Abstract
Nanoparticle based sensors are good alternatives for non-enzymatic sensing applications due to their high stability, superior photoluminescence, biocompatibility and ease of fabrication, with the only disadvantage being the cost of the synthesis process (owing to the expensive precursors and infrastructure). For the first time, we report the design of an immunosensor employing streptavidin conjugated copper nanocluster, developed at a much lower cost compared to other nanomaterials like noble metal nanoparticles and quantum dots. Using in silico tools, we have tried to establish the dynamics of conjugation of nanocluster to the streptavidin protein, based on EDC-NHS coupling. The computational simulations have successfully explained the crucial role played by the components of the immunosensor leading to an efficient design capable of high sensitivity. In order to demonstrate the functioning of the Copper Nanocluster ImmunoSensor (CuNIS), HIV-1 p24 biomarker test was chosen as the model assay. The immunosensor was able to achieve an analytical limit of detection of 23.8 pg mL−1 for HIV-1 p24 with a linear dynamic range of 27–1000 pg mL−1. When tested with clinical plasma samples, CuNIS based p24 assay showed 100% specificity towards HIV-1 p24. With the capability of multiplexed detection and a cost of fabrication 100 times lower than that of the conventional metal nanoclusters, CuNIS has the potential to be an essential low-cost diagnostic tool in resource-limited settings.
Glutathione capped copper nanoclusters were bioconjugated to streptavidin protein using EDC-NHS coupling to engineer the Copper Nanocluster Immunosensor (CuNIS), which was tested for its efficiency by using HIV-1 p24 as the model analyte.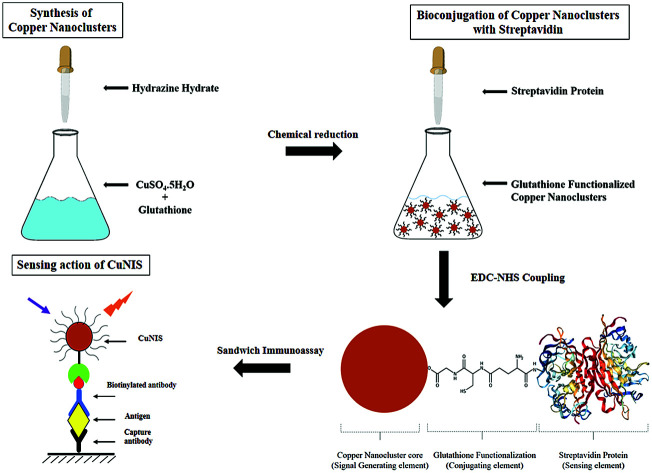
Introduction
A variety of nanomaterial-based assays have been developed for application in medical diagnostics, their key features being high specificity and sensitivity.1 Out of the various diagnostics assay configurations available, sandwich immunoassays format has been successfully modified to incorporate fluorescent nanomaterials as fluorophores.2 Consequently, a variety of nanoparticle labels ranging from carbon nanomaterials, quantum dots, noble metal nanoparticles, semiconductor nanoparticles, hybrid nanostructures, dye-doped silica nanoparticles, and metal oxide nanostructures have been deployed in the immunoassay and immunosensors as fluorescent probes.3
In the quest to develop fluorescent nanomaterials with high performance and efficiency, metal-based nanomaterials have been designed which have been successfully implemented in biodetection.4 As a result, significant attention has been focused on the utilization of metal nanoparticles in analytical chemistry as nanosized biosensing and immunosensing probes. In recent years, noble metal nanoparticles like gold, platinum, silver, etc. have been extensively used in the fabrication of novel biosensors due to their unique optical and electrocatalytic properties.5,6 However, while these noble metal nanomaterials are multifunctional, the economics of their extraction, synthesis, and application are not favorable for their application to routine and cost-effective diagnostic assays. This necessitates the quest for an inexpensive substitute which mitigates the costs involved while offering comparable detection sensitivities, similar or preferably better, compared to the currently used methods. We demonstrate in this intuitive study that a suitable alternative which satisfies all these requirements with low cost and high efficiency is copper.
Copper is one of the most widely used metals in the world. It is reported that the availability of copper in the crust is around 0.0068%.7 Copper-based nanomaterials have been widely used in different fields due to their environmentally friendly properties, economical cost of manufacturing, ease of obtainability and very high efficiency in sensing and electrocatalytic applications.8 Among the copper-based nanomaterials, the most well-known nanoparticles are Cu nanoparticles which are a prevalent fraction given their facile synthesis via the straightforward reduction of the low cost and readily available salts of copper.9 Further restriction of the size of these nanomaterials leads to the formation of atomic clusters of copper, which are called copper nanoclusters (CuNCs).10 Based on their tailored properties, copper nanoclusters are one of the most important materials derived from copper.
CuNCs are clusters of copper atoms of diameter up to 2 nm, which consist of a metallic core containing tens to hundreds of atoms protected by ligands.11 These CuNCs have exceptional physicochemical properties such as chemical stability, excellent photostability, ultra-small size and good biocompatibility which have attracted wide attention due to their applications in clinical diagnostics, nanotherapeutics, chemical catalysis, biosensing and bioimaging, luminescence-based spectroscopy and nano-optical devices.12 It is worth noting that due to quantum confinement effects, CuNCs have fascinating core size based optical and electronic properties, the most significant of them being photoluminescence which can be applied in diagnostics and monitoring of diseases.13 Combining the photoluminescence properties of CuNCs with the current need for low-cost, affordable and sensitive immunosensor, we report the development of a versatile copper nanocluster immunosensor.
In this study, we have computationally designed and experimentally tested the working of a low-cost Copper Nanocluster Immunosensor (CuNIS) in a clinical setting. The mechanistic rationale of the functioning of CuNCs vis-à-vis its conjugation was probed using simulated modeling where the factors influencing the interactions of CuNCs with streptavidin were studied using in silico tools. We attempted to rationalize the immunosensing by the CuNIS assay starting from the thermodynamics of formation of the bio-functionalized CuNC systems to their impact on the formation of streptavidin conjugated CuNCs. We also probed the role of surface functionalization by determining how glutathione functionalization affects the stability of streptavidin conjugated CuCNs. Finally, the impact of Cu nanocluster conjugated streptavidin on the interactions with biotin was evaluated. Based on the design studies, we have attempted the bioconjugation of CuNCs to the streptavidin protein which can be used for detection of any antigen or antibody using the corresponding biotin labeled antibody or antigen respectively. This immunosensor was fabricated and its potential for the detection of pathogens was evaluated with one of the most fatal diseases, AIDS which has HIV-1 as the causative virus.
In order to demonstrate the working of CuNIS in clinical settings, we chose HIV-1 p24, the biomarker for early detection of HIV, as the model analyte. A final comparison analysis of the sensitivity and cost against the other works of similar strategy highlights the significance of this endeavor. The outcome of this study will pave the way for the development of CuNC immunosensor based screening protocols with multiplexing capabilities, which can perhaps be implemented in the detection of biomarkers for any disease.
