Abstract
YAP1 and TAZ are transcriptional co-activator proteins that play fundamental roles in many biological processes, from cell proliferation and cell lineage fate determination to tumorigenesis. We previously demonstrated that Limb Expression 1 (LIX1) regulates YAP1 and TAZ activity and controls digestive mesenchymal progenitor proliferation. However, LIX1 mode of action remains elusive. Here, we found that endogenous LIX1 is localized in mitochondria and is anchored to the outer mitochondrial membrane through S-palmitoylation of cysteine 84, a residue conserved in all LIX1 orthologs. LIX1 downregulation altered the mitochondrial ultrastructure, resulting in a significantly decreased respiration and attenuated production of mitochondrial reactive oxygen species (mtROS). Mechanistically, LIX1 knock-down impaired the stability of the mitochondrial proteins PHB2 and OPA1 that are found in complexes with mitochondrial-specific phospholipids and are required for cristae organization. Supplementation with unsaturated fatty acids counteracted the effects of LIX1 knock-down on mitochondrial morphology and ultrastructure and restored YAP1/TAZ signaling. Collectively, our data demonstrate that LIX1 is a key regulator of cristae organization, modulating mtROS level and subsequently regulating the signaling cascades that control fate commitment of digestive mesenchyme-derived cells.
Keywords: Smooth muscle, Mitochondria, mtROS, Cristae, Cell fate, YAP1/TAZ, Linoleic acid, Sarcoma
Graphical abstract
Highlights
-
•
LIX1 is tightly anchored to the outer membrane of mitochondria.
-
•
LIX1 mitochondrial localization is mediated by S-palmitoylation on cysteine 84.
-
•
LIX1 knock-down reduces the stability of the mitochondrial proteins PHB2 and OPA1 and impairs cristae organization.
-
•
Redox signaling modulations regulate YAP1/TAZ activity and control fate commitment of digestive mesenchyme-derived cells.
1. Introduction
Emerging evidence suggests the implication of mitochondrial remodeling in stem cell fate decisions [[1], [2], [3], [4], [5], [6]]. Cells modulate the number and activity of mitochondria to meet the energetic and metabolic demands through biogenesis, turnover, and fusion and fission. Besides energy generation through oxidative phosphorylation (OXPHOS), mitochondria play important roles in amino acid, fatty acid and steroid metabolism, as well as in cell signaling. In recent years, mitochondrial reactive oxygen species (mtROS) have emerged as important regulators of cell differentiation [2,4,7].
Among all muscle cell types, smooth muscle cells (SMCs) in vasculature, airways, gastrointestinal tract and urogenital tract display a unique and remarkable plasticity. Unlike many other mature cell types in the adult body, SMCs do not terminally differentiate, but can reversibly modulate their phenotype, switching between a differentiated functional quiescent state and a highly proliferative mesenchymal precursor state. SMC plasticity has been extensively studied in vascular SMCs, but not in gastrointestinal SMCs. Digestive SMCs derive from the embryonic mesoderm that gives rise to the mesenchyme, which in turn differentiates into multiple tissues: submucosa, KIT-positive interstitial cells of Cajal (ICCs), and SMCs [8,9]. The differentiation of mesenchymal progenitors into SMCs includes several steps. First, they enter a determination program through the induction of MYOCD, a master regulator of SMC-restricted gene expression. Determined SMCs, defined by the early expression of the alpha and gamma isoforms of smooth muscle actin (αSMA and γSMA, respectively), will differentiate as soon as they express proteins involved in smooth muscle contractility, such as CALPONIN [10,11]. In the adult musculature, ICCs and SMCs display trans-plasticity. Indeed, inhibition of KIT activity leads to SMC differentiation, while restriction of KIT activity within these precursors leads to ICC differentiation [[12], [13], [14]]. Such plasticity is often associated with higher cancer risk [15]. For instance, gastrointestinal stromal tumors (GISTs), the most common mesenchymal neoplasm of the gastrointestinal tract [16], result from the deregulated proliferation of KIT-positive cells (ICCs or ICC/SMC mesenchymal progenitors) [14,[16], [17], [18]].
Limb Expression 1 (LIX1), a unique marker of digestive mesenchyme immaturity, controls cell fate decisions in digestive mesenchyme lineages [19]. Human LIX1 is a highly conserved gene that encodes a 282-amino acid protein with not well understood functions. LIX1 was discovered in chick embryos during a screen to identify markers of limb development [20]. An in silico prediction analysis indicated that LIX1 has a double-stranded RNA binding domain, suggesting that it could be involved in RNA processing [21]. Moreover, the arthropod homolog of LIX1, lowfat, interacts with the atypical cadherins fat and dachsous, two upstream components of the Hippo pathway that is implicated in maintaining tissue homeostasis through the regulation of the cell proliferation and differentiation balance [21,22]. In the digestive musculature, LIX1 is expressed only during fetal life. In this tissue, it stimulates the expression and activity of the Hippo effectors YAP1 and TAZ, and both LIX1 and YAP1/TAZ are key regulators of stomach mesenchymal progenitor development [19]. Moreover, LIX1 expression is high in GISTs and is associated with poor prognosis. In GISTs, LIX1 controls KIT protein level, ICC lineage specification through YAP1/TAZ, and cell proliferation [23]. Thus, although accumulating evidences indicate that LIX1 is a key regulator of muscle progenitor proliferation in vertebrates [19,24], its precise role and activity remain elusive.
In the present study, we investigated LIX1 cellular functions. We found that LIX1 is a S-palmitoylated protein with mitochondrial localization. Moreover, LIX1 regulates the commitment of digestive mesenchymal progenitors and their plasticity by controlling mitochondrial cristae shape and redox signaling.
2. Results
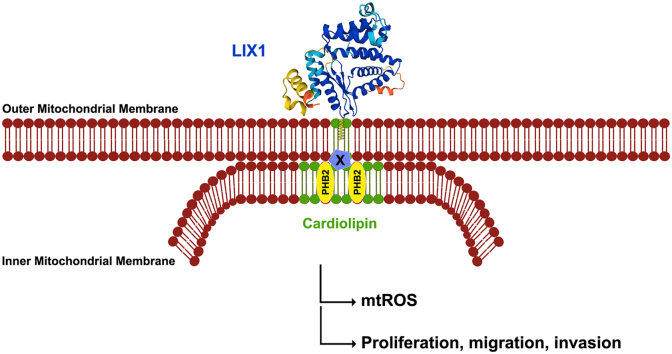
2.1. LIX1 is tightly anchored to the outer membrane of mitochondria
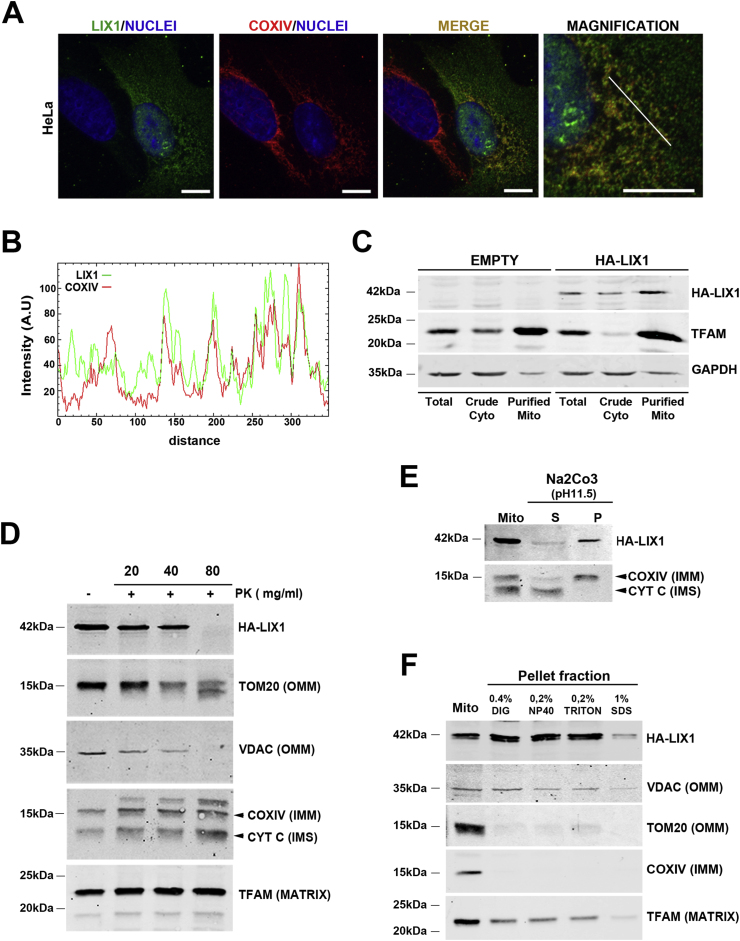
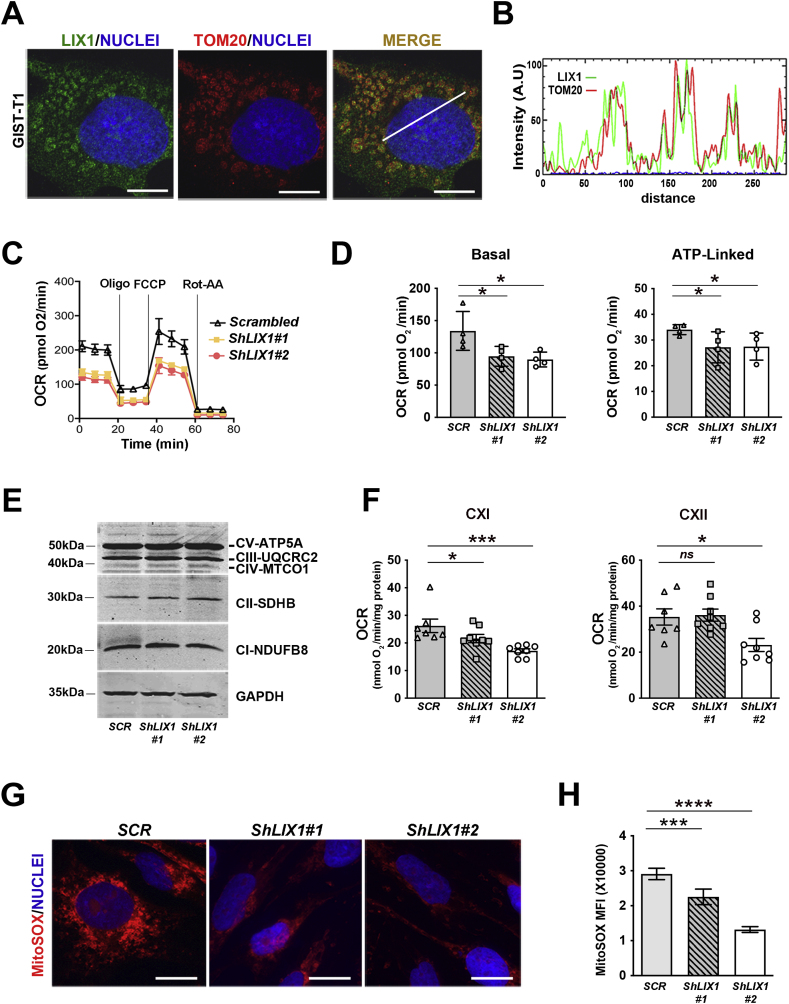
We first analyzed LIX1 localization by confocal microscopy in HeLa cells that express HA-LIX1. We found that a significant fraction of HA-LIX1 co-localized with the mitochondrial marker COXIV (Fig. 1A and B). We then performed fractionation experiments using control non-transfected cells (empty) and HA-LIX1-expressing cells to analyze LIX1 expression in the total protein extract, cytoplasmic (Crude Cyto) and mitochondrial (Purified Mito) fractions. Western blot analysis with antibodies against GAPDH (cytosol marker), TFAM (mitochondrial marker) and LIX1 showed that LIX1 is a cytoplasmic protein enriched in the mitochondrial fraction (Fig. 1C). We next determined LIX1 sub-mitochondrial localization using a proteinase K (PK) accessibility test and alkali treatments. Incubation of isolated mitochondria from HA-LIX1-expressing HeLa cells with increasing PK concentrations did not affect the levels of proteins localized in the intermembrane space (IMS) such as cytochtome C (CYT C), in the inner mitochondrial membrane (IMM), such as COXIV, and in the matrix, such as TFAM. Conversely, it strongly reduced VDAC and TOM20, two outer mitochondrial membrane (OMM) proteins, and also LIX1 levels (Fig. 1D). This suggested that LIX1 is localized at the OMM surface. To determine the strength of LIX1 interaction with the OMM, we incubated mitochondrial fractions from HA-LIX1-expressing cells with sodium carbonate (pH11.5). This treatment at high pH disrupts protein-protein and weak protein-lipid interactions, and leads to membrane integrity loss. As expected, we retrieved CYT C, a protein found in the IMS in the soluble (S) faction, whereas transmembrane proteins, such as COXIV, remained in the pellet (P)/membrane fraction as well as LIX1 (Fig. 1E). Furthermore, incubation of mitochondria purified from HA-LIX1-expressing cells with nonionic detergents (digitonin, NP-40, Triton X-100) did not affect LIX1 level, even at concentrations that extract partially or totally other mitochondrial proteins, such as TFAM (matrix), TOM20 (OMM) and COXIV that is found in the IMM (Fig. 1F). Indeed, in the Pellet fraction, we detected mitochondrial LIX1 and VDAC, a protein localized at mitochondrial detergent-resistant micro-domains. Both VDAC and LIX1 were only partially dissociated, even upon incubation with 1% SDS, a concentration that fully dissociated TOM20 and COXIV respectively from the OMM and the IMM (Fig. 1F). This indicates that LIX1 is strongly attached to the OMM, presumably in detergent-resistant micro-domains. These raft-like micro-domains, enriched in the phospholipid cardiolipin, are located at the contact sites between the IMM and OMM [25] and play important roles in mitochondrial metabolism regulation.
