Abstract
The heat shock response in alpha proteobacteria is unique in that a combination of two regulators is involved: a positive regulator, RpoH (ς32 homolog), found in the alpha, beta, and gamma proteobacteria, and a negative regulator, HrcA, widely distributed in eubacteria but not in the gamma proteobacteria. To assess the differential roles of the two regulators in these bacteria, we cloned the hrcA-grpE operon of Agrobacterium tumefaciens, analyzed its transcription, and constructed deletion mutants lacking RpoH and/or HrcA. The ΔrpoH mutant and ΔrpoH ΔhrcA double mutant were unable to grow above 30°C. Whereas the synthesis of heat shock proteins (e.g., DnaK, GroEL, and ClpB) was transiently induced upon temperature upshift from 25 to 37°C in the wild type, such induction was not observed in the ΔrpoH mutant, except that GroEL synthesis was still partially induced. By contrast, the ΔhrcA mutant grew normally and exhibited essentially normal heat induction except for a higher level of GroEL expression, especially before heat shock. The ΔrpoH ΔhrcA double mutant showed the combined phenotypes of each of the single mutants. The amounts of dnaK and groE transcripts before and after heat shock, as determined by primer extension, were consistent with those of the proteins synthesized. The cellular level of RpoH but not HrcA increased significantly upon heat shock. We conclude that RpoH plays a major and global role in the induction of most heat shock proteins, whereas HrcA plays a restricted role in repressing groE expression under nonstress conditions.
In the paradigm of bacterial heat shock response studied with Escherichia coli, ς32 (encoded by rpoH) plays a key role in controlling the transcription of all major heat shock proteins (HSPs) in the cytoplasm, including GroE and DnaK chaperone machineries and ATP-dependent proteases, such as Clp and Lon (10, 42). Upon heat shock, the cellular level of ς32 increases rapidly both by enhanced translation of rpoH mRNA and by transient stabilization of ς32, and this increase in turn activates the transcription of heat shock genes. Homologs of rpoH have been identified from some 20 species of eubacteria that belong to the alpha, beta, and gamma subgroups of proteobacteria (1, 13, 15, 20, 21, 24, 30, 31); earlier work cited in reference (21). The rpoH homologs from many gamma proteobacteria share common structural features with E. coli rpoH, a downstream box, mRNA secondary structure, and highly conserved amino acid sequence of region C, that are important for thermoregulation of rpoH translation and ς32 stability and activity in E. coli (21). In these bacteria, the RpoH homologs indeed exhibit translational induction and stabilization upon heat shock very similar to that found with E. coli (22).
In contrast, alpha and beta proteobacteria have diverged from the gamma subgroup in their modes of regulation of rpoH expression. Their rpoH genes do not contain the downstream box or mRNA secondary structure that are conserved among gamma proteobacteria. Instead, some rpoH genes from alpha proteobacteria contain an RpoH-dependent promoter that can be induced upon heat shock (24, 26, 40). Another important feature found in alpha but not in gamma proteobacteria is the presence of the CIRCE (for controlling inverted repeat of chaperone expression)-HrcA regulatory system. Recent studies of wide groups of eubacteria revealed a variety of heat shock regulatory mechanisms, either positive regulation by alternative ς factors or negative regulation by specific repressor-operator systems (see reviews in references 23 and 28). The most widely distributed system is the CIRCE-HrcA system, extensively characterized in Bacillus subtilis as the regulatory mechanism specific for the groE and dnaK operons (18, 19, 41, 43). CIRCE, with a consensus of TTAGCACTC-N9-GAGTGCTAA, represents a site for binding the HrcA repressor. HrcA could act as a stress sensor whose activity is modulated by the GroEL chaperone. This system has been found in gram-positive bacteria and proteobacteria and implicated in cyanobacteria and several other groups of eubacteria (5, 8, 9, 38, 39). Within proteobacteria, CIRCE and/or hrcA were shown to regulate groE operons of some alpha proteobacteria, including Agrobacterium tumefaciens (3, 27, 37). However a CIRCE-like sequence has not been detected in gamma proteobacteria, except for the groE operon of Chromatium vinosum (7). It seemed likely that the CIRCE-HrcA system is operative in the alpha and beta proteobacteria but not in most gamma proteobacteria. Thus, alpha and beta proteobacteria appeared to be unique in that they carry both the RpoH system, with regulatory features different from those of gamma proteobacteria, and the CIRCE-HrcA system for regulation of the heat shock response.
In this study, we used A. tumefaciens as a model to study the differential roles of the RpoH and CIRCE-HrcA systems in the heat shock response in alpha proteobacteria. The groE and dnaK operons of A. tumefaciens were previously isolated and characterized (34–37), and the CIRCE element was found in the groE, but not in the dnaK, promoter region. Although the rpoH gene was available from our previous study (21), no information on hrcA was at hand. Successful isolation of hrcA and analysis of the deletion mutants lacking rpoH and/or hrcA revealed that RpoH plays a major and global role in the induction of HSPs, whereas the role of HrcA is restricted primarily to repressing the groE operon under nonstress conditions.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
All bacterial strains used are listed in Table 1. The 4-kb SmaI fragment containing hrcA and grpE was inserted into the DraI site of pBBR122 (MoBiTec, Göttingen, Germany) to construct phrcA-grpE. For most experiments, A. tumefaciens cultures were grown at 25°C in complete medium YEB under constant aeration or on YEB agar (11). For in vivo labeling experiments, Davis minimal medium (6) supplemented with 0.2% glucose, 2 μg of thiamine/ml, 50 μg of 18 l-amino acids (excluding methionine and cysteine)/ml, and 1/100 volume of YEB were used. Luria-Bertani broth was used for growing E. coli.
TABLE 1.