Materials and methods
Computational design of the copper nanocluster immunosensor
While many groups have reported the synthesis and application of copper nanocluster in fluorometric sensing, their implementation in immunosensing has never been explored. Thus, the aim of the present work was to develop a copper nanocluster based immunosensor which can offer comparable or better sensitivity compared to other nanoparticle based systems.
To establish the design that enhances the immunosensing capabilities via in silico studies, the representative Cux structures ranging from Cu1 to Cu57 were optimized to the least energy conformation using Gaussian 09.14 The neutral Cu atom, icosahedral Cu13, tetrahedral Cu20 and the icosahedral-core modeled Cu57 cluster were evaluated for their energetics in MOE computational software using the MMFX94 force field.15 We further simulated the consequences of an increase in glutathione groups present on the surface of the Cu13 as a capping agent to study the influence of glutathione functionalization on the streptavidin–CuNC interactions via the HEX molecular docking studies.16 Cu nanoclusters of varying sizes were docked with the streptavidin protein obtained from the protein data bank without its solvent and ligands using the Hex 8.0.0 software. The different docking conditions were set where the output searched for the top 100 best energy clusters, with shape only correlation type, 3D FFT mode, grid dimension of 0.6, distance range of 40, and a translation step of 0.8. These molecular docking studies also helped determine the impact of conjugated streptavidin in its interaction with the biotin that was visualized using PyMol to capture the active site residues in the protein.17
Synthesis and characterization of copper nanoclusters
The first step in the fabrication of the CuNIS is the preparation of copper nanoclusters. The synthesis of copper nanoclusters was achieved using the chemical reduction of copper sulphate.18 Briefly, 80 mg of glutathione and 7 mg of copper sulphate were dissolved in 5 mL of distilled water which over time resulted in the formation of a supramolecular hydrogel. The next step was reduction with hydrazine hydrate. Hydrazine hydrate was diluted by a factor of 25 times in distilled water and 400 mL of the diluted hydrazine hydrate was added to the hydrogel mixture. Subsequently, the solution was further stirred at 45 °C for 15 min which led to the formation of a purple colored solution indicating the formation of nanoclusters. These nanoclusters were precipitated using isopropanol, centrifuged at 10 000 rpm, separated from the supernatant and redispersed in distilled water and stored at 4 °C for further applications in immunoassay.
To evaluate the morphology and size of the CuNC dispersion, TEM characterization was carried out using a Joel 1400 Transmission Electron Microscope operated at 80 kV. The UV-visible absorbance and photoluminescence based characterization experiments and measurements were carried out using Shimadzu 2450 UV-vis spectrophotometer and SpectraMax M5 microplate reader. The samples were accordingly diluted by 100 folds for recording the spectra.
Bioconjugation of streptavidin to glutathione functionalized copper nanoclusters
The conjugation of glutathione functionalized copper nanoclusters requires activation of the carboxyl groups of glutathione which was performed using EDC/sulfo – NHS protocol.19 The stepwise protocol is detailed below.
(1) 20 mg of CuNC was dispersed in 10 mM PBS and washed in a NanoSep centrifugal ultrafiltration device (MWCO 300 kDa).
(2) After washing, CuNCs were mixed with EDC (10 mM) and sulpho-NHS (20 mM) in PBS buffer for 30 min. This step activates the carboxyls of glutathione on CuNCs.
(3) The activated CuNCs were washed with glycine buffer. 50 μL of streptavidin protein (1 mg mL−1) prepared in carbonate buffer was added to the activated CuNCs.
(4) After an incubation period of 24 hours at 37 °C followed by multiple washes with glycine buffer, streptavidin conjugated CuNCs, which are the functional Copper Nanocluster Immunosensors (CuNIS), were obtained which were diluted to 0.1 mg mL−1 concentration in PBS. The CuNIS was kept at 4 °C for storage for future experimentation.
Post the completion of the above procedure, the next step is confirmation of conjugation. A simple way to ascertain whether the process of bioconjugation has produced conjugated nanoparticles is the fluorescence polarization (FP) method. The I∥ and I⊥ were measured for the as produced nanoclusters in the fluorescence polarization mode by SpectraMax M5 plate reader and the FP ratio was calculated. Conjugation is confirmed by the larger value of fluorescence polarization for CuNIS in comparison to the unconjugated nanoclusters.
Application of CuNIS in HIV-1 p24 biomarker detection
The CuNIS was deployed for application in HIV-1 p24 sensing which is an early biomarker for Human Immunodeficiency Virus, the causative agent of HIV. The protocol followed was as follows.
(1) 55 μL of capture antibodies of concentration 2 μg mL−1, diluted in carbonate–bicarbonate buffer, were coated in a 96 well plate. The plates were left at 4 °C for incubation for 24 hours.
(2) Post incubation step, the coated wells were washed 5 times with wash buffer. This was followed by blocking the wells with 300 μL Casein Blocking Buffer (CBB) per well. Incubation was continued for 30 minutes at 37 °C to ensure the blocking of nonspecific adsorption sites.
(3) Stock antigen p24 solution was diluted with CBB to prepare different concentrations of antigen. 100 μL of the antigen was introduced in each microwell and incubated at 37 °C with stirring for 60 minutes.
(4) After washing the microwells 5 times with wash buffer, 100 μL of biotinylated detector antibody was added per well and incubated at 37 °C for 30 minutes.
(5) To this antibody–antigen–antibody complex, 100 μL per well of CuNIS was added and the mixture was incubated at 37 °C with vigorous stirring for a period of 30 minutes.
(6) A final round of washing was performed with PBST buffer for 5 times to avoid nonspecific interactions and reduce background noise. Finally, measurements of the fluorescence signal from CuNIS were obtained via the SpectraMax microplate reader (excitation at 394 nm and emission at 598 nm).
All experiments were performed in triplicate for statistical significance. The calibration curve was obtained by plotting the measured signal intensity values against the concentration of HIV-1 p24 antigen.
Preparation of clinical samples for immunoassay
Clinical samples studies were performed to demonstrate the performance of the immunosensor in the real world context. CuNIS based p24 test was evaluated using 30 HIV positive and 30 HIV negative patient plasmas (Disclaimer – The plasma samples used in the study were obtained from the patients with their informed consent). These samples were prepared using the procedure as follows.
Preparing plasma samples
They samples were confirmed to be HIV positive or negative HIV using commercially available HIV testing kits. To prepare the patient samples, 2 μL of plasma was diluted in 188 μL of casein block buffer and 10 μL of Triton X-100 (10% aq. sol.) was added.
HIV-1 p24 spiked plasma samples
To check the efficacy of our detection system, the first step involved testing HIV negative plasma samples spiked with known concentrations of p24 antigen. These spiked plasma samples were serially diluted with PBS solution to obtain the desired concentrations. The samples were then subjected to assay and the recovered concentrations were calculated based on the calibration curve. The cross-reactivity was also evaluated with 10 samples which were HIV negative but HBV and HCV positive respectively.
