Abstract
Despite being more transmissible, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron variant only causes milder diseases in laboratory animals, often accompanied by a lower viral load compared with previous variants of concern. In this study, we report the structural basis for a robust interaction between the receptor-binding domain of the Omicron spike protein and mouse ACE2. We show that pseudovirus bearing the Omicron spike protein efficiently utilizes mouse ACE2 for entry. By comparing viral load and disease severity among laboratory mice infected by a natural Omicron variant or recombinant ancestral viruses bearing either the entire Omicron spike or only the N501Y/Q493R mutations in its spike, we find that mutations outside the spike protein in the Omicron variant may be responsible for the observed lower viral load. Together, our results imply that a post-entry block to the Omicron variant exists in laboratory mice.
Keywords: SARS-CoV-2, Omicron variant, attenuation, Balb/c mice, K18-hACE2
Graphical abstract
SARS-CoV-2 Omicron variant is known to be attenuated in laboratory animals. Liu et al. here report that the Omicron variant spike protein enables the virus to infect laboratory mice via its interaction with mouse ACE2. The attenuation of Omicron in K18-hACE2 transgenic mice, however, is attributed to mutations outside the spike region.
Introduction
The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron variant (B.1.1.529) has drastically changed the landscape of the coronavirus disease of 2019 (COVID-19) pandemic (Organization, 2021). Omicron has displaced the Delta variant in the United States and possibly the rest of the world. With 30+ changes in its spike protein, the Omicron variant has been shown to evade vaccine- or natural infection-elicited immunity (Cele et al., 2021; Pulliam et al., 2022). Within its receptor-binding domain (RBD), the Omicron variant harbors several amino acid substitutions, including S417N, T478K, E484A, Q493R, G496S, Q498R, N501Y, and Y505H, that potentially alter the species tropism of the virus. Among those, N501Y has appeared in the Alpha (B1.1.7), Beta (B1.351), and P.1 variants and has been demonstrated to enhance the binding of spike to both human and mouse ACE2 (Gu et al., 2020; Shuai et al., 2021; Xavier Montagutelli et al., 2021). The presence of N501Y along with additional mutations (K417M, E484K, Q493R, and Q498R) is associated with mouse adaptation by serial passage (Kibler et al., 2021; Lok-Yin Roy Wong et al., 2021; Muruato et al., 2021). Surprisingly, several recent studies reported an attenuated phenotype in laboratory animals (Halfmann et al., 2022; Katherine McMahan et al., 2022; Ryan et al., 2021; Abdelnabi et al., 2021; Shuai et al., 2022; Suzuki et al., 2022), raising a question of whether the Omicron variant robustly infects animal species.
Results
Omicron spike protein mediates efficient entry via ACE2 orthologs of multiple species
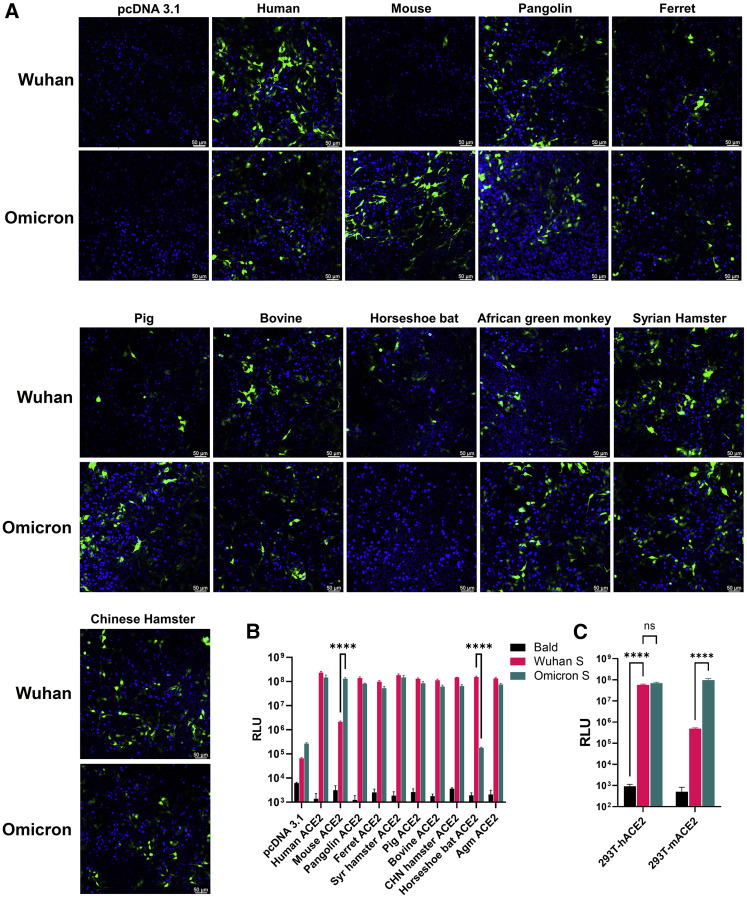
We packaged lentiviral-based pseudoviral particles that bear no spike protein (bald virus), the ancestral WA1 spike, and the Omicron spike protein. Green fluorescent protein (GFP) was the reporter gene of these pseudoviruses. We then added pseudoviruses to 293T cells that were transfected with DNA plasmids to express ACE2 of 10 species. As shown in Figure 1A, GFP-positive cells were readily visible. While the majority ACE2 homologs equally mediated WA1- or Omicron-pseudovirus infection, mouse ACE2 (mACE2) expression permitted massive infection by pseudovirus bearing the Omicron spike but not the WA1 spike. Horseshoe bat ACE2, however, only mediated entry of the pseudovirus bearing the WA1 spike. Similar observations were made when using pseudoviruses carrying the firefly luciferase (FLuc) reporter gene in transient transfections or in hACE2 and mACE2 stable cell lines (Figures 1B and 1C).
Figure 1.
Pseudovirus bearing the Omicron spike protein efficiently infects cells expressing mACE2
(A) 293T cells were transfected with ACE2 expression plasmids of 10 species and then infected by lentiviral pseudoparticles bearing the spike protein of either the Wuhan SARS-CoV-2 variant or the Omicron variant. Infected cells would be GFP positive (green). Blue dye-stained nuclei. Images shown here are representatives of two independent experiments.
(B) A similar experiment was performed as in (A), except pseudoviruses expressing firefly luciferase were employed.
(C) Infection of hACE2-and mACE2 stable 293T cells by pseudoviruses bearing the spike protein of either the Wuhan SARS-CoV-2 variant or the Omicron variant.
Results in each bar graph contain at least three technical replicates.
