Abstract
Currently an emerging human pathogenic coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), caused coronavirus disease 2019 (COVID-19) that has posed a serious threat to public health worldwide. As it is a novel severe pneumonia-type viral disease, no effective therapeutic agents are available to treat this infection to date, emphasizing an urgent need for development of effective anti-SARS-CoV-2 agents. Based on screening in computational biology and biological in vitro assays, a good number of natural compounds and their synthetic analogues have been confirmed to possess target-specific inhibitory effects against the activity of host and viral proteases, namely, cathepsin-L, TMPRSS2, Sec61, Mpro (3CL-protease), RNA-dependent RNA protease (RdRp), helicase cap-binding proteases eEF1A, eIF4A, eIF4E, which play dominant roles in progression of infection and replication of SARS-CoV-2 virus in host cells. This review paper describes the potent antiviral activity and target-specific anti-proteases activity of some natural compounds and their synthetic analogues against SARS-CoV-2 infection. It will inspire the researchers to unleash their own creativity and to design potent and safe drugs to fight the current COVID-19 pandemic.
Keywords: Natural compounds, Natural analogues, SARS-CoV-2, COVID-19, Host/virus proteases inhibitors
Abbreviations: COVID-19, coronavirus disease-2019
Graphical abstract
Abbreviations:
- ACE2
angiotensin-converting enzyme 2
- CC50
half-cytotoxic concentration
- CD-206
cluster of differentiation-206, a mannose receptor present on the surface of macrophages
- 3CLpro
3-chymotrypsin-like cysteine protease
- CMs
convoluted membranes
- CPE
cytopathic effect
- DMV
double-membrane vesicle
- EC50
concentration of the test compound necessary to inhibit 50% of viral cell growth or replication
- eEF1A
eukaryotic translation elongation factor 1A
- eIF4A
eukaryotic initiation factor 4A
- ERGIC
endoplasmic reticulum-Golgi intermediate compartment
- FRET
forster or fluorescence resonance energy transfer
- IC50
concentration necessary to inhibit 50% of target activity
- IFN
interferons
- MERS-CoV
Middle East respiratory syndrome-related coronavirus
- Mpro
main protease
- PLpro
papain-like cysteine protease
- RBD
receptor binding domain
- RBM
receptor binding motif
- RdRp,
RNA-dependent RNA polymerase
- RTC
replicase-transcriptase complex
- SRP
signal recognition particle
- SARS-CoV-2
severe acute respiratory syndrome coronavirus-2
- SI
selectivity index
- TMPRSS2
transmembrane serine protease type 2
1. Introduction
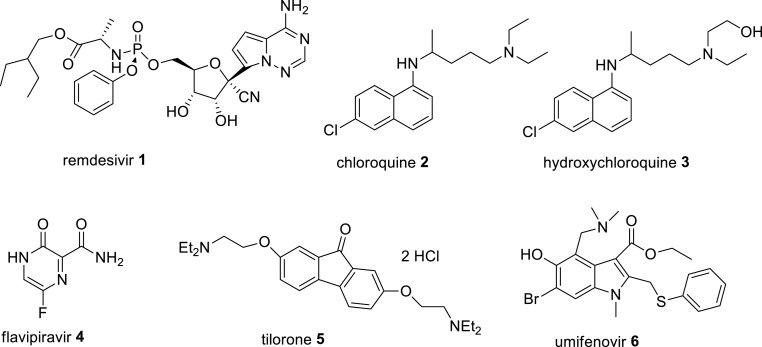
SARS-CoV-2 is a highly infectious pathogenic virus that causes the current coronavirus disease-2019 (COVID-19), a serious pneumonia-like respiratory infection, which has caused over 535 million confirmed infected cases and over 6.3 million deaths worldwide, as of 17 June 2022 [1,2]. In December 2019, this disease was originally started in the local seafood market in Wuhan city of China and then was spread across the globe rapidly in a catastrophic effect. The disease is characterized as severe respiratory disorders having flu-like symptoms, such as sore throat, fever, dry cough, shortness of breath and severe pneumonia in critical cases that lead to several organ failure and ultimately death [1}. The SARS-CoV-2 virus is highly contagious and can be transmitted through coughing and sneezing droplets of infected individuals and these virions containing droplets retained on hard surfaces for a long time and can spread to a fresh individual by direct touching to the infected surfaces [1]. The current COVID-19 pandemic is very similar to ‘Spanish Flu’ pandemic caused by a lethal influenza A virus, and resulted in nearly 100 million deaths worldwide in 1918–1919 [3]. The virus SARS-CoV-2 is a member of betacoronaviruses of family Coronaviridae, and having a positive-sense single-stranded (ss) RNA genome with 29,903 nucleotides that shares identity at the whole genome level to a bat coronavirus (CoV) (MG772933) by 96%, and sequence identity to SARS-CoV by 79.6%. Its genome consists of six major open reading frames (genes), namely replicase overlapping genes ORF1a and ORF1b, followed by structural proteins(genes), spike (S), envelope (E), membrane (M) and nucleocapsid (N) that are common to other betacoronaviruses and seven putative ORFs encoding accessory non-structural proteins that are interspersed between the structural genes, and these are arranged in order from a 5′-untranslated region (5′-UTR) to a 3′-untranslated region (3′-UTR). The replicase ORF1a/b genes code for viral functional replicase polyproteins pp1a and pp1ab in infected host cells, are further processed autoproteolytically by two cysteine proteases, papain-like protease (PLpro) and 3-chymotrypsin-like protease (3CL-pro or main protease) into 16 non-structural proteins (nsps) that play a crucial role in the formation of replicase-transcriptase complex (RTC) for viral replication and transcription [[4], [5], [6]]. This SARS-CoV-2 virus is the third highly pathogenic human coronavirus identified so far, following the earlier severe acute respiratory syndrome coronavirus (SARS-CoV) epidemic in 2003 in mainland China, and Middle East respiratory syndrome coronavirus (MERS-CoV) epidemic in 2012 in Saudi Arabia [7,8]. Available evidence on the mortality and morbidity rates on these epidemics demonstrates that the SARS-CoV-2 infected mortality rate is far lower (about 5%) than that of SARS-CoV (about 15%) and MERS-CoV (about 35%), but SARS-CoV-2 is much more contagious than the SARS-CoV and MERS-CoV viruses and affects more people over the age of 60 or those with comorbidities having weak immune system [[9], [10], [11]]. These findings indicate that SARS-CoV-2 is spreading at an alarming rate far worse than the previous human coronaviral epidemics. Till to date, there are no specific and safe antiviral drugs for treatment of SARS-CoV-2 infection and the current clinical treatments of COVID-19 by the use of some repurposed antiviral, antimalarial and immunomodulatory drugs as cocktail therapy are controversial. So far only remdesivir 1 (Fig. 1 ), a nucleoside ribose analogue, originally developed for Ebola virus infection, has been found effective in inhibition of SARS-CoV-2 infection in vitro, was approved by the FDA to treat patients with severe COVID-19. However, it appeared to be only mildly beneficial for disease recovery in randomized clinical trials in COVID-19 patients. A few patients on treatment with remdesivir faced adverse effects including nausea, elevated alanine aminotransferase level and respiratory failure [12]. Chloroquine 2 and hydroxychloroquine 3 (Fig. 1) were granted by the FDA for a while for COVID-19 treatment, but were revoked because of their serious side effects [13]. Other antiviral and immunomodulatory drugs, namely favipiravir (avigan ®) 4, tilorone (amixin ®) 5 and umifenivir (arbidol ®) 6 (Fig. 1), glucocorticoid dexamethasone and IL-6 receptor blocker monoclonal antibody tocilizumab have been approved in many European and Asian countries including Russia and China for treatment of COVID-19 patients, but their efficacy, adverse effects and mechanisms of action are not yet fully elucidated [[14], [15], [16]]. Several vaccines have been approved globally for their emergency use as preventive measure against COVID-19 virus infection via production of neutralizing antibody against viral spike S glycoprotein infection, however, their side effects, safety and efficacy are not yet fully established. Moreover, their immunizing antibody neutralization response against SARS-CoV-2 S protein, are not fully effective due to continuous mutations of RBM of the viral spike (S) glycoprotein under different geographical conditions. Some European countries have withdrawn the use of Astra Zeneca vaccine due to its adverse effects including blood clot in a few treatment subjects. Possibly, the efforts in the development of effective drugs and vaccines are hampered due to the limited knowledge on the molecular details of how SARS-CoV-2 infects host cells. For prevention of COVID-19 pandemic, the scientific community on the basis of their extensive innovative research on the cellular and molecular mechanisms of SARS-CoV-2 virus and other related human coronavirus infections in host cells, have highlighted the significance of virus-host proteins interactions and identified the major molecular determinants of the disease for the development of target-specific therapeutic interventions. The viral entry into host cells and replication of viral genome in the infected host cells are the key stages of SARS-CoV-2 life cycle. It is a complex process, which involves the action of several host and viral proteases in different stages of virus entry and replication in host cells. The study on the mechanisms of SARS-CoV-2 infection in host cells, demonstrates that some proteases of SARS-CoV-2 act as molecular connectors to host cell replication machinery to hijack the host replication machinery and to utilize it for viral RNA replication in the virus-infected host cells.
Fig. 1.
Chemical structures of some currently prescribed clinical drugs for COVID-19 treatment.
2. Major therapeutic targets in inhibition of SARS-CoV-2 infection
2.1. Targets in inhibition of viral entry into host cells
For virus entry into host cells, SARS-CoV-2 spike (S) glycoprotein requires host angiotensin-converting enzyme 2 (ACE2) or the transmembrane protein CD147 as receptor on the target cell and employs the host cellular serine proteases TMPRSS2 or cathepsin L and furin-like protease for priming, which facilitates the cleavage of S protein at the S1/S2 (multibasic PRRAR interface) site and the internal of S2 (S2′) site, and allows the fusion of the viral and host cellular membranes and this process is driven by S2 subunit. The binding of S protein to the ACE2 receptor depends on receptor binding motif (RBM), located in the receptor binding domain (RBD) of S1 unit, and cell to cell fusion depends on the activation of S2 unit. However, an extensive study on the mechanism of ACE2 and S protein interactions demonstrates that S1/S2 priming is prerequisite for subsequent TMPRSS2-mediated activation at the S2′ site, but not for S2′ site activation by cathepsin L in TMPRSS2-negative cells. In highly TMPRSS2 overexpressing cells, such as in lung Calu-3 cells, the viral entry occurs through fusion at the plasma membrane, while in cells lacking sufficientTMPRSS2, such as in human ACE2 expressing HEK-293T cells, S1/S2 priming is redundant and virus is endocytosed and fusion occurs late in acidified endo/lysosomal compartments by activation with cathepsin L. The treatment of E−64d, a blocker of cathepsin B/L activity, reduced the infection of SARS-CoV-2 pseudotyped virus in 293T cells by 90%. In such cases, S1/S2 cleavage occurs by some proteases beyond furin. It clearly indicates that the cleavage at the S2′ site by TMPRSS2 promotes plasma membrane fusion process for viral entry in the attached host cell, while cleavage of S2’ site by cathepsin L promotes clathrin-mediated endocytosis for endosomal entry of the virus. Therefore, inhibition of the activity of the proteases, TMPRSS2 or cathepsin L is a potential therapeutic strategy for inhibition of viral entry into the host cells. The utilization of host cellular serine protease TMPRSS2 or cathepsin L by the SARS-CoV-2 virus for its entry depends on the types of infected host cells and availability of these host proteases. For instance, camostat mesylate, an inhibitor of TMPRSS2 activity, on treatment in SARS-CoV-2 infected TMPRSS2 expressing Caco-2 cells, efficiently blocked the entry of the virus in Caco-2 cells, while this inhibitory effect of camostat mesylate in SARS-CoV-2 infected TMPRSS2-negative HEK-293T cells was not observed [[17], [18], [19], [20]]. Another study demonstrated that in high cathepsin L expressing Vero E6 cells, the activation for S1/S2 cleavage is not required in the infection of SARS-CoV-2 in Vero E6 cells, where S1/S2 cleavage is processed by other proteases beyond furin [21].
2.2. Inhibition of SARS-CoV-2 life cycle in infected host cells
A group of studies on the SARS-CoV-2 virus life cycle in infected human host cells identified the physical interactions of virus and human proteins and these interactions lead to paralize the normal functioning of human cells resulting in the death of the cells. Moreover, the virus proteins hijack the replication machinery of human cells for viral replication. Therefore, our adequate knowledge of the molecular details of how SARS-CoV-2 virus infects human cells, well help us to design target-specific antiviral drugs to treat COVID-19.
SARS-CoV-2 immediately after entry into human host cell releases its genomic RNA (g-RNA) in host cell cytoplasm for the onset of a complex programme of viral gene expression, and replication, which are regulated in space and time. The virus initiated the translation of the replicase genes ORF1a and ORF1b from the viral genomic RNA in infected host cells to produce two functional polyproteins pp1a and pp1ab, respectively, by recruiting host cell ribosomes. These two polyproteins are cleaved into sixteen non-structural proteins nsp1-16 by two virus cysteine proteases, one is located within nsp3, known as papain-like protease, PLpro, and another is located within nsp5, known as 3C-like protease (3CLpro), due to its similarity to the picornaviral 3C protease, or as main protease (Mpro), because it is responsible for the processing of the majority (11) of polyprotein1ab cleavage sites. These viral nsps form viral replicase-transcriptase complex (RTC), which consists of multiple enzymes, including PL protease (nsp3), main protease (nsp5), the nsp7-nsp8 primase complex, RNA-dependent RNA-polymerase (RdRp), a helicase-triphosphatase (nsp13), an exoribonuclease (nsp14), an endoribonuclease (nsp15) and N7- and 2’ O-methyltransferases (nsp10 and nsp16, respectively) [6].