Results and discussion
Computational evaluation of design and performance of the CuNIS
The immunosensor is comprised of three major elements.
(1) Copper nanoclusters which function as the signal transducers.
(2) Glutathione which function as the conjugating element.
(3) Streptavidin which function as the sensing element.
Designing and optimization of the performance of every component of the CuNIS is vital to its functioning. In order to predict the outcome of the proposed design strategy, we have computationally analyzed each element of the sensor and estimated the role played by them in the efficiency of CuNIS.
In our quest to understand the energetics involved in the fabrication of streptavidin conjugated Cu nanoclusters, we evaluated the thermodynamics of the reaction pathway involved with conjugation of streptavidin to copper nanoclusters (Scheme 1). Gaussian 09 is the software of choice to build and optimize the structures across the three steps as noted by the following sequence:Cux-gluta + EDC + NHS → Cux-gluta-EDC + NHS → Cux-gluta-NHS
Scheme 1. Schematic illustration of the reaction pathway involved in EDC-NHS activation of glutathione functionalized copper nanoclusters.
The individual reactant and product energies for the aforementioned processes were determined for their minimized least energy MMFX94 states which are tabulated in Table 1. We observed that with the increase in nanocluster size, we obtain more stable conformations. This can be reasoned based on the fact that as the size of copper nanoclusters increase, there is an accompanying increase in the negative Gibbs free energy of the reaction (as seen from Table 1), making the reaction more spontaneous. Furthermore, we note that the combined free energy (energy gain in Table 1) is positive when the number of copper atoms is less. Thus, a larger copper core makes the reaction more feasible.
Comparison of free energies of formation of various species involved in the EDC-NHS activation of copper nanoclusters.
| Process steps | ΔE0 | ΔE1 | ΔE6 | ΔE13 | ΔE20 |
|---|---|---|---|---|---|
| Cux-glutathione + EDC + NHS | 44.47 | 42.367 | 43.204 | 954.901 | 835.448 |
| Cux-glutathione-EDC | 26.93 | 15.762 | 16.69 | 901.957 | 896.757 |
| Cux-glutathione-NHS | 46.307 | 23.112 | 23.92 | 863.514 | 825.632 |
| Energy gain (step 2) | 19.377 | 7.35 | 7.23 | −38.443 | −71.125 |
| Net energy gain (step 1 + step 2) | −25.09 | −35.02 | −35.974 | −993.34 | −906.57 |
Role of copper nanoclusters as signal transducers
Role of the Cux clusters in formation of streptavidin conjugated copper nanoclusters
The role of copper nanoclusters is paramount as established in the energetics recorded above. However, the direct implication of this behavior can be best observed in the consequent binding interactions of these nanoclusters with the streptavidin protein. The best-docked poses for the various nanoclusters interacting with the active site residues in the cavity can be seen in Fig. 1. We observe that as the cluster size increases, the ligand settles deeper into the cavity, thus establishing more contact area between the electrovalent metal and the charged residues in the protein active site for interactions. As the number of interactions grow, greater binding occurs between the ligand from the nanoclusters and the target protein. This results in larger clusters having stronger binding with the protein. This is further corroborated from the observed enhancement in the binding affinity as demonstrated in the Table 2.
Fig. 1. Comparison of the best docked poses for the different sized Cux-gluta-NHS clusters with the streptavidin protein for varying cluster sizes of (a) Cu1 (b) Cu6 (c) Cu13 (d) Cu20 (e) Cu57 as determined by HEX.
Binding affinity of the various copper clusters towards streptavidin as obtained from the molecular docking studies using HEX.
| Ligand | Cu1 | Cu6 | Cu13 | Cu20 | Cu57 |
| Hex docking score | −276.49 | −275.59 | −301.06 | −344.62 | −388.80 |
Role of glutathione as the conjugating element
Role of surface functionalization of glutathione in the conjugation of copper nanocluster to streptavidin
The first step in the nanocluster-streptavidin conjugation is EDC-NHS activation which is sequentially followed by reaction with streptavidin. The interaction of EDC and NHS with the nanoclusters happens through the electron density rich surface of the nanoclusters. In this regard, the role of glutathione in bioconjugation was evaluated further by altering the number of functionalized glutathione tails on the surface of CuNCs. This is once again only a representative simulation to mimic the actual systemic state as a reaction trend. Cu13 was chosen as the model nanocluster for determining the effect of the concentration of the glutathione tails given that it has the maximum energy gain in the thermodynamic profile recorded in Table 3. We observed from the table that the growing number of the glutathione-NHS tails present on the copper nanocluster remarkably enhanced the binding affinity. This can be explained based on the increasing opportunity for each gluta-NHS tail to interact with the active site residues in the streptavidin protein pocket. We also note the impact of their steric repulsions on the interactions as the number grows given that we have chosen a relatively small cluster.
Effect of the number of glutathione molecules attached to the nanocluster surface on the binding affinity of CuNC towards streptavidin.
| No of glutathione tails | HEX docking score |
|---|---|
| Cu13-glutathione-NHS | −317.76 |
| Cu13-2 glutathione-NHS | −388.24 |
| Cu13-4 glutathione-NHS | −301.06 |
| Cu13-6 glutathione-NHS | −344.62 |
The driving force in this conjugation is the large negative total free energy that leads to the stability of the conjugated product. Therefore, to determine the stability of the conjugation, the binding affinity of the metal nanocluster interactions with streptavidin was calculated in the presence and also the absence of glutathione across various cluster sizes. The marked difference observed, as recorded in Table 4, confirms the role played by glutathione in the enhanced binding affinity. This is especially pronounced for the copper nanoclusters of smaller size as compared to the larger one. We can therefore ascertain the need of the glutathione conjugation as an important factor for the designed immunoassay.
Binding affinity of copper nanoclusters towards streptavidin in the presence and absence of glutathione.