Bacterial strains used in the study
| Strain | Genotype | Source or reference |
|---|---|---|
| A. tumefaciens | ||
| GV3101 | rpoH+hrcA+ | 12 |
| KN501 | GV3101 ΔrpoH::tetR hrcA+ | This study |
| KN613 | GV3101 rpoH+ΔhrcA | This study |
| KN201 | GV3101 ΔrpoH::tetR ΔhrcA | This study |
| E. coli | ||
| MC4100 | Δ(argF-lac)205 ara Δ139 rpsL150 thiA1 relA1 flb-5301 deoC1 ptsF25 | Laboratory stock |
| KY1454 | MC4100 grpE280 | 14 |
Construction and screening of charomid library.
Construction of the charomid library and screening by complementation of the temperature-sensitive grpE mutant was done essentially as described previously (21). The DNA inserts of the charomids obtained were subcloned into the pUC-based plasmid vector, and the nucleotide sequence was determined by standard procedures.
Construction of deletion mutants.
To construct the ΔrpoH strains, the entire rpoH coding sequence with an additional 12 bases of the 5′ noncoding region was deleted from the 7-kb BamHI fragment (21) by PCR with primers rpoH-n1r (TATCTATGGTCTGGAACGGCACCCTCTTTGG) and rpoH-r1n (GGCGACACTCTCTTAAGGAGAGCACGCCATCC) and replaced by the tetracycline resistance gene from pBR322. The BamHI fragment lacking rpoH was inserted into pK18mobsacB (32), unable to replicate in A. tumefaciens, and introduced into cells of A. tumefaciens by electrotransformation with a Gene Pulsar with Pulse Controller (Bio-Rad, Richmond, Calif.). Kanamycin-resistant clones that resulted from homologous recombination between the plasmid and the chromosomal rpoH region were selected. To obtain the clones that had lost the inserted plasmid together with the intact copy of rpoH, clones that became resistant to 10% sucrose by the loss of sacB were isolated from among the initial transformants, and those that were sensitive to kanamycin and resistant to tetracycline were selected. The deletion of rpoH was confirmed by Southern blot analysis.
Essentially the same selection procedure was used to construct the ΔhrcA strains. All of the hrcA coding region, except the four N-terminal codons and the termination codon, was deleted by PCR with primers hrcA-n1r (CCGCTGAAAAACCCATCTTGTCCGTTCTTTATCG) and hrcA-r1n (CCGCATAGGACACATCAGCAAGATCAGAGAATGCGG). The resulting fragment was used to replace the chromosomal hrcA to obtain the hrcA deletion mutants. In this case, about 50% of the sucrose-resistant clones from the second selection turned out to be ΔhrcA, as confirmed by Southern blot analysis.
Antibodies.
For raising polyclonal antibodies against RpoH and HrcA, coding sequences for each protein were inserted into the expression vector pThioHisA (Invitrogen, Carlsbad, Calif.), and the fusion protein with modified thioredoxin was expressed in E. coli, purified with Ni2+ resin, and used for immunizing rabbits. Rabbit antiserum against E. coli GroEL was previously described (17). Antisera against two other E. coli HSPs, DnaK and ClpB, were kind gifts from M. Kohiyama (University of Paris 7) and C. Squires (Tufts University), respectively.
Isolation of RNA and primer extension analysis.
Cells from 50 ml of log-phase culture were quickly chilled, harvested by centrifugation for 1 min at 13,000 × g, resuspended in 0.25 ml of lysozyme solution (104 U/ml)(Ready-Lyse Lysozyme; Epicentre Technologies, Madison, Wis.) with isohypotonic buffer, and mixed with 0.75 ml of ISOGEN-LS (Nippon Gene, Tokyo, Japan). Total RNA was isolated as described in the manufacturer's manual. The 5′ end of primer HrcA-EX1 (GCCTGATCTTTTGAAAGCGGTGC; complementary to +13 to 35 of the hrcA coding sequence) or GrpE-Ex1 (TCCGCGACGTCCGCGTCAGGTCC; complementary to +25 to 47 of the grpE coding sequence) was labeled with 32P and used for primer extension analysis of hrcA or grpE, respectively. The fluorescent primer GE1 (fluorescein isothiocyanate labeled; TGCCTAATCCCTCGATC; complementary to +70 to 86 relative to the transcription start site of groE) or DK1 (fluorescein isothiocyanate labeled; TGAAGCGAGCTGTCTGAACC; complementary to +58 to 77 relative to the transcription start site of dnaK) was used for analysis of groE or dnaK mRNA, respectively, and 16S2 (FAM labeled; TGCCACTCCCCTTGCGGGGC; complementary to +41 to 61 of the 16S rRNA) was used for analysis of 16S rRNA as an internal control. Primer extension analysis was carried out essentially as described previously (2).
Nucleotide sequence accession number.
The nucleotide sequence of the entire region around hrcA in A. tumefaciens has been deposited in GenBank under accession no. AF039940.
RESULTS
Cloning of hrcA and the neighboring genes.
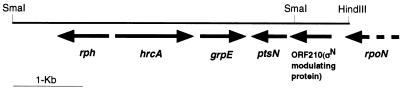
Assuming that hrcA of A. tumefaciens is located adjacent to grpE as in Caulobacter crescentus (27), we first screened a charomid library of the chromosomal DNA fragments for clones that could complement the temperature-sensitive growth of the grpE280 mutant of E. coli (KY1454). As expected, the resulting clones contained an open reading frame (ORF) which could code for a GrpE homolog. The putative GrpE protein of A. tumefaciens is 211 amino acids long and has 28.7 and 34.5% identity with the GrpEs of E. coli and C. crescentus, respectively. Sequencing of the several clones covering from about 2.5-kbp upstream to 1.5-kbp downstream of grpE revealed the presence of four other ORFs that showed homology with the known bacterial genes (Fig. 1). One of them, located directly upstream of grpE, separated by 88 bp, showed an appreciable homology with the hrcA genes of C. crescentus and other bacteria. The predicted amino acid sequence of putative HrcA was 363 amino acids and showed 47.8 and 25.4% identity with those of C. crescentus and B. subtilis, respectively. The ORF further upstream of hrcA on the opposite strand showed high sequence similarity to the rph gene encoding RNase PH of E. coli. This gene organization is identical with that found in C. crescentus (27). On the other hand, two ORFs located downstream on the opposite strand of grpE, designated ptsN and ORF210 (Fig. 1), had significant homology with ptsN, encoding phosphotransferase enzyme IIA, and ORF203, encoding a putative ς54 modulating protein of Bradyrhizobium japonicum, found at the distal end of the rpoN2 operon (16). A 3′ portion of an ORF, homologous to rpoN, was also found upstream of ORF210.
