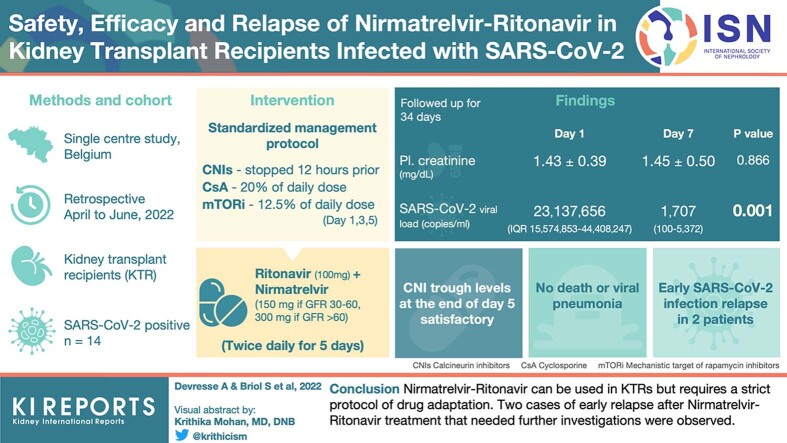
Abstract
Introduction
The efficacy of nirmatrelvir-ritonavir (NR; Paxlovid, Pfizer, New York, NY) to decrease the risk of progression to severe COVID-19 in high-risk patients has been demonstrated. However, evidence in infected kidney transplant recipients (KTRs) is lacking. Moreover, NR has significant and potentially harmful interactions with calcineurin inhibitors (CNIs).
Methods
In this single-center retrospective study, we included all KTRs treated with NR from April 28 to June 3, 2022. A standard management strategy of CNI dose adaptation (discontinuation of tacrolimus 12 hours before the start of NR and administration of 20% of the cyclosporine dose) and laboratory follow-up was applied.
Results
A total of 14 patients were included. Compared with day-0 (day before NR initiation), day-7 plasma creatinine concentrations and SARS-CoV-2 viral loads were similar (P = 0.866) and decreased (P = 0.002), respectively. CNI trough concentrations at the end of the treatment were satisfactory, nonetheless, with high individual variability. After a median follow-up time of 34 days, no death or viral pneumonia were observed. Nevertheless, 2 patients experienced early SARS-CoV-2 infection relapses (at day-10 and day-21) associated with an increase in SARS-CoV-2 viral loads.
Conclusion
NR can be used in KTRs but requires a strict protocol of drug adaptation. We observed 2 cases of early relapse after NR treatment that need further investigations.
Keywords: efficacy, kidney transplantation, nirmatrelvir-ritonavir, relapse, safety, SARS-CoV-2
Graphical abstract
Emerging new SARS-CoV-2 variants constantly modify therapeutic strategies for KTRs with COVID-19. Recently, in vitro data suggested that most monoclonal antibodies, widely used with previous variants, had lost their neutralizing ability against the BA2 variant.1 This led the United States Federal Drug Administration to recommend the use of alternative treatments in areas where the BA2 variant is endemic,2 despite low clinical evidence. Among alternatives, the safety and efficacy of NR to decrease the risk of progression to severe disease in high-risk patients has been demonstrated.3 Consequently, NR recently became one of the promising treatments for KTRs infected with SARS-CoV-2.4 However, several uncertainties remain. First, strong evidence regarding the efficacy of NR in reducing the risk of progression to severe disease in large cohorts of infected KTRs is lacking, because those patients were not represented in clinical trials.3 Second, due to the inhibitory effect of ritonavir on cytochrome (CYP) P450-3A and P-glycoprotein, NR has significant and potentially harmful interactions with many medications, including CNIs.4
The aim of the current study is to provide data regarding the management, safety, and efficacy of NR treatment of SARS-CoV-2 infection in a cohort of KTRs.
Methods
Patient Selection
NR has been available in Belgium since April 15, 2022. Since then, this treatment was applied to KTRs followed-up in our center, with the following inclusion criteria : (i) confirmed SARS-CoV-2 infection with positive polymerase-chain reaction (PCR) on nasopharyngeal swab; (ii) symptoms evolving in <5 days; (iii) baseline estimated glomerular filtration rate (by Chronic Kidney Disease Epidemiology Collaboration) >30 ml/min per 1.73 m2; (iv) no dyspnea, and oxygen saturation >95% at room air; and (v) no pregnancy or breast feeding.
This study retrospectively includes all KTRs followed in our kidney transplant center who actually received the treatment between April 28, 2022 and June 3, 2022. Patients were followed-up with until June 16, 2022 (data cutoff) for a median time of 34 days (ranging from 14 to 51 days). This work received the approval from the Ethic Committee of Cliniques universitaires Saint-Luc, Brussels, Belgium (2022/25MAI/220).