Fig. 1.
LIX1 localizes to mitochondria and is tightly anchored to the outer membrane.
(A) Confocal microscopy analysis of HA-LIX1 localization in HeLa cells. Comparison with the mitochondrial marker COXIV. Scale bars, 10 μm. (B) RGB profile of LIX1 and COXIV expression co-localization. (C) Western blot analysis of total protein extracts (Total), crude cytoplasmic (Crude Cyto) and purified mitochondria (Purified Mito) from non-transfected (Empty) and HA-LIX1-expressing HeLa cells. Membranes were probed with antibodies against the HA tag, GAPDH (cytoplasmic) and TFAM (mitochondrial marker). (D) Sub-mitochondrial localization of HA-LIX1 in isolated mitochondria from HA-LIX1-expressing HeLa cells after (+) or not (−) proteinase K (PK) incubation (accessibility test). Membranes were probed with antibodies against the HA tag, TOM20 and VDAC (Outer Mitochondrial Membrane, OMM, markers), COXIV (Inner Mitochondrial Membrane, IMM, marker), Cytochrome C (CYT C; Inter-Membrane Space, IMS, marker), and TFAM (matrix protein). (E) Western blot analysis of total isolated mitochondria (Mito) from HA-LIX1-expressing cells after incubation with Na2CO3 pH 11.5 and ultracentrifugation to separate the supernatant (S) and pellet (P) fractions. Membranes were probed with antibodies against COXIV (IMM marker) and cytochrome C (CYT C) (IMS soluble marker). (F) Western blot analysis of total isolated mitochondria (Mito) from HA-LIX1-expressing HeLa cells after solubilization (+) or not (−) with the indicated detergents and ultracentrifugation. Membranes were probed with antibodies against the HA tag, TFAM (matrix protein), VDAC and TOM20 (OMM markers), and COXIV (IMM marker).
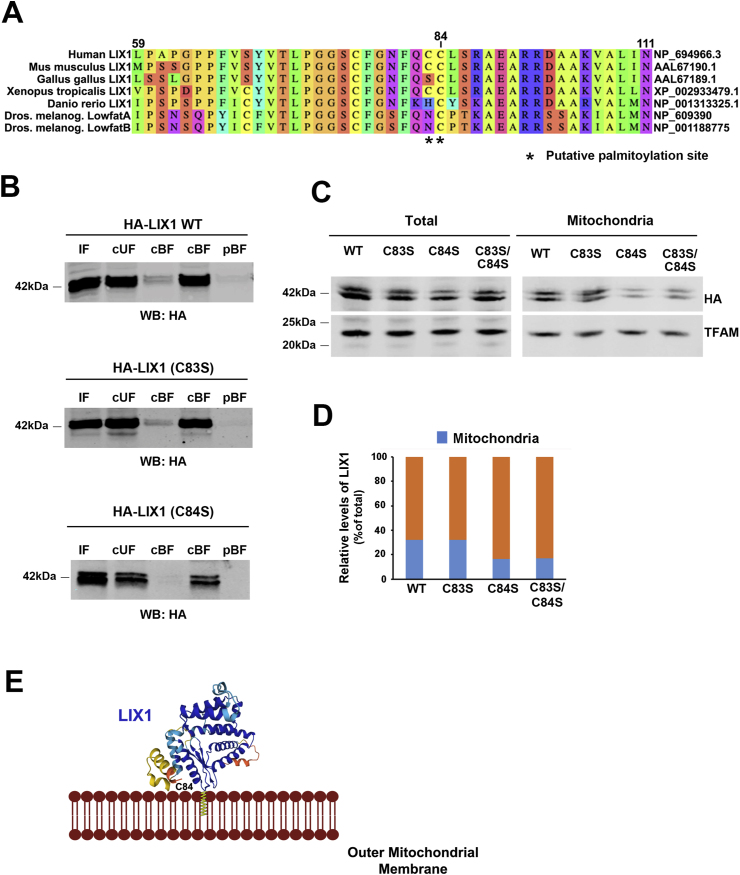
2.2. LIX1 mitochondrial localization is mediated by S-palmitoylation on cysteine 84
As LIX1 does not have any transmembrane domain, we next wanted to determine how LIX1 association with the OMM is regulated. Accumulating evidences indicate that lipid modifications can mediate protein translocation to the OMM. For instance, S-palmitoylation is required for BAX targeting to the OMM to initiate apoptosis [26]. The GPS-Lipid algorithm (http://lipid.biocuckoo.org), which allows predicting four lipid modifications (S-palmitoylation, N-myristoylation, S-farnesylation and S-geranylgeranylation) [27], identified two potential S-palmitoylation sites (cysteine 83 and 84) in the human LIX1 protein (Fig. 2A). S-palmitoylation is a dynamically regulated post-translational modification in which palmitic acid (C16.0) is reversibly attached to a cysteine residue via a thioester bond [28]. Upon S-palmitoylation, cytosolic proteins acquire a hydrophobic anchor that facilitates their docking to membranes, presumably into raft-like micro-domains [29]. To determine whether LIX1 is S-palmitoylated, we transfected HeLa cells with wild-type HA-LIX1 (LIX1 WT) or LIX1 mutants in which cysteine 83 and 84 were replaced by serine residues (LIX1 C83S, LIX1 C84S). We then tested S-palmitoylation in total protein extracts using the acyl-RAC assay [30,31]. We detected a specific signal corresponding to S-palmitoylation in the cleaved bound fraction (cBF) of LIX1 WT and LIX1 C83S samples, but not in the LIX1 C84S sample (Fig. 2B). Next, to determine whether cysteine 84 is required for LIX1 translocation to mitochondria, we analyzed by western blotting total extracts and purified mitochondrial fractions of HeLa cells that express WT HA-LIX1, LIX1 C83S, LIX1 C84S, or the double mutant LIX1 C83S/C84S. Mitochondrial targeting was strongly reduced (by ∼50%) for LIX1 C84S and LIX1 C83S/C84S compared with LIX1 WT and LIX1 C83S (Fig. 2C and D). These data indicate that LIX1 S-palmitoylation on cysteine 84 is implicated in its translocation to the OMM. The 3D structure of the LIX1 protein, established using the AlphaFold algorithm (https://alphafold.ebi.ac.uk/entry/Q8N485), revealed that cysteine 84, located between one α-helix and one β-strand, is accessible for lipid modification (Fig. 2E). Cysteine 84 is conserved in all vertebrate LIX1 orthologs and in the Drosophila homologs, but not cysteine 83 (Fig. 2A; Fig. S1).
Fig. 2.
LIX1 mitochondrial localization is controlled by S-palmitoylation at Cys84.
(A) Sequence alignment of human LIX1 and its orthologs. Asterisks indicate the position of the cysteine residues corresponding to putative S-palmitoylation sites. (B) LIX1 S-palmitoylation analysis with the Acyl RAC method. Membranes were incubated with anti-HA tag antibodies to detect LIX1. IF, input fraction; cUF, cleaved unbound fraction; cBF, cleaved bound fraction; pUF, preserved unbound fraction; pBF, preserved bound fraction. (C) Representative Western blot analysis of total extracts and isolated mitochondria from the same amount of total extract from HeLa cells that express WT HA-LIX1 (LIX1 WT), single LIX1 mutants (LIX1 C83S, LIX1 C84S) or the double mutant (LIX1 C83S/C84S) with antibodies against the HA tag and TFAM (matrix protein). (D) Quantification of the western blots showing the percentage of LIX1 (normalized to TFAM level) in mitochondria relative to LIX1 level in the total extract (normalized to TFAM level). (E) 3D structure of the LIX1 protein showing that the cysteine residue 84 is accessible for modification.