FIG. 1.
Gene arrangement around hrcA in A. tumefaciens. See the text for an explanation of each of the putative genes or ORFs.
Primer extension analysis of hrcA and grpE transcripts.
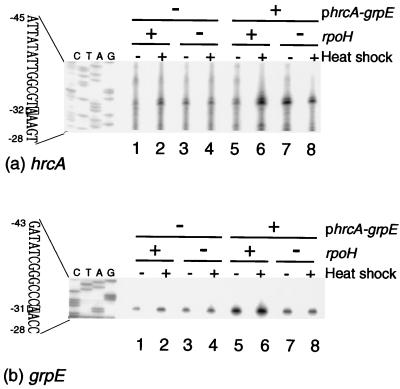
To examine transcripts of the hrcA and grpE genes, primer extension analysis was carried out with RNAs extracted from wild-type and ΔrpoH (described below) cells taken before and after a temperature shift from 25 to 37°C, using primers for the 5′ end of each gene. Since the transcripts detected were very scarce, strains harboring hrcA and grpE on a multicopy plasmid (pBBR122) were also used. A single transcript initiated at nucleotide −32 relative to the hrcA coding region was detected by using the hrcA primer, and its amount increased significantly upon heat shock (Fig. 2a). This transcript was hardly induced in the ΔrpoH mutant, suggesting possible involvement of RpoH in heat induction. Curiously, the basal level found at 25°C was higher in the ΔrpoH mutant than in the wild type. A putative promoter corresponding to this transcript showed some similarity to the heat shock promoters of A. tumefaciens and also to vegetative promoters of E. coli (Fig. 3). An inverted repeat of 12 (6 × 2) bases was found between the −10 and −35 regions of this promoter. This transcript may represent a bicistronic mRNA, since no clear hairpin structure that could implicate a transcription terminator was present between the coding regions of hrcA and grpE.
FIG. 2.
Transcription analysis of the hrcA and grpE genes. Cells were grown at 25°C in complete medium to mid-log phase and exposed to 37°C for 15 min. Samples taken before and after heat shock were used for RNA extraction. 32P-labeled primers for hrcA (a) or grpE (b) were hybridized with 10 μg of each RNA and subjected to primer extension analysis. The cDNA products were resolved by 6% sequence gels and autoradiographed. Sequence ladders produced by the same respective primers were run as markers, and the positions relative to the initiation codons are indicated. The positions of major signals are marked by boxes. Lanes: 1 and 2, GV3101 (rpoH+); 3 and 4, KN501 (ΔrpoH); 5 and 6, GV3101(phrcA-grpE); 7 and 8, KN501(phrcA-grpE). +, present; −, absent.
FIG. 3.
Promoter sequences of hrcA, grpE, and other heat shock genes in A. tumefaciens. An inverted repeat in the hrcA promoter is shown by arrows. The start sites of heat-inducible transcripts are underlined. Bases found in at least five of six possible RpoH-dependent promoters are indicated in upper case. Consensus deduced from these sequences and those of the E. coli ς32 and ς70 promoters are shown for comparison. References for the sequences are as follows: groESL, 34; dnaKJ, 35; rpoH and clpP, 19a; E. coli ς32 consensus, 10; E. coli ς70 consensus, 25.
Another transcript initiated from between hrcA and grpE, and induced upon heat shock, was detected by using the grpE primer (Fig. 2b). The heat inducibility was lost by the ΔrpoH mutation, although appreciable basal levels of this transcript remained, suggesting partial contribution of RpoH to the transcription. Consistently, a putative promoter for this transcript has some sequence similarity to other heat shock promoters whose activities depend at least partially on RpoH (Fig. 3) (see below). Thus, at least two transcripts initiated from upstream of hrcA and grpE, respectively, were shown to be involved in transcription of the two genes that may form an operon.
Construction of ΔrpoH, ΔhrcA, and the double-deletion mutants.
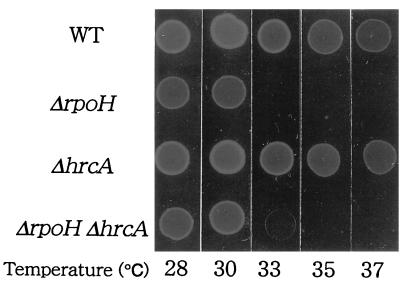
To analyze the roles of RpoH and HrcA in the heat shock response, we introduced ΔrpoH and ΔhrcA deletions into the A. tumefaciens chromosome and examined their effects, separately and in combination, on the heat shock response. The entire coding sequence of A. tumefaciens rpoH on the E. coli plasmid was replaced by the tetracycline resistance gene (tet), and ΔrpoH strains were constructed by homologous recombination between the ΔrpoH::tet fragment and rpoH on the host chromosome at 25°C. Similarly, most of the coding region of hrcA was deleted from the wild type or the ΔrpoH mutant to construct ΔhrcA or the ΔrpoH ΔhrcA double mutant, respectively. All these deletions were confirmed by Southern blotting. Immunoblotting analysis with the specific antisera revealed that the band for RpoH or HrcA, found in the wild-type extract, was not detected in the corresponding mutant extracts (data not shown). These results suggested that both the rpoH and hrcA genes exist in single copies and are expressed in the wild-type cells. The ΔrpoH mutant was able to grow at 30°C but not at higher temperatures, and growth was significantly slower even at permissive temperatures (e.g., 25°C) (Fig. 4). In contrast, the ΔhrcA mutant grew normally or slightly faster than the wild type at physiological temperatures. The double mutant lacking both RpoH and HrcA exhibited temperature-sensitive growth similar to that of the ΔrpoH mutant but grew slightly faster at the permissive temperature than the ΔrpoH single mutant.
