Abstract
Symbiotic partnerships with rhizobial bacteria enable legumes to grow without nitrogen fertilizer, because rhizobia convert atmospheric nitrogen gas into ammonia via nitrogenase. After Sinorhizobium meliloti penetrate the root nodules that they have elicited in Medicago truncatula, the plant produces a family of ca. 700 NCR (Nodule Cysteine Rich) peptides that guide differentiation of endocytosed bacteria into nitrogen fixing bacteroids. The sequences of the NCR peptides are related to the defensin class of anti-microbial peptides but have been adapted to play symbiotic roles. Using a variety of spectroscopic, biophysical and biochemical techniques we show that the most extensively characterized NCR peptide, defensin-like 24 amino acid NCR247, binds haem with nanomolar affinity. Bound haem molecules and their iron are made biologically inaccessible first by the formation of hexamers (6 haem: 6 NCR247) and then higher-order complexes. We present evidence that NCR247 is critical for an effective nitrogen-fixing symbiosis. We propose that, by sequestering haem and its bound iron, NCR247 creates a physiological state of haem deprivation. This in turn induces an iron-starvation response in rhizobia that results in iron import, which itself is required for nitrogenase activity. Using the same methods as for L-NCR247, we show that D- enantiomer of NCR247 can bind and sequester haem equivalently well. The special abilities of NCR247 and its D-enantiomer to sequester haem suggest a broad range of potential applications related to human health.
The ecologically and agriculturally important symbiosis between rhizobial bacteria and their legume hosts permits these plants to grow without nitrogen fertilizer, because the rhizobia use nitrogenase to convert nitrogen gas into ammonia. As part of this process, the bacteria penetrate the root nodules that they have elicited and are endocytosed into membrane compartments in the cytoplasm of plant cells in the interior of the nodules1. In the case of the Sinorhizobium meliloti-Medicago truncatula symbiosis, the plant expresses a family of ca. 700 defensin-related NCR (Nodule Cysteine Rich) peptides specifically in the nodules only, which guide the endocytosed bacteria into terminally differentiating into nitrogen-fixing bacteroids2. Two of these peptides NCR2113 and NCR1694 have been shown to be critical for symbiosis but their molecular mechanism is not understood. A few NCR peptides have been shown to affect membrane polarization5, while one has been shown to interact with a few bacterial proteins6. However, the molecular mechanisms of action of these NCR peptides in symbiosis is a major unanswered question in the field.
Here we set out to establish the mode of action of the smallest and best characterized of these peptides, 24 amino acid NCR247 7,8 (RNGCIVDPRCPYQQCRRPLYCRRR). Our research was strongly influenced by our unanticipated discovery that NCR247 binds and sequesters haem with nanomolar affinity. This finding was of particular interest because it suggested a possible molecular mechanism for our previous observation that the complex transcriptional response of S. meliloti cells treated with a symbiosis-relevant concentration of NCR247 in vitro included increased expression of genes that are repressed by RirA7. RirA represses transcription of at least 53 iron-responsive genes including those involved in iron uptake9. Iron is of particular importance to the nitrogen-fixing symbiosis because each nitrogenase contains about 24-32 iron atoms. We carried out a set of biochemical, biophysical and physiological experiments to test the hypothesis that haem sequestration by NCR247 is a crucial step in establishing an effective symbiosis between S. meliloti and M. truncatula. Our results have revealed the first detailed molecular mechanism of action of any NCR peptide.