Structural basis of interaction between Omicron spike protein and mACE2
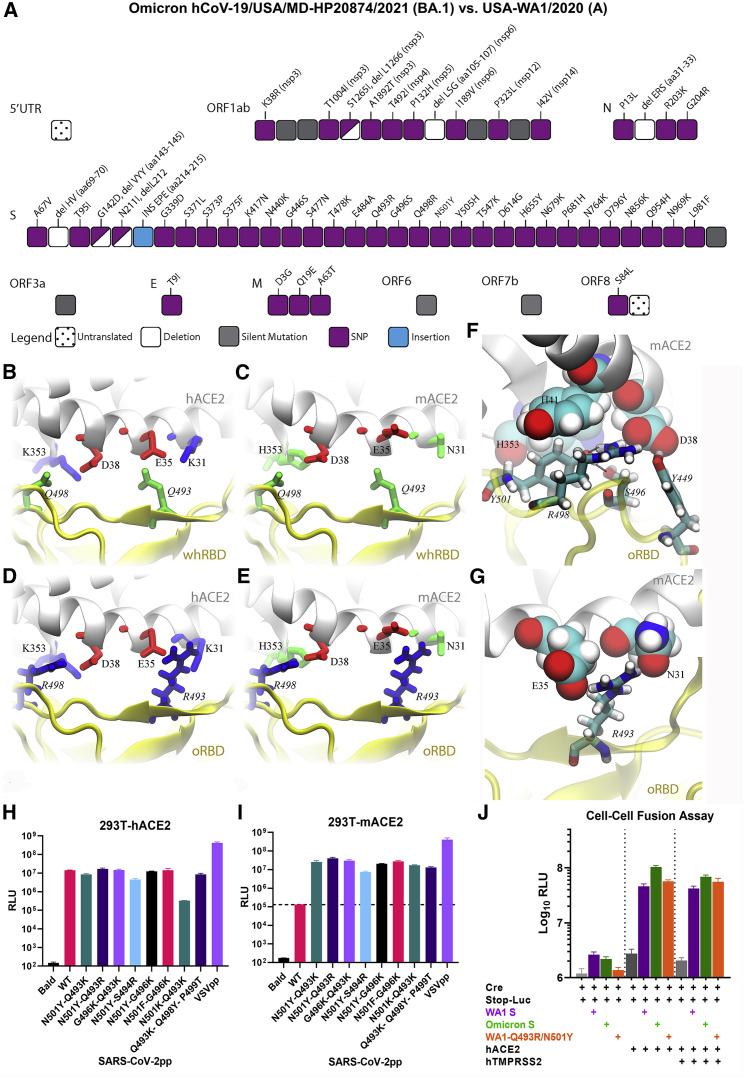
To understand the structural basis of the mACE2-Omicron RBD (oRBD) interaction, we performed all-atom molecular dynamics (MD) simulations in a physiology-like environment. Compared with the wide-type SARS-CoV-2, the Omicron variant contains 15 mutations in the RBD (Figure 2A), and many of them, including the two charged mutations Q493R and Q498R, are located inside the receptor-binding motif (RBM) that directly contacts ACE2. Generally, charged residues significantly impact the protein-protein binding interface. As shown in Figure 2B, two salt bridges (K353-D38 and E35-K31) reside at the binding surface of hACE2, while two neutral residues Q493 and Q498 are present on the Wuhan-like ancestral RBD (whRBD) surface. Besides interfacial hydrogen bonds, the two salt bridges are partially buried after the hACE2-whRBD binding, which is energetically favorable (Huynh, 2021). However, on the binding surface of the mACE2, negatively charged D38 and E35 are coordinated by two neutral residues H353 and N31, resulting in an overall charged surface (Figure 2C). When whRBD engages mACE2, two negatively charged residues (D38 and E35) will inevitably be buried at the binding interface, which is energetically unfavorable because the dielectric constant of a protein media is much less than that of water (Figure 2C). Different from whRBD, two positively charged residues (R493 and R498) are found in the RBM of oRBD. When hACE2 is bound with oRBD, as shown in Figure 2D, the net charge at the interface is +2 e because both R493 and R498 are buried (partially) at the interface. This interfacial arrangement is energetically unfavorable and may weaken the interfacial binding, but the highly favorable N501Y mutation strengthens the oRBD-hACE2 interaction. Importantly, oppositely charged oRBD and mACE2 surfaces can electrostatically attract each other, and the complex features two salt bridges (R493-E35 and R498-D38) buried at the interface with zero net charge (Figure 2E), which is energetically favorable. In support of this, a recent study reported that mACE2 binds to oRBD with a higher affinity than to RBDs of other variants (Cameroni et al., 2022).
Figure 2.
The spike protein of Omicron variant carries mutations that confer utility of mouse ACE2 for entry
(A) Illustration of all changes found in the Omicron isolate used in this study (hCoV-19/USA/MD-HP20874/2021) compared with USA-WA1/2020. Coincidentally, the Omicron spike protein in pseudoviruses carries an identical number of changes.
(B–E) Various coordinations of charged residues at the ACE2-RBD interface: (B) hACE2-wtRBD (buried charge: 0), (C) mACE2-wtRBD (buried charge: −2 e), (D) hACE2-oRBD (buried charge: +2 e), and (E) mACE2-oRBD (buried charge: 0).
(F and G) Detailed interfacial coordinations between mACE2 and oRBD.
(F) The interfacial binding near the salt bridge R498-D38.
(G) The interfacial binding near the salt bridge R493-E35.
(H and I) Pseudoviruses carrying WA1 spike with indicated combinations of mutations infected 293T-hACE2 or 293-mACE2 cell lines.
(J) Cell-cell fusion is mediated by WA1 spike or Omicron spike or the WA1-Q493R/N501Y spike protein.
Results in each bar graph contain at least three technical replicates. Data shown here are representatives of two independent experiments.
MD simulations of the mACE2-oRBD complex in the 0.15 M NaCl electrolyte also revealed an essential atomic coordination at the interface. Figure 2F illustrates that besides forming the salt bridge with D38 in mACE2, R498 also forms a hydrogen bond with Y501, which stabilizes the conformation of Y501. Additionally, S496 forms a hydrogen bond with Y501. The latter further forms the T-shape π-π stacking with H41 (mACE2) and another parallel one with H353 of mACE2. Note that the similar interaction between Y501 and H41 was also found in other ACE2-RBD complexes, such as hACE2 and the RBD of the Alpha variant (Luan et al., 2021), and is responsible for the enhanced interfacial interaction. Hence, in the case of mACE2-oRBD interaction, Y501 is in a locked conformation due to favorable interactions with H41 (mACE2), Y353 (mACE2), R498 (oRBD), and S496 (oRBD). Additionally, the salt bridge between E35 (mACE2) and R493 (oRBD) further enhances the local interfacial binding (Figure 2G).
Mutations found in Omicron spike conferred tropism in mice
The MD simulation implies that a buried charge inside the protein-protein interface reduces the binding affinity while a buried salt bridge enhances the interfacial binding affinity. To experimentally verify the hypothesis, we made several constructs expressing WA1 spike bearing the following combinations of mutations: N501Y-Q493K, N501Y-Q493R, G496K-Q493K, N501Y-S494R, N501Y-G496K, N501F-G496K, and N501K-Q493K. These mutations are expected to strengthen the RBD-mACE2 interaction through inducing salt bridges between K501/K496 of the RBD and D38 of mACE2 or between K493/R494 of RBD and E35 of mACE2. Additionally, a WA1 spike bearing Q493K/Q498Y/P499T triple mutations, which were found in a mouse-adapted virus, was included as positive control (Leist et al., 2020). Pseudoviruses bearing these spike proteins all infected 293T-mACE2 by at least two orders of magnitude compared with the WA1 spike (Figures 2H and 2I). These findings confirmed the flexibility of the RBD-ACE2 interface in dictating the species tropism. Notably, the Omicron spike protein and WA1 spike bearing Q493R/N501Y were as fusogenic as the ancestral WA1 spike in a cell-cell fusion assay (Figure 2J).
To confirm the above results in vivo, we introduced two mutations (i.e., Q493R/N501Y) into the spike protein of WA1/2020 genome using reverse genetics. The derived WA1-Q493R/N501Y was found to efficiently infect immunocompetent mice and caused weight loss in 12-month-old Blab/c mice with an inoculum of 5 × 104 plaque-forming units (PFUs) (Figures S1A and S1B). Compared wiht the ancestral WA1/2020 isolate, viral loads, measured by either quantitative real-time PCR or RNAscope, from WA1-Q493R/N501Y-infected mice were 2 to 3 logs higher (Figures S1C–S1F). Notably, the primary histopathology changes in the lungs of WA1-Q493R/N501Y-infected mice were immune cell infiltrates and limited areas of consolidation (Figures S1D–S1F). Consequently, infected animals displayed very mild disease manifestations such as ruffled fur, appearance of lethargy, and eye closure.
