Abstract
According to the Centers for Disease Control and Prevention, Acinetobacter baumannii is listed among the most threatening pathogens. A. baumannii is mainly a nosocomial pathogen with a distinctive ability to survive in multiple environments. These characteristics together with this bacterium’s ability to acquire antibiotic resistance determinants make it a notorious pathogen. The presence of human serum albumin (HSA) is associated with modification of expression levels in numerous genes. The presence of HSA in the culture medium is also correlated with a reduction in levels of the global suppressor histone-like nucleoid structure protein, H-NS. Comparative transcriptome analysis of the wild type and isogenic Δhns strains cultured in lysogeny broth (LB) in the presence or absence of HSA revealed that the expression of a subset of eleven genes are modified in the Δhns cultured in LB and the wild-type strain in the presence of HSA, pointing out these genes as candidates to be regulated by the presence of HSA through H-NS. Six and five of these genes were up- or down-regulated, respectively. Three of these genes have functions in quorum sensing (acdA, kar and fadD), one in quorum quenching (aidA), two in stress response (katE, ywrO), three in metabolism (phaC, yedL1, and yedL2), one in biofilm formation (csuAB), and one in β-oxidation of fatty acids (fadA). The regulation of these genes was assessed by: (i) transcriptional analysis and qPCR at the transcriptional level; and (ii) by determining the phenotypic characteristics of each function. The results of these studies support the hypothesis that HSA-mediated reduction of H-NS levels may be one very important regulatory circuit utilized by A. baumannii to adapt to selected environments, such as those where HSA-containing human fluids are abundant.
Subject terms: Microbiology, Pathogenesis
Introduction
Acinetobacter baumannii has emerged as a significant nosocomial pathogen as it is associated with high levels of morbidity and mortality1–5. The majority of strains recovered from patents in the hospital are multidrug resistant (MDR), a characteristic that permitted the World Health Organization (WHO) and the Centers for Disease Control and Prevention (CDC) to classify this pathogen as an “urgent threat”6,7.
A. baumannii strains are genetically diverse as a result of their extraordinary capacity to naturally acquire DNA via transformation8–13. Recent studies show that human serum albumin (HSA), a major blood protein, enhances transformation frequency and increases expression levels of type IV pilus-associated genes in A. baumannii9,14,15. Moreover, the effect of HSA is not limited to these functions; HSA also affects expression of genes involved in motility, biofilm formation, efflux pumps, metabolism, capsule synthesis, transcriptional regulation, antibiotic resistance, and pathogenesis as determined by transcriptomic analysis14. These studies also reveal that the A. baumannii h-ns (histone-like nucleoid structuring) gene expression levels decrease in cells exposed to purified HSA. An attractive hypothesis to explain a physiological role from these observations is that there is a regulatory circuit that links HSA to reduced H-NS protein intracellular concentrations, which affect expression of selected genes helping A. baumannii’s adaptation to HSA-containing human fluids14.
In A. baumannii the H-NS protein, a global transcriptional repressor in many Gram-negative bacteria16–19, plays a major role in persistence and expression of virulence-associated genes17,20, stress induced by carbapenemase expression21, as well as expression of genes associated with antibiotic resistance, biofilm formation, and quorum sensing22. H-NS may also be necessary for natural competence in A. baumannii as an hns null mutant showed significantly lower expression levels of genes related to acquisition of DNA from the environment23. To better understand the correlation of the effects produced by extracellular HSA and intracellular H-NS, we studied the effect of the presence of HSA in the milieu on the expression H-NS and other A. baumannii’s genes. Comparative transcriptomic analyses (RNA-seq) using A. baumannii AB5075 and AB5075 Δhns cultured in the presence or absence of HSA revealed significant modifications in levels of expression of specific genes. These experiments, in combination with phenotypic assays, indicate that HSA modifies expression levels of some A. baumannii genes through regulating expression of H-NS.
Results and discussion
Transcriptional effect of HSA on gene expression
Transcriptome analysis of A. baumannii AB5075 and AB5075 Δhns cultured in LB broth revealed a differential expression profile of 183 genes22. When A. baumannii AB5075 was cultured in LB broth with or without HSA there were 30 genes differentially expressed (Table S1). In both cases, an FDR-adjusted P value of < 0.05 and log2-fold change > 1 was considered as the differentially expressed-genes (DEGs). Inspection of the DEGs in both analyses showed that there were 11 genes in common (hhDEG) (Fig. 1A and Table S2). Among the eleven genes, six (acdA, kar, fadD, aidA, fadA, and phaC), and five (csuAB, katE, ywrO, yedL1, and yedL2) of these DEGs were up- or down-regulated, respectively (Fig. 1B). Three of these genes are involved in quorum sensing (acdA, kar and fadD), and one in quorum quenching (aidA)24. Other hhDEGs are associated with biofilm formation (csuAB)25, β-oxidation of fatty acid (fadA)26, stress response (katE, ywrO)27, and metabolism (phaC, yedL1, and yedL2) (Fig. 1B). Quinn et al. previous observation14, where H-NS is modified in the presence of HSA, together with genes known to be regulated by H-NS, and our present transcriptomic comparison (AB5075 vs Δhns (both strains cultured in LB broth), vs, AB5075 (cultured in LB broth) vs AB5075 (cultured in 3.5% HSA), point out these eleven genes as candidates to be regulated by the presence of HSA through modification of the intracellular H-NS concentration.
Figure 1.
