Abstract
The fungal cell wall has generated interest as a potential target for developing antifungal drugs, and the genes encoding glucan and chitin in fungal pathogens have been studied to this end. Mannoproteins, the third major component of the cell wall, contain mannose in either O- or N-glycosidic linkages. Here we describe the molecular analysis of the Candida albicans homolog of Saccharomyces cerevisiae MNN9, a gene required for the synthesis of N-linked outer-chain mannan in yeast, and the phenotypes associated with its disruption. CaMNN9 has significant homology with S. cerevisiae MNN9, including a putative N-terminal transmembrane domain, and represents a member of a similar gene family in Candida. CaMNN9 resides on chromosome 3 and is expressed at similar levels in both yeast and hyphal cells. Disruption of both copies of CaMNN9 leads to phenotypic effects characteristic of cell wall defects including poor growth in liquid media and on solid media, formation of aggregates in liquid culture, osmotic sensitivity, aberrant hyphal formation, and increased sensitivity to lysis after treatment with β-1,3-glucanase. Like all members of the S. cerevisiae MNN9 gene family the Camnn9Δ strain is resistant to sodium orthovanadate and sensitive to hygromycin B. Analysis of cell wall-associated carbohydrates showed the Camnn9Δ strain to contain half the amount of mannan present in cell walls derived from the wild-type parent strain. Reverse transcription-PCR and Northern analysis of the expression of MNN9 gene family members CaVAN1 and CaANP1 in the Camnn9Δ strain showed that transcription of those genes is not affected in the absence of CaMNN9 transcription. Our results suggest that, while the role MNN9 plays in glycosylation in both Candida and Saccharomyces is conserved, loss of MNN9 function in C. albicans leads to phenotypes that are inconsistent with the pathogenicity of the organism and thus identify CaMnn9p as a potential drug target.
Candida albicans is a dimorphic, opportunistic fungal pathogen that is responsible for the majority of fungal infections in immunocompromised hosts (33). A great deal of attention has focused on understanding host-C. albicans interactions, in particular the elements that promote the virulence of the organism (14, 47). Putative virulence factors include morphogenesis (56), proteinase production (32), phenotypic switching (57), and adherence to host cells (8, 10). A number of systems which allow C. albicans to adhere to host tissues have been identified (reviewed in references 8 and 10). In most instances, cell wall-associated mannoproteins bind to specific components on host cells (8–10, 27). Cell wall mannan has also been implicated in other pathogenicity-related processes, including immunogenicity (10). Therefore, understanding the molecular biology of glycosylation, as well as that of other processes that relate to biosynthesis of the Candida cell wall, is critical to elucidating the pathogenicity of C. albicans. In turn, such investigations may facilitate the identification of genes which encode potential targets for improved antifungal therapy. To this end, information obtained from studies of these systems in Saccharomyces cerevisiae has proven essential in facilitating such research in Candida.
The biosynthesis and structure of the mannan outer chain in S. cerevisiae have been studied extensively by Ballou and coworkers (2). Those investigations led to the isolation of a number of S. cerevisiae mannoprotein, or mnn, mutants (reviewed in references 2, 3, and 23), several of which show defects in glycosylation of secreted proteins (2–4). These mutants also exhibit phenotypes characteristic of defects in cell wall biosynthesis and/or assembly, including poor cell growth, flocculation in liquid media, clumpy growth on solid media, osmotic sensitivity, and aberrant sporulation. Of these mutants, the mnn9 strain suffers the most serious glycosylation defect. In this mutant, N-linked chains in which one α-1,6-mannose is attached to the core oligosaccharide are formed but further addition is blocked. Limited addition of α-1,2- and α-1,3-linked mannose residues results in the formation of a Man13GlcNAc2 structure (60).
The MNN9 gene has been cloned (64), and an MNN9 gene family in S. cerevisiae has been identified based on sequence homology. The other members of this family include VAN1 and ANP1. VAN1 was isolated by complementation of vanadate-resistant mutant van1-18 (28) and has been shown to be allelic to vrg7, another vanadate-resistant mutant isolated in an independent screen (5). That screen also identified vrg mutants allelic to mnn9 (vrg6), as well as other mnn mutants. Like the mnn9 strain, van1 strains underglycosylate secreted invertase (5, 29). In addition, mnn9 and van1 strains have similar growth and sporulation defects. Both strains also show sensitivity to the aminoglycoside antibiotic hygromycin B, as well as resistance to sodium orthovanadate (5, 15, 28, 29). The third member of the MNN9 gene family is ANP1, which was originally identified within a cluster of genes whose deletion resulted in sensitivity to the chloramphenicol breakdown product amino nitrophenol propanediol (39). ANP1 has since been shown to complement gem3. The gem3 mutant was isolated in a screen for mutants that mislocalize a reporter protein that should reside in an early Golgi compartment (12). The anp1 (gem3) strain also grows slowly, is osmotically sensitive, exhibits clumpy growth in liquid and solid media, is sensitive to hygromycin and resistant to vanadate, and is defective in outer-chain glycosylation (12). All three genes encode putative type II membrane proteins. Disruption of the individual genes does not result in lethality (5, 12), but deletion of both ANP1 and VAN1 renders the doubly disrupted strain inviable (12). Recently, Jungmann and Munro (26) have shown that Mnn9p, Van1p, and Anp1p colocalize in the cis Golgi apparatus subcompartment in two separate complexes, both of which contain Mnn9p (26). The separate complexes were isolated by immunoprecipitation, and both were shown to have mannosyltransferase activity. One of the isolated complexes contains Van1p and Mnn9p alone, while the other complex contains Mnn9p, Anp1p, and two other tightly associated proteins, Hoc1p and an uncharacterized protein encoded by open reading frame (ORF) YJL183w (26). Hoc1p (45) is a Golgi apparatus protein with homology to Och1p (43), the mannosyltransferase which initiates mannan backbone synthesis. However, deletion of HOC1 does not lead to defects in protein glycosylation (45). The Yjl183w protein has clear homologies to Mnn10p (16) and an α-1,2-galactosyltransferase in Schizosaccharomyces pombe (13) and has since been renamed Mnn11p (26). The enzymatic studies with the partially purified complexes show they synthesize both α-1,6- (backbone) and α-1,2 (side chain)-mannose linkages, suggesting that the complexes have multiple enzymatic activities (26). While the specific catalytic functions of each of these proteins are yet to be resolved, it is suggested that in addition to the role they play in outer-chain synthesis one or more of the enzymes in these complexes may be involved in other processes responsible for maintaining the organization and function of the secretory pathway (25, 46). Additionally, MNN9 gene family members have been identified in screens for genes involved in the cell cycle-regulated progression of polarized growth in S. cerevisiae (41, 54).
The C. albicans genome is diploid, yet no sexual cycle has been reported for this organism (31). Accordingly, genetic manipulation of this pathogen has been difficult in the past. In recent years, techniques developed for targeted gene disruption in Saccharomyces (1) have been adapted for use in Candida (18). This methodology has led to the successful disruption of many C. albicans genes, including genes involved in glycosylation (7, 53, 59), maintenance of cellular integrity (44, 48), and assembly and function of the cell wall (6, 20, 36, 40).
Here, we report the identification of an MNN9 gene family in C. albicans and describe the isolation and characterization of one gene member of that family, C. albicans MNN9. Disruption of both copies of C. albicans MNN9 results in a phenotype suggestive of severe cell wall defects, including poor growth, flocullation in liquid medium, abnormal hyphal formation, and sensitivity to β-glucanase. Additionally, C. albicans Δmnn9 strains are sensitive to hygromycin B and resistant to vanadate. These results suggest that C. albicans MNN9 encodes a protein involved in glycosylation and/or secretion of cell wall-associated mannoproteins. In turn, the severe phenotypes exhibited by the CaΔmnn9 strain and the fact that no mammalian homologs of MNN9 gene family members have been identified suggest that C. albicans MNN9 would represent an attractive target for new antifungal drugs.
MATERIALS AND METHODS
Strains and growth conditions.
Yeast and bacterial strains used in this study are listed in Table 1. C. albicans was routinely grown in YPD medium (1% yeast extract, 2% peptone, 2% glucose) or SD minimal medium (0.67% Bacto yeast nitrogen base, 2% glucose) with shaking at the appropriate temperature. C. albicans ura revertants were selected on SD agar plates containing fluoro-orotic acid (5-FOA; 1.0 mg/ml). Uridine (50 μg/ml) was added to media when ura strains were cultured. Uridine and 5-FOA were purchased from Sigma Chemical Co. (St. Louis, Mo.). Agar (2%) was added to solid media. C. albicans mnn9 strains were also propagated in liquid media supplemented with either 0.5 M KCL or 0.5 M sorbitol for osmotic stabilization.
