Agar diffusion assays with fungi and bacteria as test organisms were done on HA agar at pH 6.0 and 7.0, respectively. Malt extract peptone agar (3% malt extract, 0.3% soy peptone, 1.5% agar [pH 5.6]) was used for cultivation of C. herbarum and T. mentagrophyta.
Manipulation of DNA and cloning procedures.
Standard procedures were used for E. coli (38). Total DNA from S. tendae Tü901/8c was isolated by a large-scale method (19), and plasmid DNA was isolated by the alkaline lysis method (18). Streptomyces protoplasts were transformed as described by Bormann et al. (3).
For genetic complementation experiments, total DNA from S. tendae Tü901/8c was partially digested with BamHI, and the resulting fragments were size fractionated. Fragments of 15 to 25 kb were ligated to BglII-cleaved pIJ699 isolated from S. tendae Tü901/8c. S. tendae Tü901/NP51 was transformed with the ligation mixture, and the resulting thiostrepton-resistant transformants were grown in 10-mm2 patches on solid HA medium for 5 days. Agar plugs (6 mm in diameter) containing transformants were cut and placed onto test plates with P. variotii as the test fungus.
DNA sequencing.
The nucleotide sequence was determined by the dideoxy chain termination method using a T7 sequencing kit, deaza-dGTP reaction mixtures, [α-35S]dATP (Amersham Pharmacia Biotech Europe GmbH), and pUCK12 and subclones in pUC vectors with the M13/pUC universal and reverse primers. The CODONPREFERENCE program of the University of Wisconsin Genetics Computer Group package and the PC/GENE software package (IntelliGenetics) were used for sequence analysis. Databases were searched by using the program Ψ-BLAST (release 36) (2).
Purification and characterization of AFP1 protein.
Culture filtrate (200 ml) from S. tendae NP51(pK11) grown for 5 days in CRM medium was adjusted to pH 3.0 with 1 M HCl and stirred for 1 h at 4°C. Denatured proteins were precipitated by centrifugation at 10,000 × g for 30 min. The supernatant was desalted and buffered to 25 mM sodium acetate (pH 3.0) by chromatography on a Sephadex G-25 medium (Amersham Pharmacia Biotech) column (5 by 20 cm) previously equilibrated with the same buffer at a flow rate of 5 ml min−1. AFP1-containing fractions were fractionated on an S-Sepharose column (HR 16/10; Pharmacia) previously equilibrated with 25 mM sodium acetate buffer (pH 3.0) at a flow rate of 100 ml h−1. Most proteins did not bind to the resin. After elution of proteins by 100 ml of 0.2 M NaCl in 25 mM sodium acetate buffer (pH 3.0), AFP1 was eluted by 0.3 M NaCl in the same buffer. AFP1-containing fractions were collected, desalted by passage through Sephadex G-25 PD-10 columns (Amersham Pharmacia Biotech), and lyophilized.
AFP1 activity was determined by agar diffusion assays using P. variotii as the test organism calibrated with purified AFP1 (4 to 30 μg; Fig. 5).
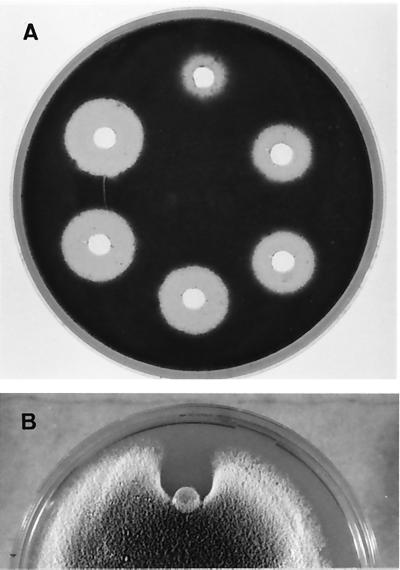
FIG. 5.
Growth inhibition of P. variotii by purified AFP1. (A) Growth of spores in response to 4, 8, 10, 15, 20, and 30 μg of AFP1 per well (clockwise beginning at the top). (B) Growth of hyphae in response to 30 μg of AFP1 applied on the paper disk, placed at the growing front of a colony and incubated for further 48 h.
The Stokes radius of purified AFP1 (100 μg) was determined by gel filtration chromatography on a Superose 12 column (HR 10/30; Amersham Pharmacia Biotech) equilibrated with 25 mM Tris-HCl buffer (pH 7)–0.1 M NaCl at a flow rate of 0.5 ml min−1. Reference proteins (Sigma) were insulin β chain (3,495.9 Da), cytochrome c (12,327 Da), carbonic anhydrase (29,000 Da), alcohol dehydrogenase from yeast (150,000 Da), and sweet potato β-amylase (200,000 Da). The molecular mass was also determined by electrospray mass spectrometry on an API III triple-quadrupole mass spectrometer (Sciex, Thornhill, Canada).
The N-terminal sequence of purified AFP1 was determined by automated Edman degradation in a pulse-liquid protein sequencer 477A (Applied Biosystems) equipped with an online analyzer 120A (Applied Biosystems) for phenylthiohydantoin-derivatized amino acids.
Determination of cysteine by using Ellmann's reagent.
To test for cysteine and cystine residues in AFP1, the reaction with Ellmann's reagent was carried out before and after treatment with sodium borohydride (17). AFP1 (20 or 40 μg in 45 μl) was incubated in the presence or absence of 2.5% sodium borohydride in water at 50°C for 1 h. After destruction of the remaining borohydride by addition of 20 μl of 1 N HCl and incubation for 30 min at room temperature, 25-μl aliquots of each sample were removed and used for agar diffusion assays with P. variotii as the test organism; 200 μl of Ellmann's reagent [0.05% 5,5′-dithio-bis(5-nitrobenzoic acid) (Sigma) in 0.22 M Tris-HCl, pH 8.2] was added to the remaining samples. After incubation for 5 min at room temperature, the absorbance at 412 nm was measured. Absorbances of the reduced AFP1 solutions (20 μg of AFP1 and 40 μg of AFP1) were 0.074 and 0.148, respectively; the absorbance of the native protein (40 μg) was 0.002. Inhibition zones toward P. variotii were not obtained for NaBH4-treated samples, while untreated AFP1 samples gave inhibition zones 12 and 14 mm in diameter.
Determination of stability.
