Abstract
Previously an Escherichia coli mutant that had acquired the ability to grow on propanediol as the sole carbon and energy source was isolated. This phenotype is the result of the constitutive expression of the fucO gene (in the fucAO operon), which encodes one of the enzymes in the fucose metabolic pathway. The mutant was found to bear an IS5 insertion in the intergenic regulatory region between the divergently oriented fucAO and fucPIK operons. Though expression of the fucAO operon was constitutive, the fucPIK operon became noninducible such that the mutant could no longer grow on fucose. A fucose-positive revertant which was found to contain a suppressor mutation in the crp gene was selected. Here we identify this crp mutation, which results in a single amino acid substitution (K52N) that has been proposed previously to uncover a cryptic activating region in the cyclic AMP receptor protein (CRP). We show that the mutant CRP constitutively activates transcription from both the IS5-disrupted and the wild-type fucPIK promoters, and we identify the CRP-binding site that is required for this activity. Our results show that the fucPIK promoter, a complex promoter which ordinarily depends on both CRP and the fucose-specific regulator FucR for its activation, can be activated in the absence of FucR by a mutant CRP that uses three, rather than two, activating regions to contact RNA polymerase. For the IS5-disrupted promoter, which retains a single CRP-binding site, the additional activating region of the mutant CRP evidently compensates for the lack of upstream regulatory sequences.
The organization of bacterial genes into operons and regulons under the control of a specific transcriptional regulator allows the cells to adapt to specific growth and survival conditions in a coordinated and parsimonious manner. On the other hand, the mechanistic linkage of gene expression results in the concealment of a considerable portion of the catalytic potential encoded in the genome, the raw material for evolutionary tinkering. We have been using the fuc regulon of Escherichia coli as a model for studying the regulatory rearrangements associated with the recruitment of an enzyme for a novel physiological function.
The fuc gene cluster, located at min 63.2 on the chromosomal map, specifies the enzymes for the utilization of l-fucose as a carbon and energy source (6, 15). The fuc genes are organized into two divergent operons, fucPIK and fucAO, under the positive control of FucR (8) (Fig. 1). FucR is activated by the effector fuculose-1-phosphate, which is the true inducer of the fuc regulon (2). Fucose metabolism is initiated by the sequential actions of a permease (encoded by fucP), an isomerase (encoded by fucI), a kinase (encoded by fucK), and an aldolase (encoded by fucA). The aldolase catalyzes the cleavage of fuculose-1-phosphate to dihydroxyacetone phosphate and l-lactaldehyde. Under aerobic respiratory conditions, l-lactaldehyde is oxidized to l-lactate by an NAD-linked aldehyde oxidoreductase of broad function (encoded by aldA at min 32). l-Lactate is then oxidized to the central metabolite pyruvate by a flavin adenine dinucleotide-dependent dehydrogenase (encoded by the lldD gene of the lldPRD operon at min 81.4; formerly the lctPRD operon). Under anaerobic fermentative conditions, however, redox balance compels the sacrifice of the lactaldehyde as a hydrogen acceptor, at the expense of NADH. This reaction is catalyzed by an NAD-linked oxidoreductase (encoded by fucO). The terminal fermentation product, l-1,2-propanediol, is then released by an exit permease (encoded by an unidentified gene) (25).
FIG. 1.
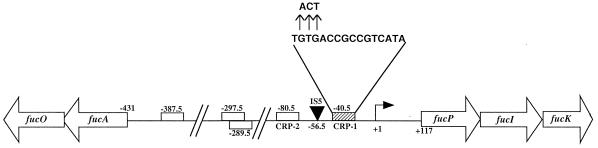
The divergent fuc operons. Shown schematically is the intergenic regulatory region between the fucAO and fucPIK operons. The bent arrow indicates the start point of transcription from the fucPIK promoter (+1). Potential CRP-binding sites are indicated by the rectangles. The sequence of CRP site 1 (hatched rectangle) is shown, as are the three base pair substitutions that were introduced to inactivate this CRP-binding site (vertical arrows). The site of the 1.3-kb IS5 insertion is also indicated (inverted triangle).
Although the fucO gene product catalyzes a reversible reaction, propanediol cannot be utilized as a sole carbon source under aerobic conditions because the compound cannot induce expression of the fuc regulon (9). Mutants that have acquired the ability to grow aerobically on propanediol, however, can be readily selected. Strain ECL3, one such mutant, was found to synthesize propanediol oxidoreductase constitutively at a level that enabled the cells to grow on propanediol almost as rapidly as on glycerol (9, 20). This mutant also synthesized the aldolase constitutively. The permease, isomerase, and kinase, on the other hand, became noninducible. As a consequence, ECL3 lost the ability to grow on fucose. Fine-structure genetic analysis revealed that strain ECL3 sustained an IS5 insertion in the intergenic region between the fucAO and fucPIK operons (the right end of IS5 on the side of fucPIK) (8). Ten other independent propanediol-positive but fucose-negative mutants were also found to have an IS5 inserted between the same base pairs and in the same orientation (15a).
Strain ECL3 was used to select fucose-positive revertants. One such revertant, strain ECL56, was found to express both the fucAO and the fucPIK operons constitutively (7, 11). Mapping experiments showed that strain ECL56 sustained a crp suppressor mutation (crp201) which caused the constitutive expression of fucPIK (24). Here we report the nature of the crp201 allele and the mechanism by which the mutant cyclic AMP receptor protein (CRP) confers the constitutive expression of fucPIK.