TABLE 1.
Strains used in this study
| Organism | Strain | Genotype | Source or reference |
|---|---|---|---|
| C. albicans | ATCC 10261 | Wild type | ATCC |
| SC5314 | Wild type | 19 | |
| CAI4 | ura3::imm434/ura::imm434; congenic to SC5314 | 18 | |
| SSCA-2 | mnn9::hisG-URA3-hisG/mnn9::hisG Δura3::imm434/Δura3::imm434 | This work | |
| SS19-4 | Δmnn9::hisG/Δmnn9::hisG Δura3::imm434/Δura3::434 | This work | |
| E. coli | DH5αmcr | K-12 (lacZYA-argF)U169 supE44 thi-1 recA1 end1 hsdR17 gyrA relA1 (φ80lacZΔM15) | 22 |
Hyphal growth was induced by use of a temperature-pH regimen in Lee's medium (34) for Northern analysis. Germ tube formation was also induced by culturing in YPD liquid medium containing fetal bovine serum (10%) at 37°C, or by culturing in RPMI 1640 medium at 37°C for experiments examining the yeast-to-hypha transition.
Escherichia coli strains were grown at 37°C in Luria-Bertani (LB) broth supplemented with ampicillin (100 μg/ml), when appropriate. Agar was added to 1.5% for LB plates.
Plasmids and genomic libraries.
The plasmid pLV4, carrying the S. cerevisiae actin gene on a 1.4-kb BamHI/NotI fragment, was kindly provided by Letty Vega (Massachusetts Institute of Technology [MIT], Cambridge, Mass.). PCR-Script (+) (Stratagene, La Jolla, Calif.) was used for subcloning and sequencing the PCR products. Construction of the C. albicans ATCC 10261 HindIII and EcoRI genomic libraries in pUC18 has been described previously (38). The methodology utilized for library screening was as previously performed (38).
Plasmid p8A was isolated after screening the genomic library and contains a 3.5-kb HindIII insert carrying C. albicans MNN9 (CaMNN9). The plasmid p8A-KpnIΔ was generated by digesting p8A to completion with KpnI. The digest was then diluted and religated before being used to transform E. coli. p8A-KpnIΔ was used for subsequent sequence analysis and construction of the mnn9 deletion construct.
The Camnn9 deletion construct was generated by digesting p8A-KpnIΔ with ClaI and EcoRV to remove 0.6 kb of the coding sequence. The ClaI site was filled in with Klenow polymerase before BglII linkers were ligated to the blunt ends. The 4.0-kb BamHI/BglII fragment from p5921 (20), carrying the hisG URA3 hisG cassette (18), was inserted into the BglII sites of the modified p8A-KpnIΔ to yield the plasmid pM9-8. Ultimately, pM9-8 was digested with HindIII and KpnI, ethanol precipitated, and used to transform the C. albicans uraΔ strain, CAI4 (18).
Recombinant DNA techniques, PCR, and DNA sequencing.
Transformation of E. coli was performed by the calcium chloride method (22). C. albicans was transformed by the lithium acetate protocol described by Ito et al. (24).
Plasmid DNA was prepared by alkaline lysis (51) or by using Miniprep columns (Qiagen, Chatsworth, Calif.). C. albicans genomic DNA for Southern analysis was isolated by standard protocols (55). Total RNA was isolated as previously described by McCreath et al. (38) or by glass bead disruption and extraction in Trizol (GIBCO/BRL, Rockville, Md.). Genomic DNA from C. albicans Miniprep columns was prepared by the spheroplast method described by Guthrie and Fink (21). Northern and Southern transfer and hybridization conditions were as previously described (38), except that Northern hybridizations were at 60°C in Church's buffer (0.5 M NaH2PO4 [pH 7.0], 1.0 mM EDTA [pH 8.0], 7% sodium dodecyl sulfate (SDS), 1% bovine serum albumin). DNA probes for hybridization were random prime labeled (17) with [α-32P]dCTP by following the manufacturer's instructions (Boehringer Mannheim, Indianapolis, Ind.). Densitometry of X-ray film images was done with a FluorS MultiImager (Bio-Rad).
PCR amplification of C. albicans genomic DNA was performed with AmpliTaq (Perkin-Elmer Cetus, Norwalk, Conn.) and carried out in an ERICOMP twin-block thermocycler. The degenerate primers used were 5′TGGGTI(C/T)(A/T)ITGGI(G/T)IGA(C/T)G(C/T)TGA3′ and 5′(C/T)(C/T)TI(G/C)C(A/G)AAI(G/C)C(C/T)TCIGT(C/T)TC3′, where I indicates inosine. The primers were designed based on the nucleotide sequence encoding conserved regions in the carboxy termini of the S. cerevisiae Mnn9, Van1, and Anp1 proteins and were synthesized by the Biopolymer Laboratory at MIT. The conditions of amplification were denaturation at 95°C for 1 min, annealing at 50°C for 1 min, and extension at 72°C for 1 min.
Double-stranded DNA sequencing was carried out by the chain termination method (52) with a cycle sequencing kit (Epicentre Technologies, Madison, Wis.). C. albicans MNN9 was progressively sequenced by extension of synthetic oligonucleotide primers (Biopolymer Laboratory, MIT) designed based on a sequence derived from previous reactions. DNA sequence analysis was performed with DNAStar. Comparison of C. albicans MNN9 nucleotide and deduced amino acid sequences to sequences present in GenBank and EMBL databases was carried out with BLAST (National Center for Biotechnology Information).
Sensitivity to hygromycin and sodium orthovanadate.
Stock solutions of 100 mM sodium orthovanadate (Sigma) and 10 mg of hygromycin B (Sigma)/ml were prepared in distilled water and filter sterilized. Fresh stock solutions were diluted into sterile YPD medium containing 2% agar for solid medium. C. albicans yeasts were grown overnight at 30°C before the cultures were equilibrated with fresh medium to an optical density at 600 nm (OD600) of 0.5 (approximately 6.0 × 106 cells/ml). Five microliters of each diluted culture was struck onto quadrants on agar plates containing various concentrations of hygromycin or sodium orthovanadate. The drug concentrations tested were 10, 50, 100, 200, 300, 350, and 400 μg of hygromycin/ml and 1, 2, 3, 4, 5, 6, 7, 10, 15, 20, and 25 mM vanadate. Growth was scored after 3 and 7 days of incubation at 30°C.
Analysis of cell wall carbohydrate.
C. albicans yeast cells (5.0-ml cultures) were labeled with UDP-[14C]glucose (7.5 μCi), and cell wall polysaccharides were fractionated and quantitated as described by Castro et al. (11).
β-Glucanase sensitivity.
β-1,6-glucanase was purified from a commercial enzyme preparation (cell wall-lysing enzyme; L-2265; Sigma) from Trichoderma harzianum by substrate affinity chromotography and preparative isoelectric focusing with the Rotofor system (Bio-Rad). The enzyme is now commercially available from BioMarin Pharmaceutical (Novato, Calif.). Both the crude enzyme preparation and pure β-1,6-glucanase were used to determine the sensitivities of C. albicans wild-type and mutant strains, as well as the release of cell wall proteins from those strains.
Sensitivities to crude β-1,3-glucanase from T. harzianum (Sigma; L-2265) and pure β-1,6-glucanase from T. harzianum were determined by a modified version of the protocol described by Ram et al. (49). Cells were harvested during logarithmic growth (approximately 1.5 × 107 to 2.0 × 107 cells/ml) and washed with 50 mM sodium acetate buffer (pH 5.0). Cells were then suspended in the same buffer containing either 1 mg of crude β-1,3-glucanase or 15 μg of purified β-1,6-glucanase and incubated at 37°C. The OD530 was measured at the onset of incubation and again 2 h later. The decrease in OD530 over time reflects the percentage of lysed cells. Cells were also incubated in the same buffer without the addition of enzyme as controls. Mutants were classified as glucanase sensitive when there was greater than 40% reduction in the measured OD530.
Isolation and analysis of cell wall proteins.