RESULTS
NCR247 elicits an iron starvation response
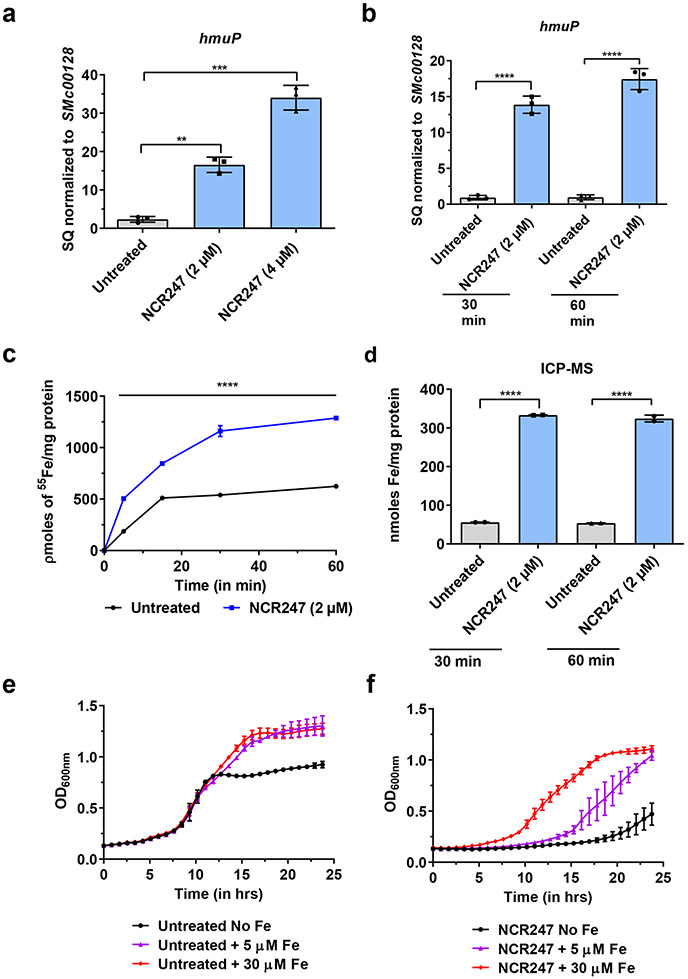
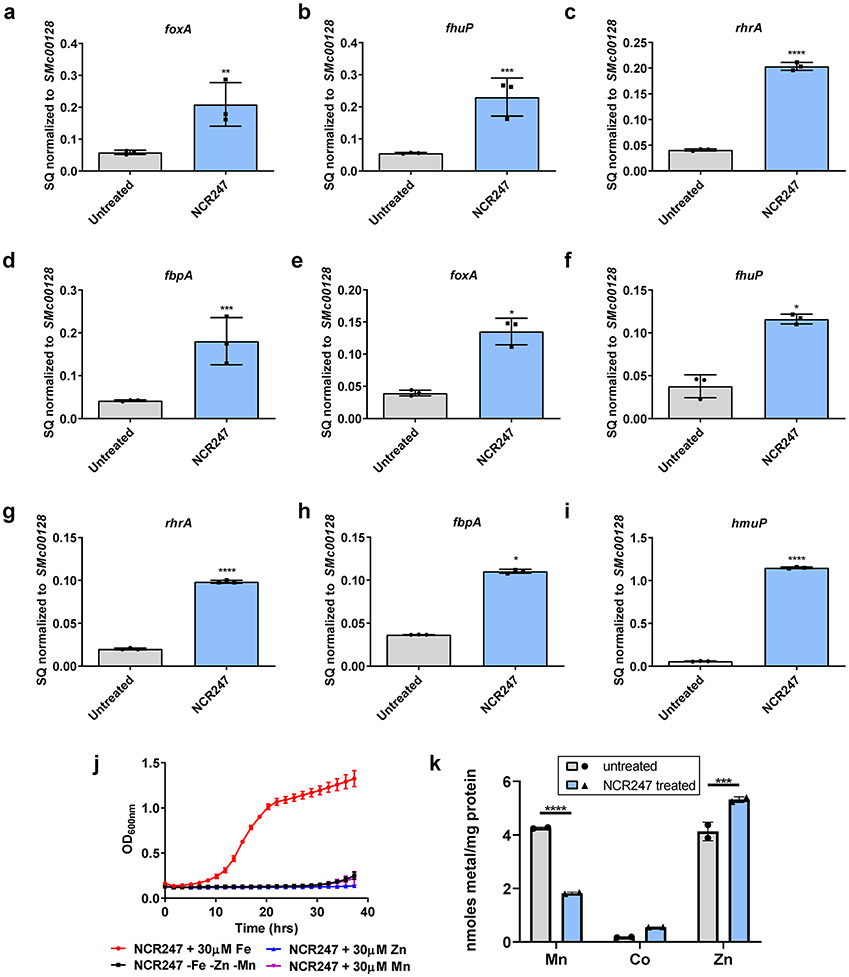
First, we tested the iron-related transcriptional response of S. meliloti to symbiotically relevant concentrations of NCR247 7,10 in minimal medium using qRT-PCR analysis. Consistent with our pervious transcriptome analysis11, many of the iron uptake genes were increased in expression (Fig. 1a). Surprisingly, we observed NCR247 treatment led to an increase in transcript levels of iron uptake genes even when S. meliloti was grown in iron-replete medium (Fig. 1b; Extended data Fig. 1a-i), a condition in which iron-uptake is normally repressed. Using radioactive 55Fe uptake assay, we found that NCR247 markedly increased the import of 55Fe (Fig. 1c) into S. meliloti even when grown in medium with sufficient iron, resulting in an increase in an elevated total iron content of the cells measured by ICP-MS analysis (Fig. 1d). NCR247 causes a reduction in growth rate of S. meliloti in a minimal medium and, based on the above observations, we hypothesized that addition of iron might suppress this phenotype. Indeed, addition of increasing concentrations of iron progressively increased the growth rate of NCR247 treated cells (Fig. 1e and 1f). Other metals we tested, including zinc and manganese, did not restore bacterial growth rates in the presence of NCR247 indicating that the effects we observed might be iron-specific responses (Extended data Fig. 1j). The slight alterations in Mn and Zn content of S. meliloti may be due to alteration in iron homeostatic mechanisms of S. meliloti (Extended data Fig. 1k). Our data support the hypothesis that NCR247 causes an iron starvation response in S. meliloti even under iron-replete conditions, but the molecular mechanism responsible for this effect was not clear.
Fig.1∣. NCR247 induces iron starvation response and drives iron import into S. meliloti.
a, Increase in expression of a gene involved in iron import (hmuP) in S. meliloti upon NCR247 treatment for 30 mins, when grown in minimal media -Fe, as quantitated by qRT-PCR analysis. b, Increase in expression of hmuP upon NCR247 (2 μM) treatment when S. meliloti was grown in iron sufficient medium (5 μM). In a and b, the data are expressed as starting quantities (SQ) of respective mRNAs normalized to the control gene SMc00128 and are presented as an average of three technical replicates ± s.d. c, Increase in uptake of 55Fe upon treatment with 2 μM NCR247 when compared to untreated S. meliloti. d, Increase in total iron content of 2 μM NCR247 treated S. meliloti, measured by ICP-MS analysis. e, Growth pattern of untreated cells in iron-depleted (-Fe), iron sufficient (5 μM), and iron-replete media (30 μM). f, Rescue of growth by Fe in 2 μM NCR247 treated cells. In c-f, data are presented as mean of three biological replicates ± s.d. In a-d, **P=0.0035, ***P=0.0002 and ****P<0.0001 vs untreated sample; two-way analysis of variance (ANOVA) with multiple comparisons.
