Abstract
Little is known about the molecular mechanism by which histone-like nucleoid-structuring (H-NS) protein and cyclic AMP-catabolite activator protein (CAP) complex control bacterial motility. In the present paper, we show that crp and hns mutants are nonmotile due to a complete lack of flagellin accumulation. This results from a reduced expression in vivo of fliA and fliC, which encode the specific flagellar sigma factor and flagellin, respectively. Overexpression of the flhDC master operon restored, at least in part, motility in crp and hns mutant strains, suggesting that this operon is the main target for both regulators. Binding of H-NS and CAP to the regulatory region of the master operon was demonstrated by gel retardation experiments, and their DNA binding sites were identified by DNase I footprinting assays. In vitro transcription experiments showed that CAP activates flhDC expression while H-NS represses it. In agreement with this observation, the activity of a transcriptional fusion carrying the flhDC promoter was decreased in the crp strain and increased in the hns mutant. In contrast, the activity of a transcriptional fusion encompassing the entire flhDC regulatory region extending to the ATG translational start codon was strongly reduced in both hns and crp mutants. These results suggest that the region downstream of the +1 transcriptional start site plays a crucial role in the positive control by H-NS of flagellum biosynthesis in vivo. Finally, the lack of complementation of the nonmotile phenotype in a crp mutant by activation-deficient CAP mutated proteins and characterization of cfs, a mutation resulting in a CAP-independent motility behavior, demonstrate that CAP activates flhDC transcription by binding to its promoter and interacting with RNA polymerase.
The structure and the function of the flagellum in Escherichia coli and Salmonella typhimurium have been extensively studied for many years. Its biosynthesis seems to play a crucial role in adaptation to various environmental conditions and is affected by numerous adverse conditions (51). Furthermore, motility has frequently been associated with virulence and/or inflammatory response in various microorganisms, such as Bordetella bronchiseptica (1), Vibrio cholerae (19), and S. typhimurium (13). Finally, it has recently been shown that motility is critical for colonization and/or biofilm formation, e.g., in Vibrio fischeri (23) and in E. coli (45).
In E. coli, numerous mutations are known to alter motility, especially those affecting synthesis of bacterial membrane components, such as porins (27) and lipopolysaccharide (44). Moreover, several regulators have been shown to be involved in the control of swarming behavior in E. coli and S. typhimuium, in particular, the cyclic AMP (cAMP)-catabolite activator protein (CAP) complex (54, 61) and the histone-like nucleoid-structuring (H-NS) protein (7, 26). The former positively or negatively regulates a vast number of genes involved in various functions in E. coli (12). The latter controls expression of many genes regulated by environmental parameters, such as pH, temperature, and osmolarity (3). Mutations in crp or in hns, the structural genes of CAP and H-NS, respectively, result in a complete loss of motility in E. coli (7, 54). However, little is known about the molecular mechanism by which these regulatory proteins control flagellar gene expression.
In E. coli and S. typhimurium, flagellum biosynthesis requires expression of nearly 50 genes clustered at several places on the chromosome. Their transcription forms an ordered cascade in which the expression of one gene located at a given level requires the transcription of another one at higher level (35). At the top of the hierarchy, the flhD and flhC genes constitute the master operon which controls the expression of all other flagellar genes. In E. coli, the FlhD and FlhC proteins have been shown to act as positive regulators of the flagellar regulon (33). The fliA gene, one of those located at the second level of the cascade, encodes the flagellar sigma factor ς28. This protein is required for the expression of most genes located at the third level, e.g., fliC, the flagellin structural gene (35).
In the present study, we investigated the mechanism by which H-NS and the cAMP-CAP complex regulate flagellum biosynthesis. By gel retardation and footprinting experiments, we demonstrated the abilities of both regulators to bind to the flhDC regulatory region. Despite a similar complete loss of flagellin accumulation in hns and crp mutants, in vitro transcription assays showed that CAP activates expression of the master operon while H-NS represses it. Using transcriptional fusions, we demonstrated that the positive control by H-NS of flagellar gene expression in vivo requires the region downstream of the +1 transcriptional start site.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Strains carrying the hns-1001 (7) and crp::Sm (29a) mutations were constructed by P1 transduction with phage P1vir as previously described (39). The strains were grown in Luria-Bertani or tryptone medium supplemented with 0.4% (wt/vol) sodium succinate as a carbon source. Tryptone swarm plates containing 1% Bacto-Tryptone, 0.5% NaCl, and 0.3% Bacto Agar were used to test bacterial motility as previously described (6). When required, antibiotics were added at the following concentrations: chloramphenicol, 20 μg/ml; kanamycin, 20 μg/ml; ampicillin, 50 μg/ml; and streptomycin, 50 μg/ml. All experiments were performed in accordance with the European regulation requirements concerning the contained use of Genetically Modified Organisms of Group-I (agreement no. 2735).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotypea | Reference or source |
|---|---|---|
| Strain | ||
| FB8 | Wild type | 10 |
| MG1655 | Wild type | CGSC6300 |
| BE1815 | MG1655 crp::Sm | 29a |
| BE1816 | MG1655 hns-1001 | This study |
| BE1817 | MG1655 crp::Sm hns-1001 | This study |
| PS2209 | Wild type | 7 |
| PS2652 | PS2209 hns-1001 | 7 |
| BE1818 | PS2209 crp::Sm | This study |
| BE1919 | PS2209 hns-1001 crp::Sm | This study |
| JLV70-2 | Δcrp cfs | 57 |
| Plasmid | ||
| pKK232-8 | Cloning vector for promoter analysis; Apr | Pharmacia |
| pSR | pBR322 derivative carrying the gal promoter upstream of a λ oop terminator | 46a |
| pJCD01 | pUC19 derivative containing a polylinker flanked by the divergent terminators rpoCt and rrnBT1T2 | 36 |
| pDIA525 | pBR322 derivative carrying the flhDC promoter (nt −213 to +78) upstream of a λ oop terminator | This study |
| pDIA528 | pKK232-8 derivative carrying the flhDC promoter (nt −213 to +78) | This study |
| pDIA545 | pKK232-8 derivative carrying the flhDC regulatory region (nt −213 to +205) | This study |
| pDIA546 | pJCD01 derivative carrying the flhDC regulatory region (nt −213 to +202) | This study |
| pDIA551 | pKK232-8 derivative carrying the promoter and part of the coding sequence of the flhDC operon (nt −213 to +826) | This study |
| pDIA559 | pKK232-8 derivative carrying the fliC promoter region | This study |
| pPM61 | ColE1 derivative cloning multicopy vector carrying the flhDC operon under its native promoter | 5 |
| pDCRP | pBR322 derivative containing the wild-type crp coding sequence | 58 |
| pDCRP-H159L | pDCRP derivative containing mutation H159L | 58 |
| pDCRP-T158A | pDCRP derivative containing mutation T158A | 58 |
nt, nucleotide.
Two-dimensional electrophoresis.
Protein extracts were analyzed by two-dimensional electrophoresis according to the procedure previously described (31). The purified protein was subjected to internal amino acid sequencing by the Laboratoire de Microséquençage des Protéines, Institut Pasteur, Paris, France.
RNA preparation.
Total RNA was extracted from 4 ml of culture grown to an optical density at 600 nm (OD600) of 0.4 to 0.5 with the High Pure RNA isolation kit (Boehringer Mannheim). The RNA concentration and purity were determined by OD260 and OD280 measurements.
Probe labelling.
A 651-bp DNA probe corresponding to part of the fliC coding region was generated by PCR amplification with oligonucleotides 5′-CATTAATACCAACAGCCTCTCGC-3′ and 5′-ATTGAAGCTGGGTTAGTTCCGCC-3′ and the PCR DIG Probe synthesis kit (Boehringer Mannheim) according to the manufacturer's instructions.