Yeast cells from 100-ml cultures were harvested during logarithmic growth (1.25 × 107 to 2.5 × 107 cells/ml) and washed three times with 10 mM sodium acetate buffer (pH 5.0) containing 1 mM phenylmethylsulfonyl fluoride (PMSF; Sigma). The cells were then resuspended in 1.0 ml of the same buffer before the addition of glass beads (0.45 mm in diameter) to the meniscus. Cells were disrupted by vortexing three times for 1 min each, interspersed with cooling on ice for 30 s. The cell lysate was separated from the glass beads by centrifugation and collected. The glass beads were then washed two times with 1 M NaCl, and the washes were pooled with the lysate. Cell walls were pelleted by centrifugation at 1,000 × g for 10 min, washed two times with 1 M NaCl containing PMSF, and then stored in PMSF at −20°C.
Noncovalently bound proteins were extracted from cell wall preparations by incubation in Tris-HCl (pH 7.8) containing 2% SDS, 100 mM EDTA, and 40 mM β-mercaptoethanol for 5 min at 100°C. The cell wall fraction was pelleted by centrifugation for 5 min at 10,000 × g, and the supernatant containing SDS-soluble proteins was collected.
Glucanase-extractable proteins were isolated from the SDS-extracted cell wall fractions as follows. SDS-treated cell walls were washed several times with 1 mM PMSF to remove residual SDS. Cell walls were then incubated with pure β-1,6-glucanase from T. harzianum (0.4 g wet weight of cell walls) in 50 mM sodium acetate buffer (pH 5.0) containing 1 mM PMSF at 37°C for 2 h. As a control, cell walls were incubated in the same buffer without the addition of enzyme. The reaction mixture was centrifuged for 5 min at 10,000 × g, and the supernatant was analyzed for protein release after glucanase treatment. SDS- and glucanase-extractable proteins were separated by linear-gradient (4 to 20%) SDS-polyacrylamide gel electrophoresis (PAGE) and visualized by silver staining (Daiichi silver stain; Integrated Separation Systems, Natick, Mass.).
Photomicroscopy.
Cells were mounted on glass slides and photographed with a ×40 objective with Nomarski optics on a Nikon Diaphot microscope.
Electrophoretic karyotyping.
C. albicans chromosomes were prepared and separated by pulsed-field electrophoresis as previously described (63). The chromosomal blot was kindly provided by Joy Sturtevant (Georgetown University Medical Center, Washington, D.C.). The chromosomal blot was hybridized with the internal ClaI/EcoRV fragment of CaMNN9 derived from p8A-KpnIΔ. Hybridization was performed in Church's buffer at 60°C before the filter was washed in 2× SSC (1× is 0.15 M NaCl plus 0.015 M sodium citrate) containing 0.1% SDS at 60°C for 15 min.
RT-PCR.
Total RNA was isolated from wild-type and Camnn9Δ strains grown as yeast. Cultures (50 ml) were centrifuged, and the collected cells were washed with sterile water and resuspended in Trizol (GIBCO/BRL) at 1.0 ml per 2.0 g of wet cells. Cells were disrupted by being vortexed in the presence of glass beads (0.45 nm in diameter), and RNA was extracted in accordance with the manufacturer's instructions. RNA samples were precipitated with 0.5 M NaCl and 2 volumes of ethanol before RNA concentrations were determined by measuring the OD260. Total RNA samples (1.0 to 2.0 μg) were treated with DNase (amplification grade; GIBCO/BRL) by following the manufacturer's instructions. Total DNase reaction mixtures (10 μl) were subjected to first-strand cDNA synthesis with a Superscript II kit (GIBCO/BRL) and internal 3′ primers specific for either C. albicans MNN9, VAN1 (5′GCGCTCGAGATCACCATTACGATCCGG3′ [57a]), or ANP1 (5′AGGAGGTTTACAATTGGC3′ [57a]). Reverse transcription (RT) reaction mixtures (2 to 5 μl) were utilized directly as templates in PCR mixtures (100 μl) containing 1× PCR buffer, 25 mM MgCl2, 0.8 mM dNTP mixture, 1.0 μM concentrations of each 3′ (above) and 5′ gene-specific primer (CaVAN1, 5′GCGCCATGGCCATCTTATATTGAAAGCG3′, and CaANP1, 5′AAATCACATAATGAGGTAACC3′ [57a]), and 4 U of BIO-X-ACT DNA polymerase (ISC:Bioexpress, Kaysville, Utah). PCR consisted of denaturation at 95°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1 min. Amplification products were separated by electrophoresis on 1.2% Separide (GIBCO/BRL) agarose gels, stained with ethidium bromide, and visualized under UV light.
Nucleotide sequence accession number.
The C. albicans MNN9 nucleotide sequence has been deposited in the GenBank and EMBL databases under the accession no. U63642.
RESULTS
Isolation and analysis of the C. albicans MNN9 gene.
Degenerate primers were designed based on conserved regions present in the S. cerevisiae MNN9, VAN1, and ANP1 nucleotide sequences (Fig. 1). The primers were used to amplify a 420-bp product from C. albicans ATCC 10261 genomic DNA. The amplification product was cloned into PCR-Script (+), and several individual clones were sequenced. Sequence analysis revealed the presence of two sequences which, based on sequence similarity with the catalogued S. cerevisiae MNN9 and VAN1 sequences in the GenBank database, were identified as portions of the putative C. albicans MNN9 and VAN1 genes. The mixture of amplification products was used to probe digests of genomic DNA derived from three strains of C. albicans and hybridized to 3.5- and 4.5-kb HindIII fragments and 2.4- and 6.5-kb EcoRI fragments in all strains analyzed (data not shown). The PCR mixture was subsequently used to screen C. albicans ATCC 10261 HindIII and EcoRI genomic libraries constructed in pUC18 (38). Several positive clones were isolated from each library. Restriction analysis of the positive clones isolated from the respective libraries revealed HindIII inserts of 3.5 kb or 2.4-kb EcoRI inserts (data not shown). Initial sequence analysis of the clone containing the 2.4-kb EcoRI fragment showed it to carry the 3′ portion of the C. albicans VAN1 gene (which will be described in detail in a different study). The clone containing the 3.5-kb HindIII insert was subcloned to a 2.1-kb HindIII/KpnI fragment in pUC18 (plasmid p8A-KpnIΔ; see Fig. 4), and both DNA strands were sequenced. Sequence analysis revealed an ORF of 1,107 bp predicted to encode a protein of 368 amino acids. The deduced amino acid sequence showed 65.6% overall identity with the S. cerevisiae Mnn9 protein, strongly suggesting that the C. albicans homolog had been isolated. A relevant feature observed in the deduced amino acid sequence is the presence of a region (amino acids 18 to 34) predicted to be a membrane-spanning domain. A similar domain is present in the same region of the S. cerevisiae MNN9 amino acid sequence (64). Alignment of the C. albicans MNN9 gene product sequence with those of the S. cerevisiae MNN9, VAN1, and ANP1 genes indicated significant homology within the carboxy regions of all predicted proteins (Fig. 1). A putative C. albicans homolog of S. cerevisiae ANP1 has been identified by PCR amplification and Southern analysis (see above; data not shown), indicating the presence of a similar MNN9 gene family in Candida.
FIG. 1.
Amino acid alignments of the CaMNN9 product with protein sequences encoded by members of the S. cerevisiae MNN9 gene family. Boxes indicate amino acid identity. The putative membrane-spanning domains present in the CaMNN9 and ScMNN9 proteins are overlined. Arrows indicate sequences used to design primers for amplification of C. albicans genomic DNA.
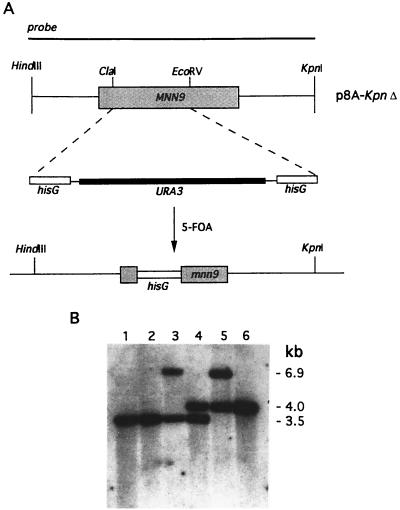
FIG. 4.
