Abstract
Three VirB proteins (VirB1*, VirB2, and VirB5) have been implicated as putative components of the T pilus from Agrobacterium tumefaciens, which likely mediates binding to plant cells followed by transfer of genetic material. Recently, VirB2 was indeed shown to be its major component (E.-M. Lai and C. I. Kado, J. Bacteriol. 180:2711–2717, 1998). Here, the influence of other Vir proteins on the stability and cellular localization of VirB1*, VirB2, and VirB5 was analyzed. Solubility of VirB1* and membrane association of VirB2 proved to be inherent features of these proteins, independent of virulence gene induction. In contrast, cellular levels of VirB5 were strongly reduced in the absence of other Vir proteins, indicating its stabilization by protein-protein interactions. The assembly and composition of the T pilus were analyzed in nopaline strain C58(pTiC58), its flagellum-free derivative NT1REB(pJK270), and octopine strain A348(pTiA6) following optimized virulence gene induction on solid agar medium. In all strains VirB2 was the major pilus component and VirB5 cofractionated during several purification steps, such as ultracentrifugation, gel filtration, and sucrose gradient centrifugation. VirB5 may therefore be directly involved in pilus assembly, possibly as minor component. In contrast, secreted VirB1* showed no association with the T pilus. In-frame deletions in genes virB1, virB2, virB5, and virB6 blocked the formation of virulence gene-dependent extracellular high-molecular-weight structures. Thus, an intact VirB machinery as well as VirB2 and VirB5 are required for T-pilus formation.
Agrobacterium tumefaciens is a natural genetic engineer capable of transferring a segment of DNA (T DNA) on its Ti (tumor-inducing) plasmid to plant cells, where it stably integrates into the nuclear genome. This process depends on the action of several virulence proteins for production of a single-stranded copy of T DNA (VirC1, VirC2, VirD1, and VirD2), protection from nucleolytic cleavage (VirE2), entry into the plant nucleus and integration (VirD2 and VirE2), and transfer into plant cells (VirB1 to VirB11 and VirD4) (8, 19, 23, 37, 46, 47). Transfer of the T strand in a complex with Vir proteins (T complex) from A. tumefaciens to plant cells resembles conjugative DNA transfer between bacteria in many aspects (29, 41). First, sequence comparison shows significant similarities of components of the transfer machinery, i.e., VirB1 to VirB11 and VirD4, to constituents of several bacterial conjugation and protein secretion systems, suggesting that a common mechanism was adapted in evolution for trafficking of different substrates (8, 27, 43). Second, structural predictions suggest membrane association and export of several of these proteins. A multimeric transmembrane complex, the T-complex transfer machinery (see reference 8 and citations therein), may correspond to the electron-dense regions at contact sites between donor and recipient bacteria during conjugation observed by electron microscopy (11). Third, electron microscopy shows pili on the surface of virulence gene-induced A. tumefaciens, suggesting that Vir proteins, e.g., components of the membrane-associated VirB complex, may play a role during pilus assembly or constitute its structural components (15, 16).
Bacterial pili play crucial roles in cell-cell interactions, for example, by conferring adhesion of pathogenic bacteria to host tissues (20) or by initiating cell-cell contact leading to transfer of genetic information (14). Analogous to pilus-dependent conjugative transfer of the F episome, the A. tumefaciens T pilus may bind plant cells as a first step leading to T-complex transfer (30). To analyze the mechanism of pilus assembly and plant cell binding, pilus structural components need to be identified. Based on different lines of evidence, three VirB proteins, i.e., VirB1*, VirB2, and VirB5, were assigned potential roles as pilus components. VirB1 shows similarity to bacterial transglycosylases in its N-terminal domain and may facilitate assembly of the T-complex transfer machinery by localized lysis of the murein cell wall (6, 28, 32), and it undergoes processing followed by secretion of the C-terminal portion of VirB1* (3). VirB1* is the only soluble VirB component and partly localizes to the extracellular space, and electron microscopic studies detected VirB1-cross-reactive material on the surface of virulence gene-induced agrobacteria (9). These results suggested that it may play a role in plant cell interaction, possibly as a pilus component (3, 5, 30). Based on its weak sequence similarity to TraA, the major component of the F pilus, VirB2 was predicted to be a pilus component (21, 38). Lai and Kado (26) showed recently that VirB2, which proved to be a cyclic peptide (12), is a major component of the A. tumefaciens T pilus, but other constituents were not identified. A potential role of VirB5 as a pilus component derived from studies on TraC, its homolog from the incompatibility N (IncN) group plasmid pKM101. Conjugative transfer of pKM101 derivatives with transposon insertions in traC (pKM101traC::Tn5) is strongly reduced compared to that of the wild-type plasmid, but addition of a helper strain expressing a functional DNA transfer machinery strongly increases conjugative transfer (44). This extracellular complementation supports an exterior localization of TraC, possibly as part of a pilus structure, allowing transfer of the protein from the helper strain to pKM101traC::Tn5-carrying cells (43). Analysis of pKM101-carrying Escherichia coli showed the association of TraC with an exterior high-molecular-weight structure, further supporting its role either in assembly or as a structural component of the conjugative pilus (36).
So far, additional components have not been identified in conjugative pili, although indirect evidence suggests that they might exist in case of the F pilus (2, 13). In contrast, adhesive pili typically contain one major pilin and one or more minor components, which determine specific pilus functions such as cell adhesion (34, 39). To address the presence of minor pilus components, we isolated T pili and performed compositional analysis with all VirB protein-specific antisera. In addition to VirB2, VirB5 consistently copurified with T pili isolated from nopaline strains C58(pTiC58) and NT1REB(pJK270), as well as from octopine strain A348(pTiA6). Thus, VirB5 may be a minor component of the T pilus, in contrast to VirB1*, which showed no association with extracellular high-molecular-weight structures. Analyses of virB mutants suggest that the stability of cell-bound VirB5 and its extracellular association with VirB2-containing T pili are mediated by an interaction(s) with components of the membrane-associated T-complex transfer machinery.
MATERIALS AND METHODS
Bacterial growth and virulence gene induction.