MATERIALS AND METHODS
Bacterial strains, growth media, and reagents.
Relevant genotypes and sources of the bacterial strains used in this study are given in Table 1. Luria-Bertani and MacConkey media were prepared as described by Miller (16). MacConkey lactose medium and MacConkey base medium were purchased from Difco Laboratories and prepared as directed. Minimal M9 medium was prepared as described in “Current protocols in Molecular Biology” (1). When used, supplements were added to the following concentrations: 0.2% l-fucose, 0.4% dl-propanediol, 75 μg of carbenicillin/ml, 30 μg of chloramphenicol/ml, 30 μg of kanamycin/ml, and 100 μg of streptomycin/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or characteristic | Source or reference |
|---|---|---|
| Strains | ||
| ECL1 | HfrC fuc+ crp+ | 7 |
| ECL56 | HfrC fucAO(Con) IS5-fucPIK crp201 | 24 |
| CSH100 | F′ lac proA+B+ (lacIqlacPL8)/araD (gpt-lac)5 | Cold Spring Harbor Laboratory |
| CSH100 fucAO(Con) IS5-fucPIK | fucAO(Con) IS5-fucPIK from ECL56 | This study |
| FW102 | F−/araD (gpt-lac)5 Strr | 21 |
| JCB43Δcrp39 | F−/Δ(gpt-proAB-arg-lac)XIII Δcrp39 | 13 |
| JCB43Δcrp39[F′ pfuc-lacZ] | fucPIK promoter-lacZ fusion on F′ | This study |
| JCB43Δcrp39[F′ IS5 pfuc-lacZ] | IS5-containing fucPIK promoter-lacZ fusion on F′ | This study |
| JCB43Δcrp39[F′ crp1 pfuc-lacZ] | CRP site 1 in cis to fucPIK promoter inactivated | This study |
| JCB43Δcrp39[F′ IS5-crp1 pfuc-lacZ] | CRP site 1 in cis to IS5-containing promoter inactivated | This study |
| ECL733 | F−/Δfuc ΔlacU169 | 25 |
| ECL733Δcrp39 | Δcrp39 from JCB43Δcrp39 | This study |
| ECL733Δcrp39[F′ pfuc-lacZ] | fucPIK promoter-lacZ fusion on F′ | This study |
| ECL733Δcrp39[F′ IS5 pfuc-lacZ] | IS5-containing fucPIK promoter-lacZ fusion on F′ | This study |
| ECL733Δcrp39[F′ crp1 pfuc-lacZ] | CRP site 1 in cis to fucPIK promoter inactivated | This study |
| ECL733Δcrp39[F′ IS5-crp1 pfuc-lacZ] | CRP site 1 in cis to IS5-containing promoter inactivated | This study |
| Plasmids | ||
| pHA7Ea | Directs expression of crp+ | 12 |
| pHA7-K52Na | Directs expression of crpK52N | 12a;1. |
| pHA7-H159La | Directs expression of crpH159L | 12a |
| pHA7-K52N,H159La | Directs expression of crpK52N,H159L | 12a |
| pFW11-nullb | Contains promoterless lacZ fragment | 21 |
Provides resistance to carbenicillin.
Provides resistance to chloramphenicol.
Identification of the crp201 mutation.
To define the crp201 mutation, a crp+ fucAO(Con) IS5-fucPIK strain [CSH100 fucAO(Con) IS5-fucPIK] was first constructed. Bacteriophage P1 was used to transduce the fucAO(Con) IS5-fucPIK allele from strain ECL56 to strain CSH100 (crp+). Transductants were selected for the ability to grow on propanediol as the sole carbon and energy source and purified on the same agar. They were then scored for inability to grow on fucose (24). The genotype of strain CSH100 fucAO(Con) IS5-fucPIK, a crp+ fucAO(Con) IS5-fucPIK isolate, was verified by plating the cells on MacConkey-propanediol agar (which gave rise to slightly red colonies) and on MacConkey-fucose agar (which gave rise to pale colonies). Second, the plasmid pHA7E bearing the crp+ allele was used to transform strain ECL56 to allow homologous recombination of the host crp201 and plasmid crp+ alleles. A pool of plasmid DNA was then prepared from the transformants and used to transform strain CSH100 fucAO(Con) IS5-fucPIK. A red transformant colony on MacConkey-fucose agar was then isolated. To verify that this phenotype was plasmid linked, plasmid DNA was isolated from this transformant and used to retransform strain CSH100 fucAO(Con) IS5-fucPIK. All of the resulting transformants manifested the fucose-positive phenotype, suggesting that the crp201 mutation had been transferred to the plasmid.
The plasmid-borne crp201 mutation was then mapped to a HindIII-EagI restriction fragment encompassing codons 1 to 140 of crp. The DNA contained in this restriction fragment was fully sequenced, and a single base pair substitution, specifying the amino acid replacement of lysine 52 by asparagine (K52N), was uncovered.
Construction of fucPIK promoter-lacZ fusion strains.