2.3. Endogenous LIX1 is enriched in mitochondria of gastric mesenchymal progenitors and GIST cells
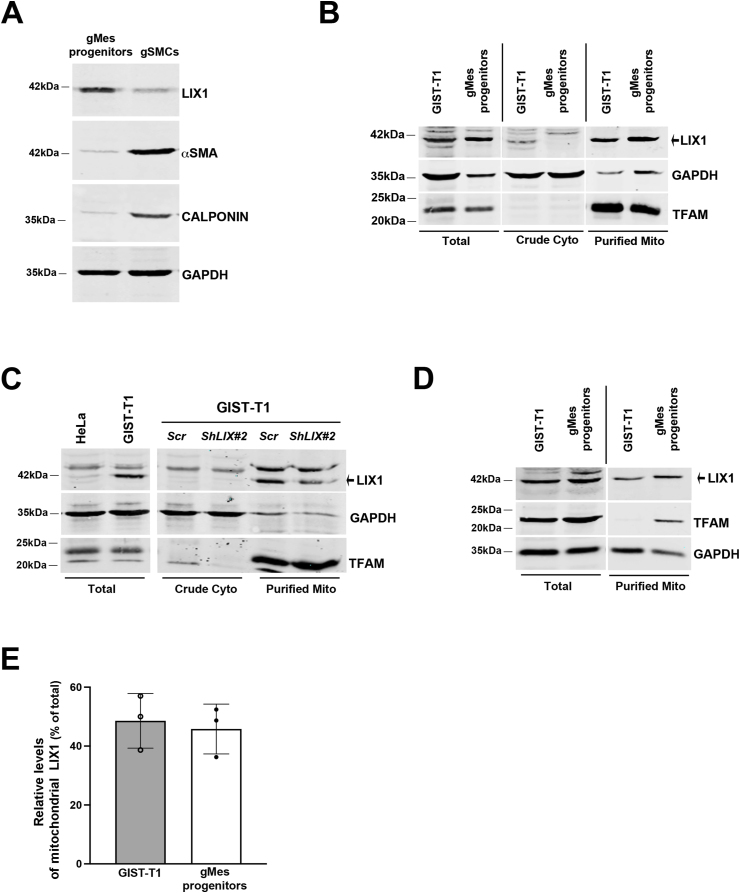
We next examined whether endogenous LIX1 co-fractionates with mitochondria. During development, LIX1 expression is detected in digestive mesenchymal progenitors [19]. Accordingly, we found that LIX1 was highly expressed in human gastric mesenchymal (gMes) progenitors, whereas it was barely detectable in differentiated gastric SMCs that are characterized by the expression of αSMA and CALPONIN (Fig. 3A). LIX1 was also expressed in the GIST-T1 cell line established from a metastatic human GIST (Fig. 3B) [23]. We then performed fractionation experiments using gMes progenitors and GIST-T1 cells. LIX1 was enriched in the mitochondrial fraction in both cell types (Fig. 3B). The signal detected for LIX1 in mitochondria was specific because it was strongly reduced in GIST-T1 cells that stably express ShRNAs against LIX1 (GIST-T1-ShLIX1#2) (Fig. 3C). In both cell types, endogenous mitochondrial LIX1 represented approximately 50% of total LIX1 (Fig. 3D and E). Our data demonstrate that endogenous LIX1 is enriched in mitochondria in gastric mesenchymal progenitors and in GIST cells derived from such progenitors.
Fig. 3.
Endogenous LIX1 is enriched in mitochondria of GIST-T1 cells.
(A) Representative Western blot of total extracts from gastric mesenchymal progenitor cells (gMes progenitors) and fully differentiated gastric SMCs (gSMCs). Membranes were probed with antibodies against LIX1, αSMA (marker of determined SMCs), and CALPONIN (marker of differentiated SMCs). Equal loading was verified by GAPDH expression. (B) Western blot analysis of total extracts, crude cytoplasmic (Crude Cyto) and purified mitochondria (Purified Mito) fractions (equal amounts) from GIST-T1 and gMes progenitors. Membranes were probed with antibodies against LIX1, GAPDH (cytoplasmic) and TFAM (mitochondrial marker). (C) Western blot analysis of total extracts from GIST-T1 and HeLa (negative control) cells, and of cytoplasmic (Crude Cyto) and mitochondrial (Purified Mito) fractions from GIST-T1 cells that stably express shRNAs against LIX1 (ShLIX1#2) or scramble shRNAs (Scr). Membranes were probed with antibodies against LIX1, GAPDH (cytoplasmic) and TFAM (mitochondrial marker). (D) Western blot analysis of total extracts and purified mitochondria (Purified Mito) fractions GIST-T1 cells and gMes progenitors. Membranes were probed with antibodies against LIX1, GAPDH (cytoplasmic), and TFAM (mitochondrial marker). (E) Quantification of the amount of endogenous LIX1 (normalized to TFAM level) in mitochondria relative to the total extract. Values are the mean ± SD of three independent experiments is shown.
2.4. LIX1 promotes mitochondrial respiration in GIST cells
Confocal analysis of GIST-T1 cells using the mitochondrial markers TOM20 and COXIV confirmed the mitochondrial localization of a significant fraction of endogenous LIX1 (Fig. 4A and B; Fig. S2). The mitochondrial localization of LIX1 prompted us to investigate its role in respiration. To this aim, we used GIST cell lines that stably express GIST-T1-Scrambled (negative control shRNA) and shRNAs against LIX1 (GIST-T1-ShLIX1#1 and GIST-T1-ShLIX1#2) [23]. We assessed the impact of LIX1 downregulation on the oxygen consumption rate (OCR). Extracellular flux (Seahorse) analysis showed that basal OCR (baseline minus non-mitochondrial respiration) and ATP-linked OCR (difference between OCR at baseline and respiration following oligomycin addition) were significantly lower in GIST-T1-ShLIX1 cells than in control GIST-T1-Scrambled cells (Fig. 4C and D). This indicated lower reliance on OXPHOS for energy production in GIST cells in which LIX1 was downregulated. The decreased OCR was not likely due to changes in electron transport chain proteins because the expression levels of several subunits were comparable in silenced and control cells (Fig. 4E). On the other hand, OCR quantification in permeabilized GIST-T1 cells demonstrated a significantly lower complex I (CXI)-driven oxygen consumption in both GIST-T1-ShLIX1 cell lines (Fig. 4F). CXII-driven oxygen consumption was moderately decreased only in GIST-T1-ShLIX1#2 cells, possibly due to the higher efficacy of this shRNA [23]. Mitochondria are bioenergetic, biosynthetic and signaling organelles [32]. A consequence of mitochondrial oxidative metabolism is the generation of copious amounts of reactive oxygen species (ROS) by the electron transport chain. High mtROS levels activate signaling pathways that promote cancer cell proliferation [[33], [34], [35], [36]]. Complex I is responsible for the majority of mtROS produced by the respiratory chain [37,38]. We found that mtROS production, measured by the level of MitoSOX immunofluorescence, was significantly decreased in GIST-T1-ShLIX1 cells compared with controls (Fig. 4G and H). These data indicate that LIX1 downregulation attenuates CXI-driven respiration, leading to decreased mtROS production.
Fig. 4.
LIX1regulatesmitochondrialrespiration and ROS production in GIST-T1 cells.
(A) Confocal microscopy analysis of endogenous LIX1 localization in GIST-T1 cells. Comparison with the mitochondrial marker TOM20. (B) LIX1/TOM20 RGB profile analyzed at the level of the white line. (C) Oxygen consumption rate (OCR) in GIST-T1-Scrambled, GIST-T1-ShLIX1#1 and GIST-T1-ShLIX1#2 cells incubated with oligomycin (Oligo), FCCP and rotenone/antimycin A (Rot-AA) using a Seahorse XF96 Extracellular Flux analyzer. Data are the mean ± SEM of 4 independent experiments. Cells were left for 4–6 h in the plates before starting the injection of drugs. (D) Quantification of the basal and ATP-linked OCR values from the Seahorse analysis. Non-mitochondrial OCR was determined after the addition of rotenone/antimycin A (Rot-AA) and subtracted from all the other values before calculating the respiratory parameters. Values are the mean ± SD of n=4 experiments using GIST-T1-Scrambled, GIST-T1-ShLIX1#1 and -ShLIX1#2 cells. *P < 0.05 (two-tailed Mann–Whitney tests). (E) Representative Western blot of total protein extracts from GIST-T1-Scrambled, GIST-T1-ShLIX1#1 and GIST-T1-ShLIX1#2 cells. Membranes were probed with antibodies against the indicated OXPHOS proteins. (F) ETC-CXI- and ETC-CXII-driven respiration measured using Oxygraph-2k high-resolution respirometry. Values are the mean ± SEM of GIST-T1-Scrambled (n=7), GIST-T1-ShLIX1#1 (n=8) and GIST-T1-ShLIX1#2 (n=8) cells. *P < 0.05, ***P < 0.001 (one-tailed Mann–Whitney test). (G) MitoSOX staining of GIST-T1-Scrambled, GIST-T1-ShLIX1#1 and -ShLIX1#2 cells. Nuclei were visualized with Hoechst. Scale bars, 20 μm. (H) Quantification of the MitoSOX signal. Values are the mean ± SEM of GIST-T1-Scrambled (n=34), GIST-T1-ShLIX1#1 (n=39), and GIST-T1-ShLIX1#2 (n=35) cells from three independent experiments. ***P < 0.001; ****P < 0.0001 (two-tailed Mann–Whitney test).