Omicron spike protein mediated efficient infection of laboratory mice
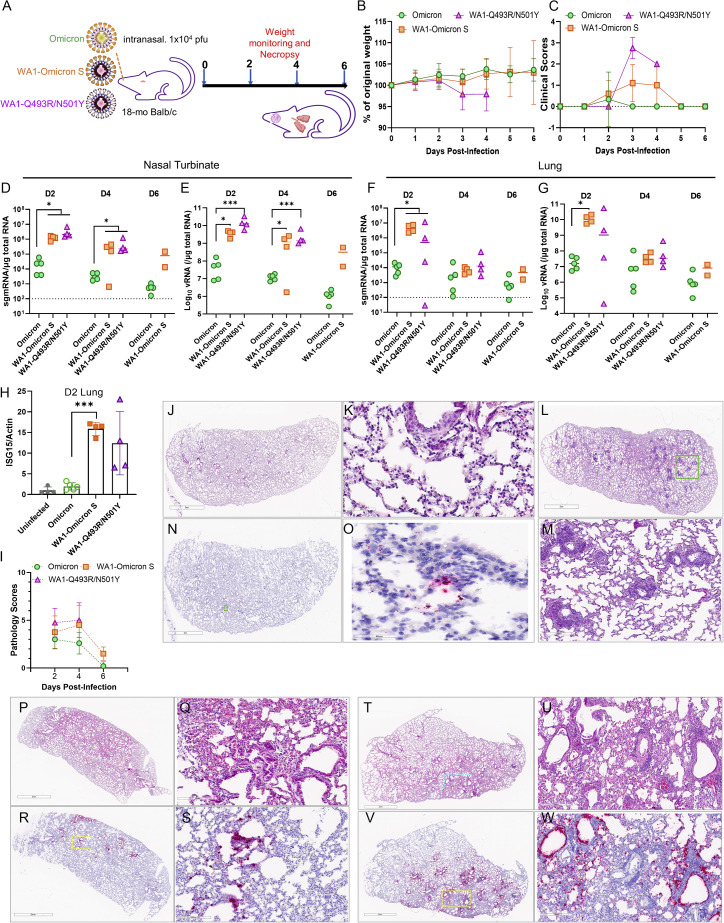
We subsequently characterized the infection of 18-month-old Balb/c mice with the Omicron variant (isolate hCoV-19/USA/MD-HP20874/2021). Because of reported attenuation of Omicron in Syrian hamster and K18-transgenic mice (Halfmann et al., 2022; Katherine McMahan et al., 2022; Shuofeng Yuan et al., 2022), we also included a recombinant virus (WA1-Omicron-S) in which the spike protein sequence of WA1/2020 ancestral virus was replaced with that of the Omicron variant. For comparison, WA1-Q493R/N501Y was included as a positive control virus (Figure 3A). WA1-Omicron-S contains the WA1 backbone with the Omicron spike protein. Therefore, WA1-Omicron-S should enter cells in the same manner as Omicron does. If the natural Omicron variant fails to infect cells in the lung due to defective entry, as implied in several studies (Meng et al., 2022; Thomas et al., 2021), infection by the WA1-Omicron-S will similarly result in low viral load in the lung because the same entry blockage should exist. If the attenuated phenotype of the natural Omicron variant is due to poor replication or rapid elimination by host innate immune response, one might expect the WA1-Omicron-S virus would robustly infect mouse lungs. Notably, our natural Omicron variant stock had a relatively low infectious titer (∼2 × 105 PFUs/mL); we therefore chose 1 × 104 PFUs in 50 μL for intranasal inoculation in this study. Under this challenge dose, the only group of mice that had modest weight loss was those inoculated with the WA1-Q493R/N501Y virus (Figure 3B). WA1-Omicron-S- and Q493R/N501Y-infected mice nonetheless had clinical signs of illness at 3 and 4 days post-infection (DPI) (Figure 3C). The presence of subgenomic RNA (sgmRNA) of the E gene and total viral RNA was readily detectable in both the nasal turbinates and the lungs at 2 and 4 DPI in all three groups, although viral loads detected from WA1-Omicron-S- and WA1-Q493R/N501Y- infected mice were at least two logs higher than those from the natural variant infected animals (Figures 3D–3G). The infection by WA1-Omicron-S and WA1-Q493R/N501Y also markedly upregulated the interferon-stimulated gene 15 (ISG15) (Figure 3H) and induced pathology in the lungs (Figure 3I). Interestingly, histopathology examination of the natural Omicron variant-infected lungs found that most infected animals had a low degree of immune cell infiltration (Figures 3J and 3K). Occasionally, mice infected with the natural Omicron variant displayed more pronounced perivascular infiltrates (Figures 3L–3M), despite very little viral RNA detected in the lung by RNAscope (Figures 3N–3O). Mice infected by WA1-Omicron-S and WA1-Q493R/N501Y exhibited more diffuse peribronchiolar, perivascular, and alveolar inflammatory infiltrates and with clear presence of viral RNA as detected by RNAscope using a probe that specifically targets the S gene (Figures 3P–3W). The viral RNA was detected not only along bronchial epithelia but also in the lung parenchyma from WA1-Omicron-S- and WA1-Q493R/N501Y-infected mice. At 7 DPI, the histopathological changes have mostly resolved in the lungs of all three groups.
Figure 3.
Spike-independent attenuation of the Omicron variant in Balb/c mice
(A) Overall study design.
(B) Weight loss profile.
(C) Clinical scores of all three groups of mice overall the period of study.
(D–G) Subgenomic RNA (sgmRNA) and total viral RNA levels in nasal turbinates (D and E) and lungs (F and G).
(H) Induction of ISG15 at 2 DPI in mouse lungs.
(I) Summed pathology scores of all three groups of mice overall the period of study.
(J–M) Representative H&E images of the natural Omicron-infected mice. (K) is a closeup image from (J), and (M) is a closeup from (L).
(N) An RNAscope image of the natural Omicron variant-infected mouse.
(O) A closeup from (N). Blue, nuclei; red, viral RNA.
(P) A representative H&E image of the WA1-Omicron S-infected mouse.
(Q) A closeup from (P).
(R) A representative RNAscope image of the WA1-Omicron S-infected mouse.
(S) A closeup from (R).
(T) A representative H&E image of the WA1-Q493R/N501Y-infected mouse.
(U) A closeup from (T).
(V) A presentative RNAscope image of the WA1-Q493R/N501Y-infected mouse.
(W) A closeup from (V).
Results shown in this figure were derived from one experiment (n = 10–15 for each group). Each solid shape represents one animal in (D)–(H).
Omicron spike-independent attenuation in human ACE2 transgenic mice
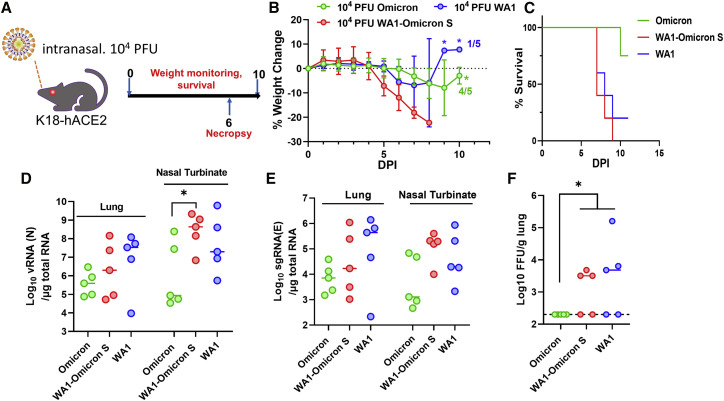
Since SARS-CoV-2 infection in immunocompetent mice generally leads to very mild disease, even for the WA1-Q493R/N501Y virus, we subsequently conducted an experiment in K18-hACE2 mice. Infection of K18-hACE2 by SARS-CoV-2 is lethal (Bao et al., 2020; Golden et al., 2020; Oladunni et al., 2020; Rathnasinghe et al., 2020a, 2020b; Winkler et al., 2020; Yinda et al., 2020), which makes this model useful in assessing pathogenic differences between variants. For this purpose, we inoculated K18-hACE2 mice with 104 PFUs of WA1, WA1-Omicron-S, and the natural Omicron variant (n = 10 for each group) and monitored weight loss and survival. Five mice from each group were sacrificed on day 6 after infection for viral load analyses (Figure 4A). Both WA1- and WA1-Omicron-S-infected animals displayed severe weight loss starting from day 5 onward (Figure 4B). All WA1-Omicron-S-infected mice succumbed to infection, and only one in the WA1-infected group survived (Figure 4C). By contrast, most natural Omicron variant-infected mice displayed minimal weight loss, and only one succumbed to the infection at 10 DPI. Viral loads from the natural Omicron variant-infected mice, including total viral RNA or single guide RNA (sgRNA) or infectious virus titers, were consistently lower than those from WA1- or WA1-Omicron-S-infected animals (Figures 4D–4F).