Quantitative analysis of mRNA.
RNA (500 ng) was denatured in 300 μl of RNA dilution buffer (water, 20× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], and formaldehyde in the ratio 5:3:2) at 65°C for 15 min. The 20× SSC solution (3 M NaCl–0.3 M trisodium citrate adjusted to pH 7) was treated with diethylpyrocarbonate. It was then applied to Hybond N+ nylon filters (Amersham) with a PR600 SlotBlot applicator (Hoefer Scientific). The RNAs were covalently cross-linked to the membrane by UV cross-linking at 0.51 J/cm2. DIG-labeled probe (20 μl) was hybridized to the immobilized RNA at 50°C for 16 h with DIG Easy Hyb buffer (Boehringer Mannheim). The membrane was washed two times with 2× SSC–0.1% sodium dodecyl sulfate at room temperature and then two times with 0.1× SSC–0.1% sodium dodecyl sulfate at 68°C. The labeled probe was visualized with the CSPD chemiluminescence detection system (Boehringer Mannheim) and Hyperfilm-MP X-ray film (Amersham). Bands were scanned with a JX-330 SHARP scanner and quantified with PDI software PDQuest based on a SUN computer system.
DNA manipulations.
Plasmid pDIA525 was constructed by PCR amplification as previously described (6). Briefly, a DNA fragment containing the flhDC promoter region was generated with the primers O1 (5′-GGAATTCTGCGCAACATCCC-3′) and O2 (5′-CCCAAGCTTGCAGAACCACC-3′) (see Fig. 2) and genomic DNA from E. coli FB8. The 308-bp PCR fragment was purified with the High Pure PCR purification kit (Boehringer Mannheim) and cloned into the pSR plasmid, restricted by EcoRI and HindIII sites to remove the gal promoter located upstream of a λ oop terminator. Plasmid pDIA528 was constructed by cloning a similar DNA fragment into the pKK232-8 vector (Pharmacia) after PCR amplification with the oligonucleotides O1′ (5′-CGGGATCCTGCGCAACATCC-3′) and O2 (5′-CCCAAGCTTGCAGAACCACC-3′). The O1′ primer differed from the O1 primer by the presence of a BamHI site instead of an EcoRI site. Plasmid pDIA545 was constructed by PCR amplification of the flhDC regulatory region with the primers O1′ (5′-CGGGATCCTGCGCAACATCC-3′) and O3 (5′-CCCAAGCTTAGGTATGCATTATTCCCACCC-3′). The resulting 435-nucleotide fragment was cloned into the plasmid pKK232-8 (Pharmacia). Plasmid pDIA546 was constructed by cloning a 423-bp BamHI/NsiI fragment containing the flhDC regulatory region of pDIA545 into plasmid pJCD01 (36). The fragment was purified from agarose gel with the JETsorb kit (GENOMED). Plasmid pDIA551 containing the promoter region and part of the coding sequence of the flhDC operon was constructed by PCR amplification of a 1,048-nucleotide fragment with primers O1′ (5′-CGGGATCCTGCGCAACATCC-3′) and O4 (5′-CCCAAGCTTGCCATTACACAAACCGG-3′). The resulting DNA fragment was cloned into the BamHI and HindIII sites of plasmid pKK232-8. Similarly, plasmid pDIA559 was constructed by PCR amplification of the fliC promoter with the primers 5′-GGGATCCGTAAAACGAATACCGGG-3′ and 5′-CCCAAGCTTGGTATTAATGACTTGTGCC-3′. The 276-bp fragment was cloned into the BamHI and HindIII sites of plasmid pKK232-8.
FIG. 2.
Regulatory region of the flhDC master operon. The nucleotides are numbered relative to the transcriptional start site (+1), indicated by a solid bent arrow. The unique CAP binding site consensus sequence is indicated by a box. Regions protected by H-NS are underlined with solid lines. Downstream of the +1 site, DNase footprinting assays were performed on the sole noncoding strand. The positions of the −10 and −35 sequences are indicated in italics. Nucleotide substitutions identified in the flhDC promoter of a cfs strain are indicated by arrowheads. The ATG translation initiation codon identified by mass spectrometry and a putative ribosome binding site (RBS) are indicated in boldface and by a dotted line, respectively. Dashed arrows numbered O1/O1′ to O3 represent oligonucleotides used in PCR amplification and/or +1 mapping. O1/O1′-to-O2 and O1′-to-O3 PCR fragments were used to construct plasmids pDIA525 and pDIA528 and plasmids pDIA545 and pDIA546, respectively.
Protein purification.
CAP, H-NS, and RNA polymerase were prepared as described previously (references 20, 60, and 24, respectively).
Gel retardation experiments.
Plasmid pDIA525, containing the flhDC promoter region, was cleaved by EcoRI, HindIII, NdeI, and SspI. DNA fragments (100 ng) were incubated with H-NS for 15 min at room temperature in the reaction mixture previously described (22). Protein-DNA complexes were resolved on 3% MetaPhor agarose gel with Tris-borate-EDTA as the running buffer. Similar experimental conditions were used with CAP except that the reaction mixture and running buffer both contained 200 μM cAMP.
Oligonucleotide labeling.
The oligonucleotides used in footprinting analysis and primer extension experiments were end labeled with phage T4 polynucleotide kinase and [γ-32P]ATP (3,000 Ci/mmol) according to standard procedures (48).
Primer extension.
The reaction was performed according to standard procedures (4), with some modifications. Ten picomoles of end-labeled O3 oligonucleotide complementary to the region including the translational start site of flhDC mRNA (see Fig. 2) was precipitated with ammonium acetate and ethanol at −20°C, washed with 70% ethanol, dried, and resuspended in 40 μl of diethyl pyrocarbonate-treated 10 mM Tris–1 mM EDTA (pH 7.6) buffer to a concentration of 2.5 ng/μl. Five nanograms of primer was annealed with 10 μg of total RNA in avian myeloblastosis virus reverse transcriptase reaction buffer (Boehringer Mannheim) and 1 mM deoxynucleoside triphosphate at 65°C for 10 min. The reaction was kept going while the temperature slowly decreased to 30°C. RNasin (20 U) (Promega) was added, and the reaction was performed with 40 U of avian myeloblastosis virus reverse transcriptase (Boehringer Mannheim) at 42°C for 90 min. One microliter of 0.5 M EDTA (pH 8.0) and 1 μl of DNase-free pancreatic RNase (Boehringer Mannheim) were added, and the reaction was further incubated at 37°C for 30 min. The reaction mixture was precipitated with ammonium acetate and ethanol, washed with 70% ethanol, and resuspended in formamide loading buffer. As a reference, sequencing reactions were performed with the Thermosequenase radiolabeled terminator cycle sequencing kit from Amersham with the same primer used in primer extension experiments.
Footprinting analysis.
DNase I footprinting experiments were performed as previously described (36) with some modifications. A 308-nucleotide fragment containing the flhDC promoter region and a 435-nucleotide fragment containing the entire regulatory region extending to the ATG translational start codon were generated by PCR amplification with a combination of the labeled and unlabeled oligonucleotides O1 (5′-GGAATTCTGCGCAACATCCC-3′)-O2 (5′-CCCAAGCTTGCAGAACCACC-3′) and O1-O3 (5′-CCCAAGCTTAGGTATGCATTATTCCCACCC-3′), respectively. Complexes with the labeled fragment were formed in 14 μl of binding buffer (36) with purified CAP and H-NS for 30 min at room temperature or at 30°C when RNA polymerase was present in the mixture. Then, 3 μl of DNase I solution at 0.45 μg/ml was added and incubated at 30°C for 15 s for samples without proteins, 20 s for the complexes with CAP, and 30 s when RNA polymerase was present in the mixture. The reaction was stopped by the addition of 40 μl of phenol, followed by vortexing and addition of 183 μl of stop solution. Protected bands were identified by comparison with the migration of the same fragment treated for A+G sequencing reactions by the method of Maxam and Gilbert (38).