FIG. 4.
Growth of the ΔrpoH and ΔhrcA mutants. Overnight cultures grown in complete medium (YEB) at 25°C were diluted 10-fold, and 5-μl samples were spotted on the YEB plates and incubated overnight at the indicated temperatures. WT, GV3101; ΔrpoH, KN501; ΔhrcA, KN613; ΔrpoH ΔhrcA, KN201.
Effects of ΔrpoH and ΔhrcA mutations on HSP synthesis.
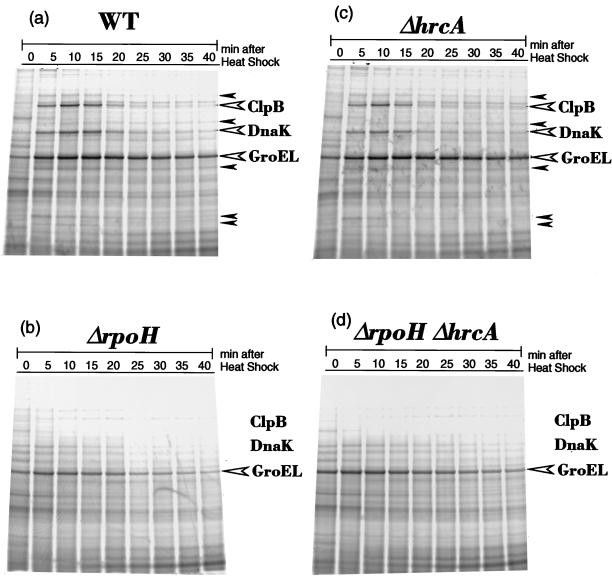
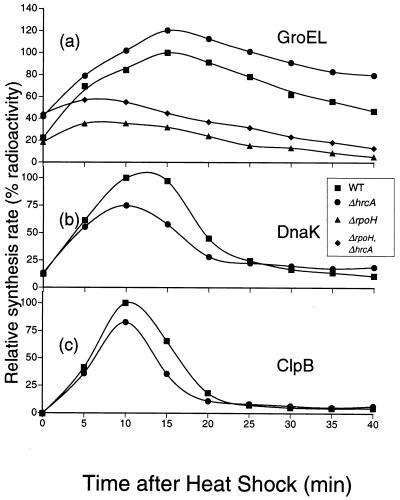
The set of mutants described above were examined for the synthesis of HSPs before and after temperature upshift. When wild-type cells grown at 25°C were shifted to 37°C, the synthesis of several HSPs was rapidly and transiently induced (Fig. 5a). Among them, the most abundant proteins, of about 90, 70, and 60 kDa, were identified as ClpB, DnaK, and GroEL homolog, respectively, by immunoprecipitation with specific antisera for the respective proteins of E. coli (data not shown). In the ΔrpoH mutant lacking RpoH, none of these HSPs except GroEL appeared to be induced significantly; GroEL was synthesized apparently normally at 25°C but was only modestly enhanced upon heat shock (Fig. 5b). These results suggested that RpoH is responsible for the heat induction of most, if not all, HSPs in A. tumefaciens. On the other hand, the ΔhrcA deletion hardly affected the synthesis of HSP, except that GroEL synthesis was slightly higher than in the wild type both before and after temperature upshift (Fig. 5c), indicating the role of HrcA in repressing GroE expression. The synthesis patterns in the double mutant were very similar to those in the ΔrpoH single mutant except for the higher rates of GroEL expression throughout (Fig. 5d); thus, the ΔrpoH and ΔhrcA mutations exhibited additive effects on the synthesis of these proteins.
FIG. 5.
Sodium dodecyl sulfate (SDS)-polyacrylamide gel patterns of HSP synthesis in mutants. The cells were grown in enriched minimal medium at 25°C and shifted to 37°C at time zero. Samples were taken at intervals, pulse labeled with [35S]methionine for 2 min, and treated with trichloroacetic acid. Whole cell proteins were analyzed by SDS-polyacrylamide gel (7.5% gel) electrophoresis and visualized with a phosphorimager. The bands of the most abundantly synthesized HSPs (GroEL, DnaK, and ClpB) are marked with open arrowheads, and other possible HSPs are marked with closed arrowheads. (a) WT, GV3101; (b) ΔrpoH, KN501; (c) ΔhrcA, KN613; (d) ΔhrcA ΔrpoH, KN201.
The synthesis rates of three major HSPs were quantified to further evaluate the effects of the deletions on the kinetics and extent of heat shock response. The GroEL synthesis was indeed higher in the ΔhrcA mutant than in the wild type at all periods tested, but especially higher rates of derepression were observed before heat shock and during the shutoff periods, not at the peak of heat induction (Fig. 6a). A slight but significant induction of GroEL which peaks at about 10 min was observed in the ΔrpoH mutant and even in the ΔhrcA ΔrpoH mutant. This induction in the double-deletion mutant may be due to specific processing and stabilization of the groEL mRNA (36), since the amount of mRNA immediately downstream of the 5′ end of the groE operon did not change significantly upon heat shock (see below). On the other hand, induction of DnaK and ClpB in the ΔhrcA mutant was similar to that in the wild type, except for the slightly lower extents of induction for both proteins (Fig. 6b and c). Synthesis of DnaK and ClpB in the ΔrpoH mutant, which could not be accurately determined due to the background, was found by immunoprecipitation to be less than 6 or 4%, respectively, of the maximum synthesis rates in the wild type (data not shown).
FIG. 6.
Kinetics of HSP induction upon temperature upshift. Radioactivity associated with the protein bands of the three major HSPs shown in Fig. 3 were quantified to analyze the kinetics of induction. (a) GroEL; (b) DnaK; (C) ClpB. The radioactivity of each band was plotted after normalization to the maximum synthesis rate in the wild type, set at 100. DnaK and ClpB in the ΔrpoH and ΔhrcA ΔrpoH mutants were not plotted, as they were below the background levels. ■, GV3101 (wild type); ▴, KN501 (ΔrpoH); ●, KN613 (ΔhrcA); ⧫, KN201 (ΔhrcA ΔrpoH).