| Nanocluster | Binding affinity in absence of glutathione-NHS tail (kcal mol−1) | Binding affinity in presence of glutathione-NHS (kcal mol−1) |
|---|---|---|
| Cu1 | −37.70 | −276.49 |
| Cu6 | −114.95 | −275.59 |
| Cu13 | −137.85 | −301.06 |
| Cu20 | −185.36 | −344.62 |
| Cu57 | −294.07 | −388.80 |
Role of streptavidin as the sensing element
Role of streptavidin conjugated CuNCs in interaction with biotin
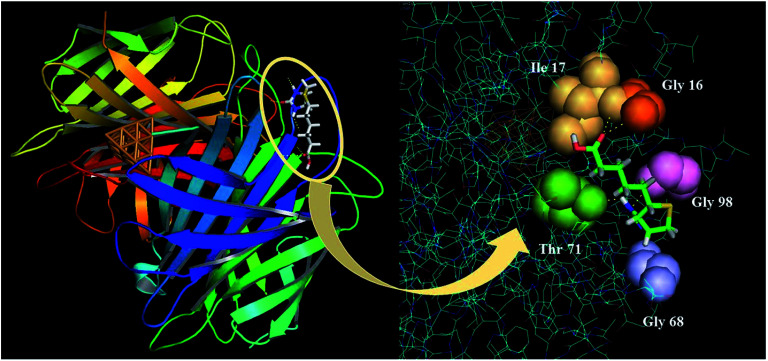
As compiled in Table 5, we observe that there is a significant contribution from metal nanocluster towards the interactions of streptavidin protein with biotin. From the Cu0 to the Cu1, we observe a major increase in the binding affinity of streptavidin by almost 40 units. The interacting amino acid residues in the active sites of the protein are largely glycine and threonine. Active sites of Gly 16, Gly 68, Gly 98 and Thr 71 interact with the carboxylic group in the biotin chain. We can clearly see that the optimal size of the nanocluster with highest interactions is Cu20, after which the interactions again return to the native state. This can be attributed to the steric size of the bigger metal nanocluster. The highest binding score could be explained by the presence of the additional interactions with the Ile 17 solely for this protein conformation. The best-docked pose for the interactions of the biotin with the streptavidin-Cu20 is captured using PyMol as shown in Fig. 2. The inset shows the binding interactions of biotin with the specific amino acid residues demonstrated as spheres.
The binding score of the interactions of the biotin and its corresponding active site residues for the best-docked poses of various Cux-streptavidin protein.
| Nanocluster | BA_biotin (kcal mol−1) | Interacting active site residues |
|---|---|---|
| Cu0 | −224.65 | Gly 16, Glu 98 |
| Cu1 | −264.78 | Gly 16, Gly 68, Gly 98, Thr 71 |
| Cu6 | −265.58 | Gly 16, Gly 68, Gly 98, Thr 71 |
| Cu13 | −254.13 | Gly 16, Gly 98, Thr 71 |
| Cu20 | −273.37 | Gly 16, Ile 17, Gly 68, Gly 98, Thr 71 |
| Cu57 | −234.20 | Gly 16, Glu 98 |
Fig. 2. The best docked pose for the interactions of the biotin with the streptavidin-Cu20 simulated through Pymol.
Based on this study, we can predict that as the particle size increases, the interactions also increase between streptavidin and the biotin ligand.
Synthesis and characterization of the glutathione functionalized copper nanoclusters
The synthesis of copper nanocluster required a technique that does not have much infrastructural requirement and can be performed with cost-effective reagents in a short reaction duration. This is achieved with the chemical reduction method. In this method, the copper salts are reduced using a reducing agent to create zero-valent copper atoms.20 In our set up, copper sulphate pentahydrate was the salt of choice. Glutathione was chosen as the capping agent.21
The mechanism of formation of copper nanoclusters involves hydrazine. The copper salt and glutathione are allowed to react first to form the hydrogel. Following this, hydrazine is added to the mixture and stirred for 15 minutes at 45 °C to reduce the hydrogel to generate the desired copper nanoclusters.18 The stirring which redistributes the heat in the system is vital, for it supports the formation of stable nanoclusters. The reaction is depicted in the equation below: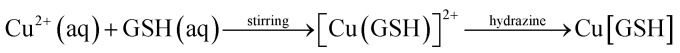
The TEM characterization of the copper nanocluster is presented in the ESI Fig. S1,† while the UV-vis absorbance and photoluminescence spectroscopy is presented in ESI Fig. S2 in the ESI.†
Experimental evaluation of conjugation of copper nanoclusters to streptavidin
The design of an immunosensor involves a sensing reaction which is essentially an interaction between the sensing elements on the sensor and analyte (directly or indirectly).22 In CuNC immunosensor, streptavidin was chosen as the sensing element which interacts with the biotin on the detector antibody. So the most important step of fabricating CuNC immunosensor is the conjugation of streptavidin to CuNCs.
The process of conjugation of streptavidin to copper nanocluster was carried out using the 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC)-N-hydroxysuccinimide (NHS) coupling mechanism which is well established.23 EDC reacts and activates the carboxylic acid groups present in glutathione leading to the formation of a reactive O-acylisourea intermediate. When the amino groups on the streptavidin attack the nanocluster, the intermediate is easily displaced allowing the primary amine groups to form an amide bond with the carboxyl groups form glutathione. The intermediate is an unstable by-product which doesn't undergo any further reaction with non-amine groups, thus ensuring that no other moieties interfere in this reaction.
The EDC-NHS methodology was chosen as the preferred method of bioconjugation due to two main reasons:
(1) The EDC-NHS method is a very well-known method which can be easily repeated in any setting given the protocol is followed. This allows for standardization of the protocol to a great extent allowing a minimal variation in sensitivity of the assay with every new batch of nanoclusters.
(2) The EDC-NHS establishes a very strong and stable amide bond between the conjugating entities. In the course of the conjugation, EDC-NHS are eliminated from the end product. This also helps in keeping the chemical identity of the conjugating molecules intact.
The as-obtained product of the coupling reaction can be ascertained for conjugation using the method of fluorescence polarization (FP). Fluorescent polarization is defined as a phenomenon where a fluorophore radiates light in different axes of polarization with different intensities. This anisotropy is dependent on the size and motion of the fluorophore, which can be tuned by the conjugation and surface functionalization.24 Thus, fluorescence polarization can be used to confirm the conjugation of biomolecules to fluorophores.
We, therefore, adopted the FP method to test the conjugation of streptavidin to copper nanocluster. The FP measurements were made in SpectraMax M5 microplate reader in the fluorescence polarization mode.
The change in the FP value corresponds to the increase in the size of the fluorophore after conjugation. The binding of the protein to the nanoclusters increases its dimensions, which slows down its molecular rotation compared to the nanoclusters which are unconjugated.25,26 This causes the FP value to be higher for streptavidin conjugated nanoclusters compared to unconjugated nanoclusters. I∥ and I⊥ values were obtained from the spectrophotometer. The calculation of the FP values was done based on the formula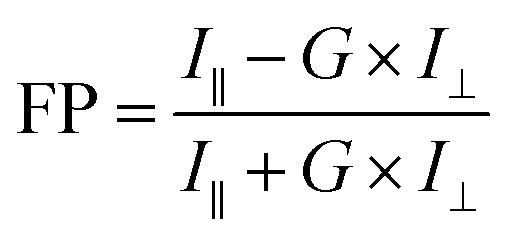
The corresponding data is tabulated in Table 6. As it can be inferred, the FP value increases with size of the particles. Thus, FP becomes an easy tool to monitor the functionalization of any moiety where an increase in the size is expected. The FP value of the copper nanocluster conjugated to streptavidin was higher than unconjugated copper nanocluster (CuNCs), which confirmed the conjugation of streptavidin to CuNCs.