2.5. Mitochondrial remodeling regulates YAP1/TAZ and drives GIST cell identity
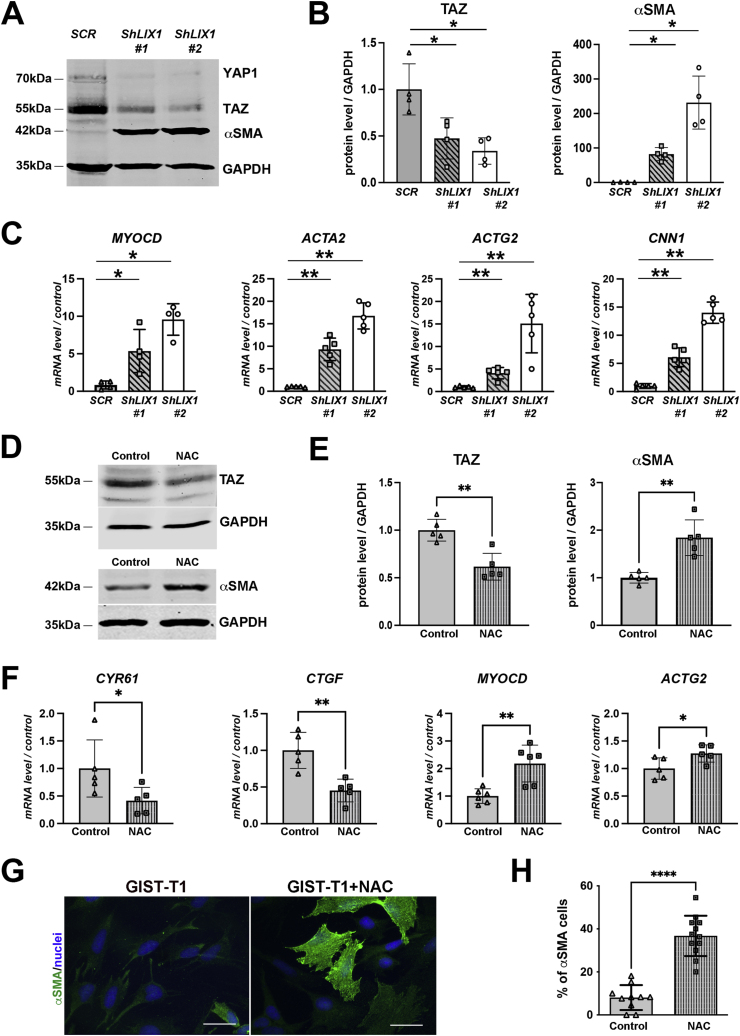
LIX1 promotes GIST cell proliferation in vitro and in vivo and controls cell invasion in vitro [23]. In more than 80% of cases, GISTs are driven by oncogenic KIT mutations leading to constitutive or ligand-independent activation of this tyrosine kinase receptor. The resulting activation of intracellular signaling cascades promotes proliferation, survival, and tumor growth [39]. Upon LIX1 inactivation in GIST cells, YAP1 and TAZ expression levels were reduced and their activity (monitored through the level of its transcriptionally targets CYR61 and CTGF) was decreased (Fig. 5A and B) [23]. In addition, reducing YAP1 or TAZ expression led to a marked decrease of KIT expression [23]. Conversely, LIX1 inactivation induced the upregulation of MYOCD, a master regulator of SMC-restricted gene expression and of SMC-restricted contractile genes, such as ACTA2 and ACTG2 (that encode αSMA and γSMA, respectively) and CNN1 (that encodes CALPONIN) in GIST-T1-ShLIX1 cells (Fig. 5C). As LIX1 silencing induced a decrease in mtROS production (Fig. 4G and H) and reprograms KIT-positive GIST cells to the SMC lineage [23] (Fig. 5A–C), we hypothesized that changes in GIST cell identity could be mediated through mtROS modulation. Therefore, we evaluated the functional importance of mtROS modulation on YAP1/TAZ expression/activity and SMC marker induction. Reducing mtROS levels in GIST-T1 cells using N-acetyl cysteine (NAC), a ROS scavenger, or by inducing the targeted expression of CATALASE in mitochondria [40,41], to mimic the LIX1 knock-down phenotype, led to a marked decrease in TAZ level and activity, as shown by CYR61 and CTGF downregulation, and to a strong increase in SMC markers (Fig. 5D–H; Fig. S3). These findings highlight the pivotal role of redox signaling in regulating digestive mesenchymal cell fate and GIST tumorigenesis.
Fig. 5.
Mitochondrial remodeling controls GIST cell identity.
(A) Representative Western blot showing YAP1/TAZ and αSMA levels in GIST-T1-Scrambled, GIST-T1-ShLIX1#1 and -ShLIX1#2 cells. Equal loading was verified by GAPDH expression. (B) Graph represents the quantification of TAZ and αSMA levels in four independent experiments. Data were normalized to GAPDH and converted to fold change. Values are the mean ± SEM of n = 4 samples. *P < 0.05 (two-tailed Mann–Whitney test). (C) RT-qPCR analysis of MYOCD, ACTA2, ACTG2 and CNN1 relative mRNA expression in GIST-T1-Scrambled cells vs GIST-T1-ShLIX1#1 and -ShLIX1#2 cells. Data were normalized to the mean HBMS and YWHAZ expression, and converted to fold change. Values are the mean ± SEM of n = 4 samples for MYOCD expression and n = 5 samples for ACTA2, ACTG2 and CNN1 expression. *P < 0.05 and **P < 0.01 (two-tailed Mann–Whitney test). (D) Representative Western blot showing TAZ and αSMA levels in GIST-T1 cells incubated or not (control) with 3 mM N-acetyl cysteine (NAC) for 18 h. Equal loading was verified by GAPDH expression. (E) Quantification of TAZ and αSMA levels normalized to GAPDH and converted to fold changes. Values are the mean ± SEM of n = 5 samples. **P < 0.01 (two-tailed Mann–Whitney test). (F) RT-qPCR analysis of CYR61, CTGF, MYOCD and ACTG2 relative mRNA expression in GIST-T1- cells vs NAC-treated GIST-T1 cells. Data were normalized to the mean HBMS and YWHAZ expression, and converted to fold changes. Values are the mean ± SEM of n = 5 samples. *P < 0.05 and **P < 0.01 (two-tailed Mann–Whitney test). (G) Immunofluorescence analysis of GIST-T1 and GIST-T1 + NAC cells with anti- αSMA antibodies. Nuclei were visualized with Hoechst. Scale bar, 50 μm. (H) Quantification of the percentage of αSMA-positive cells. Values are the mean ± SEM of n =136 GIST-T1-Scrambled and n=125 for GIST-T1+NAC. ****P < 0.0001 (two-tailed Mann–Whitney test).
2.6. LIX1 regulates the mitochondrial network morphology and the inner membrane architecture in GIST cells
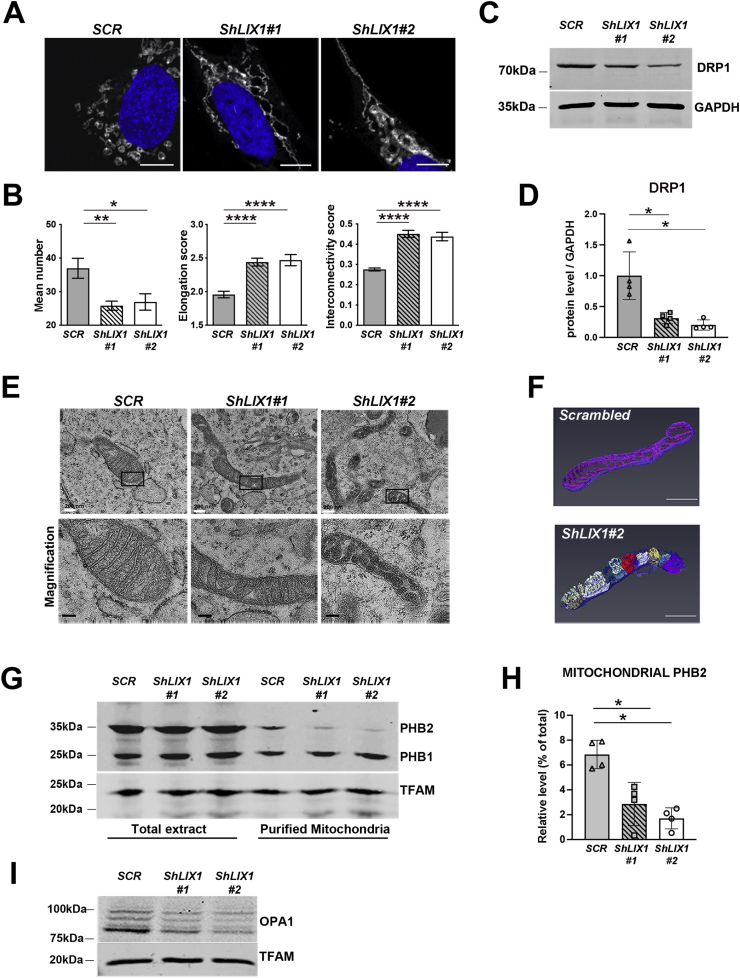
As mitochondrial bioenergetics is influenced by the mitochondrial morphology and ultrastructure [42], we next evaluated LIX1 implication in the regulation of mitochondrial morphology. MitoTracker Red staining, we observed fragmented mitochondria in GIST-T1-Scrambled cells, and an elongated network in GIST-T1-ShLIX1 cells (Fig. 6A). Quantification of these findings showed that the number of mitochondria was reduced in LIX1-silenced compared with control cells (Fig. 6B). Again, the mitochondrial mass did not appear to be affected by LIX1 silencing (Fig. 4E). The decreased number of mitochondria in GIST-T1-ShLIX1 cells was associated with a slight increase in the mean size, elongation and interconnectivity of mitochondria, compared with control cells (Fig. 6B). This phenotype was associated with a reduction in the level of dynamin-related protein 1 (DRP1) that promotes mitochondrial fission (Fig. 6C and D). Converselly, HA-LIX1 expression in HeLa cells induced mitochondrial fragmentation, a phenotype that led to a significant increase in mitochondrial number and a decrease in mitochondrial elongation and interconnectivity (Figs. S4A and B). As the electron transport chain is in the IMM, we next analyzed the IMM architecture by transmission electron microscopy (TEM). Compared with GIST-T1-Scrambled cells, the IMM and cristae were altered in GIST-T1-ShLIX1 cells (Fig. 6E; Fig. S5). These morphological changes were more important in GIST-T1-ShLIX1#2 cells in which LIX1 downregulation was more efficient. Specifically, lamellar cristae were almost completely lost and replaced by vesicle-like cristae without cristae junctions. We confirmed the remodeling of cristae in GIST-T1-ShLIX1#2 cells by three-dimensional reconstructions from serial TEM images (Fig. 6F; Supplementary Movie 1 and Movie 2). Such aberrant balloon-like cristae were previously described in prohibitin 2 (PHB2)-deficient mitochondria, where this phenotype was caused by the loss of the long OPA1 isoform, a dynamin-like GTPase required for mitochondrial fusion and cristae morphogenesis [[43], [44], [45]]. Analysis of PHB1 and PHB2 expression in total protein extracts and purified mitochondria from GIST-T1-Scrambled and GIST-T1-ShLIX1 cells showed that their expression in total extracts was not affected by LIX1 silencing, whereas only PHB2 was decreased in mitochondria of GIST-T1-ShLIX1 cells (Fig. 6G and H). This was associated with a reduction of all OPA1 isoforms in mitochondria (Fig. 6I). Our data demonstrate that LIX1 silencing in GIST-T1 cells impairs the stability of proteins required for proper cristae morphology and alters the mitochondrial membrane ultrastructure, resulting in decreased respiration and mtROS production.
Fig. 6.
LIX1 regulates mitochondrial network morphology and inner membrane architecture in GIST-T1 cells.