In vitro transcription assays.
In vitro transcription experiments were performed with pDIA525 containing the flhDC promoter region and pDIA546 containing the entire flhDC regulatory region as previously described (36) with the following modifications. Seven microliters of the reaction mixture containing H-NS at 334 nM and/or CAP at 50 nM was incubated at 30°C for 10 min with RNA polymerase at 120 nM final concentration. Then, 3.5 μl of a mixture containing nucleoside triphosphates and heparin was added to perform the polymerization at 30°C for 10 min. Quantitative data were obtained with a PhosphorImager (Molecular Dynamics).
Chloramphenicol acetyltransferase assay.
Strains were grown in tryptone medium to an OD600 of 0.15 to 0.3, 15 ml of the culture was centrifuged for 10 min at 6,000 × g, and the pellet was resuspended in 500 μl of 100 mM Tris-HCl (pH 7.8). The cells were disrupted by sonication (five cycles of 40 s at +4°C), and cell debris was removed by centrifugation for 10 min at 13,000 × g. The total-protein concentration in the cell extract was determined by the method of Bradford (9). Chloramphenicol acetyltransferase activity was measured by the method of Shaw (49) at 405 nm and 30°C for 1 min with a spectrophotometer equipped with a temperature-controlled chamber. One unit was defined as 1 μmol of chloramphenicol acetylated per min per μg of protein.
RESULTS
The reduced expression of the flhDC master operon in crp and hns mutants results in a lack of flagellin biosynthesis.
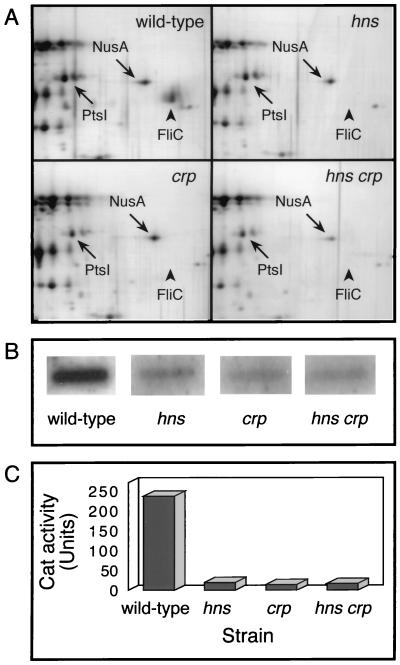
In E. coli, CAP- and H-NS-deficient strains are known to be completely nonmotile (7, 54). To evaluate the flagellin content in hns and crp mutants, total-protein extracts were analyzed by two-dimensional gel electrophoresis. In the wild-type strain, a protein was resolved as a single spot of pI 4.7 and an apparent mass of 53 kDa, in agreement with the theoretical values computed from the FliC amino acid sequence. This protein was undetectable in extracts of crp and/or hns mutants compared to extracts of the wild type (Fig. 1A). An internal fragment of the purified polypeptide was subjected to microsequencing. The amino acid sequence obtained matched the sequence of the fliC gene product (data not shown). This result provides evidence that the loss of motility associated with hns and crp mutations results from a complete lack of flagellin, in agreement with previous examination of such mutant strains by electron microscopy (7, 54).
FIG. 1.
Effect of hns and/or crp mutations on flagellin synthesis. (A) Protein extracts of the MG1655 wild-type strain and of hns, crp, and hns crp derivatives were resolved by two-dimensional electrophoresis, and the gels were silver stained. Only the region in the vicinity of FliC is shown. FliC is indicated by an arrowhead; NusA and PtsI are indicated as landmarks to facilitate the comparison. (B) The fliC mRNA level was analyzed by slot blot hybridization with a 651-bp probe specific to the fliC gene. Quantitation was performed with the Bio-Rad Multi-Analyst system. (C) fliC-cat transcriptional fusion activity was measured in early exponential growth phase. The data are the mean values from three independent assays that differed by less than 10%.
To ascertain whether the lack of flagellin accumulation in crp and hns mutants resulted from a reduced expression of the fliC gene located at the third level in the flagellar-gene cascade, we performed a comparative analysis of flagellin-encoding mRNA in crp and/or hns strains. In contrast to the wild-type strain, fliC mRNA was not significantly different from the background level in the mutant strains (Fig. 1B), in agreement with the absence of flagellin observed in these mutants (Fig. 1A). The role of CAP and H-NS in the initiation of fliC transcription was investigated by measuring the activity of a cat-fliC transcriptional fusion in crp and/or hns strains. The strains were grown to mid-exponential phase, which corresponds to the highest level of flagellar-gene expression (46). More than a 10-fold decrease in fliC transcription was observed in crp and hns strains, compared to the level measured in the wild-type strain (Fig. 1C).
FliC synthesis requires the expression of fliA, encoding the specific flagellar sigma factor, located at the second level in the flagellar hierarchy. Therefore, to study the role of H-NS and CAP in flagellar-gene expression, we constructed transcriptional fusions between the cat gene and the fliA gene, encoding the specific flagellar sigma factor, as well as fliL and flhB, two genes also located at the second level in the ordered cascade and regulated by the FlhDC regulatory proteins (33). In comparison with the wild-type strain, more than a 10-fold decrease in cat activity was measured in the presence of hns and/or crp mutations (data not shown). This suggests that both regulators mainly control flagellar-gene expression by affecting flhDC located at the first level of the cascade. To test this hypothesis, we examined motile behavior in crp and hns mutants in the presence of a plasmid overexpressing in trans the flhDC master operon (Table 2). Compared with their mutant counterpart, motility was restored, at least in part, within both mutant strains in the presence of plasmid pPM61, a ColE1 derivative. Such a reversion of the motility defect by increasing the gene dosage of the flhDC operon has been observed in a dnaK mutant (52) or under adverse conditions (51). Taken together, these observations suggest that flhDC constitutes the major target of CRP and H-NS, with regard to regulation in flagellar-gene expression.
TABLE 2.
Motility of E. coli strains on semisolid agar plates
| Strain genotype | Plasmid (protein) | Motilitya |
|---|---|---|
| Wild-type | None | 30 ± 3 |
| hns-1001 | None | 3 ± 1 |
| hns-1001 | pPM61 (FlhD and FlhC) | 28 ± 3 |
| crp::Sm | None | 3 ± 1 |
| crp::Sm | pPM61 (FlhD and FlhC) | 8 ± 1 |
Expressed as the diameter of the swarming ring (in millimeters) after 15 h at 30°C. The data are the mean values ± standard deviations of three independent experiments.
Determination of the flhDC transcriptional start site.
The location of the flhDC transcription initiation site has been previously investigated by primer extension analysis, but multiple transcriptional start sites were identified. These observations did not allow the unambiguous identification of any promoter sequence (53). Moreover, FlhD synthesis could initiate at a GTG or at an ATG located 3 codons downstream (5). To clarify these points, and in particular, to determine the translational initiation codon precisely, purified FlhD protein was analyzed by mass spectrometry (41a). The result, i.e., an experimental molecular mass of 13.317 kDa, provides evidence that flhD translation initiates at the ATG. A putative ribosome binding site was identified upstream from this translational start codon (Fig. 2).