Comparison of fluorescence polarization value of the unconjugated CuNCs with the conjugated CuNCs.
| Sample | I ∥ | I ⊥ | FP |
|---|---|---|---|
| Unconjugated CuNCs | 94 | 78 | 0.093 |
| Conjugated CuNCs (CuNIS) | 132 | 99 | 0.143 |
The as-obtained structure post-EDC-NHS based streptavidin conjugation protocol is the functional Copper Nanocluster ImmunoSensor (CuNIS) which can be tested for implementation in various immunodiagnostic applications.
Deployment of copper nanocluster immunosensor (CuNIS) in HIV diagnostics CuNIS based sandwich immunoassay for HIV-1 p24 detection
Acquired immune deficiency syndrome (AIDS) is one of the most severe health issues that humanity has come across.27 Since, there is no stipulated treatment that can cure this disease as yet, the only way to control and further avoid the transmission of this disease is by spreading social awareness at a community level and early detection of new infections at a clinical level.28 Clinical diagnostics utilize either RNA based PCR techniques or HIV-1 p24 antigen-based molecular biology techniques.29 In this study, we have chosen AIDS as the disease of the choice to test the performance of CuNIS. HIV-1 p24 biomarker has been chosen as the target analyte due to its abundant availability in the bloodstream when the infection is in early stages and concentrations of HIV antibodies is low.30
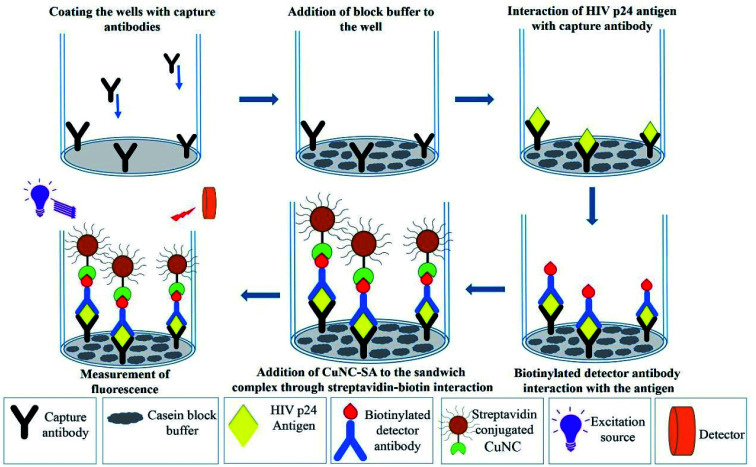
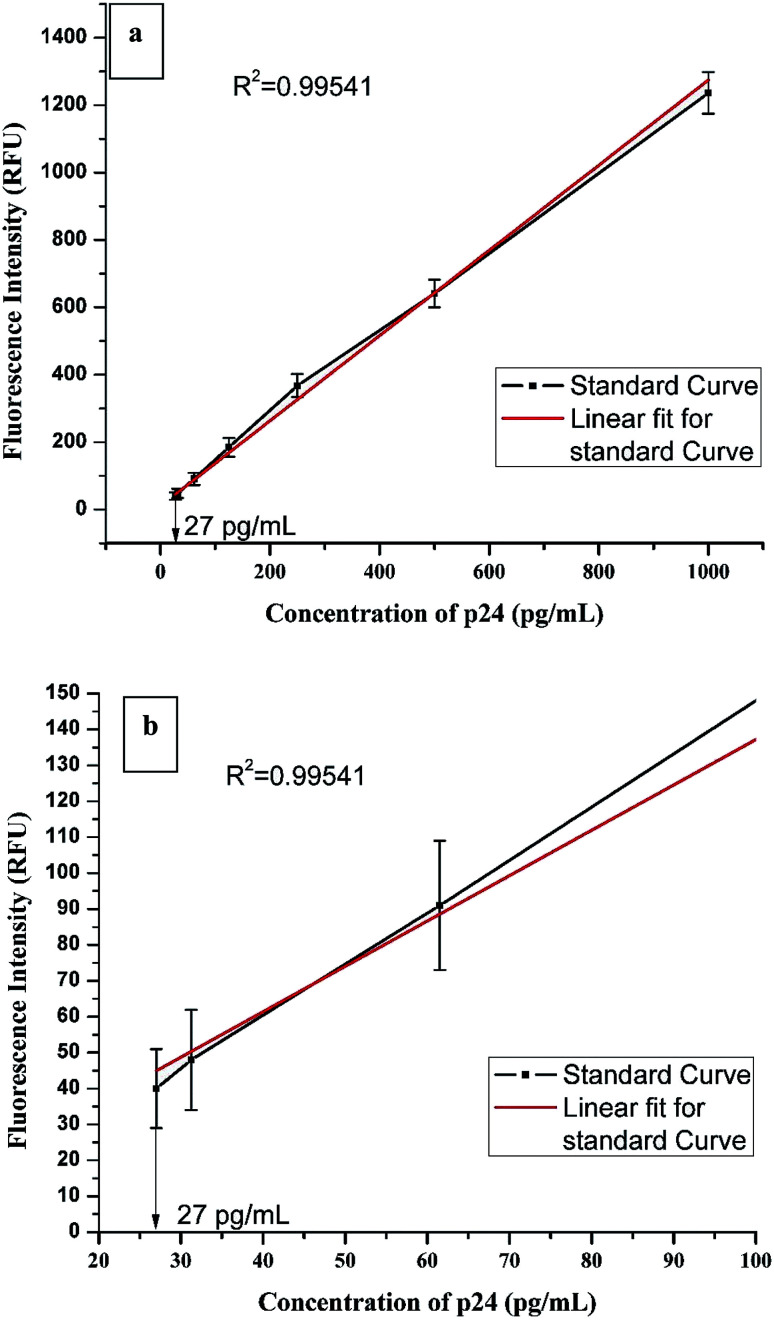
HIV-1 p24 biomarker test was chosen as the model assay to study the real world performance of CuNIS. The HIV-1 p24 immunoassay that we have adopted is a sandwich format assay wherein a sandwich of antibody–antigen–antibody complex is formed. The detector antibodies are biotinylated which interact with the streptavidin from CuNIS in the detection step (Scheme 2). The strong interactions between biotin and streptavidin attach the CuNIS to the sandwich complex which emits when excited with the excitation energy. The signal intensity measured using the SpectraMax M5 microplate reader is dependent on HIV-1 p24 concentration in the sample. The measured signal intensity was plotted against the concentration of p24 antigen to obtain the calibration curve as depicted in Fig. 3(a) and (b).
Scheme 2. Schematic illustration of CuNIS application for detection of HIV-1 p24 antigen.
Fig. 3. (a) Calibration curve of CuNIS based HIV-1 p24 assay (b) the calibration curve of CuNIS based HIV-1 p24 assay with resolved axis showing the lower end of the linear dynamic range.