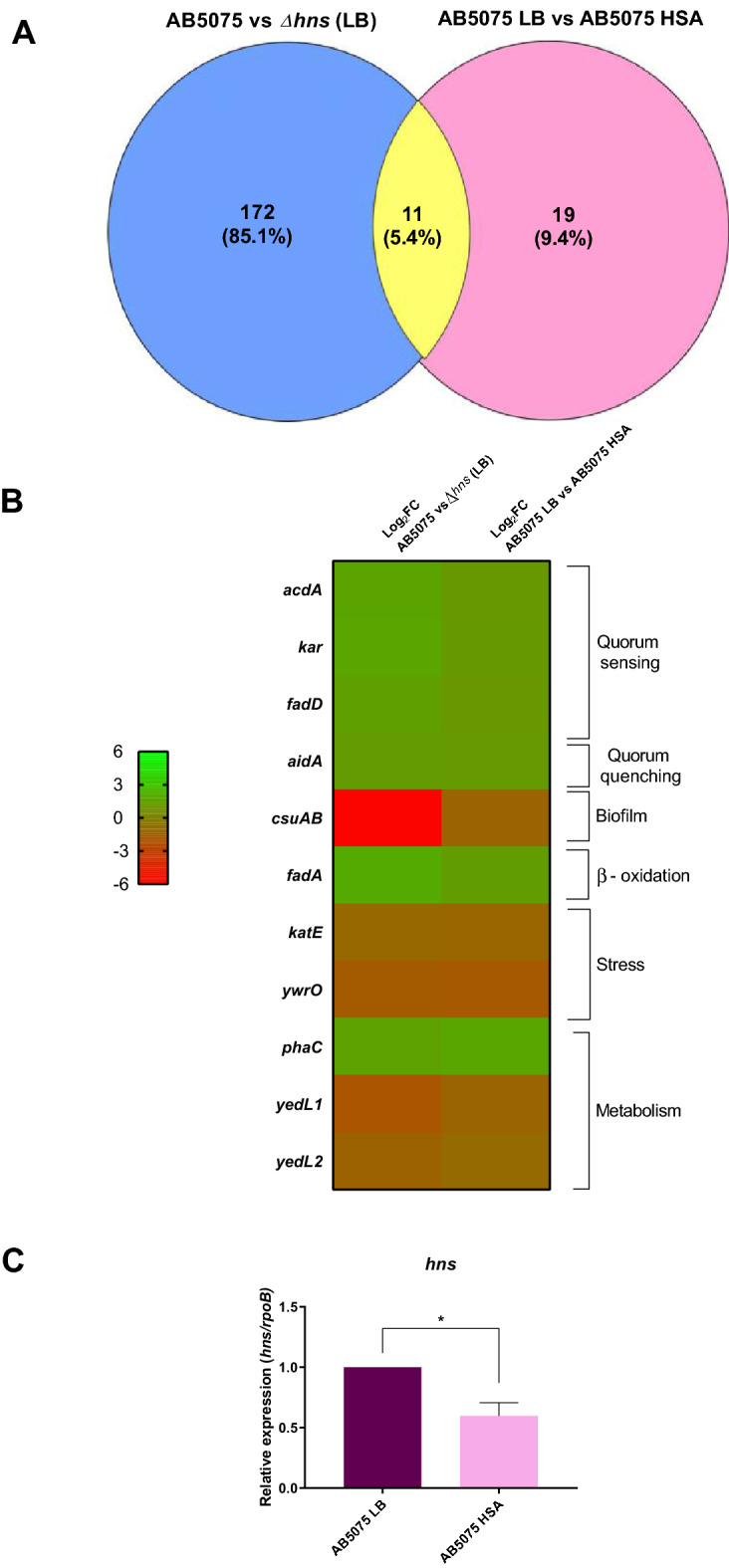
(A) Venn diagram shown the transcriptional analysis comparison between A. baumannii AB5075 vs A. baumannii Δhns (both strains cultured in LB broth) and AB5075 (cultured in LB broth) vs A. baumannii AB5075 (cultured in LB broth supplemented with 3.5% HSA). (B) Heat-map shown the total of 11 DEGs (hhDGEs) found in common between the two analyses. The scale goes from − 6 to 6. (C) qRT-PCR analysis of hns of A. baumannii AB5075 cultured in LB broth vs AB5075 cultured in LB broth supplemented with 3.5% HSA. Fold changes were calculated using double ΔCt analysis. At least three independent samples were used, and four technical replicates were performed from each sample. Student’s t test analysis was performed using GraphPad Prism (GraphPad software, San Diego, CA, USA). A P < 0.05 was considered significant. Data are presented as mean ± SD.
Cultures of the multidrug resistant, hypervirulent A. baumannii AB5075 model strain in the presence or absence of a physiological concentration of HSA were next subjected to quantitative RT-PCR (qRT-PCR) using total RNA to confirm that the presence of HSA in the growth medium results in a reduction of the H-NS synthesis (Fig. 1C).