The virus nsp1 protein C-terminal binds to cellular 40S and 80S ribosomal subunits and obstructs the mRNA entry tunnel and thereby suppresses host cap-dependent protein translation and gene expression, as well as host innate immune response for clearance of the infection. The shutdown of the key parts of host immune system possibly facilitates efficient viral replication and immune evasion. Therefore, inhibition of the activity of SARS-CoV-2 or nsp1 is a potential target for inhibition of COVID-19 infection. As PL protease generates nsp1 by the cleavage of polyprotein 1a, the inhibition of the activity of PLpro is a potential drug target for prevention of SARS-CoV-2 infection [22].
The main protease (Mpro) is responsible for the processing of polyprotein1ab for release of nsps that are responsible for viral replication via formation of RTC. Therefore, inhibition of the Mpro activity is a potential antiviral drug target. Thus, the compounds that block the activity of Mpro in cell culture assays, could be useful in the development of potential anti-SARS-CoV-2 agents [23].
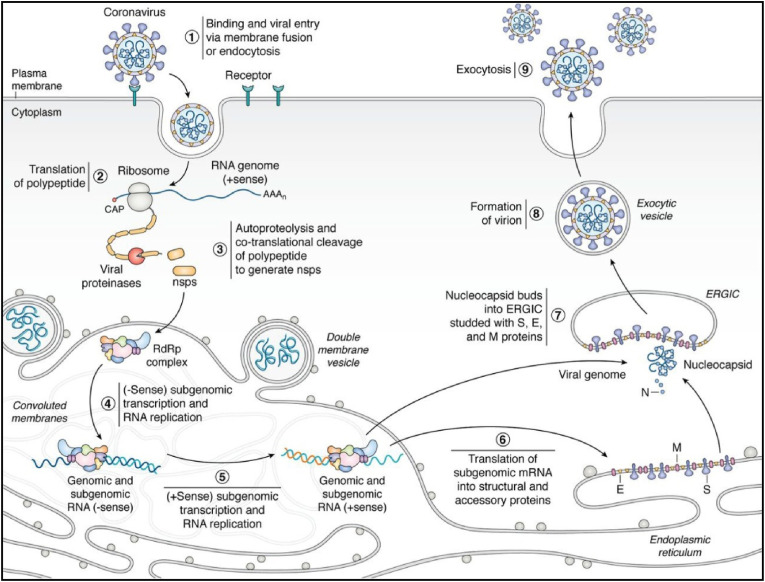
The non-structural proteins nsp2-16 are involved to form viral replicatase-transcriptase complex (RTC), also known as RNA polymerase or RdRp complex for viral RNA synthesis. Among them, nsp2-11 are believed to provide supporting functions to accommodate viral RTC in host cell via modulation of intracellular membrane, host immune evasion and functioning as cofactors in replication [24]. While nsp12-16 are involved in RNA synthesis, RNA proofreading and RNA modification. The nsp12, known as RNA-dependent RNA polymerase (RdRp) is the key enzyme responsible for RNA synthesis with the help of its two cofactors nsp7 and nsp8 as primase complex. The nsp13, known as helicase, unwinds double-stranded (ds) RNA into single stranded negative RNA in a 5′- to 3′- direction for its use as a template for viral RNA synthesis by RdRp. Moreover, it acts as RNA 5′-triphosphatase (RTPase) that may be involved in viral mRNA capping. The nsp14 provides a 3′-5′ exonuclease activity that assists RNA proofreading function. The nsp16 performs the function of 2′-O-methyltransferase activity in RNA modification. The interactions of these nsps with host cell factors determine the efficacy of viral RNA synthesis from RTC. The viral RTC undergoes a series of molecular events for the synthesis of full-length negative-sense genomic RNA copies, which function as templates for generation of new positive-sense genomic RNAs. These new genomic RNAs are packaged into new virions with the help of structural nucleocapsid (N) protein. The newly generated enveloped virions are exported from the infected host cell into adjacent host cell by cxocytosis [24]. The mechanism of viral replication and infection in host cells by SARS-CoV-2 and other coronaviruses is depicted in Fig. 2 .
Fig. 2.
The mechanism of infection and replication (life-cycle) of SARS-CoV-2 and other coronaviruses in host cells. (1) The viral entry into host cell on binding to host cell ACE2 receptor followed by membrane fusion or endocytosis and deposition of viral genomic RNA (g-RNA) into host cell cytoplasm. (2) The virus replicase genes ORF1a and ORF1b from (+)-sense- g-RNA are translated by host translation machinery into functional polyproteins pp1a and pp1ab, respectively. (3) The polyproteins 1a and 1 ab are cotranslationally cleaved by two virus cysteine proteases, papain-like protease (PLpro) and 3-chymotrypsin-like protease (3CLpro) or main protease (Mpro) into 16 non-structural proteins (nsp1-16) and to form viral replicase-transcriptase complex (RTC) or RdRp complex. (4) The RTC uses the viral g-RNA as a template to generate (−)-sense subgenomic (sg) and genome-length (g) RNAs. (5) These (−)-sense sg- and g-RNAs are used as templates for synthesis of (+)- sense full-length progeny genomes and sg- RNAs. (6) The components of RTC carry out transcription and replication of virus in CMs adjacent to DMVs that are both derived from host cell rough endoplasmic reticulum (ER). (7) The (+)-sense g-RNA is bound by viral nucleocapsid (N) protein and buds into ERGIC and these nucleocapsid buds are decorated with structural proteins S, E, and M translated from (+)-sense sg-RNAs to form enveloped virions. (8) and (9) The newly formed enveloped virions are exported from the infecred cell into adjacent host cell by exocytosis for infection and replication. [Adapted from Ref. [24]].
The multifunctional SARS-CoV-2 N protein on localizing in RTC, promotes viral RNA synthesis and translation by recruiting host factors and promotes RNA template switching from discontinuous to continuous transcription. Moreover, it protects the virus from the host immune response and interacts with many host proteins, namely binds to stress granule proteins G3BP1 and G3BP2 and to other host mRNA binding proteins including the mTOR-regulated translational repressor LARP1, two subunits of casein kinase 2, and mRNA decay factors UPF1 and MOV10, suppresses their expression and activity in SARS-CoV-2 infected cells to increase viral mRNA translation. Available evidence demonstrates that the blocking of stress granule (SG) formation by reduction of the levels of SG marker proteins G3BP1/2 significantly increases viral replication in coronavirus-infected cells. Moreover, the mammalian target of repamycin complex 1 (mTORC1), a critical regulator of protein synthesis, increases the activity of La-related protein 1 (LARP1) that binds strongly to the terminal oligopyrimidine (TOP)-mRNA and suppresses the binding interaction of eIF4G to TOP-mRNA and thereby negatively influences the translation of viral mRNA in infected cells. All coronavirus mRNAs rely on their cap-dependent translation by producing proteins eIF4F complex, consisting of eIF4E, eIF4G and the DEAD-box helicase eIF4A, in a process enhanced in trans by SARS-CoV N protein. The inhibitors of eIF4F/eIF4A have been reported to increase G3BP proteins aggregation and to reduce viral translation. Possibly, N protein on phosphorylation by host GSK-3β, recruits host helicase DDX1 to bind viral RTC and assists the viral proteins in 3′- to 5′- unwinding for long range continuous translation. Further, host protein arginine methyltransferase 1 (PRMT1) methylates SARS-CoV-2 N protein at residues R95 and R177 in RGG/RG motifs, for viral packaging and virion production.Therefore, inhibition of the activity of virus cap-binding protein complex eIF4F or the expression of N protein or the PRMT-1 activity may be a potential therapeutic target in inhibition of SARS-CoV-2 infection [[25], [26], [27], [28], [29], [30]].
Host cells produce variety of antiviral factors to create an antiviral state and targets for inhibition of viral infection. For instance, host epigenetic regulator histone deacetylase 2 (HDAC2) protein expression is upregulated to stimulate the expression of interferon-related gene, viperin in virus-infected cells for inhibition of viral infection. The SARS-CoV-2 nsp5 interacts with host HDAC2 and inhibits its transport into the nucleus and thereby potentially inhibits the ability of HDAC2 in induction of host innate antiviral response [31]. Further, host cellular tRNA methyltransferase 1 (TRMT1) gene is responsible for catalyzing the dimethylguanosine (m2,2G) in both nuclear and mitochondrial tRNAs for maintenance of cellular redox homeostasis and growth via protein synthesis. The mutation of TRMT1 in viral and cytotoxic infections results in the defects in RNA binding and loss of cellular stress sensitivity [32]. An interaction of SARS-CoV-2 nsp5 with host TRMT1 leads to the removal of the zinc finger from TRMT1, and thereby results in an exclusive mitochondrial localization and prevents its function in cellular translation and protein synthesis [30]. Therefore, inhibition of the activity of virus main protease, nsp5 is a potential therapeutic target against SARS-CoV-2 infection.
The formation of viral RTC is crucial for virus replication and hence a promising target for antiviral drugs against SARS-CoV-2. The RdRp is the key enzyme in RTC, responsible for viral RNA synthesis. Therefore, the inhibitors of RdRp enzyme activity in cell culture assays in low nano or micromolar concentration ranges could be useful in development of potential anti-SARS-CoV-2 drugs.
A group of accumulating evidence demonstrates that SARS-CoV-2 and other positive-sense RNA viruses induce modification of the ER membrane by co-opting the ER-associated protein biosynthesis machinery and creating characteristic membrane structures, known as double membrane vesicles (DMVs) or replication organelles (ROs) or replication factories, the sites for viral RNA synthesis. These DMVs provide a way to shield the replicative materials, which would otherwise be recognized by cellular defenses and spark an innate immune response for its destruction. A SARS-CoV-2 virus and human protein interaction map reveals that virus nsp8 protein interacts with three signal recognition particles (SRPs), namely SRP19, SRP54 and SRP72 to utilize them in viral protein synthesis via their RNA GTPase activity [33] and other host protein, Sec 61 translocon, an essential machinery of the ER for nascent proteins transport and processing, for manipulation of ERAD (ER-associated degradation) pathway and use them in manufacturing these DMVs and in synthesis of viral RNAs. The inhibitors of Sec61 have been reported to reduce viral replication. The virus nsp8 showed good interactions with host EDEM3 (ER-degradation enhancing α-mannosidase like protein 3), OS-9, an ER-resident lectin that targets misfolded glycoproteins in ERAD pathway and two ERAD (ER-associated degradation)-related proteins that are abundant in coatomer complex II-independent vesicles. These proteins are known as ‘EDEMoses’ and are the key elements in cellular proteostasis adjustment via controlling the ERAD components. These EDEMosomes are used by SARS-CoV-2 in the formation of DMVs and for viral proteostasis as well as to protect the viral RTC from the sensors of cytosol and host IFNs. Therefore, inhibition of the activity of Sec 61 in virus-infected cells, is a potential therapeutic target in inhibition of SARS-CoV-2 infection [30,34,35].
The SARS-CoV-2 virus nsp3 and nsp4 generate the DMVs, whereas nsp6, through oligomerization, colocalyzes within the ER-reporter protein Cb5 (the C-tail of cytochrome b5) and acts as a filter in communication between the replication organelle (RO) and the ER, and allows the lipid flow but restricts the access of ER luminal proteins or ER membrane proteins into the DMVs. This ER zippering activity of nsp6 depends on the adaptive evolution of SARS-CoV-2 variants of concern (VOC). Six variants of SARS-CoV-2, namely Alpha, Beta, Gamma, Eta, Iota and Lambda have a three amino acid residues deletion event (ΔSGF, in positions 106–108) during evolution, in the predicted second and longest nsp6 luminal loop, and have higher zippering activity, which in turn increases transmissibility, infectivity and immune escape of the virus. Moreover, viral nsp6 acts as an organizer of DMV clusters and mediates a contact with lipid droplets (LDs) through LD-tethering complex DFCP1-RAB18 [36]. Available evidence demonstrates that the RO and the LD contacts are important players in the development and progression of viral infection via utilization of released fatty acids from LD generated by lipophagy, as an energy source for morphogenesis of progeny virions [37]. Further, SARS-CoV-2 nsp6 suppresses host IFN-1 signaling more efficiently than SARS-CoV and MERS-CoV by inhibiting TBK1 and IRF3 activation and promotes viral pathogenesis [38]. In addition, SARS-CoV-2 nsp6 directly interacts with host sigma-1 receptor (Sig-1R) to promote viral replication. Under cellular stress, Sig-1R resides in the ER reticular network and plasma membrane and regulates ER stress responses, Ca2+ homeostasis, lipid remodelling, protein degradation and autophagy for cell survival [30,39]. Therefore, the inhibition of the activity of SARS-CoV-2 nsp6 is a promising drug target against COVID-19 infection.
SARS-CoV-2 virus envelope (E) protein plays an important role in viral life cycle and infection. It interacts with virus M protein to maintain the shape of viral particles (virions). Moreover, it plays a key role in the intracellular transport of virions via its viroporin or cation ion channel activity. The E viroporin maintains Ca2+ homeostasis by increasing Ca2+ permeability in a voltage-dependent manner. Other virus proteins orf3a and orf8a also generate ion channels, the activity of ion permeability of E protein ion channel is higher than that of orf3a and orf8a. The ion channel proteins induce inflammasome activity and upregulate the expression of inflammatory cytokines including IL-1β. Suppression of E protein expression of SARS-CoV-2 causes significant reduction in viral titer and maturity, suggesting the important role of E protein in virus production and maturation. Moreover, blockade of E protein channel activity significantly reduces the viral pathogenicity. Therefore, suppression of E protein expression is a potential antiviral and vaccine target [40].