Local Standard Protocol
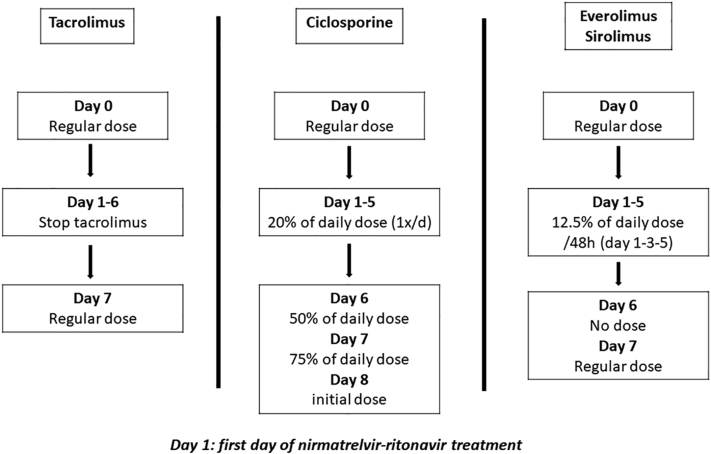
Before the first use of nirmatrelvir or ritonavir, we implemented a standardized management protocol for all KTRs with suspicion of SARS-CoV-2 infection (symptoms and/or positive antigenic test and/or high-risk contact) still ongoing. The patient is first examined in our SARS-CoV-2 outpatient clinic and a PCR on a nasopharyngeal swab is performed. If eligible to treatment with nirmatrelvir or ritonavir, all patient’s concurrent medications (immunosuppressive drugs and others) are carefully reviewed and are adapted by the nephrologist together with the clinical pharmacologist according to recommendations recently published by the French Society of Pharmacology and Therapeutics.5 Detailed management of both CNIs and mechanistic target of rapamycine inhibitors following NR prescription, is described in Figure 1.
Figure 1.
Protocol of calcineurin inhibitors and mechanistic target of rapamycine inhibitors dose adpatation during nirmatrelvir-ritonavir treatment.
According to our local protocol, the antimetabolite drug is interrupted for 7 days and steroid dose remains unchanged. Written recommendations are provided to the patient with treatment adaptation. NR is then prescribed (100 mg of ritonavir combined with 150 mg of nirmatrelvir [if baseline estimated glomerular filtration rate is 30–60 ml/min per 1.73 m2] or with 300 mg of nirmatrelvir [if estimated glomerular filtration rate is >60 ml/min per 1.73 m2] twice a day for 5 days).
During and after NR treatment, standard biological monitoring was performed, including serum creatinine and CNI or mechanistic target of rapamycine inhibitors trough levels at several time points (day [D] 0 [D1= first day of treatment], D2, D5, D7 and D9), assessment of anti-SARS-CoV2 serology on D0 and SARS-CoV-2 viral load on D0 and D7). Finally, patients were clinically re-evaluated on D9 at the outpatient clinic. Patients were advised to reach the nephrologist if they experienced any side effect at anytime.
Microbiology
The vaccine-induced humoral response was measured using an immunoassay detecting antibody against the spike protein receptor-binding domain (Elecsys anti-SARS-CoV-2, Roche Diagnostics GmbH, Mannheim, Germany - positive threshold >0.8 BAU/ml). The detection of SARS-CoV-2 on nasopharyngeal swab was performed using the Alinity m SARS-CoV-2 assay (Abbott Molecular Inc., Des Plaines, IL). This real-time reverse transcription polymerase reaction test is a dual target assay for the RdRp and N genes.
Positive samples presenting Ct values <25 were then sequenced starting from the same Nasopharyngeal swabs. The technique of sequencing is detailed in the Supplementary Materials and Methods.
Statistics
Paired-samples t test and Wilcoxon test were applied for comparison of serum creatinine and viral loads between the different time points. All tests were 2-sided and P ≤ 0.05 were considered significant. Statistical analysis was performed using SPSS v27 (IBM, New York, NY) and Prism v9 (GraphPad, San Diego).
Results
Baseline Characteristics
A total of 14 patients were eligible for treatment with NR during the study period. Baseline characteristics of the cohort are detailed in Table 1. Briefly, the median age was 60 (ranges 33-79) years, 9 (64%) were male, and 3 (24%) were patients with diabetes. Immunosuppressive therapy mainly consisted on a combination of tacrolimus (86%), mycophenolate mofetil (79%), and steroids (100%). None of the patients had a history of prior Sars-CoV-2 infection. All but 1 received anti SARS-CoV-2 vaccine (4 doses [last dose a median time of 69 days before] and 3 doses [last dose a median time of 210 days before], in 10 and 3 KTRs, respectively) before infection. Eight patients mounted a robust humoral response (anti-receptor-binding domain antibody titers >250 BAU/ml), 2 patients had a moderate humoral response (0–250 BAU /ml), and 4 had no humoral response after vaccination. Sequencing (available for 13 patients) revealed that all were infected by Omicron variants at D0. Several sublineages were identified: 8 patients with a BA.2 (Nextstrain clade 21L), 1 patient with a BA.2.3 (Nextstrain clade 21L), 2 patients with a BA.2.9 (Nextstrain clade 21L), 1 patient with a BA.2.36 (Nextstrain clade 21L) and 1 patient with a BA.2.12.1 (Nextstrain clade 22C) (Table 2).