The strains used are listed in Table 1. Growth of E. coli for cloning procedures was performed at 37°C in Luria-Bertani medium, whereas agrobacteria were grown at 28°C in YEB (0.5% beef extract, 0.1% yeast extract, 0.5% peptone, 0.5% sucrose, 2 mM MgSO4) liquid medium or in petri dishes containing medium solidified with 2% agar. For plasmid propagation, media were supplemented with streptomycin (50 μg/ml for E. coli and 100 μg/ml for A. tumefaciens), spectinomycin (50 μg/ml for E. coli and 300 μg/ml for A. tumefaciens), ampicillin (100 μg/ml), and chloramphenicol (20 μg/ml).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Agrobacterium strains | ||
| C58 | Wild type, pTiC58 | 42 |
| CB1001 | pTiC58 carrying in-frame deletion of virB1 | This study |
| CB1005 | pTiC58 carrying in-frame deletion of virB5 | 36 |
| NT1REB(pJK270) | Flagellum-free derivative of C58 | 26 |
| A348 | Wild type, pTiA6NC | P. Christie |
| PC1001 | pTiA6NC carrying in-frame deletion of virB1 | 7 |
| PC1002 | pTiA6NC carrying in-frame deletion of virB2 | 7 |
| PC1005 | pTiA6NC carrying in-frame deletion of virB5 | 7 |
| PC1006 | pTiA6NC carrying in-frame deletion of virB6 | 7 |
| E. coli JM109 | Cloning host | 45 |
| Plasmids | ||
| pTrc200 | Strr Spcr, pVS1 derivative, lacIq, trc promotor expression vector | 36 |
| pTrcB1 | Strr Spcr, virB1 PCR fragment cloned downstream of the trc promotor of pTrc200 | This study |
| pTrcB2 | Strr Spcr, virB2 PCR fragment cloned downstream of the trc promotor of pTrc200 | This study |
| pTrcB5 | Strr Spcr, virB5 PCR fragment cloned downstream of the trc promotor of pTrc200 | 36 |
| pGVO310 | Apr, pBR322 derivative carrying the virA-virB region and part of virG from pTiC58 | 10 |
| pVirAB4 | Cmr, pBCSK+.Nde carrying a 2.6-kb ApaI/HindIII fragment from pGVO310 carrying genes virB1, virB2, and virB3 | This study |
A. tumefaciens virulence genes were induced by growth in AB minimal medium (10 g of glucose per liter, 4 g of MES [morpholineethanesulfonic acid] per liter, 2 g of NH4Cl per liter, 0.3 g of MgSO4 · 7H2O per liter, 0.15 g of KCl per liter, 0.01 g of CaCl2 per liter, 0.0025 g of FeSO4 · 7H2O per liter, and 1 mM potassium phosphate [pH 5.5]) by the addition of acetosyringone (AS) at a final concentration of 200 μM. For isolation of pili, cells were induced for 3 or 4 days at 20°C on AB medium solidified with 2% agar. For induction of the LacI-repressed trc promotor in pTrc200 constructs, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.5 mM.
DNA modification procedures.
Standard methods were used for DNA preparation, modification, and cloning, with enzymes purchased from MBI Fermentas and New England Biolabs (31). DNA sequences of PCR clones and in-frame deletion constructs were confirmed by sequencing on an ABI Prism 377 sequencer. DNA fragments were PCR amplified with Goldstar DNA polymerase (Eurogentec) from 1 ng of plasmid template by using the following cycle conditions: denaturation (one time), 2 min at 95°C; cycling (30 times), 44°C for 1 min, 72°C for 2 min, and 95°C for 30 s; strand completion (one time), 44°C for 1 min and 72°C for 5 min; and termination at 4°C.
To generate constructs for AS-independent gene expression, pTrc200 (36) was cleaved with NcoI and SmaI or BamHI and treated with calf intestine phosphatase. Genes were PCR amplified from target plasmid pGVO310, cleaved with NcoI or AflIII at the 5′ end and with ScaI or BglII at the 3′ end (see underlined sequence below), and ligated with pTrc200. Oligonucleotides used for amplification are as follows: for virB1, B1Afl (5′GCCACATGTTGAAGGCAACAGGG-3′) and B13 (5′-GGGAGATCTTTCAAAGCATCG-3′), and for virB2, B25 (5′-GGGGCCATGGGATGCTTTGAAAGATACC-3′) and B23 (5′-GAAAGTACTTAGCCACCTCCAGTC-3′).
Construction of virB1 deletion strain CB1001.
Strain CB1001, carrying an in-frame deletion of virB1 on the Ti plasmid of strain C58, was constructed essentially as described previously (7, 36). Oligonucleotides dB1-1 (5′-AGCTTGGGGAGATGGGGAATGCGATGCTTTGAAAGA-3′) and dB1-2 (5′-TCTTTCAAAGCATCGCATTCCCCATCTCCCCAAGCT-3′) were used to construct an in-frame deletion of virB1 in plasmid pVirAB4, which was then introduced into the Ti plasmid of strain C58 by double recombination.
Subcellular fractionation.
Total cell lysate, soluble protein, and membrane proteins were isolated from A. tumefaciens grown on AB agar medium as described previously (3).
Isolation and characterization of T pili.
Cells were grown on AB agar in 15-cm-diameter plates, suspended in 10 ml of buffer P (50 mM potassium phosphate, pH 5.5), and then centrifuged at 10,000 rpm in an SS34 rotor in an RC-5B centrifuge (Sorvall) for 60 min. Cell pellets were suspended in 1 ml of buffer P, passed eight times through a 26-gauge needle to remove surface-associated high-molecular-weight structures, and then centrifuged in a microcentrifuge for 60 min at 15,000 rpm. The supernatant was subjected to high-speed centrifugation at 40,000 rpm for 90 min in a 70.1 Ti rotor in an OTD 50B centrifuge (Sorvall) to separate high-molecular-weight structures, like flagella and pili, in the pellet from soluble constituents removed from the cells by shearing.
To assess the composition and molecular weight of pili, pellets obtained by high-speed centrifugation were suspended in 300 μl of buffer P and applied to a Superdex 200 column (Pharmacia), followed by chromatography with a flow rate of 0.5 ml/ml. Reference proteins for calibration of the column are indicated in the legend to Fig. 6.
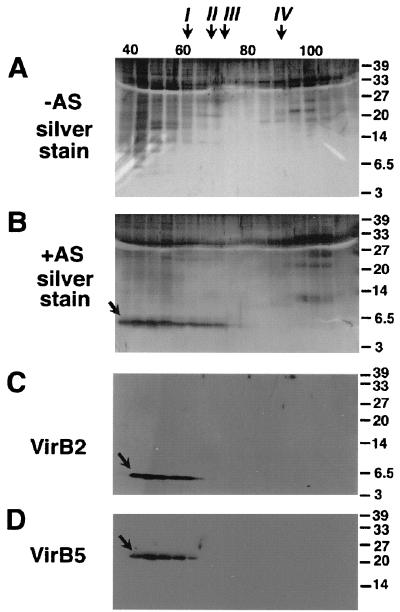
FIG. 6.