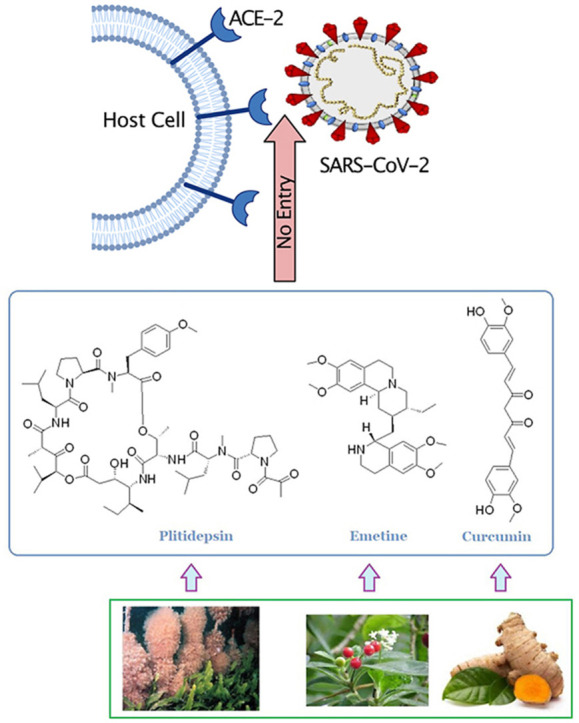
3. Promising natural compounds and their synthetic analogues to fight against SARS-CoV-2 and other viral infections
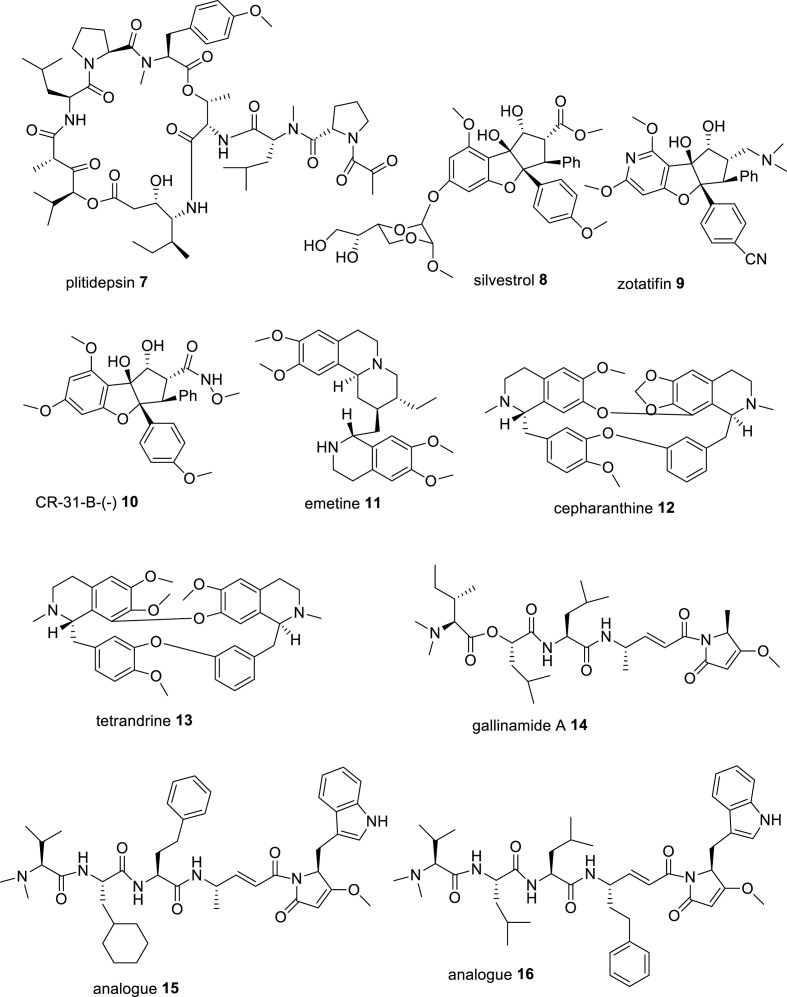
Plitidepsin, also known as dehydrodidemmin B 7 (Fig. 3 ), a cyclic depsipeptide, isolated from Mediterranean marine tunicate Aplidium albicans, later on synthesized and commercialized under the trade name aplidin® and is developed for treatment of various types of tumors including multiple myeloma, has been found to exhibit potential antiviral activity against SARS-CoV-2 virus. It inhibited the infection of SARS-CoV-2 in Vero E6 cells with an IC50 of 0.70 nM (IC90, 1.76 nM) and in human ACE2-expressing 293T cells with an IC50 of 0.73 nM (IC90, 0.88 nM) and showed about 27.5 fold more efficacy than remdesivir (IC90 of 2.25 μM and 24.20 nM in Vero E6 cells and hACE2-293T cells, respectively). The CC50 of plitidepsin in Vero E6 and hACE2-293T cells was 1.99 nM and >200 nM, respectively. Moreover, Plitidepsin inhibited SARS-CoV-2 virus replication in human pneumocyte-like cells with an IC90 of 3.14 nM (IC50, 1.62 nM) and SI of 40.4, suggesting its potent antiviral activity in primary human lung cells. The study of the molecular mechanism of anti-SARS-CoV-2 activity revealed that plitidepsin inhibited viral replication through inhibition of the activity of host eukaryotic translation elongation factor 1A (eEF1A) protein, which is essential for viral translation and gene expression [41]. In vivo, pretreatment of plitidepsin in SARS-CoV-2 infected mice expressing human ACE2 at a dose of 0.3 mg or 1.0 mg/kg for 3 days, significantly reduced the viral load in the lungs of the mice as compared with the vehicle group [41]. Available evidence demonstrates that the translation factor eEF1A promotes replication of many RNA viruses through various mechanisms. In the respiratory syncytial virus (RSV) life cycle, eEF1A interacts with nucleocapsid (N), phosphoprotein (P) and matrix (M) protein in the formation of RTC, where N is the strongest binding partner. It indicates that the interaction of eEF1A with N protein facilitates viral genomic RNA synthesis and virion production [42]. In support of this evidence, plitidepsin in SARS-CoV-2 infected Vero E6 cells significantly reduced viral replication and N protein expression levels [41]. In a preclinical study, plitidepsin was found to distribute preferentially to lung over plasma with almost similar potency against several SARS-CoV-2 variants. In Australia, plitidepsin plus dexamethasone has been approved for the therapy in patients with refractory/relapsed multiple myeloma disease. In an open-label, randomized phase I/II clinical trials (NCT04382066) conducted by a Spanish group, PharmaMar, Madrid, in 10 Spanish hospitals, plitidepsin on iv treatment to the hospitalized COVID-19 patients (n = 46), at the doses of 1.5 mg (n = 15), 2.0 mg (n = 16) and 2.5 mg (n = 15) once daily for 3 days along with dexamethasone phosphate (8 mg/d) showed significant reduction of viral load (70%) and lung inflammation after 15 days, along with increased lymphocyte counts and decreased neutrophil counts, and improved discharge rate of the patients. However, treatment-related adverse events including hypersensitivity, diarrhea, nausea and vomiting were found in more than 5% of the patients and 3 patients died. A few patients were advised to receive additional treatment. The patients did not face any cardiac severity problems. Overall, plitidepsin showed a favourable safety profile in COVID-19 patients. A phase III clinical study to demonstrate the efficacy of plitidepsin in hospitalized COVID-19 patients, who require oxygen therapy is in progress (NEPTUNO: NCT04784559) [43]. Although it is administered intravenously similar to remdesivir, its toxicity profile and side effects remain to be fully elucidated before its commercial application as an alternative of remdesivir in COVID-19 treatment.
Fig. 3.
Chemical structures of some natural compounds and their analogues having potent in vitro anti-SARS-CoV-2 and anti-proteases activities.
Silvestrol 8 (Fig. 3), a natural rocaglate, also known as flavagline having a cyclopenta [b] benzofuran moiety and a dioxan side chain, isolated from South Asian plants, Aglaia silvestris and Aglaia foveolata, has potential antiviral activity against a variety of pathogenic viruses including Ebola virus, poliovirus type 1, Chikungunya virus, picorna virus, hepatitis E virus, Zika virus, influenza A virus, MERS-CoV, HCoV-229E and SARS-CoV-2 in micromolar concentration ranges via inhibition of the activity of eukaryotic initiation factor 4A (eIF4A) protein, essential for viral mRNA translation. It potently reduced the replication of two coronaviruses, HCoV-229E and MERS-CoV in human embryonic lung fibroblast MRC-5 cells with EC50 values of 3.0 and 1.3 nM (EC90 of 27 and 12 nM), respectively, and infection of picornavirus poliovirus type-1 (PV-1) in MRC-5 cells with an EC50 value of 20 nM as well as completely abolished the replication of SARS-CoV-2 in normal human bronchial epithelial cells at a concentration of 100 nM. The study of antiviral mechanism of silvestrol reveals that silvestrol strongly inhibits the expression levels of coronavirus structural and non-structural proteins N and nsp8, and the formation of viral RTC [[44], [45], [46]]. The study of molecular mechanism of cap-binding protein eIF4A reveals that eukaryotic initiation factor 4F (eIF4F), a heterotrimeric complex, composed of helicase eIF4A, cap-binding subunit eIF4E and the scaffold protein eIF4G is required for recruitment of 40S ribosomal subunit to capped mRNAs. The helicase eIF4A unwinds a number of messenger RNAs (mRNAs) in their 5′-untranslated region (5′-UTR) to initiate the binding of 43S preinitiation complex [47]. Silvestrol also modulated host immune system by down-regulating the expression of pro-inflammatory cytokines (IL-6, IL-8 and CCL-2), and amplified the anti-inflammatory potential of M2-macrophages by increasing the expression of anti-inflammatory surface markers CD-206 and TREM2 (triggering receptor expressed on myeloid cells-2) on human monocyte-derived macrophages and dendritic cells to increase phagocytosis via down-regulation of STAT-1 expression in macrophages and STAT-3 in dendritic cells [48]. A synthetic analogue of silvestrol, zotatifin 9 (Fig. 3), was developed by the Effector therapeutic company as a potent inhibitor of eIF4A, and it inhibited the replication of SARS-CoV-2 in Vero E6 cells with an IC90 of 37 nM, suggesting its potency to treat COVID-19 [30]. It is in the process of phase I clinical trials in COVID-19 patients by the Effector group [NCT04632381] [49]. Another synthetic analogue of silvestrol, CR-31-B- (−) 10 (Fig. 3) was developed as a specific inhibitor of eIF4A, and it potently reduced the replication of SARS- CoV-2 in Vero E6 cells with an EC50 of 1.82 nM and SI of >50 and of the replication of HCoV-229E and MERS-CoV in Vero E6 cells with EC50 values of about 2.9 nM and about 1.9 nM, respectively. Moreover, compound 10 almost.
Completely reduced the expression levels of SARS-CoV-2 replication (RTC) and N protein in SARS-CoV-2 infected normal human bronchial epithelial (NHBE) cells, collected from the donors of COVID-19 patients, but this effect was not observed with its (+)-enantiomer [45]. Thus, these rocaglates are promising drug candidates for clinical trials against SARS-CoV-2 infection in COVID-19 patients.