Table 1.
Baseline characteristics at SARS-CoV-2 infection of the cohort
| Variable | Population N = 14 |
|---|---|
| Age, years, median (ranges) | 60 (33–79) |
| Male, n (%) | 9 (64) |
| Time since transplantation, months (ranges) | 116 (10–332) |
| Treated hypertension, n (%) | 10 (71) |
| Treated diabetes, n (%) | 3 (21) |
| Overweight-obesity, n (%) | 8 (57) |
| Treated dyslipidemia, n (%) | 11 (79) |
| Prior anti-SARS-CoV-2 vaccine, n (%) | |
| 4 doses | 10 (71) |
| 3 doses | 3 (21) |
| 0 dose | 1 (8) |
| Anti-RBD antibody level prior infection, n (%) | |
| >250 BAU/ml | 8 (57) |
| 0-250 BAU/ml | 2 (14) |
| 0 | 4 (29) |
| Prior COVID-19 infection, n | 0 |
| CYP3A5 genotype, n (%) | |
| 3∗/3∗ | 8 (57) |
| 1∗/3∗ | 2 (14) |
| CYP3A4 genotype, n (%) | |
| 1∗/1∗ | 7 (50) |
| Immunosuppression at admission, n (%) | |
| Immediate release tacrolimus | 1 (7) |
| Slow-release tacrolimus | 11 (79) |
| Mycophenolate mofetil | 11 (79) |
| Cyclosporine | 2 (14) |
| Azathioprine | 1 (7) |
| Steroids | 14 (100) |
| Nirmatrelvir-ritonavir dose, n (%) | |
| 300/100 mg | 5 (36) |
| 150/100 | 9 (64) |
CYP, cytochrome P450; RBD, receptor-binding domain.
Table 2.
Baseline and relapse nextstrain clade, pangolin sublineage assignment and GISAID submission references
| Day 0 |
Relapse |
|||||
|---|---|---|---|---|---|---|
| Patients | Nextclade | Pangolin | GISAID | Nextclade | Pangolin | GISAID |
| Patient 1 | 21L | BA.2.3 | EPI_ISL_12587374 | 21L | BA.2.3 | EPI_ISL_13017215 |
| Patient 2 | 21L | BA.2 | EPI_ISL_12587390 | |||
| Patient 3 | 21L | BA.2 | EPI_ISL_12660432 | |||
| Patient 4 | 21L | BA.2.9 | EPI_ISL_13017301 | 21L | BA.2.9 | EPI_ISL_13017221 |
| Patient 5 | 21L | BA.2 | EPI_ISL_13017302 | |||
| Patient 6 | 21L | BA.2 | EPI_ISL_13017212 | |||
| Patient 7 | 21L | BA.2.9 | EPI_ISL_13017210 | |||
| Patient 8 | NA | NA | NA | |||
| Patient 9 | 21L | BA.2.36 | EPI_ISL_13204423 | |||
| Patient 10 | 21L | BA.2 | EPI_ISL_13204424 | |||
| Patient 11 | 21L | BA.2 | EPI_ISL_13204422 | |||
| Patient 12 | 21L | BA.2 | EPI_ISL_13424237 | |||
| Patient 13 | 21L | BA.2 | EPI_ISL_13424225 | |||
| Patient 14 | 22C | BA.2.12.1 | EPI_ISL_13424238 | |||
GISAID, Global Initiative on Sharing Avian Influenza Data; NA, not available
Evolution of Laboratory Parameters During and After Treatment With NR
D0 and D7 mean (± SD) plasma creatinine concentrations were comparable (1.43 [±0.39] mg/dl vs. 1.45 [±0.50] mg/dl, P = 0.866) (Figure 2). Two patients experienced acute kidney injury respectively at D0 (patient 7) and D9 (patient 3) due to urinary infection requiring hospitalization and acute diarrhea, respectively. In both cases, plasma creatinine rapidly returned to baseline values after hydration and resolution of clinical symptoms. Compared with D0, median SARS-CoV-2 viral load assessed on D7 dramatically dropped (23,137,656 (interquartile range: 15,574,853–44,408,247) copies/ml vs. 1707 (interquartile range: 100–5372) copies/ml, P = 0.001) (Figure 3).
Figure 2.