VirB2 and VirB5 elute in a high-molecular-weight structure from a Superdex 200 gel filtration column. Surface structures isolated by high-speed centrifugation from agrobacteria grown under virulence gene-inducing (+AS) and noninducing (−AS) conditions were subjected to gel filtration chromatography. Column fractions were subjected to SDS-PAGE followed by silver staining (A and B) or Western blotting with specific antisera for VirB2 (C) or VirB5 (D). VirB2 and VirB5 are indicated by arrows. Molecular weights and elution volumes of reference proteins for calibration of the gel filtration column: I, ferritin (440,000 and 63 ml); II, aldolase (158,000 and 70.4 ml); III, bovine serum albumin (68,000 and 76.8 ml); IV, cytochrome c (12,000 and 92.8 ml). Numbers on the right are molecular weights in thousands.
Pellets obtained by high-speed centrifugation of surface-exposed macromolecules from eight AB agar plates of strain NT1REB(pJK270) were suspended in 200 μl of buffer P, applied to an isopycnic 30 to 70% sucrose gradient in buffer P, and centrifuged for 8 h in an AH627 rotor in an OTD 50 B centrifuge (Sorvall).
Electron microscopy.
For negative staining, a drop of the pellets obtained by high-speed centrifugation was placed on a carbon-coated copper grid, removed with a pipette after 2 min, air dried, and stained for 1 min with 1% phosphotungstic acid–0.01% glucose, pH 7.0. The specimens were examined in a Zeiss EM 912 transmission electron microscope operated with the OMEGA energy filter in the zero loss mode.
Protein analysis.
VirB proteins in cell lysates were detected after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described previously (25, 35), followed by silver staining or Western blotting, incubation with VirB-specific polyclonal antisera, and detection with horseradish peroxidase-conjugated antirabbit secondary antibody (Bio-Rad), using standard protocols of the BM chemiluminescence detection system (Roche). The following reference proteins (1 μg/lane) were used for determination of molecular masses: T7 RNA polymerase (97 kDa), fructose 6-phosphate kinase (85 kDa), glutamate dehydrogenase (55 kDa), aldolase (39 kDa), lactate dehydrogenase (33 kDa), triosephosphate isomerase (27 kDa), trypsin inhibitor (20 kDa), lysozyme (14 kDa), aprotinin (6.5 kDa), and insulin (3 kDa).
Generation of VirB2 protein-specific antiserum.
A specific antiserum was generated by injection of 500 μg of purified inclusion bodies of phage T7 gene 10 protein fused to amino acids 9 to 121 of VirB2 in New Zealand White rabbits for immunization. To reduce nonspecific cross-reactions, antisera were affinity purified by binding to polyvinylidene difluoride membrane-fixed antigen and elution at acidic pH values by standard procedures (18).
Image processing.
Fluorographs and gels were scanned with a UMAX UC840 MaxVision, processed with Adobe Photoshop 2.5 software, and printed on an Epson Stylus Photo printer.
RESULTS
Cellular localization of candidate pilus components.
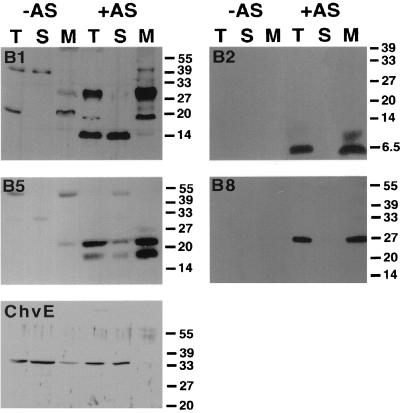
Three different VirB proteins (VirB1*, VirB2, and VirB5) were proposed as putative components of the extracellular T pilus. VirB2, VirB5, and full-length VirB1 cofractionate with membranes of agrobacteria after virulence gene induction in liquid culture, whereas the C-terminal processing product VirB1* localizes to the soluble fraction. However, the previously used induction at 28°C in liquid culture proved to be inappropriate for pilus assembly and/or function of the T-complex transfer machinery (15, 16). Since our goal was to isolate the T pilus and analyze its content, the cellular localization of putative pilus components was reassessed after virulence gene induction under optimized conditions on agar medium at 20°C (15), which led to increased levels of VirB proteins in nopaline Ti plasmid pTiC58-carrying strain C58 compared to induction in liquid culture at 28°C. Agrobacterial virulence genes were induced on acidified AB agar with AS and lysed by passage through a French press, and subcellular fractions (total lysate, soluble protein, and membrane fractions) were analyzed for localization of VirB1, VirB2, VirB5, VirB8, and the periplasmic sugar-binding protein ChvE. VirB8 was included as a control for an intrinsically membrane-bound protein, and activity assays of the inner membrane enzyme NADH oxidase confirmed the correct separation of subcellular fractions (not shown). Figure 1 shows that, as in liquid-induced cells, full-length VirB1, VirB2, VirB5, and VirB8 fractionated predominantly or exclusively with the membranes, whereas the C-terminal processing product VirB1* and ChvE were predominantly soluble in strain C58. Deletion of individual virB genes (virB1, virB2, virB5, and virB6) did not detectably affect subcellular localization of VirB1, VirB2, and VirB5 (not shown); however, we observed reduced levels of VirB5 in a virB6 deletion mutant (see below). Molecular mass determination showed that VirB2 is a 5.5-kDa protein, which constantly migrates below the 6.5-kDa molecular mass marker protein aprotinin. In contrast, VirB2 was previously reported to be a 7.2-kDa protein (26, 38), perhaps due to different preparation of gels used for SDS-PAGE analysis.
FIG. 1.
Cellular localization of VirB1, VirB2, VirB5, VirB8, and ChvE in strain C58. Western blot analysis with different specific antisera after SDS-PAGE of subcellular fractions from agrobacteria grown on AB minimal medium plates under virulence gene-inducing (+AS) and noninducing (−AS) conditions is shown. Lanes: T, total cell lysate; S, soluble fraction; M, membrane proteins. ChvE is not encoded by the virulence regulon and therefore is detected in lysates from cells grown under both conditions. Numbers on the right are molecular weights in thousands.