Emetine 11 (Fig. 3), a tetrahydroisoquinoline alkaloid, found in high concentration in the roots of Brazilian plant, Psychotria ipecacuanha Stokes syn. Cephaelis ipecuanha Rich (Rubiaceae) and in other plants of families, Alangiaceae and Icacinaceae, is a FDA approved drug to treat amebic dysentery, caused by Entamoeba histolytica [50]. Several groups reported its potent antiviral activities against several human pathogenic viruses, including Dengue viruses, cytomegaloviruses, HIV-1, Ebola virus, rabies virus, herpes simplex viruses, enterovirus (EV)-A71, EVD68, echnovirus-6, coxsackievirus A16, coxsackie B virus, Zika virus, and several coronaviruses, such as HCoV-OC43, HCoV-NL63, MERS-CoV and SARS-CoV-2 in nanomolar concentration ranges. It potently inhibited the replication of HCoV-OC43, HCoV-NL63, MERS-CoV and MHV-A59 (mouse hepatitis virus) with EC50 values of 0.30, 1.43, 0.34 and 0.12 μM, respectively in virus infected BHK-21, LLC-MK2, Vero E6 and DBT cells, respectively [51,52]. Several groups evaluated the in vitro antiviral efficacy of emetine against SARS-CoV-2. Choy et al. reported that emetine hydrochloride potently inhibited the replication of SARS-CoV-2 in Vero E6 cells with an EC50 of 0.46 μM by reduction of RNA copy numbers and showed better activity than remdesivir (EC50 of 23.15 μM) [53]. Wang et al. reported that emetine efficiently suppressed the replication of SARS-CoV-2 in Vero E6 cells with an EC50 of 0.007 μM and SI of 280 and was about 30-fold more effective than remdesivir (EC50 of 0.24 μM). The study of molecular mechanism of antiviral activity of emetine revealed that emetine significantly decreased the level of virus N protein and on pretreatment in SARS-CoV-2-infected Vero E6 cells blocked the entry of the virus with an EC50 of 0.019 μM, possibly disrupted the binding interaction of virus spike RBD with host ACE2 receptor, while remdesivir did not block the virus entry and only inhibited viral replication [54]. Kumar et al. reported that emetine potently suppressed the replication of SARS-CoV-2 in Vero E6 cells at a low nanomolar concentration with an EC50 of 0.147 nM, CC50 of 1603.8 nM and SI of 10910.4. This group suggested that emetine inhibited the activity of cap-binding protein eukaryotic translation factor 4E (eIF4E) in viral protein translation. Molecular docking and molecular dynamics stimulation studies suggested that emetine binds to the cap-binding pocket of eIF4E in a similar conformation as m7-GTP of viral mRNA binds. They suggested that emetine possibly disrupted the ERK/MNK1/eIF4A signaling of SARS-CoV-2 for viral replication in host cells [55]. Luo et al. has shown that emetine suppressed the number of RNA copies in SARS-CoV-2 infected 293T cells with an IC50 of 0.2729 μM by inhibiting the activity of virus RdRp enzyme, orf6 and nucleocapsid N proteins. The orf6 protein provides a trans-stimulation of viral replication and transcription by blocking of host interferon-α/β (IFN-α/β) signaling and STAT-1 activation. A study on the role of SARS-CoV-2 orf6 protein in viral replication, indicates that orf6 on interaction with host proteins Rae1 and Nup98 that are involved in mRNA nuclear export, reduces their expression to disrupt host mRNA export and immune response against viral infection and preferentially translates viral transcripts and thereby improves viral replication [30,56,57]. The surface plasmon resonance (SPR)-based binding affinity study of emetine-SARS-CoV-2 RdRp complex revealed that emetine binds to RdRp with a dissociation constant KD of 25.7 μM and another alkaloid cephaeline binds with a KD of 19.6 μM. Both emetine and cephaeline showed strong binding interaction with SARS-CoV-2 N protein with KD values of 53.76 and 58.24 μM, respectively and thus prevented the maturation of the virus. Ren et al. suggested that emetine and cephaeline inhibited the replication of SARS-CoV-2 in Vero E6 cells with EC50 values of 0.000117 and 0.0123 μM, respectively in qRT-PCR assay [58]. Therefore, emetine inhibited SARS-CoV-2 replication targeting S protein, RdRp, eIF4A, N and orf6 proteins. In a clinical trial by a Chinese group (registration no; ChiCTR2000030022), treatment of emetine in mild COVID-19 patients at a dose of 3.6 mg per OS, 3 times a day for 10 days showed significant reduction of viral load and negative SARS-CoV-2 RNA in RT-PCR testing without any apparent adverse effects, particularly no severity of cardinal symptoms. Thus, emetine at low dose was much effective in the treatment of COVID-19 patients [59].
Cepharanthine (CEP) 12 (Fig. 3), a natural bis-benzylisoquinline alkaloid isolated from Japanese herb Stephania cephalantha Hayata, and its natural analogue tetrandrine (TET) 13, isolated from Chinese herb Stephania tetrandra S. Moore, potently inhibited the infection of SARS-CoV-2 in high TMPRSS2 expressing Vero E6 cells with IC50 values of 1.90 μM and 10.37 μM, respectively, and IC90 of 4.46 μM and 14.80 μM, respectively, as compared with remdesivir (IC50 of 0.99 μM). The study of the molecular mechanism of anti-SARS-CoV-2 activity revealed that both CEP and TET inhibited viral entry by inhibiting the activities of lysosomal membrane proteins, Niemann-Pick type c intracellular cholesterol transporter-1 (NPC-1) by CEP, and two-pore segment channel-2 (TPC-2) by TET. Accumulating evidence indicates that NPC-1 is a cholesterol transporter protein and acts in preserving cholesterol in the endosomal or lysosomal compartments and facilitates the entry of the virus into the endosome of the host cells, while TPC-2 is an endosomal cation channel and acts in trafficking low density lipoprotein (LDL)-cholesterol molecules and helps in virus entry through endosome into host cells. In silico docking analysis of the conformations of these compounds suggested that the diphenyl ester moiety of these molecules was a putative pharmacophore to inhibit the channel activities of these cholesterol transporter proteins. Possibly, the ammonium cations of these molecules interact with the channel proteins to inhibit their activity [60]. Another study reported that CEP at 10 μM concentration, almost completely abolished the viral RNA level (only 0.08% remained) in SARS-CoV-2 infected Vero E6 cells. Moreover, CEP inhibited SARS-CoV-2 spike (S) protein-mediated pseudovirus entry in ACE2 expressing 293T cells with an EC50 value of 0.315 μM, but in presence of 20 μM BAPTA-AM, a selective Ca2+ ion cheletor, the inhibitory effect on pseudovirus entry of CEP was decreased highly with an EC50 of 3.25 μM. These findings suggested that Ca2+ ion is essential for virus entry into the host cells and S-ACE2-mediated cell fusion is promoted by host calcium channels [61]. These alkaloids showed antiviral activity against other viruses, CEP inhibited the replication of HIV with an IC50 value of 26.4 nM and TET inhibited the replication of Ebola virus with an IC50 value of 55 nM, and both CEP and TET inhibited the infection of HCoV-OC43 in human lung MRC-5 cells with IC50 values of 0.83 μM and 0.33 μM, respectively, and significantly reduced the replication and expression levels of viral S and N proteins in MRC-5 cells [62]. These alkaloids could be useful in clinical study in SARS-CoV-2 infected animals.
Gallinamide A 14 (Fig. 3), also known as symplostatin 4, a natural modified depsipeptide, isolated from marine cyanobacteria of genus Schizothrix and Symploca, collected from the Caribbean coast near Panama and Florida, respectively, has potent antimalarial effect in a murine model [63,64]. Gallinamide A and its two synthetic analogues 15 and 16 (Fig. 3) have been shown to inhibit the infection of SARS-CoV-2 in cathepsin L (Cat L) overexpressing Vero E6 cells in nanomolar concentration ranges with EC50 values of 28, 168 and 920 nM, respectively, and CC50 values of >100 μM. Moreover, these compounds 14–16 strongly in vitro inhibited the activity of human recombinant cathepsin L (Cat L) activity with IC50 values of 17.6, 5.6 and 17.0 pM, respectively. Further, synthetic analogues 15 and 16 completely inhibited the cytopathic effect (CPE) of SARS-CoV-2 in human alveolar basal epithelial ACE2 expressing A549 cells at a concentration of 310 nM, while the natural molecule 14 did it at a concentration of 625 nM. These compounds showed no host cell cytotoxicity up to 100 μM concentration and also safe in animal models. In addition, gallinamide A 14 and its synthetic analogue 15 completely neutralized the entry of SARS-CoV-2 in TMPRSS2 overexpressing human fetal lung fibroblast MRC-5/ACE2/TMPRSS2 cells at a concentration of 32 nM, as compared with TMPRSS2 inhibitor nafamostat mesylate, which completely blocked the entry of virus in the said cells at 4 μM concentration. These results suggested that Cat L is still important for viral entry in TMPRSS2 overexpressing cells. Although, both Cat L and TMPRSS2 are highly expressed in lung tissue and other tissues, however, their preferential expression depends on the cell type and organ system. For instance, both TMPRSS2 and Cat L protease pathways are operational and important for induction of infection of SARS-CoV-2 virus in MRC-5/ACE2/TMPRSS2 cells, but Cat L is the predominant pathway in Vero E6 cells. In a study on the antiviral activity of gallinamide A in inhibition of SARS-CoV-2 infection in Vero E6 cells, gallinamide A was added to various time points of the Vero cell culture from 30 min pre-infection to 120 min post-infection and virus load was examined after 24 h post-infection, the inhibitory effect was maximum when 14 was added prior to or at the time of infection and this effect was substantially reduced when added at 60 min or later post-infection. The TMPRSS2 inhibitor nafamostat mesylate showed potent antiviral effect against SARS-CoV-2 infection in HEK-293T-/ACE2/TMPRSS2 cells, but not effective in Vero E6 cells. Moreover, a combination of gallinamide A and nafamostat mesylate showed a better antiviral effect against SARS-CoV-2 in HEK-293T-/ACE2/TMPRSS2 cells by their synergistic effect. The compounds 14–16 were inactive against SARS-CoV-2 Mpro and PLpro enzymes. These compounds could be potential Cat L inhibitors in COVID-19 treatment. However, their short-term treatment may be fruitful to develop host innate and adaptive responses against viral infection, but long-term treatment may cause adverse effects including bone and heart defects [65].
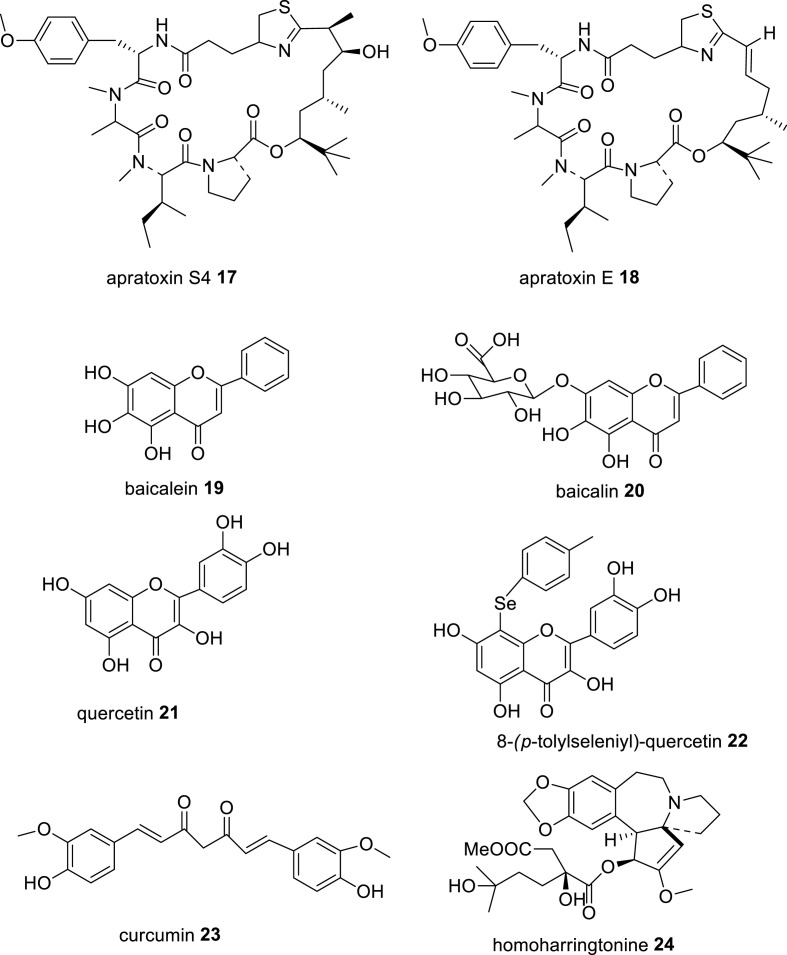
Apratoxin S4 17 (Fig. 4 ), a synthetic analogue of natural heteromeric depsipeptide apratoxin E 18 (Fig. 3), was developed as potent antitumor drug against ocular angiogenic disease and potent inhibitor of host protein biosynthesis machinery Sec61 and thereby prevents cotranslational translocation of secretory proteins, receptor tyrosine kinases into tumor cells for their growth, leading to anticancer and anti-angiogenic activities. Apratoxin E isolated from marine cyanobacterium, Lyngbya bouillonii, have potential cytotoxic effect against various cancer cells including colon, cervix and bone cancers [66,67]. Apratoxin S4 (Apra S4) showed potent antiviral effect against SARS-CoV-2 as compared with currently used RdRp inhibitor remdesivir. Apra S4 potently reduced the replication of SARS-CoV-2 in monkey Vero E6 cells and human ACE2-transfected HeLa cells in a dose-dependent manner with IC50 values of 170 nM and 0.71 nM, respectively, and SI of >58 and >1400, respectively, and have about 20- and 50-fold higher potency as compared to remdesivir in Vero E6 and HeLa-hACE2 cells, respectively. It also inhibited the replication of flaviviruses, Zika virus, Dengue virus and West Nile virus in virus-infected Huh-7.5 cells with IC50 values of 15 nM, 3.3 nM and 14 nM, respectively, and SI of >67, >300 and >71, respectively, as well as potently inhibited the replication of influenza A virus in human A549 cells with an IC50 of 0.46 nM and SI of >2200. The study of the molecular mechanism of antiviral activity revealed that Apra S4 inhibited the activity of host Sec61 in suppression of viral replication. Apra S4 at 1 μM concentration on treatment in SARS-CoV-2 infected Vero CCL81 cells, specially designed improved host cells for virus propagation, after 16 h post-infection, markedly reduced the formation of stacked double membrane vesicles, ds-RNA, marker of viral replication, trafficking of spike protein and virions production, while remdesivir at 10 μM concentration was less effective under the same experimental conditions. Possibly, Apra S4 by inhibiting the activity of Sec61, prevented the SARS-CoV-2 virus in the utilization of host protein processing machinery in viral replication process. Its efficacy in SARS-CoV-2 infected animal models could be useful to consider this molecule in clinical trials in COVID-19 patients as an alternative to remdesivir in COVID-19 pandemic [68].
Fig. 4.
Chemical structures of some natural compounds and their analogues having potent in vitro anti-SARS-CoV-2 and anti-proteases activities.