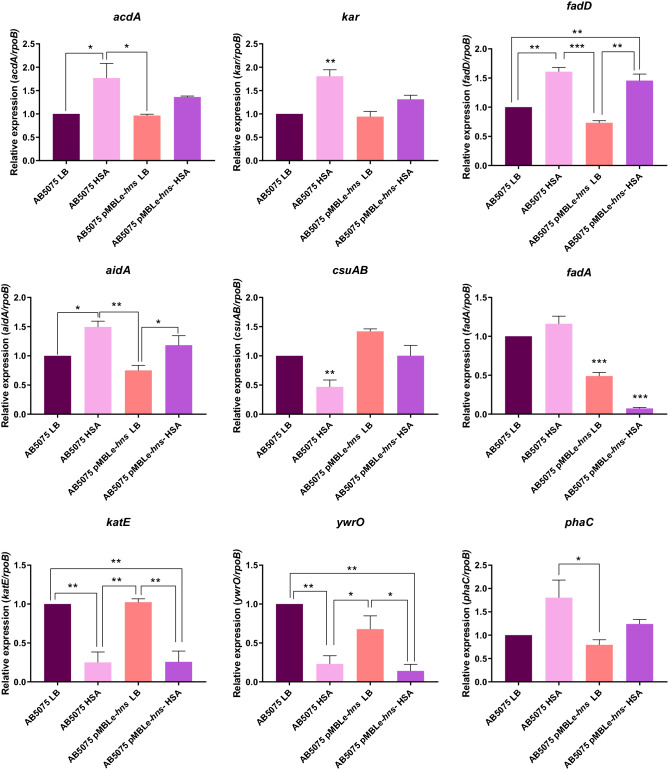
We next determined the levels of H-NS in A. baumannii AB5075 pMBLe-hns, a strain that carries a plasmid that overexpresses H-NS. Figure S1 shows that expression was elevated in cells grown in LB medium, but the H-NS levels were reduced fourfold when the medium was supplemented with 3.5% HSA. This strain and the wild type A. baumannii AB5075 were used to assess the expression of nine selected hhDEGs by qRT-PCR using RNA obtained from cultures carried out in LB broth supplemented with and without 3.5% HSA. Expression of acdA, kar, fadD, and aidA were upregulated about twofold in A. baumannii AB5075 grown in the presence of HSA. However, it was of interest that increasing the concentration of H-NS did not modify the expression of these genes (compare bar AB5075LB with AB5075 pMBLe-hns in Fig. 2). Expression of csuAB was downregulated twofold in A. baumannii AB5075 growing in the presence of HSA.
Figure 2.
Genetic analysis of genes of A. baumannii AB5075 or AB5075 pMBLe-hns cultured in LB broth or LB broth supplemented with 3.5% HSA. qRT-PCR of genes associated with quorum sensing, acdA, kar, and fadD, with quorum quenching, aidA, with biofilm, csuAB, with β-oxidation, fadA, with stress, katE and ywrO and metabolism phaC expressed in LB or LB supplemented with HSA. Fold changes were calculated using double ΔCt analysis. At least three independent samples were used, and four technical replicates were performed from each sample. The A. baumannii AB5075 cultured in LB was used as reference. Data are presented as mean ± SD. Statistical significance (P < 0.05) was determined by ANOVA followed by Tukey’s multiple-comparison test, one asterisks: P < 0.05; two asterisks: P < 0.01 and three asterisks: P < 0.001. This figure was performed using GraphPad Prism version number 9 (GraphPad software, San Diego, CA, USA, https://www.graphpad.com/).
In contrast, the fadA gene behaved unexpectedly; significant changes were not observed in the presence or absence of HSA in A. baumannii AB5075 and a tenfold reduction in expression was observed in the overproducing strain that was driven by the presence of HSA, i.e., when overexpression of H-NS was reversed (Fig. 2). The molecular basis of these modifications and changes are unknown at this time. The stress-response related genes katE and ywrO were downregulated around threefold in the presence of HSA and differences were not noted when H-NS was overexpressed (Fig. 2). The metabolic gene phaC a key enzyme in the polymerization of polyhydroxyalkanoates (PHAs), showed a significant twofold up-regulation in both strains, but significant modifications to levels of expression when H-NS was overexpressed was not seen (Fig. 2).
Since biofilm and quorum sensing are intimately related to A. baumannii’s virulence (and we identified genes involved in these two processes that are regulated by HSA and H-NS), we expanded the qRT-PCR analysis to two other genes associated with these processes. The expression of csuE, associated with biofilm formation, was downregulated about twofold when A. baumannii AB5075 was cultured in the presence of HSA. In A. baumannii AB5075 pMBLe-hns was overexpressed; but a significant downregulation was observed when HSA was present in the culture medium (Fig. S2). In the case of abaI, which codes for the acyl homoserine lactone (AHL) synthase, the presence of HSA in the milieu produced a significant threefold reduction in expression. A. baumannii AB5075 pMBLe of H-NS growing in the presence or absence of HSA expressed the gene at low levels (Fig. S2).
HSA plays a role in modulating the biofilm formation, the quorum sensing network, and the oxidative stress through H-NS
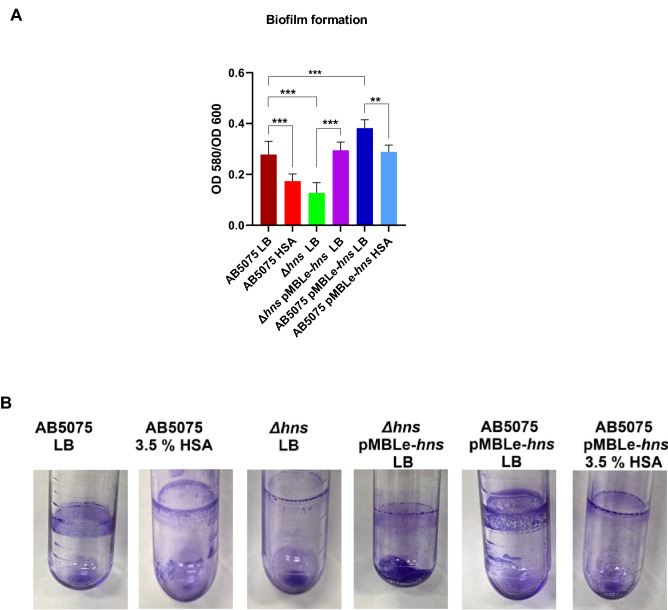
A. baumannii AB5075 grown in the presence of HSA and the hns deficient mutant strain produced a reduced mass of biofilm in comparison to the wild type when the biofilm production was quantified using a previously described method in polystyrene wells (Fig. 3A). Similar results were observed when the biofilm formation was assessed in tubes (Fig. 3B). The A. baumannii AB5075 pMBLe-hns strain as well as the mutant complemented by the plasmid pMBLe-hns cultured in the absence of HSA showed elevated levels of biofilm production. In A. baumannii AB5075 pMBLe-hns a regulatory effect was noted in HSA-containing medium. Taken together, these results support a regulatory role of HSA through modifying H-NS expression.