The wild-type fucPIK promoter and the mutant derivative rendered uninducible by the IS5 insertion were each fused to a lacZ reporter gene. This was done by using the PCR to amplify the relevant promoter regions from the chromosomal DNA of strains ECL1 and ECL56, respectively. Oligonucleotide primers were designed to introduce a BamHI restriction site (underlined) near the start of the fucA gene (5′-CTACCTCTCTCGGATCCAAAACAG-3′) and a SalI restriction site (underlined) near the start of the fucP gene (5′-CCTCTTAGGTCGACAAGCTTAAGCAC-3′). The PCR-amplified DNAs were digested with BamHI and SalI, and the resulting fragments were inserted upstream of a promoterless lacZ gene fragment carried on pFW11-null (21) to generate plasmids pFW11-pfuc-lacZ and pFW11-IS5 pfuc-lacZ. The pFW11-null vector was specially designed to facilitate the transfer of promoter-lacZ fusions onto an F′ episome by homologous recombination (21). Accordingly, the pFW11 derivatives were transformed into strain CSH100 to permit the promoter-lacZ fusions to be recombined onto the F′ episome present in this strain. pFW11 bears a kanamycin resistance gene which is transferred to the F′ episome during the recombination event, thus permitting recombinant episomes to be selected (for details, see reference 21). The recombinant F′ episomes were isolated by mating the CSH100 transformants with F− recipient strain FW102, thus generating strains FW102[F′ pfuc-lacZ] and FW102[F′ IS5 pfuc-lacZ]. Strains FW102[F′ pfuc-lacZ] and FW102[F′ IS5 pfuc-lacZ] were then used as donor strains for the transfer of the F′ episomes into a Δcrp strain (JCB43Δcrp39) to generate a pair of reporter strains designated JCB43Δcrp39[F′ pfuc-lacZ] and JCB43Δcrp39[F′ IS5 pfuc-lacZ].
Derivatives of the two promoter-lacZ fusions (pfuc-lacZ and the IS5 pfuc-lacZ) bearing mutations designed to inactivate CRP-binding site 1 within the fuc regulatory region were constructed (Fig. 1). The mutations (three base pair substitutions) were introduced into plasmids pFW11-pfuc-lacZ and pFW11-IS5 pfuc-lacZ by site-directed mutagenesis using oligonucleotide primers 5′-AAGTACTACCGCCGTCATA-3′ and 5′-CCTAATAAGTACTACCGCCG-3′, respectively (substituted base pairs underlined). The resulting pFW11 derivatives were then used to transfer the mutated promoter-lacZ fusions to an F′ episome as described above. These F′ episomes were again mated from intermediate recipient strain FW102 into strain JCB43Δcrp39, thus generating a pair of reporter strains designated JCB43Δcrp39[F′ crp1 pfuc-lacZ] and JCB43Δcrp39[F′ IS5-crp1 pfuc-lacZ].
The F′ episomes bearing each of the four reporter constructs described above were also transferred to a strain bearing a deletion of the chromosomal fucR gene as well as the crp gene. This ΔfucRΔcrp strain was constructed by using bacteriophage P1 to transduce the Δcrp39 allele from strain JCB43Δcrp39 to recipient strain ECL733 (a ΔfucR strain). Streptomycin-resistant transductants were selected and then screened for resistance to infection by bacteriophage λ (to identify those that had acquired both the streptomycin resistance allele and the linked crp deletion). They were also rechecked for the inability to grow on fucose as the sole carbon source. The resulting strain, ECL733Δcrp39, was used as the recipient for the transfer of each F′ episome from FW102 donor cells. The final four ΔfucRΔcrp reporter strains are designated ECL733Δcrp39[F′ pfuc-lacZ], ECL733Δcrp39[F′ IS5 pfuc-lacZ], ECL733Δcrp39[F′ crp1 pfuc-lacZ], and ECL733Δcrp39[F′ IS5-crp1 pfuc-lacZ].
β-Galactosidase assays.
Sodium dodecyl sulfate-CHCl3-permeabilized cells were assayed for β-galactosidase activity essentially as described previously (16), except that the cells were grown in Luria broth. Each set of assays was performed at least three times in duplicate on separate occasions, with similar results. Shown are the averaged values from a single representative assay; duplicate measurements differed by less than 10%.
DNase I protection experiment.
DNase I protection experiments were performed essentially as described previously (11a), except that the reaction mixtures contained 100 μM cyclic AMP. The restriction fragments bearing the wild-type and the IS5-disrupted fucPIK promoters were derived, respectively, from plasmids pFW11-pfuc-lacZ and pFW11-IS5 pfuc-lacZ after digestion with HindIII and NciI. The promoter-bearing fragments were 3′ end labeled at the HindIII end (∼100 bp downstream of the transcription start site) with [α-32P]dATP and Klenow fragment. The radiolabeled DNA templates were then purified by gel electrophoresis.
RESULTS
To identify the nature of the crp201 allele in strain ECL56, we transferred the mutation to a plasmid-borne crp gene by homologous recombination. A plasmid directing expression of the crp gene was propagated in strain ECL56 to allow the plasmid-borne gene to recombine with the chromosomal crp allele. The plasmid DNA was then purified and transformed into strain CSH100 fucAO(Con) IS5-fucPIK (see Materials and Methods). Because CSH100 fucAO(Con) IS5-fucPIK cells cannot be induced to express the fucPIK operon, they are unable to form colonies on minimal media containing fucose. We therefore plated the transformants on minimal fucose medium to select for those that could express the fucPIK operon due to the acquisition of the crp201 allele that confers the constitutive phenotype in strain ECL56.