Baicalein 19 (Fig. 4), a major flavonoid constituent of Chinese herb, Scutellaria baicalensis Georgi roots and found in other plants including Oroxylum indicum bark, Thymus vulgaris leaves and Scutellaria lateriflora roots, has been widely used in TCM to treat hepatitis and upper respiratory infection. Baicalein showed potent antiviral activity against SARS-CoV-2 by inhibiting the replication of SARS-CoV-2 in Vero cells with an EC50 value of 2.94 μM and SI of >172, as well as inhibiting the activity of SARS-CoV-2 3CL protease in vitro with an IC50 value of 0.39 μM. A natural glycoside of baicalein, baicalin 20 (Fig. 4), showed a weak inhibitory effect against SARS-CoV-2 3CL protease with an IC50 value of 83.4 μM. The docking analysis of the complex of baicalein with SARS-CoV-2 3CL protease showed a strong binding interaction of baicalein with the catalytic dyad residues His41 and Cys145 of the main protease by the 6- and 7-hydroxyl groups and carbonyl group of baicalein [69]. Another group reported that baicalein and baicalin inhibited the activity of SARS-CoV-2 3CL protease with IC50 values of 0.94 μM and 6.41 μM, respectively, in a FRET-based protease assay. Moreover, baicalein and baicalin inhibited the infection (cytopathic effect, CPE) of SARS-CoV-2 in Vero E6 cells with EC50 values of 2.94 μM and 27.87 μM, respectively, and CC50 s of >200 μM, and SI values of >68 and >7, respectively [70]. Both baicalein and baicalin showed better anti-SARS-CoV-2 effect in human lung epithelial Calu-3 cells with EC50 values of 1.2 μM and 8.0 μM, respectively, and CC50 values of 91 and >100 μM, respectively, as compared to remdesivir (EC50, 0.14 μM). Moreover, both baicalein and baicalin inhibited the activity of SARS-CoV-2 RdRp enzyme in RNA synthesis, and baicalein showed better activity in a biochemical assay using RNA polymerase complex of the virus annealed to a 14-mer RNA template-[α-32P]-GTP as visualization agent of the RNA product. Molecular docking analysis of these flavonoids with SARS-CoV-2 RdRp complex showed that the calculated free binding energy of baicalein and baicalin were −8.7 and −7.8 kcal/mol, respectively as compared with remdesivir (−6.5 kcal/mol). In silico data suggested that baicalein possibly binds to His133 and Asn705 amino acid residues of RdRp. In a thermal shift assay using a protein thermal shift software, baicalein caused an elevation of melting temperature, ΔTf of 3.9 °C of SARS-CoV-2 nsp12, suggesting a strong binding to the main component of RdRp [71]. Baicalein also inhibited the SARS-CoV-2 nsp13 associated unwinding activity in viral translation with an IC50 value of 2.90 μM. The docking analysis suggested that baicalein binds to both orthosteric and allosteric binding sites of helicase nsp13 to prevent the unwinding activity [72]. Several studies on the antiviral activities of natural flavonoids reported that baicalein showed antiviral activity against a wide range of enveloped RNA viruses including influenza A-, Dengue-, Japanese encephalitis-, chikungunya-, human immunodeficiency- and Zika-viruses and non-enveloped RNA viruses, such as enteroviruses. Baicalein potently inhibited the in vitro replication of influenza A pandemic HINI virus with an IC50 of 0.018 μM. Currently baicalein is in a phase II clinical trial for treatment of influenza virus infected patients by a Chinese group, CSPC ZhongQi Pharm. Technology Co Ltd [73]. In.
An animal model, baicalein in its crystal form β having absolute bioavailability of 47.40%, on oral treatment at a dose of 200 mg/kg/d, for 5 days to h-ACE2-transgenic mice-infected with SARS-CoV-2, significantly inhibited body weight loss, replication of the virus and reduced viral load and lesions in the lung tissue of the mice. In LPS-induced acute lung injury in mice, baicalein improved respiratory function and decreased inflammatory cell infiltration in lung tissue and the elevated levels of serum IL-1β and TNF-α in mice [73]. It indicates that baicalein could be a potential natural drug for treatment of COVID-19 patients.
Quercetin 21 (Fig. 4), another plant flavonoid, found in several common dietary fruits, vegetables and grains, and in medicinal plants, has been used in food supplement, due to its potential antioxidant, anti-inflammatory, immunomodulating and antiviral activities. It has been reported to exhibit antiviral activity against many RNA and DNA viruses including respiratory syncytial virus (RSV), polio type 1 virus, para-influenza type 3 virus, herpes simplex virus-1 (HSV-1), cytomegalovirus, dengue virus type 2 (DENV-2), coronaviruses- SARS-CoV and SARS-CoV-2 [74]. Quercetin inhibited the SARS-CoV-2 replication in Vero E6 cells in relatively high concentrations, with an IC50 value of 192 μM, while its synthetic analogue, 8-(p-tolylselenyl)-quercetin 22, inhibited the replication of SARS-CoV-2 in Vero E6 cells in relatively low concentrations, with an IC50 value of 8 μM, and quercetin and this analogue were non-toxic to Vero E6 cells up to the concentration of 500 μM and 100 μM, respectively. Moreover, quercetin and its analogue 22 inhibited the activity of the SARS-CoV-2 main protease (Mpro) with IC50 values of 21 μM and 11 μM, respectively, and with inhibition constant (Ki) of 7.4 μM and 3.8 μM, respectively. Possibly, poor bioavailability of quercetin into the cellular compartment is the major reason for its poor anti-SARS-CoV-2 effect. The molecular docking analysis of the complex of analogue 22 with SARS-CoV-2 Mpro showed a strong binding affinity with a free binding energy of −8.2 kcal/mol and binding interaction of quercetin moiety with the catalytic dyad residues Cys145 and His41 as well as a hydrogen bond between the selenium atom and side chain residue Gln189 of the Mpro [75]. Quercetin potently inhibited the SARS-CoV-2 nsp13 helicase RNA unwinding activity with an IC50 value of 0.53 μM and another flavonoid myricetin showed a better activity with an IC50 value of 0.41 μM. Molecular docking analysis showed that both quercetin and myricetin have good binding affinity for 5′-RNA-binding site of the virus RNA [72]. In a clinical trial, quercetin phytosome ® (QP), a lecithin-based novel bioavailable form of quercetin, with 200 mg quercetin in 500 mg capsule, on treatment (1 capsule thrice a day for 7 days followed by 2 capsules per day for next 7 days) to subjects with mild COVID-19, significantly reduced viral load (negative SARS-CoV-2 in RT-PCR test) and improved the blood parameters in reduction of LDH, ferritin, CRP and D-dimer levels in the patients [76].
Curcumin 23 (Fig. 4), the main polyphenolic constituent of Asian traditional spice turmeric, Curcuma longa L., has attracted a significant attention for the treatment of COVID-19 patients because of its versatile biological activities, such as anti-inflammatory, antioxidant, immunomodulatory, antifungal, antiviral and antitumor activities. It showed potent antiviral activities against a broad-range of viruses including Dengue virus, Zika virus, chikungunya virus, enterovirus, HIV, respiratory syncytial virus, hepatitis B virus (HBV), HCV, influenza A virus and SARS-coronaviruses- SARS-CoV and SARS-CoV-2 [77]. It potently inhibited the infection of SARS-CoV-2 D614G and Delta strains in Vero E6 cells with EC50 values of 4.06 μM and 1.14 μM, respectively, in pre-postinfection treatment, and SI of 4.06 and 14.5, respectively, and CC50 of 16.5 μM. Moreover, at a concentration of 10 μg/ml, it reduced the infectivity of these tested SARS-CoV-2 strains by more than 99% in Vero E6 cells. Further, on pretreatment (10 μg/ml) in SARS-CoV-2 infected peripheral blood mononuclear cells (PBMCs), significantly decreased the elevated expression levels of pro-inflammatory cytokines, IL-1β, IL-6 and IL-8 [78]. Accumulating evidence demonstrates that SARS-CoV-2 D614G strain plays a dominant role in global COVID-19 pandemic because of its spike protein evolution in composition of amino acid residues and fitness advantage in different environments increases its efficient replication and transmission, which leads to greater infectivity and higher viral load in upper respiratory tract, but not in disease severity in COVID-19 patients [79] and Delta variant (sub-type B.1.617.2) is believed to spread faster because its spike (S) protein increases the immune evasion potently due to its diverse mutations in the N-terminal domain (NTD) and RBD as compared to other variants. The antibody neutralization property of Pfizer or Astra-Zeneca vaccine is about five-fold lower efficient against the Delta variant than against the Alpha-variant (B.1.1.7) [80]. The study of molecular mechanism of antiviral effect revealed that curcumin inhibited the activity of the SARS-CoV-2 3CL protease with an IC50 value of 11.9 μM in a FRET-based protease assay [81]. Moreover, curcumin inhibited the activity of host cellular protease TMPRSS2, which is one of the major activating proteases of host cell receptor ACE2 for the entry of SARS-CoV-2 in host cells [82]. Molecular docking analysis reported that curcumin showed good binding interaction to the SARS-CoV-2 spike protein and host ACE2 receptor with BE of −7.9 kcal/mol and −7.8 kcal/mol, respectively [83]. In addition, curcumin inhibited the activity of vacuolar-ATPase expression, which acts as a proton pump during acidification of endosomal vesicles and is essential for activation of cathepsin L for the viral entry via endosomal membrane fusion, as well as for maturation of the virions in the endosome for its transport to lysosomes for replication [84,85]. Curcumin on treatment in influenza A virus (IAV)-infected mice significantly improved the survival rate of mice by reducing the lung inflammatory cytokines levels, viral load and fibrosis and improving the pulmonary function via activating the Nrf2 signal and suppressing the IAV-induced activation of TLR2/4, p38/JNK MAPK and NF-ĸB signaling pathways [86]. Pulmonary fibrosis is a devasting outcome of COVID-19 infection. SARS-CoV-2 infects upper and lower respiratory tracts, results in the induction of TLR signals for activation of inflammasome and generation of inflammatory cytokines including IL-1β and IL-6, which mediate pulmonary inflammation and fibrosis [87]. Therefore, curcumin is effective in suppression of pulmonary inflammation and fibrosis in COVID-19 patients. In a clinical trial (CTRI/2020/05/025482), curcumin formulation (curcumin C3 complex ® (Sami Direct, India) containing curcumin 525 mg and piperine 2.5 mg in tablet) on supplementation as adjunct therapy in mild to severe COVID-19 patients at a dose of 2 tablets daily for 14 days, significantly improved general symptoms (reduction of fever, cough, sore throat and breathlessness) and pulmonary inflammation and duration of hospitalization and deaths [88].
Homoharringtonine (HHT) 24 (Fig. 4), a natural alkaloid isolated from the plant Cephalotaxus harringtonii, has been prescribed in TCM for treatment of chronic myeloid leukemia and other tumor diseases for more than 30 years. HHT showed potent antiviral activity against diverse DNA and RNA viruses including vesicular stomatitis virus (VSV), new castle disease virus (NDV), herpes simplex virus type 1 (HSV-1), porcine epidemic diarrhea virus (PEDV), hepatitis B virus (HBV), echovirus-1 and coronaviruses in the nanomolar concentrations. For instance, it inhibited the replication of HSV-1 and PEDV in Vero cells with IC50 values of 139 nM and 112 nM, respectively, and SI of 40 and 50, respectively. The study of molecular mechanism of antiviral activity of HHT revealed that it reduced the phosphorylation level of the endogenous and exogenous eukaryotic initiation factor 4E (p-eIF4E), an essential protein that regulates the selective translation of specific mRNA in a cap-binding way of DNA and RNA viruses [89]. HHT potently inhibited the infection of SARS-CoV-2 virus, collected from COVID-19 patient, in Vero E6 cells with an EC50 value of 2.55 μM at 48 h post-infection, and reduced the viral RNA copy numbers in Vero E6 cells with an EC50 value of 2.14 μM and CC50 of 59.75 μM. Possibly HHT inhibited the viral replication via inhibition of the activity of viral protein translation [59]. Another study reported that HHT reduced the expression of TMPRSS2 in TMPRSS2 expressing primary human bronchial epithelial (BEAS-2B) cells with an IC50 value of 61 nM and inhibited the entry of SARS-CoV-2 pseudotyped virus in TMPRSS2 expressing human intestinal epithelial Caco-2 cells with an IC50 value of 30 nM. Moreover, HHT at 30 nM concentration on treatment for 18 h in human lung cancer Calu-3 cells, significantly reduced the expression of TMPRSS2 in the Calu-3 cells. These findings suggest that HHT prevents the infection of SARS-CoV-2 in Calu-3 cells by inhibition of viral entry via suppression of TMPRSS2 expression and activity. Hence this natural alkaloid could be a potential therapeutic agent against COVID-19 virus [90].
The major therapeutic targets of the promising anti-SARS-CoV-2 natural products and their analogues that are discussed above, as well as their efficacy in inhibition of SARS-CoV-2 infection in host cells with EC50 or IC50 values are documented in Table-1 .
Table 1.