Figure 4.
WA1-Omicron-S is lethal in K18-hACE2 mice
(A) Overall study design. n = 10 per group.
(B) Weight loss profile. Note, after 9 DPI there was only one WA1-infected mouse alive. All WA-Omicron-S-infected mice succumbed to infection by day 8.
(C) Survival curves.
(D) Viral RNA (D) and sgmRNA (E) levels in nasal turbinates (NBs) and the lungs at 6 DPI.
(F) Infectious viral titers from lung homogenates at 6 DPI. ∗p < 0.05.
Results shown in this figure were derived from one experiment. Each solid shape represents one animal in (D)–(F).
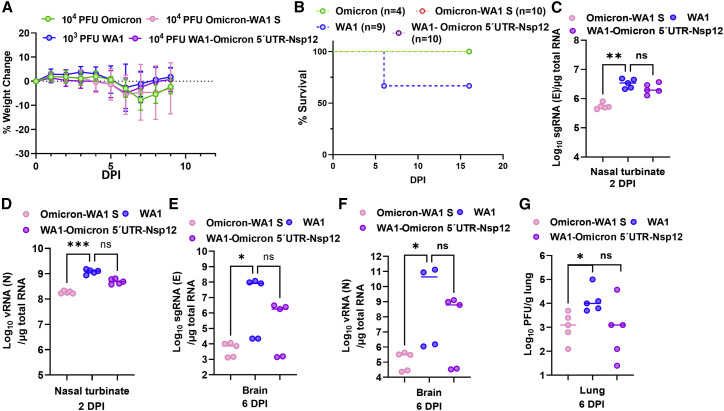
To further explore the Omicron spike-independent attenuation in human ACE2 transgenic mice, we generated two additional chimeric viruses: Omicron-WA1 S, in which the Omicron BA.1 variant (sequence based on isolate hCoV-19/USA/MD-HP20874/2021) now contains the WA1/2020 spike protein, and WA1-Omicron 5′ UTR-Nsp12, in which the WA1 virus contains mutations of the Omicron variant ranging from 5′ UTR to Nsp12 × 104 PFUs, viruses were given to K18-hACE2 mice intranasally. Because often >90% mice inoculated with 104 PFU WA1 become moribund by day 6 or 7, making it difficult to assess tissue viral loads at 6 DPI, we chose to inoculate K18-hACE2 with 103 PFU WA1 so that enough outwardly healthy animals remain at 6 DPI for comparison. As shown in Figure 5A, K18-hACE2 mice infected by 104 PFUs of Omicron-WA1 S or WA1-Omicron 5′ UTR-Nsp12 lost no more than 5% of their original body weight by 6 DPI and then started regaining weight afterward. By 9 DPI, infected mice fully recovered their body weight, and none succumbed to infection. By contrast, infection with only 103 PFUs of WA1 led to 3 out of 9 mice dead by day 9. Viral loads from Omicron-WA1 S-infected mice, including total viral RNA or sgRNA in nasal turbinates and brains and infectious virus titers in lungs, were significantly lower than those from WA1-infected animals. WA1-Omicron 5′ UTR-Nsp12-infected mice also yielded lower viral load, although the difference did not reach statistical significance (Figures 5D–5G).
Figure 5.
Omicron bearing WA1 spike is attenuated in K18-hACE2 mice
(A) Body weight loss of K18 mice after infection with 104 PFUs of Omicron-WA1 S, WA1-Omicron 5′ UTR-Nsp12, or Omicron or 103 PFUs WA1.
(B) Survival curves after infection. Numbers of mice in each group are indicated in the legend.
(C and D) sgmRNA (C) and viral RNA (D) levels in NBs at 2 DPI.
(E and F) sgmRNA (E) and viral RNA (F) levels from brains at 6 DPI.
(G) Infectious virus titers from lungs at 6 DPI. ∗p < 0.05.
Results shown in this figure were derived from one experiment. Each solid shape represents one animal in (C)–(G).
Discussion
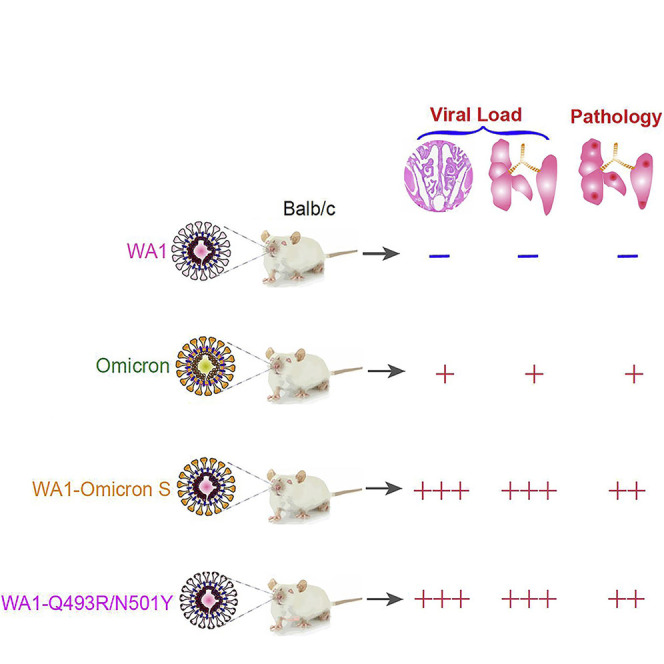
We present evidence that the spike protein of Omicron variant has obtained multiple mutations that enable efficient usage of mACE2 for cellular entry both in vitro and in vivo. The mutations found in the Omicron spike protein may potentially alter the species tropism of SARS-CoV-2 variants. This is especially interesting regarding the usage of mouse and horseshoe bat ACE2. Omicron contrasts with the ancestral WA1 isolate in that the former gained the ability to engage mACE2 but then lost usage of horseshoe bat ACE2. A striking observation is that despite an efficient usage of mACE2 by the Omicron variant, laboratory mice infected by Omicron only exhibited mild disease, accompanied by low viral loads, as has been reported by others (Halfmann et al., 2022). It was recently proposed that the Omicron spike protein is not efficiently cleaved at S1/S2, and hence the virus may shift away from TMPRSS2-expressing cells, such as type II pneumocytes, and become less pathogenic (Meng et al., 2022). In support, the Omicron spike is less fusogenic compared with that of the Delta variant (Cong Zeng et al., 2021; Zhao et al., 2021). We, however, found that the Omicron spike protein mediates cell-cell fusion as efficiently as does the ancestral spike protein (Figure 2J). Our findings further suggest that genetic changes outside the Spike protein in the natural Omicron variant may contribute to the observed attenuation in laboratory mice because WA1-Omicron-S is neither attenuated in Balb/c nor in K18-hACE2 mice. Therefore, the low viral load detected in Omicron-infected mice is unlikely caused by an inability of the variant to enter mouse tissues, despite that the disease severity may still be influenced by mutations within the spike protein. One limitation of the current study is the relatively low inoculum used. Nonetheless, findings from this study directly challenge the notion that somehow the SARS-CoV-2 Omicron variant fails to reach mouse lungs. Instead, the observed lower viral load and attenuation in laboratory mice appear to be a result of post-entry blockage linked to mutations outside the spike protein in the Omicron variant. This blockage could be an intrinsic replicative defect of Omicron in mice or a heightened susceptibility to rodent innate immunity. As illustrated in Figure 2A, there are 17 amino acid changes outside the spike protein in Omicron. Interestingly, although the reduction of viral load in WA1-Omicron 5′ UTR-Nsp12-infected mice did not reach statistical significance, a cloned Omicron virus bearing the WA1 spike protein was indeed attenuated in the K18-hACE2 mice. A thorough mutational analysis will be required to pinpoint what mutation(s) contribute to the attenuated phenotype of Omicron in laboratory mice.