We purified the plasmid DNA from a clone that grew on minimal fucose medium and verified that the fucose+ phenotype was plasmid linked. We then mapped the mutation to a restriction fragment encompassing codons 1 to 140 of the crp gene (and the 5′-untranslated region). DNA sequence analysis of this restriction fragment revealed a single base pair substitution resulting in amino acid substitution K52N. Interestingly, this very same mutation has been isolated previously as an intragenic suppressor of a positive control mutation (resulting in the substitution H159L) that specifically impairs the ability of CRP to activate transcription (3). Analysis of this suppression by Williams and coworkers suggested that the K52N substitution results in the creation of an additional activating region on CRP that can be utilized only when the CRP-binding site is positioned suitably close to the binding determinants for RNA polymerase (RNAP) (22, 23). Specifically, it was found that the K52N mutation suppresses the effect of the H159L mutation when CRP activity was assayed at a so-called class II promoter which bears a single CRP-binding site near position −41 relative to the start point of transcription. No suppression occurs at a class I promoter which bears a single CRP-binding site farther upstream (3, 22).
To facilitate analysis of the effect of CRP with the K52N mutation (CRPK52N) on transcription from the fucPIK promoter with or without the IS5 element in the upstream regulatory region, we fused each promoter to the lacZ gene. We used the PCR to amplify a DNA fragment extending from just upstream of the fucP gene to just upstream of the divergently transcribed fucA gene (Fig. 1). The PCR product derived from either the wild-type or the mutant (containing the IS5 element) promoter was cloned upstream of a promoterless lacZ gene. The fucPIK promoter-lacZ fusions were first assembled on a plasmid vector and then transferred to an F′ episome by homologous recombination as described by Whipple (21).
We introduced each of the resulting F′ episomes into a Δlac Δcrp strain, thus generating a pair of reporter strains (JCB43Δcrp39[F′ pfuc-lacZ] and JCB43Δcrp39[F′ IS5 pfuc-lacZ]). We transformed each of the strains with a set of plasmids encoding either wild-type CRP (CRP+), CRPK52N, CRPH159L, or CRPH159L,K52N. We then performed β-galactosidase assays to quantify the levels of lacZ expression in the presence and absence of fucose. The results of these assays are shown in Table 2. Cells containing CRP+ and harboring the wild-type promoter expressed lacZ at a low level when grown in the absence of fucose. In the presence of fucose, the expression was induced roughly 40-fold. On the other hand, cells containing CRP+ and harboring the IS5-disrupted promoter were unable to be induced, in agreement with the results in previous studies (8). By contrast, cells containing CRPK52N and harboring either the wild-type or the IS5-disrupted promoter expressed lacZ constitutively (i.e., expressed lacZ at a high level in the absence of fucose). When these cells were grown in the presence of fucose, the activity of the wild-type promoter increased about threefold but that of the IS5-disrupted promoter increased only modestly. The levels of lacZ expression in cells containing CRPH159L and harboring either promoter were similar to those in cells containing CRP+. Finally, the uninduced level of lacZ expression in cells containing CRPH159L,K52N and harboring either promoter was partially constitutive, and this expression was dramatically superinduced in the presence of fucose in cells harboring the wild-type (but not the IS5-disrupted) promoter (see Discussion).
TABLE 2.
Effects of different crp alleles on the activities of wild-type and IS5 promoters in the presence or absence of inducer
| Plasmid-encoded form of CRP | β-Galactosidase activity (Miller units) with:
|
|||
|---|---|---|---|---|
| Wild-type promotera
|
IS5 promoterb
|
|||
| With fucose | Without fucose | With fucose | Without fucose | |
| CRP+ | 130 | 3.2 | 1.9 | 1.7 |
| CRPK52N | 280 | 90 | 210 | 140 |
| CRPH159L | 100 | 2.3 | 1.2 | 1.2 |
| CRPK52N,H159L | 1,600 | 62 | 35 | 24 |
Strain JCB43Δcrp39[F′ pfuc-lacZ] cells containing the wild-type fucPIK promoter fused to the lacZ gene were transformed with pHA7 derivatives encoding the indicated forms of CRP.
Strain JCB43Δcrp39[F′ IS5 pfuc-lacZ] cells containing the IS5-disrupted fucPIK promoter fused to the lacZ gene were transformed with pHA7 derivatives encoding the indicated forms of CRP.
Based on the results of Busby and coworkers (3, 22), we anticipated that the CRPK52N protein likely exerted its effect when bound to a CRP recognition site centered ∼41.5 bp upstream of the start point of fucPIK transcription for both the wild-type and IS5-disrupted promoters. To find out if this was the case, we first determined the transcription start points for each of these promoters. This was done by primer extension analysis (data not shown), which revealed the same start site located 116 bp upstream of the start of the fucP gene in each case (Fig. 1). As anticipated, a putative CRP-binding site (one of several previously identified in the intergenic regulatory region) is located at position −40.5 relative to the transcription start site. DNA sequence analysis of the IS5-disrupted promoter localized the site of the 1.3-kb insertion to position −56.5, just upstream of this CRP-binding site (hereafter designated CRP site 1) (Fig. 1). Thus, the IS5 element separates the fucPIK core promoter and CRP site 1 from putative regulatory sequences located further upstream.