Analysis of the dnaK and groE transcripts with the mutants.
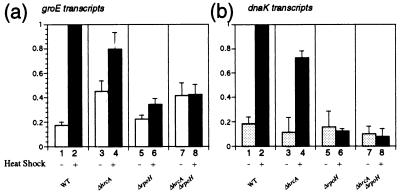
To clarify the roles of RpoH and HrcA in the transcriptional induction of HSP genes, we chose groE and dnaK operons that had been analyzed previously and examined the amounts of the respective mRNAs by primer extension (Fig. 7). The groE major transcript markedly increased upon heat shock in the wild type, as reported previously (36). This induction was strikingly affected by the ΔrpoH mutation, the mRNA level after heat shock being less than 40% that of the wild type (Fig. 7a, compare lanes 2 and 6). However, the basal-level expression before heat shock was hardly affected by the lack of RpoH (compare lanes 1 and 5). A similar effect was observed in the ΔhrcA background (compare lanes 4 and 8 and lanes 3 and 7). These results indicated that RpoH contributes greatly to the heat induction but not to the basal-level transcription of groE mRNA. Apparently, the basal transcription depends mostly on a ς factor(s) other than RpoH.
FIG. 7.
Transcription analyses of the groE and dnaK genes. Cells were grown at 25°C in complete medium to mid-log phase and exposed to 37°C for 15 min. Samples taken before and after heat shock were used for RNA extraction. The amount of heat shock transcripts was determined by primer extension analysis. The extension reaction was carried out with each of the groE (a) or dnaK (b) primers complementary to the region downstream of the known transcription start sites of each operon and with 16S rRNA primer labeled with different fluorescent dyes. The cDNA products were analyzed on 8% sequence gels. Four experiments with two separate RNA samples were carried out. The transcripts were quantified by using 16S rRNA as an internal reference and then normalized to the value for the heat-shocked wild-type cells, and average values are presented with standard errors. Open and shaded bars, RNA from nonstressed cells; solid bars, RNA from heat-shocked cells.
On the other hand, the ΔhrcA deletion caused about a twofold increase in the groE transcript under nonstress conditions in both the rpoH+ and ΔrpoH strains (Fig. 7a, compare lanes 1 and 3 and lanes 5 and 7) but had little effect on the transcript level obtained after heat shock (compare lanes 2 and 4 and lanes 6 and 8). Thus, the repression by HrcA is exerted efficiently during steady-state growth but much less so upon temperature upshift. This was consistent with the previous Northern analyses of transcripts from the groE operon with deletions or mutations in the CIRCE sequence (37). These results are also in good agreement with findings with other bacteria (27, 33, 41).
When both rpoH and hrcA were deleted, no induction of groE transcription was observed upon heat shock, in contrast to the partial induction observed with either of the single mutants. It appears, therefore, that the two regulatory factors, RpoH and HrcA, work independently and that their activities are mostly responsible for heat induction of groE transcription in A. tumefaciens.
Transcriptional regulation of dnaK which does not contain CIRCE appeared simpler. The major transcript, initiated from the single known start site, markedly increased upon heat shock in the wild type and in the ΔhrcA mutant, whereas no induction was detected in the ΔrpoH mutant or the ΔrpoH ΔhrcA double mutant (Fig. 7b), indicating that dnaK transcriptional induction upon heat shock depends solely on RpoH. On the other hand, the ΔhrcA mutation caused significant reduction in dnaK transcript both before and after heat shock. This reduction, consistent with the reduced rate of DnaK synthesis (Fig. 6b), may be attributed to the increased level of GroE which might somehow reduce the activity of RpoH.
Increased expression of RpoH but not HrcA during heat shock response.
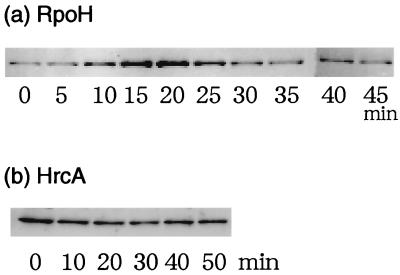
To analyze the expression of RpoH and HrcA in A. tumefaciens, we raised antisera against recombinant RpoH and HrcA and used them to determine the changes in protein levels during the heat shock response by immunoblotting. A significant level of RpoH protein was detected in cells grown at 25°C, and its level was markedly enhanced (by four- to fivefold) upon the shift to 37°C. After reaching the maximum at about 15 min, the RpoH level gradually decreased to near the preshift level after about 30 min (Fig. 8a). In spite of the increase of hrcA transcript observed, the HrcA protein was not induced by the heat shock and the HrcA level did not change significantly for at least 50 min after the shift to 37°C (Fig. 8b).
FIG. 8.
The cellular levels of RpoH and HrcA during the heat shock response. Cells of wild-type A. tumefaciens (GV3101) were grown to mid-log phase in complete medium at 25°C and shifted to 37°C at time zero. Samples were taken at the indicated times, mixed with an equal volume of 2× sodium dodecyl sulfate (SDS) loading buffer, and boiled for 5 min. Equal amounts of protein adjusted by the optical density (in Klett units) of the culture were loaded on the SDS-polyacrylamide gels (12.5% gels), blotted onto a nitrocellulose membrane (Hybond ECL; Amersham Life Science), and detected with rabbit antiserum against RpoH (a) or HrcA (b) by chemiluminescence techniques.
DISCUSSION
Previous work indicated the involvement of the CIRCE-HrcA system in regulation of the groE operons in alpha proteobacteria, including A. tumefaciens (3, 27, 37). These studies showed that the repression of groE transcription is mediated by the CIRCE-HrcA system, particularly under nonstress conditions. However, the role of this system in the heat induction of groE could not be directly examined, because heat shock induction occurred even when hrcA or CIRCE had been deleted, presumably by contribution of the heat shock ς factor, RpoH. Here we have shown that both HrcA and RpoH contribute independently to the heat shock response of groE transcription in A. tumefaciens, since the single-deletion mutants lacking either hrcA or rpoH could still respond to heat shock, albeit weakly, and enhanced groE transcripts significantly whereas the double deletions virtually abolished the ability to respond to high temperature.