Limitations of the study
A potential caveat of the work is that the Omicron BA.1 isolate tested in this study may not be representative of all Omicron variants. Additionally, the mechanism of attenuation of the Omicron variant may differ between unmodified laboratory mice and K18-hACE2 transgenic mice, with the latter mouse model displaying wilder expression of human ACE2 as a transgene. As Omicron spike protein interacts with both human and mACE2, the variant infects K18-hACE2 mice via both receptors, whereas infection of genetically unmodified mice is only mediated by endogenous mACE2.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| SARS-Coronavirus 2, Isolate USA-WA1/2020 | BEI Resources | Cat#NR-52281 |
| SARS-Coronavirus 2, Isolate hCoV-19/USA/MD-HP20874/2021 | BEI Resources | Cat#NR-56461 |
| SARS-Coronavirus 2, WA1-Omicron-S | This manuscript | N/A |
| SARS-Coronavirus 2, WA1-Q493R/N501Y | This manuscript | N/A |
| Endura™ Chemically Competent Cells | Lucigen | Cat#60240-2 |
| TransforMax™ EPI300™ Competent Cells | Lucigen | Cat#C300C105 |
| Critical commercial assays | ||
| Lipofectamine-3000 Transfection Reagent | ThermoFisher | Cat#L3000015 |
| Luciferase Assay System | Promega | Cat#E1501 |
| SuperScript IV Reverse Transcriptase | ThermoFisher Scientific | Cat#18090010 |
| Phusion® High-Fidelity DNA Polymerase | New England Biolabs | Cat#M0530L |
| mMESSAGE mMACHINE™ T7 Transcription Kit | ThermoFisher Scientific | Cat#AM1344 |
| Experimental models: Cell lines | ||
| Lenti-X™ 293T Cell Line | Takara | Cat#632180 |
| Vero (C1008) E6 | ATCC | Cat#CRL-1586 |
| Oligonucleotides | ||
| Primer M13 F (GTAAAACGACGGCCAGT) | Facility for Biotechnology Resources, FDA | N/A |
| Primer SARS-2S-PRRA F (AGACAAACTC CCGGTCTGTGGCAAGCCAGTC) |
Facility for Biotechnology Resources, FDA | N/A |
| Primer Q493R F (TTGTTACTTTCCTTTAA GATCATATGGTTTCCAACCCACT) |
Facility for Biotechnology Resources, FDA | N/A |
| Primer Q493R R (GAAACCATATGATCTT AAAGGAAAGTAACAATTAA) |
Facility for Biotechnology Resources, FDA | N/A |
| Primer N501Y F (GTTTCCAACCCACTT ATGGTGTTGGTTACCAACCATA) |
Facility for Biotechnology Resources, FDA | N/A |
| Primer N501Y R (ACCAACACCATAA GTGGGTTGGAAACCATATGA) |
Facility for Biotechnology Resources, FDA | N/A |
| Primer BglII R (CAGCATCTGCAAGTGTCACT) | Facility for Biotechnology Resources, FDA | N/A |
| Recombinant DNA | ||
| pcDNA-human-ACE2 | GenScript | N/A |
| pcDNA-mouse-ACE2 | GenScript | N/A |
| pcDNA-ferret-ACE2 | GenScript | N/A |
| pcDNA-Syrian hamster-ACE2 | GenScript | N/A |
| pcDNA-Grivet (African green monkey)-ACE2 | GenScript | N/A |
| pcDNA-horseshoe bat-ACE2 | GenScript | N/A |
| pcDNA-pig-ACE2 | GenScript | N/A |
| pcDNA-bovine-ACE2 | GenScript | N/A |
| pcDNA-Chinese hamster-ACE2 | GenScript | N/A |
| pcDNA- Malayan pangolin -ACE2 | GenScript | N/A |
| pcDNA- SARS-CoV-2 Wuhan Spike | This manuscript | N/A |
| pcDNA- SARS-CoV-2 Omicron Spike | This manuscript | N/A |
| pcDNA- SARS-CoV-2 Spike N501Y-Q493K | This manuscript | N/A |
| pcDNA- SARS-CoV-2 Spike N501Y-Q493R | This manuscript | N/A |
| pcDNA- SARS-CoV-2 Spike G496K-Q493K | This manuscript | N/A |
| pcDNA- SARS-CoV-2 Spike N501Y-S494R | This manuscript | N/A |
| pcDNA- SARS-CoV-2 Spike N501Y-G496K | This manuscript | N/A |
| pcDNA- SARS-CoV-2 Spike N501F-G496K | This manuscript | N/A |
| pcDNA- SARS-CoV-2 Spike N501K-Q493K | This manuscript | N/A |
| pcDNA- SARS-CoV-2 Spike Q493K-Q498Y-P499T | This manuscript | N/A |
| TMPRSS2 | Addgene | Cat#53887 |
| psPAX2 | Addgene | Cat#12260 |
| pCMV-CREM | Addgene | Cat#8395 |
| pSV-STOP-luc | Addgene | Cat#8390 |
| RNAscope reagents | ||
| RNAscope 2.5 HD RED kit | ACD | Cat#322373 |
| V-SARS-Cov-2-S | ACD | Cat#854841 |
| Mm PPIB probe (positive control) | ACD | Cat#313911 |
| dapB probe (negative control) | ACD | Cat#310043 |
| Software and algorithms | ||
| Prism 9.0 software | GraphPad | N/A |
| SnapGene | GSL Biotech LLC | N/A |
Resource availability
Lead contact
Further information and requests for reagents may be directed to and will be fulfilled by Lead Contact Tony Wang (Tony.Wang@fda.hhs.gov).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Experimental model and subject details
Viruses and cells
Vero E6 cell line (Cat # CRL-1586, gender unknown) cell line was purchased from American Type Culture Collection (ATCC) and cultured at 37°C with 5% CO2in eagle’s minimal essential medium (MEM) supplemented with 10% fetal bovine serum (Invitrogen) and 1% penicillin/streptomycin and L-glutamine. Calu-3 cell line (Cat # HTB-55, obtained from a male, lung adenocarcinoma patient) was obtained from ATCC and maintained in EMEM+20%FBS at 37°C with 5% CO2.
The SARS-CoV-2 isolate hCoV-19/USA/MD-HP20874/2021 was obtained from BEI Resources, NIAID, NIH, and had been tittered on Vero E6 cells by plaque assay.
Mice
Aged male and female Balb/c mice and K18-hACE2 were previously purchased from the Jackson laboratory and held at FDA vivarium. Littermates of the same sex were randomly assigned to experimental groups. All experiments were performed within the biosafety level 3 (BSL-3) suite on the White Oak campus of the U.S. Food and Drug Administration. The study protocol details were approved by the White Oak Consolidated Animal Care and Use Committee and carried out in accordance with the PHS Policy on Humane Care & Use of Laboratory Animals.
Method details
SARS-CoV-2 pseudovirus production and infection assay
Human codon-optimized cDNA encoding SARS-CoV-2 S glycoprotein of the Wuhan-Hu-1 isolate (NC_045512) or the B.1.1.529 variant (sequence available upon request) was synthesized by GenScript and cloned into eukaryotic cell expression vector pcDNA 3.1 between the BamHI and XhoI sites. Pseudovirions were produced by co-transfection of Lenti-X 293T cells with psPAX2, pTRIP-luc or pTRIP-GFP, and SARS-CoV-2 S expressing plasmid using Lipofectamine 3000. The supernatants were harvested at 48 and 72 h post-transfection and filtered through 0.45-μm membranes. Infection was done as previously described (Liu et al., 2021).
SARS-CoV-2 spike-mediated cell–cell fusion assay
To assess the Spike protein-mediated membrane fusion, we have previously established a quantitative cell-cell fusion assay in which 293T acceptor cells, containing a loxP-flanked STOP cassette that blocks transcription of the downstream luciferase reporter gene, were cocultured with Cre-expressing donor cells (Liu et al., 2021). In this assay, fusion between donor and acceptor cell membranes removes the STOP cassette and hence permits luciferase production. Various spike constructs were transfected into donor 293T cells and then the hACE2 plasmid was transfected into the acceptor 293T cells. Twenty-four hours later, recipient cells were mixed at a 1:1 ratio with 293T cells expressing Cre and CoV S (donor cells) to initiate cell–cell fusion. Luciferase activity was measured 48 h thereafter.