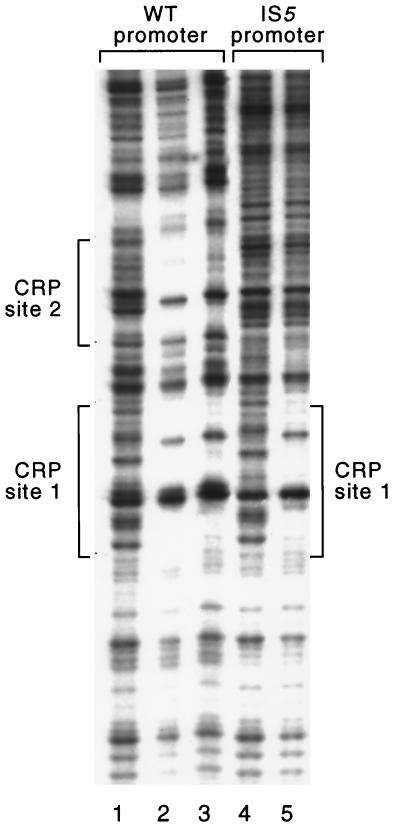
DNase I footprint analysis revealed that purified CRP bound to CRP site 1 on both the wild-type and the IS5 templates (Fig. 2). The wild-type template also contains CRP site 2 (Fig. 1), and the footprint showed that CRP bound to this site as well. The concentration of CRP required to occupy these sites was similar to that required to occupy the CRP-binding site associated with the wild-type lac promoter (data not shown).
FIG. 2.
CRP-binding sites associated with wild-type (WT) and IS5-disrupted promoters. Shown are the results of a DNase I protection experiment performed with purified CRP. The reaction mixtures loaded in lanes 1 and 4 contained no CRP, that loaded in lane 2 contained 320 nM CRP dimer, and those loaded in lanes 3 and 5 contained 64 nM CRP dimer. CRP site 1 is present on both templates, whereas CRP site 2 is present on the wild-type template only (see text).
We then sought to determine if CRP site 1 was required for the ability of CRPK52N to activate both the IS5-disrupted and the wild-type fucPIK promoters under noninducing conditions. To this end, we used site-directed mutagenesis to inactivate CRP site 1 in cis to both the wild-type and IS5-disrupted promoters. Thus modified, the promoters were again transferred to F′ episomes, and a new pair of reporter strains was generated (JCB43Δcrp39[F′ crp1 pfuc-lacZ] and JCB43Δcrp39[F′ IS5-crp1 pfuc-lacZ]). We transformed each of these strains with the same set of four plasmids encoding either CRP+ or a mutant derivative. Table 3 shows the activity levels of β-galactosidase in these new strains grown in the presence or absence of fucose. The salient results are as follows. First, cells containing CRPK52N and harboring either promoter expressed lacZ at a low level in the absence of fucose, indicating that CRP site 1 is required for CRPK52N to direct constitutive expression of lacZ under the control of either the wild-type or the IS5-disrupted promoter. Second, cells containing CRP+ or one of the mutant derivatives and harboring the wild-type crp1 promoter were inducible for lacZ expression to different degrees in the presence of fucose. The highest level of lacZ expression was achieved with CRPH159L, and, intriguingly, the inactivation of CRP site 1 completely abrogated the superinduction mediated by CRPH159L,K52N (compare Tables 2 and 3 [1,600 versus 31 Miller units]). Third, cells harboring the IS5 crp1 promoter expressed lacZ at a low level in all cases.
TABLE 3.
Effects of different crp alleles on the activities of crp1 and IS5-crp1 promoters in the presence or absence of inducer
| Plasmid-encoded form of CRP | β-Galactosidase activity (Miller units) with:
|
|||
|---|---|---|---|---|
|
crp1 promotera
|
IS5-crp1 promoterb
|
|||
| With fucose | Without fucose | With fucose | Without fucose | |
| CRP+ | 33 | 0.9 | 0.8 | 0.7 |
| CRPK52N | 18 | 2.6 | 1.7 | 1.5 |
| CRPH159L | 79 | 2.1 | 1.1 | 1.0 |
| CRPK52N,H159L | 31 | 4.6 | 1.9 | 2.0 |
Strain JCB43Δcrp39[F′ crp1 pfuc-lacZ] cells containing the crp1 fucPIK promoter fused to the lacZ gene were transformed with pHA7 derivatives encoding the indicated forms of CRP.
Strain JCB43Δcrp39[F′ IS5-crp1 pfuc-lacZ] cells containing the IS5-disrupted crp1 fucPIK promoter fused to the lacZ gene were transformed with pHA7 derivatives encoding the indicated forms of CRP.
We then wanted to confirm that in the presence of a functional CRP site 1, CRPK52N exerted its constitutive effect on lacZ transcription independently of the fucose-specific regulator, FucR. For this purpose we took advantage of a strain bearing a deletion that removes the fucR gene. We constructed a Δcrp Δlac derivative of this strain and then introduced the F′ episomes bearing the wild-type and IS5-disrupted promoters. The resulting pair of reporter strains (ECL733Δcrp39[F′ pfuc-lacZ] and ECL733Δcrp39[F′ IS5 pfuc-lacZ]) was again transformed with each of the four CRP plasmids. Table 4 shows that lacZ expression in these strains was about the same in the presence of fucose as in its absence and, in particular, that cells containing CRP+ could no longer be induced for the expression of lacZ, as expected. Furthermore, CRPK52N remained active on both promoters in this genetic background, confirming that it functions independently of FucR.
TABLE 4.
Effects of different crp alleles on the activities of wild-type and IS5 promoters in a background lacking the specific regulator FucR
| Plasmid-encoded form of CRP | β-Galactosidase activity (Miller units) with:
|
|||
|---|---|---|---|---|
| Wild-type promotera
|
IS5 promoterb
|
|||
| With fucose | Without fucose | With fucose | Without fucose | |
| CRP+ | 1.9 | 2.3 | 1.2 | 1.1 |
| CRPK52N | 68 | 60 | 120 | 97 |
| CRPH159L | 2.4 | 2.7 | 1.7 | 1.4 |
| CRPK52N,H159L | 44 | 59 | 15 | 18 |
Strain ECL733Δcrp39[F′ pfuc-lacZ] cells containing the wild-type fucPIK promoter fused to the lacZ gene were transformed with pHA7 derivatives encoding the indicated forms of CRP.