Figure 3.
Phenotypic analysis of biofilm formation. Biofilm assays performed with the AB5075 strain grown in LB or LB supplemented with 3.5% HSA, Δhns and Δhns pMBLe-hns strains cultured in LB, and AB5075 pMBLe-hns culture in LB or LB supplemented with 3.5% HSA. (A) Quantification of the radio of biofilm to total biomass in polystyrene wells. The mean ± SD is informed of three independent experiments. Statistical significance (P < 0.05) was determined by ANOVA followed by Tukey’s multiple comparison test, one asterisks: P < 0.05; two asterisks: P < 0.01 and three asterisks: P < 0.001. (B) Tubes of biofilm experiment after the stained with 1% crystal violet y removed the excess. A representative experiment is shown. This figure was performed using GraphPad Prism version number 9 (GraphPad software, San Diego, CA, USA, https://www.graphpad.com/).
Phenotypic modifications in quorum sensing were assessed determining levels of acyl-homoserine lactone (AHL) using the Agrobacterium tumefaciens-based solid plate assays28,29. Rodgers et al. demonstrated that the supernatants from A. baumannii AB5075 or AB5075 Δhns produced similar intensity of color, likely caused by an increase in both AHL synthesis (quorum sensing) and lactonase activity (quorum quenching) in AB5075 Δhns strain22. To study the effect of HSA on AHL secretion into the surrounding medium, A. baumannii AB5075, AB5075 Δhns, and the strains complemented with pMBLe-hns were cultured in different conditions before assessing the AHL concentration in the growth medium. A. baumannii AB5075 produced much lower amount of AHL when it was cultured in the presence of HSA (Fig. S3). A. baumannii AB5075 pMBLe-hns growing in the presence or absence of HSA and A. baumannii AB5075 Δhns pMBLe-hns cultured in LB broth, all produced low levels of AHL. The low AHL concentration in the growth medium could be explained by cancelling modifications in expression of genes associated with AHL synthesis (acdA, kar, fadD, and abaI) and lactonase, which is responsible for quorum quenching activity (aidA) (Figs. 1, 2, and S2).
Hydrogen peroxide is a disinfectant with potent bactericidal activity that is used in vaporized form to control outbreaks of multi-resistant A. baumannii infections30–32. Since A. baumannii is catalase-positive and the katE gene was a hhDEG, down-regulated in the presence of HSA, we assessed the production of catalase activity, as the decrease in absorbance at 240 nm resulting from the consumption of H2O2 (Fig. 4). Addition of HSA to the growth medium resulted in a reduction of catalase activity in the wild type strain. Equally low activity was produced by A. baumannii AB5075 Δhns growing in the absence of added HSA, which suggest that the action of HSA occurs through reduction of H-NS synthesis (Fig. 4). These results agreed with both RNA-seq analyses and RT-qPCR results. As expected, the complemented A. baumannii AB5075 Δhns pMBLe-hns produced higher catalase activity when compared to A. baumannii AB5075 (Fig. 4). Surprisingly, overexpressing hns showed reduced catalase activity when compared to the wild type strain; we still do not know the molecular bases for this unexpected results (Fig. 4).
Figure 4.
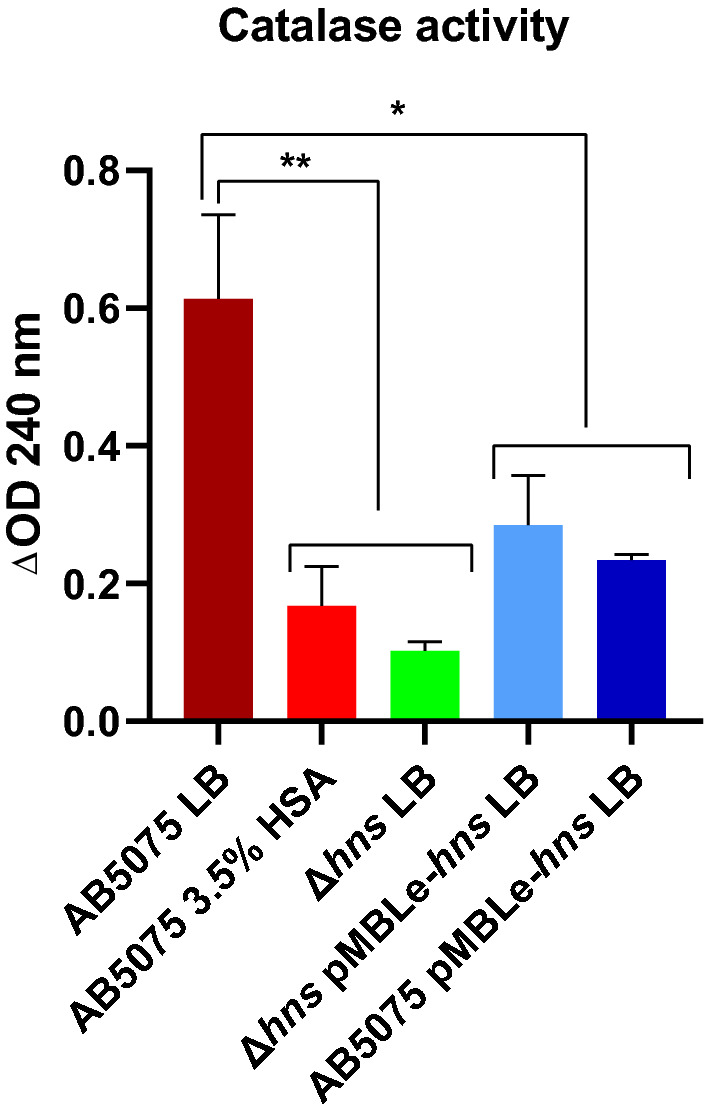
Phenotypic analysis of catalase activity. Monitoring the decrease in absorbance at 240 nm of crude extract of A. baumannii AB5075 and derivate strains cultured in LB or LB supplemented with 3.5% HSA. The mean ± SD is informed of three independent experiments. Statistical significance (P < 0.05) was determined by ANOVA followed by Tukey’s multiple comparison test, one asterisks: P < 0.05; two asterisks: P < 0.01 and three asterisks: P < 0.001. This figure was performed using GraphPad Prism version number 9 (GraphPad software, San Diego, CA, USA, https://www.graphpad.com/).