List of some promising natural compounds and their analogues that are discussed in the text is documented along with their potent in vitro anti-SARS-CoV-2 activity in virus-infected host cells and their major therapeutic targets against SARS-CoV-2 via inhibition of the activity of host/virus proteases.
| Natural compound/synthetic analogue | In vitro inhibition of SARS-CoV-2 infection/replication in host cells | Major therapeutic targets against SARS-CoV-2 via inhibition of host/virus protease activity | Reference (s) |
|---|---|---|---|
| Plitidepsin 7 | IC90 of 1.76 nM in Vero E6 cells and 0.88 nM in hACE2/293T cells as compared to remdesivir (IC90 of 2.25 μM and 24.20 nM, resp.) | Inhibits the activity of host eEF1A protease | [41] |
| Silvestrol 8 | Abolishes replication completely at 100 nM in human bronchial epithelial cells | Inhibits the activity of eIF4A protease, N and nsp8 proteins | [45,46] |
| Zotatifin 9 | IC90 of 37 nM in Vero E6 cells | Inhibits the activity of host eIF4A protease | [30] |
| CR-31-B-(−) 10 | EC50 of 1.82 nM in Vero E6 cells | Inhibits the activity of EIF4A and N proteins | [45] |
| Emetine 11 | EC50 of 0.147 nM in Vero E6 cells and SI of 10910.4 as compared to remdesivir (EC50 of 0.24 μM); IC50 of 0.2729 μM in 293T cells in suppression of virus RNA copies | Inhibits the activity of S, eIF4E, RdRp, N and orf6 proteins | [33,[53], [54], [55], [56], [57], [58]] |
| Cepharanthine 12 | IC50 of 1.90 μM in Vero E6 cells and at 10 μM completely abolished the viral replication in Vero E6 cells | Inhibits the activity of S and NPC-1 proteins | [60] |
| Tetrandrine 13 | IC50 of 10.37 μM in Vero E6 cells | Inhibits the activity of S and TPC-2 proteins | [60] |
| Gallinamide A 14 | EC50 of 28 nM in Vero E6 cells and CC50 of >100 μM | Inhibits the activity of cathepsin L with IC50 of 17.6 pM | [65] |
| Synthetic analogue 15 | EC50 of 168 nM in Vero E6 cells and CC50 of >100 μM; completely inhibits the virus infection in ACE2/A549 cells at 310 nM concn | Inhibits the activity of cathepsin L with IC50 of 5.6 pM | [65] |
| Synthetic analogue 16 | EC50 of 920 nM in Vero E6 cells; completely inhibits the CPE in ACE2/A549 cells at 310 nM concn | Inhibits the activity of cathepsin L with IC50 of 17.0 pM | [65] |
| Apratoxin S4 18 | IC50 of 170 nM and 0.71 nM in Vero E6 and hACE2/HeLa cells, resp. and SI of >58 and > 1400 resp.; completely inhibits the viral replication in hACE2/HeLa cells at 2 nM concn | Inhibits the activity of host Sec61 protease | [68] |
| Baicalein 19 | EC50 of 2.94 μM in Vero E6 cells and SI of >172; EC50 of 1.2 μM in Calu-3 cells and CC50 of 91 μM | Inhibits the activity of virus Mpro with IC50 of 0.39 μM; of virus RdRp and helicase nsp13 | [[69], [70], [71], [72]] |
| Quercetin 21 | IC50 of 192 μM in Vero E6 cells and CC50 of 500 μM | Inhibits the activity of virus Mpro with an IC50 of 21 μM and of virus helicase unwinding activity with IC50 of 0.53 μM | [72,75] |
| Quercetin analogue 22 | IC50 of 8 μM in Vero E6 cells and CC50 of 100 μM | Inhibits the activity of virus Mpro with an IC50 of 11 μM | [75] |
| Curcumin 23 | EC50 of 4.06 μM and 1.14 μM against virus D614G and Delta strains in Vero E6 cells, resp; completely inhibits the CPE of these strains at 10 μg/ml concn | Inhibits the activity of virus Mpro with an IC50 of 11.9 μM | [78,81] |
| Homoharringtonine 24 | EC50 of 2.14 μM in Vero E6 cells; IC50 of 30 nM in pseudovirus entry in Caco-2 cells | Inhibits the activity of host TMPRSS2 and eIF4E proteases | [53,89,90] |
4. Concluding remarks
COVID-19 from SARS-CoV-2 virus infection is the third highly pathogenic human coronavirus disease to date. Although it is less deadly than earlier SARS-CoV and MERS-CoV, the rapid spreading of this disease has posed the severest threat to global health in this century. This SARS-CoV-2 outbreak has lasted for more than two and half year now, and is likely to coexist with us for a long time. Till to date no effective drug is available for treatment of this viral infection. Currently prescribed the viral RdRp inhibitors, remdesivir, favipiravir, and molnupiravir and Mpro inhibitors ribavirin,and lopinavir/ritonavir, viral entry inhibitors hydroxychloroquine and umifenovir along with immunosuppressives dexamethasone and tocilizumab in multidrug regimen treatment have been found effective in the reduction of infectious viral loads and general symptoms in severe COVID-19 patients, however, their synergistic efficacy, definite ratio for optimal efficacy and adverse effects including mutagenic effects in patients are not yet fully investigated [91]. Several clinically approved vaccines are widely available and used for antibody neutralization of this viral infection in many countries, however, their efficacy is restricted due to mutations of spike protein of this virus. The natural compounds play a key role in the discovery of drugs against various diseases. Indeed, about half of the approved drugs in global market during the period, between 1981 and 2014, were derived from or mimicked a natural compound [92]. Moreover, in the current COVID-19 pandemic, many patients prefer complementary or traditional medicinal therapies, albeit using them almost exclusively in conjunction with western medicine. For instance, one study reported that almost 92% of 135 hospitalized COVID-19 patients in northeast Chongqing (China) received traditional Chinese medicine in addition to western medicine (antiviral kaletra ® (lopinavir/ritonavir) and interferon) for treatment [93]. Indeed, it is hard to separate the potential effects of, and the interactions between traditional Chinese medicine and western medicine in the treatment of the disease via their anti-proteases targets.
In this review, we have highlighted the potent anti-SARS-CoV-2 and anti-proteases activities of 16 natural compounds including their synthetic analogues in nanomolar concentration ranges for their use in drug design for effective drugs against current COVID-19 pandemic. Among them, 11 compounds, namely plitidepsin, silvestrol and its analogues zotatifin and CR-31-B-(−), emetine, gallinamide A and its two synthetic analogues, apratoxin S4, baicalein and curcumin may be considered as lead molecules for design of effective drugs for COVID-19 infection. In phase I/II clinical trials, the treatments of host eEF1A inhibitor plitidepsin and multi-proteases inhibitor emetine in hospitalized COVID-19 patients showed better efficacy than remdesivir, and hence these compounds could be designed for their balanced clinical doses to get optimal efficacy and minimal toxicity and side effects for treatment in different stages of the disease. As these are clinically approved drugs for treatment of tumor and amebic dysentery, their pharmacokinetics and safety profile have already been evaluated. A recent report on the host DNA mutagenesis by the initial metabolite of viral RdRp inhibitor molnupiravir, β-D-N4-hydroxycytidine, raised a question on the use this drug as well as use of other nucleoside analogues as viral RdRp inhibitors in COVID-19 treatment. The mutagenesis of host DNA mutation may contribute to the development of cancer and birth defects of the offspring [94]. In clinical trials, zotatifin and curcumin on treatment in hospitalized COVID-19 patients showed good efficacy in reduction of viral load and infiltrates in lung tissues. Silvestrol may be utilized for design of more potent anti-SARS-CoV-2 drug than zotatifin. Baicalein, a non-nucleoside viral RdRp inhibitor as well as potential Mpro inhibitor, showed potent efficacy in influenza A virus-infected patients in a phase II clinical trial, could be a promising drug for clinical trial in COVID-19 infection. Apratoxin S4, a synthetic analogue of natural apratoxin E, showed potent in vitro anti-SARS-CoV-2 activity via inhibition of viral DMV and ds RNA formation, protein synthesis and trafficking secretory spike protein, could be a potential candidate for clinical trial against COVID-19 infection. It is highly effective in inhibition of viral life cycle in infected host cells. Gallinamide A and its two analogues have potent inhibitory activity against host protease cathepsin L, responsible for viral entry into host cells. These compounds are the prospective drug candidates for clinical trials in SARS-CoV-2 infected human ACE2-expressing animal models. Homoharringtonine, a potential inhibitor of host TMPRSS2 protease, could be a potential drug candidate for clinical trials in SARS-CoV-2 infected animals. Dietary quercetin and its syntheic Se-derivative could be processed in nanoformulations or liposomes to increase their bioavailability in the disease-targeted lung epithelial tissues for improvement of their efficacy and reduction of host cell toxicity. The alkaloids, cepharanthine and tetrandrine, potent inhibitors of viral entry in endocytosis, could be utilized in the design of effective anti-SARS-CoV-2 drugs in inhibition of viral entry into the host cells. Indeed, it is a great challenge and social commitment of the researchers and pharmaceutical groups to develop effective drugs against SARS-CoV-2 at the earliest to save the people from the present COVID-19 pandemic threat. A multi-disciplinary and global collaborative approach may achieve this goal.
Funding
This work did not receive any specific grant from the funding agencies in the public, commercial or non-profit sectors.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
MD is thankful to the University of Virginia for her PDF support from a grant of NIH, United States in the group of Prof. J. S. Smith, Department of Biochemistry and Molecular Genetics.
References
- 1.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-Cov-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coronavirus disease (COVID-19) pandemic. http://www.who.int./emergencies/diseases/novel-coronavirus-2019/ad group survey Available at: Accessed.
- 3.Aassve A., Alfani G., Gandolfi F., LeMoglie M. Epidemics and trust: the case of the Spanish Flu. Health Econ. 2021;30:840–857. doi: 10.1002/hec.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Shi H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan R.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W., Cheng G., Jiang T. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27(3):325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan J.F., Kok K.H., Zhu Z., Chu H., To K.K., Yuan S., Yuen K.Y. Genome characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan, Emerg. Microbes Infect. 2020;9(1):221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C.T., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustiand K., Oisen-Rasmussen M., Fouchi R., Gunther S., Osterhaus A.D.M.E., Drosten C., Pallensch M.A., Anderson L.J., Bellim W.J. Chaterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300(5624):1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 8.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 9.Tang B., Bragazzi N.L., Li Q., Tang S., Xiao Y., Wu J. An updated estimation of the risk of transmission of the novel coronavirus (2019-nCoV) Infect. Dis. Model. 2020;5:248–255. doi: 10.1016/j.idm.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peiris J.S.M., Guan Y., Yuen K.Y. Severe acute respiratory syndrome. Nat. Med. 2004;10:S88–S97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Middle East respiratory syndrome coronavirus (MERS-CoV) https://www.who.int./emergencies/mers-cov/en Available at: Accessed.