Strain ECL733Δcrp39[F′ IS5 pfuc-lacZ] cells containing the IS5-disrupted fucPIK promoter fused to the lacZ gene were transformed with pHA7 derivatives encoding the indicated forms of CRP.
We also constructed ΔfucR Δcrp Δlac strains harboring the crp1 derivative of each promoter (ECL733Δcrp39[F′ crp1 pfuc-lacZ] and ECL733Δcrp39[F′ IS5-crp1 pfuc-lacZ]). With these strains, the level of lacZ expression was low irrespective of the form of CRP introduced (Table 5), thus showing that the partial induction of lacZ expression seen in cells harboring the wild-type promoter in cis to the inactivated CRP site 1 was dependent on FucR.
TABLE 5.
Effects of different crp alleles on the activities of crp1 and IS5-crp1 promoters in a background lacking the specific regulator FucR
| Plasmid-encoded form of CRP | β-Galactosidase activity (Miller units) with:
|
|||
|---|---|---|---|---|
|
crp1 promotera
|
IS5-crp1 promoterb
|
|||
| With fucose | Without fucose | With fucose | Without fucose | |
| CRP+ | 0.7 | 0.7 | 0.4 | 0.4 |
| CRPK52N | 3.5 | 3.1 | 1.3 | 1.0 |
| CRPH159L | 1.6 | 1.7 | 1.0 | 1.0 |
| CRPK52N,H159L | 6.9 | 6.3 | 1.7 | 1.7 |
Strain ECL733Δcrp39[F′ crp1 pfuc-lacZ] cells containing the crp1 fucPIK promoter fused to the lacZ gene were transformed with pHA7 derivatives encoding the indicated forms of CRP.
Strain ECL733Δcrp39[F′ IS5-crp1 pfuc-lacZ] cells containing the IS5-disrupted crp1 fucPIK promoter fused to the lacZ gene were transformed with pHA7 derivatives encoding the indicated forms of CRP.
DISCUSSION
We have characterized a crp mutation that restores a fucose-positive phenotype to an E. coli mutant strain that cannot be induced by fucose to express the fucPIK operon because of the presence of an IS5 insertion element in the intergenic regulatory region between the divergent fucPIK and fucAO operons. This crp mutation, leading to constitutive expression of the fucPIK operon, specifies a single amino acid substitution, K52N. Interestingly, such a mutation has been found previously as a second-site change that restored the ability of CRPH159L to activate transcription from a class II promoter (3). We found that CRPK52N activated transcription from both the wild-type fucPIK promoter and the IS5-containing derivative in an essentially fucose-independent manner. This is in contrast to CRP+, which effectively activated the wild-type fucPIK promoter only in the presence of fucose but failed to activate the IS5-disrupted promoter in the presence or absence of fucose, as previously reported (8).
We then sought to identify the site of action of the CRPK52N protein. Determination of the transcription start point for both the wild-type and the IS5-disrupted promoters revealed the presence of a CRP-binding site (CRP site 1) centered at position −40.5 (a class II position). We also found that the 1.3-kb IS5 element was inserted directly upstream of this CRP recognition site, presumably separating it from other essential regulatory elements located further upstream (additional CRP-binding sites and presumably also one or more yet to be located FucR-binding sites). These findings indicate that, whereas the binding of wild-type CRP to site 1 is not sufficient to activate transcription from the fucPIK promoter, binding of CRPK52N is sufficient, at least for the IS5-disrupted promoter.
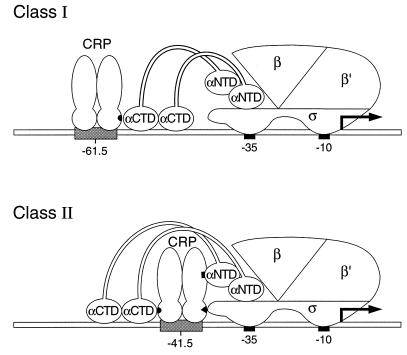
CRP has the potential to make at least three different contacts with RNAP (reviewed in reference 5). When bound at a class I promoter (at some distance upstream of the promoter −35 region, as for the lac promoter), the CRP dimer makes a single contact with RNAP by using residues in a solvent-exposed loop known as activating region 1 (AR1) in the promoter-proximal subunit to contact the C-terminal domain of the α subunit (α-CTD) (Fig. 3) (reviewed in reference 4). However, when bound at a class II promoter (immediately adjacent to the promoter −35 region, as for the gal promoter), wild-type CRP makes two productive contacts with RNAP (17) (Fig. 3): (i) it uses a second activating region (AR2) in the promoter-proximal subunit to contact the N-terminal domain of the α subunit (α-NTD) and (ii) it uses AR1 in the promoter-distal subunit to contact the α-CTD (which in turn associates with the DNA upstream of the DNA-bound CRP dimer). Inactivation of AR1 (by the H159L substitution, for example) ordinarily prevents CRP from activating transcription from both class I and class II promoters. Evidence from the previous CRPH159L suppression work suggests that the K52N substitution exposes a cryptic activating region (AR3) (22, 23), which may interact productively with the ς subunit of RNAP (5, 14). In the context of the present work, the K52N substitution presumably renders CRP a more potent activator that can function independently to activate a promoter whose activity ordinarily depends on multiple activators working in concert. CRPH159L,K52N provides for a significantly lower level of constitutive expression from the IS5-disrupted promoter than does CRPK52N (Table 2, 210 versus 35 Miller units), consistent with the idea that CRPK52N is utilizing at least two (and probably all three) of its activating regions to stimulate transcription from the mutant promoter.