The results described in this section support the hypothesis that some A. baumannii genes are regulated by H-NS, whose expression is modified by the presence of HSA in the milieu. H-NS is a nucleoid-associated protein that binds DNA in a relatively nonspecific manner (it shows preference for AT-rich and curved regions) and alters its topology, which modifies levels of transcription. The effects of H-NS in A. baumannii have not been thoroughly studied17,20,22. An early analysis found that an A. baumannii mutant containing a disrupted hns gene was modified in the expression of several virulence genes such as those associated with the autotransporter Ata, the type VI secretion system, a type I pilus cluster, the acetoin metabolism, and phenylacetic acid degradation17. This strain also showed altered adherence to biotic surfaces and increase virulence as determined using the Caenorhabditis elegans infection model system17. A previous study showed that exposure of the model A. baumannii A118 to HSA was accompanied by a reduction of expression of H-NS and a wide variety of genes related to antibiotic resistance and stress response as well as genes that encode functions associated with natural competence14,22,23,33.
The global nature of H-NS as transcriptional regulator makes it challenging to definitely associate genes that are regulated by H-NS through variations in levels of expression of the regulator when the cells are exposed to HSA. However, despite these limitations, the results described in this work strongly suggest that there is a group of genes whose expression is indirectly regulated at the transcriptional level by HSA through reducing H-NS expression. We conclude that HSA-mediated reduction of H-NS levels may be one regulatory circuit utilized by A. baumannii to adapt to selected environments such as those where HSA-containing human fluids are abundant.
Conclusions
H-NS is a highly abundant intracellular protein that functions as a nucleoid organizer and a transcriptional silencer. In this work we gathered evidence that supports the role of HSA and H-NS in regulating quorum sensing and quorum quenching, biofilm, β-oxidation, stress, and metabolism related genes. Future analysis planned using chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq), in wild-type or an h-ns mutant, in the presence or absence of HSA will contribute to new insights into the molecular mechanisms governing the roles of HSA and H-NS in regulating the genes identified in this work and most probably other genes involved in the mechanisms of persistence and pathogenicity of A. baumannii.
Materials and methods
Bacterial strains
The multidrug and hypervirulent AB5075 strain34, its isogenic hns mutant (AB5075Δhns)35 and AB5075 ΔhnspMBLe-hns22 were used in the present study.
Electroporation
Electro-competent A. baumannii AB5075 cells were prepared and mixed with pMBLe-hns plasmid DNA (containing apramycin resistance) followed by electroporation with a Bio-RadGene Pulser instrument as described previously21. The electroporated cells were placed in recovery in a shaking incubator followed by culturing overnight at 37 °C on LB agar containing15 μg/ml apramycin (aac(3)-IV). At least 10 colonies were picked to confirm the presence of the different plasmids. To confirm their presence, plasmid extraction followed by gel electrophoresis analysis.
RNA extraction and RNA-seq analysis
A. baumannii AB5075 and derivative strains were cultured in LB broth and incubated with agitation for 18 h at 37 °C. Overnight cultures were then diluted 1:10 in fresh LB broth or LB broth supplemented with HSA and incubated with agitation for 7 h at 37 °C. The Direct-zol RNA Kit (Zymo Research, Irvine, CA, USA) was used to perform the RNA extraction in triplicates. RNA samples were DNase treated (Thermo Fisher Scientific, Waltham, MA, USA) following manufacturer’s instruction. Samples were confirmed to have no DNA contamination through PCR amplification of the 16S rDNA gene. RNA sequencing was outsourced to Novogene (Novogene Corporation, Sacramento, CA, USA). Ribosomal RNA-depletion was done using the Ribo-Zero kit (Illumina) and the construction of the cDNA library was performed with the TruSeq Stranded Total RNA Library Prep kit (Illumina) from three independent replicates per sample. Analysis of the quality of the Illumina reads, trimming of low-quality bases and removal of Illumina adapters was performed as described previously36. Reads were aligned to the genome of A. baumannii AB5075 using Burrows-Wheeler Alignment (BWA) software (v0.7.17) BWA and visualized using the Integrative Genomics Viewer (IGV). Read counts per gene were calculated using FeatureCounts37, and differential expression analysis was performed using DEseq2.Differentially expressed genes (DEGs) were defined as those displaying an FDR adjusted P value of < 0.05 and log2 fold change > 1. In the present work, previously published RNA-seq reads (GEO accession No GSE167117) corresponding to A. baumannii AB5075 vs AB5075 Δhns incubated in LB broth22 or A. baumannii AB5075 grown in LB broth vs grown in 3.5% HSA were analyzed.
qRT-PCR was performed to confirm and analyze the expression genes. cDNAwas prepared using the iScript Reverse Transcription Supermix for qRT-PCR (BioRad, Hercules, CA, USA) and quantitative PCR was performed using iQ SYBR Green Supermix (BioRad, Hercules, CA, USA) per the manufacturer’s recommendations, respectively. Results were analyzed using the 2−ΔΔCt method38 in which rpoB was used as the control gene. rpoB genes was confirmed to be a suitable reference gene showing a stable expression under the tested conditions using four other additional genes (16S rRNA, recA, secA and gyrB) per comparison. Experiments were performed in technical and biological triplicates. The results from experiments performed were statistical analyzed (ANOVA followed by Tukey’s multiple comparison test) using GraphPad Prism (GraphPad software, San Diego, CA, USA). A P value < 0.05 was considered significant.