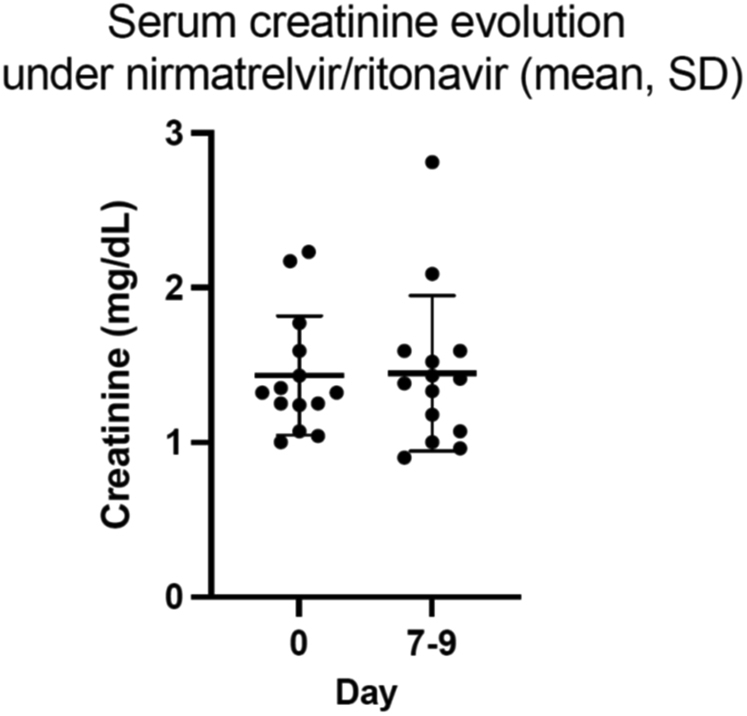
Comparison of plasma creatinine concentrations before (day 0) and 7 days after (day 7) nirmatrelvir-ritonavir initation. Day 0 and day 7 plasma creatinine concentrations were comparable (1.43 [1–2.23] mg/dl vs 1.45 [0.9–2.81] mg/dl, P = 0.866, paired-samples t test). Means ± SDs of plasma creatinine values are represented.
Figure 3.
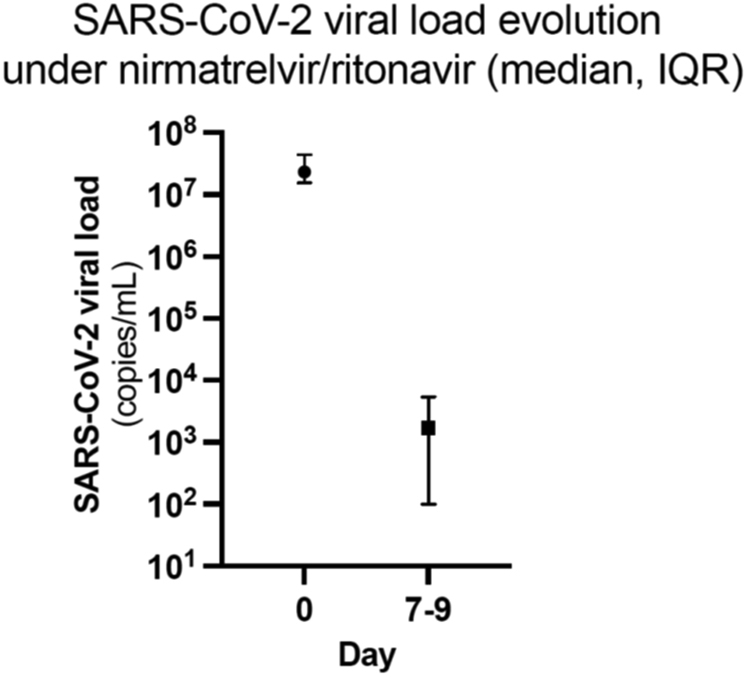
Evolution of SARS-CoV-2 viral load values. Comparison before (day 0) and 7 days after (day 7) nirmatrelvir-ritonavir initation. Compared with day 0, median SARS-CoV-2 viral load assessed at day 7 dramatically dropped (23,137,655 copies/ml vs. 1746 copies/ml, Wilcoxon Signed rank test; P = 0.002). Medians and IQRs of SARS-CoV-2 viral loads are represented. IQR, interquartile range.