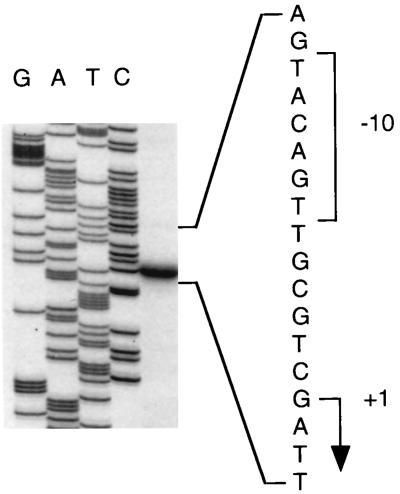
To determine the transcription initiation site, primer extension experiments were performed with RNA isolated from a wild-type strain expressing part of the flhDC operon from plasmid pDIA551 (Table 1) and primer O3 containing the translational start site. A major band for transcription initiation was detected (Fig. 3), which indicates that transcription of flhDC arises from the G residue located 198 nucleotides upstream from the ATG start codon (Fig. 2). Hexamers (−35 and −10) showing 50 and 67% similarity, respectively, with the ς70 consensus, were identified upstream from the transcriptional start site. These two boxes are separated by a 17-bp spacer (Fig. 2). The sizes of transcripts obtained by in vitro transcription experiments and primer extension experiments performed from these transcripts further confirmed the location of the +1 site (see below). Finally, the location of the transcriptional start site was in full agreement with that of a single-stranded region within the open complex. A characteristic permanganate reactivity with single-stranded thymine residues was indeed observed at positions −11 and −9 at the noncoding strand (data not shown).
FIG. 3.
Identification of the flhDC transcriptional start site. Primer extension analysis was performed with RNA extracted from a wild-type strain carrying plasmid pDIA551. As a reference, a DNA sequencing ladder is shown (lanes GATC). The sequence is complementary to the strand shown to the right and was obtained with the same primer used for primer extension. The −10 box and the transcription start point (+1) are indicated by a bracket and an arrow, respectively.
Identification of CAP and H-NS binding sites in the flhDC promoter region.
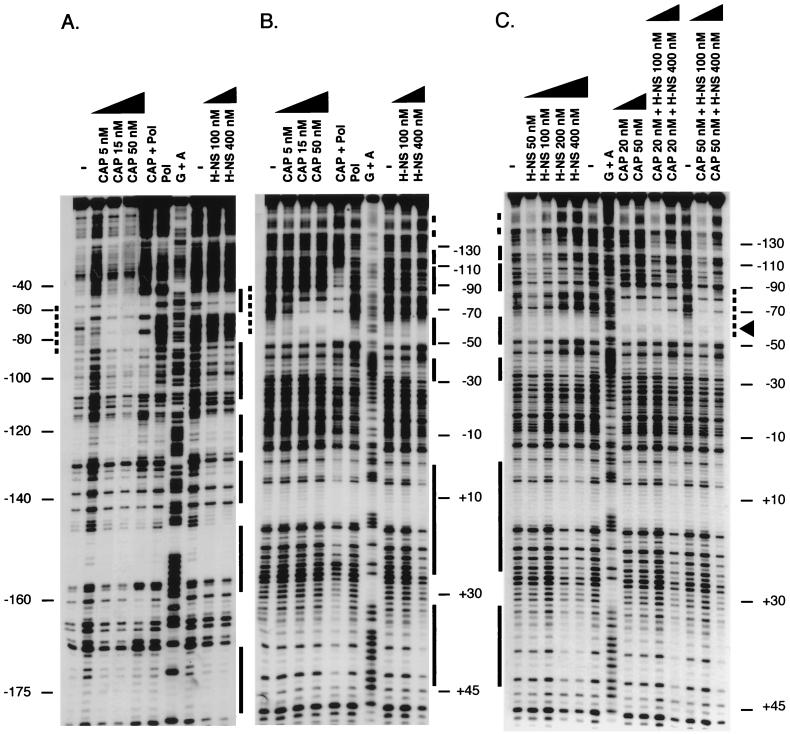
To determine the mechanism of regulation by the cAMP-CAP complex and H-NS, and their abilities to bind the flhDC promoter region, gel retardation experiments were performed with purified proteins. Plasmid pDIA525 carrying the flhDC promoter was digested by different restriction enzymes to generate various DNA fragments used as competitors for binding to both regulators. A preferential binding of H-NS to the flhDC promoter was observed when the concentration of H-NS reached 2 μM (Fig. 4). In addition, the 191-bp DNA fragment corresponding to the bla promoter was also retarded by H-NS, in agreement with previous results (6, 34, 62). Similarly, the 301-bp fragment corresponding to the flhDC promoter region was found to be specifically retarded in the presence of the cAMP-CAP complex (Fig. 4). Indeed, a 15 nM concentration of CAP protein was sufficient to promote a significant retardation in the electrophoretic mobility of this fragment. At a 50 nM concentration of CAP, a full retardation of the flhDC promoter region was observed. A competitive gel shift assay was also performed with a PCR-amplified DNA fragment encompassing the flhDC promoter (positions −213 to +14 with respect to the transcriptional start site) and the region extending downstream of the +1 transcriptional start site (position +14 with respect to the transcriptional start site to the ATG codon), respectively (Fig. 2). After amplification, this fragment was restricted by the DdeI restriction enzyme to generate two DNA fragments of 200 and 235 bp. The sole DNA fragment corresponding to the flhDC promoter (positions −213 to +14 with respect to the +1 site) was specifically shifted in the presence of CAP. The electrophoretic mobility of this DNA-protein complex was further retarded in the presence of H-NS. This suggests that both regulators were able to bind together to the flhDC promoter region. Strikingly, in the presence of H-NS alone, a similar retardation was observed for both DNA fragments (data not shown), suggesting that H-NS could also bind to a region downstream of the +1 site. These results are in agreement with those obtained by footprinting experiments (see below).
FIG. 4.
Competitive gel retardation assay with H-NS or CAP and restriction fragments derived from plasmid pDIA525. This plasmid, containing the promoter region of flhDC, was cleaved by EcoRI, HindIII, NdeI, and SspI, and DNA fragments were incubated with the indicated concentrations of each protein. After protein-DNA complex formation, the fragments were resolved on a 3% MetaPhor agarose gel. The upper arrow indicates the position of the 301-bp fragment containing the flhDC promoter; the lower arrow indicates the position of the fragment containing the 191-bp bla promoter; the third DNA fragment serves as a negative control in the experiment. In the presence of CAP, the reaction mixture and running buffer both contained 200 μM cAMP. The left-hand lanes contain the 1-kb DNA ladders (Gibco BRL).
To determine the precise location of CAP and H-NS binding sites in the flhDC regulatory region, DNase I footprint experiments were performed on the flhDC promoter (residues −213 to +78 with respect to the +1 transcriptional start site). The cAMP-CAP complex protected a region between positions −56 and −83 from DNase I cleavage (Fig. 5A and B). This region contains a CAP binding site consensus sequence (positions −61 to −82) centered at position −71.5 with respect to the transcription start site. The same DNA fragment was used in footprinting experiments with H-NS. Unlike CAP, several binding sites were identified on the flhDC promoter, i.e., −178 to −170, −158 to −148, −139 to −130, −126 to −116, −110 to −85, −64 to −50, −40 to −27, −1 to +26, and +32 to +44 with respect to the +1 site (Fig. 5). The region extending from −64 to −50 was protected by H-NS alone and in part by CRP alone (Fig. 5C). In contrast, in the presence of both regulators, a new pattern was observed. Indeed, bands −58 and −59 remained visible while bands −54, −55, and −56 were no longer detected. This indicates that the two proteins together are able to bind the same DNA fragment, in agreement with what we observed in gel retardation experiments (data not shown). Such a modification in the protection pattern has been reported as a consequence of the simultaneous binding of RNA polymerase and cAMP-CAP to the lacUV5 promoter region (29). Moreover, the binding of CAP to its site did not alter H-NS binding to the sites identified in the flhDC regulatory region, as shown by H-NS footprinting observed downstream of the CAP binding site, i.e., −40 to −27, −1 to +26, and +32 to +44 (Fig. 5C). Finally, DNase footprint experiments performed on the region extending from the +1 site to the ATG start codon revealed that H-NS is able to bind to several sites, i.e., +75 to +84, +104 to +119, +134 to +144, and +162 to +174 (Fig. 2).