The optimization of the concentration of CuNIS has been presented in the ESI through ESI Fig. S3.†
Analytical performance of CuNIS in HIV-1 p24 immunoassay
Statistically, the immunoassay employing CuNIS had a fluorescence cut-off value of 10.15 RFU (Relative Fluorescence Units).31 The limit of detection (LOD) for this assay was recorded to be 23.8 pg mL−1 in the dynamic range of 27–1000 pg mL−1. The signal to cutoff ratio was set to be 2 : 1 for positive samples.32 The linear correlation equation was as followsFluorescence intensity = 1.26 × concentration of p24 + 10.71
A good correlation was observed between p24 antigen concentrations and the signal count from CuNIS. The value of the coefficient of correlation was r2 = 0.99541, which leads to the inference that this assay can be explained as a linear dose dependent-model. Thus, it can be reported from this study that the CuNIS based assays could achieve detection sensitivity at the picogram per milliliter level. To our knowledge, this is one of the highest sensitivity reported using copper nanoclusters in biochemical sensing. This high value of sensitivity can be attributed to the utilization of highly specific streptavidin–biotin interaction as the sensing reaction. Table 7 compares the detection sensitivities of other copper nanocluster based detection methodologies with CuNIS based detection protocol.
Comparison of sensitivities of various other copper nanocluster based sensors.
Clinical serum sample analysis
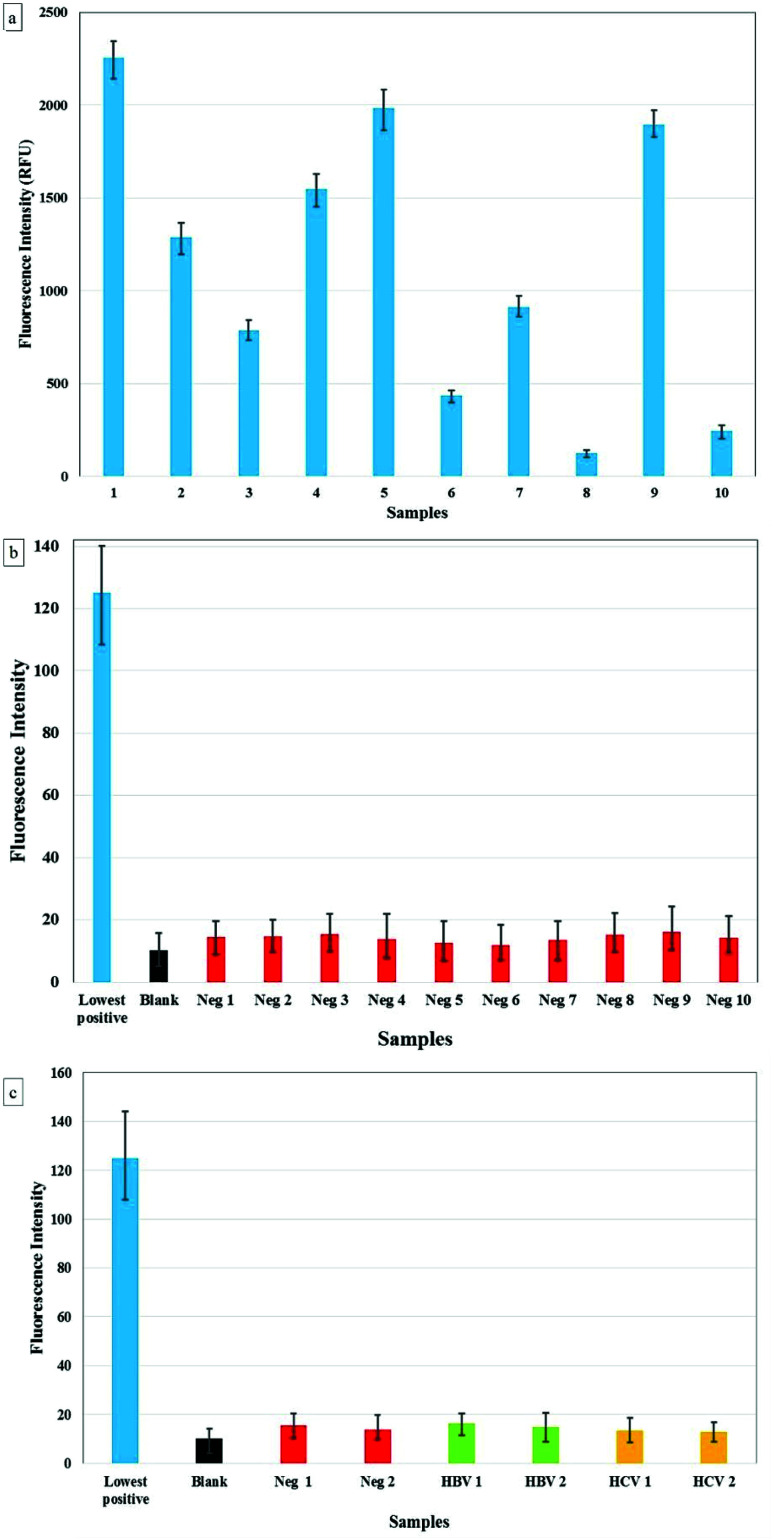
Furthermore, we tried to establish the effectiveness of CuNIS by testing its specificity. The testing with interfering biomolecules showed no effect on the performance of CuNIS, as depicted in ESI Fig. S4.† In order to test the recovery rate of the assays, HIV negative samples clinical plasma samples were spiked with HIV-1 p24 concentrations and the assay was performed to calculate the recovered concentrations based on the calibrations curve. The data of the study is presented in Table S1 of the ESI.†
The specificity was further assessed by performing the assay on samples from HIV positive individuals. In this regard, the plasma samples were first diluted 100 times prior to the analysis. We tested 30 samples confirmed as HIV positive and 30 samples confirmed as HIV negative (tested by 4th gen commercial ELISA kits) using CuNIS and no false negatives or false positive were observed. We tested the performance of immunosensor for cross-reactivity with 10 HBV positive and 10 HCV positive plasma samples, all of which were confirmed as HIV negative. We observed that the signal intensity from these samples was as low as the negative control. The comparison of the signal intensities from the samples is presented in Fig. 4(a–c). This allowed us to conclude that the CuNIS based p24 assay is very specific to p24 antigen and is not susceptible to interference from other pathogen biomarkers. As it is clearly evident, the CuNIS has very high specificity with negligible cross-reactivity.
Fig. 4. (a) Results for CuNIS based p24 assay for (a) 10 HIV +ve samples (b) 10 HIV −ve samples (c) HIV −ve, HBV +ve/HIV −ve, and HCV +ve/HIV −ve.
Comparison of cost of fabrication of various metal nanocluster based immunosensor
The key highlight of this research endeavor was the design and fabrication of a robust, selective and specific low-cost immunosensor. The other metal nanoclusters of gold, silver, and platinum are known to be expensive which limits their application in developing low-cost assays. But copper is economically priced, affordable to all margins of society and there are a wide range of precursors that are available to synthesize copper nanoclusters. Table 8 demonstrates that using copper nanoclusters can reduce the cost of the immunosensor by almost 10 times in comparison to silver and almost 100 times in comparison to the gold nanocluster, even while their performance is comparable. This could be an enormous gain, especially, when we are looking at a global disease. Thus, CuNIS could be the preferred choice for deployment of these diagnostic immunoassays especially in the resource-limited settings that are the crucial areas of the disease prevalence.