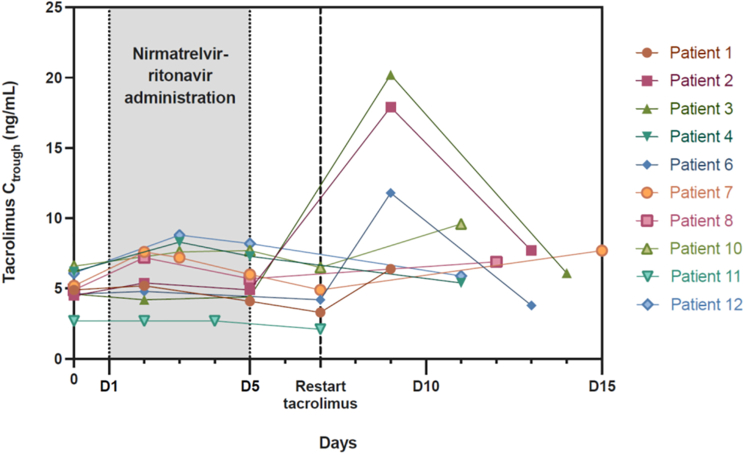
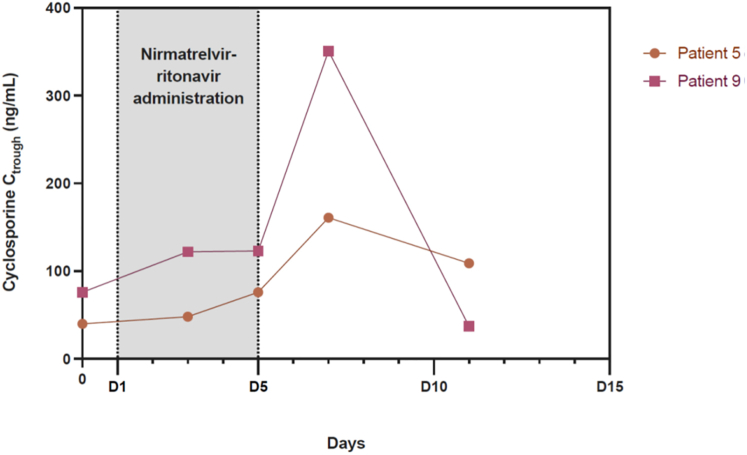
Therapeutic drug monitoring data were analyzable for the first 12 patients (Figures 4 and 5). CNI trough concentrations at the end of the treatment (D5 or D7) were satisfactory. Nevertheless, there was a large variability in tacrolimus half-life during antiviral treatment (range from 173 hours to 619 hours). Four patients (#2, 3, 6, and 9) presented CNI accumulation from D9 with trough concentrations increasing >10 ng/ml for tacrolimus and 300 ng/ml for cyclosporine.
Figure 4.
Evolution of tacrolimus trough concentrations (Ctrough) during and after nirmatrelvir-ritonavir administration. A total of 10 patients received nirmatrelvir-ritonavir 12 hours after the last dose of tacrolimus for a period of 5 days (D1–D5). Tacrolimus was restarted at the initial dose on D7. Tacrolimus Ctrough increased >10 ng/ml for 3 patients (#2, 3, and 6), so the dosage was adjusted after D9 for these patients. #, number; D, day.
Figure 5.
Evolution of cyclosporine trough concentrations (Ctrough) during and after nirmatrelvir-ritonavir administration. Cyclosporine was administered to 2 patients at 20% of the usual daily dosage during nirmatrelvir-ritonavir treatment (D1–D5) and then gradually increased from D6 to D8 (D6 : 50%, D7 : 75%, D8 : 100% of the initial dose). Cyclosporine Ctrough remained stable during nirmatrelvir-ritonavir administration. On D7, the patient 9’s cyclosporine trough level rose >350 ng/ml, warranting a subsequent dose reduction. D, day.
Clinical Outcome
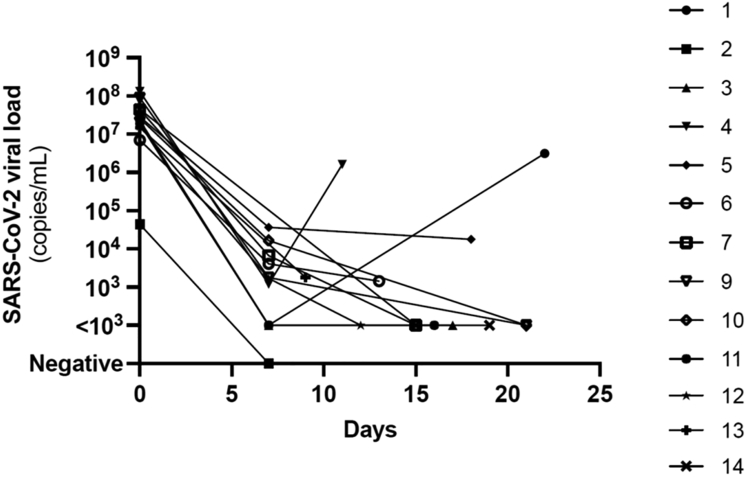
NR was well tolerated with no treatment discontinuation. No clinical biopsy-proven acute rejection was diagnosed during the study period. Two patients required hospitalization, respectively on D11 (patient 4) and D2 (patient 7) for acute pyelonephritis of the graft, which was rapidly resolved with antibiotics (IV ceftriaxone and then oral ciprofloxacine). No patient died during the study period and none presented with viral pneumonia. However, early relapses of COVID-19 symptoms occurred in 2 patients (patient 1 and 4; Figure 6) together with an increase in viral load.
Figure 6.
Individual evolution overtime of SARS-CoV-2 viral loads. Early relapses of COVID-19 symptoms occurred in 2 patients (patient 1 and 4) together with a relapse in viral load. SARS-CoV-2 viral load data were not available for one patient (patient 8).