FIG. 5.
Analysis of CAP and H-NS binding to the flhDC promoter by DNase I footprinting assays. (A) The labeled DNA fragment represents the coding strand; (B) DNA fragment labeled at the noncoding strand; (C) binding to the noncoding strand was examined in the presence of both regulators. The CAP binding sites are indicated by dotted lines. The regions protected by H-NS are marked by solid lines. The region where both regulators interact is highlighted by an arrowhead. Lanes G+A contain the products of a sequencing reaction used to determine the coordinates (to the left of panel A and to the right of panels B and C) of the regions protected against DNase I cleavage relative to the transcription start site. Above the lanes, the CAP or H-NS concentrations used in the experiment are indicated. Pol, RNA polymerase was present in the reaction mixture; −, no protein was present.
The region downstream of the +1 transcriptional start site is required in activation by H-NS but not by CRP.
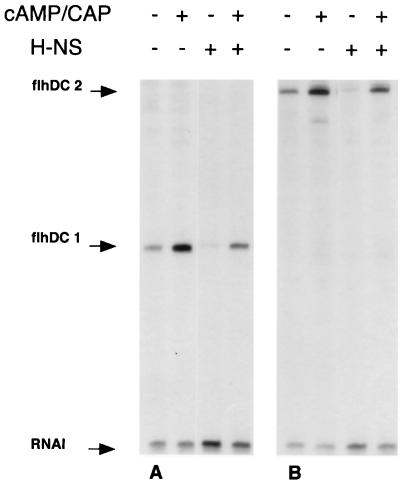
In vitro transcription experiments were performed with plasmid pDIA525 (Table 1) carrying the flhDC promoter region. A 169-bp transcript was observed (Fig. 6A), in agreement with the position of the transcriptional start site (Fig. 3). In the presence of 50 nM cAMP-CAP complex, in vitro transcription of the master operon was increased more than threefold. In contrast, the presence of 334 nM H-NS in the reaction mixture resulted in a sevenfold decrease in flhDC transcription. When H-NS and CAP were both present, the in vitro transcriptional level was close to that observed in the absence of both regulators (Fig. 6A). Similar results were obtained with plasmid pDIA546 synthesizing a 271-bp transcript encompassing the whole flhDC regulatory region extending to the ATG start codon (Fig. 6B).
FIG. 6.
In vitro transcription assay by RNA polymerase from flhDC promoter. Supercoiled DNA of plasmids pDIA525 and pDIA546 was incubated with RNA polymerase in the presence (+) or the absence (−) of cAMP-CAP complex at 50 nM and/or H-NS protein at 334 nM. Samples were subjected to electrophoresis on 7% sequencing gels. The flhDC1 and flhDC2 transcripts originating from the flhDC promoter (169 and 271 nucleotides, respectively) and from the RNA-I promoter located on the same plasmids (108 nucleotides) are indicated by arrows. This result is representative of three independent experiments.
To investigate the discrepancy between the repressor effect of H-NS in vitro (Fig. 6) and its activator effect in vivo (Fig. 1), we constructed two transcriptional fusions by using the cat reporter gene. The flhDC promoter (residues −213 to +78 with respect to the +1 transcriptional start site) and the whole regulatory region (residues −213 to +205) were cloned into pKK232-8, giving pDIA528 and pDIA545, respectively. While a fivefold decrease in enzymatic activity was observed from plasmid pDIA528 in the crp mutant (Table 3), more than a twofold increase in cat activity was measured in the hns strain, suggesting that H-NS represses the activity of the flhDC promoter in vivo. Moreover, the cat activity was restored to a level close to that in the wild type in an hns crp double mutant. These results are in agreement with those obtained in vitro (Fig. 6A). In contrast, up to a 100-fold decrease in cat activity was measured from the fusion carried by plasmid pDIA545 in a crp mutant and in an hns crp double mutant. More importantly, a threefold decrease in activity was observed in the hns strain, suggesting that the in vivo flhDC activation by H-NS requires the entire regulatory region encompassing both the flhDC promoter and the region extending downstream of the +1 transcriptional start site to the ATG translational codon. Similar results were obtained with a flhDC-lacZ transcriptional fusion located on the chromosome (data not shown).
TABLE 3.
Effect of hns and/or crp mutations on flhDC-cat transcriptional fusion activity
| Transcriptional fusion [gene (regulatory region)b and plasmid] | CATa activity (Uc) in strain (genotype):
|
|||
|---|---|---|---|---|
| PS2209 (wild type) | PS2652 (hns-1001) | BE1818 (crp::Sm) | BE1819 (hns crp) | |
| flhDC (nt −213 to +78), pDIA528 | 2,124 ± 82 | 4,943 ± 144 | 399 ± 17 | 1,629 ± 159 |
| flhDC (nt −213 to +205), pDIA545 | 3,260 ± 188 | 1,158 ± 67 | 30 ± 4 | 48 ± 2 |
CAT, chloramphenicol acetyltransferase.
nt, nucleotide.
U = 1 μmol of chloramphenicol acetylated per min per μg of protein.
Activation by cAMP-CAP complex of the flhDC master operon results from a direct interaction with RNA polymerase.
Promoter mutations, in particular, those in the lac promoter, are known to release transcription from the requirement for cAMP and CAP. Mutant strains, known as cfs (constitutive flagellar synthesis), synthesize flagella in a CAP-independent manner (54). To determine whether this phenotype was associated with a mutation located in the flhDC promoter region, we examined its nucleotide sequence in a cfs mutant strain (57). We observed two nucleotide substitutions, i.e., an A→C transversion close to the CAP binding site and a C→T transition in the −10 box. This substitution increases the homology of the −10 region with the Pribnow box consensus sequence (Fig. 2). All attempts to isolate similar mutants showing an H-NS-independent swarming in hns strains were unsuccessful (unpublished results).
In most cases, the cAMP-CAP complex activates transcription by recruiting RNA polymerase through the activating region I (residues 156 to 162) (12). We wanted to confirm that the CAP-dependent activation of the flhDC operon resulted from a similar mechanism. Therefore, we complemented the loss of motility in a crp mutant by wild-type and two mutated CAP proteins carrying amino acid substitution T158A or H159L in activating region I (15). In contrast to the wild-type CAP, both mutated proteins were unable to complement the nonmotile phenotype in the crp mutant (Table 4). Taken together, these results suggest that the cAMP-CAP complex positively regulates expression of the flagellar master operon by interaction with RNA polymerase.
TABLE 4.
Motility of E. coli strains expressing wild-type or mutated CAP protein on semisolid agar plates
| Strain genotype | Plasmid (protein) | Motilitya |
|---|---|---|
| Wild-type | None | 30 ± 3 |
| crp::Sm | None | 3 ± 1 |
| crp::Sm | pDCRP (CAP) | 21 ± 2 |
| crp::Sm | pDCRP-T158A (mutated CAP) | 3 ± 1 |
| crp::Sm | pDCRP-H159L (mutated CAP) | 3 ± 1 |
Expressed as the diameter of the swarming ring (in millimeters) after 15 h at 30°C. The data are the mean values ± standard deviations of three independent experiments.