Temperature stability was investigated by incubating purified AFP1 (1 mg ml−1 in 25 mM MES-NaOH (pH 6) in sealed vessels at 40 to 100°C. Samples (20 μl) were removed between 0 and 60 min. pH stability of purified AFP1 (1 mg ml−1) was investigated at pH 1.5 to 12 at 4 and 24°C. Samples (20 μl) were taken after 0, 1, 2, 3, 4, 5, and 8 days. The following buffers (25 mM) were used: glycine-HCl, pH 1.5 and 2; sodium acetate-NaOH, pH 3; citric acid-Na2HPO4, pH 4 and 5; MES-NaOH, pH 6; TES (N-tris[hydroxymethyl]methyl-2-aminoethanesulfonic acid)-NaOH, pH 7; HEPES-NaOH, pH 8; TAPS (N-tris[hydroxymethyl]methyl-3-aminopropanesulfonic acid)-NaOH, pH 9; and glycine-NaOH, pH 10, 11, and 12. Stability toward proteases was assayed by incubating 7 μg of AFP1 and 10 μg of pepsin, proteinase K, pronase, or trypsin (all from Sigma) in 0.1 M citric acid–0.2 M Na2PO4 (at pH 4 and 5 for pepsin and at pH 7 for the other enzymes) for 3 and 12 h at 37°C. Antifungal activity of the samples was determined by agar diffusion assays using P. variotii as the test organism.
PAGE.
Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 16% polyacrylamide gels containing 6 M urea in the Tris-Tricine (N-tris[hydroxymethyl]glycine) buffer system described by Schägger and von Jagow (39), which allowed a good separation of proteins with molecular masses of <10 kDa. Prestained molecular mass standards (Gibco BRL, Life Technologies) were insulin α and β chain (3,000 Da), bovine trypsin inhibitor (6,200 Da), lysozyme (14,300 Da), β-lactoglobulin (18,400 Da), carbonic anhydrase (29,000 Da), and ovalbumin (43,000 Da). Proteins in gels were detected by staining with Coomassie blue G250.
Determination of antifungal activity.
Agar diffusion assays were performed to determine the antifungal activity of AFP1-containing agar or solutions and to characterize the spectrum of biological activity of AFP1. Test plates (85 mm in diameter) contained 17.5 ml of HA agar seeded with test organisms, either spores of filamentous fungi or cells of bacteria and yeasts grown overnight in liquid HA medium, at a concentration of (105). AFP1-containing solutions were applied to paper disks (6 mm in diameter) or in wells (6 mm in diameter) punched into the agar. Test plates were incubated for at least 24 h at 37°C for E. coli, M. miehei, A. fumigatus, and P. variotii, at 24°C for C. herbarum, and at 27°C for the remaining test organisms. All results deduced from agar diffusion assays were calculated from three independent experiments. To assay for inhibition of hyphal growth, an agar plug containing mycelia of the test fungus was placed in the center of a petri dish containing 17.5 ml of HA agar and incubated until the colony reached a diameter of 3 to 4 cm. A paper disk containing 20 or 30 μg of AFP1 was placed at the growing front of the hyphae, and the test plates were incubated further.
High-pressure liquid chromatography (HPLC) analysis was used to check culture filtrates for nikkomycins (10).
Labeling of AFP1.
AFP1 (1 mg) was labeled with 0.236 mg of 5(6)-carboxyfluorescein-N-hydroxysuccinimide ester according to the specifications of the fluorescein labeling kit supplied by Boehringer (Mannheim, Germany).
Assays.
Chitin azur (Sigma), p-nitrophenyl β-d-N,N′,N"-triacetylchitotriose (Sigma), laminarin (Sigma), and whole mycelia of P. variotii were used as substrates to assay AFP1 for chitinase, β-1,3-glucanase, and cell wall-degrading activity, respectively. P. variotii mycelia grown in liquid HA medium were homogenized in an Ultraturrax blender for 15 s. Cell debris was sedimented by centrifugation at 20,000 × g for 20 min, washed three times with deionized water, lyophilized, and used for estimation of sugar liberation. The reaction mixture consisted of 0.5 ml of substrate suspension (4 mg ml−1) in 0.1 M citric acid–0.2 M Na2HPO4 buffer (pH 7) or in 0.1 M ammonia acetate buffer (pH 5) and 0.1 ml of AFP1 (50 μg in distilled water) and was incubated at 37°C with shaking for 12 to 48 h. Undissolved substrates were removed by centrifugation. Degradation of chitin azur was determined by measuring the absorbance at 570 nm. The amount of reduced sugars released from laminarin or fungal mycelia was assayed with the low-alkalinity copper reagent of Somogyi (41) and arsenomolybdate chromogen of Nelson (32). With p-nitrophenyl β-d-N,N′,N"-triacetylchitotriose as the substrate, the reaction mixture consisted of 0.12 ml of substrate (2 mg ml−1) in 0.1 M citric acid–0.2 M Na2HPO4 buffer (pH 7.0) and 100 μg of AFP1. After incubation for 4 h at 37°C, absorbance was determined at 410 nm. Positive control reactions were performed with chitinase from S. griseus (Sigma) and laminarinase from mollusk (Sigma).
Binding assays were performed with Avicel PH-101 (Fluka BioChemika), xylan from birchwood (Sigma), curdlan from Alcaligenes faecalis (Sigma), purified crab shell chitin (Sigma), chitosan from crab shells (Sigma), and cell walls of P. variotii. Fluorescein isothiocyanate (FITC)-labeled AFP1 (10 μg) was incubated with 5 mg of polyglycan and 1 mg of cell walls in 100 μl of 100 mM Tris-HCl (pH 7.6) overnight on a rotator. After three washes with the same buffer containing 150 mM NaCl, the samples were inspected by microscopy.
Microscopy.
For fluorescence microscopy, samples were mounted in 16.7% (wt/vol) Moviol (Hoechst)–33.3% (vol/vol) glycerol in 66.7 mM Tris-HCl (pH 8 (37). A Zeiss Axiophot 2 microscope, the FITC filter to visualize FITC-labeled AFP1, and Kodak T-Max films (ASA 100 and ASA 1000) were used for photographs.
Nucleotide accession number.
The nucleotide sequence data described in this report have been deposited at the EMBL nucleotide sequence database under accession no. AJ242827.
RESULTS
Cloning of the AFP1-encoding gene.
The mutant S. tendae Tü901/NP51, which does not synthesize biologically active nikkomycins (4), was transformed with a genomic library of S. tendae Tü901 in pIJ699, constructed as described in Materials and Methods. To screen for antifungally active transformants, agar plugs containing thiostrepton-resistant transformants grown in patches on complex agar medium were cut and incubated on plates containing spores of P. variotii. Among approximately 2,000 transformants, one caused a clear zone of growth inhibition about 10 mm in diameter. The recombinant plasmid pAE11 isolated from this transformant carried a 14.5-kb insert (Fig. 1). When pAE11 was reintroduced into mutant NP51, about 70% of the new transformants grown on solid medium caused zones of growth inhibition of P. variotii.