FIG. 3.
Interactions between CRP and RNAP at a class I and a class II promoter. When bound at a class I promoter, CRP uses AR1 (small black semicircle) in the promoter-proximal subunit to contact the α-CTD. When bound at a class II promoter, wild-type CRP makes two contacts with RNAP by using AR1 in the promoter-distal subunit to contact the α-CTD and AR2 (small black rectangle) in the promoter-proximal subunit to contact the α-NTD. The CRPK52N mutant possesses a third activating region, designated AR3 (small black triangle), that has been proposed to contact the ς subunit (see text).
Why does CRP site 1 not mediate activation by wild type CRP at the IS5-disrupted fucPIK promoter? Our results suggest that this is not likely to be a consequence of poor site occupancy. Rather, we suggest that the failure of wild-type CRP to activate the IS5-disrupted promoter may be an inherent property of the fucPIK core promoter. We do not know what feature of the fucPIK core promoter might distinguish it from other class II promoters that can be activated by a single DNA-bound CRP dimer. We note, however, that unlike a number of well-characterized CRP-dependent class II promoters, the fucPIK promoter does not have an extended −10 region (3a). Perhaps this or other features of the fucPIK core promoter help account for the fact that the wild-type fucPIK promoter cannot be activated by CRP in the absence of FucR. Many other bacterial promoters whose activities depend on two transcriptional activators have been described (10; see, for example, reference 19). In our case, the dependence of the fucPIK promoter on a specific complex of regulatory proteins for its activity can apparently be overcome in the presence of a mutant CRP protein with three, rather than two, activating regions. At promoters such as the semisynthetic one studied by Bell and colleagues (3), two activating regions are sufficient for CRP to stimulate transcription and AR3 can substitute functionally for AR1. However, at the IS5-disrupted fucPIK promoter, AR3 compensates not for a defective activating region but for the lack of upstream regulatory sequences.
Interestingly, the inactivation of AR1 by the H159L substitution does not impair the ability of CRP to activate transcription from the wild-type promoter under inducing conditions (i.e., in the presence of fucose). This suggests that at the complex fucPIK promoter, any interactions between CRP and the α-CTD do not contribute significantly to promoter activation. One possibility is that at the wild-type promoter, the role of CRP in the activation process is indirect. By analogy with the regulation of the divergent mal operons in E. coli (18), CRP might, for example, function as an architectural element to facilitate the formation of a higher-order complex involving one or more FucR molecules and RNAP. Another possibility is that one CRP dimer (that bound at site 1) interacts with RNAP by using AR2 but that the arrangement of the upstream regulatory elements precludes productive interaction between AR1 and the α-CTD. In this regard, it is particularly interesting that transcription from the wild-type promoter was superinduced in cells containing the doubly substituted CRPH159L,K52N. That is, inactivation of AR1 in the context of CRPK52N dramatically improved its ability to function together with FucR to stimulate transcription under inducing conditions. This superinduction was dependent on CRP site 1, suggesting that under these conditions the CRP dimer bound at site 1 uses AR3 (and presumably also AR2) to exert a direct effect on transcription. The stimulatory effect of the H159L substitution suggests that, when functional, AR1 may mediate an inhibitory interaction between CRP and the α-CTD, disruption of which promotes the formation of additional productive interactions, presumably involving FucR. Such an inhibitory interaction might involve the CRP dimer bound at site 1 and/or one or more additional CRP dimers bound further upstream. Further analysis of the fuc intergenic regulatory region will be required to elucidate the mechanisms by which both wild-type CRP and the CRPK52N,H159L mutant cooperate with FucR to stimulate transcription from the wild-type promoter.
ACKNOWLEDGMENTS
This work was supported by grants GM44025 (to A.H.) and GM30693 (to E.C.C.L.).