The RNA-seq data analysed during the current study are available in the NCBI repository with the GEO accession No GSE167117.
Catalase activity measurements
Crude extract of A. baumannii AB5075 and derivate strains cultured in LB or LB supplemented with 3.5% HSA were utilized to determinate the catalase activity spectrophotometrically by monitoring the decrease in absorbance at 240 nm resulting from the consumption of H2O2 using a UV visible spectrophotometer (SpectraMax M3)39.
Biofilm assay
A. baumannii AB5075 and derivative strains were cultured in fresh LB medium, or LB supplemented with 3.5% HSA with agitation for 18 h at 37 °C. Tubes were emptied, washed three times with 1X phosphate-buffered saline (PBS) and stained with 1% crystal violet (CV) for 15 m. Excess CV was removed by washing three more with 1X PBS. In addition, quantification of biofilm production in polystyrene wells were carried out using a protocol from previously described method14. Experiments were performed in triplicate, with at least three technical replicates per biological replicate.
N-acyl homoserine lactone (AHL) detection
Agrobacterium tumefaciens-based solid plate assays were carried out to detect N-Acyl Homoserine Lactone (AHL) production40 as described in previous work28.Initially, 500 μL of the homogenate were loaded in a central well of 0.7% LB agar plates supplemented with 40 μg of 5-bromo-3-indolyl-β-D-galactopyranoside (X-Gal) per mL and 250 μL (OD = 2.5) of the overnight culture of Agrobacterium tumefaciens biosensor. The presence of AHL was determined by the development of blue coloring29. As a positive control, 100 μL of N-decanoyl-dl-homoserine lactone (C10-AHL) 12.5 mg/mL was utilized. Quantification of 5,5′-dibromo-4,4′-dichloro-indigo production in different conditions was determined using ImageJ software (NIH) by measuring the intensity of each complete plate, and subtracting the intensity measured in the negative control. The values were normalized to the positive control, which received the arbitrary value of 100.
Statistical analysis
Experiments performed at least in triplicates were statistically analyzed by ANOVA followed by Tukey’s multiple comparison tests using GraphPad Prism (GraphPad software, San Diego, CA, USA). A P value < 0.05 was considered significant.
All procedures performed in this study were in accordance with the CSUF Institutional Biosafety Committee Approval plan (DBH117-01) and follow the NIH, CDC, OSHA and other environmental and occupational regulations.
Supplementary Information
Author contributions
J.E., B.N., M.R.T., T.S., C.P., N.G., R.S., R.A.B, M.E.T. and M.S.R. conceived the study and designed the experiments. J.E., B.N., M.R.T., T.S., C.P., N.G., R.S., and M.S.R. performed the experiments and genomics and bioinformatics analyses. J.E., B.N., M.R.T., T.S., C.P., N.G., R.S., R.A.B., M.E.T. and M.S.R. analyzed the data and interpreted the results. R.A.B., M.E.T. and M.S.R. contributed reagents/materials/analysis tools. M.R.T., T.S., R.S., R.A.B., M.E.T. and M.S.R. wrote and revised the manuscript. All authors read and approved the final manuscript.
Funding
The authors’ work was supported by NIH SC3GM125556 to MSR, R01AI100560 to RAB, R01AI063517, R01AI072219 to RAB and 2R15 AI047115 to MET. JE and CP were supported by Grant MHRT 2T37MD001368from the National Institute on Minority Health and Health Disparities, National Institute of Health. This study was supported in part by funds and/or facilities provided by the Cleveland Department of Veterans Affairs, Award Number 1I01BX001974 to RAB from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development and the Geriatric Research Education and Clinical Center VISN 10 to RAB. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Department of Veterans Affairs. MRT and TS are recipient of a postdoctoral fellowship from CONICET. R.S. is a staff member from CONICET.
Data availability
The datasets generated and analyzed during the current study are available in the Gene Expression Omnibus (GEO) repository, (GEO accession No GSE167117).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jenny Escalante and Brent Nishimura.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-19012-y.
References
- 1.Ramirez MS, Bonomo RA, Tolmasky ME. Carbapenemases: transforming Acinetobacter baumannii into a yet more dangerous menace. Biomolecules. 2020 doi: 10.3390/biom10050720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paterson DL, Bonomo RA. Multidrug-resistant gram-negative pathogens: The urgent need for ‘old’ polymyxins. Adv. Exp. Med. Biol. 2019;1145:9–13. doi: 10.1007/978-3-030-16373-0_2. [DOI] [PubMed] [Google Scholar]
- 3.Spellberg B, Bonomo RA. The deadly impact of extreme drug resistance in Acinetobacter baumannii. Crit. Care Med. 2014;42:1289–1291. doi: 10.1097/CCM.0000000000000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spellberg B, Bonomo RA. “Airborne assault”: A new dimension in Acinetobacter baumannii transmission*. Crit. Care Med. 2013;41:2042–2044. doi: 10.1097/CCM.0b013e31829136c3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 2007;51:3471–3484. doi: 10.1128/AAC.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.CDC. Antibiotic resistance threats in the United States. Atlanta, GA: US Department of Health and Human Services, CDC (2019).
- 7.(WHO). WHO. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics (2017).