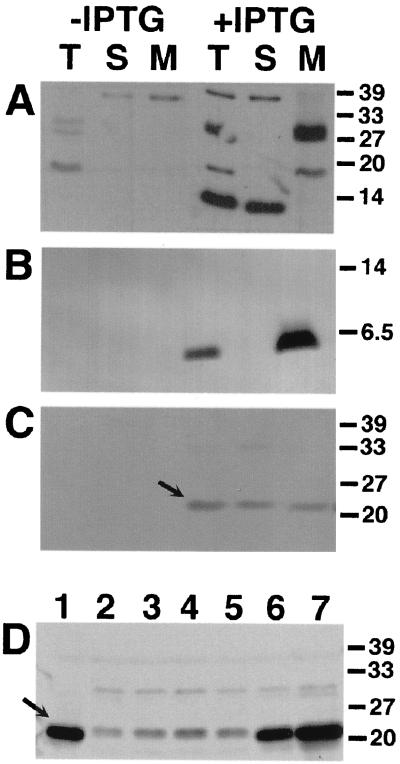
Whereas protein sequence analysis predicts membrane-spanning regions in VirB2 and VirB8, and thus intrinsic localization to membranes, VirB1 and VirB5 do not contain significant stretches of hydrophobic amino acids. This led to the hypothesis that protein-protein interactions with other VirB proteins may mediate their association with membranes. To directly test this hypothesis, genes virB1, virB2, and virB5 were cloned in pTrc200 for expression from the trc promotor, and the resulting plasmids pTrcB1, pTrcB2, and pTrcB5 were individually transformed into avirulent A. tumefaciens strains carrying deletions in the corresponding genes on their Ti plasmid. Complementation experiments showed that expression of nopaline VirB2 and VirB5 completely restored the virulence of octopine as well as nopaline strains carrying deletions of virB2 and virB5, respectively (not shown). Complementation of strains carrying deletions in virB1, which show attenuated virulence, was not quantified. Transformed cells were grown on AB agar plates without AS in the presence of IPTG (0.5 mM) to induce expression from the trc promotor, followed by cell lysis and preparation of subcellular fractions. Analysis of VirB protein content by SDS-PAGE and Western blotting with specific antisera showed that hydrophobic VirB2 as well as apparently hydrophilic VirB1 and VirB5 cofractionated with the membranes even in the absence of other VirB proteins (Fig. 2A to C). Compared to the wild type, however, VirB5 protein levels were strongly reduced and a larger portion was detected in the soluble fraction, whereas VirB1 and VirB2 accumulated to wild-type levels. To analyze the importance of Vir proteins for accumulation of VirB5 in the cell, CB1005(pTrcB5) was grown in the presence of IPTG for induction of the trc promotor as described above, using various concentrations of AS to achieve different levels of virulence gene induction. Figure 2D shows that the amount of cell-bound VirB5 parallels increased virulence gene induction, indicating that Vir proteins exert a stabilizing effect.
FIG. 2.
Cellular localization of VirB1, VirB2, and VirB5 in the absence of Vir proteins. Western blot analysis with specific antisera after SDS-PAGE of cell lysates from strains CB1001(pTrcB1) (A), PC1002(pTrcB2) (B), and CB1005(pTrcB5) (C) grown on AB minimal medium plates in the presence of IPTG (0.5 mM) for induction is shown. Lanes: T, total cell lysate; S, soluble fraction; M, membrane proteins. (D) Cellular levels of VirB5 in strain C58 (lane 1) (with AS) and in strain CB1005(pTrcB5) grown in the presence of IPTG and the following concentrations (micromolar) of AS for virulence gene induction: 0 (lane 2), 0.01 (lane 3), 0.1 (lane 4), 1 (lane 5), 10 (lane 6), and 200 (lane 7). Detection of VirB5 is indicated by arrows. Numbers on the right are molecular weights in thousands.
Isolation of the T pilus from agrobacteria induced on agar medium.
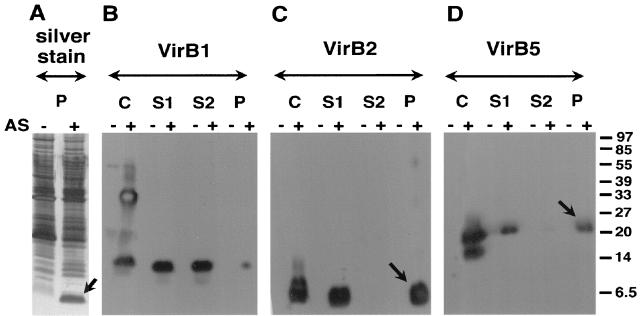
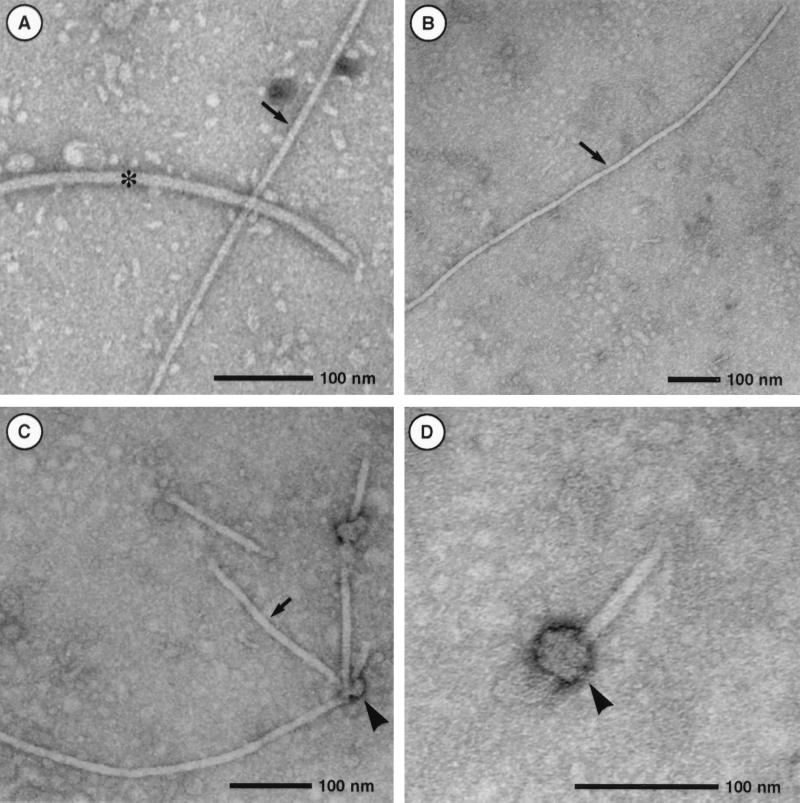
We next assessed the protein components of the T pilus in nopaline strain C58. Shearing forces applied to bacteria, such as repeated passing through a thin needle, remove multimeric structures like pili and flagella from the surface. To isolate the T pilus from A. tumefaciens, we followed a procedure described for the Hrp pilus from phytopathogenic Pseudomonas syringae (33). Cells were grown on agar plates in the presence of AS, washed from the surface, and repeatedly sheared through a 26-gauge needle. After an initial low-speed centrifugation to remove cells, the total supernatant was fractionated by high-speed centrifugation into soluble components and high-molecular-weight components that sedimented in the pellet. Samples from the pellet fraction, expected to contain flagella and pili, were separated by SDS-PAGE followed by silver staining of proteins. Figure 3A shows several proteins in a broad molecular mass range in the pellet fractions isolated from induced and noninduced cells. The major bands in the 32-kDa range probably correspond to flagellins, and they do not appear in fractions isolated from a flagellum-free strain (see below). However, large amounts of a protein of 5.5 kDa are present only in the fraction containing high-molecular-mass surface structures from virulence gene-induced cells, which probably corresponds to VirB2, the major component of the T pilus (26). Different fractions isolated from the cell surface were analyzed by SDS-PAGE and Western blotting with specific antisera (anti-VirB1, anti-VirB2, anti-VirB3, anti-VirB4, anti-VirB5, anti-VirB8, anti-VirB9, anti-VirB11, and anti-VirE2). Significant detection occurred only for candidate pilus proteins VirB1*, VirB2, and VirB5 (Fig. 3B to D). VirB1* was predominantly soluble, whereas VirB2 and VirB5 sedimented during high-speed centrifugation and thus are part of a high-molecular-mass structure. Based on an apparent molecular mass of 5.5 kDa, VirB2 corresponds to the abundant protein detected in silver-stained gels. In contrast to our previous hypothesis, VirB1* is not a pilus component. Fractions containing high-molecular-mass structures isolated by shearing and ultracentrifugation were analyzed by transmission electron microscopy, and Fig. 4A and B show dimensions of T pili and flagella similar to those reported by others (26). In addition, we frequently observed T pili with terminal sacculus-like structures (Fig. 4C and D), which may represent either terminal knobs at the pilus tip or pieces of the cell membrane removed by shearing.