The antifungal activity of the transformants was obviously not derived from genetic complementation of mutant NP51 to nikkomycin synthesis since nikkomycins (at concentrations >3 mg/liter) were not detected by HPLC of culture filtrates of transformants grown in nikkomycin production medium. Furthermore, when the culture filtrate was passed through a membrane filter with an exclusion limit of 10 kDa, which would not retain nikkomycins (approximately 600 Da), the antifungally active compound was retained on the filter. To localize the encoding gene, designated afp1 (antifungal protein 1), subclones of pAE11 were constructed. The presence of the deletion plasmid pK11, which lacked three KpnI fragments and contained a 2.8-kb insert, or plasmid pK112, which carried the 1.1-kb XhoI-BamHI fragment from pK11 (Fig. 1), led to expression of the antifungal protein in S. tendae NP51 and S. lividans TK23.
DNA sequence analysis.
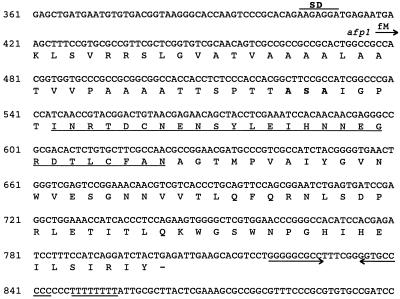
The nucleotide sequence of the 2,087-bp BamHI-XbaI fragment cloned from pK11 into pUC19 (forming pUCK12) was determined and analyzed. A complete open reading frame, afp1, composed of 384 bp, and the 5′ end of a putative second open reading frame, orf2, were identified (Fig. 1 and 2). The afp1 gene has two potential ATG start codons, at nucleotides (nt) 411 and 417; the latter is preceded by a potential ribosome-binding site (GAGG; nt 407 to 410) and therefore is the proposed translational start site. The afp1 gene terminates at a TGA stop codon at nt 801 and could encode a protein of 128 amino acids. The G+C content of the afp1 gene is 63.3%, much lower than that of the total Streptomyces genome (73%). The region upstream of the afp1 gene (nt 1 to 416) also has a low G+C content (60.7%). In the distal region of the 3′ end of the afp1 gene are found a 9-bp, GC-rich inverted repeat region and an adjacent poly(I) region of 8 nt, which are rare for potential Streptomyces transcription terminators. Database searches revealed limited sequence similarity (25 to 33% identity, 41 to 53% similarity) between 40 to 77 amino acid residues of AFP1 and N-acetylmuramoyl-l-alanine amidase from Synechocystis sp., cellulase CelE from Pseudomonas fluorescens, and endo-1,4-β-xylanases from Bacillus and Ruminococcus spp. (GenBank accession no. D90909, X86798, X59059, and U43089). The region of sequence similarity within CelE constitutes a part of the 100-amino-acid cellulose-binding domain, which is characterized by several highly conserved tryptophan residues in addition to glycine and asparagine residues and the lack of charged amino acids (12a). The region of similarity between AFP1 and the xylanases is located in the catalytic domain of the enzymes. Ψ-Blast iterations revealed similarity between AFP1 and cellulose-binding domains of other cellodextrinases, xylanases, and esterase D from Pseudomonas fluorescens (Fig. 3) and to a sequence in the catalytic domain of various xylanases.
FIG. 2.
Nucleotide sequence of the S. tendae Tü901 afp1 gene and the predicted amino acid sequence. A putative ribosome-binding site (SD) is overlined. The translational start site of the afp1 gene is indicated by an arrow, and the termination codon is marked by a dash. Amino acids determined experimentally by N-terminal sequencing are underlined. The proposed cleavage site for signal peptidase is in boldface. →← indicates an inverted repeat sequence, and the poly(T) region is underlined.
FIG. 3.
BLASTP sequence alignment of amino acid residues 3 to 75 of the extracellular AFP1 with segments of N-acetylmuramoyl-l-alanine amidase from Synechocystis sp. (AmiA) and cellulase CelE (CelE), endoglucanase A (EGA), cellodextrinase (EGC), xylanase A (XylA), and esterase D (EstD) from Pseudomonas fluorescens. The sequences were obtained from GenBank (AFP1, this work; CelE, X86798; AmiA, D90909; CelE, X86798; EGA, X12570; EstD, X15429), from the PIR database (EGC, S19652), and from Swiss-Prot (XylA, P14768). CBD indicates residues of cellulose-binding domains which are identical or similar in the aligned sequences of CelE, EGA, EGC, XylA, and EstD. Aromatic amino acid residues involved in protein-sugar binding (34) are boxed. Amino acid residues that are identical in six of seven sequences are in boldface.
The potential open reading frame orf2 could begin at the ATG start codon at nt 1249 or at nt 1285, each of which is preceded by a potential ribosome-binding site (AGGG, nt 1239 to 1242; GGGA, nt 1279 to 1282). The corresponding incomplete orf2 has a G+C content of 70.9%. The encoded N-terminal portion of the protein, composed of 277 or 266 amino acids, respectively, did not reveal any similarity to protein sequences in databases.
Purification of the AFP1 protein.
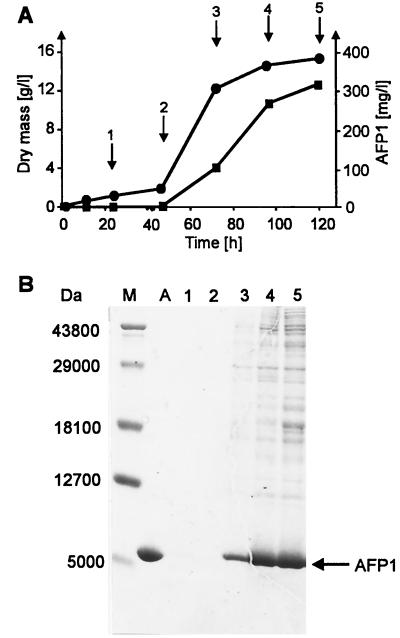
To purify AFP1, S. tendae NP51 carrying pK11 was first cultivated in various media and culture filtrates were analyzed for antifungal activity by agar diffusion assays. Antifungal activity was of a similar high value when SP or CRM medium was used. CRM medium, which contains less proteinaceous compounds that could interfere with the purification procedure, was used for further experiments. Culture filtrates were collected throughout the growth of S. tendae NP51(pK11) and assayed by agar diffusion tests and SDS-PAGE (Fig. 4). Antifungal activity of culture filtrates increased to a maximum level corresponding to approximately 300 mg of AFP1 liter−1 at 120 h in the stationary growth phase and remained constant when mycelia were cultivated for a further 24 h. SDS-PAGE analysis revealed that AFP1 appeared as the major protein of antifungally active culture filtrates and migrated near the 5,000-Da marker protein.
FIG. 4.