- 8.Vesel N, Blokesch M. Pilus production in Acinetobacter baumannii is growth phase dependent and essential for natural transformation. J. Bacteriol. 2021 doi: 10.1128/JB.00034-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Traglia GM, Quinn B, Schramm ST, Soler-Bistue A, Ramirez MS. Serum Albumin and Ca2+ Are Natural Competence Inducers in the Human Pathogen Acinetobacter baumannii. Antimicrob. Agents Chemother. 2016;60:4920–4929. doi: 10.1128/AAC.00529-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Touchon M, Cury J, Yoon EJ, Krizova L, Cerqueira GC, Murphy C, Feldgarden M, Wortman J, Clermont D, Lambert T, Grillot-Courvalin C, Nemec A, Courvalin P, Rocha EP. The genomic diversification of the whole Acinetobacter genus: Origins, mechanisms, and consequences. Genome Biol. Evol. 2014;6:2866–2882. doi: 10.1093/gbe/evu225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramirez MS, Merkier AK, Quiroga MP, Centron D. Acinetobacter baumannii is able to gain and maintain a plasmid harbouring In35 found in Enterobacteriaceae isolates from Argentina. Curr. Microbiol. 2012;64:211–213. doi: 10.1007/s00284-011-0052-9. [DOI] [PubMed] [Google Scholar]
- 12.Martinez J, Liu C, Rodman N, Fernandez JS, Barberis C, Sieira R, Perez F, Bonomo RA, Ramirez MS. Human fluids alter DNA-acquisition in Acinetobacter baumannii. Diagn. Microbiol. Infect. Dis. 2018;93(3):183–187. doi: 10.1016/j.diagmicrobio.2018.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Domingues S, Rosario N, Candido A, Neto D, Nielsen KM, Da Silva GJ. Competence for natural transformation is common among clinical strains of resistant Acinetobacter spp. Microorganisms. 2019;7:30. doi: 10.3390/microorganisms7020030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quinn B, Rodman N, Jara E, Fernandez JS, Martinez J, Traglia GM, Montana S, Cantera V, Place K, Bonomo RA, Iriarte A, Ramirez MS. Human serum albumin alters specific genes that can play a role in survival and persistence in Acinetobacter baumannii. Sci. Rep. 2018;8:14741. doi: 10.1038/s41598-018-33072-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le C, Pimentel C, Tuttobene MR, Subils T, Nishimura B, Traglia GM, Perez F, Papp-Wallace KM, Bonomo RA, Tolmasky ME, Ramirez MS. Interplay between meropenem and human serum albumin on expression of carbapenem resistance genes and natural competence in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2021;65(10):e01019. doi: 10.1128/AAC.01019-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H, Ayala JC, Benitez JA, Silva AJ. RNA-seq analysis identifies new genes regulated by the histone-like nucleoid structuring protein (H-NS) affecting Vibrio cholerae virulence, stress response and chemotaxis. PLoS ONE. 2015;10:e0118295. doi: 10.1371/journal.pone.0118295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eijkelkamp BA, Stroeher UH, Hassan KA, Elbourne LD, Paulsen IT, Brown MH. H-NS plays a role in expression of Acinetobacter baumannii virulence features. Infect. Immun. 2013;81:2574–2583. doi: 10.1128/IAI.00065-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prosseda G, Fradiani PA, Di Lorenzo M, Falconi M, Micheli G, Casalino M, Nicoletti M, Colonna B. A role for H-NS in the regulation of the virF gene of Shigella and enteroinvasive Escherichia coli. Res. Microbiol. 1998;149:15–25. doi: 10.1016/S0923-2508(97)83619-4. [DOI] [PubMed] [Google Scholar]
- 19.Hurtado-Escobar GA, Grepinet O, Raymond P, Abed N, Velge P, Virlogeux-Payant I. H-NS is the major repressor of Salmonella Typhimurium Pef fimbriae expression. Virulence. 2019;10:849–867. doi: 10.1080/21505594.2019.1682752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deveson Lucas D, Crane B, Wright A, Han ML, Moffatt J, Bulach D, Gladman SL, Powell D, Aranda J, Seemann T, Machado D, Pacheco T, Marques T, Viveiros M, Nation R, Li J, Harper M, Boyce JD. Emergence of high-level colistin resistance in an Acinetobacter baumannii Clinical isolate mediated by inactivation of the global regulator H-NS. Antimicrob. Agents Chemother. 2018 doi: 10.1128/AAC.02442-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang F, Fitchett N, Razo-Gutierrez C, Le C, Martinez J, Ra G, Lopez C, Gonzalez LJ, Sieira R, Vila AJ, Bonomo RA, Ramirez MS. The H-NS regulator plays a role in the stress induced by carbapenemase expression in Acinetobacter baumannii. mSphere. 2020;5:e00793. doi: 10.1128/mSphere.00793-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodgers D, Le C, Pimentel C, Tuttobene MR, Subils T, Escalante J, Nishimura B, Vescovi EG, Sieira R, Bonomo RA, Tolmasky ME, Ramirez MS. Histone-like nucleoid-structuring protein (H-NS) regulatory role in antibiotic resistance in Acinetobacter baumannii. Sci. Rep. 2021;11:18414. doi: 10.1038/s41598-021-98101-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le C, Pimentel C, Tuttobene MR, Subils T, Escalante J, Nishimura B, Arriaga S, Rodgers D, Bonomo RA, Sieira R, Tolmasky ME, Ramírez MS. Involvement of the histone-like nucleoid structuring protein (H-NS) in Acinetobacter baumannii’s natural transformation. Pathogens. 2021;10:1083. doi: 10.3390/pathogens10091083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez M, Mayer C, Fernandez-Garcia L, Blasco L, Muras A, Ruiz FM, Bou G, Otero A, Tomas M, Geih G. Quorum sensing network in clinical strains of A. baumannii: AidA is a new quorum quenching enzyme. PLoS ONE. 2017;12:e0174454. doi: 10.1371/journal.pone.0174454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomaras AP, Flagler MJ, Dorsey CW, Gaddy JA, Actis LA. Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology. 2008;154:3398–3409. doi: 10.1099/mic.0.2008/019471-0. [DOI] [PubMed] [Google Scholar]