Patient 1 was a 47-year-old male transplanted 10 years ago. His immunosuppressive regimen consisted of a combination of tacrolimus, mycophenolate mofetil, and steroids. After 5 days of symptoms (runny nose, sore throat, cough, chills, and muscle pain), he was treated with 150 mg of nirmatrelvir and 100 mg of ritonavir twice a day for 5 days. Symptoms rapidly resolved. SARS-CoV-2 viral loads on D0 and D7 were measured at 18,941,591 and <1000 copies/ml respectively. On D21, the patient experienced a new onset of symptoms (runny nose, fatigue, headaches) and a repeated PCR (D22) showed a viral load relapse at 3,148,427 copies/ml. Patient 4 was a 68-year-old female transplanted 1 year earlier. Her immunosuppressive regimen consisted of a combination of tacrolimus, mycophenolate mofetil, and steroids. After 5 days of symptoms (runny nose and cough), she was treated with 150 mg of nirmatrelvir and 100 mg of ritonavir twice a day for 5 days. Symptoms rapidly resolved, though a mild cough persisted. SARS-CoV-2 viral loads on D0 and D7 were measured at 131,344,593 and 1,189 copies/ml, respectively. On D10, the patient presented new symptoms (malaise, fatigue, and nausea) and a repeated PCR (D11) showed a relapse in the viral load at 1,616,064 copies/ml.
Both patients had rapid resolution of symptoms within few days without further intervention and a negative PCR test was subsequently obtained on D26 (patient 1) and D35 (patient 4), respectively.
For patient 1, although the viral strains sequenced on D0 and at relapse are Omicron variants with the same sublineage assignment BA.2.3 (or Nextstrain clade: 21L) (Table 2), the strain present at the time of relapse (D22) shows additional variations: one amino acid substitution in the Spike protein compared with the starting strain S:R408S as well as the deletions ORF1a:G82-, ORF1a:G83-, ORF1a:G84-, ORF1a:G85-, ORF1a:G86-, and S:Y144-. For patient 4, the viral strains sequenced on D0 and at relapse (D11) are Omicron variants with the same sublineage assignment BA.2.9 (or Nextstrain clade: 21L) (Table 2). In this second case, no differences could be demonstrated between the strains present at the beginning of the infection and at the time of relapse.
To assess whether patient 1 was infected by another SARS-CoV-2 strain, a phylogenic analysis was performed (Supplementary Figure S1) showing that both strains (the one of the initial infection compared with the one during relapse) were very close, making the hypothesis of an infection by a new strain unlikely.
Discussion
Rapidly emerging new variants of concern of SARS-CoV-2 present a constant challenge requiring frequent adaptations in therapeutic strategies for KTRs infected with SARS-CoV-2. To date, the following 3 treatments are indicated to treat SARS-CoV-2 infection in areas where BA.2 variant is prevalent: (i) remdesivir (only i.v. administration), (ii) molnupiravir (oral administration, not currently available in our country outside medical need programs), and (iii) NR (oral administration).4 However, evidence regarding safety and efficacy of all these treatments in KTRs is very low, because of the exclusion, or the very low rate of inclusion, of transplanted patients in clinical trials. Consequently, there is an urgent need to collect available data on this subject. That is even more relevant for NR use because of the high risk of potentially harmful drug interactions associated with its use. We here report the largest case series of KTRs treated with NR.
Treatment with NR was effective in our patients to prevent severe COVID-19. Indeed, none of them required hospitalization for viral pneumonia and none of them died. In addition, viral load massively decreased on D7 in all patients. Noteworthy, 2 patients were hospitalized for urinary sepsis, but the causal relationship with the treatment is unlikely because it was not reported in the phase 3 trial.3 This is in line with previous case series on the use of NR in transplanted patients.6, 7, 8 Salerno et al.6 reported that among 25 solid organ transplant patients (including 5 KTRs), 4 required hospitalization (one for diarrhea and 3 for COVID-19 symptoms), and none died, within 30 days.