DISCUSSION
One of the most striking features of microorganisms is their ability to grow under a wide range of osmolarity, temperature, or nutrient availability conditions. To survive under detrimental conditions, bacteria have acquired the ability to adapt their structures and physiologies rapidly. These mechanisms are based on the existence of multiple regulatory networks in which genes are regulated in a coordinate manner in response to environmental stimuli. Motility and chemotaxis are processes known to be affected by numerous factors, e.g., salts and temperature (32). This suggests that flagellum biosynthesis is a complex process involving multiple controls on gene expression.
In the present study, we attempted to determine more precisely the role of H-NS and cAMP-CAP on flagellar gene expression and, in particular, on the flhDC master operon. We showed that mutations affecting hns or crp resulted in up to a 100-fold decrease in flhDC-cat expression (Table 3). Although the decrease in cat activity was less in the hns mutant than in the crp strain, it is still sufficient to explain the complete lack of motility associated with hns mutation in E. coli (7, 14, 55). First, the absence of flagellin synthesis in hns and crp mutants resulted from a similar strongly reduced expression in fliC, the flagellin structural gene (Fig. 1), and in fliA, encoding the flagellar sigma factor (data not shown). These data are in agreement with a previous electron microscopy observation showing that E. coli mutant strains deficient in H-NS or CAP lack flagella (7, 54). Second, it has been previously demonstrated that mutations affecting the level of either the heat shock protein DnaK (52) or the initiating factor of chromosomal DNA replication, DnaA (40), or mutations resulting in a modification of the membrane phospholipid content (50), as well as some adverse conditions such as temperature (51), can lead to a strong reduction in fliA and/or fliC transcription despite a moderate reduction in flhDC expression. Third, a high level of flhDC expression from a multicopy plasmid was able to overcome the moderate repression of the master operon in the hns strain (Table 3), resulting in a restoration of motility in this mutant (Table 2). Although, it has been shown that H-NS can interact with FliG, a protein of the flagellar motor (14, 37), our results provide evidence that the altered swarming behavior in such an hns mutant mainly results from the control by H-NS of flagellum synthesis rather than of its functioning.
To investigate the molecular mechanism by which H-NS protein and the cAMP-CAP complex control the flhDC operon, we performed gel shift assays and DNase I footprinting experiments. These experiments demonstrated that both proteins are able to bind to the regulatory region of the master operon in vitro. A unique CAP binding site was identified centered at position −71.5 upstream from the +1 transcription start site (Fig. 5). Its location relative to the +1 site and its strong homology with the CAP binding site consensus sequence suggest that the binding of the cAMP-CAP complex to this site leads to flhDC activation (12, 53). On the other hand, several H-NS binding sites were identified in the flhDC regulatory region (Fig. 5). Such multiple binding sites have been previously observed with H-NS on the promoter region of different E. coli genes, such as hns (17), proU (34), lac (47), rrnB (56), and virF (16).
H-NS was shown to positively affect the synthesis of flagella (Fig. 1), although it has usually been described as a general repressor of transcription (59). Therefore, it was of interest to know whether the positive effect of H-NS on flhDC expression was a direct consequence of its binding to the promoter region of the master operon. In vitro transcription experiments performed with H-NS showed that this regulator represses in vitro expression of the master operon (Fig. 6). In contrast, we demonstrated that CAP is able to promote flhDC activation (Fig. 6). Such a positive regulatory role is further supported by characterization of a cfs mutation. The nucleotide substitutions observed in the promoter region of the cfs mutant, known to synthesize flagella in a CAP-independent manner (Fig. 2), are somewhat reminiscent of the mutations in the lacUV5 promoter, known to restore promoter activity in the absence of the cAMP-CAP complex. Moreover, CAP proteins carrying mutations in activating region I, a region involved in interactions between CAP and the C-terminal part of the RNA polymerase α subunit, were unable to complement the loss of motility in crp mutant strains (Table 4). This provides evidence that the positive control of CAP results from a direct interaction between the cAMP-CAP complex and RNA polymerase.
Recently, interactions between H-NS and various regulators have been demonstrated. For example, H-NS is known to interfere with ςs and Lrp in transcription of the osmC gene (8) and with FNR and CAP in the regulation of cai and fix operons (11). In some cases, the role of the activator, e.g., CAP in the regulation of bgl and pap operons (18, 41) and ςs in the regulation of csgBA (2) has been thought to relieve, at least in part, the repression mediated by H-NS. Using in vivo (Table 3) and in vitro (Fig. 6) assays, we demonstrated that CAP relieves the repression mediated by H-NS on the activity of the sole flhDC promoter region. However, this mechanism is not sufficient to explain the complex regulation affecting the flhDC master operon, as suggested by our previous observation that cfs mutants synthesizing flagella in a cAMP-CAP-independent manner remain nonflagellate in an hns background (7). First, gel retardation and footprinting experiments demonstrated that H-NS and CAP are able to bind together to the flhDC promoter fragment, without altering significantly the binding of the other regulator (Fig. 5). Moreover, an hns crp double mutant was completely deficient in flagellin accumulation and in fliC mRNA synthesis (Fig. 1), in agreement with the lack of motility recently observed in a similar mutant in S. typhimurium (30). Finally, H-NS exerted a positive control in vivo of the full-length flhDC regulatory region extending to the translational ATG start codon (Table 3).
During the last decade, the regulation of the proU operon has been extensively studied, in particular, with regard to H-NS (21). However, it has recently been demonstrated that the mechanism of proU repression by H-NS cannot be explained solely by the binding of the regulator to the promoter region (28). Similarly, our data provide evidence that the in vitro binding of H-NS to the flhDC regulatory region (Fig. 6) is not sufficient to explain its clear positive control observed in vivo on flagellar-gene expression (Fig. 1 and Table 3). One hypothesis would be that H-NS acts indirectly on flhDC by regulating the synthesis of, or by interacting with another protein required for full expression of, the master operon. The observation that the positive control by CAP was also modulated by the region extending from the +1 transcriptional start site to the ATG translational codon (Table 3) further supports the involvement of an ancillary factor in the regulation of flhDC expression in vivo. In any case, an understanding of the mechanism by which H-NS affects expression of the flagellar master operon, the first example of positive control studied so far at the molecular level, will require further investigation.
The function of the flagellum-chemotaxis regulon seems to play an important role in adaptation to stressful environmental conditions. In this respect, the master operon flhDC constitutes a good example of stress-responsive genes. The regulation of its expression requires a complex network involving several regulators. In addition to CAP and H-NS, HU, Fis, and/or Lrp have been suggested to affect the flagellum-chemotaxis regulon in E. coli (42), S. typhimurium (43), and Proteus mirabilis (25). Such a multiple control of motility in enterobacteria could be the basis of a fine tuning of flagellar-gene expression in response to environmental challenges.
ACKNOWLEDGMENTS
We are grateful to H. Buc for critical reading of the manuscript and to T. Pugsley for helpful advice. We thank A. Campos and P. Matsumura for providing us with purified FlhD protein. We also thank A. Namane for analysis of FlhD protein by mass spectrometry.
Financial support came from the Institut Pasteur and the Centre National de la Recherche Scientifique (URA 1129).