NCR247 binds to haem
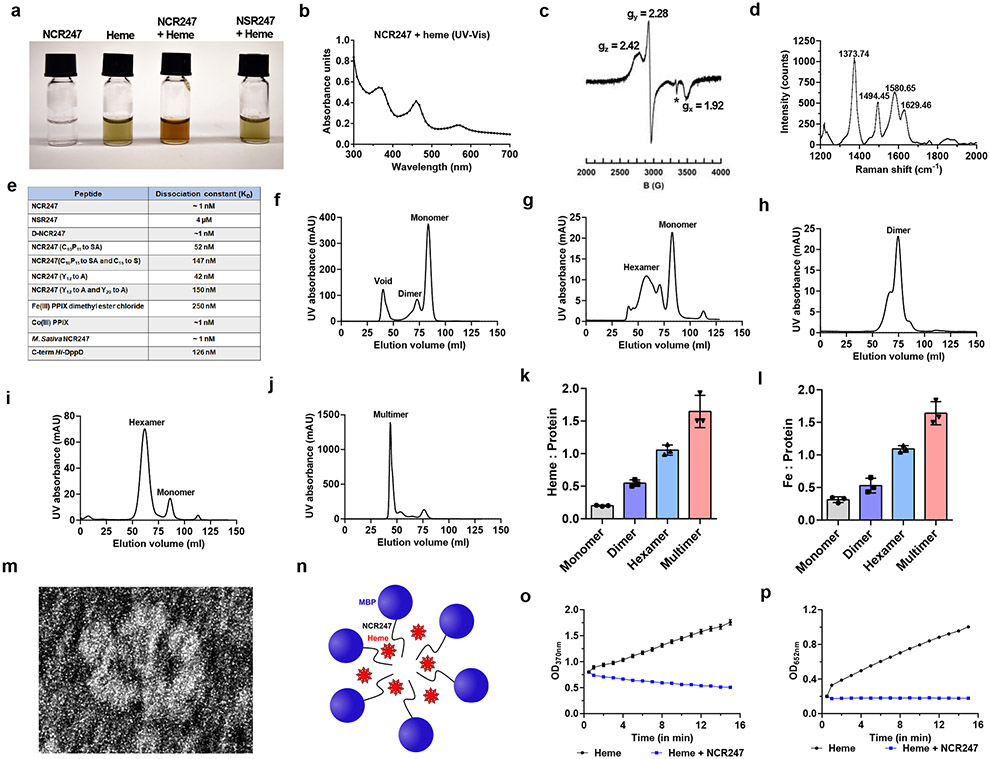
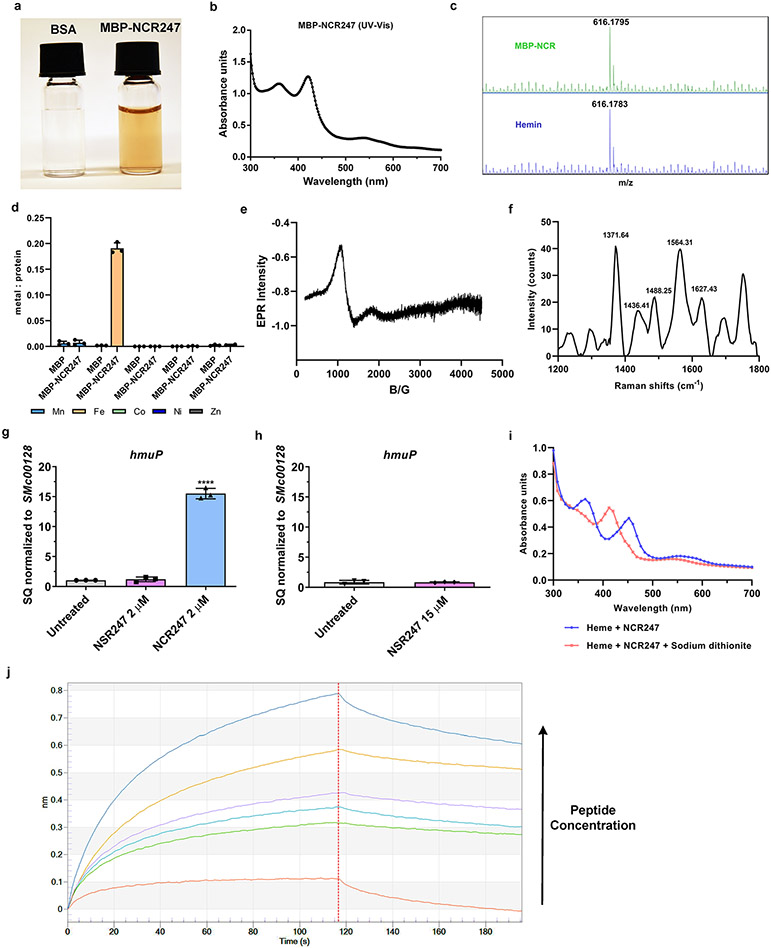
We made a serendipitous discovery when we observed that the Maltose Binding Protein (MBP)-tagged NCR247 purified as a reddish protein from E. coli (Extended data Fig. 2a). Several observations indicated that the colour of this chimeric protein was due to haem, including the UV-Vis absorption spectrum, which had a curve typical of haem-binding proteins (Extended data Fig. 2b); the direct detection of haem by mass spectrometry (Extended data Fig. 2c); the presence of porphyrin (porphyrin/protein ~0.20), and; the presence of iron (Extended data Fig. 2d).
In order to test if the chemically synthesized peptide used in the previous experiments as well as in many others, is also capable of binding haem, we added haem in a 1:1 ratio and it bound within seconds, as evidenced by a visible colour change (Fig. 2a). Moreover, we observed a UV-Vis absorption spectrum with maxima at 366, ~450, and 560 nm (Fig. 2b). An atypical hyper porphyrin (split-Soret) spectrum (similarly intense peaks at 366 and 450 nm) of NCR247-haem complex is very similar to that of only two other hemoproteins, both of which use two cysteines as axial ligands - DGCR812 and the N-terminal fragment of CD7413. Similar to haem-bound DGCR8, the EPR spectrum of NCR247-haem complex clearly revealed the presence of low spin Fe(III) ferric haem (Fig. 2c), and Raman spectroscopy identified the presence of ferric iron and a six-coordinate, low spin b-type haem (Fig. 2d).
Fig.2∣. NCR247 binds and sequester haem.
a, Chemically synthesized NCR247 shows reddish-brown color upon haem binding. NSR247 with haem shows no color change. b, UV-Vis spectrum of heme bound NCR247 with split-Soret peaks at 366 nm, ~450 nm, and 560 nm. c, EPR spectrum of NCR247-heme complex showing rhombic signal with g values indicating a low spin ferric heme. *shows an imperfection in the cavity. d, Resonance-Raman spectrum of NCR247-heme complex with prominent ν peaks indicative of a Fe3+, six-coordinate, low spin (6cLS) b-type heme. e, Dissociation constant (KD) of NCR247 and variants to haem as determined from Biolayer interferometry using biotinylated heme as ligand. For Fe(III) PPIX dimethyl ester Cl and Co(III) PPIX, biotinylated NCR247 was used as ligand. f and g, Size-Exclusion chromatograms of native MBP-NCR247 from E. coli grown without ALA (predominant monomer) (f), and with ALA (mixed multimers) (g). h-j, Size-Exclusion chromatograms after addition of half molar equivalent (predominant dimer) (h), equimolar (predominant hexamer) (i), and excess (predominant multimer) (j) heme to the purified monomer fraction of MBP-NCR247. k and l, Heme content (Heme assay kit) (k), and iron content (ICP-MS analysis) (l) of fractions isolated from size exclusion chromatography. m, Representative negative staining image of the hexamer fraction of MBP-NCR247. n, Current model for haem sequestration by NCR247. o and p, Peroxidase activity of heme and NCR247-heme complex on a chromogenic substrate TMB, measured by absorption at 370 nm (o) and 652nm (p). In a-j, representative data from three independent experiments is shown. In k, l, o and p, data are presented as mean of three replicates ± s.d.