FIG. 3.
Isolation of T pili from the surface of nopaline strain C58. High-molecular-weight structures were isolated by shearing from the surface of agrobacteria grown on AB minimal medium plates under virulence gene-inducing (+AS) and noninducing (−AS) conditions. The resulting samples C (cells), S1 (supernatant after shearing), S2 (supernatant after high-speed centrifugation), and P (pellet after high-speed centrifugation) were subjected to SDS-PAGE and analyzed by silver staining (A) or Western blotting with specific antisera for VirB1 (B), VirB2 (C), or VirB5 (D). VirB components detected in the pellet after ultracentrifugation are indicated by arrows. Numbers on the right are molecular weights in thousands.
FIG. 4.
Ultrastructural analysis of extracellular high-molecular-weight structures. Transmission electron microscopic analysis of T pili and flagella isolated from strain C58 is shown. (A) Comparison of flagellum (asterisk) and T pilus (arrow); (B) low-magnification image of long T-pilus fragment; (C) bundle of T pili with terminal sacculi (arrowhead); (D) higher magnification of sacculus-like structure.
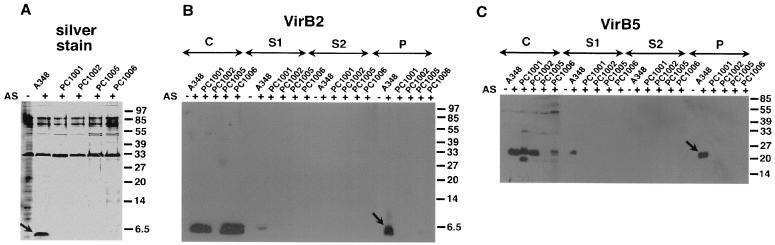
Octopine Ti plasmid-carrying strain A348 virulence genes were induced on AB agar plates with AS, followed by isolation of surface structures, high-speed centrifugation, and analysis of the different fractions by SDS-PAGE, Western blotting, and detection with VirB protein-specific antisera. Figure 5A shows that the sediment obtained after high-speed centrifugation of supernatants from virulence gene-induced cells contains a protein with a molecular mass of 5.5 kDa (detected by silver staining), which corresponds to VirB2 detected by Western blotting (Fig. 5B). In addition, VirB5-specific antibodies detected a protein of the predicted molecular weight in the pilus-containing fractions isolated from A348-carrying cells (Fig. 5C), but VirB5 was never detected in silver-stained gels. Thus, VirB2 and VirB5 cofractionate in T-pilus preparations from strains carrying two different Ti plasmids.
FIG. 5.
Isolation of the T pili from the surface of octopine strain A348 and virB deletion mutants PC1001 (virB1 deletion), PC1002 (virB2 deletion), PC1005 (virB5 deletion), and PC1006 (virB6 deletion). High-molecular-weight structures were isolated by shearing from the surface of agrobacteria grown on AB minimal medium plates under virulence gene-inducing (+AS) and noninducing (−AS) conditions. The resulting samples C (cells), S1 (supernatant after shearing), S2 (supernatant after high speed centrifugation), and P (pellet after high-speed centrifugation) were subjected to SDS-PAGE. Pellet fractions were analyzed by silver staining (A). Western blotting with specific VirB2 (B) or VirB5 (C) antisera was used to monitor T-pilus purification. VirB components detected in the pellet after ultracentrifugation are indicated by arrows. Numbers on the right are molecular weights in thousands.
Surface structures were isolated from derivatives of strain A348 carrying in-frame deletions in individual virB genes on the Ti plasmid: PC1001, carrying a deletion in the virB1 gene; PC1002, carrying a deletion in the virB2 gene; PC1005, carrying a deletion in the virB5 gene; and PC1006, carrying a deletion in the virB6 gene. Figure 5 shows that strains PC1002 and PC1005, which are defective for synthesis of VirB2 and VirB5, respectively, do not assemble T pili on their surfaces. Analysis of PC1001, which is defective in a putative transglycosylase involved in assembly of the VirB transmembrane structure, and of PC1006, which is defective in a transmembrane protein which may constitute the pore for T-complex transfer, gave similar results. Thus, assembly of functional T pili depends on pilus subunits as well as on other VirB components of the machinery for genetic transformation of plants. To further characterize the association of VirB2 and VirB5, T pili were further purified by different methods.
Gel filtration chromatography confirms that the T pilus constitutes a high-molecular-weight structure.