(A) MitoTracker staining of GIST-T1-Scrambled, GIST-T1-ShLIX1#1 and -ShLIX1#2 cells. Nuclei were visualized with Hoechst. Scale bars, 10 μm. (B) Quantification of the MitoTracker data with the Mito-Morphology Macro in Image J. Values are the mean ± SEM of GIST-T1-Scrambled (n=25), GIST-T1-ShLIX1#1 (n=28), and GIST-T1-ShLIX1#2 (n=28) cells. *P < 0.05; **P < 0.001; ****P < 0.0001 (two-tailed Mann–Whitney test). (C) Representative Western blot showing DRP1 levels in GIST-T1-Scrambled, GIST-T1-ShLIX1#1 and GIST-T1-ShLIX1#2GIST-T1 cells. Equal loading was verified by GAPDH expression. (D) Quantification of DRP1 level normalized to GAPDH and converted to fold change. Values are the mean ± SEM of n = 4 samples for each condition. *P < 0.05 (two-tailed Mann–Whitney test). (E) Ultrastructure of mitochondria in GIST-T1-Scrambled, GIST-T1-ShLIX1#1, and -ShLIX1#2 cells. Scale bars, 100 nm. Lower panels: magnifications of the areas in the black boxes in the upper panels. (F) Three-dimensional reconstructions from serial transmission electron microscopy sections of GIST-T1-Scrambled and GIST-T1-ShLIX1#2 cells. Scale bars, 500 nm. The vesicle-like cristae are shown in different colors. (G) Western blot analysis of total protein extracts and purified mitochondria from GIST-T1-Scrambled, GIST-T1-ShLIX1#1, and -ShLIX1#2 cells. Membranes were probed with anti-PHB and -TFAM (mitochondrial marker) antibodies. (H) Quantification of PHB2 level normalized to GAPDH and converted to fold change. Values are the mean ± SEM of n = 4 samples for each condition. *P < 0.05 (two-tailed Mann–Whitney test). (I) Representative Western blot of total protein extracts from GIST-T1-Scrambled, GIST-T1-ShLIX1#1, and -ShLIX1#2 cells. Membranes were probed with anti-OPA1 and -TFAM (mitochondrial marker) antibodies.
Supplementary video related to this article can be found at https://doi.org/10.1016/j.redox.2022.102431
The following are the supplementary data related to this article:
2.7. Supplementation with linoleic acid abrogates the effect of LIX1 downregulation on mitochondrial morphology and ultrastructure in GIST cells and restores YAP/TAZ levels
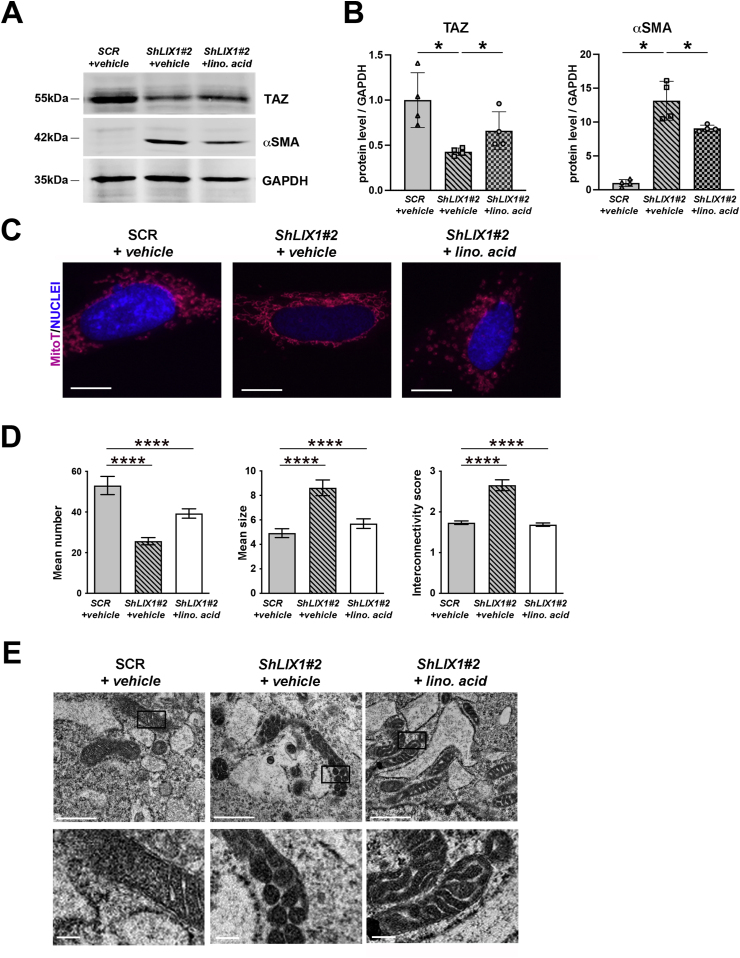
Gene interaction data have linked the function of PHB complexes to cardiolipin, a dimeric phospholipid with four fatty acyl chains found mainly in the IMM and composed predominantly of the tetra-linoleic form in muscle cells [46]. Cardiolipin levels are significantly reduced in mitochondria of cells lacking PHB2 [47], but they can be restored by supplementing the growth medium with linoleic acid [48]. As PHB2 levels were reduced in mitochondria of cells in which LIX1 was silenced (Fig. 6G and H), we supplemented GIST-T1-ShLIX1 and control cells with linoleic acid or vehicle (EtOH) for 12 h. Linoleic acid supplementation induced a marked increase in TAZ level and activity and a strong decrease in SMC markers (Fig. 7A, B; Fig. S6). Analysis of the mitochondrial morphology and cristae organization showed that untreated (vehicle) GIST-T1-ShLIX1 cells displayed an elongated mitochondrial network, whereas linoleic acid-supplemented GIST-T1-ShLIX1 cells harbored more fragmented mitochondria, as observed in untreated GIST-T1-Scrambled cells (Fig. 7C and D). Supplementation with linoleic acid slightly increased the number of mitochondria in GIST-T1-ShLIX1 cells, and decreased the mean elongation and interconnectivity scores (Fig. 7D). Furthermore, analysis of the IMM architecture by TEM showed that untreated GIST-T1-ShLIX1 cells harbored vesicle-like cristae without junctions, whereas linoleic acid-supplemented GIST-T1-ShLIX1 cells displayed lamellar cristae, similar to control cells (Fig. 7E). Thus, linoleic acid supplementation significantly attenuated the mitochondrial defects caused by LIX1 silencing and restored YAP1/TAZ expression. Altogether, our data highlight the pivotal role of mitochondrial metabolism and redox signaling modulations in LIX1-driven mesenchymal cell fate commitment in the digestive musculature and GIST tumorigenesis.
Fig. 7.
The effect of LIX1 silencing on mitochondrial morphology in GIST-T1 cells is reversed by linoleic acid.
(A) Representative Western blot showing TAZ and αSMA levels in GIST-T1-Scrambled (+ETOH vehicle), GIST-T1-ShLIX1#2 (+EtOH vehicle) and -ShLIX1#2 (+50 μM of linoleic acid) cells. Equal loading was verified by GAPDH expression. (B) Quantification of the Western blot bands to determine TAZ and αSMA expression levels relative to GAPDH in the different experimental conditions. Normalized expression levels were converted into fold change. Values are the mean ± SEM of n = 3 samples for each condition. *P < 0.05 (two-tailed Mann–Whitney test). (C) MitoTracker staining of GIST-T1-Scrambled (+EtOH vehicle), GIST-T1-ShLIX1#2 (+EtOH vehicle), and -ShLIX1#2 cells (+50 μM of linoleic acid). Nuclei were visualized with Hoechst. Scale bars, 10 μm. (D) Quantification of the MitoTracker data with the Mito-Morphology Macro in Image J. Values are the mean ± SEM of GIST-T1-Scrambled (+DMSO vehicle) (n=25), GIST-T1-ShLIX1#2 (+DMSO vehicle) (n=28), and GIST-T1-ShLIX1#2 cells (+50 μM of linoleic acid) (n=28); ****P < 0.0001 (two-tailed Mann–Whitney test). (E) Ultrastructure of mitochondria in GIST-T1-Scrambled (+EtOH vehicle), GIST-T1-ShLIX1#2 (+EtOH vehicle), and -ShLIX1#2 (+50 μM of linoleic acid) cells. Lower panels: magnification of the areas in the black boxes in the upper panels.
3. Discussion
LIX1 regulates YAP1/TAZ signaling levels and plasticity in the digestive musculature between the ICC and SMC lineages [19,23]. The human LIX1 gene encodes a 282-amino acid protein the function of which remains to be determined.
Here, we identified a cellular function of LIX1. We found that endogenous LIX1 is a S-palmytoylated protein that localizes in the mitochondria of gastric mesenchymal progenitors and GIST cells, which are derived from such progenitors. LIX1 is tightly anchored to the OMM where it may be localized in detergent-resistant lipid domains. These raft-like micro-domains are enriched in the phospholipid cardiolipin that is present mainly in the IMM and at contact sites between the IMM and OMM [[49], [50], [51], [52], [53], [54]]. Interestingly, cysteine 84, which is required for LIX1 S-palmitoylation, is conserved in all vertebrate LIX1 orthologs and in the Drosophila homologs, suggesting a conserved cellular regulatory mechanism to modulate LIX1 functions.
Then, we investigated LIX1 role in mitochondria in the context of GISTs. Altered mitochondrial metabolism is considered a cancer hallmark [35]. Here, we found that LIX1 downregulation in GIST cells resulted in the generation of mitochondria with an elongated mitochondrial network and defective cristae as well as in a decrease of respiration and mtROS production. As elongation and interconnectivity of mitochondria were slightly increased in LIX1-silenced cells, we hypothesized that this phenotype could be a consequence of mtROS production decrease because ROS-mediated mechanisms have been involved in mitochondrial remodeling [55]. We found that LIX1 is required to control the mitochondrial stability of PHB2, a protein involved in cristae organization. As observed upon LIX1 silencing, PHB2 ablation destabilizes the long OPA1 isoform and disrupts cristae structure, a phenotype that leads to decreased cell proliferation [43,56]. This effect on the IMM ultrastructure, where the respiratory chain is localized and the activity of which is crucial for ATP and mtROS production to ensure uncontrolled proliferation of cancer cells [[33], [34], [35], [36]], confers to LIX1 a pivotal role in GIST tumorigenesis. Due to the sequence similarity with lipid raft-associated proteins [[57], [58], [59]], PHBs might function as IMM scaffolds to control the lateral distribution of proteins and lipids, and thereby define functional platforms [60,61]. The de novo synthesis of cardiolipin in mitochondria is followed by a remodeling process, in which cardiolipin undergoes cycles of deacylation and acylation mediated by the Taffazin protein. In muscle cells, remodeled cardiolipin is predominantly composed of the tetra-linoleic form [46]. As cardiolipin and OPA1 levels are reduced in mitochondria lacking PHB2 [43,47], PHB2 might segregate cardiolipin into specialized membrane domains where it could bind to and stabilize proteins required for its remodeling [47] and also to proteins involved in cristae organization (e.g. OPA1 and its regulatory protease OMA1) [47,56,62,63]. Therefore, LIX1 might act as a scaffold to control PHB2 stability and consequently cardiolipin distribution or remodeling at mitochondrial contact sites.