Based on these analyses, we ascertained that the Fe(III) in the haem is double-cysteine ligated in the NCR247-haem complex14,15. Consistent with this, an NCR247 derivative in which all four cysteines were mutated to serine (NSR247), which unlike NCR247, did not change colour in response to haem addition (Fig. 2a), had only one high spin species corresponding to free haem in EPR and Raman spectra (Extended data Fig 2e and 2f), and did not increase expression of iron uptake genes, even at concentrations of 15 uM of the peptide (Extended data Fig. 2g and 2h). NCR247 also binds sodium dithionite-reduced Fe(II) ferrous haem (Extended data Fig. 2i). Biolayer interferometry using biotinylated haem16 revealed that NCR247 binds haem with a KD of ~1 nM (Fig. 2e and Extended data Fig. 2j). Since haem is not a chiral molecule, the D-enantiomer of NCR247 bound heme similarly, as predicted (Fig. 2e). The NSR247 derivative bound heme ca. 4000x less well (Fig. 2e), consistent with cysteines having an important role in haem binding.
Due to the importance of CP motif in some haem binding proteins17,18, we tested whether C10P11 motif of NCR247 contributes to the tight binding to haem. Mutating the CP motif to SA decreased binding affinity, while additionally mutating C15 to S caused a further decrease. Other amino acids are important as well. For example, since tyrosines have been known to contribute to haem binding 19,20, we mutated Y12 to A or both Y12 and Y20 to A’s and also observed decreased haem binding affinity (Fig. 2e).
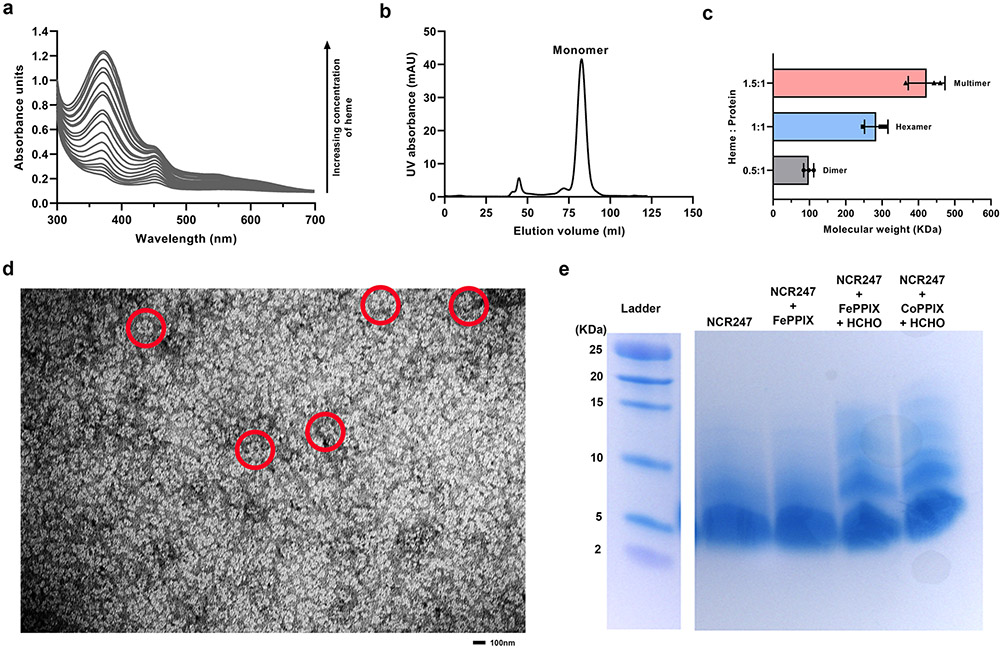
We hypothesized that the two axial cysteines that coordinate the haem iron likely come from different peptides, because altering the NCR247-haem ratio by titrating in haem increased the magnitude of absorption of both the 450 nm and 560 nm peaks until the ratio reached 2:1 (Extended data Fig 3a). However, absorbance at 366 nm increased even after the other peaks were saturated, suggesting that, as for DGCR8, this could be due to higher-order structures forming as more haem is added12. Consistent with this notion, we used size exclusion chromatography to analyse MBP-NCR247 purified from normally grown E. coli and identified a major peak corresponding to the size of a monomer (haem/protein <0.2) and a minor peak corresponding to a dimer (Fig. 2f), whereas MBP-NCR247 purified from E. coli, grown on medium supplemented with 5-aminolevulinic acid (ALA) to increase the level of intracellular haem, migrated as a mixture of higher molecular weight species (Fig. 2g). MBP-NSR247 purified as a monomer, even after addition of ALA, implying that haem binding triggers multimerization (Extended Fig. 3b). Addition of a half molar equivalent of haem to purified MBP-NCR247 monomer results in a major dimer peak (Fig 2h) but addition of equimolar haem yields a hexamer (Fig. 2i) and a small monomeric peak. Addition of even more haem leads to higher molecular weight multimers (Fig. 2j). Interestingly, the haem (and iron) to MBP-NCR247 ratio of monomer, dimer, hexamer, and multimer were ca. <0.2:1, 1:2,1:1 and >1.3:1 respectively (Fig. 2k and 2l). Mass photometry results upon addition of increasing concentration of haem to MBP-NCR247 monomers also indicated species with molecular weights corresponding to dimer, hexamer, or multimer (Extended data Fig. 3c).
Interestingly, negative staining of the hexameric MBP-NCR247- haem species revealed numerous examples of a flower-like structure with six petals (Fig. 2m; Extended data Fig. 3d). Taken together, these observations led us to propose a simple model, in which two NCR247 peptides initially bind a single haem on opposite sides. The resulting dimers then interact with additional haem at dimer-dimer interfaces to form hexamers (Fig. 2n), which can then form higher-order structures. As expected, NCR247 peptide can form multimers upon addition of haem and iron does not catalyze the formation of multimers (Extended data Fig. 3e). NCR247’s special mode of interacting with haem would be expected to make both the iron and the porphyrin ring chemically inaccessible, an expectation supported by our observation that the intrinsic peroxidase activity of haem is quenched upon NCR247 binding (Fig. 2o and 2p). It is unclear whether the recent report on the ability of some anti-microbial peptides to bind haem ~1000 fold less tightly than NCR247 and forming undefined assemblages is related to the haem binding properties of NCR24721.