- 12.Goldman J.D., Lye D.C.B., Hui D.S., Marks K.M., Bruno R., Montejano R., Spinner C.D., Galli M., Ahn M.Y., Nahass R.G., Chen Y.S., SenGupta D., Hyland R.H., Osinusi A.O., Cao H., Blair C., Wei X., Gaggar A., Brainard D.M., Towner W.J., Munoz J., Mullane K.M., Marty F.M., Tashima K.T., Diaz G., Subramanian A. Remdesivir for 5 or 10 days in patients with severe COVID-19. N. Engl. J. Med. 2020;383(19):1827–1837. doi: 10.1056/NEJMoa2015301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khuroo M.S. Chloroquine and hydroxychloroquine in coronavirus disease 2019 (COVID- 19), facts, fiction, and hype: a critical appraisal. Int. J. Antimicrob. Agents. 2020;56(3) doi: 10.1016/j.ijantimicag.2020.106101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai Q., Yang M., Liu D., Chen J., Shu D., Xia J., Liao X., Gu Y., Cai Q., Yang Y., Shen C., Li X., Peng L., Huang D., Zhang J., Zhang S., Wang F., Liu J., Chen L., Chen S., Wang Z., Zhang Z., Cao R., Zhong W., Liu Y., Liu L. Experimental treatment with favipiravir for COVID-19: an open-label control study. Engineering. 2020;6(10):1192–1198. doi: 10.1016/j.eng.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ekins S., Lane T.R., Madrid P.B. Tilorone: a broad-spectrum antiviral invented in the USA and commercialized in Russia and beyond. Pharm. Res. 2020;37:71. doi: 10.1007/s11095-020-02799-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang D., Yu H., Wang T., Yang H., Yao R., Liang Z. Efficacy and safety of umifenovir for coronavirus disease 2019 (COVID-19): a systematic review and meta-analysis. J. Med. Virol. 2021;93(1):481–490. doi: 10.1002/jmv.26256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS- CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang K., Chen W., Zhang Z., Deng Y., Lian J.Q., Du P., Wei D., Zhang Y., Sun X.X., Gong L., Yang X., He L., Zhang L., Yang Z., Geng J.J., Chen R., Zhang H., Wang B., Zhu Y.M., Nan G., Jiang J.L., Li L., Wu J., Lin P., Huang W., Xie L., Zheng Z.H., Zhang K., Miao J.L., Cui H.Y., Huang M., Zhang J., Fu L., Yang X.M., Zhao Z., Sun S., Gu H., Wang Z., Wang C.F., Lu Y., Liu Y.Y., Wang Q.Y., Bian H., Zhu P., Chen Z.N. CD-147 spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target Ther. 2020;5(1):283. doi: 10.1038/s41392-020-00426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bayati A., Kumar R., Francis V., McPherson P.S. SARS-CoV-2 infects cells following viral entry via clathrin-mediated endocytosis. J. Biol. Chem. 2021;296 doi: 10.1016/j.jbc.2021.100306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson C.B., Farzan M., Chen B., Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022;23:3–20. doi: 10.1038/s41580-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang T., Jaimes J.A., Bidon M.K., Straus M.R., Daniel S., Whittaker G.R. Proteolytic activation of SARS-CoV-2 spike at the S1/S2 boundary: potential role of protease beyond furin. ACS Infect. Dis. 2021;7(20):264–272. doi: 10.1021/acsinfecdis.0c00701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thoms M., Buschauer R., Ameismeier M., Koepke L., Denk T., Hirschenberger M., Kratzat H., Hayn M., Mackens-Kiani T., Cheng J., Stran J.H., Sturzel C.M., Frohlich T., Berninghausen O., Becker T., Kirchhoff F., Sparrer K.M.J., Beckmann R. Structural basis for translational shutdown and immune evasion by the nsp1 protein of SARS-CoV-2. Science. 2020;369(6508):1249–1255. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ullrich S., Nitsche C. The SARS-CoV-2 main protease as drug target. Bioorg. Med. Chem. Lett. 2020;30(17) doi: 10.1016/j.bmcl.2020.127377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartenian E., Nandakumar D., Lari A., Lv M., Tucker J.M., Glaunsinger B.A. The molecular virology of coronaviruses. J. Biol. Chem. 2020;295(37):12910–12934. doi: 10.1074/jbc.REV120.013930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu C.H., Chen P.T., Yeh S.H. Nucleocapsid phosphorylation and RNA helicase DDX1 recruitment enables coronavirus transition from discontinuous to continuous transcription. Cell Host Microbe. 2014;16(4):462–472. doi: 10.1016/j.chom.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu S., Ye Q., Singh D., Cao Y., Diedrich J.K., Yates J.R., III, Villa E., Cleveland D.W., Corbett K.D. The SARS-CoV-2 nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein. Nat. Commun. 2021;12:502. doi: 10.1038/s41467-020-20768-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai T., Yu Z., Wang Z., Liang C., Richard S. Arginine methylation of SARS-CoV-2 nucleocapsid protein regulates RNA binding, its ability to suppress stress granule formation, and viral replication. J. Biol. Chem. 2021;297(1) doi: 10.1016/j.jbc.2021.100821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakagawa K., Narayanan K., Wada M., Makino S. Inhibition of stress granule formation by Middle East respiratory syndrome coronavirus 4a accessory protein facilitates viral translation, leading to efficient viral replication. J. Virol. 2018;92(20):e00902–e00918. doi: 10.1128/JVI.00902-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fonseca B.D., Zakaria C., Jia J.J., Graber T.E., Svitkin Y., Tahmasebi S., Healy D., Hoang H.D., Jensen J.M., Diao I.T., Lussier A., Dajadian C., Padmanabhan N., Wang W., Matta- Camacho E., Hearnden J., Smith E.M., Tsukumo Y., Yanagiya A., Morita M., Petroulakis E., Gonzalez J.L., Hernandez G., Alain T., Damgaad C.K. La-related protein 1 (LARP1) represses terminal oligopyrimidine (TOP) mRNA translation downstream of mTOR complex 1 (mTORC1) J. Biol. Chem. 2015;290(26):15996–16020. doi: 10.1074/jbc.M114.621730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gordon D.E., Jang G.M., Bonhaddon M., Xu J., Obenier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.l., Tummino T.A., Huttenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Kain A.M., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Menon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkatareman S., Liboy-Lugo J., Lin Y., Huang X.P., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O'Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu D., Wang H.Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Naack J., hubert M., Stroud R.M., Frankel A.D., rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., Zastrow M.V., Verdin E., Ashworth A., Schwartz O., d'Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.R., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., Garcia-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagesh P.T., Hussain M., Galvin H.D., Husain M. Histone deacetylase 2 is a component of influenza A virus-induced host antiviral response. Front. Microbiol. 2017;8:1315. doi: 10.3389/fmicb.2017.01315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dewe J.M., Fuller B.L., Lentini J.M., Kellner S.M., Fu D. TRMT1-catalyzed tRNA modifications are required for redox homeostasis to ensure proper cellular proliferation and oxidative stress survival. Mol. Cell. Biol. 2017;37(21):e00214–e00217. doi: 10.1128/MCB.00214-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Politz J.C., Yarovoi S., Kiroy S.M., Gowda K., Zwieb C., Pederson T. Signal recognition particle components in the nucleolus. Proc. Natl. Acad. Sci. USA. 2000;97(1):55–60. doi: 10.1073/pnas.97.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sicari D., Chatziioannou A., Koutsandreas T., Sitia R., Chevet E. Role of the early secretory pathway in SARS-CoV-2 infection. J. Cell Biol. 2020;219(9) doi: 10.1083/jcb.202006005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ravindran M.S., Bagchi P., Cunningham C.N., Tsai B. Opportunistic intruders: how viruses orchestrate ER functions to infect cells. Nat. Rev. Microbiol. 2016;14:407–420. doi: 10.1038/nrmicro.2016.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ricciardi S., Guarino A.M., Giaquinto L., Polishchuk E.V., Santoro M., Tullio G.D., Wilson C., Panariello F., Soares V.C., Dias S.S.G., Santos J.C., Souza T.M.L., Fusco G., Viscardi M., Brandi S., Bozza P.T., Polishchuk R.S., Venditti R., De Matteis M.A. The role of NSP6 in the biogenesis of the SARS-CoV-2 replication organelle. Nature. 2022;606(7915):761–768. doi: 10.1038/s41586-022-04835-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herker E., Vieyres G., Beller M., Krahmer N., Bohnert M. Lipid droplet contact sites in health and disease. Trends Cell Biol. 2021;31(5):345–358. doi: 10.1016/j.tcb.2021.01.004. [DOI] [PubMed] [Google Scholar]
- 38.Xia H., Cao Z., Xia X., Zhang X., Chen J.Y.C., Wang H., Menachery V.D., Rajsbaun R., Shi P.Y. Evasion of type 1 interferon by SARS-CoV-2. Cell Rep. 2020;33(1) doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vela J.M. Repurposing sigma-1 receptor ligands for COVID-19 therapy. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.582310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao Y., Yang R., Lee I., Zhang W., Sun J., Wang W., Meng X. Characterization of the SARS-CoV-2 E protein: sequence, structure, viroporin, and inhibitors. Protein Sci. 2021;30(6):1114–1130. doi: 10.1002/pro.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.White K.M., Rosales R., Yildiz S., Kehrer T., Miorin L., Morino E., Jangra S., Uccellini M.B., Rathnasinghe R., Coughlan L., Martinez-Romero C., Batra J., Rojc A., Bouhaddou M., Fabius J.M., Obernier K., Dejosez M., Guillen M.J., Losada A., Aviles P., Schotsaert M., Zwaka T., Vignuzzi M., Shokat K.M., Krogan N.J., Garcia-Sastre A. Plitidepsin has potent preclinical efficacy against SARS-CoV-2 by targeting the host protein eEF1A. Science. 2021;371:926–931. doi: 10.1126/science.abf4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei T., Li D., Marcial D., Khan M., Lin M.H., Snape N., Ghildyal R., Harrich D., Spann K. The eukaryotic elongation factor 1A is critical for genome replication of the paramyxovirus respiratory syncytial virus. PloS One. 2014;9(12) doi: 10.1371/journal.pone.0114447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varona J.F., Landete P., Lopez-Martin J.A., Estrada V., Paredes R., Guisado-Vasco P., de Oreuta L.F., Torralba M., Fortun J., Vates R., Barberan J., Clotet B., Ancochea J., Carnevali D., Cabella N., Porras L., Gijon P., Monereo A., Abad D., Zuniga S., Sola I., Rodon J., Vergara-Alert J., Izquierdo-Useros N., Fudio S., Pontes M.J., de Rivas B., de Velasco P.G., Nieto A., Gomez J., Aviles P., Lubomirov R., Belgrano A., Sopesen B., White K.M., Rosale R., Yildiz S., Reuschi A.K., Thorne L.G., Jolly C., Towers G.J., Zuliani-Alvarez L., Bouhaddou M., Obernier K., McGovern B.L., Rodriguez M.L., Enjuanes L., Fernandez-Sousa J.M., Krogan N.J., Jimeno J.M., Garcia-Sastre A. Preclinical and randomized phase I studies of plitidepsin in adults hospitalized with COVID-19. Life Sci. Alliance. 2022;5(4) doi: 10.26508/lsa.202101200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nebigil C.G., Moog C., Vagner S., Benkirane-Jessel N., Smith D.R., Desanbry L. Flavaglines as natural products targeting eIF4A and prohibitins: from traditional Chinese medicine to antiviral activity against coronaviruses. Eur. J. Med. Chem. 2020;203 doi: 10.1016/j.ejmech.2020.112653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muller C., Obermann W., Karl N., Wendel H.G., Taroncher-Oldenburg G., Pleschka S., Hartmann R.K., Grunweller A., Ziebuhr J. The rocaglate CR-31-B- (-) inhibits SARS-CoV-2 replication at non-toxic, low nanomolar concentrations in vitro and ex-vivo. Antiviral Res. 2021;186 doi: 10.1016/j.antiviral.2021.105012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muller C., Schulte F.W., Lange-Grunweller K., Obermann W., Madhugiri R., Pleschka S., Ziebuhr J., Hartmann R.K., Grunweller A. Broad-spectrum antiviral activity of the eIF4A inhibitor silvestrol against corona- and picornaviruses. Antiviral Res. 2018;150:123–129. doi: 10.1016/j.antiviral.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malka-Mahieu H., Newman M., Desaubry L., Robert C., Vagner S. Molecular pathways: the eIF4A translation initiation complex-new opportunities for cancer treatment. Clin. Cancer Res. 2016;23(1):21–25. doi: 10.1158/1078-0432.CCR-14-2362. [DOI] [PubMed] [Google Scholar]
- 48.Blum L., Geisslinger G., Parnham M.J., Grunweller A., Schiffmann S. Natural antiviral compound silvestrol modulates human monocyte-derived macrophages and dendritic cells. J. Cell. Mol. Med. 2020;24(12):6988–6999. doi: 10.1111/jcmm.15360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harrison C. Drug researchers pursue new lines of attack against COVID-19. Nat. Biotechnol. 2020;38:659–662. doi: 10.1038/d41587-020-00013-z. [DOI] [PubMed] [Google Scholar]
- 50.Wiegrebe W., Kramer W.J., Shamma M. The emetine alkaloids. J. Nat. Prod. 1984;47(3):397–408. [Google Scholar]