REFERENCES
- 1.Akerley B J, Cotter P A, Miller J F. Ectopic expression of the flagellar regulon alters development of the Bordetella-host interaction. Cell. 1995;80:611–620. doi: 10.1016/0092-8674(95)90515-4. [DOI] [PubMed] [Google Scholar]
- 2.Arnqvist A, Olsén A, Normark S. ςS-dependent growth-phase induction of the csgBA promoter in Escherichia coli can be achieved in vivo by ς70 in the absence of the nucleoid-associated protein H-NS. Mol Microbiol. 1994;13:1021–1032. doi: 10.1111/j.1365-2958.1994.tb00493.x. [DOI] [PubMed] [Google Scholar]
- 3.Atlung T, Ingmer H. H-NS: a modulator of environmentally regulated gene expression. Mol Microbiol. 1997;24:7–17. doi: 10.1046/j.1365-2958.1997.3151679.x. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Short protocols in molecular biology: a compendium of methods from current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1992. [Google Scholar]
- 5.Bartlett D H, Frantz B B, Matsumura P. Flagellar transcriptional activators FlbB and FlaI: gene sequences and 5′ consensus sequences of operons under FlbB and FlaI control. J Bacteriol. 1988;170:1575–1581. doi: 10.1128/jb.170.4.1575-1581.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertin P, Benhabiles N, Krin E, Laurent-Winter C, Tendeng C, Turlin E, Thomas A, Danchin A, Brasseur R. The structural and functional organization of H-NS-like proteins is evolutionarily conserved in Gram-negative bacteria. Mol Microbiol. 1999;31:319–330. doi: 10.1046/j.1365-2958.1999.01176.x. [DOI] [PubMed] [Google Scholar]
- 7.Bertin P, Terao E, Lee E H, Lejeune P, Colson C, Danchin A, Collatz E. The H-NS protein is involved in the biogenesis of flagella in Escherichia coli. J Bacteriol. 1994;176:5537–5540. doi: 10.1128/jb.176.17.5537-5540.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouvier J, Gordia S, Kampmann G, Lange R, Hengge-Aronis R, Gutierrez C. Interplay between global regulators of Escherichia coli: effect of RpoS, Lrp and H-NS on transcription of the gene osmC. Mol Microbiol. 1998;28:971–980. doi: 10.1046/j.1365-2958.1998.00855.x. [DOI] [PubMed] [Google Scholar]
- 9.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 10.Bruni C B, Colantuoni V, Sbordone L, Cortese R, Blasi F. Biochemical and regulatory properties of Escherichia coli K-12 his mutants. J Bacteriol. 1977;130:4–10. doi: 10.1128/jb.130.1.4-10.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buchet A, Eichler K, Mandrand-Berthelot M A. Regulation of the carnitine pathway in Escherichia coli: investigation of the cai-fix divergent promoter region. J Bacteriol. 1998;180:2599–2608. doi: 10.1128/jb.180.10.2599-2608.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Busby S, Kolb A. The CAP modulon. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. R. C. New York, N.Y: Landes; 1996. pp. 255–279. [Google Scholar]
- 13.Ciacci-Woolwine F, Blomfield I C, Richardson S H, Mizel S B. Salmonella flagellin induces tumor necrosis factor alpha in a human promocytic cell line. Infect Immun. 1998;6:1127–1134. doi: 10.1128/iai.66.3.1127-1134.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donato G M, Kawula T H. Enhanced binding of altered H-NS protein to flagellar rotor protein FliG causes increased flagellar rotational speed and hypermotility in Escherichia coli. J Biol Chem. 1998;273:24030–24036. doi: 10.1074/jbc.273.37.24030. [DOI] [PubMed] [Google Scholar]
- 15.Ebright R H. Transcription activation at class I CAP-dependent promoters. Mol Microbiol. 1993;8:797–802. doi: 10.1111/j.1365-2958.1993.tb01626.x. [DOI] [PubMed] [Google Scholar]
- 16.Falconi M. Thermoregulation of Shigella and Escherichia coli EIEC pathogenicity. A temperature-dependent structural transition of DNA modulates accessibility of virF promoter to transcriptional repressor H-NS. EMBO J. 1998;17:7033–7043. doi: 10.1093/emboj/17.23.7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falconi M, Higgins N P, Spurio R, Pon C L, Gualerzi C O. Expression of the gene encoding the major bacterial nucleoid protein H-NS is subject to transcriptional auto-repression. Mol Microbiol. 1993;10:273–282. doi: 10.1111/j.1365-2958.1993.tb01953.x. [DOI] [PubMed] [Google Scholar]
- 18.Forsman K, Sondén B, Görannson M, Uhlin B E. Antirepression function in Escherichia coli for the cAMP-cAMP receptor protein transcriptional activator. Proc Natl Acad Sci USA. 1992;89:9880–9884. doi: 10.1073/pnas.89.20.9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gardel C L, Mekalanos J J. Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect Immun. 1996;64:2246–2255. doi: 10.1128/iai.64.6.2246-2255.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghosaini L R, Brown A M, Sturtevant J M. Scanning calorimetric study of the thermal unfolding of catabolite activator protein from Escherichia coli in the absence and presence of cyclic mononucleotides. Biochemistry. 1988;27:5257–5261. doi: 10.1021/bi00414a046. [DOI] [PubMed] [Google Scholar]
- 21.Gowrishankar J, Manna D. How is osmotic regulation of transcription of the Escherichia coli proU operon achieved? A review and a model. Genetica. 1996;97:363–378. doi: 10.1007/BF00055322. [DOI] [PubMed] [Google Scholar]
- 22.Goyard S, Bertin P. Characterization of BpH3, an H-NS like protein in Bordetella pertussis. Mol Microbiol. 1997;24:815–823. doi: 10.1046/j.1365-2958.1997.3891753.x. [DOI] [PubMed] [Google Scholar]
- 23.Graf J, Dunlap P V, Ruby E G. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J Bacteriol. 1994;176:6986–6991. doi: 10.1128/jb.176.22.6986-6991.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hager D A, Jin D J, Burgess R R. Use of Mono Q high-resolution ion exchange chromatography to obtain highly pure and active Escherichia coli RNA polymerase. Biochemistry. 1990;29:7890–7894. doi: 10.1021/bi00486a016. [DOI] [PubMed] [Google Scholar]
- 25.Hay N A, Tipper D J, Gygi D, Hughes C. A nonswarming mutant of Proteus mirabilis lacks the Lrp global transcriptional regulator. J Bacteriol. 1997;179:4741–4746. doi: 10.1128/jb.179.15.4741-4746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinton J C D, Santos D S, Seirafi A, Hulton C J, Pavitt G D, Higgins C F. Expression and mutational analysis of the nucleoid-associated protein H-NS of Salmonella typhimurium. Mol Microbiol. 1992;6:2327–2337. doi: 10.1111/j.1365-2958.1992.tb01408.x. [DOI] [PubMed] [Google Scholar]
- 27.Ingham C, Buechner M, Adler J. Effect of outer membrane permeability on chemotaxis in Escherichia coli. J Bacteriol. 1990;172:3577–3583. doi: 10.1128/jb.172.7.3577-3583.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jordi B J A M, Fielder A, Burns C M, Hinton J C D, Dover N, Ussery D W, Higgins C F. DNA binding is not sufficient for H-NS-mediated repression of proU expression. J Biol Chem. 1997;272:12083–12090. doi: 10.1074/jbc.272.18.12083. [DOI] [PubMed] [Google Scholar]
- 29.Kolb A, Igarashi K, Ishihama A, Lavigne M, Buckle M, Buc H. E. coli RNA polymerase, deleted in C-terminal part of its α-subunit, interacts differently with the cAMP-CRP complex at the lac P1 and the gal P1 promoter. Nucleic Acids Res. 1993;21:319–326. doi: 10.1093/nar/21.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a.Krin, E. Unpublished data.