We previously demonstrated that LIX1 regulates the proliferative and invasive capacities of GIST cells upstream of YAP1/TAZ [23]. More than 80% of GISTs harbor a gain of function mutation in KIT that results in the constitutive activation of the KIT receptor and signaling pathways, leading to spontaneous tumor cell proliferation and uncontrolled tumor growth [39]. We found that reducing YAP1/TAZ protein level or activity markedly decreases KIT expression. This indicates that LIX1 regulates KIT protein level upstream of YAP1/TAZ [23]. Pervasive activation of YAP1/TAZ has been observed in many tumor types [64]. Supplementation with unsaturated fatty acids counteracts the effects of LIX1 knock-down on mitochondrial morphology and ultrastructure and restores YAP1/TAZ signaling. It is tempting to speculate that LIX1-mediated mitochondrial remodeling controls YAP1/TAZ levels and subsequent GIST malignancy. In line with this hypothesis, it was shown that YAP1 activity is influenced by the mitochondrial morphology and ROS levels [65,66].
In summary, this study brings insights into LIX1 cellular function. We found that LIX1 contributes to the maintenance of the cristae organization and mitochondrial function, which are essential for cell proliferation and differentiation. Furthermore, we showed that mtROS fine-tuning is a key event in the control of YAP1/TAZ signaling activity. Identifying the regulatory mechanisms by which YAP1/TAZ control stem/progenitor cell differentiation into the various major gastrointestinal lineages should provide valuable information to develop new therapeutic strategies for regenerative medicine and for cancers.
4. Methods
4.1. Cell culture and reagents
The GIST-T1 cell line was obtained from Cosmo Bio (Japan). This cell line was established from a metastatic human GIST that harbors a heterozygous deletion of 57 bases in exon 11 of KIT [67]. HeLa cells were a kind gift of V. Baldin and A. Debant (CRBM, Montpellier France). Human gastric SMCs, provided by Innoprot Innovative (Spain), were grown at low density to maintain a synthetic undifferentiated phenotype, or at high density to induce their differentiation. All cells were cultured in Dulbecco's Modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. GIST-T1 stable cell lines (GIST-T1-Scrambled and GIST-T1-ShLIX1#1 and GIST-T1-ShLIX1#2) were generated as previously reported [23]. For linoleic acid supplementation, stable GIST-T1 cell lines were incubated with EtOH (vehicle) or with 50 μM linoleic acid for 12 h. For NAC experiments, GIST-T1 cells were incubated with 3 mM NAC for 18 h. All cell lines were routinely tested for the absence of mycoplasma contamination (VenorGeM OneStep Test, BioValley). For mitochondrial CATALASE overexpression, GIST-T1 cells were transfected with the pcDNA3-mitoCat plasmid using lipofectamine (Invitrogen), as previously described [40,41]. CATALASE-overexpressing GIST-T1 cells were selected using G418 (250 μg/ml) (Invitrogen).
4.2. DNA plasmids and constructs
The LIX1 expressing plasmid was from Origene (#SC108332, USA). The cDNA encoding human LIX1 was subcloned in the pCS2HA plasmid to generate the HA-LIX1 fusion protein in which the HA-tag is at the N-terminus of LIX1. The pCS2HA- HA-LIX1 plasmid was used as template to generate the LIX1 variants in which cysteine 83 and 84 were substituted by serine residues with the QuikChange Site-Directed Mutagenesis Kit (Stratagene), according to the manufacturer's protocol, and the primers listed in Supplementary Table 1.
4.3. Subcellular fractionation
Subcellular fractionation was performed as previously reported [45]. Briefly, cells were resuspended in Mitochondria Isolation Buffer (MIB, 200 mM sucrose, 10 mM Tris/MOPS, 1 mM EGTA/Tris, and protease inhibitors), and then lysed using a motor-driven homogenizer operating at 1600 rpm. An aliquot of the resulting extract was used as whole cell lysate (Total). The remaining lysate was centrifuged at 600 g, 4 °C for 10 min to remove any contaminants coming from the nuclear fraction. This supernatant (crude cytoplasmic fraction) was then centrifuged again at 7000 g, 4 °C, for 10 min to pellet mitochondria. The mitochondrial fraction was washed twice in MIB and resuspended in MIB. For the protease protection assay, purified mitochondria were resuspended in MIB and incubated or not with increasing concentrations of PK on ice for 30 min. PMSF (2 mM) was then added to inhibit PK. For alkaline sodium carbonate extraction, 100 mg of mitochondrial fraction was re-suspended in 100 mM Na2CO3 (pH 11.5) and incubated on ice for 30 min. After centrifugation at 100,000g for 30 min, the supernatant (soluble proteins) and the pellet (membranes) fractions were harvested. Samples were then separated by SDS-PAGE followed by Western blot analysis. For the detergent assay, purified mitochondria were resuspended in MIB and incubated or not with 1/10 of 4% digitonin, 2% NP40, 2% Triton X-100, or 10% SDS. Samples were then centrifuged at 16,000g and the pellet (membranes) fractions were harvested. Samples were separated by SDS-PAGE followed by Western blot analysis.
4.4. Acyl-RAC assay
LIX1 S-palmitoylation was assessed using the acyl-resin assisted capture (Acyl-RAC) method and the CAPTUREome™ S-Palmitoylated Protein Kit (Badrilla, K010-310; Leeds, UK), as previously described [31]. Briefly, free thiol groups were first blocked. The remaining palmitate groups were then treated with a thioester cleavage reagent (cleaved fraction) or with a preservation reagent (control, preserved fraction). Then, proteins with newly released thiol groups were incubated with the capture resin. Both bound and unbound fractions were analyzed. For the experiment, 1 mg of total cell lysates was used. Samples were separated by SDS–PAGE followed by Western blot analysis.
4.5. Cell fixation, immunofluorescence microscopy, quantification
Cells were seeded on fibronectin-coated (50 μg/ml per coverslips) coverslips, fixed with 4% paraformaldehyde in PBS containing 0.01% Triton X-100 for 10 min, blocked with 1% goat serum for 1 h before incubation with primary (Supplementary Table 2) and Alexa 350-, 488-, and 555-conjugated secondary antibodies (Life Science) in 0.1% goat serum. Nuclei were labeled with Hoechst (Invitrogen). Cells were imaged using a Zeiss AxioVision fluorescence microscope. Mitochondria were imaged using a Zeiss LSM780 confocal microscope. Live cell imaging was performed using a Leica SP8-UVconfocal microscope. For staining mitochondria, cells were incubated with 200 nM of MitoTracker™ Deep Red FM (Molecular Probes™) at 37 °C for 30 min. Cells were then washed three times with DMEM, rinsed in PBS twice, and processed for immunofluorescence. Quantification was performed with a macro designed by Dagda et al. (2009) in ImageJ [68]. Mitochondrial superoxide ions were detected as described by Arena et al.(2018) [69]. Cells cultured on glass coverslips were incubated with DMEM and 5 μM MitoSOX™ Red Mitochondrial Superoxide Indicator (Molecular Probes™) at 37 °C for 10 min. Cell counting and pixel quantification were performed with ImageJ. Antibodies are listed in Supplementary Table 2.
4.6. Seahorse analysis and high-resolution oxygraph respirometry
Oxygen consumption rates (OCR) were measured using the XFe-96 Extracellular Flux Analyzer (Seahorse Bioscience). 0.5 x105 GIST cells (50–60% of confluence) were seeded on poly-d-lysine-coated XF96 plates in XF medium (non-buffered DMEM containing 10 mM glucose and 2 mM l-glutamine). Cells were left for 4–6 h before starting the injection of drugs. OCRs were measured using the mitochondrial stress test in basal conditions and in response to oligomycin (1 μM), FCCP (1 μM), rotenone (100 nM), and antimycin A (1 μM; Sigma).
OCRs were measured also by high-resolution Oxygraph respirometry (Oroboros). The respiratory rates of 2-3x106 cells were recorded at 37 °C in 2 ml glass chambers. Cells were resuspended in respiratory buffer (0.5 mM EGTA, 3 mM MgCl2, 60 mM potassium lactobionate, 20 mM taurine, 10 mM KH2PO4, 20 mM HEPES, 110 mM sucrose, and 1 mg/mL BSA at pH 7.1) and permeabilized by incubation with digitonin (15 μg/106 cells). Malate (5 mM) and pyruvate (5 mM) were added to provide NADH to complex I (CXI). The addition of succinate (10 mM) and rotenone (10 mM) allowed measuring respiration driven by complex II (CXII).
4.7. Transmission electron microscopy
GIST-T1 cells were fixed in 2.5% glutaraldehyde in PHEM buffer, pH7.2, at room temperature for 1 h, and post-fixed in 0.5% osmic acid in the dark and room temperature for 2 h, dehydrated in a graded series of ethanol solutions (30–100%), and embedded in EmBed 812 using an Automated Microwave Tissue Processor for Electronic Microscopy (Leica EM AMW). Thin sections (70 nm; Leica-Reichert Ultracut E) collected at different levels of each block were counterstained with 1.5% uranyl acetate in 70% ethanol and lead citrate and observed using a Tecnai F20 transmission electron microscope at 200 KV at the CoMET MRI facilities, INM, Montpellier France. Three-dimensional reconstructions were performed with the AMIRA software.
4.8. Western blotting
Cells were resuspended in lysis buffer (20 mM Tris pH8, 50 mM NaCl, 1% NP40, cOmplete EDTA-free Protease Inhibitor Cocktail (Roche)) or in MIB (to analyze mitochondria). Protein lysates were boiled in SDS-PAGE sample buffer, separated by SDS-PAGE, and transferred to nitrocellulose membranes. Membranes were incubated with primary antibodies (Supplementary Table 2).
4.9. Reverse transcription and quantitative polymerase chain reaction (RT-qPCR)
Total RNA extraction, reverse transcription and qPCR was performed as previously described [23]. PCR primers (Supplemental Table 3) were designed using the LightCycler Probe Design 2.0 software. Expression levels were determined with the LightCycler analysis software (version 3.5) relative to standard curves. Data are the mean level of gene expression relative to the expression of the reference genes HBMS and YWHAZ calculated using the 2−ΔΔCT method.
5. Statistical analysis
Data were analyzed with two-tailed Mann-Whitney tests and the GraphPad Prism 6 software. Results were considered significant when P < 0.05 (*), P < 0.01 (**), P < 0.001 (***), or P < 0.0001 (****).
Author s’ contribution
A.G. performed confocal imaging and biochemical analyses. C.A. performed High-Resolution Oxygraphy experiment and analyzed the data; S.K. performed Seahorse experiment and analyzed the data. C.C. performed electron microscopy experiments. S.F. and P.d.S.B. conceived the study. S.F., P.d.S.B., A.L., A.M., N.T. designed experiments with contributions from C.A., J.F., M.R., S.K. S.F. wrote the manuscript with inputs from A.G. and P.d.S.B. All authors read and approved the manuscript.