Production of SARS-CoV-2 recombinant virus
SARS-CoV-2 recombinant viruses containing 2019-nCoV/USA_WA1/2020 sequence or substitutions (Omicron S or Q493R/N501Y) in the Spike protein of WA1 was generated using a 7-plasmid reverse genetic system which was based on the virus strain (2019-nCoV/USA_WA1/2020) isolated from the first reported SARS-CoV-2 case in the U.S. (Xie et al., 2020). The initial 7 plasmids were generous gifts from Dr. P-Y Shi (UTMB). Upon receipt, fragment 4 was subsequently subcloned into a low-copy plasmid pSMART LCAmp (Lucigen) to increase stability. Standard molecular biology technique was employed to create mutations. In vitro transcription and electroporation were carried following procedures that were detailed elsewhere (Shufeng Liu et al., 2021; Xie et al., 2021). Recombinant viruses were further deep sequenced to confirm the presence of mutations. To generate the Omicron-WA1-S and WA1-Omicron 5′UTR-Nsp12, plasmids containing omicron mutations were constructed by using RT-PCR and overlap PCR methods. Reverse transcription viral RNA template was extracted from the supernatant of SARS-Related Coronavirus 2, Isolate hCoV-19/USA/MD-HP20874/2021 (BEI, Cat# NR-56461) using QIAamp Viral RNA Mini Kit (Qiagen, Cat# 52906) as per the manufacturer’s protocol. Reverse transcription was performed by using SuperScript IV reverse transcriptase (ThermoFisher Scientific, Cat# 18090010) with sequence specific primers and Omicron viral RNA template. The cDNA templates were amplified by PCR using Phusion® High-Fidelity DNA Polymerase (NEB, Cat# M0530L) with the following primers:
pBACf: ccgttcttcttcgtcataa
C241Tf: gcacatctaggttttgtccgggtgtgaccgaaagg
C241Tr: cggtcacacccggacaaaacctagatgtgctgatga
KasIr: gactttagatcggcgccgta
KasIf: tagttacggcgccgatctaaa
PacIr: acacaagtgtaactttaattaactgctt
PacIf: aagcagttaattaaagttacacttgtgt
XmaIr: ttaaaccctgacccgggtaagtggt
XmaIf: accacttacccgggtcagggtttaa
MluIr: agttggcgtatacgcgtaat
MluIf: ccagatatattacgcgtatacgccaact
BamHIr: agaggatccggtctcaactcacagtgtcaacaatttcagca
Nsp14f: atataggcggccgccgtctcagagtgctttggtttatgataa
NheIr: tatagccacggaacctccaa
25000f: tcattcaaggaggagttaga
BamHIr: caaatttgcagcaagatccacaagaacaacagc
BamHIf: tcttgtggatcttgctgcaaatttgatgaa
29000r: ttcttagtgacagtttggcc
29000f: ggccaaactgtcactaagaa
M13r: caggaaacagctatgac
Mouse infection experiments
Aged male and female Balb/c mice and K18-hACE2 were previously purchased from the Jackson laboratory and held at FDA vivarium. All experiments were performed within the biosafety level 3 (BSL-3) suite on the White Oak campus of the U.S. Food and Drug Administration. The study protocol details were approved by the White Oak Consolidated Animal Care and Use Committee and carried out in accordance with the PHS Policy on Humane Care & Use of Laboratory Animals.
For infection studies, mice were first anesthetized by 3-5% isoflurane. Intranasal inoculation was done by pipetting 104 PFU SARS-CoV-2 in 50 μL volume dropwise into the nostrils of the mouse. Mice were weighed and observed daily. For tissue collections, mice were euthanized by CO2 overdose on days 2, 4, 6 as necessary. Categories included in clinical scoring include weight loss (0–1), posture and appearance of fur (0–1), appearance of lethargy (0–2), and eye closure (0–1).
RNA isolation from tissues
RNA was extracted from 0.1-gram tissue homogenates using Trizol and the RNeasy Mini kit (Qiagen) and eluted with 50 μL of water. One microgram RNA was used for each reaction in real-time RT-PCR.
Real-time PCR assay of SARS-CoV-2 viral and subgenomic RNA
Quantification of SARS-CoV-2 viral RNA (vRNA) was conducted using the SARS-CoV-2 (2019-nCoV) CDC qPCR Probe Assay (IDTDNA) using iTaq Universal Probes One-Step Kit (Bio-Rad). The standard curve was generated using 2019-nCoV_N_Positive Control (IDTDNA). The detection limit of the vRNA was determined to be 100 copies/reaction. Quantification of SARS-CoV-2 E gene subgenomic mRNA (sgmRNA) was conducted using Luna Universal Probe One-Step RT-qPCR Kit (New England Biolabs) on a Step One Plus Real-Time PCR system (Applied Biosystems). The primer and probe sequences were: SARS2EF: CGATCTCTTGTAGATCTGTTCT; PROBE: FAM-ACACTAGCCATCCTTACTGCGCTTCG- BHQ-1; SARS2ER: ATATTGCAGCAGTACGCACACA. To generate a standard curve, the cDNA of SARS-CoV-2 E gene sgmRNA was cloned into a pCR2.1-TOPO plasmid. The copy number of sgmRNA was calculated by comparing to a standard curve obtained with serial dilutions of the standard plasmid. The limit of quantification (LoQ) of the sgmRNA was determined to be 100 copies/reaction. Values below LoQ were mathematically extrapolated based on the standard curves for graphing purpose. When graphing the results in Prism 9, values below LoQ were arbitrarily set to half of the LoQ values.
Histopathology analyses
Tissues (lungs and nasal turbinates) were fixed in 10% neural buffered formalin for 2–7 days and then processed for paraffin embedding. The 5-μm sections were stained with hematoxylin and eosin for histopathological examinations. Images were scanned using an Aperio ImageScope and scored under the following categories: consolidation, alveolar wall thickening, alveolar airway infiltrates, perivascular Infiltrates, perivascular edema, peribronchiolar infiltrates, type II pneumocyte hyperplasia, necrosis (alveoli and bronchiole), bronchiole mucosal hyperplasia, bronchiole airway infiltrates, proteinaceous fluid, hemorrhage, vasculitis. Grading scale: 0 = none, 1 = mild, 2 = moderate, 3 = severe.
In-situ hybridization
To detect SARS-CoV-2 genomic RNA in FFPE tissues, ISH was performed using the RNAscope 2.5 HD RED kit, a single plex assay method (Advanced Cell Diagnostics; Catalog 322373) according to the manufacturer’s instructions. Briefly, Mm PPIB probe detecting peptidylprolyl isomerase B gene (housekeeping gene) (catalog 313911, positive-control RNA probe), dapB probe detecting dihydrodipicolinate reductase gene from Bacillus subtilis strain SMY (a soil bacterium) (catalog 310043, negative-control RNA probe) and V-SARS-Cov-2-S (catalog 854841) targeting SARS-CoV-2 positive-sense (genomic) RNA. Tissue sections were deparaffinized with xylene, underwent a series of ethanol washes and peroxidase blocking, and were then heated in kit-provided antigen retrieval buffer and digested by kit-provided proteinase. Sections were exposed to ISH target probes and incubated at 40°C in a hybridization oven for 2 h. After rinsing, ISH signal was amplified using kit-provided pre-amplifier and amplifier conjugated to alkaline phosphatase and incubated with a fast-red substrate solution for 10 min at room temperature. Sections were then stained with 50% hematoxylin solution followed by 0.02% ammonium water treatment, dried in a 60°C dry oven, mounted, and stored at 4°C until image analysis.