Strategy used for disruption of both alleles of the C. albicans MNN9 gene. (A) Restriction map of a genomic fragment containing CaMNN9. The 600-bp ClaI/EcoRV fragment was replaced with the 4.0-kb BglII/BamHI fragment carrying the hisG URA3 hisG cassette (see Materials and Methods). (B) Southern analysis of genomic DNA from strains obtained during the deletion process. DNA was digested with HindIII and separated by agarose gel electrophoresis. After transfer to a nylon membrane, the blot was probed with the 2.1-kb HindIII/KpnI fragment containing CaMNN9. Lane 1, SC5314 (wild type); lane 2, CAI4 (wild type, Ura−); lane 3, SS2+ (MNN9/mnn9, Ura+); lane 4, SS2− (MNN9/mnn9, Ura−); lane 5, SSCA-2 (mnn9/mnn9, Ura+); lane 6, SS19-4 (mnn9/mnn9, Ura−). The 3.5-kb band represents the wild-type MNN9 locus. The 6.9- and 4.0-kb bands correspond to the Camnn9::hisG-URA3-hisG and Camnn9::hisG loci, respectively.
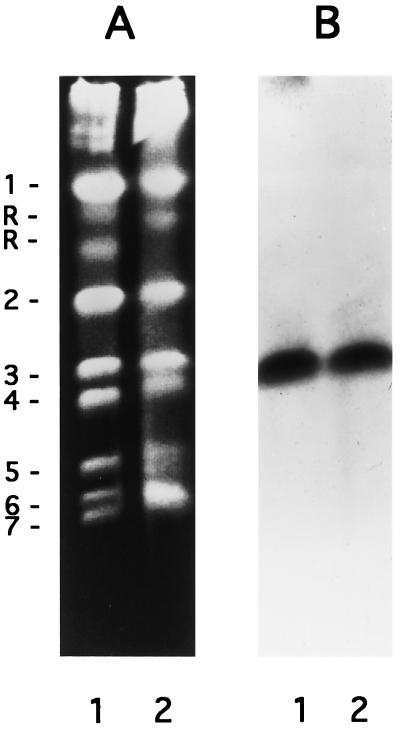
Analysis of chromosomes isolated from two strains of C. albicans showed that MNN9 resides on chromosome 3 in both strains (Fig. 2). The same chromosomal blot was stripped and reprobed with the C. albicans URA3 gene (derived from plasmid p5921), which has been shown to be located on chromosome 3 (19, 37) for verification of chromosomal mapping (data not shown). These results, as well as those obtained from restriction mapping and Southern analysis (data not shown), suggest that there is only one copy of CaMNN9 present in the C. albicans genome.
FIG. 2.
Chromosomal analysis of CaMNN9. Chromosomes isolated from C. albicans CAI4 and ATCC 10261 (A, lanes 1 and 2, respectively) were probed with the internal 600-bp ClaI/EcoRV fragment from p8A-KpnIΔ (B). Chromosomal designations are on the left. CaMNN9 resides on chromosome 3 in both strains.
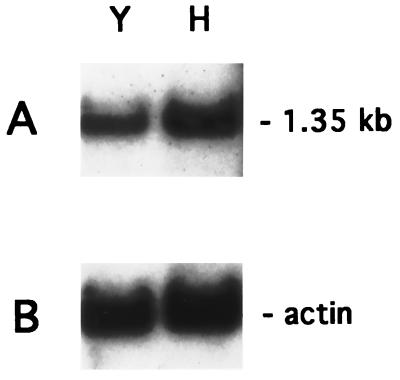
Northern analysis of total RNA obtained from C. albicans ATCC 10261 growing in either yeast or hyphal phases identified an mRNA of approximately 1.35 kb that hybridized with the HindIII/KpnI fragment from p8A-KpnIΔ carrying CaMNN9 (Fig. 3). The level of MNN9 transcript, as measured by densitometry, was only slightly elevated during mycelial growth (1.4 times the yeast phase signal) when normalized to values from signals obtained after hybridization with the S. cerevisiae actin gene (Fig. 3).
FIG. 3.
Analysis of expression of CaMNN9. Shown is a Northern analysis of total RNA (10 μg) isolated from C. albicans ATCC 10261 growing in either yeast (lane Y) or hyphal (lane H) growth phases. The blot was probed with the internal ClaI/EcoRV fragment of CaMNN9 (A), stripped, and reprobed with the S. cerevisiae actin gene (B).
Deletion of the MNN9 gene in C. albicans.
A homozygous mnn9Δ strain was constructed by the method devised by Fonzi and Irwin (18) in order to determine the phenotypes associated with disruption of the C. albicans MNN9 gene. Approximately 600 bp of the CaMNN9 ORF was replaced with the hisG URA3 hisG cassette (see Materials and Methods), and the resulting construct was transformed into CAI4, a C. albicans ura3Δ strain (18). More than 30 Ura+ transformants were obtained, and Southern analysis of three isolates showed that homologous recombination had occurred (Fig. 4). One of the Ura+ MNN9/mnn9 isolates, SS2+, was plated on SD medium containing uridine and 5-FOA to select for Ura− revertants that would arise from excision of the URA3 gene as a result of recombination of the flanking hisG repeats (18). Two of the three isolates analyzed by Southern hybridization had undergone the desired event (Fig. 4), resulting in strains that were MNN9/mnn9 heterozygotes, auxotrophic for uracil.
In order to obtain a homozygous mnn9 deletion strain, the aforementioned heterozygotes, SS2− and SS6−, were transformed with the same deletion construct and Ura+ transformants were once again selected. Over 60 transformants were checked by Southern analysis before one homozygous disruptant was identified (Fig. 4). Ultimately, five Ura+ mnn9/mnn9 strains were isolated (all showed the same hybridization pattern, suggesting that the same recombination event had occurred in each strain), and one such strain, SSCA-2, was used for further studies. Ura− auxotrophic strain SS19-4 was derived from SSCA-2 by following the same protocol as that described above. Northern analysis of total RNA obtained from each of the null mutants (Ura+ and Ura−) verified that CaMNN9-specified transcripts were not produced in those strains (data not shown). In most instances, the Ura+ strains SC5314 and SSCA-2 were used for the analyses described below, since their respective Ura− derivatives (CAI4 and SS19-4) tended to grow more slowly. Often, experiments were repeated with the other independently isolated homozygous deletion strains to verify that the phenotypes were the same. When SS19-4 was used in studies, it was compared to Ura− strain CAI4.
Morphology and growth phenotype of mnn9Δ cells.
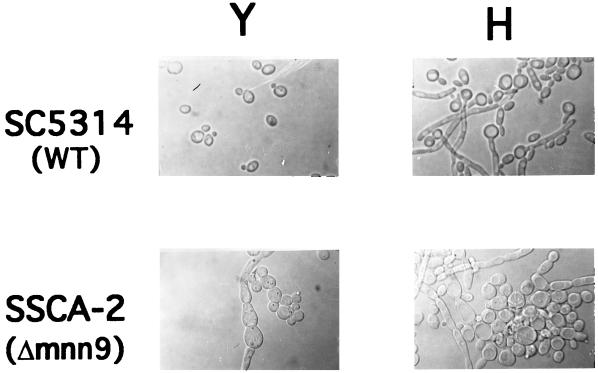
The C. albicans mnn9/mnn9 double disruptants exhibited many of the same growth phenotypes as those observed for their S. cerevisiae mnn9 strain counterparts (2, 64). Specifically, Camnn9Δ strains grew more slowly than the wild-type strain, exhibited a small, dry-colony phenotype on solid media, and grew as large clumps in liquid media. Addition of osmotic stabilizers, such as sorbitol and KCl, to media provided some reparation of growth defects but did not restore normal growth rates or phenotypes. Disruption of CaMNN9 also resulted in an impairment in conversion of yeast cells to hyphal growth forms. Strains SSCA-2 and SS19-4 were induced to form germ tubes in either 10% serum or RPMI 1640 medium, and in both instances the frequency of germ tube formation was greatly reduced compared to those for strains SC5314 and CAI4, respectively (data not shown). Hyphal formation also appeared abnormal in that many pseudohyphae were observed among yeast and hyphal cells in large aggregates. When cells were viewed under a microscope with Normarski optics, the differences in the cell morphology of both yeast and hyphal cells were found to be quite pronounced (Fig. 5). These results are indicative of a defect that affects the biosynthesis and/or assembly of the C. albicans cell wall, as would be expected in strains defective in protein glycosylation or secretion processes. In turn, poor growth and the inability to properly form hyphae suggest that the mnn9 strains might lack the appropriate adherence and/or invasive properties necessary to establish infection.
FIG. 5.