T pili isolated from the surface of virulence gene-induced agrobacteria sedimented during high-speed centrifugation, indicating a high-molecular-weight structure, which was further assessed by gel filtration chromatography. Surface structures were isolated from A. tumefaciens C58 (virulence gene induced and uninduced); pellets obtained after high-speed centrifugation were resuspended in buffer P and subjected to gel filtration on a Superdex 200 column followed by SDS-PAGE. Silver staining identified a 5.5-kDa protein eluting at 40 to 70 ml from the gel filtration column in samples from virulence gene-induced cells, whereas no prominent band in this molecular mass range was detected in samples from noninduced cells (Fig. 6A and B). Comparison with the elution volumes of reference proteins suggests that the major portion eluted with a structure with a molecular mass significantly larger than 440 kDa, the molecular mass of the ferritin reference protein, which is well in accord with a multimeric structure such as a pilus. The broad range of elution from the column likely indicates different degrees of fragmentation of the pilus conferred by shearing forces during its purification. Vir protein-specific antibodies were then used for compositional analyses (Figs. 6C and D), and VirB2 and VirB5 antigens eluted at similar positions from the column as the 5.5-kDa protein detected by silver staining. Thus, gel filtration chromatography adds independent proof that VirB2 and VirB5 associated with high-molecular-weight structures of similar size. Due to its abundance, the major pilus component VirB2 was detected by silver staining in samples isolated from virulence gene-induced cells, but the presence of multiple proteins in the 20- to 30-kDa molecular mass range, which may correspond to flagellar components, hindered detection of VirB5.
Sucrose gradient fractionation of T pili shows cofractionation of VirB2 and VirB5.
To isolate T pili without contaminating flagellar components, and to further substantiate the association of VirB2 and VirB5, high-molecular-weight structures were isolated from virulence gene-induced and uninduced cells of a flagellum-free derivative of strain C58 [NT1REB (pJK270)] (24) by shearing and ultracentrifugation as described above. The pellets were resuspended and subjected to centrifugation in a linear 30 to 70% sucrose gradient, and the fractions were collected and analyzed by SDS-PAGE. T-pilus-containing fractions were identified by silver staining of the major component VirB2, but VirB5 was never detected. However, Western blot analyses detected copurification of VirB2 and VirB5, suggesting that both proteins associate with the T pilus (not shown).
DISCUSSION
VirB2 was identified as major component of the T pilus (12, 26), but different lines of evidence had suggested that VirB1* and VirB5 may also be part of that structure (43, 47). To directly analyze pilus composition, extracellular high-molecular-weight structures were isolated from different A. tumefaciens strains by shearing and high-speed centrifugation, followed by further purification by gel filtration chromatography and sucrose gradient centrifugation. A complete set of antisera specific for all nopaline VirB proteins demonstrated that VirB5 copurified with T pili during different purification steps. This confirms earlier reports on VirB2 as the major subunit of the strain C58 T pilus. The fact that the T pilus of strain A348 has the same major subunit as the T pilus of strain C58 suggests that VirB2 and its homologs in related bacterial conjugation systems, e.g., TraM from the IncN plasmid pKM101, TrwL from the IncW plasmid pR388, and TrbC from the IncP plasmid pRP4, may constitute major components of their conjugative pili. This gives a clear direction for future studies directed towards analysis of the transfer mechanism of this large group of broad-host-range plasmids.
VirB5 was not detected in silver-stained gels analyzing purified T pili. However, compared to antisera used for detection of the major component VirB2 by Western blotting, anti-VirB5 sera are highly specific, demonstrating its copurification with T pili from different A. tumefaciens strains. This suggests its association with these surface-exposed structures as a minor constituent. Alternatively, VirB5 may constitute a different extracellular high-molecular-weight structure, but there is no evidence for the existence of additional cell appendages. Whereas copurification suggests association of VirB5 with the T pilus, our present data do not reveal its exact localization therein. VirB5 may be part of an assembly complex at the pilus base which is partly removed from the cells by shearing. The pilus-associated sacculi observed by electron microscopy could correspond to that structure. Alternatively, VirB5 may be a minor component which localizes at the pilus tip, mediating specific adhesion to recipient cells. However, our present data do not exclude an indirect interaction or association of VirB5 with the pilus base in the periplasm. Chromosomally encoded proteins are necessary for plant cell attachment (19), and future studies using isolated T pili will assess whether they contribute to specific binding. Studies on the F episome-determined pilus suggest that in addition to the major VirB2-homologous subunit TraA, a minor component may play a role in recipient-cell binding, but there is no direct evidence for such a protein (2, 13). Similarly, a role of VirB5 as a pilus component was proposed based on conjugation experiments suggesting exterior localization of its homolog TraC from the IncN plasmid pKM101 conjugation machinery (43, 44). TraC was indeed isolated as part of an extracellular high-molecular-weight complex from pKM101-carrying cells (36). Further studies, e.g., by immunoelectron microscopy, are needed to analyze the localization of VirB5 and TraC in conjugative pili to understand their role in DNA transfer.
VirB1*, the C-terminal processing product of VirB1, was hypothesized to be part of the T pilus because of its predominantly soluble nature and secretion (3), and electron microscopic studies further support its exterior localization (9). However, we showed here that VirB1* localizes to the supernatant after shearing of virulence gene-induced cells, like VirB2 and VirB5, but that it does not associate with the pellet containing high-molecular-weight structures after high-speed centrifugation. Thus, VirB1* is likely not a structural component of the T pilus. The VirB1-cross-reactive material detected by immunoelectron microscopy may correspond to secreted protein which remains loosely attached to the cell exterior. However, this does not exclude a role in genetic transformation of plants. Expression of VirB1* alone in a virB1 deletion strain partly compensates for its reduced tumorigenicity, suggesting that VirB1* may play another role in T-complex transfer, possibly as a soluble virulence factor mediating plant cell interactions (30a).
The present analysis does not rule out the presence of other, minor components in the pilus, which may have evaded detection due to their low abundance or low titers of specific antibodies. Also, the outer membrane anchor linking the pilus to the cell has not been identified, but VirB3 is a prime candidate for this function. VirB3 is an essential virulence protein (7), localizes predominantly to the outer membrane (22), and has homologs in related macromolecular transfer systems, including the IncF plasmid conjugation machinery. Outer membrane association of the heterodimeric VirB7-VirB9 complex depends on the N-terminal lipoprotein modification of VirB7 and is required for the stability of many VirB proteins (1, 4, 40). Cross-linking suggests an interaction of VirB9 with VirB1*, and its localization in the outer membrane indicates that it may be closely juxtaposed to the base of the pilus (3). Interactions between VirB proteins probably mediate assembly of the T pilus, and VirB2 and VirB5 associate with the membranes in virulence gene-induced wild-type cells. AS-independent expression of VirB2 from the trc promotor affects neither its stability nor its cellular localization. In contrast, AS-independent expression results in strongly reduced amounts of cell-bound VirB5, which may reflect its susceptibility to degradation by periplasmic proteases. However, when virulence gene expression is induced in such a strain by addition of AS, the amount of cell-bound VirB5 reaches the wild-type level. This indicates interaction of VirB5 with other virulence gene-induced proteins, which may confer protection from degradation in the periplasm and association with the T pilus.