In B. subtilis and perhaps other low-G+C gram-positive bacteria, the CIRCE-HrcA system plays a key regulatory role in expression of both the groE and dnaK operons encoding major chaperones (33, 41). In contrast, CIRCE is found in many species of alpha proteobacteria but only in the groE operons (3, 4, 37, 38), implying that groE is probably the only target regulated by the CIRCE-HrcA system in this group. The present study revealed that HrcA is indeed involved in the regulation of groE transcription of A. tumefaciens by repressing it under nonstress conditions and releasing the repression upon heat stress. However, HrcA contributes only modestly to the temperature regulation of groE transcription, which is also controlled by RpoH. In addition, the heat shock regulation of GroE in this bacterium is not confined to transcriptional control but also involves specific processing and stabilization of groEL mRNA (36): thus, the contribution of HrcA to the heat shock response of GroE is further limited. The specific stabilization of groEL mRNA may explain the slight induction of GroEL synthesis in the ΔrpoH ΔhrcA double mutant and the higher rate of GroEL synthesis in the ΔhrcA mutant upon heat shock (Fig. 6a), neither of which was observed at the transcript level as determined for the groES segment (Fig. 7a).
As for the heat induction of other HSPs, no appreciable effect was observed from the lack of HrcA, although induction of both ClpB and DnaK was slightly reduced (Fig. 6b and c). Since no direct effect of HrcA on dnaK transcription is expected, this may be an indirect effect of the increased GroE level in the ΔhrcA mutant, which might diminish the stress caused by heat shock and reduce the RpoH-mediated heat shock response.
The hrcA transcript of A. tumefaciens, determined by semiquantitative primer extension, was significantly induced after heat shock. Since the putative promoter has some similarity to other heat shock promoters, contribution of RpoH to this promoter was expected, as in the case of C. crescentus (27). Indeed, the possible contribution of RpoH to this transcription was suggested, because heat induction observed with the wild type was not found in the ΔrpoH mutant. In addition, the level of this transcript seen under nonstress condition was higher in the ΔrpoH mutant than in the wild type, suggesting possible involvement of another regulatory mechanism for the transcription. Currently, the nature of the mechanism remains unknown; an inverted repeat found between the −35 and −10 regions might play a role in this or other regulation of HrcA expression. In this connection, an interesting paradox is that the heat-induced transcription of hrcA did not result in significant enhancement of HrcA protein upon heat shock. The transcriptional induction might produce untranslatable RNA, or HrcA might be destabilized at high temperature. Further work is needed to solve these problems.
In contrast to the restricted role of HrcA, RpoH appears to be responsible for heat induction of most HSPs, acting as a global regulator of the heat shock response, like ς32 in E. coli. When produced in E. coli, RpoH of A. tumefaciens can correctly recognize the dnaK and groE heat shock promoters of E. coli (20). Since most of the HSP genes so far examined in A. tumefaciens, groESL, dnaKJ, grpE, rpoH, and clpP, contain heat-inducible promoters similar to the heat shock promoters of E. coli (Fig. 3), RpoH was anticipated to be responsible for transcription of these promoters. As expected, the deletion of rpoH resulted in a complete or partial loss of heat-induced transcription from the dnaK or groE promoter, respectively (Fig. 7). Furthermore, all the heat shock proteins detected by pulse labeling were markedly affected in the ΔrpoH mutant, clearly implying that induction of most if not all HSPs in A. tumefaciens is primarily controlled by RpoH.
In spite of a major role of RpoH in the synthesis of HSP, the ΔrpoH mutant of A. tumefaciens showed a relatively mild temperature sensitivity, the inability to grow only above 30°C, compared with the E. coli ΔrpoH mutant, which cannot grow above 20°C. This may be related to the relatively high basal expression of GroEL in A. tumefaciens in the absence of RpoH (Fig. 5 to 7), in view of the findings with the E. coli ΔrpoH mutant and its temperature-resistant revertants that the cellular level of GroE primarily determines the upper limit of growth temperature (17). The expression of GroEL in the ΔrpoH mutant depends on the transcript initiated apparently from the same start site used in the rpoH+ strain. We don't know which sigma factor(s) is responsible for this transcription in the ΔrpoH mutant, but it seems likely that the same sigma is responsible for basal level transcription of groE, grpE, and other heat shock genes in the rpoH+ strain. Such a sigma factor would not be activated by heat shock, as judged by the groE transcription in the ΔrpoH ΔhrcA double mutant or by dnaK transcription in the ΔrpoH mutants. The existence of a multiple RpoH-like ς factor(s), as was found in B. japonicum, seems unlikely, because neither proteins that can cross-react with anti-RpoH polyclonal antibodies nor a DNA fragment that hybridizes with an rpoH fragment even with very low stringencies was found in cell extracts or genomic DNA of ΔrpoH cells, respectively. However a very distant relative of RpoH might exist and be responsible for basal transcription of heat shock genes, including groE. Alternatively, the vegetative sigma factor SigA, a ς70 homolog, could recognize the groE promoter, which has some similarity to ς70 promoters, especially at its −35 region (34). Also, the region upstream of −35 might be important for the high basal activity of the groE promoter, since deletion of bases −37 to −53 greatly reduces transcription from this promoter (37); an AT stretch present in this region may act like an UP element (29). In any event, the potentially high basal activity specifically seen with the groE promoter may provide a basis for negative control by the CIRCE-HrcA system even in the absence of activation by RpoH, leading to efficient and versatile regulation by the combination of two systems.