Morphology of yeast and hyphal growth forms of C. albicans wild-type and mnn9Δ strains. Yeast cells (Y) were grown in YPD to an OD600 of 1.0. The dimorphic transition to hyphal growth was induced by the addition of serum to 10% and shifting the culture to 37°C. Hyphal samples (H) were observed after 4 h of incubation. Cells were viewed under a microscope with Nomarski optics.
Effect of MNN9 deletion on sensitivities to hygromycin and sodium orthovanadate.
Yeast mutants with defects in Golgi apparatus-specific glycoprotein processing, including all members of the S. cerevisiae MNN9 gene family, have shown resistance to sodium orthovanadate and sensitivity to the aminoglycoside antibiotic hygromycin B (5, 15). Thus, the C. albicans mnn9Δ strains were tested to determine sensitivity to these compounds. Both Ura+ and Ura− derivative Camnn9Δ strains, as well as the respective Ura+ and Ura− wild-type Candida strains, were scored for growth on YPD plates containing various amounts of hygromycin B or sodium orthovanadate (Table 2). As anticipated, the mnn9Δ strains displayed resistance to sodium orthovanadate, being able to grow on plates containing 20 mM vanadate. In contrast, the wild-type parental strains were completely inhibited on plates containing 15 mM vanadate. In turn, the Candida mnn9 mutants were sensitive to hygromycin. Both wild-type strains were able to grow on medium supplemented with 300 μg of hygromycin per ml, while the mnn9 strains were completely growth inhibited at 100 μg of hygromycin/ml. Since hygromycin sensitivity is due, at least in part, to defects in glycosylation (15), these results suggested that disruption of C. albicans MNN9 results in a glycosylation defect similar to that observed in S. cerevisiae mnn9 strains (3, 64).
TABLE 2.
Phenotypic effects of disruption of CaMNN9
| Strain | Drug sensitivitya
|
% [14C]glucose incorporatedb into:
|
Glucanase sensitivityc
|
||||
|---|---|---|---|---|---|---|---|
| Vanr | Hygs | Alkali-insoluble fraction | Alkali-soluble β-glucan | Mannan | Crude β-1,3 | Pure β-1,6 | |
| SC5314 (WT)d | − | + | 52 | 16 | 14 | − | + |
| SSCA-2 (Δmnn9) | + | − | 56 | 24 | 7 | − | − |
Resistance to sodium orthovanadate (Van) and sensitivity to hygromycin B (Hyg) were measured as growth on YPD medium supplemented with various concentrations of drug (see Materials and Methods). +, phenotype present; −, phenotype absent.
Results are percentages of incorporation of radioactivity from [14C]glucose into different cell wall polysaccharide fractions. Experiments were done in triplicate.
Crude glucanase preparation from T. harzianum (Sigma) contains β-1,3-glucanase, β-1,6-glucanase, and chitinase activities. +, sensitive; −, not sensitive.
WT, wild type.
mnn9Δ strain cell walls contain reduced mannan levels.
The results of drug studies suggested that disruption of CaMNN9 resulted in an inability to properly glycosylate secreted and/or cell wall-associated mannoproteins. Such a defect should directly result in a reduction of the mannan content of the mnn9 strain cell wall. Thus, mutant and wild-type strains were grown in YPD medium containing radioactive glucose to label the various cell wall carbohydrate components. Cell walls were then isolated, and the amounts of radioactivity incorporated into the alkali-insoluble, alkali-soluble β-glucan, and mannan cell wall fractions were determined (Table 2). As anticipated, the mnn9 deletion strain cells exhibited a significant decrease (50%) in the mannan portion of their cell walls compared to the wild-type strain. In cell walls extracted from strain SSCA-2 mannan comprised only 7% of total cell wall carbohydrate, compared to 14% for wild-type strain SC5314. Interestingly, the incorporation of radioactive glucose into the alkali-soluble β-glucan fraction was clearly increased in the disrupted strains. This might reflect an attempt by the mutant cells to compensate for the mannan synthesis defect.
Effect of MNN9 deletion on sensitivity to glucanase.
Sensitivity to glucanase is often used as a parameter for changes in cell wall composition in yeast (35, 49). The C. albicans mnn9Δ strains were tested for sensitivity to a crude β-1,3-glucanase and a purified β-1,6-glucanase preparation. Both Ura+ and Ura− derivative Camnn9Δ strains were found to be sensitive to β-1,3-glucanase compared to the Ura+ and Ura− wild-type Candida strains. In contrast, both the mutant and wild-type strains (Ura+ and Ura−) were insensitive to lysis by treatment with pure β-1,6-glucanase (Table 2). Control cells incubated under similar conditions without the addition of either enzyme showed no lysis.
Analysis of cell wall-associated proteins.
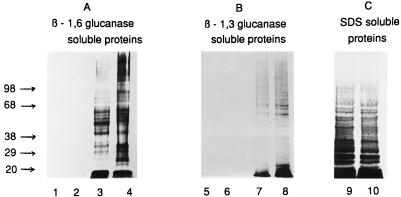
The S. cerevisiae mnn9 strain has recently been used to study the anchorage, structure, and types of cell wall proteins released after treatment with glucanases (30, 42, 61). Therefore, it was of interest to determine if disruption of CaMNN9 altered the profile of proteins released by this method. Strains SC5314 and SSCA-2 were grown to exponential phase, and cell walls were prepared (see Materials and Methods). Proteins released from cell walls after treatment with SDS were separated by SDS-PAGE and detected by silver staining. No obvious differences between the profiles of proteins released from the cell walls of mnn9Δ strain cells and wild-type strain cells were observed (Fig. 6). Cell wall fractions remaining after extraction with SDS were digested with either a purified β-1,6-glucanase from Trichoderma or a crude Trichoderma β-1,3-glucanase to release covalently bound mannoproteins. SDS-PAGE analysis of the resulting protein extracts showed the release of β-1,6-glucosylated mannoproteins from both mutant and wild-type strains (Fig. 6). Indeed, more high-molecular-weight proteins were released from the mnn9Δ strain cell walls than from the wild-type cell walls. It is possible that the reduction in the mannan content of the mutant cell wall facilitates the release of those glucan-linked proteins by providing better enzyme access. Treatment with the crude β-1,3-glucanase resulted in the release of a low level of proteins detectable by silver staining from SDS-extracted cell walls derived from either the mutant or wild-type strains (Fig. 6).
FIG. 6.
SDS-PAGE analysis of β-1,6-glucanase-soluble (A), β-1,3-glucanase-soluble (B), and SDS-soluble (C) proteins extracted from C. albicans SSCA-2 (Δmnn9) and SC5314 (wild type) cell walls. Controls are represented as untreated wild-type (lanes 1 and 5) and Δmnn9 (lanes 2 and 6) strain cell walls. Proteins isolated from wild-type (odd-numbered lanes) and mutant (even-numbered lanes) strain cell walls after treatment with either β-1,6-glucanase (lanes 3 and 4), β-1,3-glucanase (lanes 7 and 8), or SDS (lanes 9 and 10) were separated on 4- to 20% gradient gels and visualized by silver staining. Molecular mass standards (in kilodaltons) are shown on the left.
Analysis of MNN9 gene family transcript levels in mnn9Δ strain cells.
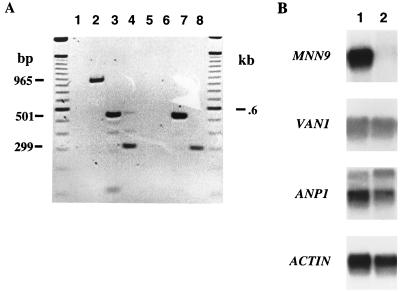
The proteins encoded by the Saccharomyces MNN9 gene family have been shown to interact physically and might have overlapping and/or redundant enzymatic activities (26). Thus, experiments were performed to determine if expression of the Candida family member genes was affected in a mnn9Δ strain background. PCR was initially performed with genomic DNAs obtained from both wild-type C. albicans SC5314 and the mnn9Δ strain SSCA-2 as the templates. Primers were designed on the basis of known sequences of C. albicans MNN9, VAN1, and ANP1 (57a). Amplification products of the expected sizes were obtained for all template-primer combinations, except for MNN9 with mnn9Δ DNA as the template, as expected (data not shown). In addition, equivalent amounts of DNA were amplified for each gene in both strains (data not shown). Each of the gene-specific primers was then used in RT-PCR, and the predicted products were obtained (Fig. 7). Little or no product was obtained when the control reaction mixture with no reverse transcriptase was used as the template with ANP1-specific primers or when MNN9 gene-specific primers were used to amplify the SSCA-2 cDNA reaction. The expected 500- and 350-bp products were amplified from the respective VAN1 and ANP1 cDNAs derived from the wild-type and mnn9Δ strains, with no apparent significant differences in product amount. In turn, the results of Northern blot experiments show no substantial difference in ANP1 or VAN1 transcript levels between wild-type and mnn9Δ strains (Fig. 7). Total RNA (10 μg) obtained from both SC5314 and SSCA-2 was Northern blotted and probed (sequentially, stripping the blot after each hybridization) with either the 501-bp VAN1 PCR product, the 299-bp ANP1 product, or the 965-bp MNN9 product. The relative transcript levels were similar in all cases to levels of actin transcript obtained after stripping the blot and reprobing with the S. cerevisiae actin gene (Fig. 7). As expected, no transcript was identified when the MNN9 PCR product was used to probe RNA derived from strain SSCA-2. Jungmann and Munro previously observed a lower level of Anp1 protein immunoprecipitated from an S. cerevisiae mnn9Δ strain (26). Our results suggest that the decrease observed is due to posttranslational processing (most probably degradation of uncomplexed Anp1p) rather than changes at the transcriptional level.