Time course of AFP1 production in S. tendae NP51(pK11) cultivated in CRM medium. (A) AFP1 (■) in the culture filtrate determined by agar diffusion assays with P. variotii as the test fungus compared to growth as determined by dry weight (●). (B) SDS-PAGE of culture filtrates from S. tendae NP51(pK11) and purified AFP1. Lanes 1 to 5, culture filtrate (40 μl) taken at time points (1 to 5) indicated by arrows in panel A; lane A, purified AFP1 (5 μg); lane M, molecular mass markers. Proteins were stained with Coomassie blue. AFP1 is marked by an arrow.
AFP1 was purified to apparent homogeneity by a two-step purification based on the stability and solubility of AFP1 at acidic pH; AFP1 has a calculated pI of 5.3. After precipitation of most proteins from culture filtrates at pH 3.0, the remaining proteins were separated by cation-exchange chromatography using S-Sepharose equilibrated in 25 mM acetate buffer (pH 3.0). After elution of contaminating proteins with 0.2 M NaCl in 25 mM acetate buffer (pH 3.0), AFP1 was eluted with 0.3 M NaCl in the same buffer and appeared to be homogeneous on SDS-polyacrylamide gels (Fig. 4). Final recovery of AFP1 was 30%.
Properties of purified AFP1.
The N-terminal amino acid sequence of AFP1 determined by Edman degradation revealed that 42 amino acid residues were cleaved from the N terminus of the afp1-encoded protein (Fig. 2). The molecular mass of AFP1 was determined by electrospray mass spectrometry to be 9,862 ± 2.7 Da, which coincided with that deduced from the nucleotide sequence (9,860 Da). The molecular mass determined by SDS-PAGE after AFP1 was reduced by dithiothreitol (Fig. 4) and gel filtration gave a considerably lower value of 6,100 Da.
Native AFP1 did not react with Ellman's reagent, which detects free thiol groups, but gave a positive reaction after treatment with sodium borohydride, which indicated that Cys-6 and Cys-24 of AFP1 form a disulfide bridge (Fig. 2). The reduced form of AFP1 did not exhibit antifungal activity in agar diffusion assays.
AFP1 was stable over the pH range of 1.5 to 12 and resisted almost completely digestion by pepsin at pH 4 and 5 and by proteinase K, pronase, or trypsin at pH 7 (data not shown). Heating purified AFP1 at temperatures between 70 and 100°C for 60 min reduced antifungal activity to 50%.
Biological activity.
The antimicrobial activity of AFP1 was determined by growth inhibition assay on agar plates seeded with various microorganisms (Table 1). AFP1 inhibited growth of specific ascomycetes; P. variotii was the most sensitive fungus tested. In contrast, AFP1 had no effect on the growth of E. coli, Micrococcus luteus, S. lividans TK23, Mucor species, and yeasts. Growth of P. variotii was inhibited at AFP1 concentrations of >4 μg per well (Fig. 5). In addition, AFP1 applied to the growing front of a colony directly inhibited hyphal growth of fungi that were affected in agar diffusion assays (shown for P. variotii in Fig. 5). Growth inhibition zones caused by AFP1 were stable for weeks when test plates were further incubated at room temperature.
TABLE 1.
Biological activity of AFP1 in agar diffusion assays
| Test organism | Inhibition
zonea (mm)
|
|||
|---|---|---|---|---|
| AFP1
|
Nik (5 μg) | AFP1 (20 μg) + Nik (5 μg) | ||
| 20 μg | 40μg | |||
| Escherichia coli | 0 | 0 | NT | NT |
| Micrococcus luteus | 0 | 0 | NT | NT |
| Streptomyces lividans TK23 | 0 | 0 | NT | NT |
| Mucor miehei, M. hiemalis | 0 | 0 | NT | NT |
| Candida albicans | 0 | 0 | 0/−13 | 0/−13 |
| Saccharomyces cerevisiae | 0 | 0 | NT | NT |
| Yarrowia lipolytica | 0 | 0 | NT | NT |
| Aspergillus fumigatus | 10/−11 | 13/−15 | 0/−10 | 15/−23 |
| A. niger | 0 | 7 | 0 | 12b |
| A. terreus | 9 | 11 | 0 | 13/−18 |
| Cladosporium herbarum | 0 | 0 | 0/−40 | 0/−40 |
| Mycotypha africana | 0 | 0 | 32 | 32 |
| Paecilomyces variotii | 18 | 20 | 0/−47 | 22/−47 |
| Penicillium chrysogenum | 0 | 0 | 0 | 12b |
| Trichophyton mentagrophyta | 0/−11 | 11 | 0 | 0/−11 |
Diameter of complete growth inhibition/− diameter of incomplete growth inhibition. NT, not tested.
Diameter of sporulation inhibition. Nik, nikkomycin Z.
Combinations of AFP1 and the chitin synthetase inhibitor nikkomycin were tested against fungi affected by AFP1 to detect possible synergistic effects between the antifungal agents. Synergism was clearly observed against Aspergillus species, especially A. fumigatus (Table 1).
Test for enzyme activities and polysaccharide binding.
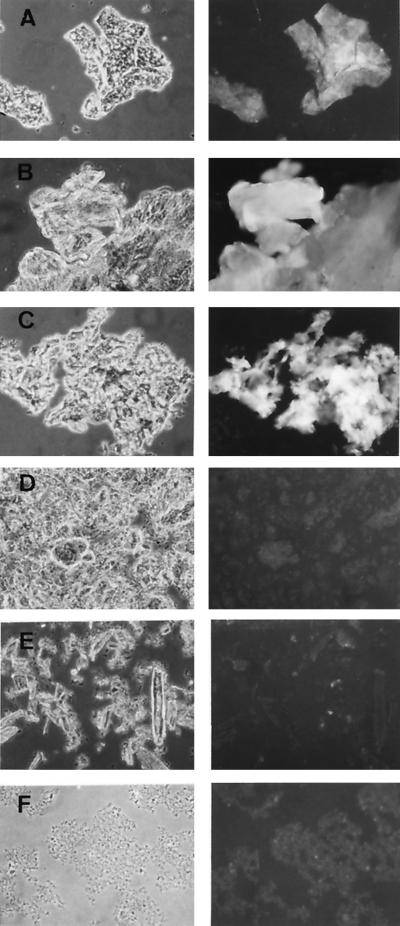
As the biological activity of AFP1 was exclusively directed against ascomycetes, AFP1 was assayed for enzyme activities directed against chitin and β-1,3-glucan, the main components of their cell wall (50). However, neither chitinase and β-1,3-glucanase activity nor the release of reducing sugars from cell walls of P. variotii could be detected. Since AFP1 had limited sequence similarity to cellulose-binding domains, binding of fluorescein-labeled AFP1 to various polysaccharides and the cell wall of P. variotii was studied. Microscopy observations revealed that AFP1 binds to crab shell chitin, chitosan, and P. variotii cell wall but does not to β-1,3-glucan (curdlan), crystalline cellulose (Avicel), and xylan (Fig. 6).