Physiological effects of NCR247- haem binding
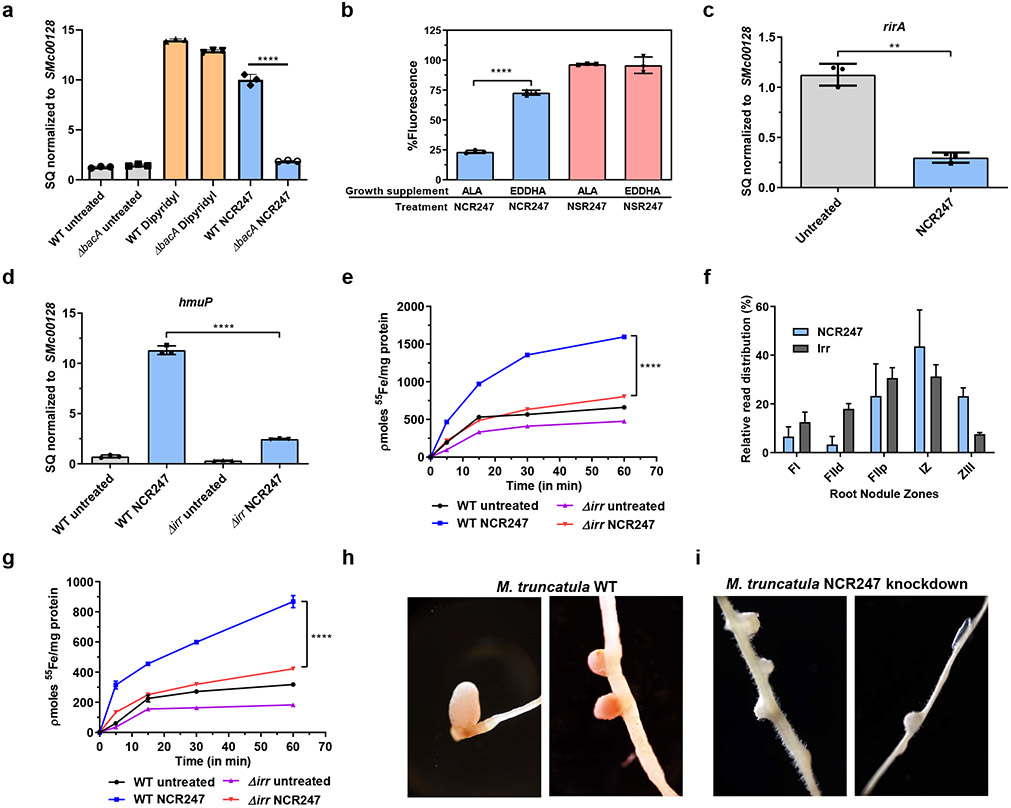
BacA is an inner-membrane transporter that has a crucial role in uptake of NCR247 into the cytoplasm of S. meliloti 22,23,24. A ΔbacA strain responds to iron starvation by increasing the expression of hmuP (a gene involved in iron uptake), similar to wildtype. However, there is very little change in expression of hmuP upon treatment with NCR247 (Fig. 3a), indicating that the iron starvation response caused by NCR247 is mediated in the cytoplasm of S. meliloti.
Fig.3∣. NCR247 binds intracellular haem and drives iron uptake by modulating Irr mediated iron regulation.
a, Diminished expression of gene involved in iron uptake (hmuP) in NCR247 treated ΔbacA compared to NCR247 treated wildtype and iron chelator (dipyridyl) treated wildtype and ΔbacA strain as measured by qRT-PCR analysis. b, Increased quenching of fluorescence of N-FITC labeled NCR247 by cellular extracts from S. meliloti grown with ALA when compared to S. meliloti grown with EDDHA. N-FITC labeled NSR247 is used as a control. c, Decrease in expression of rirA upon treatment with NCR247 as measured by qRT-PCR analysis d, Decrease in expression of hmuP in NCR247 treated Δirr when compared to NCR247 treated wildtype S. meliloti. In a, c and d, 2 μM NCR247 was treated for 30 mins and the data are expressed as starting quantities (SQ) of respective mRNAs normalized to the control gene SMc00128 and are presented as average of three technical replicates ± s.d. e, Decreased uptake of 55Fe in NCR247 treated Δirr when compared to NCR247 treated Wildtype S. meliloti. f, Relative expression levels of NCR247 and irr along the symbiotic process represented by distinct nodule sections. NCR247 and irr are expressed at higher levels in the interzone (IZ) where bacteroids are differentiating and preparing for nitrogen fixation. FI-meristematic zone, FIId (distal), FIIp (proximal fraction) of Zone II-infection and differentiation zone, ZIII-nitrogen fixation zone. Data is obtained from Symbimics website curated from previous publication and represents a mean of three technical replicates80. The individual data points are not available. g, Decreased uptake of 55Fe in NCR247 treated bacteroids from nodules of inoculated with Δirr when compared to NCR247 treated bacteroids from wildtype S. meliloti inoculated plants. h and i, Representative image of 12-day nodules of Wildtype (h) and NCR247 knockdown (i) of M. truncatula (A17) inoculated with wildtype S. meliltoi (Rm1021). In b, e and g, data are presented as mean of three biological replicates ± s.d. In b, ****P<0.0001 EDDHA vs ALA treated sample; c, **P=0.001 NCR247 Vs Untreated sample; a, d, e and g, ****P<0.0001 NCR247 treated WT vs ΔbacA/Δirr samples; two-way analysis of variance (ANOVA) with multiple comparisons. In h-i, representative image from 8 roots with each genotype is shown.
To demonstrate the ability of NCR247 to bind haem inside cells, we exploited the finding that haem quenches green fluorescence25 by showing that the green fluorescence of FITC-labelled NCR247 can be effectively quenched by haem. As expected, the fluorescence of FITC-labelled NSR247 remains unquenched even after addition of excess haem (Extended data Fig. 4a). Cellular extracts made from S. meliloti grown in an ALA-supplemented medium (to raise intracellular haem) quenched the fluorescence of FITC-NCR247 more than the extracts made from cells grown with the iron chelator EDDHA (to lower intracellular haem). The fluorescence of FITC-labelled NSR247 remains the same in both conditions (Fig. 3b).