FIG. 7.
RT-PCR (A) and Northern (B) analysis of expression of MNN9 family genes in C. albicans wild-type and Δmnn9 strains. RT-PCR was performed with RNA extracted from wild-type strain SC5314 (lanes 1 to 4) and Δmnn9 strain SSCA-2 (lanes 5 to 8). cDNA synthesis and subsequent amplification were done with primers specific for the C. albicans MNN9 (lanes 2 and 6), VAN1 (lanes 3 and 7), and ANP1 (lanes 4 and 8) genes. Control reactions (lanes 1 and 5) utilized ANP1-specific primers, and reaction mixtures contained no reverse transcriptase. Northern blot analysis was performed with total RNA (10 μg) isolated from strains SC5314 (lane 1) and SSCA-2 (lane 2), and the purified wild-type MNN9, VAN1, and ANP1 RT-PCR products were used as probes. The blot was stripped sequentially between hybridizations and was probed with the S. cerevisiae actin gene as a quantitative control for gel loading.
DISCUSSION
We have isolated and characterized the MNN9 gene from the fungal pathogen C. albicans and examined some of the phenotypic effects of disruption of this gene. Analysis of the gene sequence revealed an intron-free ORF of 1,107 bp encoding a protein of 368 amino acids. As expected, CaMNN9 has a high degree of homology with its S. cerevisiae counterpart at the deduced amino acid sequence level (65.6%), with several blocks of strong homology within the predicted carboxy termini. A predicted transmembrane domain is present between residues 18 and 34 of the amino acid sequence. Similar regions have been identified in the amino termini of S. cerevisiae MNN9 gene family member proteins (12, 64). Additionally, electrophoretic karyotype analysis mapped CaMNN9 to chromosome 3 in both strains examined. A single mRNA of approximately 1.35 kb was identified in both the yeast and hyphal morphological growth phases of C. albicans. In addition, no significant difference in transcript levels was observed, which suggests that the CaMNN9 gene is not growth phase regulated. Both chromosomal copies of CaMNN9 were disrupted by the “URA blaster” method (18). Many of the phenotypes exhibited by the mutant were indicative of cell wall defects, including osmotic instability, slow growth, severe clumping in liquid media, and abnormal colony formation on solid media. The addition of osmotic stabilizers did not have a pronounced effect on cell growth or stability. Hyphal formation was also impaired both in terms of frequency of conversion of yeast cells to hyphal forms and the elaboration of defective hyphae and many pseudohyphae. An inability to correctly synthesize cell wall components would certainly influence proper expansion of the cell wall during yeast-to-mycelium conversion. Indeed, examination of radioactively labeled cell wall carbohydrate components showed Camnn9Δ strain cell walls to contain half the amount of mannose present in parental strain cell walls. In addition, a greater amount of alkali-soluble β-glucans was present in mutant cell walls than in wild-type cell walls, suggesting an attempt to compensate for the mannan defect. Like all strains carrying mutant members of the S. cerevisiae MNN9 gene family, Camnn9Δ strains were shown to be resistant to sodium orthovanadate and sensitive to the aminoglycoside antibiotic hygromycin B. Camnn9Δ strains also proved to be sensitive to lysis by a crude β-1,3-glucanase preparation, while the parental strains were resistant. Since increased sensitivity to β-1,3-glucanases is indicative of altered cell wall composition (35, 49) and since sensitivity to hygromycin has been shown to correlate with defects in glycosylation (15), it is assumed that deletion of CaMNN9 directly affects the mannosylation of cell wall-associated mannoproteins. Treatment of SDS-extracted cell walls with a purified T. harzianum β-1,6-glucanase resulted in the release of more covalently bound, high-molecular-weight proteins from mutant cell walls than from wild-type cell walls. These proteins probably represent glycosyl phosphatidylinositol (GPI)-anchored proteins that are heavily O-mannosylated. Examination of secreted chitinase from S. cerevisiae MNN9 gene family knockout strains showed that the processes of O glycosylation remain intact in these mutant strains and that only N-linked glycosylation is affected (57a). It could be inferred that the overall reduction in the mannan content of the Camnn9 mutant cell wall would facilitate the release of these proteins linked to glucan. Indeed, the S. cerevisiae mnn9 strain has been utilized by others in similar experiments to gain a clearer understanding of the types of proteins that are cell wall associated and how, and to what cell wall components, they are anchored (30, 42, 61).
The shared phenotypes of S. cerevisiae mnn9 gene family mutants suggest that Mnn9p, Anp1p, and Van1p might perform redundant enzymatic activities. Certain proteins encoded by members of the MNN9 gene family have also been shown to physically interact with each other (26) (see below), suggesting either overlapping or coordinate functions. It was of interest to determine if the loss of function of one gene family member in C. albicans would affect the transcriptional activity of either of the other family member genes. The results of RT-PCR and Northern analysis of CaVAN1 and CaANP1 expression in a Camnn9Δ strain showed that neither of these genes is up or down regulated in the absence of the CaMNN9 transcript. It had been observed that the steady-state level of S. cerevisiae Anp1p was reduced in an S. cerevisiae mnn9Δ strain (26). Our results suggest that Anp1p is probably degraded in the absence of its complex partner, Mnn9p.
Recently, Jungmann and Munro (26) have provided greater insight into the role S. cerevisiae MNN9 gene family members play in the synthesis of the outer chain of cell wall mannan. The model they propose suggests that a Mnn9p-Van1p complex is responsible for the synthesis of a short α-1,6-mannose chain onto the initiating mannose residue, placed by Och1p. The mannose polymer is then extended to its full length by the action of the Mnn9p-Anp1p complex, which also catalyzes the addition of the initial α-1,2-mannose branches. Mnn2p and Mnn5p have recently been identified as the α-1,2-mannosyltransferases responsible for the initiation and extension of additional α-1,2-mannose branches, respectively (50). Ultimately, the chains are terminated by the Mnn1p (64)-catalyzed addition of an α-1,3-linked mannose, and phosphomannose is incorporated by Mnn6p (62).
While proteins encoded by MNN9 gene family members are obviously directly involved in yeast glycosylation, acting either as mannosyltransferases themselves or by organizing and directing the appropriate mannosyltransferases, there is a great deal of evidence which suggests that they are involved in other cellular processes. Loss of Anp1p results in mislocalization of various Golgi apparatus resident proteins (12, 46). Anp1p has also been shown to interact genetically with Bet5p, a protein involved in transport from the endoplasmic reticulum to the Golgi apparatus (25). Additionally, ANP1 was identified in a screen for genes of mutants defective in polarized growth, but with no defects in actin cytoskeleton structure (41). Interestingly, the mutation of ANP1 was recently shown to be synthetically lethal in conjunction with cmd1A, a temperature-sensitive yeast calmodulin mutant which confers a defect in actin organization (54). In turn, both anp1 and cmd1A proved to be synthetically lethal with a specific mutation of MYO2, a gene that encodes a calmodulin-binding class V myosin (54). These results implicate Anp1p either directly, or indirectly by virtue of its role in glycosylation, in the regulation of actin organization.