FIG. 6.
Binding of AFP1 to chitin, chitosan, and fungal cell walls. Fluorescein-labeled AFP1 was incubated overnight with crab shell chitin (A), crab shell chitosan (B), P. variotii cell wall (C), β-1,3-glucan (curdlan; D), Avicel (E), and xylan (F). After three washes with 100 mM Tris-HCl (pH 7.6)–150 mM NaCl, samples were analyzed by phase-contrast (left) and fluorescence (right) microscopy.
Effect of AFP1 on fungal morphology and its localization.
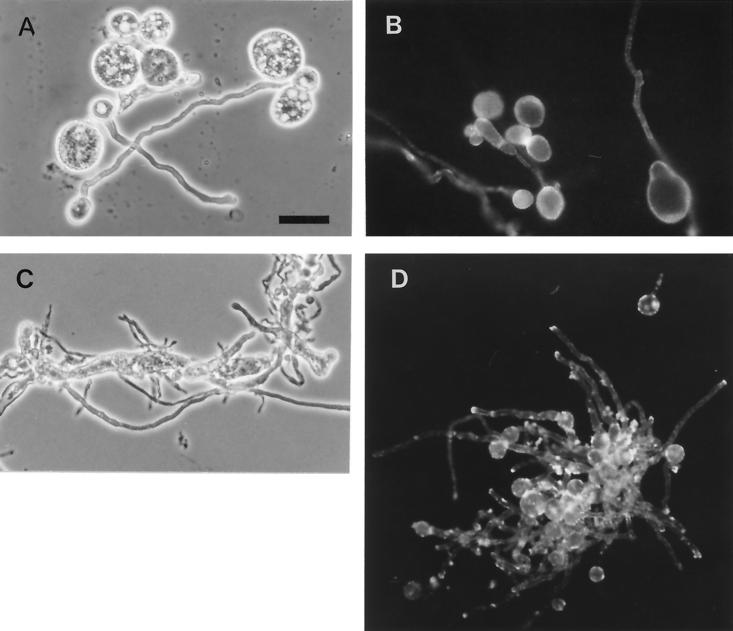
The phenotypes of P. variotii and A. fumigatus affected by AFP1 were determined by inoculating 200 μl of complete medium (HA) containing various concentrations of AFP1 with conidia and cultivating the plates for 12 h. At 50 μg of AFP1 ml−1, approximately 80% of P. variotii conidia (4 to 6 μm) grew isotropically to large, round cells up to 25 μm in diameter. After washing and plating onto solid medium, the large germ spheres grew to form colonies. The remaining conidia germinated normally and grew to germ spheres 11 μm in diameter and formed a germ tube (Fig. 7A). At higher concentrations of AFP1 (>75 μg ml−1), in addition to the large round cells, hyphal swelling was observed (data not shown). In contrast, large spheres were not observed for A. fumigatus. At concentrations of 50 μg of AFP1 ml−1, A. fumigatus germinated and grew normally (Fig. 7D); at concentrations of >75 μg of AFP1 ml−1, aberrant branching of hyphae and increased hyphal width were observed (Fig. 7C). Mycelia lysed easily upon mechanical force.
FIG. 7.
Effect of AFP1 on the morphology of P. variotii and A. fumigatus and its localization. P. variotii (A and B) and A. fumigatus (C and D) were grown in the presence of fluorescein-labeled AFP1 (A, B, and D, 50 μg ml−1; C, 75 μg ml−1) for 12 h and analyzed by phase-contrast (left) and fluorescence (right) microscopy. Scale bar represents 20 μm.
Fluorescein-labeled AFP1 (50 μg ml−1) was used to localize AFP1 on both fungi (Fig. 7). With P. variotii, most of the fluorescence appeared on the surface of the large spheres, and fluorescence was also detected along hyphae. With A. fumigatus, strong fluorescence appeared at hyphal tips and at the wall of germinated conidia that had formed a germ tube.
DISCUSSION
The gene for the extracellular AFP1 from S. tendae Tü901 was cloned on a multicopy vector in a nikkomycin-nonproducing mutant. Overexpression was a prerequisite for detection of the gene by the screening method used. Since AFP1 was produced in large amounts by the transformants and remained stable and soluble at acid pH, it was rapidly purified to homogeneity from culture filtrate. Mature AFP1 consists of 86 amino acid residues according to the N terminus defined by Edman degradation and the molecular mass obtained by electrospray mass spectrometry. The estimation of an apparent lower molecular mass for AFP1 by SDS-PAGE and gel filtration might be due its relatively small size and an unusual molecular shape caused by the presence of a disulfide bridge, which could influence the binding of SDS (5) and lead to deviations from the ideal behavior in gel filtration. A significantly lower apparent molecular mass has also been determined for the S. tendae 4158 α-amylase inhibitor tendamist, which consists of 74 amino acids and contains two disulfide bridges (46).
The afp1 gene encodes a typical signal peptide required for secretion with three positively charged amino-terminal amino acid residues (two Arg and one Lys) and a hydrophobic core region. However, extracellular AFP1 does not appear to be the direct product of cleavage by signal peptidase I, as the residues upstream of the determined N terminus of extracellular AFP1 (Gly-Pro-Thr) do not conform to the −1,−3 rule, which states that proline must be absent from positions −3 to +1 relative to the cleavage site of signal peptidase I (48). Two sites that follow the −1,−3 rule (Ala-Ala-Ala and Ala-Ser-Ala) and that are preceded by a turn-promoting proline at distances of 2 and 3 residues, respectively, are located 13 and 4 amino acid residues upstream of the N terminus. Since purified AFP1 from S. lividans(pK11) consisted of a small proportion (10 to 20%) of the protein with five additional amino acids (Ala-Ile-Gly-Pro-Thr-) at the N terminus (data not shown), AFP1 isolated from S. tendae NP51(pK11) might be the result of extracellular processing subsequent to the initial signal peptidase cleavage. Similar phenomena have been reported for protein protease inhibitors from S. lividans and S. longisporus overproduced in S. lividans; the protein revealed amino-terminal amino acid heterogeneity in relation to the length of cultivation (44).