Declaration of competing interest
The authors disclose no potential conflicts of interest.
Acknowledgements
This work was supported by the Association contre les Myopathies (AFM N°23800 to SF), by Inserm Transfert (CoPoc N°MAT-PI-13315-A-02 to SF), and by Ligue Régionale Contre le Cancer Languedoc-Roussillon (2020 to P.d.SB). Work was also supported by institutional funds by the University of Montpellier, Inserm and CNRS. AG is a recipient of an AFM-Téléthon PhD studentship. We thank Dr. Ewa Gurgul-Convey for kindly providing the pcDNA3-mitoCat plasmid. We deeply thanks Alexandre de Santa Barbara his help in source code and ImageJ quantification. We are grateful to members of Sandrine Faure and Pascal de Santa Barbara's team for critical reading of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2022.102431.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Mandal S., Lindgren A.G., Srivastava A.S., Clark A.T., Banerjee U. Mitochondrial function controls proliferation and early differentiation potential of embryonic stem cells. Stem Cell. 2011;29:486–495. doi: 10.1002/stem.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wanet A., Arnould T., Najimi M., Renard P. Connecting mitochondria, metabolism, and stem cell fate. Stem Cell. Dev. 2015;24:1957. doi: 10.1089/scd.2015.0117. –1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khacho M., Clark A., Svoboda D.S., Azzi J., MacLaurin J.G., Meghaizel C., Sesaki H., Lagace D.C., Germain M., Harper M.-E., Park D.S., Slack R.S. Mitochondrial dynamics impacts stem cell identity and fate decisions by regulating a nuclear transcriptional program. Cell Stem Cell. 2016;19:232–247. doi: 10.1016/j.stem.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 4.Khacho M., Slack R.S. Mitochondrial dynamics in the regulation of neurogenesis: from development to the adult brain. Dev. Dynam. 2018;247:47–53. doi: 10.1002/dvdy.24538. [DOI] [PubMed] [Google Scholar]
- 5.Bahat A., Gross A. Mitochondrial plasticity in cell fate regulation. J. Biol. Chem. 2019;294:13852–13863. doi: 10.1074/jbc.REV118.000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hinge A., He J., Bartram J., Javier J., Xu J., Fjellman E., Sesaki H., Li T., Yu J., Wunderlich M., Mulloy J., Kofron M., Salomonis N., Grimes H.L., Filippi M.-D. Asymmetrically segregated mitochondria provide cellular memory of hematopoietic stem cell replicative history and drive HSC attrition. Cell Stem Cell. 2020;26:420–430. doi: 10.1016/j.stem.2020.01.016. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hock M.B., Kralli A. Transcriptional control of mitochondrial biogenesis and function. Annu. Rev. Physiol. 2009;71:177–203. doi: 10.1146/annurev.physiol.010908.163119. [DOI] [PubMed] [Google Scholar]
- 8.Wallace A.S., Burns A.J. Development of the enteric nervous system, smooth muscle and interstitial cells of Cajal in the human gastrointestinal tract. Cell Tissue Res. 2005;319:367–382. doi: 10.1007/s00441-004-1023-2. [DOI] [PubMed] [Google Scholar]
- 9.Le Guen L., Marchal S., Faure S., de Santa Barbara P. Mesenchymal-epithelial interactions during digestive tract development and epithelial stem cell regeneration. Cell. Mol. Life Sci. 2015;72:3883–3896. doi: 10.1007/s00018-015-1975-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faure S., McKey J., Sagnol S., de Santa Barbara P. Enteric neural crest cells regulate vertebrate stomach patterning and differentiation. Development. 2015;142:331–342. doi: 10.1242/dev.118422. [DOI] [PubMed] [Google Scholar]
- 11.Bourret A., Chauvet N., de Santa Barbara P., Faure S. Colonic mesenchyme differentiates into smooth muscle before its colonization by vagal enteric neural crest-derived cells in the chick embryo. Cell Tissue Res. 2017;368:503–511. doi: 10.1007/s00441-017-2577-0. [DOI] [PubMed] [Google Scholar]
- 12.Torihashi S., Ward S.M., Sanders K.M. Development of c-Kit-positive cells and the onset of electrical rhythmicity in murine small intestine. Gastroenterology. 1997;112:144–155. doi: 10.1016/s0016-5085(97)70229-4. [DOI] [PubMed] [Google Scholar]
- 13.Klüppel M., Huizinga J.D., Malysz J., Bernstein A. Developmental origin and Kit-dependent development of the interstitial cells of cajal in the mammalian small intestine. Dev. Dynam. 1998;211:60–71. doi: 10.1002/(SICI)1097-0177. 199801. 211:1<60::AID-AJA6>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 14.Torihashi S., Nishi K., Tokutomi Y., Nishi T., Ward S., Sanders K.M. Blockade of kit signaling induces transdifferentiation of interstitial cells of cajal to a smooth muscle phenotype. Gastroenterology. 1999;117:140–148. doi: 10.1016/s0016-5085(99)70560-3. [DOI] [PubMed] [Google Scholar]
- 15.Vicente-Dueñas C., Pérez-Caro M., Abollo-Jiménez F., Cobaleda C., Sánchez-García I. Stem-cell driven cancer: “hands-off” regulation of cancer development. Cell Cycle. 2009;8:1314–1318. doi: 10.4161/cc.8.9.8217. [DOI] [PubMed] [Google Scholar]
- 16.Corless C.L., Barnett C.M., Heinrich M.C. Gastrointestinal stromal tumours: origin and molecular oncology. Nat. Rev. Cancer. 2011;11:865–878. doi: 10.1038/nrc3143. [DOI] [PubMed] [Google Scholar]
- 17.Ordog T., Zörnig M., Hayashi Y. Targeting disease persistence in gastrointestinal stromal tumors. Stem Cells Transl Med. 2015;4:702–707. doi: 10.5966/sctm.2014-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hapkova I., Skarda J., Rouleau C., Thys A., Notarnicola C., Janikova M., Bernex F., Rypka M., Vanderwinden J.-M., Faure S., Vesely J., de Santa Barbara P. High expression of the RNA-binding protein RBPMS2 in gastrointestinal stromal tumors. Exp. Mol. Pathol. 2013;94:314–321. doi: 10.1016/j.yexmp.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 19.McKey J., Martire D., de Santa Barbara P., Faure S. LIX1 regulates YAP1 activity and controls the proliferation and differentiation of stomach mesenchymal progenitors. BMC Biol. 2016;14:34. doi: 10.1186/s12915-016-0257-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swindell E.C., Moeller C., Thaller C., Eichele G. Cloning and expression analysis of chicken Lix1, a founding member of a novel gene family. Mech. Dev. 2001;109:405–408. doi: 10.1016/s0925-4773(01)00535-4. [DOI] [PubMed] [Google Scholar]
- 21.Bando T., Hamada Y., Kurita K., Nakamura T., Mito T., Ohuchi H., Noji S. Lowfat, a mammalian Lix1 homologue, regulates leg size and growth under the Dachsous/Fat signaling pathway during tissue regeneration. Dev. Dynam. 2011;240:1440–1453. doi: 10.1002/dvdy.22647. [DOI] [PubMed] [Google Scholar]
- 22.Mao Y., Kucuk B., Irvine K.D. Drosophila lowfat, a novel modulator of Fat signaling. Development. 2009;136:3223–3233. doi: 10.1242/dev.036152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guérin A., Martire D., Trenquier E., Lesluyes T., Sagnol S., Pratlong M., Lefebvre E., Chibon F., de Santa Barbara P., Faure S. LIX1 regulates YAP activity and controls gastrointestinal cancer cell plasticity. J. Cell Mol. Med. 2020;24:9244–9254. doi: 10.1111/jcmm.15569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi M.-C., Ryu S., Hao R., Wang B., Kapur M., Fan C.-M., Yao T.-P. HDAC4 promotes Pax7-dependent satellite cell activation and muscle regeneration. EMBO Rep. 2014;15:1175–1183. doi: 10.15252/embr.201439195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sorice M., Manganelli V., Matarrese P., Tinari A., Misasi R., Malorni W., Garofalo T. Cardiolipin-enriched raft-like microdomains are essential activating platforms for apoptotic signals on mitochondria. FEBS Lett. 2009;583:2447–2450. doi: 10.1016/j.febslet.2009.07.018. [DOI] [PubMed] [Google Scholar]
- 26.Fröhlich M., Dejanovic B., Kashkar H., Schwarz G., Nussberger S. S-palmitoylation represents a novel mechanism regulating the mitochondrial targeting of BAX and initiation of apoptosis. Cell Death Dis. 2014;5:e1057. doi: 10.1038/cddis.2014.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie Y., Zheng Y., Li H., Luo X., He Z., Cao S., Shi Y., Zhao Q., Xue Y., Zuo Z., Ren J. GPS-Lipid: a robust tool for the prediction of multiple lipid modification sites. Sci. Rep. 2016;6 doi: 10.1038/srep28249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang X., Chatterjee V., Ma Y., Zheng E., Yuan S.Y. Protein palmitoylation in leukocyte signaling and function. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.600368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Charollais J., Van Der Goot F.G. Palmitoylation of membrane proteins (Review) Mol. Membr. Biol. 2009;26:55–66. doi: 10.1080/09687680802620369. [DOI] [PubMed] [Google Scholar]
- 30.Forrester M.T., Hess D.T., Thompson J.W., Hultman R., Moseley M.A., Stamler J.S., Casey P.J. Site-specific analysis of protein S-acylation by resin-assisted capture. J. Lipid Res. 2011;52:393–398. doi: 10.1194/jlr.D011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chytła A., Gajdzik-Nowak W., Biernatowska A., Sikorski A.F., Czogalla A. High-level expression of palmitoylated MPP1 recombinant protein in mammalian cells. Membranes. 2021;11:715. doi: 10.3390/membranes11090715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chandel N.S. Mitochondria as signaling organelles. BMC Biol. 2014;12:34. doi: 10.1186/1741-7007-12-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weinberg F., Hamanaka R., Wheaton W.W., Weinberg S., Joseph J., Lopez M., Kalyanaraman B., Mutlu G.M., Budinger G.R.S., Chandel N.S. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc. Natl. Acad. Sci. U.S.A. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vyas S., Zaganjor E., Haigis M.C. Mitochondria and cancer. Cell. 2016;166:555–566. doi: 10.1016/j.cell.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeBerardinis R.J., Chandel N.S. Fundamentals of cancer metabolism. Sci. Adv. 2016;2 doi: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kong H., Chandel N.S. Regulation of redox balance in cancer and T cells. J. Biol. Chem. 2018;293:7499–7507. doi: 10.1074/jbc.TM117.000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lenaz G., Tioli G., Falasca A.I., Genova M.L. Complex I function in mitochondrial supercomplexes. Biochim. Biophys. Acta. 2016;1857:991–1000. doi: 10.1016/j.bbabio.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 38.Herrero A., Barja G. Localization of the site of oxygen radical generation inside the complex I of heart and nonsynaptic brain mammalian mitochondria. J. Bioenerg. Biomembr. 2000;32:609–615. doi: 10.1023/a:1005626712319. [DOI] [PubMed] [Google Scholar]