We found that the amount of RpoH increases markedly upon heat shock and reaches a maximum at about 15 min. This induction seems to be autoregulated primarily at the level of transcription initiated from an RpoH-dependent promoter (19a), as reported in C. crescentus (26, 40). If that is the case, an additional mechanism(s) which not only triggers but also terminates the positive-feedback circuit should exist for modulating RpoH induction. Indeed, the induction of DnaK and ClpB peaks within 10 min after temperature upshift, when the cellular level of RpoH still continues to increase, suggesting that a certain mechanism controlling RpoH activity is involved. Further work on the regulation of RpoH in A. tumefaciens should reveal the nature of both conserved and divergent strategies in the regulation of RpoH and of the heat shock response in proteobacteria.
ACKNOWLEDGMENTS
We are grateful to M. Kohiyama and C. Squires for providing antibodies and to A. Oka for helpful discussion. We also thank Masako Nakayama and Seiji Takahara for technical assistance.
REFERENCES
- 1.Andersson S G, Zomorodipour A, Andersson J O, Sicheritz-Ponten T, Alsmark U C, Podowski R M, Naslund A K, Eriksson A S, Winkler H H, Kurland C G. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley; 1987. [Google Scholar]
- 3.Babst M, Hennecke H, Fischer H M. Two different mechanisms are involved in the heat-shock regulation of chaperonin gene expression in Bradyrhizobium japonicum. Mol Microbiol. 1996;19:827–839. doi: 10.1046/j.1365-2958.1996.438968.x. [DOI] [PubMed] [Google Scholar]
- 4.Baldini R L, Avedissian M, Gomes S L. The CIRCE element and its putative repressor control cell cycle expression of the Caulobacter crescentus groESL operon. J Bacteriol. 1998;180:1632–1641. doi: 10.1128/jb.180.7.1632-1641.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ballard S A, Go M, Segers R, Adler B. Molecular analysis of the dnaK locus of Leptospira interrogans serovar copenhageni. Gene. 1998;216:21–29. doi: 10.1016/s0378-1119(98)00329-1. [DOI] [PubMed] [Google Scholar]
- 6.Davis B D. Isolation of biochemically deficient mutants of bacteria by means of penicillin. Proc Natl Acad Sci USA. 1949;35:1–10. doi: 10.1073/pnas.35.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferreyra R G, Soncini F C, Viale A M. Cloning, characterization, and functional expression in Escherichia coli of chaperonin (groESL) genes from the phototrophic sulfur bacterium Chromatium vinosum. J Bacteriol. 1993;175:1514–1523. doi: 10.1128/jb.175.5.1514-1523.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glatz A, Horvath I, Varvasovszki V, Kovacs E, Torok Z, Vigh L. Chaperonin genes of the Synechocystis PCC 6803 are differentially regulated under light-dark transition during heat stress. Biochem Biophys Res Commun. 1997;239:291–297. doi: 10.1006/bbrc.1997.7463. [DOI] [PubMed] [Google Scholar]
- 9.Grandvalet C, Rapoport G, Mazodier P. hrcA, encoding the repressor of the groEL genes in Streptomyces albus G, is associated with a second dnaJ gene. J Bacteriol. 1998;180:5129–5134. doi: 10.1128/jb.180.19.5129-5134.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gross C A. Function and regulation of the heat shock proteins. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C.: ASM Press; 1996. pp. 1382–1399. [Google Scholar]
- 11.Hirayama T, Muranaka T, Ohkawa H, Oka A. Organization and characterization of the virCD genes from Agrobacterium rhizogenes. Mol Gen Genet. 1988;213:229–237. doi: 10.1007/BF00339586. [DOI] [PubMed] [Google Scholar]
- 12.Holsters M, Silva B, Van Vliet F, Genetello C, De Block M, Dhaese P, Depicker A, Inze D, Engler G, Villarroel R. The functional organization of the nopaline A. tumefaciens plasmid pTiC58. Plasmid. 1980;3:212–230. doi: 10.1016/0147-619x(80)90110-9. [DOI] [PubMed] [Google Scholar]
- 13.Huang L H, Tseng Y H, Yang M T. Isolation and characterization of the Xanthomonas campestris rpoH gene coding for a 32-kDa heat shock sigma factor. Biochem Biophys Res Commun. 1998;244:854–860. doi: 10.1006/bbrc.1998.8367. [DOI] [PubMed] [Google Scholar]
- 14.Ishiai M, Wada C, Kawasaki Y, Yura T. Mini-F plasmid mutants able to replicate in Escherichia coli deficient in the DnaJ heat shock protein. J Bacteriol. 1992;174:5597–5603. doi: 10.1128/jb.174.17.5597-5603.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karls R K, Brooks J, Rossmeissl P, Luedke J, Donohue T J. Metabolic roles of a Rhodobacter sphaeroides member of the ς32 family. J Bacteriol. 1998;180:10–19. doi: 10.1128/jb.180.1.10-19.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kullik I, Fritsche S, Knobel H, Sanjuan J, Hennecke H, Fischer H M. Bradyrhizobium japonicum has two differentially regulated, functional homologs of the ς54 gene (rpoN) J Bacteriol. 1991;173:1125–1138. doi: 10.1128/jb.173.3.1125-1138.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kusukawa N, Yura T. Heat shock protein GroE of Escherichia coli: key protective roles against thermal stress. Genes Dev. 1988;2:874–882. doi: 10.1101/gad.2.7.874. [DOI] [PubMed] [Google Scholar]
- 18.Mogk A, Homuth G, Scholz C, Kim L, Schmid F X, Schumann W. The GroE chaperonin machine is a major modulator of the CIRCE heat shock regulon of Bacillus subtilis. EMBO J. 1997;16:4579–4590. doi: 10.1093/emboj/16.15.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mogk A, Volker A, Engelmann S, Hecker M, Schumann W, Volker U. Nonnative proteins induce expression of the Bacillus subtilis CIRCE regulon. J Bacteriol. 1998;180:2895–2900. doi: 10.1128/jb.180.11.2895-2900.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.Nakahigashi, K. Unpublished results.