AFP1 is a chitin-binding protein that strongly interacted with crab shell chitin, chitosan, and fungal cell walls. Binding of proteins to carbohydrates is mediated by aromatic residues, primarily tryptophan and tyrosine, through hydrogen-bonding and hydrophobic interactions (42). Three tryptophan residues of bacterial cellulose-binding domains to which AFP1 shows sequence similarity have been shown to be exposed to one side of the domain and to be involved in protein-sugar binding (34, 52). AFP1 has aromatic amino acid residues at the relevant positions: histidine, tyrosine, and tryptophan at positions 15, 36, and 40, respectively. Tryptophan, tyrosine, and histidine are also found at analogous positions in cellulose-binding domains of distinct families of proteins (45). Bacterial cellulose-binding domains are found in many cellulases, xylanases, and other hemicellulases that have a modular structure (13). These enzymes appear to have evolved from a limited number of ancestral sequences by domain fusion with subsequent modifications of the domains. A significant transfer of xylanase genes that fall into two distinct families seems to have occurred between bacteria and eukaryotic microorganisms. Since the afp1 gene has features that are extraordinary for Streptomyces (a low G+C content of DNA and a poly[T] region and stem-loop structure downstream of the gene), it is tempting to speculate that it may have evolved from a progenitor polysaccharide-binding domain of microorganisms having a G+C content lower than that of Streptomyces. Many streptomycetes, including S. tendae Tü901, utilize chitin as sole carbon and nitrogen source and can be easily enriched from soil on plates containing chitin as the only nutrient. Whether AFP1 is part of the chitinolytic system that facilitates degradation of chitin in a way similar to that proposed for cellulose-binding domains in microbial plant cell wall hydrolases (26) is now being investigated.
AFP1 represents the first chitin-binding protein from bacteria that exhibits antifungal activity. As antifungal activity has also been demonstrated for AFP1 isolated to homogeneity from E. coli BL21(DE3) cell extracts carrying the portion of the afp1 gene for the mature AFP1 protein in the T7 RNA polymerase-based pET11a (data not shown), there is no doubt that antifungal activity is caused by AFP1 alone. The chitin-binding proteins, CHB1 and CHB2, recently isolated from Streptomyces also bind to α-chitin and fungal hyphae but have no antifungal activity (40). AFP1 does not show sequence similarity to CHBs, which have a higher molecular mass (18.7 kDa) and an isoelectric point at alkaline pH (9.01). The antifungal property of AFP1 might be related to its small size, which would allow it to penetrate the fungal wall. An estimate of fungal wall pore size predicts that proteins larger than 15 to 20 kDa will not be able to pass through the fungal wall (29). Chitin-binding proteins from plants that have antifungal effects (e.g., Ac-AMPs from seeds of amaranth, nettle lectin, and hevein) are of small size (reference 35 and references herein). Members of the RP-4 family have molecular masses of 13 to 15 kDa (22). The most potent chitin-binding lectins are the 6-kDa Ac-AMPs that act against a broad spectrum of fungal pathogens at concentrations of 1 to 10 μg ml−1. The 8.5-kDa nettle lectin inhibits germ tube growth of seven fungi at concentrations of 20 to 150 μg ml−1, which is the range required for antifungal activity of AFP1, while the 5-kDa hevein, which causes swelling of hyphae, is three to five times less active.
The mechanism of growth inhibition by the plant chitin-binding proteins is not known. Microscopy studies revealed that fluorescein-labeled AFP1 preferentially binds to sites where cell wall synthesis takes place: at conidia which grow isotropically by adding new wall material uniformly in every direction and to hyphal tips (50). Therefore, it might be speculated that AFP1 interferes with cell wall synthesis by binding to nascent chitin, eventually cross-linking the polymer and interfering with growth polarity. The susceptibility of chitin synthetase for the antibiotic nikkomycin appeared to be enhanced in Aspergillus species, which are less sensitive to nikkomycin alone. Similar synergism has been reported for zeamatin and thaumatin-like proteins, which belong to the RP-5 family and facilitate penetration of nikkomycin by interaction with the fungal membrane (16, 36). In fungi, wall synthesis is coupled to exocytosis of vesicles containing wall-synthesizing enzymes and wall material, a process that determines growth polarity. The transport of vesicles is assumed to be mediated by a cytoskeleton, of which actin is the most important component. Actin is concentrated in areas of growth, and linkages between actin and plasmalemma have been postulated (reference 15 and references therein). The factors that establish polarity and subsequent cell wall synthesis of filamentous fungi and that maintain polarity are relatively unknown. Recently, eight Aspergillus nidulans mutants were isolated and characterized. These mutants have defects in one of these processes, and their germinated conidia and hyphae are misshapen (28).
The predominant effect of AFP1 on P. variotii occurred during spore germination. AFP1 interfered with polarized growth and prevented the emergence of a normal germ tube but allowed nonpolar growth, leading to large spherical cells with a rigid wall. In addition, AFP1 interfered with polarized growth at hyphal tips, leading to abnormal branching and swollen hyphae with weakened walls that did not resist internal turgor pressure upon mechanical stress. The different effects of AFP1 on P. variotii and A. fumigatus, where AFP1 appeared only to interfere with growth polarity of hyphae and the selectivity of AFP1 toward ascomycetes, may be due to the cell wall-associated proteins, which might act as a physical barrier for AFP1 and determine accessibility of the target. Cell wall proteins have been shown to be determinants of resistance of Saccharomyces cerevisiae toward tobacco osmotin and are presumed to mask binding sites in the plasma membrane that mediate osmotin action (55). AFP1 may be a valuable tool for studying chitin synthesis in filamentous fungi and its coupling to processes involved in determination of growth polarity and for investigating determinants of resistance.
ACKNOWLEDGMENTS
We thank S. Stevanovic and J. Metzger (Institute of Organic Chemistry, University of Tübingen) for analytical support, B. Lattermann (Max-Planck-Institut für Entwicklungsbiologie, Tübingen, Germany) for introduction to photomicroscopy, P. L. Huynh and G. Scheer for making photographs, and K. A. Brune for editing the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB 323, A4, and C2).