A cost comparison of different metal nanocluster immunosensors.
| S. No | Nanocluster | Precursor | Costa per milligramb ($ per US) |
|---|---|---|---|
| 1 | Platinum | Chloroplatinic acid hexahydrate | 1.33 |
| 2 | Gold | Tetrachloroauric(III) acid | 1.07 |
| 3 | Silver | Silver nitrate | 0.175 |
| 4 | Copper | Copper(ii) sulphate pentahydrate | 0.012 |
All the costs have been based on the commercial prices for the chemicals from Sigma-Aldrich.
The equivalent masses were calculated taking into account the amount of precursor used for each reaction.
Discussion
Over that last couple of decades, there have been many sensitive nanoparticle based biosensors which have been designed and proposed by research groups all over the world.39–41 Most of them demonstrated high sensitivity and excellent stability. But the cost, deployment and the shortcomings of the specific nanoparticle system remained a key challenge, making such nanoparticle based biosensors inadequate in many applications and preventing their widespread uses.42 Copper nanocluster based immunosensor has clearly demonstrated the ability to overcome these challenges. With an easy synthesis protocol, requiring bare minimum infrastructure, and a straightforward bioconjugation technique, CuNIS definitely opens the door to the world of low cost immunosensing.
The important facet of CuNIS is that it could achieve pg mL−1 sensitivity without the need for any highly specialized instruments or expensive chemicals and reagents, like those required in PCR based methods. Based on the performance of the CuNIS, it can be said that CuNIS is a robust and sensitive immunosensor because of its good signal strength and high signal to noise ratio. Also, there are no requirements for sophisticated equipments and specific storage conditions as CuNIS is not an enzyme-based immunosensor. Due to the high specificity of CuNIS, it could be developed into a multifunctional biosensor applicable in any detection protocol where biotinylated biomolecules are in use. This feature of CuNIS makes it possible to develop it as a universal label which can be used for detection of any pathogen just by changing the capture antibodies and detector antibodies specific to the disease biomarker. This allows for multiplexed detection of diseases as specific antibodies can be used specifically to detect a disease biomarker. This kind of multiplexed platform for clinical diagnostics can greatly benefit in reducing the wait time for an assay as multiple diseases can be detected simultaneously. Interestingly, there is no need for specific training of technicians to employ CuNIS in immunoassays as it is similar to traditional ELISA which is widely practiced across various laboratories and clinics.
One important observation is that the CuNIS, with a limit of detection of 23.8 pg mL−1 for HIV-1 p24, has a sensitive comparable to the current generation ELISA.43 The matter of fact is that this experiment was a pilot study to demonstrate the working of the conjugation and the immunosensing strategy of CuNIS. We have not really focused on optimization of the parameters to the best possible conditions. We believe that on further optimization of conditions such as the method of synthesis, bioconjugation protocol and optimization of concentrations of components involved, CuNIS can actually break the 5 pg mL−1 barrier. With further application of plasmonic enhancers and microfluidic platforms, we foresee that it can even detect the analyte at sub-picogram level. Presently, these modifications are being implemented by our group to enhance the sensitivity of CuNIS to the highest possible level.
Thus, we predict that on optimization and simplification, CuNIS could be developed into a rapid and ultra-sensitive immunodiagnostic tool for real-world diagnostics which can even be deployed in resource-constrained regions.
Conclusions
Fluorescent copper nanoclusters were synthesized to engineer the streptavidin labeled fluorescent copper nanoclusters. They were designed and evaluated for immunosensing via methods that highlighted the impact of conjugated copper nanocluster systems on the formation of streptavidin conjugated CuNC. It also established the role played by surface functionalization in the interaction of streptavidin conjugated CuCNs with biotin. The performance of CuNIS was tested by deployment in HIV-1 p24 assay, with HIV-1 p24 being the biomarker for AIDS-causing HIV virus. 100% specificity was observed in CuNIS based assay when tested with HIV negative plasma samples and those infected with other viruses such as HBV and HCV. Results also indicated that positive plasma samples were accurately detected as positive by CuNIS. The CuNIS was found to work efficiently in detecting p24 in the linear working range of 27 pg mL−1 to 1000 pg mL−1 with a limit of detection of 23.8 pg mL−1. This dynamic range observed here is very crucial for the detection of p24 antigen concentration within days after exposure up to 3 weeks, after which owing to seroconversion, the antigen becomes undetectable.
The presented results in this study pave the way for new methodologies that can be applied for the detection of various disease-causing agents. Based on our findings, we conclude that CuNIS can offer high sensitivity and specificity which can be deployed for improved point-of-care diagnostics at minimal fabrication cost.
Disclaimer
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Food and Drug Administration, U.S. Department of Health and Human Services.
Funding
No Applicable source of funding.
Data and materials availability
All data needed to evaluate the conclusions in the paper are present in the paper and/or the ESI.† Additional data related to this paper may be requested from the authors.
Conflicts of interest
The authors declare that they have no competing interests.
Supplementary Material
Acknowledgments
All authors are grateful to Bhagawan Sri Sathya Sai Baba for his constant inspiration and guidance. Aditya Kurdekar thanks DST for their support through the DST-INSPIRE Fellowship program, Ministry of Science and Technology, Government of India. The authors thank the Sri Sathya Sai Institute of Higher Medical Sciences for providing us the plasma samples for clinical analysis.