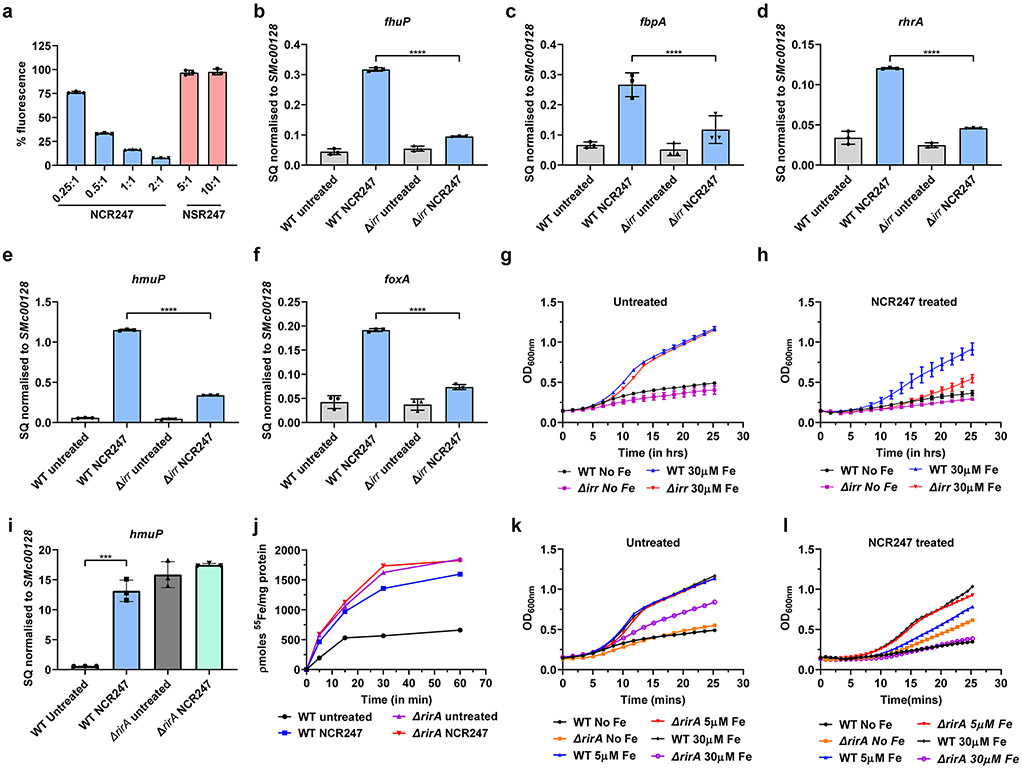
Because haem has a crucial role in maintaining iron homeostasis in rhizobial bacteria26, the capacity of NCR247 to interact with free/labile intracellular haem and render it biologically inaccessible could account for the ability of NCR247 to induce a state of iron starvation. Iron homeostasis in Sinorhizobium and some related bacteria including Rhizobium, Agrobacterium, Brucella, and Bartonella27,28,29 is controlled by two transcriptional regulators, Irr and RirA (Extended data Fig. 5)26. During iron-replete conditions, haem binding causes Irr to lose its DNA binding ability, thus causing de-repression of target genes involved in haem synthesis, iron storage, export, and, importantly, rirA30. After RirA is metalated to its fully active [4Fe-4S] bound form, it binds DNA and represses iron uptake genes to prevent further iron uptake31,9. We hypothesized that intracellular haem sequestration by NCR247 causes Irr to stay active even under iron-replete conditions and repress the transcription of rirA, thus leading to an increase in transcription of iron import genes. Consistent with this hypothesis we found that rirA transcription was reduced upon NCR247 treatment (Fig 3c); that NCR247-mediated increase in transcription of iron uptake genes (Fig. 3d and Extended data Fig 4b-4f) and uptake of 55Fe were diminished in an irr deletion mutant (Δirr) compared to wild type S. meliloti (Fig. 3e); and that iron addition could only partially rescue the growth of NCR247-treated Δirr mutant (Extended data Fig. 4g and 4h). The slight increase in expression of iron uptake genes in NCR247-treated Δirr compared to untreated Δirr may result from iron being redirected to replenish low levels of haem caused by haem sequestration rather than to metalate the Fe-S cluster of RirA and cause repression. In support of this model, a ΔrirA mutant had increased expression of hmuP and increased 55Fe uptake activity (Extended data Fig. 4i and 4j) similar to NCR247-treated wildtype. Upon NCR247 treatment, a ΔrirA mutant showed improved growth when compared to wildtype in low iron and iron-sufficient conditions. However, at high iron conditions a ΔrirA mutant shows reduced growth rate and this worsens upon NCR247 treatment (Extended data Fig. 4k and 4l) probably due to oxidative stress caused by excess iron.