REFERENCES
- 1.Abad L R, D'Urzo M P, Liu D, Narasimhan M L, Reuveni M, Zhu J K, Niu X, K. S N, Hasegawa P M, Bressan R A. Antifungal activity of tobacco osmotin has specificity and involves plasma membrane permeabilization. Plant Sci. 1996;118:11–23. [Google Scholar]
- 2.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bormann C, Aberle K, Fiedler H-P, Schrempf H. Genetic complementation of Streptomyces tendaedeficient in nikkomycin production. Appl Microbiol Biotechnol. 1990;32:424–430. doi: 10.1007/BF00903777. [DOI] [PubMed] [Google Scholar]
- 4.Bormann C, Mattern S, Schrempf H, Fiedler H P, Zähner H. Isolation of Streptomyces tendaemutants with an altered nikkomycin spectrum. J Antibiot. 1989;42:913–918. doi: 10.7164/antibiotics.42.913. [DOI] [PubMed] [Google Scholar]
- 5.Broekaert W F, Marien W, Terras F R G, De Brolle M F C, Proost P, Van Damme J, Dillen L, Claeys M, Rees S B, Vanderleyden J, Cammue B P A. Antimicrobial peptides from Amaranthus caudatusseeds with sequence homology to the cysteine/glycine-rich domain of chitin-binding proteins. Biochemistry. 1992;31:4308–4314. doi: 10.1021/bi00132a023. [DOI] [PubMed] [Google Scholar]
- 6.Dempsey D M A, Silva H, Klessig D F. Engineering disease and pest resistance in plants. Trends Microbiol. 1998;6:54–61. doi: 10.1016/S0966-842X(97)01186-4. [DOI] [PubMed] [Google Scholar]
- 7.Doran J L, Leskiw B K, Aippersbach S, Jensen S E. Isolation and characterization of a β-lactamase-inhibitory protein from Streptomyces clavuligerusand cloning and analysis of the corresponding gene. J Bacteriol. 1990;172:4909–4918. doi: 10.1128/jb.172.9.4909-4918.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fehlbaum P, Bulet P, Michaut L, Lagueux M, Broekaert W F, Hetru C, Hoffmann J A. Insect immunity. Septic injury of Drosophila induces the synthesis of a potent antifungal peptide with sequence homology to plant antifungal peptides. J Biol Chem. 1994;269:33159–33163. [PubMed] [Google Scholar]
- 9.Fiedler H-P. Biosynthetic capacities of actinomycetes. 1. Screening for secondary metabolites by HPLC and UV-visible absorbance spectral libraries. Nat Prod Lett. 1993;2:119–128. [Google Scholar]
- 10.Fiedler H-P. Screening for new microbial products by high-performance liquid chromatography using a photodiode array detector. J Chromatogr. 1984;316:487–494. doi: 10.1016/s0021-9673(00)96176-4. [DOI] [PubMed] [Google Scholar]
- 11.Fiedler H-P, Kulik A, Schüz T C, Volkmann C, Zeeck A. Biosynthetic capacities of Actinomycetes. 2. Juglomycin Z, a new naphthoquinone antibiotic from Streptomyces tendae. J Antibiot. 1994;47:1116–1122. doi: 10.7164/antibiotics.47.1116. [DOI] [PubMed] [Google Scholar]
- 12.Fiedler H-P, Schüz T, Decker H. An overview of nikkomycins: history, biochemistry, and applications. In: Rippon J W, Fromtling R A, editors. Cutaneous agents. New York, N.Y: Marcel Dekker Inc.; 1993. pp. 325–352. [Google Scholar]
- 12a.Hazlewood G P, Gilbert H J. Xylan and cellulose utilization by the clostridia. Bio/Technology. 1993;25:311–341. [PubMed] [Google Scholar]
- 13.Gilkes N R, Henrissat B, Kilburn D G, Miller R C, Jr, Warren R A J. Domains in microbial β-1,4-glycanases: sequence conservation, function, and enzyme families. Microbiol Rev. 1991;55:303–315. doi: 10.1128/mr.55.2.303-315.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goto A, Matsui Y, Ohyama K, Arai M, Murao S. Purification and characterization of an alpha-amylase inhibitor (Haim) produced by Streptomyces griseosporeusYM-25. J Agric Biol Chem. 1983;47:83–88. [Google Scholar]
- 15.Heath I B. The role of actin in tip growth of fungi. Int Rev Cytol. 1990;123:95–127. [Google Scholar]
- 16.Hejgaard J, Jacobson S, Svendsen I. Two antifungal thaumatin-like proteins from barley grain. FEBS Lett. 1991;291:127–131. doi: 10.1016/0014-5793(91)81119-s. [DOI] [PubMed] [Google Scholar]
- 17.Henschen A. Analysis of cyst(e)ine residues, disulfide bridges, and sulfhydryl groups in proteins. In: Wittmann-Liebold B, Salnikow J, Erdmann V A, editors. Advanced methods in protein microsequence analysis. Berlin, Germany: Springer-Verlag; 1986. pp. 244–255. [Google Scholar]
- 18.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomycetes. Norwich, United Kingdom: The John Innes Foundation; 1985. [Google Scholar]
- 19.Hunter I S. Gene cloning in Streptomyces. Biochem Soc Trans. 1985;12:19–44. [Google Scholar]
- 20.Iijima R, Kurata S, Natori S. Purification, characterization, and cDNA cloning of an antifungal protein from hemolymph of Sarcophaga peregrina(flesh fly) larvae. J Biol Chem. 1993;268:12055–12061. [PubMed] [Google Scholar]
- 21.Kieser T, Melton R E. Plasmid pIJ699, a multi-copy positive-selection vector for Streptomyces. Gene. 1988;65:83–19. doi: 10.1016/0378-1119(88)90419-2. [DOI] [PubMed] [Google Scholar]
- 22.Kombrink E, Somssich I E. Pathogenesis-related proteins and plant defense. In: Esser K, Lemke P A, editors. The Mycota. 5A. Berlin, Germany: Springer-Verlag; 1997. pp. 107–128. [Google Scholar]
- 23.König W A, Hagenmaier H, Dähn U. Stoffwechselprodukte von Mikroorganismen, 146. Massenspektrometrische Identifizierung von 3-Cyclohexenylgylcin im Kulturfiltrat von Streptomyces tendae. Z Naturforsch Teil B. 1975;30:626–628. [Google Scholar]
- 24.Linthorst H J M. Pathogenesis-related proteins of plants. Crit Rev Plant Sci. 1991;10:123–150. [Google Scholar]
- 25.Marx F, Haas H, Reindl M, Stöffler G, Lottspeich F, Redl B. Cloning, structural organization and regulation of expression of the Penicillium chrysogenum pafgene encoding an abundantly secreted protein with antifungal activity. Gene. 1995;167:167–171. doi: 10.1016/0378-1119(95)00701-6. [DOI] [PubMed] [Google Scholar]
- 26.Millward-Sadler S J, Poole D M, Henrissat B, Hazlewood G P, Clarke J H, Gilbert H J. Evidence for a general role for high-affinity non-catalytic cellulose binding domains in microbial plant cell wall hydrolases. Mol Microbiol. 1994;11:375–382. doi: 10.1111/j.1365-2958.1994.tb00317.x. [DOI] [PubMed] [Google Scholar]