MD simulations
We carried out all-atom MD simulations for a complex of the mouse’s ACE2 and the RBD of Omicron’s spike protein using the NAMD2.13 package (Phillips et al., 2005) running on the IBM Power Cluster. We obtained the secondary structure of the mouse’s ACE2 (mACE2) through the homology modeling according to the structure of human ACE2 (PDB code: 6M0J) using SWISS-MODEL (Waterhouse et al., 2018). The atomic structure of the Omicron RBD (oRBD) was adopted from the recently resolved crystal structure (PDB code: 7T9L) (Mannar et al., 2022). The complex of mACE2 and oRBD was assembled by the structural alignment to the known complex structure of hACE2 and oRBD (PDB code: 7T9L). After solvating the mACE2-oRBD complex in a 0.15 NaCl electrolyte, we performed all-atom molecular dynamics simulations to equilibrate the built complex in its physiology-like environment. The final simulation system comprises 209,460 atoms.
We used the CHARMM36 m force field (Huang et al., 2017) for proteins, the TIP3P model(Jorgensen et al., 1983; Neria et al., 1996) for water, the standard force field (Beglov and Roux, 1994) for Na+ and Cl−. The periodic boundary conditions (PBC) were applied in all three dimensions. Long-range Coulomb interactions were computed using particle-mesh Ewald (PME) full electrostatics with the grid size of about 1 Å in each dimension. The pairwise van der Waals (vdW) energies were calculated using a smooth (10–12 Å) cutoff. The temperature T was kept at 300 K by applying the Langevin thermostat (Allen and Dominic, 1987),while the pressure was maintained constant at 1 bar using the Nosé-Hoover method (Martyna et al., 1994). With the SETTLE algorithm (Miyamoto and Kollman, 1992) enabled to keep all bonds rigid, the simulation time-step was 2 fs for bonded and non-bonded (including vdW, angle, improper and dihedral) interactions, and the time-step for Coulomb interactions was 4 fs, with the multiple time-step algorithm (Tuckerman et al., 1992).
Quantification and statistical analysis
The number of technical or biological replicates of each experiment is indicated in related figure legends. Unless specified, bar graphs were plotted to show mean ± standard deviation (SD). Statistical analyses were performed using One-way ANOVA through GraphPad Prism (9.1.2) software for Windows, GraphPad Software, San Diego, California USA. p < 0.05 is deemed statistically significant.
Acknowledgments
The following reagent was obtained through BEI Resources, NIAID, NIH: SARS-related coronavirus 2, isolate hCoV-19/USA/MD-HP20874/2021 (lineage B.1.1.529; Omicron variant), NR-56461, contributed by Andrew S. Pekosz. We are grateful to Dr. P. Shi (UTMB) for the original 7 plasmids to perform SARS-CoV-2 reverse genetics. We thank Drs. K. Erlandson and C. Florence for assistance in obtaining reagents and Dr. C. Weiss for critical reading of the manuscript. The work described in this manuscript was supported by the US Food and Drug Administration (FDA) intramural research fund and partially by the US FDA, Office of the Chief Scientist, Medical Countermeasures Initiative, under OCET Intramural Program ID# 2022-1821. The article reflects the views of the authors and does not represent views or policies of the FDA. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
S.L., P.S., K.S., and T.T.W. conducted the experiments; B.L. performed the MD simulation; S.L., P.S., and T.T.W. analyzed the data; B.L. and T.T.W. designed the experiments and wrote the paper. T.T.W. is the lead contact.
Declaration of interests
The authors declare no competing interests.
Published: September 13, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111359.
Supplemental information
Data and code availability
-
•
Original data may be directed to and will be fulfilled by lead contact Tony Wang (Tony.Wang@fda.hhs.gov).
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abdelnabi R., Foo C.S., Zhang X., Lemmens V., Maes P., Slechten B., Raymenants J., André E., Weynand B., Dallemier K., Neyts J. The omicron (B.1.1.529) SARS-CoV-2 variant of concern does not readily infect Syrian hamsters. bioRxiv. 2021 doi: 10.1101/2021.12.24.474086. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen M.P., Dominic J. Oxford University Press; 1987. Computer Simulation of Liquids. [Google Scholar]
- Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F., et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- Beglov D., Roux B. Finite representation of an infinite bulk system: solvent boundary potential for computer simulations. J. Chem. Phys. 1994;100:9050–9063. [Google Scholar]
- Cameroni E., Bowen J.E., Rosen L.E., Saliba C., Corti D., Zepeda S.K., Culap K., Pinto D., VanBlargan L.A., De Marco A., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2022;602:664–670. doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cele S., Jackson L., Khoury D.S., Khan K., Moyo-Gwete T., Tegally H., San J.E., Cromer D., Scheepers C., Amoako D.G., et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2021;602:654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Zeng J.P.E., Qu P., Faraone J., Zheng Y.-M., Carlin C., Bednash J.S., Zhou T., Lozanski G., Mallampalli R., Saif L.J., et al. Neutralization and stability of SARS-CoV-2 omicron variant. bioRxiv. 2021 doi: 10.1101/2021.12.16.472934. Preprint at. [DOI] [Google Scholar]
- Golden J.W., Cline C.R., Zeng X., Garrison A.R., Carey B.D., Mucker E.M., White L.E., Shamblin J.D., Brocato R.L., Liu J., et al. Human angiotensin-converting enzyme 2 transgenic mice infected with SARS-CoV-2 develop severe and fatal respiratory disease. JCI Insight. 2020;5:142032. doi: 10.1172/jci.insight.142032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H., Chen Q., Yang G., He L., Fan H., Deng Y.Q., Wang Y., Teng Y., Zhao Z., Cui Y., et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369:1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann P.J., Iida S., Iwatsuki-Horimoto K., Maemura T., Kiso M., Scheaffer S.M., Kawaoka Y., Darling T.L., Joshi A., Loeber S., et al. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature. 2022;603:687–692. doi: 10.1038/s41586-022-04441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Rauscher S., Nawrocki G., Ran T., Feig M., de Groot B.L., Grubmüller H., MacKerell A.D., Jr. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods. 2017;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh T. Insights into SARS-CoV-2’s mutations for evading human antibodies: sacrifice and survival. J. Med. Chem. 2021;65:2820–2826. doi: 10.1021/acs.jmedchem.1c00311. [DOI] [PubMed] [Google Scholar]
- Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- Katherine McMahan V.G., Giffin V., Tostanoski L.H., Chung B., Siamatu M., Suthar M.S., Halfmann P., Kawaoka Y., Piedra-Mora C., Martinot A.J., et al. Reduced pathogenicity of the SARS-CoV-2 omicron variant in hamsters. bioRxiv. 2022 doi: 10.1101/2022.01.02.474743. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibler K.V., Szczerba M., Douglas L., Roeder A.J., Rahman M., Hogue B.G., Roy Wong L.-Y., Perlman S., Li Y., Bertram L., Jacobs Intranasal immunization with a vaccinia virus vaccine vector expressing pre-fusion stabilized SARS-CoV-2 spike fully protected mice against lethal challenge with the heavily mutated mouse-adapted SARS2-N501YMA30 strain of SARS-CoV-2. bioRxiv. 2021 doi: 10.1101/2021.12.06.471483. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leist S.R., Dinnon K.H., 3rd, Schäfer A., Tse L.V., Okuda K., Hou Y.J., West A., Edwards C.E., Sanders W., Fritch E.J., et al. A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell. 2020;183:1070–1085.e12. doi: 10.1016/j.cell.2020.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Selvaraj P., Lien C.Z., Nunez I.A., Wu W.W., Chou C.K., Wang T.T. The PRRA insert at the S1/S2 site modulates cellular tropism of SARS-CoV-2 and ACE2 usage by the closely related Bat RaTG13. J. Virol. 2021;95 doi: 10.1128/JVI.01751-20. JVI.01751-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok-Yin Roy Wong J.Z., Kevin W., Li K., Ortiz N.J.S., Pezzulo A.A., Szachowicz P.J., Klumpp K., Aswad F., Rebo J., Narumiya S., Murakami M., et al. Eicosanoid signaling as a therapeutic target in middle-aged mice with severe COVID-19. bioRxiv. 2021 doi: 10.1101/2021.04.20.440676. Preprint at. [DOI] [Google Scholar]