- 51.Valipour M. Different aspects of emetine's capabilities as a highly potent SARS-CoV-2 inhibitor against COVID-19. ACS Pharmacol. Transl. Sci. 2022;5(6):387–399. doi: 10.1021/acsptsci.2c00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen L., Niu J., Wang C., Huang B., Wang W., Zhu N., Deng Y., Wang H., He F., Cen S., Tan W. High-throughput screening and identification of potent broad-spectrum inhibitors of coronaviruses. J. Virol. 2019;93(12) doi: 10.1128/JVI.00023-19. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choy K.T., Wong A.Y.L., Kaewpreedee P., Sia S.F., Chen D., Hui K.P.Y., Chu D.K.W., Chan M.C.W., Cheung P.P.H., Huang X., Peiris M., Yen H.L. Remdesivir, lopinavir, emetine and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antiviral Res. 2020;178 doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang A., Sun Y., Liu Q., Wu H., Liu J., He J., Yu J., Chen Q.Q., Ge Y., Zhang Z., Hu C., Chen C., Qi Z., Zou F., Liu F., Hu J., Zhao M., Huang T., Wang B., Wang L., Wang W., Wang W., Ren T., Liu J., Sun Y., Fan S., Wu Q., Liang C., Sun L., Su B., We W., Liu Q. Low dose of emetine as potential anti-SARS-CoV-2 virus therapy: preclinical in vitro inhibition and in vivo pharmacokinetic evidences. Mol. Biomed. 2020;1(1):14. doi: 10.1186/s43556-020-00018-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumar R., Afsar M., Khandelwal N., Chander Y., Riyesh T., Dedar R.K., Gulati B.R., Pal Y., Barua S., Tripathi B.N., Hussain T., Kumar N. Emetine suppresses SARS-CoV-2 replication by inhibiting interaction of viral mRNA with eIF4E. Antiviral Res. 2021;189 doi: 10.1016/j.antiviral.2021.105056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luo Y., Yu F., Zhou M., Liu Y., Xia B., Zhang X., Liu J., Zhang Z., Du Y., Li R., Wu L., Zhang X., Pan T., Guo D., Peng T., Zhang H. Engineering a reliable and convenient SARS-CoV-2 replicon system for analysis of viral RNA synthesis and screening of antiviral inhibitors. mBio. 2021;12(1) doi: 10.1128/mBio.02754-20. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Addetia A., Lieberman N.A.P., Phung Q., Hsiang T.Y., Xie H., Roychoudhury P., Shrestha L., Loprieno M.A., Huang M.L., Gale M., Jr., Jerome K.R., Greninger A.L. SARS-CoV-2 ORF6 disrupts bidirectional nucleocytoplasmic transport through interactions with Rae1 and Nup98. J. Virol. 2021;12(2) doi: 10.1128/mBio.00065-21. 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ren P.X., Shang W.J., Yin W.C., Ge H., Wang L., Zhang X.L., Li B.Q., Li H.L., Xu Y.C., Xu E.H., Jiang H.L., Zhu L.L., Zhang N.K., Bai F. A multi-targeting drug design strategy for identifying potent anti-SARS-CoV-2 inhibitors. Acta Pharmacol. Sin. 2022;43(2):483–493. doi: 10.1038/s41401-021-00668-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fan S., Zhen Q., Chen C., Wang W., Wu Q., Ma H., Zhang C., Zhang L., Lu B., Ge H., Yong L., Li B., Yu Y., Chen W., Mao Y., Qu G., Su L., Wang A., Ding Z., Li H., Zhang J., Wang Y., Gao Y., Xu X., Zhu Z., Chen J., Zhang L., Liang H., Wu S., Huang M., Xia Q., Li P., Sun Y., Liang C., Wei W., Liu Q., Sun L. Clinical efficacy of low-dose emetine for patients with COVID-19: a real world study. J. BioX. Res. 2021;4(2):53–59. doi: 10.1097/JBR.0000000000000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hijikata A., Shionyu-Mitsuyama C., Nakae S., Shionyu M., Ota M., Kanaya S., Hirokawa T., Nakajima S., Watashi K., Shirai T. Evaluating cepharanthine analogues as natural drugs against SARS-CoV-2. FEBS Open Bio. 2022;12(1):285–294. doi: 10.1002/2211-5463.13337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.He C.L., Huang L.Y., Wang K., Gu C.J., Hu J., Zhang G.J., Xu W., Xie Y.H., Tang N., Huang A.L. Identification of bis-benzylisquinoline alkanoids as SARS-CoV-2 entry inhibitors from a library of natural products, Signal Transduct. Target. Ther. 2021;6:131. doi: 10.1038/s41392-021-00531-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim D.E., Min J.S., Jang M.S., Lee J.Y., Shin Y.S., Park C.M., Song J.H., Kim H.R., Kim S., Jin Y.H., Kwon S. Natural bis-benzylisoquinoline alkanoids-tetrandrine, fangchinoline, and cepharanthine, inhibit human coronavirus OC43 infection of MRC-5 human lung cells. Biomolecules. 2019;9(11):696. doi: 10.3390/biom9110696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Linington R.G., Clark B.R., Trimble E.E., Almanza A., Urena L.D., Kyle D.E., Gerwick W.H. Antimalarial peptides from marine cyanobacteria: isolation and structural elucidation of gallinamide A. J. Nat. Prod. 2009;72(1):14–17. doi: 10.1021/np8003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Conroy T., Guo J.T., Elias N., Cergol K.M., Gut J., Legac J., Khatoon L., Liu Y., McGowan S., Rosenthal P.J., Hunt N.H., Payne R.J. Synthesis of gallinamide A analogues as potent falcipain inhibitors and antimalarials. J. Med. Chem. 2014;57(24):10557–10563. doi: 10.1021/jm501439w. [DOI] [PubMed] [Google Scholar]
- 65.Ashhurst A.S., Tang A.H., Fajtova P., Yoon M.C., Aggarwal A., Bedding M.J., Stoye A., Beretta L., Pwee D., Drelich A., Skinner D., Li l., Meek T.D., Mckerrow J.H., Hook V., Tseng C.T., Larance M., Turville S., Gerwick W.H., Donoghue A.J.O., Payne R.J. Potent anti-SARS-CoV-2 activity by the natural product gallinamide A and analogues via inhibition of cathepsin L. J. Med. Chem. 2022;65(4):2956–2970. doi: 10.1021/acs.jmedchem.1c01494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mathew S., Schupp P.J., Luesch H., Apratoxin E. A cytotoxic peptolide from a Guamanian collection of marine cyanobacterium Lyngbya bouillonii. J. Nat. Prod. 2008;71(6):1113–1116. doi: 10.1021/np700717s. [DOI] [PubMed] [Google Scholar]
- 67.Chen Q.Y., Liu Y., Cai W., Luesch H. Improved total synthesis and biolgical evaluation of potent apratoxin S4 based anticancer agents with different stability and further enhanced activity. J. Med. Chem. 2014;57(7):3011–3029. doi: 10.1021/jm4019965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pohl M.O., Martin-Sancho L., Ratnayake R., White K.M., Riva L., Chen Q.Y., Lieber G., Busnadiego I., Yin X., Lin S., Pu Y., Pache L., Rosales R., Dejosez M., Qin Y., De Jesus P.D., Beall A., Yoh S., Hale B.G., Zwaka T.P., Matsunaga N., Garcia-Sastre A., Stertz S., Chanda S.K., Luesch H. Sec 61 inhibitor apratoxin S4 potently inhibits SARS-CoV-2 and exhibits broad-spectrum antiviral activity. ACS Infect. Dis. 2022;8(7):1265–1279. doi: 10.1021/acsinfecdis.2c00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu H., Ye F., Sun Q., Liang H., Li C., Li S., Lu R., Huang B., Tan W., Lai L. Scutellaria baicalensis extract and baicalein inhibit replication of SARS-CoV-2 and its 3C-like protease in vitro. J. Enzyme Inhib. Med. Chem. 2021;36(1):497–503. doi: 10.1080/14756366.2021.1873977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Su H.X., Yao S., Zhao W.F., Li M.J., Liu J., Shang W.J., Xie H., Ke C.Q., Hu H.C., Gao M.N., Yu K.Q., Liu H., Shen J.S., Tang W., Zhang L.K., Xiao G.F., Ni L., Wang D.W., Zuo Z.P., Jiang H.L., Bai F., Wu Y., He Y., Xu Y.C. Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients. Acta Pharmacol. Sin. 2020;41:1167–1177. doi: 10.1038/s41401-020-0483-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zandi K., Musali K., Oo A., Cao D., Liang B., Hassandarvish P., Lan S., Slack R.L., Kirby K.A., Bassit L., Amblard F., Kim B., AbuBakar S., Sarafianos S.G., Schnazi R.F. Baicalein and baicalin inhibit SARS-CoV-2 RNA-dependent RNA polymerase. Microorganisms. 2021;9(5):893. doi: 10.3390/microorganisms9050893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Corona A., Wycisk K., Talarico C., Manelfi C., Milia J., Cannalire R., Esposito F., Gibbon P., Zaliani A., Iaconis D., Beccari A.R., Summa V., Nowotny M., Tramontano E. Natural compounds inhibit SARS-CoV-2 nsp13 unwinding and ATPase enzyme activities. ACS Pharmacol. Transl. Sci. 2022;5(4):226–239. doi: 10.1021/acsptsci.1c00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Song J., Zhang L., Xu Y., Yang D., Zhang L., Yang S., Zhang W., Wang J., Tian S., Yang S., Yuan T., Liu A., Lv Q., Li F., Liu H., Hou B., Peng X., Lu Y., Du G. The comprehensive study on the therapeutic effects of baicalein for the treatment of COVID-19 in vivo and in vitro. Biochem. Pharmacol. 2021;183 doi: 10.1016/j.bcp.2020.114302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Biancatelli R.M.L.C., Berrill M., Catravas J.D., Marik P.E. Quercetin and vitamin C: an experimental, synergistic therapy for the prevention and treatment of SARS-CoV-2 related disease (COVID-19) Front Immunol. 2020;11:1451. doi: 10.3389/fimmu.2020.01451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mangiavacchi F., Botwina P., Menichetti E., Bagnoli L., Rosati O., Marini F., Fonseca S.F., Abenante L., Alves D., Dabrowska A., Kula-Pacurar A., Ortega-Alarcn D., Jimenez-Alsanco A., Ceballos-Laita L., Vega S., Rizuti B., Abian O., Lenardao E.J., Velazquez-Campoy A., Pyrc K., Sancineto L., Santi C. Seleno-functionalization of quercetin improves the non-covalent inhibition of Mpro and its antiviral activity in cells against SARS-CoV-2. Int. J. Mol. Sci. 2021;22(13):7048. doi: 10.3390/ijms22137048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pierro F.D., Iqtadar S., Khan A., Mumtaz S.U., Chaudhury M.M., Bertuccioli A., Derosa G., Maffioli P., Togni S., Riva A., Allegrini P., Khan S. Potential clinical benefits of quercetin in the early stage of COVID-19: results of a second, pilot, randomized, controlled and open-label clinical trial. Int. J. gen. Med. 2021;14:2807–2816. doi: 10.2147/IJGM.S318949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mathew D., Hsu W.L. Antiviral potential of curcumin. J. Funct. Foods. 2018;40:692–699. [Google Scholar]
- 78.Marin-Palma D., Tabares-Guevara J.H., Zapata-Cardona M.I., Florez-Alvarez L., Yepes L.M., Rugeles M.T., Zapata-Builes W., Hernandez J.C., Taborda N.A. Curcumin inhibits in vitro SARS-CoV-2 inhibition in Vero E6 cells through multiple antiviral mechanisms. Molecules. 2021;26(22):6900. doi: 10.3390/molecules26226900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827. doi: 10.1016/j.cell.2020.06.043. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Parrot F., Robillard N., Puech J., Prot M., Gallais F., Gantner P., Valey A., Guen J.L., Kassis-Chikhani N., Edriss D., Belee L., Seve A., Courtellemont L., Pere H., Hocqueloux L., Fafi-Kremer S., Prazuck T., Mouquet H., Bruel T., Simon-Loriere E., Rey F.A., Schwartz O. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 81.Bahun M., Jukic M., Oblak D., Kranjc L., Bagc G., Butala M., Bozovicar K., Bratkovic T., Podlipnik C., Ulrih N.P. Inhibition of the SARS-CoV-2 3CL pro main protease by plant polyphenols. Food Chem. 2022;373B doi: 10.1016/j.foodchem.2021.131594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thangapazham R.L., Shaheduzzaman S., Kim K.H., Passi N., Tadese A., Vahey M., Dobi A., Srivastava S., Maheshwari R.K. Androgen responsive and refractory prostate cancer cells exhibit distinct curcumin regulated transcriptome. Cancer Biol. Ther. 2008;7(9):1427–1435. doi: 10.4161/cbt.7.9.6469. [DOI] [PubMed] [Google Scholar]
- 83.Jena A.B., Kanungo N., Nayak V., Chainy G.B.N., Dandapat J. Catechin and curcumin interact with S protein of SARS-CoV-2 and ACE2 of human cell membrane: insights from computational studies. Sci. Rep. 2021;11:2043. doi: 10.1038/s41598-021-81462-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nagahama K., Utsumi T., Kumano T., Mackawa S., Oyama N., Kawakami J. Discovery of a new function of curcumin which enhances its anticancer therapeutic potency. Sci. Rep. 2016;6 doi: 10.1038/srep30962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu J., Cao R., Xu M., Wang X., Zhang H., Hu H., Li Y., Hu Z., Zhong W., Wang M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discover. 2020;6:16. doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dai J., Gu L., Su Y., Wang Q., Zhao Y., Chen X., Deng H., Li W., Wang G., Li K. Inhibition of curcumin on influenza A virus infection and influenzal pneumonia via oxidative stress, TLR2/4, p38/JNK MAPK and NF-ĸB pathways. Int. Immunopharmacol. 2018;54:177–187. doi: 10.1016/j.intimp.2017.11.009. [DOI] [PubMed] [Google Scholar]
- 87.Conti P., Ronconi G., Caraffa A., Gallenga C.E., Ross R., Frydas I., Kritas S.K. Induction of proinflammatory cytokines (IL-1 and IL-6) and lung inflammation by coronavirus-19 (COVI-19 or SARS-CoV-2): anti-inflammatory strategies. J. Biol. Regul. Homeost. Agents. 2020;34(2):327–331. doi: 10.23812/CONTI-E. [DOI] [PubMed] [Google Scholar]
- 88.Pawar K.S., Mastud R.N., Pawar S.K., Pawar S.S., Bhoite R.S., Bhoite R.R., Kulkarni M.V., Deshpande A.R. Oral curcumin with piperine as adjuvant therapy for the treatment of COVID-19: a randomized clinical trial. Front Pharmacol. 2021;12 doi: 10.3389/fphar.2021.669362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dong H.J., Wang Z.H., Meng W., Li C.C., Hu Y.X., Zhou L., Wang X.J. The natural compound homoharringtonine presents broad antiviral activity in vitro and in vivo. Viruses. 2018;10(11):601. doi: 10.3390/v10110601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen Y., Lear T.B., Evankovich J.W., Larsen M.B., Lin B., Alfaras I., Kennerdell J.R., Salminen L., Camarco D.P., Lockwood K.C., Tuncer F., Liu J., Myerburg M.M., McDyer J.F., Liu Y., Finkel T., Chen B.B. A high-throughut screen forTMPRSS2 expression identifies FDA-approved compounds that can limit SARS-CoV-2 entry. Nat. Commun. 2021;12:3907. doi: 10.1038/s41467-021-24156-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kumar S., Caliskan D.M., Janowski J., Faist A., Conrad B.C.G., Lange J., Ludwig S., Brunotte L. Beyond vaccines: clinical status of prospective COVID-19 therapeutics. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.752227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Newman D.J., Cragg G.M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016;79(3):629–661. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- 93.Wan S., Xiang Y., Fang W., Zhang Y., Li B., Hu Y., Lang C., Huang D., Sun Q., Xiong Y., Huang X., Lv J., Luo Y., Shen L., Yang H., Huang G., Yang R. Clinical features and treatment of COVID-19 patients in northeast Chongqing. J. Med. Virol. 2020;92(7):797–806. doi: 10.1002/jmv.25783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhou S., Hill C.S., Sarkar S., Tse L.V., Woodburn B.M.D., Schinazi R.F., Sheahan T.P., Baric R.S., Heise M.T., Swanstrom R. β-D-N4-Hydroxycytidine inhibits SARS-CoV-2 through lethal mutagenesis but is also mutagenic to mammalian cells. J. Infect. Dis. 2021;224(3):415–419. doi: 10.1093/infdis/jiab247. [DOI] [PMC free article] [PubMed] [Google Scholar]