- 39.Nakahara M., Isozaki K., Hirota S., Miyagawa J., Hase-Sawada N., Taniguchi M., Nishida T., Kanayama S., Kitamura Y., Shinomura Y., Matsuzawa Y. A novel gain-of-function mutation of c-kit gene in gastrointestinal stromal tumors. Gastroenterology. 1998;115:1090–1095. doi: 10.1016/s0016-5085(98)70079-4. [DOI] [PubMed] [Google Scholar]
- 40.Gurgul E., Lortz S., Tiedge M., Jörns A., Lenzen S. Mitochondrial catalase overexpression protects insulin-producing cells against toxicity of reactive oxygen species and proinflammatory cytokines. Diabetes. 2004;53:2271–2280. doi: 10.2337/diabetes.53.9.2271. [DOI] [PubMed] [Google Scholar]
- 41.Gurgul-Convey E., Mehmeti I., Lortz S., Lenzen S. Cytokine toxicity in insulin-producing cells is mediated by nitro-oxidative stress-induced hydroxyl radical formation in mitochondria. J. Mol. Med. (Berl.) 2011;89:785–798. doi: 10.1007/s00109-011-0747-1. [DOI] [PubMed] [Google Scholar]
- 42.Galloway C.A., Lee H., Yoon Y. Mitochondrial morphology-emerging role in bioenergetics. Free Radic. Biol. Med. 2012;53:2218–2228. doi: 10.1016/j.freeradbiomed.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Merkwirth C., Dargazanli S., Tatsuta T., Geimer S., Löwer B., Wunderlich F.T., von Kleist-Retzow J.-C., Waisman A., Westermann B., Langer T. Prohibitins control cell proliferation and apoptosis by regulating OPA1-dependent cristae morphogenesis in mitochondria. Genes Dev. 2008;22:476–488. doi: 10.1101/gad.460708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olichon A., Baricault L., Gas N., Guillou E., Valette A., Belenguer P., Lenaers G. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J. Biol. Chem. 2003;278:7743–7746. doi: 10.1074/jbc.C200677200. [DOI] [PubMed] [Google Scholar]
- 45.Frezza C., Cipolat S., Martins de Brito O., Micaroni M., Beznoussenko G.V., Rudka T., Bartoli D., Polishuck R.S., Danial N.N., De Strooper B., Scorrano L. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell. 2006;126:177–189. doi: 10.1016/j.cell.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 46.Semba R.D., Moaddel R., Zhang P., Ramsden C.E., Ferrucci L. Tetra-linoleoyl cardiolipin depletion plays a major role in the pathogenesis of sarcopenia. Med. Hypotheses. 2019;127:142–149. doi: 10.1016/j.mehy.2019.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richter-Dennerlein R., Korwitz A., Haag M., Tatsuta T., Dargazanli S., Baker M., Decker T., Lamkemeyer T., Rugarli E.I., Langer T. DNAJC19, a mitochondrial cochaperone associated with cardiomyopathy, forms a complex with prohibitins to regulate cardiolipin remodeling. Cell Metabol. 2014;20:158–171. doi: 10.1016/j.cmet.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 48.Valianpour F., Wanders R.J.A., Overmars H., Vaz F.M., Barth P.G., van Gennip A.H. Linoleic acid supplementation of Barth syndrome fibroblasts restores cardiolipin levels: implications for treatment. J. Lipid Res. 2003;44:560–566. doi: 10.1194/jlr.M200217-JLR200. [DOI] [PubMed] [Google Scholar]
- 49.Daum G. Lipids of mitochondria. Biochim. Biophys. Acta. 1985;822:1–42. doi: 10.1016/0304-4157(85)90002-4. [DOI] [PubMed] [Google Scholar]
- 50.Ardail D., Privat J.P., Egret-Charlier M., Levrat C., Lerme F., Louisot P. Mitochondrial contact sites. Lipid composition and dynamics. J. Biol. Chem. 1990;265:18797–18802. [PubMed] [Google Scholar]
- 51.Hovius R., Lambrechts H., Nicolay K., de Kruijff B. Improved methods to isolate and subfractionate rat liver mitochondria. Lipid composition of the inner and outer membrane. Biochim. Biophys. Acta. 1990;1021:217–226. doi: 10.1016/0005-2736(90)90036-n. [DOI] [PubMed] [Google Scholar]
- 52.Simbeni R., Pon L., Zinser E., Paltauf F., Daum G. Mitochondrial membrane contact sites of yeast. Characterization of lipid components and possible involvement in intramitochondrial translocation of phospholipids. J. Biol. Chem. 1991;266:10047–10049. [PubMed] [Google Scholar]
- 53.Hoch F.L. Cardiolipins and biomembrane function. Biochim. Biophys. Acta. 1992;1113:71–133. doi: 10.1016/0304-4157(92)90035-9. [DOI] [PubMed] [Google Scholar]
- 54.Schlame M., Rua D., Greenberg M.L. The biosynthesis and functional role of cardiolipin. Prog. Lipid Res. 2000;39:257–288. doi: 10.1016/s0163-7827(00)00005-9. [DOI] [PubMed] [Google Scholar]
- 55.Deheshi S., Dabiri B., Fan S., Tsang M., Rintoul G.L. Changes in mitochondrial morphology induced by calcium or rotenone in primary astrocytes occur predominantly through ros-mediated remodeling. J. Neurochem. 2015;133:684–699. doi: 10.1111/jnc.13090. [DOI] [PubMed] [Google Scholar]
- 56.Anderson C.J., Kahl A., Fruitman H., Qian L., Zhou P., Manfredi G., Iadecola C. Prohibitin levels regulate OMA1 activity and turnover in neurons. Cell Death Differ. 2020;27 doi: 10.1038/s41418-019-0469-4. 1896–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tavernarakis N., Driscoll M., Kyrpides N.C. The SPFH domain: implicated in regulating targeted protein turnover in stomatins and other membrane-associated proteins. Trends Biochem. Sci. 1999;24:425–427. doi: 10.1016/s0968-0004(99)01467-x. [DOI] [PubMed] [Google Scholar]
- 58.Langhorst M.F., Reuter A., Stuermer C.a.O. Scaffolding microdomains and beyond: the function of reggie/flotillin proteins. Cell. Mol. Life Sci. 2005;62:2228–2240. doi: 10.1007/s00018-005-5166-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morrow I.C., Parton R.G. Flotillins and the PHB domain protein family: rafts, worms and anaesthetics. Traffic. 2005;6:725–740. doi: 10.1111/j.1600-0854.2005.00318.x. [DOI] [PubMed] [Google Scholar]
- 60.Osman C., Haag M., Potting C., Rodenfels J., Dip P.V., Wieland F.T., Brügger B., Westermann B., Langer T. The genetic interactome of prohibitins: coordinated control of cardiolipin and phosphatidylethanolamine by conserved regulators in mitochondria. J. Cell Biol. 2009;184:583–596. doi: 10.1083/jcb.200810189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Christie D.A., Lemke C.D., Elias I.M., Chau L.A., Kirchhof M.G., Li B., Ball E.H., Dunn S.D., Hatch G.M., Madrenas J. Stomatin-like protein 2 binds cardiolipin and regulates mitochondrial biogenesis and function. Mol. Cell Biol. 2011;31:3845–3856. doi: 10.1128/MCB.05393-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Merkwirth C., Martinelli P., Korwitz A., Morbin M., Brönneke H.S., Jordan S.D., Rugarli E.I., Langer T. Loss of prohibitin membrane scaffolds impairs mitochondrial architecture and leads to tau hyperphosphorylation and neurodegeneration. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1003021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ban T., Ishihara T., Kohno H., Saita S., Ichimura A., Maenaka K., Oka T., Mihara K., Ishihara N. Molecular basis of selective mitochondrial fusion by heterotypic action between OPA1 and cardiolipin. Nat. Cell Biol. 2017;19:856–863. doi: 10.1038/ncb3560. [DOI] [PubMed] [Google Scholar]
- 64.Fullenkamp C.A., Hall S.L., Jaber O.I., Pakalniskis B.L., Savage E.C., Savage J.M., Ofori-Amanfo G.K., Lambertz A.M., Ivins S.D., Stipp C.S., Miller B.J., Milhem M.M., Tanas M.R. TAZ and YAP are frequently activated oncoproteins in sarcomas. Oncotarget. 2016;7:30094–30108. doi: 10.18632/oncotarget.8979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.von Eyss B., Jaenicke L.A., Kortlever R.M., Royla N., Wiese K.E., Letschert S., McDuffus L.-A., Sauer M., Rosenwald A., Evan G.I., Kempa S., Eilers M. A MYC-driven change in mitochondrial dynamics limits YAP/TAZ function in mammary epithelial cells and breast cancer. Cancer Cell. 2015;28:743–757. doi: 10.1016/j.ccell.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 66.Ji F., Shen T., Zou W., Jiao J. UCP2 regulates embryonic neurogenesis via ROS-mediated yap alternation in the developing neocortex. Stem Cell. 2017;35:1479–1492. doi: 10.1002/stem.2605. [DOI] [PubMed] [Google Scholar]
- 67.Taguchi T., Sonobe H., Toyonaga S., Yamasaki I., Shuin T., Takano A., Araki K., Akimaru K., Yuri K. Conventional and molecular cytogenetic characterization of a new human cell line, GIST-T1, established from gastrointestinal stromal tumor. Lab. Invest. 2002;82:663–665. doi: 10.1038/labinvest.3780461. [DOI] [PubMed] [Google Scholar]
- 68.Dagda R.K., Cherra S.J., Kulich S.M., Tandon A., Park D., Chu C.T. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J. Biol. Chem. 2009;284:13843–13855. doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arena G., Cissé M.Y., Pyrdziak S., Chatre L., Riscal R., Fuentes M., Arnold J.J., Kastner M., Gayte L., Bertrand-Gaday C., Nay K., Angebault-Prouteau C., Murray K., Chabi B., Koechlin-Ramonatxo C., Orsetti B., Vincent C., Casas F., Marine J.-C., Etienne-Manneville S., Bernex F., Lombès A., Cameron C.E., Dubouchaud H., Ricchetti M., Linares L.K., Le Cam L. Mitochondrial MDM2 regulates respiratory complex I activity independently of p53. Mol. Cell. 2018;69:594–609. doi: 10.1016/j.molcel.2018.01.023. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.