- 26.Jiang JH, Hassan KA, Begg SL, Rupasinghe TWT, Naidu V, Pederick VG, Khorvash M, Whittall JJ, Paton JC, Paulsen IT, McDevitt CA, Peleg AY, Eijkelkamp BA. Identification of novel Acinetobacter baumannii host fatty acid stress adaptation strategies. MBio. 2019;10:e02056. doi: 10.1128/mBio.02056-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun D, Crowell SA, Harding CM, De Silva PM, Harrison A, Fernando DM, Mason KM, Santana E, Loewen PC, Kumar A, Liu Y. KatG and KatE confer Acinetobacter resistance to hydrogen peroxide but sensitize bacteria to killing by phagocytic respiratory burst. Life Sci. 2016;148:31–40. doi: 10.1016/j.lfs.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pimentel C, Le C, Tuttobene MR, Subils T, Martinez J, Sieira R, Papp-Wallace KM, Keppetipola N, Bonomo RA, Actis LA, Tolmasky ME, Ramirez MS. Human pleural fluid and human serum albumin modulate the behavior of a hypervirulent and multidrug-resistant (MDR) Acinetobacter baumannii representative strain. Pathogens. 2021;10:471. doi: 10.3390/pathogens10040471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cha C, Gao P, Chen YC, Shaw PD, Farrand SK. Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol. Plant Microbe Interact. 1998;11:1119–1129. doi: 10.1094/MPMI.1998.11.11.1119. [DOI] [PubMed] [Google Scholar]
- 30.Lomovskaya O, Nelson K, Rubio-Aparicio D, Tsivkovski R, Sun D, Dudley MN. The impact of intrinsic resistance mechanisms on potency of QPX7728, a new ultra-broad-spectrum beta-lactamase inhibitor of serine and metallo beta-lactamases in Enterobacteriaceae, Pseudomonas aeruginosa, and Acinetobacter baumannii. Antimicrob. Agents Chemother. 2020;64(6):e00552. doi: 10.1128/AAC.00552-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ray A, Perez F, Beltramini AM, Jakubowycz M, Dimick P, Jacobs MR, Roman K, Bonomo RA, Salata RA. Use of vaporized hydrogen peroxide decontamination during an outbreak of multidrug-resistant Acinetobacter baumannii infection at a long-term acute care hospital. Infect. Control Hosp. Epidemiol. 2010;31:1236–1241. doi: 10.1086/657139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chmielarczyk A, Higgins PG, Wojkowska-Mach J, Synowiec E, Zander E, Romaniszyn D, Gosiewski T, Seifert H, Heczko P, Bulanda M. Control of an outbreak of Acinetobacter baumannii infections using vaporized hydrogen peroxide. J. Hosp. Infect. 2012;81:239–245. doi: 10.1016/j.jhin.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 33.Martinez J, Fernandez JS, Liu C, Hoard A, Mendoza A, Nakanouchi J, Rodman N, Courville R, Tuttobene MR, Lopez C, Gonzalez LJ, Shahrestani P, Papp-Wallace KM, Vila AJ, Tolmasky ME, Bonomo RA, Sieira R, Ramirez MS. Human pleural fluid triggers global changes in the transcriptional landscape of Acinetobacter baumannii as an adaptive response to stress. Sci. Rep. 2019;9:17251. doi: 10.1038/s41598-019-53847-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacobs AC, Thompson MG, Black CC, Kessler JL, Clark LP, McQueary CN, Gancz HY, Corey BW, Moon JK, Si Y, Owen MT, Hallock JD, Kwak YI, Summers A, Li CZ, Rasko DA, Penwell WF, Honnold CL, Wise MC, Waterman PE, Lesho EP, Stewart RL, Actis LA, Palys TJ, Craft DW, Zurawski DV. AB5075, a highly virulent isolate of Acinetobacter baumannii, as a model strain for the evaluation of pathogenesis and antimicrobial treatments. MBio. 2014;5:e01076–e1114. doi: 10.1128/mBio.01076-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gallagher LA, Ramage E, Weiss EJ, Radey M, Hayden HS, Held KG, Huse HK, Zurawski DV, Brittnacher MJ, Manoil C. Resources for genetic and genomic analysis of emerging pathogen Acinetobacter baumannii. J. Bacteriol. 2015;197:2027–2035. doi: 10.1128/JB.00131-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodman Nyah MJ, Sammie F, Jun N, Myers AL, Harris CM, Emily D, Fernandez JS, Christine L, Mendoza AM, Veronica J, Nikolas N, Brennan CA, Bonomo RA, Rodrigo S, Soledad RM. Human pleural fluid elicits pyruvate and phenylalanine metabolism in Acinetobacter baumannii to Enhance cytotoxicity and immune evasion. Front. Microbiol. 2019;10:1581. doi: 10.3389/fmicb.2019.01581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Sartorio MG, Repizo GD, Cortez N. Catalases of the polyextremophylic Andean isolate Acinetobacter sp. Ver 3 confer adaptive response to H. FEBS J. 2020;287:4525–4539. doi: 10.1111/febs.15244. [DOI] [PubMed] [Google Scholar]
- 40.Paulk Tierney AR, Rather PN. Methods for detecting N-acyl homoserine lactone production in Acinetobacter baumannii. Methods Mol. Biol. 2019;1946:253–258. doi: 10.1007/978-1-4939-9118-1_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during the current study are available in the Gene Expression Omnibus (GEO) repository, (GEO accession No GSE167117).