- Luan B., Wang H., Huynh T. Enhanced binding of the N501Y-mutated SARS-CoV-2 spike protein to the human ACE2 receptor: insights from molecular dynamics simulations. FEBS Lett. 2021;595:1454–1461. doi: 10.1002/1873-3468.14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannar D., Saville J.W., Zhu X., Srivastava S.S., Berezuk A.M., Tuttle K.S., Marquez A.C., Sekirov I., Subramaniam S. SARS-CoV-2 Omicron variant: antibody evasion and cryo-EM structure of spike protein-ACE2 complex. Science. 2022;375:760–764. doi: 10.1126/science.abn7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martyna G.J., Tobias D.J., Klein M.L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994;101:4177–4189. [Google Scholar]
- Meng B., Abdullahi A., Ferreira I.A.T.M., Goonawardane N., Saito A., Kimura I., Yamasoba D., Gerber P.P., Fatihi S., Rathore S., et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature. 2022;603:706–714. doi: 10.1038/s41586-022-04474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S., Kollman P.A. SETTLE: an analytical version of the SHAKE and RATTLE algorithm for rigid water molecules. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- Muruato A., Vu M.N., Johnson B.A., Davis-Gardner M.E., Vanderheiden A., Lokugamage K., Schindewolf C., Crocquet-Valdes P.A., Langsjoen R.M., Plante J.A., et al. Mouse-adapted SARS-CoV-2 protects animals from lethal SARS-CoV challenge. PLoS Biol. 2021;19:e3001284. doi: 10.1371/journal.pbio.3001284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neria E., Fischer S., Karplus M. Simulation of activation free energies in molecular systems. J. Chem. Phys. 1996;105:1902–1921. [Google Scholar]
- Oladunni F.S., Park J.G., Pino P.A., Gonzalez O., Akhter A., Allué-Guardia A., Olmo-Fontánez A., Gautam S., Garcia-Vilanova A., Ye C., et al. Lethality of SARS-CoV-2 infection in K18 human angiotensin-converting enzyme 2 transgenic mice. Nat. Commun. 2020;11:6122. doi: 10.1038/s41467-020-19891-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organization W.H. Classification of omicron (B.1.1.529): SARS-CoV-2 variant of concern, 27th November 2021. 2021. https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern
- Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kalé L., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulliam J.R.C., van Schalkwyk C., Govender N., von Gottberg A., Cohen C., Groome M.J., Dushoff J., Mlisana K., Moultrie H. Increased risk of SARS-CoV-2 reinfection associated with emergence of the Omicron variant in South Africa. Science. 2022;376:eabn4947. doi: 10.1126/science.abn4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathnasinghe R., Strohmeier S., Amanat F., Gillespie V.L., Krammer F., Garcia-Sastre A., Coughlan L., Schotsaert M., Uccellini M. Comparison of transgenic and adenovirus hACE2 mouse models for SARS-CoV-2 infection. bioRxiv. 2020 doi: 10.1101/2020.07.06.190066. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathnasinghe R., Strohmeier S., Amanat F., Gillespie V.L., Krammer F., García-Sastre A., Coughlan L., Schotsaert M., Uccellini M.B. Comparison of transgenic and adenovirus hACE2 mouse models for SARS-CoV-2 infection. Emerg. Microb. Infect. 2020;9:2433–2445. doi: 10.1080/22221751.2020.1838955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K.A., Watson R.J., Bewley K.R., Burton C., Carnell O., Cavell B.E., Challis A., Coombes N.S., Emery K., Fell R., et al. Convalescence from prototype SARS-CoV-2 protects Syrian hamsters from disease caused by the Omicron variant. bioRxiv. 2021 doi: 10.1101/2021.12.24.474081. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai H., Chan J.F.W., Yuen T.T.T., Yoon C., Hu J.C., Wen L., Hu B., Yang D., Wang Y., Hou Y., et al. Emerging SARS-CoV-2 variants expand species tropism to murines. EBioMedicine. 2021;73:103643. doi: 10.1016/j.ebiom.2021.103643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai H., Chan J.F.-W., Hu B., Chai Y., Yuen T.T.-T., Yin F., Huang X., Yoon C., Hu J.-C., Liu H., et al. Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature. 2022;603:693–699. doi: 10.1038/s41586-022-04442-5. [DOI] [PubMed] [Google Scholar]
- Shufeng Liu T.H., Huynh T., Stauft C.B., Wang T.T., Luan B. Structure-Function analysis of resistance to bamlanivimab by SARS-CoV-2 variants kappa, delta, and lambda. J. Chem. Inf. Model. 2021;61:5133–5140. doi: 10.1021/acs.jcim.1c01058. [DOI] [PubMed] [Google Scholar]
- Shuofeng Yuan Z.-W.Y., Ye Z.W., Liang R., Tang K., Zhang A.J., Lu G., Poon V.K.-M., Chan C.C.S., Mok B.W.Y., et al. The SARS-CoV-2 Omicron (B.1.1.529) variant exhibits altered pathogenicity, transmissibility, and fitness in the golden Syrian hamster model. bioRxiv. 2022 doi: 10.1101/2022.01.12.476031. Preprint at. [DOI] [Google Scholar]
- Suzuki R., Yamasoba D., Kimura I., Wang L., Sato K., Kishimoto M., Ito J., Morioka Y., Nao N., Nasser H., et al. Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature. 2022;603:700–705. doi: 10.1038/s41586-022-04462-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P., Peacock J.C.B., Zhou J., Thakur N., Newman J., Kugathasan R., Sukhova K., Kaforou M., Bailey D., Barclay W.S. the SARS-CoV-2 variant, Omicron, shows rapid replication in human primary nasal epithelial cultures and efficiently uses the endosomal route of entry. bioRxiv. 2021 doi: 10.1101/2021.12.31.474653. Preprint at. [DOI] [Google Scholar]
- Tuckerman M., Berne B.J., Martyna G.J. Reversible multiple time scale molecular dynamics. J. Chem. Phys. 1992;97:1990–2001. [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler E.S., Bailey A.L., Kafai N.M., Nair S., McCune B.T., Yu J., Fox J.M., Chen R.E., Earnest J.T., Keeler S.P., et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 2020;21:1327–1335. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier Montagutelli M.P., Levillayer L., Baquero Salazar E., Jouvion G., Conquet L., Beretta M., Donati F., Albert M., Gambaro F., Behillil S., et al. Variants with the N501Y mutation extend SARS-CoV-2 host range to mice, with contact transmission. bioRxiv. 2021 doi: 10.1101/2021.03.18.436013. Preprint at. [DOI] [Google Scholar]
- Xie X., Muruato A., Lokugamage K.G., Narayanan K., Zhang X., Zou J., Liu J., Schindewolf C., Bopp N.E., Aguilar P.V., et al. An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe. 2020;27:841–848.e3. doi: 10.1016/j.chom.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Lokugamage K.G., Zhang X., Vu M.N., Muruato A.E., Menachery V.D., Shi P.Y. Engineering SARS-CoV-2 using a reverse genetic system. Nat. Protoc. 2021;16:1761–1784. doi: 10.1038/s41596-021-00491-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yinda C.K., Port J.R., Bushmaker T., Owusu I.O., Avanzato V.A., Fischer R.J., Schulz J.E., Holbrook M.G., Hebner M.J., Rosenke R., et al. K18-hACE2 mice develop respiratory disease resembling severe COVID-19. bioRxiv. 2020 doi: 10.1101/2020.08.11.246314. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Lu L., Peng Z., Chen L.L., Meng X., Zhang C., Ip J.D., Chan W.M., Chu A.W.H., Chan K.H., et al. SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with delta variant in TMPRSS2-expressed cells. Emerg. Microb. Infect. 2021;11:277–283. doi: 10.1080/22221751.2021.2023329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Original data may be directed to and will be fulfilled by lead contact Tony Wang (Tony.Wang@fda.hhs.gov).
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.