In conclusion, since MNN9 gene structure and function seem to be well conserved between C. albicans and S. cerevisiae and since gene families have been identified in both genera, the fundamental role this gene plays in C. albicans cellular processes should be further investigated. Disruption of CaMNN9 results in phenotypes that are inconsistent with pathogenicity, including poor growth, aberrant hypha formation, and altered cell wall composition. Since the Candida cell wall is essential for the biology and pathogenicity of the organism it is likely that mutation of MNN9 will render the strain avirulent. In fact, recent studies have shown that disruption of genes that encode O-mannosyltransferases in C. albicans does alter the virulence capabilities of the knockout strains (7, 59). Thus, we are currently assessing the adherence capabilities of the Camnn9Δ strains and ultimately will test the virulence of these strains in animal models of infection. Unfortunately, the severe clumping phenotype and osmotic instability of the deletion mutant present obstacles to these studies and are being addressed at this time. Ultimately, the results of those experiments will define members of this family as potential drug targets.
ACKNOWLEDGMENTS
We thank Carlos Hirschberg (Boston University Medical Center) for critical reading of this manuscript, W. A. Fonzi, June Zhao, and Joy Sturtevant (Georgetown University Medical Center) for strains, plasmids, and the karyotype blot, respectively, and Letty Vega (MIT) for the S. cerevisiae actin gene. We also thank Paul Awald (MIT) for help with the cell wall carbohydrate analysis and Stu Levitz (Boston University Medical Center) for use of the Nikon Diaphot microscope.
This work was supported by grants from the National Institutes of Health, GM45188 (to P.W.R.) and CA14051 (to R. O. Hynes).
REFERENCES
- 1.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballou C E. Yeast cell wall and cell surface. In: Strathern J N, Jones E W, Broach J R, editors. The molecular biology of the yeast Saccharomyces. Metabolism and gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. pp. 335–360. [Google Scholar]
- 3.Ballou C E. Isolation, characterization and properties of Saccharomyces mnn mutants with non-conditional protein glycosylation defects. Methods Enzymol. 1990;185:440–476. doi: 10.1016/0076-6879(90)85038-p. [DOI] [PubMed] [Google Scholar]
- 4.Ballou L, Cohen R E, Ballou C E. Saccharomyces cerevisiae mannoprotein mutants that make mannoproteins with a truncated outer chain. J Biol Chem. 1980;255:5986–5991. [PubMed] [Google Scholar]
- 5.Ballou L, Hitzeman R A, Lewis M S, Ballou C E. Vanadate-resistant yeast are defective in protein glycosylation. Proc Natl Acad Sci USA. 1991;88:3209–3212. doi: 10.1073/pnas.88.8.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bulawa C E, Miller D W, Henry L K, Becker J W. Attenuated virulence of chitin-deficient mutants of Candida albicans. Proc Natl Acad Sci USA. 1995;92:10570–10574. doi: 10.1073/pnas.92.23.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buurman E T, Westwater C, Hube B, Brown A J, Odds F C, Gow N A. Molecular analysis of CaMnt1p, a mannosyl transferase important for adhesion and virulence of Candida albicans. Proc Natl Acad Sci USA. 1998;95:7670–7675. doi: 10.1073/pnas.95.13.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calderone R A, Braun P C. Adherence and receptor relationships of Candida albicans. Microbiol Rev. 1991;55:1–20. doi: 10.1128/mr.55.1.1-20.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calderone R A, Fukayama M. Virulence-associated mannoproteins of Candida albicans. In: Sutcliffe J A, Georgeopapadakou N H, editors. Emerging targets in antibacterial and antifungal chemotherapy. New York, N.Y: Chapman and Hall; 1992. pp. 524–545. [Google Scholar]
- 10.Calderone R A. The recognition of host cells by the pathogenic yeast, Candida albicans. Jpn J Med Mycol. 1994;35:9–18. [Google Scholar]
- 11.Castro C, Ribas J C, Valdivieso M H, Varona R, Del Rey F, Duran A. Papulacandin B resistance in budding and fission yeast: isolation and characterization of a gene involved in (1,3)β-d-glucan synthesis in Saccharomyces cerevisiae. J Bacteriol. 1995;177:5732–5739. doi: 10.1128/jb.177.20.5732-5739.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chapman R E, Munro S. The functioning of the yeast Golgi apparatus requires an ER protein encoded by ANP1, a member of a new family of genes affecting the secretory pathway. EMBO J. 1994;13:4896–4907. doi: 10.1002/j.1460-2075.1994.tb06817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chappell T, Hajibagheri N, Ayscough K, Pierce M, Warren G. Localization of an α-1,2 galactosyltransferase activity to the Golgi apparatus of Schizosaccharomyces pombe. Mol Biol Cell. 1994;5:519–528. doi: 10.1091/mbc.5.5.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cutler J E. Putative virulence factors of Candida albicans. Annu Rev Microbiol. 1991;45:187–218. doi: 10.1146/annurev.mi.45.100191.001155. [DOI] [PubMed] [Google Scholar]
- 15.Dean N. Yeast glycosylation mutants are sensitive to aminoglycosides. Proc Natl Acad Sci USA. 1995;92:1287–1291. doi: 10.1073/pnas.92.5.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dean N, Poster J B. Molecular and phenotypic analysis of the S. cerevisiae MNN10 gene identifies a family of related glycosyltransferases. Glycobiology. 1996;6:73–81. doi: 10.1093/glycob/6.1.73. [DOI] [PubMed] [Google Scholar]
- 17.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 18.Fonzi W A, Irwin M Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gillum A M, Tsay E Y, Kirsch D R. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet. 1984;198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- 20.Gow N A R, Robbins P W, Lester J W, Brown A J P, Fonzi W A, Chapman T, Kinsman O. A hyphal-specific chitin synthase gene (CHS2) is not essential for growth, dimorphism or virulence of Candida albicans. Proc Natl Acad Sci USA. 1994;91:6216–6220. doi: 10.1073/pnas.91.13.6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guthrie C, Fink G R. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:169–182. [PubMed] [Google Scholar]
- 22.Hanahan D. Techniques for transformation of E. coli. In: Glover D M, editor. DNA cloning. Oxford, United Kingdom: IRL Press; 1988. pp. 109–135. [Google Scholar]
- 23.Herscovics A, Orlean P. Glycoprotein synthesis in yeast. FASEB J. 1993;7:540–548. doi: 10.1096/fasebj.7.6.8472892. [DOI] [PubMed] [Google Scholar]
- 24.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang Y, Scarpa A, Zhang L, Stone S, Feliciano E, Ferro-Novick S. A high-copy suppressor screen reveals genetic interactions between BET3 and a new gene: evidence for a novel complex in ER-to-Golgi transport. Genetics. 1998;49:833–841. doi: 10.1093/genetics/149.2.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jungmann J, Munro S. Multi-protein complexes in the cis Golgi of S. cerevisiae with α-1,6 mannosyltransferase activity. EMBO J. 1998;17:423–434. doi: 10.1093/emboj/17.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanbe T, Han Y, Redgrave B, Rieselman M, Cutler J. Evidence that mannans of Candida albicans are responsible for adherence of yeast forms to spleen and lymph node tissue. Infect Immun. 1993;61:2578–2584. doi: 10.1128/iai.61.6.2578-2584.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanik-Ennulat C, Neff N. Vanadate-resistant mutants of Saccharomyces cerevisiae show alterations in protein phosphorylation and growth control. Mol Cell Biol. 1990;10:898–909. doi: 10.1128/mcb.10.3.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanik-Ennulat C, Montalvo E, Neff N. Sodium orthovanadate-resistant mutants of Saccharomyces cerevisiae show defects in Golgi-mediated protein glycosylation, sporulation and detergent resistance. Genetics. 1995;140:933–943. doi: 10.1093/genetics/140.3.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kapteyn J C, Montijn R C, Vink E, de la Cruz J, Shimoi H, Lipke P N, Klis F M. Retention of Saccharomyces cerevisiae cell wall proteins through a phosphodiester-linked β-1,3-/β-1,6-glucan heteropolymer. Glycobiology. 1996;6:337–345. doi: 10.1093/glycob/6.3.337. [DOI] [PubMed] [Google Scholar]