Future studies will address the protein-protein interactions leading to stabilization of VirB5 and T-pilus assembly. To this end, VirB5 was purified and found to bind a virulence gene-induced protein in a gel overlay assay. This protein may stabilize VirB5 and/or mediate its assembly into a shear-sensitive complex (24a). Alternatively, cross-linking agents may be used to identify interacting partners by immunoprecipitation of cross-linked complexes followed by analysis with VirB protein-specific antisera. Additional clues towards achievement of this goal may result from cross-linking analysis of A. tumefaciens strains carrying in-frame deletion mutations in single virB genes, which do not assemble T pili (references 15 and 26 and this study). Cross-linking analysis of VirB proteins in such strains may reveal factors linking the pilus to the membranes, identify interacting partners of VirB5, or clarify whether VirB2, like TraC, its homolog from the IncP plasmid pRP4 conjugation system, requires processing after removal of the signal peptide for its function in T-complex transfer (12, 17).
ACKNOWLEDGMENTS
We thank Peter Christie and Stephen C. Winans for gifts of strains and plasmids and August Böck for support and discussions. Yasunori Machida kindly provided ChvE-specific antiserum.
This study was supported by grants from the Deutsche Forschungsgemeinschaft (DFG) to C.B. (BA 1416/2-1) and from the National Science Foundation (IBN-9507782) to P.C.Z.
REFERENCES
- 1.Anderson L B, Vogel Hertzel A, Das A. Agrobacterium tumefaciens VirB7 and VirB9 form a disulfide-linked protein complex. Proc Natl Acad Sci USA. 1996;93:8889–8894. doi: 10.1073/pnas.93.17.8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anthony K G, Sherbourne C, Sherburne R, Frost L S. The role of the pilus in recipient cell recognition during bacterial conjugation mediated by F-like plasmids. Mol Microbiol. 1994;13:939–953. doi: 10.1111/j.1365-2958.1994.tb00486.x. [DOI] [PubMed] [Google Scholar]
- 3.Baron C, Llosa M, Zhou S, Zambryski P C. VirB1, a component of the T-complex transfer machinery of Agrobacterium tumefaciens, is processed to a C-terminal secreted product, VirB1*. J Bacteriol. 1997;179:1203–1210. doi: 10.1128/jb.179.4.1203-1210.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baron C, Thorstenson Y R, Zambryski P C. Biochemical analysis of the complex between the lipoprotein VirB7 and VirB9 in the membranes of Agrobacterium tumefaciens. J Bacteriol. 1997;179:1211–1218. doi: 10.1128/jb.179.4.1211-1218.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baron C, Zambryski P C. Plant transformation: a pilus in Agrobacterium T-DNA transfer. Curr Biol. 1996;6:1567–1569. doi: 10.1016/s0960-9822(02)70773-2. [DOI] [PubMed] [Google Scholar]
- 6.Bayer M, Eferl R, Zellnig G, Terferle K, Dijkstra A, Koraimann G, Högenauer G. Gene 19 of plasmid R1 is required for both efficient conjugative DNA transfer and bacteriophage R17 infection. J Bacteriol. 1995;177:4279–4288. doi: 10.1128/jb.177.15.4279-4288.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger B R, Christie P J. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J Bacteriol. 1994;176:3646–3660. doi: 10.1128/jb.176.12.3646-3660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christie P J. Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J Bacteriol. 1997;179:3085–3094. doi: 10.1128/jb.179.10.3085-3094.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chumakov M I, Kurbanova I V. Localization of the protein VirB1 involved in contact formation during conjugation among Agrobacterium cells. FEMS Microbiol Lett. 1998;168:297–301. doi: 10.1111/j.1574-6968.1998.tb13287.x. [DOI] [PubMed] [Google Scholar]
- 10.Depicker A, DeWilde M, De Vos G, De Vos R, Van Montagu M, Schell J. Molecular cloning of overlapping segments of the nopaline Ti plasmid of pTiC58 as a means to restriction endonuclease mapping. Plasmid. 1980;31:193–211. doi: 10.1016/0147-619x(80)90109-2. [DOI] [PubMed] [Google Scholar]
- 11.Dürrenberger M B, Villiger W, Bächi T. Conjugational junctions: morphology of specific contacts in conjugating Escherichia coli bacteria. J Struct Biol. 1991;107:146–156. doi: 10.1016/1047-8477(91)90018-r. [DOI] [PubMed] [Google Scholar]
- 12.Eisenbrandt R, Kalkum M, Lai E M, Lurz R, Kado C I, Lanka E. Conjugative pili of IncP plasmids and the Ti plasmid T pilus are composed of cyclic subunits. J Biol Chem. 1999;274:22548–22555. doi: 10.1074/jbc.274.32.22548. [DOI] [PubMed] [Google Scholar]
- 13.Frost L S, Lee J, Scraba D, Paranchych W. Two monoclonal antibodies specific for different epitopes within the amino-terminal region of F pilin. J Bacteriol. 1986;168:192–198. doi: 10.1128/jb.168.1.192-198.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frost L S, Ippen-Ihler K, Skurray R A. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol Rev. 1994;58:162–210. doi: 10.1128/mr.58.2.162-210.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fullner K J, Lara J L, Nester E W. Pilus assembly by Agrobacterium T-DNA transfer genes. Science. 1996;273:1107–1109. doi: 10.1126/science.273.5278.1107. [DOI] [PubMed] [Google Scholar]
- 16.Fullner K J, Nester E W. Temperature affects the T-DNA transfer machinery of Agrobacterium tumefaciens. J Bacteriol. 1996;178:1498–1504. doi: 10.1128/jb.178.6.1498-1504.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haase J, Lanka E. A specific protease encoded by the conjugative DNA transfer systems of IncP and Ti plasmids is essential for pilus synthesis. J Bacteriol. 1997;179:5728–5735. doi: 10.1128/jb.179.18.5728-5735.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harlow E, Lane D, editors. Antibodies: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 19.Hooykaas P J J, Beiersbergen A G M. The virulence system of Agrobacterium tumefaciens. Annu Rev Phytopathol. 1994;32:157–179. [Google Scholar]
- 20.Hultgren S J, Abraham S, Caparon M, Falk P, St. Geme III J W, Normark S. Pilus and nonpilus bacterial adhesins: assembly and function in cell recognition. Cell. 1993;73:887–901. doi: 10.1016/0092-8674(93)90269-v. [DOI] [PubMed] [Google Scholar]
- 21.Jones A L, Lai E-M, Shirasu K, Kado C I. VirB2 is a processed pili-like protein encoded by the Agrobacterium tumefaciens Ti plasmid. J Bacteriol. 1996;178:5706–5711. doi: 10.1128/jb.178.19.5706-5711.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones A L, Shirasu K, Kado C I. The product of the virB4 gene of Agrobacterium tumefaciens promotes accumulation of VirB3 protein. J Bacteriol. 1994;176:5255–5261. doi: 10.1128/jb.176.17.5255-5261.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kado C I. Promiscuous DNA transfer system of Agrobacterium tumefaciens: role of the virB operon in sex pilus assembly and synthesis. Mol Microbiol. 1994;12:17–22. doi: 10.1111/j.1365-2958.1994.tb00990.x. [DOI] [PubMed] [Google Scholar]
- 24.Kao J C, Perry K L, Kado C I. Indoleacetic acid complementation and its relation to host range specifying genes on the Ti plasmid of Agrobacterium tumefaciens. Mol Gen Genet. 1982;188:425–432. doi: 10.1007/BF00330044. [DOI] [PubMed] [Google Scholar]
- 24a.Krall, L., and C. Baron. Unpublished data.