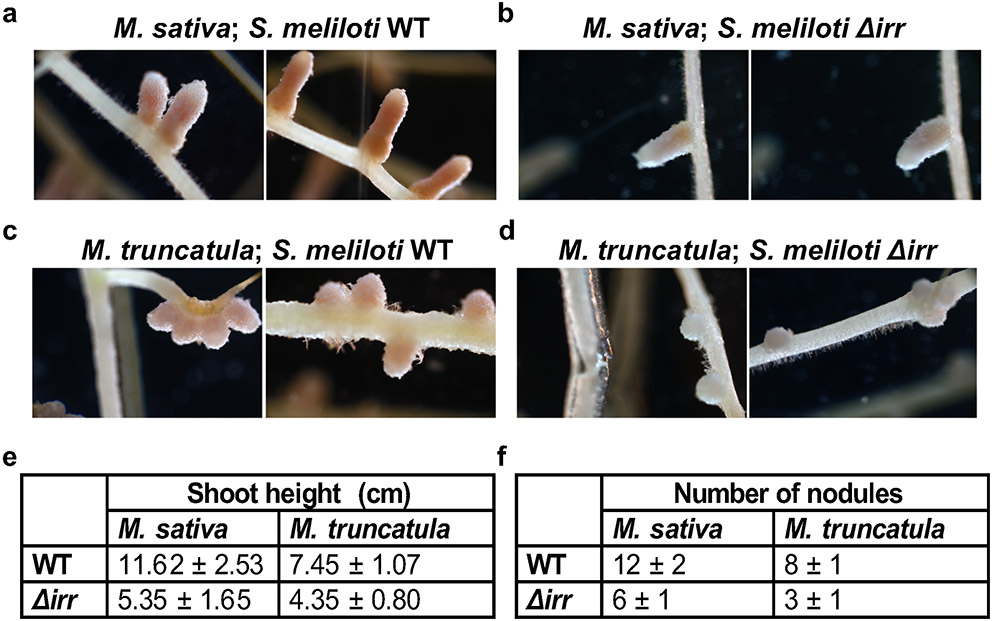
Rhizobia-Legume symbiosis is a highly iron-requiring process32,33 because each molecule of the nitrogen-fixing enzyme nitrogenase requires 24-32 iron atoms34,35. Iron is also an essential component of ferrodoxin and cytochromes, both of which are involved in respiration32. The iron uptake systems of roots are activated during nodulation36, but how developing bacteroids increase iron import as they prepare to fix nitrogen has not been reported previously. In order to evaluate this further, we reanalysed a dataset from a laser-capture microdissection study coupled with RNA sequencing37. We found that the expression pattern of S. meliloti irr mRNA coincides with the expression pattern of NCR247 in various zones of the nodules (Fig. 3f). Additional support for our hypothesis comes from the observation that S. meliloti bacteroids isolated from M. truncatula nodules increase uptake of Fe55 upon NCR247 treatment when compared with untreated bacteroids, and that this NCR247-mediated response was diminished in Δirr bacteroids isolated from nodules (Fig. 3g). Consistent with this, M. sativa and M. truncatula inoculated with Δirr S. meliloti had pale-coloured, small nodules, decreased shoot height and reduced numbers of nodules, when compared with plants inoculated with the wild-type S. meliloti implying ineffective symbiosis (Extended data Fig. 6a-d)). Interestingly, since an active Irr can repress haem synthesis38, the iron that is brought in could be directly utilized for incorporation into nitrogenase rather than for haem synthesis. Taken together, these observations suggest that NCR247 plays an important role in boosting iron import into the bacteroids during symbiosis by sequestering haem to override the usual iron homeostasis machinery of the bacterium.
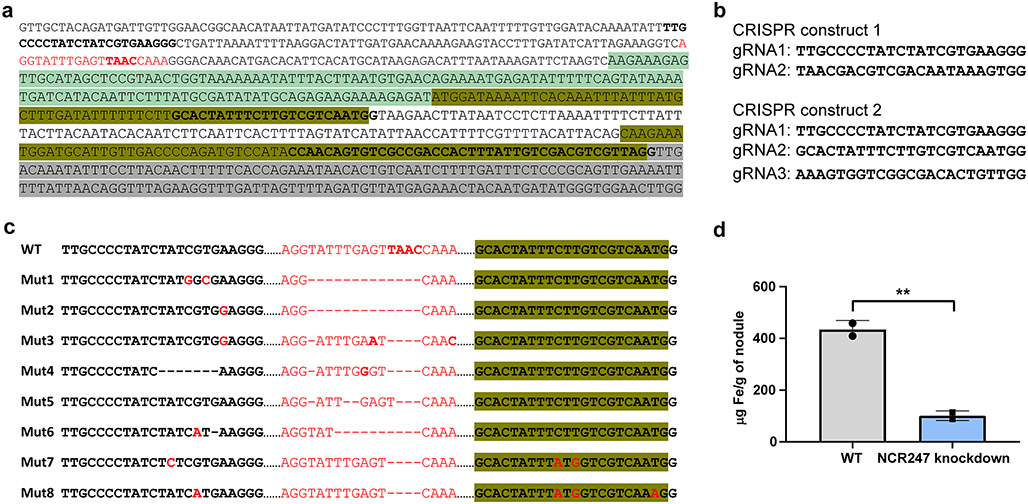
Finally, we made a CRISPR knockdown of NCR247 in M. truncatula (A17) using Agrobacterium rhizogenes mediated hairy root transformation. 8 /100 roots obtained from transforming two CRISPR constructs contained deletions in the NCR247 promoter region and 2/8 roots contained an additional substitution mutation in the putative peptidase recognition sequence (ALFLVV to ALFMVV) (Extended data Fig 7a-c). All 8 roots had small, white nodules, indicating that NCR247 is required for effective symbiosis (Fig 3h, 3i). The rest of the 92 roots contained wildtype or heterogenic sequence and elicited pink nodules. These data indicate that the expression of NCR247 in plant nodules is critical to establish an effective symbiosis. Consistent with our model for NCR247 function, these small white nodules had a 4-fold reduction in iron content compared with normal nodules (Extended Fig. 7d).