The strategy of CNI adaptation during NR treatment we applied was based on the strategy proposed by the French Society of Pharmacology and Therapeutics (Figure 1).5 Ritonavir is well known to significantly increase CNI concentrations, which could lead to drug toxicity.9 Lange et al.10 proposed another, yet rather similar, strategy consisting of immediate holding of tacrolimus or 80% dose reduction of cyclosporine on initiation of nirmatrelvir or ritonavir, until the end of the 5-day course treatment. Measuring the CNI level on D3, D6, and D7 after nirmatrelvir or ritonavir initiation was also recommended to assist with timing of CNI resumption. This strategy was tested in 4 KTRs by Wang et al.8 The authors found high variability in tacrolimus concentration evolution between patients but they anticipated that tacrolimus might be reintroduced partially or in full dose between D8 and D10. Salerno et al. further evaluated the strategy on their larger cohort (25 solid organ transplant) and obtained similar tacrolimus concentrations before and after treatment.6 However, 4 patients experienced a supratherapeutic tacrolimus concentration after restarting tacrolimus. In our study, therapeutic drug monitoring data were available for 12 patients. Using the proposed recommendations5 of discontinuation 12 hour before the start of NR for tacrolimus and administering 20% of the usual daily dosage for cyclosporine, CNI trough concentrations at the end of the treatment were mostly satisfactory. However, we also observed important individual variability. These data suggest that CNI clearance might present a large inter-individual variability among transplant recipients and that some patients may display a longer CYP3A4 and/or P-glycoprotein inhibition leading to increased exposure on restarting immunosuppressive treatment. Therapeutic drug monitoring is thus particularly recommended in this population of patients. The D7 to D10 period appears particularly at risk and should be closely monitored. Though the number of patients included is limited, no risk factor (CYP3A5 or CYP3A4 genotype, concentration over dose ratio) for higher or lower CNI clearance during NR treatment or for longer CNI metabolism and/or transport inhibition have been found. The identification of such factors would be particularly relevant to identify patients at risk of rapid CNI elimination and therefore at risk for rejection or in contrast, patients at risk of CNI accumulation and toxicity. Importantly, kidney function remained remarkably stable during the study period (with the exception of 2 patients affected by concomittant sepsis and diarrhea), suggesting that this rather simple strategy seems safe regarding the graft function.
In our experience, the management of NR was much more demanding than the management with previous treatments, such as sotrovimab.11 Indeed, it requires manpower and important logistics to control tacrolimus concentrations. Moreover and most importantly, it requires an active participation of the patient because of the modifications made to the immunosuppressive treatment as well as the temporary discontinuation of some other drugs. We think that providers should be very selective in prescribing NR only to patients who can understand and strictly adhere to drug modifications, and also in centers having the logistics to manage the treatment.
Emerging data have demonstrated that NR can be associated with early relapse.12, 13, 14 A recent US retrospective study that included 11,270 people treated with NR showed 7-day and 30-day relapse rates of 3.53% and 5.40% for COVID-19 infection, 2.31% and 5.87% for COVID-19 symptoms, and 0.44% and 0.77% for hospitalizations, respectively.14 In our study, we observed 2 cases (14% of the cohort) who experienced early relapse of COVID-19 symptoms associated with proven increase in SARS-CoV-2 viral load. The 2 patients did not differ in baseline characteristics compared with those who did not experience early relapse. Both patients reported no deviation in the prescribed posology of NR, thus making the hypothesis of low adherence unlikely. For 1 patient (patient 4), SARS-CoV-2 strains present at the beginning of the infection and at the time of relapse were similar. However, we detected additional variations in the strain at the time of relapse, compared with the initial strain, in the Spike protein and in the ORF-1 gene in the other patient (patient 1). Phylogenic analysis revealed that these variations were most likely acquired during or after NR treatment. Nevertheless, the clinical relevance of this finding in explaining the COVID-19 relapse experienced by the patient is uncertain and remains to be demonstrated.15, 16, 17, 18 Lower blood concentrations of nirmatrelvir (not routinely available) or other, here undetected, acquired genetic mutations are other hypothesis to explore on larger cohorts.
Our study has limitations. It is a small cohort with a retrospective design, making it subject to bias, and a short follow-up. It also lacks a control group that could help determine the drug’s true efficacy in a predominantly vaccinated population. Nevertheless, we administered the current recommended treatment to our vulnerable population and were able to closely monitor and follow-up our patients.
In conclusion, NR can be used in KTRs infected with SARS-CoV-2. It seems effective in preventing the occurrence of severe COVID-19. Drug adaptation is feasible and mandatory to avoid CNI toxicity. We observed 2 cases of early relapse after NR treatment that need urgent further investigations.
Disclosure
FL received research grants (paid to institution) from Chiesi, Astellas Pharma, and Sandoz; and fees to attend meetings from Chiesi and Sandoz. All the other authors declared no competing interests.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author on reasonable request.
Author Contributions
AD, SB, and NK undertook the following: research idea, study design, data analysis, and writing of the manuscript; SB and JDG performed data acquisition; AS and BK performed serologic analysis; JDG performed statistical analysis; LB, VH, and FL performed pharmacologic analysis; KDP, RM, and BB performed microbiologic analysis (PCR and genetic); AD, SB, JDG, FL, LB, VH, AS, BK, JCY, LB, TD, AB, KDP, RM, BB, EG, and NK took care of the patients. All authors discussed and reviewed the manuscript.
Footnotes
Supplementary Materials and Methods. SARS-CoV-2 variants sequencing.
Figure S1. Phylogeny of the 14 patients at day 0 and of the 2 patients (patient 1 and 4) at relapse.
Supplementary Reference.
Supplementary Material
Supplementary Materials and Methods. SARS-CoV-2 variants sequencing.
Figure S1. Phylogeny of the 14 patients at day 0 and of the 2 patients (patient 1 and 4) at relapse.