- 25.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Lai E-M, Kado C I. Processed VirB2 is the major subunit of the promiscuous pilus of Agrobacterium tumefaciens. J Bacteriol. 1998;180:2711–2717. doi: 10.1128/jb.180.10.2711-2717.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lanka E, Wilkins B M. DNA processing reactions in bacterial conjugation. Annu Rev Biochem. 1995;64:141–169. doi: 10.1146/annurev.bi.64.070195.001041. [DOI] [PubMed] [Google Scholar]
- 28.Lehnherr H, Hansen A-M, Ilyina T. Penetration of the bacterial cell wall: a family of lytic transglycosylases in bacteriophages and conjugative plasmids. Mol Microbiol. 1998;30:454–457. doi: 10.1046/j.1365-2958.1998.01069.x. [DOI] [PubMed] [Google Scholar]
- 29.Lessl M, Balzer D, Pansegrau W, Lanka E. Sequence similarities between the RP4 Tra2 and the Ti VirB region strongly support the conjugation model for T-DNA transfer. J Biol Chem. 1992;267:20471–20480. [PubMed] [Google Scholar]
- 30.Llosa M, Zambryski P C. On the origin and function of pili. Trends Microbiol. 1998;6:98–99. doi: 10.1016/s0966-842x(98)01222-0. [DOI] [PubMed] [Google Scholar]
- 30a.Llosa, M., J. Zupan, and P. C. Zambryski. Unpublished data.
- 31.Maniatis T A, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 32.Mushegian A R, Fullner K J, Koonin E V, Nester E W. A family of lysozyme-like virulence factors in bacterial pathogens. Proc Natl Acad Sci USA. 1996;93:7321–7326. doi: 10.1073/pnas.93.14.7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roine E, Wei W, Yuan J, Nurmiaho-Lassila E L, Kalkkinen N, Romantschuk M, He S Y. Hrp pilus: an hrp-dependent bacterial surface appendage produced by Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci USA. 1997;94:3459–3464. doi: 10.1073/pnas.94.7.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakellaris H, Balding D P, Scott J R. Assembly proteins of CS1 pili of enterotoxigenic Escherichia coli. Mol Microbiol. 1996;21:529–541. doi: 10.1111/j.1365-2958.1996.tb02562.x. [DOI] [PubMed] [Google Scholar]
- 35.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range of 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 36.Schmidt-Eisenlohr H, Domke N, Baron C. TraC of IncN plasmid pKM101 associates with membranes and extracellular high-molecular-weight structures in Escherichia coli. J Bacteriol. 1999;181:5563–5571. doi: 10.1128/jb.181.18.5563-5571.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sheng J, Citovsky V. Agrobacterium-plant cell DNA transport: have virulence proteins, will travel. Plant Cell. 1996;8:1699–1710. doi: 10.1105/tpc.8.10.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shirasu K, Kado C I. Membrane localization of the Ti plasmid VirB proteins involved in the biosynthesis of a pilin-like structure in Agrobacterium tumefaciens. FEMS Microbiol Lett. 1993;111:287–294. doi: 10.1111/j.1574-6968.1993.tb06400.x. [DOI] [PubMed] [Google Scholar]
- 39.Soto G E, Hultgren S J. Bacterial adhesins: common themes and variations in architecture and assembly. J Bacteriol. 1999;181:1059–1071. doi: 10.1128/jb.181.4.1059-1071.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spudich G M, Fernandez D, Zhou X-R, Christie P J. Intermolecular disulfide bonds stabilize VirB7 homodimers and VirB7/VirB9 heterodimers during biogenesis of the Agrobacterium tumefaciens T-complex transport apparatus. Proc Natl Acad Sci USA. 1996;93:7512–7515. doi: 10.1073/pnas.93.15.7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stachel S E, Timmermann B, Zambryski P C. Generation of single-stranded T-DNA molecules during the initial stages of T-DNA transfer from Agrobacterium tumefaciens to plant cells. Nature. 1986;322:706–712. [Google Scholar]
- 42.van Larebeke N, Engler G, Holsters M, van den Elsacker S, Zaenen I, Schilperoort R A, Schell J. Large plasmids in Agrobacterium tumefaciens essential for crown gall-inducing ability. Nature. 1974;252:169–170. doi: 10.1038/252169a0. [DOI] [PubMed] [Google Scholar]
- 43.Winans S C, Burns D L, Christie P J. Adaptation of a conjugal system for the export of pathogenic macromolecules. Trends Microbiol. 1996;4:64–68. doi: 10.1016/0966-842X(96)81513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winans S C, Walker G C. Conjugal transfer system of the N incompatibility plasmid pKM101. J Bacteriol. 1985;161:402–410. doi: 10.1128/jb.161.1.402-410.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC18 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 46.Zambryski P C. Chronicles from the Agrobacterium-plant cell DNA transfer story. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:465–490. [Google Scholar]
- 47.Zupan J R, Ward D, Zambryski P C. Assembly of the VirB transport complex for DNA transfer from Agrobacterium tumefaciens to plant cells. Curr Opin Microbiol. 1998;1:649–655. doi: 10.1016/s1369-5274(98)80110-0. [DOI] [PubMed] [Google Scholar]