- 20.Nakahigashi K, Kanemori M, Morita M, Yanagi H, Yura T. Conserved function and regulation of ς32 homologues in Gram-negative bacteria. J Biosci. 1998;23:407–414. [Google Scholar]
- 21.Nakahigashi K, Yanagi H, Yura T. Isolation and sequence analysis of rpoH genes encoding ς32 homologs from gram negative bacteria: conserved mRNA and protein segments for heat shock regulation. Nucleic Acids Res. 1995;23:4383–4390. [PMC free article] [PubMed] [Google Scholar]
- 22.Nakahigashi K, Yanagi H, Yura T. Regulatory conservation and divergence of ς32 homologs from Gram-negative bacteria: Serratia marcescens, Proteus mirabilis, Pseudomonas aeruginosa, and Agrobacterium tumefaciens. J Bacteriol. 1998;180:2402–2408. doi: 10.1128/jb.180.9.2402-2408.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Narberhaus F. Negative regulation of bacterial heat shock genes. Mol Microbiol. 1999;31:1–8. doi: 10.1046/j.1365-2958.1999.01166.x. [DOI] [PubMed] [Google Scholar]
- 24.Narberhaus F, Krummenacher P, Fischer H M, Hennecke H. Three disparately regulated genes for ς32-like transcription factors in Bradyrhizobium japonicum. Mol Microbiol. 1997;24:93–104. doi: 10.1046/j.1365-2958.1997.3141685.x. [DOI] [PubMed] [Google Scholar]
- 25.Record M T, Jr, Reznikoff W S, Craig M L, McQuade K L, Cshlax P J. Escherichia coli RNA polymerase (Eς70), promoters, and the kinetics of the steps of transcription initiation. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C.: ASM Press; 1996. pp. 792–821. [Google Scholar]
- 26.Reisenauer A, Mohr C D, Shapiro L. Regulation of a heat shock ς32 homolog in Caulobacter crescentus. J Bacteriol. 1996;178:1919–1927. doi: 10.1128/jb.178.7.1919-1927.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts R C, Toochinda C, Avedissian M, Baldini R L, Gomes S L, Shapiro L. Identification of a Caulobacter crescentus operon encoding hrcA, involved in negatively regulating heat-inducible transcription, and the chaperone gene grpE. J Bacteriol. 1996;178:1829–1841. doi: 10.1128/jb.178.7.1829-1841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ron E Z, Segal G, Robinson M, Graur D. Control elements in the regulation of bacterial heat shock response. In: Rosenberg E, editor. Microbial ecology and infectious disease. Washington, D.C.: ASM Press; 1998. pp. 143–152. [Google Scholar]
- 29.Ross W, Gosink K K, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse R L. A third recognition element in bacterial promoters: DNA binding by the α subunit of RNA polymerase. Science. 1993;262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- 30.Sahu G K, Chowdhury R, Das J. The rpoH gene encoding ς32 homolog of Vibrio cholerae. Gene. 1997;189:203–207. doi: 10.1016/s0378-1119(96)00849-9. [DOI] [PubMed] [Google Scholar]
- 31.Sato S, Ishikawa H. Expression and control of an operon from an intracellular symbiont which is homologous to the groE operon. J Bacteriol. 1997;179:2300–2304. doi: 10.1128/jb.179.7.2300-2304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schafer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Puhler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 33.Schulz A, Schumann W. hrcA, the first gene of the Bacillus subtilis dnaK operon, encodes a negative regulator of class I heat shock genes. J Bacteriol. 1996;178:1088–1093. doi: 10.1128/jb.178.4.1088-1093.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Segal G, Ron E Z. Heat shock transcription of the groESL operon of Agrobacterium tumefaciens may involve a hairpin-loop structure. J Bacteriol. 1993;175:3083–3088. doi: 10.1128/jb.175.10.3083-3088.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Segal G, Ron E Z. The dnaKJ operon of Agrobacterium tumefaciens: transcriptional analysis and evidence for a new heat shock promoter. J Bacteriol. 1995;177:5952–5958. doi: 10.1128/jb.177.20.5952-5958.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Segal G, Ron E Z. The groESL operon of Agrobacterium tumefaciens: evidence for heat shock-dependent mRNA cleavage. J Bacteriol. 1995;177:750–757. doi: 10.1128/jb.177.3.750-757.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Segal G, Ron E Z. Heat shock activation of the groESL operon of Agrobacterium tumefaciens and the regulatory roles of the inverted repeat. J Bacteriol. 1996;178:3634–3640. doi: 10.1128/jb.178.12.3634-3640.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Segal G, Ron E Z. Regulation and organization of the groE and dnaK operons in Eubacteria. FEMS Microbiol Lett. 1996;138:1–10. doi: 10.1111/j.1574-6968.1996.tb08126.x. [DOI] [PubMed] [Google Scholar]
- 39.Tan M, Wong B, Engel J N. Transcriptional organization and regulation of the dnaK and groE operons of Chlamydia trachomatis. J Bacteriol. 1996;178:6983–6990. doi: 10.1128/jb.178.23.6983-6990.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu J, Newton A. The Caulobacter heat shock sigma factor gene rpoH is positively autoregulated from a ς32-dependent promoter. J Bacteriol. 1997;179:514–521. doi: 10.1128/jb.179.2.514-521.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan G, Wong S L. Isolation and characterization of Bacillus subtilis groE regulatory mutants: evidence for orf39 in the dnaK operon as a repressor gene in regulating the expression of both groE and dnaK. J Bacteriol. 1995;177:6462–6468. doi: 10.1128/jb.177.22.6462-6468.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yura T, Nagai H, Mori H. Regulation of the Escherichia coli heat-shock response. Annu Rev Microbiol. 1993;47:321–350. doi: 10.1146/annurev.mi.47.100193.001541. [DOI] [PubMed] [Google Scholar]
- 43.Zuber U, Schumann W. CIRCE, a novel heat shock element involved in regulation of heat shock operon dnaK of Bacillus subtilis. J Bacteriol. 1994;176:1359–1363. doi: 10.1128/jb.176.5.1359-1363.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]