Electronic supplementary information (ESI) available: Characterization of copper nanoclusters, optimization of the CuNIS concentration, the effects of interfering biomolecules and recovery concentration study. See DOI: 10.1039/c9na00503j
References
- Burgess R. Stud. Health Technol. Inform. 2009;149:257–283. [PubMed] [Google Scholar]
- de la Escosura-Muñiz A. Parolo C. Merkoçi A. Mater. Today. 2010;13:24–34. doi: 10.1016/S1369-7021(10)70125-5. [DOI] [Google Scholar]
- Pei X. Zhang B. Tang J. Liu B. Lai W. Tang D. Anal. Chim. Acta. 2013;758:1–18. doi: 10.1016/j.aca.2012.10.060. [DOI] [PubMed] [Google Scholar]
- Doria G. Conde J. Veigas B. Giestas L. Almeida C. Assunção M. Rosa J. Baptista P. V. Sensors. 2012;12:1657–1687. doi: 10.3390/s120201657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Li G. Zang W. Wang L. Zhang X. Catal. Sci. Technol. 2014;4:3290. doi: 10.1039/C4CY00580E. [DOI] [Google Scholar]
- Lee K.-S. El-Sayed M. A. J. Phys. Chem. B. 2006;110:19220–19225. doi: 10.1021/jp062536y. [DOI] [PubMed] [Google Scholar]
- Technical data for Copper, http://periodictable.com/Elements/029/data.html
- Tamilvanan A. Balamurugan K. Ponappa K. Kumar B. M. Int. J. Nanosci. 2014;13:1430001. doi: 10.1142/S0219581X14300016. [DOI] [Google Scholar]
- Khan A. Rashid A. Younas R. Chong R. Int. Nano Lett. 2016;6:21–26. doi: 10.1007/s40089-015-0163-6. [DOI] [Google Scholar]
- Chen C.-Y. Chen C.-L. Wang C.-M. Liao W.-S. Nanomaterials. 2018;8:97. doi: 10.3390/nano8020097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. Chen B. Rogach A. L. Nanoscale Horiz. 2017;2:135–146. doi: 10.1039/C7NH00013H. [DOI] [PubMed] [Google Scholar]
- Feng J. Chen Y. Han Y. Liu J. Ma S. Zhang H. Chen X. ACS Omega. 2017;2:9109–9117. doi: 10.1021/acsomega.7b01052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y. Wei W. Chen W. Chin. Sci. Bull. 2012;57:41–47. doi: 10.1007/s11434-011-4896-y. [DOI] [Google Scholar]
- Frisch F. M. J., Trucks G. W., Schlegel H. B., Scuseria G. E., Robb M. A., Cheeseman J. R., Scalmani G., Barone V., Petersson G. A., Nakatsuji H., Li X., Caricato M., Marenich A., Bloino J., Janesko B. G., Gomperts R., Mennucci B., Hratchian H. P., Ort J. V. and Fox D. J., Gaussian 09 (Revision A.02), Gaussian Inc., Pittsburgh, PA, 2016 [Google Scholar]
- Molecular Operating Environment, https://www.chemcomp.com/MOE-Molecular_Operating_Environment.htm, accessed 9 September 2016
- Hex Protein Docking, http://hex.loria.fr/, accessed 6 September 2016
- Schrödinger L. L. C., The {PyMOL} Molecular Graphics System, Version∼1.8, 2015
- Wang C. C. Wang C. C. Xu L. Cheng H. Lin Q. Zhang C. Nanoscale. 2014;6:1775–1781. doi: 10.1039/C3NR04835G. [DOI] [PubMed] [Google Scholar]
- Hermanson G. T., Bioconjugate Techniques, Academic Press, Boston, 2013, p. xvii [Google Scholar]
- Wang C. Ling L. Yao Y. Song Q. Nano Res. 2015;8:1975–1986. doi: 10.1007/s12274-015-0707-0. [DOI] [Google Scholar]
- Iravani S. Korbekandi H. Mirmohammadi S. V. Zolfaghari B. Res. Pharm. Sci. 2014;9:385–406. [PMC free article] [PubMed] [Google Scholar]
- Luppa P. B. Sokoll L. J. Chan D. W. Clin. Chim. Acta. 2001;314:1–26. doi: 10.1016/S0009-8981(01)00629-5. [DOI] [PubMed] [Google Scholar]
- Fischer M. J. E., ed. J. N. Mol and E. M. J. Fischer, Amine Coupling Through EDC/NHS: A Practical Approach BT - Surface Plasmon Resonance: Methods and Protocols, Humana Press, Totowa, NJ, 2010, pp. 55–73 [Google Scholar]
- Moerke N. J., in Current Protocols in Chemical Biology, John Wiley & Sons, Inc., Hoboken, NJ, USA, 2009 [Google Scholar]
- Lea W. A. Simeonov A. Expert Opin. Drug Discovery. 2011;6:17–32. doi: 10.1517/17460441.2011.537322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluorescence Polarization, https://www.thermofisher.com/in/en/home/references/molecular-probes-the-handbook/technical-notes-and-product-highlights/fluorescence-polarization-fp.html, accessed 9 February 2016
- Colvin C. J. Glob. Health. 2011;7:31. doi: 10.1186/1744-8603-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley S. L. P & T : a peer-reviewed journal for formulary management. 2018;43(1):40–57. [PMC free article] [PubMed] [Google Scholar]
- Onen C. L., in AIDS in Africa, Kluwer Academic Publishers, Boston, 2007, pp. 297–321 [Google Scholar]
- Gray E. R. Bain R. Varsaneux O. Peeling R. W. Stevens M. M. McKendry R. A. AIDS. 2018;32:2089–2102. doi: 10.1097/QAD.0000000000001982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltzman C. L. Herzenberg J. E. Phillips W. A. Oncology. 1988;2:15. [PubMed] [Google Scholar]
- Shrivastava A. Gupta V. Chron. Young Sci. 2011;2:21. doi: 10.4103/2229-5186.79345. [DOI] [Google Scholar]
- Chen P.-C. Li Y.-C. Ma J.-Y. Huang J.-Y. Chen C.-F. Chang H.-T. Sci. Rep. 2016;6:24882. doi: 10.1038/srep24882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. Zhang H. Xu X. Zhou J. Lu F. Zhang Z. Hu Z. Luo J. Spectrochim. Acta, Part A. 2018;202:65–69. doi: 10.1016/j.saa.2018.05.030. [DOI] [PubMed] [Google Scholar]
- Zhou D.-L. Huang H. Wang Y. Anal. Methods. 2017;9:5668–5673. doi: 10.1039/C7AY02035J. [DOI] [Google Scholar]
- Wang H.-B. Li Y. Bai H.-Y. Liu Y.-M. Anal. Lett. 2018;51:2868–2877. doi: 10.1080/00032719.2018.1454457. [DOI] [Google Scholar]
- Momeni S. Ahmadi R. Safavi A. Nabipour I. Talanta. 2017;175:514–521. doi: 10.1016/j.talanta.2017.07.056. [DOI] [PubMed] [Google Scholar]
- Mao X. Liu S. Yang C. Liu F. Wang K. Chen G. Anal. Chim. Acta. 2016;909:101–108. doi: 10.1016/j.aca.2016.01.009. [DOI] [PubMed] [Google Scholar]
- Sanvicens N. Pastells C. Pascual N. Marco M.-P. TrAC, Trends Anal. Chem. 2009;28:1243–1252. doi: 10.1016/j.trac.2009.08.002. [DOI] [Google Scholar]
- Holzinger M. Le Goff A. Cosnier S. Front. Chem. 2014;2:63. doi: 10.3389/fchem.2014.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farka Z. Juřík T. Kovář D. Trnková L. Skládal P. Chem. Rev. 2017;117:9973–10042. doi: 10.1021/acs.chemrev.7b00037. [DOI] [PubMed] [Google Scholar]
- Sin M. L. Y. Mach K. E. Wong P. K. Liao J. C. Expert Rev. Mol. Diagn. 2014;14:225–244. doi: 10.1586/14737159.2014.888313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystryak S. Santockyte R. JAIDS, J. Acquired Immune Defic. Syndr. 2015;70:109–114. doi: 10.1097/QAI.0000000000000726. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.