- 30.Kutsukake K. Autogenous and global control of flagellar master operon, flhD, in Salmonella typhimurium. Mol Gen Genet. 1997;254:440–448. doi: 10.1007/s004380050437. [DOI] [PubMed] [Google Scholar]
- 31.Laurent-Winter C, Ngo S, Danchin A, Bertin P. Role of Escherichia coli histone-like nucleoid-structuring protein in bacterial metabolism and stress response. Eur J Biochem. 1997;244:767–773. doi: 10.1111/j.1432-1033.1997.00767.x. [DOI] [PubMed] [Google Scholar]
- 32.Li C, Louise C J, Shi W, Adler J. Adverse conditions which cause lack of flagella in Escherichia coli. J Bacteriol. 1993;175:2229–2235. doi: 10.1128/jb.175.8.2229-2235.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X, Matsumura P. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J Bacteriol. 1994;176:7345–7351. doi: 10.1128/jb.176.23.7345-7351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lucht J M, Dersch P, Kempf B, Bremer E. Interactions of the nucleoid-associated DNA-binding protein H-NS with the regulatory region of the osmotically controlled proU operon of Escherichia coli. J Biol Chem. 1994;269:6578–6586. [PubMed] [Google Scholar]
- 35.Macnab R M. Flagella and motility. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 1. Washington, D.C.: American Society for Microbiology; 1996. pp. 123–145. [Google Scholar]
- 36.Marschall C, Labrousse V, Kreimer M, Weichart D, Kolb A, Hengge-Aronis R. Molecular analysis of the regulation of csiD, a carbon starvation-inducible gene in Escherichia coli, that is exclusively dependent on ςs and requires activation by cAMP-CRP. J Mol Biol. 1998;276:339–353. doi: 10.1006/jmbi.1997.1533. [DOI] [PubMed] [Google Scholar]
- 37.Marykwas D L, Schmidt S A, Berg H C. Interacting components of the flagellar motor of Escherichia coli revealed by two-hybrid system in yeast. J Mol Biol. 1996;256:564–576. doi: 10.1006/jmbi.1996.0109. [DOI] [PubMed] [Google Scholar]
- 38.Maxam A M, Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci USA. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 40.Mizushima T, Koyanagi R, Katayama T, Miki T, Sekimizu K. Decrease in expression of the master operon of flagellin synthesis in a dnA46 mutant of Escherichia coli. Biol Pharm Bull. 1997;20:327–331. doi: 10.1248/bpb.20.327. [DOI] [PubMed] [Google Scholar]
- 41.Mukerji M, Mahadevan M. Characterization of the negative elements involved in silencing the bgl operon of Escherichia coli: possible role for DNA gyrase, H-NS, and CRP-cAMP in regulation. Mol Microbiol. 1997;24:617–627. doi: 10.1046/j.1365-2958.1997.3621725.x. [DOI] [PubMed] [Google Scholar]
- 41a.Namane, A. Unpublished data.
- 42.Nishida S, Mizushima T, Miki T, Sekimizu K. Immotile phenotype of an Escherichia coli mutant lacking the histone-like protein HU. FEMS Microbiol Lett. 1997;150:297–301. doi: 10.1111/j.1574-6968.1997.tb10384.x. [DOI] [PubMed] [Google Scholar]
- 43.Osuna R, Lienau D, Hughes K T, Johnson R C. Sequence, regulation, and functions of fis in Salmonella typhimurium. J Bacteriol. 1995;177:2021–2032. doi: 10.1128/jb.177.8.2021-2032.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parker C T, Kloser A W, Schnaitman C A, Stein M A, Gottesman S, Gibson B W. Role of the rfaG and rfaP genes in determining the lipopolysaccharide core structure and cell surface properties of Escherichia coli K-12. J Bacteriol. 1992;174:2525–2538. doi: 10.1128/jb.174.8.2525-2538.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pratt L A, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 46.Prüß B M, Matsumura P. Cell cycle regulation of flagellar genes. J Bacteriol. 1997;179:5602–5604. doi: 10.1128/jb.179.17.5602-5604.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46a.Rimsky, S. Unpublished data.
- 47.Rimsky S, Spassky A. Sequence determinants for H1 binding on Escherichia coli lac and gal promoters. Biochemistry. 1990;29:3765–3771. doi: 10.1021/bi00467a024. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook J, Fritsch E F, Maniatis T, editors. Molecular cloning. A laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 49.Shaw W V. Chloramphenicol acetyltransferase from chloramphenicol resistant bacteria. Methods Enzymol. 1975;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- 50.Shi W, Bogdanov M, Dowhan W, Zusman D R. The pss and psd genes are required for motility and chemotaxis in Escherichia coli. J Bacteriol. 1993;175:7711–7714. doi: 10.1128/jb.175.23.7711-7714.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi W, Li C, Louise C J, Adler J. Mechanism of adverse conditions causing lack of flagella in Escherichia coli. J Bacteriol. 1993;175:2236–2240. doi: 10.1128/jb.175.8.2236-2240.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi W, Zhou Y, Wild J, Adler J, Gross C A. DnaK, DnaJ, and GrpE are required for flagellum synthesis in Escherichia coli. J Bacteriol. 1992;174:6256–6263. doi: 10.1128/jb.174.19.6256-6263.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shin S, Park C. Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J Bacteriol. 1995;177:4696–4702. doi: 10.1128/jb.177.16.4696-4702.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Silverman M, Simon M. Characterization of Escherichia coli flagellar mutants that are insensitive to catabolite repression. J Bacteriol. 1974;120:1196–1203. doi: 10.1128/jb.120.3.1196-1203.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Timchenko T, Bailone A, Devoret R. Btcd, a mouse protein that binds to curved DNA, can substitute in Escherichia coli for H-NS, a bacterial nucleoid protein. EMBO J. 1996;15:3986–3992. [PMC free article] [PubMed] [Google Scholar]
- 56.Tippner D, Afflerbach H, Bradaczek C, Wagner R. Evidence for a regulatory function of the histone-like Escherichia coli protein H-NS in ribosomal RNA synthesis. Mol Microbiol. 1994;11:589–604. doi: 10.1111/j.1365-2958.1994.tb00339.x. [DOI] [PubMed] [Google Scholar]
- 57.Vogler A P, Lengeler J W. Indirect role of adenylate cyclase and cyclic AMP in chemotaxis to phosphotransferase system carbohydrates in Escherichia coli K-12. J Bacteriol. 1987;169:593–599. doi: 10.1128/jb.169.2.593-599.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.West D, Williams R, Rhodius V, Bell A, Sharma N, Zou C, Fujita N, Ishihama A, Busby S. Interactions between the Escherichia coli cyclic AMP receptor protein and RNA polymerase at Class II promoters. Mol Microbiol. 1993;10:789–797. doi: 10.1111/j.1365-2958.1993.tb00949.x. [DOI] [PubMed] [Google Scholar]
- 59.Williams R M, Rimsky S. Molecular aspects of the E. coli nucleoid protein, H-NS: a central controller of gene regulatory networks. FEMS Microbiol Lett. 1997;156:175–185. doi: 10.1111/j.1574-6968.1997.tb12724.x. [DOI] [PubMed] [Google Scholar]
- 60.Williams R M, Rimsky S, Buc H. Probing the structure, function and interactions of the Escherichia coli H-NS and StpA proteins using dominant negative derivatives. J Bacteriol. 1996;178:4335–4343. doi: 10.1128/jb.178.15.4335-4343.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yokota T, Gots J. Requirement of adenosine 3′,5′-cyclic monophosphate for flagellation in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1970;103:513–516. doi: 10.1128/jb.103.2.513-516.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zuber F, Kotlars D, Rimsky S, Buc H. Modulated expression of promoters containing upstream curved DNA sequences by the Escherichia coli nucleoid protein H-NS. Mol Microbiol. 1994;12:231–240. doi: 10.1111/j.1365-2958.1994.tb01012.x. [DOI] [PubMed] [Google Scholar]