- 31.Kurtz M B, Kelly R, Kirsch D R. Molecular biology of Candida albicans. In: Kirsch D R, Kelly R, Kurtz M B, editors. The genetics of Candida albicans. Boca Raton, Fla: CRC Press, Inc.; 1990. pp. 21–74. [Google Scholar]
- 32.Kwon-Chung K J, Lehman D, Good C, Magee P T. Genetic evidence for role of extracellular proteinase in virulence of Candida albicans. Infect Immun. 1985;49:571–575. doi: 10.1128/iai.49.3.571-575.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwon-Chung K J, Bennett J E. Medical mycology. Philadelphia, Pa: Lea and Febiger; 1992. pp. 280–336. [Google Scholar]
- 34.Lee K L, Buckley H R, Campbell C C. An amino acid liquid synthetic medium for development of mycelial and yeast forms of Candida albicans. Sabouraudia. 1975;13:148–153. doi: 10.1080/00362177585190271. [DOI] [PubMed] [Google Scholar]
- 35.Lussier M, White A M, Sheraton J, diPaolo T, Treadwell J, Southard S B, Horenstein C I, Chen-Weiner J, Ram A R J, Kapteyn J C, Roemer T, Vo D H, Bondoc D C, Hall J, Zhong W W, Sidcu A M, Davies J, Klis F M, Robbins P W, Bussey H. Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics. 1997;147:435–450. doi: 10.1093/genetics/147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lussier M, Sidcu A M, Shahinian S, Bussey H. The Candida albicans KRE9 gene is required for cell wall β-1,6-glucan synthesis and is essential for growth on glucose. Proc Natl Acad Sci USA. 1998;95:9825–9830. doi: 10.1073/pnas.95.17.9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Magee B B, Koltin Y, Gorman J A, Magee P T. Assignment of cloned genes to the seven electrophoretically separated Candida albicans chromosomes. Mol Cell Biol. 1988;8:4721–4726. doi: 10.1128/mcb.8.11.4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCreath K J, Specht C A, Robbins P W. Molecular cloning and characterization of chitinase genes from Candida albicans. Proc Natl Acad Sci USA. 1995;92:2544–2548. doi: 10.1073/pnas.92.7.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKnight G L, Cardillo T S, Sherman F. An extensive deletion causing overproduction of yeast iso-2-cytochrome c. Cell. 1981;25:409–419. doi: 10.1016/0092-8674(81)90059-3. [DOI] [PubMed] [Google Scholar]
- 40.Mio T, Adachi-Shimizu M, Tachibana Y, Tabuchi H, Inoue S B, Yabe T, Yamada-Okabe T, Arisawa M, Watanabe T, Yamada-Okabe H. Cloning of the Candida albicans homolog of Saccharomyces cerevisiae GSC1/FKS1 and its involvement in β-1,3-glucan synthesis. J Bacteriol. 1997;179:4096–4105. doi: 10.1128/jb.179.13.4096-4105.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mondesert G, Clarke D J, Reed S I. Identification of genes controlling growth polarity in the budding yeast Saccharomyces cerevisiae: a possible role of N-glycosylation and involvement of the exocyst complex. Genetics. 1997;147:421–434. doi: 10.1093/genetics/147.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Montijn R C, van Rinsum J, van Schagen F A, Klis F M. Glucomannoproteins in the cell wall of Saccharomyces cerevisiae contain a novel type of carbohydrate side chain. J Biol Chem. 1994;269:19338–19342. [PubMed] [Google Scholar]
- 43.Nakayama K, Nagusa T, Shimma Y, Kuromitu J, Jigami Y. OCH1 encodes a novel membrane bound mannosyltransferase: outer chain elongation of asparagine-linked oligosaccharides. EMBO J. 1992;11:2511–2519. doi: 10.1002/j.1460-2075.1992.tb05316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Navarro-Garcia F, Sanchez M, Pla J, Nombela C. Functional characterization of the MKC1 gene of Candida albicans, which encodes a mitogen-activated protein kinase homolog related to cell integrity. Mol Cell Biol. 1995;15:2197–2206. doi: 10.1128/mcb.15.4.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neiman A M, Mhaiskar V, Manus V, Galibert F, Dean N. Saccharomyces cerevisiae HOC1, a suppressor of pkc1, encodes a putative glycosyltransferase. Genetics. 1997;145:637–645. doi: 10.1093/genetics/145.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nothwehr S F, Bryant N J, Stevens T H. The newly identified yeast GRD genes are required for retention of late-Golgi membrane proteins. Mol Cell Biol. 1996;16:2700–2707. doi: 10.1128/mcb.16.6.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Odds F C. Candida species and virulence. ASM News. 1994;60:313–318. [Google Scholar]
- 48.Paravicini G, Mendiza A, Antonsson B, Cooper M, Losberger C, Payton M. The Candida albicans PKC1 gene encodes a protein kinase C homolog necessary for cellular integrity, but not for dimorphism. Yeast. 1996;12:741–756. doi: 10.1002/(sici)1097-0061(19960630)12:8<741::aid-yea967>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 49.Ram A F J, Wolters A, Hoopen R T, Klis F M. A new approach for isolating cell wall mutants in Saccharomyces cerevisiae by screening for hypersensitivity to Calcofluor white. Yeast. 1994;10:1019–1030. doi: 10.1002/yea.320100804. [DOI] [PubMed] [Google Scholar]
- 50.Rayner J C, Munro S. Identification of the MNN2 and MNN5 mannosyltransferases required for forming and extending the mannose branches of the outer chain mannans of Saccharomyces cerevisiae. J Biol Chem. 1998;273:26836–26843. doi: 10.1074/jbc.273.41.26836. [DOI] [PubMed] [Google Scholar]
- 51.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 52.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sarthy A V, McGonigal T, Coen M, Frost D J, Meulbroek J A, Goldman R C. Phenotype in Candida albicans of a disruption of the BGL2 gene encoding a 1,3-β-glucosyltransferase. Microbiology. 1997;143:367–376. doi: 10.1099/00221287-143-2-367. [DOI] [PubMed] [Google Scholar]
- 54.Sekiya-Kawasaki M, Botstein D, Ohya Y. Identification of functional connections between calmodulin and the yeast actin cytoskeleton. Genetics. 1998;150:43–48. doi: 10.1093/genetics/150.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1986. [Google Scholar]
- 56.Sobel J D, Muller G, Buckley H R. Clinical role of germ-tube formation in the pathogenesis of candidal vaginitis. Infect Immun. 1984;44:576–580. doi: 10.1128/iai.44.3.576-580.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soll D R, Langtimm C J, McDowell J, Hicks J, Galask R. High-frequency switching in Candida strains from vaginitis patients. J Clin Microbiol. 1987;25:1611–1622. doi: 10.1128/jcm.25.9.1611-1622.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57a.Southard, S. Unpublished data.
- 58.Thompson J R, Douglas C M, Li W, Jue C K, Premanik B, Yuan X, Rude T H, Tofaletti D L, Perfect J R, Kurtz M. A glucan synthase FKS1 homolog in Cryptococcus neoformans is single copy and encodes an essential function. J Bacteriol. 1999;181:444–453. doi: 10.1128/jb.181.2.444-453.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Timpel C, Strahl-Bolsinger S, Ziegelbauer S, Ernst J F. Multiple functions of Pmt1p-mediated O-mannosylation in the fungal pathogen Candida albicans. J Biol Chem. 1998;273:20837–20846. doi: 10.1074/jbc.273.33.20837. [DOI] [PubMed] [Google Scholar]
- 60.Tsai P K, Frevert P, Ballou C E. Carbohydrate structure of Saccharomyces cerevisiae mnn9 mannoprotein. J Biol Chem. 1984;259:3805–3811. [PubMed] [Google Scholar]
- 61.van der Vaart J M, Caro L H P, Chapman J W, Klis F M, Verrips C T. Identification of three mannoproteins in the cell wall of Saccharomyces cerevisiae. J Bacteriol. 1995;177:3104–3110. doi: 10.1128/jb.177.11.3104-3110.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang X H, Nakayama K, Shimma Y, Tanaka A, Jigami Y. MNN6, a member of the KRE2/MNT1 family, is the gene responsible for mannosylphosphate transfer in Saccharomyces cerevisiae. J Biol Chem. 1997;272:18117–18124. doi: 10.1074/jbc.272.29.18117. [DOI] [PubMed] [Google Scholar]
- 63.Wickes B L, Golin J E, Kwon-Chung K J. Chromosomal rearrangement in Candida stellatoidea results in a positive effect on phenotype. Infect Immun. 1991;59:1762–1771. doi: 10.1128/iai.59.5.1762-1771.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yip C L, Welch S K, Klebel F, Gilbert T, Seidel P, Grant F J, O'Hara P J, MacKay V L. Cloning and analysis of the S. cerevisiae MNN9 and MNN1 genes required for complex glycosylation of secreted proteins. Proc Natl Acad Sci USA. 1994;91:2723–2727. doi: 10.1073/pnas.91.7.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]