- 27.Möhrle V, Roos U, Bormann C. Identification of cellular proteins involved in nikkomycin production in Streptomyces tendae Tü901. Mol Microbiol. 1995;15:561–571. doi: 10.1111/j.1365-2958.1995.tb02269.x. [DOI] [PubMed] [Google Scholar]
- 28.Momany M, Westphal P J, Abramowsky G. Aspergillus nidulans swomutants show defects in polarity establishment, polarity maintenance and hyphal morphogenesis. Genetics. 1999;151:557–567. doi: 10.1093/genetics/151.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Money N P. Measurement of pore size in hyphal cell wall of Achyla bisexualis. Exp Mycol. 1990;14:234–242. [Google Scholar]
- 30.Murao S, Sato S, Muto N. Isolation of alkaline protease inhibitor from Streptomyces albogriseolusS-3253. Agric Biol Chem. 1972;36:1737–1744. [Google Scholar]
- 31.Nakaya K, Omata K, Okanashi I, Nakamura Y, Kolkenbrock H, Ulbrich N. Amino acid sequence and disulfide bridges of an antifungal protein isolated from Aspergillus giganteus. Eur J Biochem. 1990;139:31–38. doi: 10.1111/j.1432-1033.1990.tb19300.x. [DOI] [PubMed] [Google Scholar]
- 32.Nelson N. A photometric adaption of the Somogyi method for the determination of glucose. J Biol Chem. 1944;153:375–380. [Google Scholar]
- 33.Peczynska-Czoch W, Modarski M. Actinomycete enzymes. In: Goodfellow M, Williams S T, Modarski M, editors. Actinomycetes in biotechnology. San Diego, Calif: Academic Press, Inc.; 1988. pp. 219–283. [Google Scholar]
- 34.Poole D M, Hazlewood G P, Huskisson N S, Virden R, Gilbert H J. The role of conserved tryptophan residues in the interaction of a bacterial cellulose binding domain with its ligand. FEMS Microbiol Lett. 1993;106:77–84. doi: 10.1111/j.1574-6968.1993.tb05938.x. [DOI] [PubMed] [Google Scholar]
- 35.Raikhel N V, Lee H-I. Structure and function of chitin-binding proteins. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:591–615. [Google Scholar]
- 36.Roberts W K, Selitrennikoff C P. Zeamatin, an antifungal protein from maize with membrane-permeabilizing activity. J Gen Microbiol. 1990;136:1771–1778. [Google Scholar]
- 37.Rodriguez J, Deinhardt F. Preparation of a semipermanent mounting medium for fluorescent antibody studies. J Virol. 1960;12:316–317. doi: 10.1016/0042-6822(60)90205-1. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Schägger H, von Jagow G. Tricine sodium dodecyl sulphate-polyacrylamide gel electrophoresis for the separation of proteins in the range of 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 40.Schnellmann J, Zeltins A, Blaak H, Schrempf H. The novel lectin-like CHB1 is encoded by a chitin-inducible Streptomyces olivaceoviridisgene and binds specifically to crystalline a-chitin of fungi and other organisms. Mol Microbiol. 1994;13:807–819. doi: 10.1111/j.1365-2958.1994.tb00473.x. [DOI] [PubMed] [Google Scholar]
- 41.Somogyi M. Notes on sugar determination. J Biol Chem. 1952;195:19–23. [PubMed] [Google Scholar]
- 42.Spurlino J C, Lu G-Y, Quiocho F A. The 2.3-A resolution structure of the maltose- or maltodextrin-binding protein, a primary receptor of the bacterial active transport and chemotaxis. J Biol Biochem. 1991;266:5202–5219. doi: 10.2210/pdb1mbp/pdb. [DOI] [PubMed] [Google Scholar]
- 43.Stinzi A, Heitz T, Prasad V, Wiedemann-Merdinoglu S, Kaufmann S, Geoffroy P, Legrand M, Fritig B. Plant pathogenesis-related proteins and their role in defense against pathogens. Biochimie. 1993;75:687–706. doi: 10.1016/0300-9084(93)90100-7. [DOI] [PubMed] [Google Scholar]
- 44.Strickler J E, Berka T R, Gorniak J, Fornwald J, Keys R, Rowland J J, Rosenberg M, Taylor D P. Two novel Streptomycesprotein protease inhibitors. Purification, activity, cloning, and expression. J Biol Chem. 1992;267:3236–3241. [PubMed] [Google Scholar]
- 45.Tormo J, Lamed R, Chirino A J, Morag E, Bayer E A, Shoam Y, Steitz T A. Crystal structure of a bacterial family-III cellulose-binding domain: a general mechanism for attachment to cellulose. EMBO J. 1996;15:5739–5751. [PMC free article] [PubMed] [Google Scholar]
- 46.Vértesy L, Oeding V, Bender R, Zepf K, Nesemann G. Tendamistat (HOE 467), a tight-binding α-amylase inhibitor from Streptomyces tendae4158. Eur J Biochem. 1984;141:505–512. doi: 10.1111/j.1432-1033.1984.tb08221.x. [DOI] [PubMed] [Google Scholar]
- 47.Vigers A J, Wiedemann S, Roberts W K, Legrand M, Selitrennikoff C P, Fritig B. Thaumatin-like pathogenesis-related proteins are antifungal. Plant Sci. 1992;83:155–161. [Google Scholar]
- 48.von Heinje G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ward J M, Janssen G R, Kieser T, Bibb M J, Bibb M J. Construction and characterization of a series of multicopy promoter-probe plasmid vectors for Streptomyces using the aminoglycoside phosphotransferase gene from Tn5as indicator. Mol Gen Genet. 1986;203:468–478. doi: 10.1007/BF00422072. [DOI] [PubMed] [Google Scholar]
- 50.Wessels J G H. Developmental regulation of fungal cell wall formation. Annu Rev Phytopathol. 1994;32:413–437. [Google Scholar]
- 51.Woloshuk C P, Meulenhoff J S, Sela-Buurlage M, van den Elzen P J M, Cornelissen B J C. Pathogen-induced proteins with inhibitory activity toward Phytophthora infestans. Plant Cell. 1991;3:619–628. doi: 10.1105/tpc.3.6.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu G Y, Ong E, Gilkes N R, Kilburn D G, Muhandiram D R, Harris-Brandts M, Carver J P, Kay L E, Harvey T S. Solution of a cellulose-binding domain from Cellulomonas fimiby nuclear magnetic resonance spectroscopy. Biochemistry. 1995;34:6993–7009. [PubMed] [Google Scholar]
- 53.Yanish-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 54.Yun D-J, Bressan R A, Hasegawa P M. Plant breeding, rev. 14. New York, N.Y: Wiley; 1997. Plant antifungal proteins; pp. 39–88. [Google Scholar]
- 55.Yun D-J, Zhao Y, Pardo J M, Narasimhan M L, Damsz B, Lee H, Abad L R, Paino D'Urzo M, Hasegawa P M, Bressan R A. Stress proteins on the yeast cell surface determine resistance to osmotin, a plant antifungal protein. Proc Natl Acad Sci USA. 1997;94:7082–7087. doi: 10.1073/pnas.94.13.7082. [DOI] [PMC free article] [PubMed] [Google Scholar]