Supplementary Reference
References
- 1.Takashita E., Kinoshita N., Yamayoshi S., et al. Efficacy of antiviral agents against the SARS-CoV-2 omicron subvariant BA.2. N Engl J Med. 2022;386:1475–1477. doi: 10.1056/NEJMc2201933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.FDA updates Sotrovimab emergency use authorization Food and Drug Administration. https://www.fda.gov/drugs/drug-safety-and-availability/fda-updates-sotrovimab-emergency-use-authorization Updated May 4. 2022.
- 3.Hammond J., Leister-Tebbe H., Gardner A., et al. Oral nirmatrelvir for high-risk, nonhospitalized adults with Covid-19. N Engl J Med. 2022;386:1397–1408. doi: 10.1056/NEJMoa2118542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fishbane S., Hirsch J.S., Nair V. Special considerations for Paxlovid treatment among transplant recipients with SARS-CoV-2 infection. Am J Kidney Dis. 2022;79:480–482. doi: 10.1053/j.ajkd.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lemaitre F., Grégoire M., Monchaud C., et al. Management of drug-drug interactions with nirmatrelvir/ritonavir in patients treated for Covid-19: guidelines from the French Society of Pharmacology and Therapeutics (SFPT) Therapie. Published online April 20. 2022 doi: 10.1016/j.therap.2022.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salerno D.M., Jennings D.L., Lange N.W., et al. Early clinical experience with nirmatrelvir/ritonavir for the treatment of COVID-19 in solid organ transplant recipients. Am J Transplant. 2022;22:2083–2088. doi: 10.1111/ajt.17027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Radcliffe C., Palacios C.F., Azar M.M., Cohen E., Malinis M. Real-world experience with available, outpatient COVID-19 therapies in solid organ transplant recipients during the omicron surge. Am J Transplant. Published May. 2022 doi: 10.1111/ajt.17098. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang A.X., Koff A., Hao D., Tuznik N.M., Huang Y. Effect of nirmatrelvir/ritonavir on calcineurin inhibitor levels: early experience in four SARS-CoV-2 infected kidney transplant recipients. Am J Transplant. 2022;22:2117–2119. doi: 10.1111/ajt.16997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morelle J., Goffin E., Wallemacq P., et al. Extended release tacrolimus and antiretroviral therapy in a renal transplant recipient: so extended. Transpl Int. 2010;23:1065–1067. doi: 10.1111/j.1432-2277.2010.01098.x. [DOI] [PubMed] [Google Scholar]
- 10.Lange N.W., Salerno D.M., Jennings D.L., et al. Nirmatrelvir/ritonavir use: managing clinically significant drug-drug interactions with transplant immunosuppressants. Am J Transplant. 2022;22:1925–1926. doi: 10.1111/ajt.16955. [DOI] [PubMed] [Google Scholar]
- 11.Fernandes G., Devresse A., Scohy A., et al. Monoclonal antibody therapy in kidney transplant recipients with delta and omicron variants of SARS-CoV-2: a single-center case series. Kidney Med. 2022;4:100470. doi: 10.1016/j.xkme.2022.100470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rubin R. From positive to negative to positive again-the mystery of why COVID-19 rebounds in some patients who take Paxlovid. JAMA. 2022;327:2380–2382. doi: 10.1001/jama.2022.9925. [DOI] [PubMed] [Google Scholar]
- 13.COVID-19 rebound after Paxlovid treatment CDC health ADVISORY. Centers for Disease Control and Prevention. https://emergency.cdc.gov/han/2022/pdf/CDC_HAN_467.pdf Updated May 25. 2022. Accessed June 29, 2022.
- 14.Wang L., Berger N.A., Davis P.B., et al. COVID-19 rebound after Paxlovid and Molnupiravir during January–June 2022 [Preprint]. medRxiv. Posted online June 21. 2022 doi: 10.1101/2022.06.21.22276724. [DOI] [Google Scholar]
- 15.Chakraborty C., Bhattacharya M., Sharma A.R. Emerging mutations in the SARS-CoV-2 variants and their role in antibody escape to small molecule-based therapeutic resistance. Curr Opin Pharmacol. 2022;62:64–73. doi: 10.1016/j.coph.2021.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Velazquez-Salinas L., Zarate S., Eberl S., et al. Positive selection of ORF1ab, ORF3a, and ORF8 genes drives the early evolutionary trends of SARS-CoV-2 during the 2020 COVID-19 pandemic. Front Microbiol. 2020;11:550674. doi: 10.3389/fmicb.2020.550674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shirato K., Kakizaki M., Tomita Y., et al. Detection of the ORF1 gene is an indicator of the possible isolation of severe acute respiratory syndrome coronavirus 2. Pathogens. 2022;11:302. doi: 10.3390/pathogens11030302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ambike S., Cheng C.C., Feuerherd M., et al. Targeting genomic SARS-CoV-2 RNA with siRNAs allows efficient inhibition of viral replication and spread. Nucleic Acids Res. 2022;50:333–349. doi: 10.1093/nar/gkab1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author on reasonable request.