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 2.Bartkus J M, Mortlock R P. Isolation of a mutation resulting in constitutive synthesis of l-fucose catabolic enzymes. J Bacteriol. 1986;165:710–714. doi: 10.1128/jb.165.3.710-714.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell A, Gaston K, Williams R, Chapman K, Kolb A, Buc H, Minchin S, Williams J, Busby S. Mutations that alter the ability of the Escherichia coli cyclic AMP receptor protein to activate transcription. Nucleic Acids Res. 1990;18:7243–7250. doi: 10.1093/nar/18.24.7243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Brown J A, Barne K A, Minchin S D, Busby S J W. Extended −10 promoters. In: Eckstein F, Lilley D, editors. Nucleic acids and molecular biology. Vol. 11. Berlin, Germany: Springer-Verlag; 1997. pp. 41–52. [Google Scholar]
- 4.Busby S, Ebright R H. Promoter structure, promoter recognition, and transcription activation in prokaryotes. Cell. 1994;79:743–746. doi: 10.1016/0092-8674(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 5.Busby S, Ebright R H. Transcription activation at class II CAP-dependent promoters. Mol Microbiol. 1997;23:853–859. doi: 10.1046/j.1365-2958.1997.2771641.x. [DOI] [PubMed] [Google Scholar]
- 6.Chakrabarti T, Chen Y-M, Lin E C C. Clustering of genes for l-fucose dissimilation by Escherichia coli. J Bacteriol. 1984;157:984–986. doi: 10.1128/jb.157.3.984-986.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y-M, Chakrabarti T, Lin E C C. Constitutive activation of l-fucose genes by an unlinked mutation in Escherichia coli. J Bacteriol. 1984;159:725–729. doi: 10.1128/jb.159.2.725-729.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y-M, Lu Z, Lin E C C. Constitutive activation of the fucAO operon and silencing of the divergently transcribed fucPIK operon by an IS5 element in Escherichia coli mutants selected for growth on l-1,2-propanediol. J Bacteriol. 1989;171:6097–6105. doi: 10.1128/jb.171.11.6097-6105.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cocks G T, Aguilar J, Lin E C C. Evolution of the l-1,2-propanediol catabolism in Escherichia coli by recruitment of enzymes for l-fucose and l-lactate metabolism. J Bacteriol. 1974;118:83–88. doi: 10.1128/jb.118.1.83-88.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collado-Vides J, Magasanik B, Gralla J D. Control site location and transcriptional regulation in Escherichia coli. Microbiol Rev. 1991;55:371–394. doi: 10.1128/mr.55.3.371-394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hacking A J, Lin E C C. Disruption of the fucose pathway as a consequence of genetic adaptation to propanediol as a carbon source in Escherichia coli. J Bacteriol. 1976;126:1166–1172. doi: 10.1128/jb.126.3.1166-1172.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Hochschild A. Long-range cooperativity in protein-DNA interactions. Methods Enzymol. 1991;208:343–361. doi: 10.1016/0076-6879(91)08019-e. [DOI] [PubMed] [Google Scholar]
- 12.Joung J K, Chung E H, King G, Yu C, Hirsh A S, Hochschild A. Genetic strategy for analyzing specificity of dimer formation: Escherichia coli cyclic AMP receptor protein mutant altered in its dimerization specificity. Genes Dev. 1995;9:2986–2996. doi: 10.1101/gad.9.23.2986. [DOI] [PubMed] [Google Scholar]
- 12a.Joung, J. K., and A. Hochschild. Unpublished data.
- 13.Joung J K, Koepp D M, Hochschild A. Synergistic activation of transcription by bacteriophage λ cI protein and E. coli cAMP receptor protein. Science. 1994;265:1863–1866. doi: 10.1126/science.8091212. [DOI] [PubMed] [Google Scholar]
- 14.Lonetto M A, Rhodius V, Lamberg K, Kiley P, Busby S, Gross C. Identification of a contact site for different transcription activators in region 4 of the Escherichia coli RNA polymerase sigma70 subunit. J Mol Biol. 1998;284:1353–1365. doi: 10.1006/jmbi.1998.2268. [DOI] [PubMed] [Google Scholar]
- 15.Lu Z, Lin E C C. The nucleotide sequence of Escherichia coli genes for L-fucose dissimilation. Nucleic Acids Res. 1989;17:4883–4884. doi: 10.1093/nar/17.12.4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Lu, Z., and E. C. C. Lin. Unpublished data.
- 16.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 17.Niu W, Younggyu K, Tau G, Heyduk T, Ebright R H. Transcription activation at class II CAP-dependent promoters: two interactions between CAP and RNA polymerase. Cell. 1996;87:1123–1134. doi: 10.1016/s0092-8674(00)81806-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richet E, Vidal-Ingigliardi D, Raibaud O. A new mechanism for coactivation of transcription initiation: repositioning of a transcriptional activator triggered by the binding of a second activator. Cell. 1991;66:1185–1195. doi: 10.1016/0092-8674(91)90041-v. [DOI] [PubMed] [Google Scholar]
- 19.Scott S, Busby S, Beacham I. Transcriptional co-activation at the ansB promoters: involvement of the activating regions of CRP and FNR when bound in tandem. Mol Microbiol. 1995;18:521–531. doi: 10.1111/j.1365-2958.1995.mmi_18030521.x. [DOI] [PubMed] [Google Scholar]
- 20.Sridhara S, Wu T T, Chused T M, Lin E C C. Ferrous-activated nicotinamide adenine dinucleotide-linked dehydrogenase from a mutant of Escherichia coli capable of growth on 1,2-propanediol. J Bacteriol. 1969;98:87–95. doi: 10.1128/jb.98.1.87-95.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whipple F W. Genetic analysis of prokaryotic and eukaryotic DNA-binding proteins in Escherichia coli. Nucleic Acids Res. 1998;26:3700–3706. doi: 10.1093/nar/26.16.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams R, Bell A, Sims G, Busby S. The role of two surface exposed loops in transcription activation by the Escherichia coli CRP and FNR proteins. Nucleic Acids Res. 1991;19:6705–6712. doi: 10.1093/nar/19.24.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams R M, Rhodius V, Bell A I, Kolb A, Busby S J W. Orientation of functional activating regions in the Escherichia coli CRP protein during transcription activation at class II promoters. Nucleic Acids Res. 1996;24:1112–1118. doi: 10.1093/nar/24.6.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu Y, Lin E C C. A mutant crp allele that differentially activates the operons of the fuc regulon in Escherichia coli. J Bacteriol. 1988;170:2352–2358. doi: 10.1128/jb.170.5.2352-2358.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu Y, Lin E C C. l-1,2-Propanediol exits more rapidly than l-lactaldehyde from Escherichia coli. J Bacteriol. 1989;171:862–867. doi: 10.1128/jb.171.2.862-867.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]