Sequence features of NCR247 enable haem binding
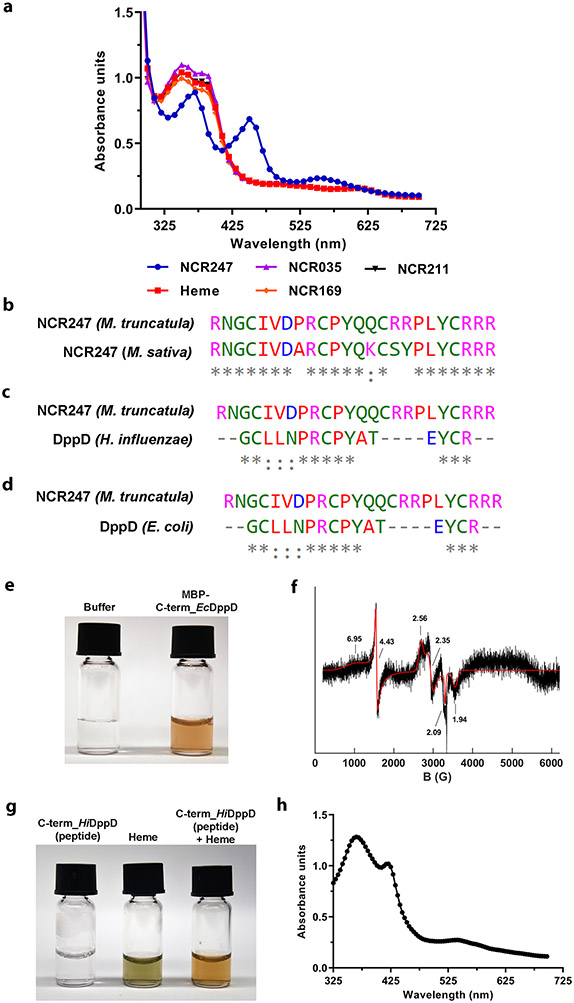
The features of the NCR247 sequence that enable it to bind and sequester haem so effectively seem to be rare, at least according to bioinformatics analysis. Sequences homologous to NCR247 were not found in the ca. 700 other NCR peptides of M. truncatula suggesting that the ability of NCR247 to bind haem with high-affinity may be unique among these peptides. We tested two NCR peptides that are reported to be essential for symbiosis (NCR2113 and NCR1694) and the cationic antimicrobial peptide NCR0352 for haem binding using UV-Vis spectrometry. None of these three peptides were able to bind haem as shown in Extended data Fig. 8a, indicating that they exert their symbiotic roles by different molecular mechanisms. We then performed a NCBI BLAST search with a cutoff of e-value less than 5 but did not find any related sequences in eukaryotes. We did identify one eukaryotic sequence by analysing M. truncatula’s close relative, M. sativa (alfalfa) whose sequences were not in the NCBI database. This revealed that M. sativa encodes an NCR247 ortholog, which has a 4-amino acid change compared to M. truncatula NCR24739 (Extended data Fig. 8b). We show that a chemically synthesized M. sativa (alfalfa) NCR247 ortholog, binds haem with the same affinity using biolayer interferometry (Fig. 2e). Within the bacterial kingdom, our NCBI Blast search revealed that NCR247 has 83% similarity to a sequence within the C-terminal end of the protein DppD (Extended data Fig. 8c and 8d). DppD is the ATPase subunit of the inner membrane component of a haem ABC transporter that is required for haem iron utilization in E. coli, Haemophilus influenzae and Mycobacterium tuberculosis40,41,42. MBP-fused to a 20 amino acid peptide from C-terminal of E. coli DppD that is homologous to NCR247 purified as a reddish protein, and had signals corresponding to the presence of haem according to EPR (Extended data Fig. 8e and 8f). Moreover, chemically synthesized peptide corresponding to the homologous region of DppD of H. influenzae readily bound haem in solution and exhibited a UV-Vis spectrum with Soret bands that were consistent with a haem binding protein (Extended data Fig. 8g and 8h). However, this NCR247 related DppD peptide bound haem ~120 times less strongly than NCR247 (Fig. 2e).
Potential therapeutic applications of NCR247s haem sequestering property
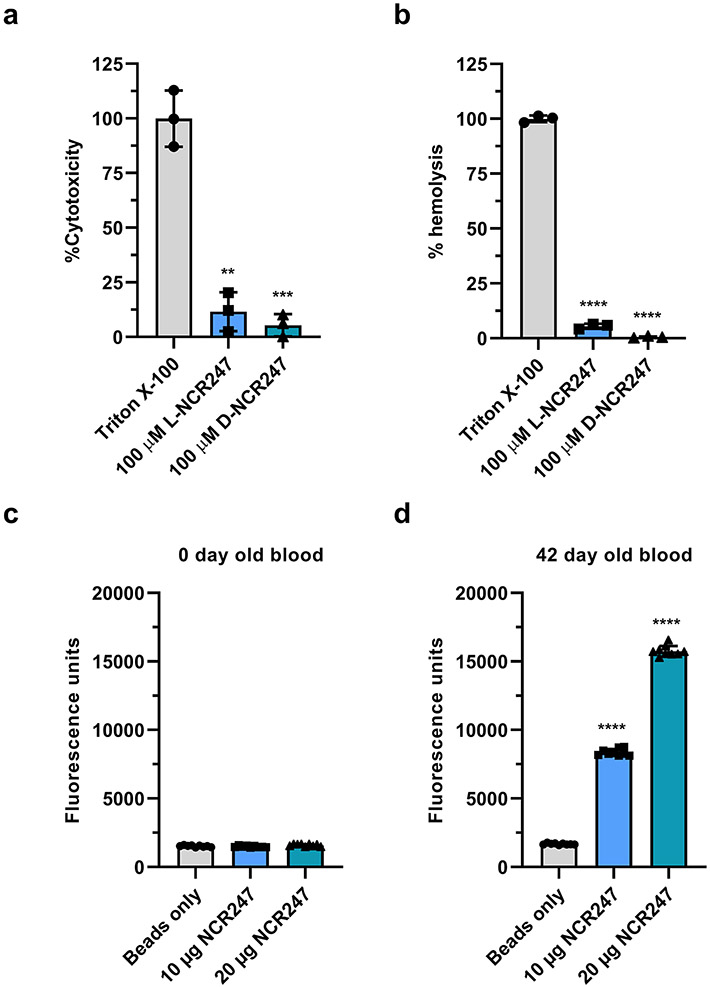
Several features of NCR247’s chemical and functional properties are interesting from a translational perspective. NCR247 binds haem with a KD of ca. 1 nM (Fig. 2e), which means that NCR247 can bind free haem, or labile haem loosely bound to proteins such as serum albumin (KD 40 μM)43, without removing haem from the important haem -binding proteins hemoglobin (KD 0.01 pM) and the haem scavenger haempexin (KD < 1 pM)44. Also, the interaction of NCR247 cysteines with both axial positions of the iron in haem, followed by NCR247/ haem oligomers forming aggregates, renders haem unreactive (Fig. 2o and 2p). Further, the D-enantiomer of NCR247 binds haem as well as the L-enantiomer (Fig. 2e), but is resistant to proteolytic degradation. NCR247 is highly water soluble and is small which means that it can be synthesized chemically.45 We tested L and D-NCR247 and found negligible cytotoxicity in mammalian cell line HEK-293 and did not detect haemolytic activities in a standard haemolysis assay (Extended data Fig. 9a and 9b). Interestingly, as discussed in detail previously 46, the sequences of NCR peptides including NCR247 are related to antimicrobial peptides47,48 and share similarities such as pairs of cysteines with some conotoxins49,50. Certain anti-microbial peptides and conotoxins have been used clinically.
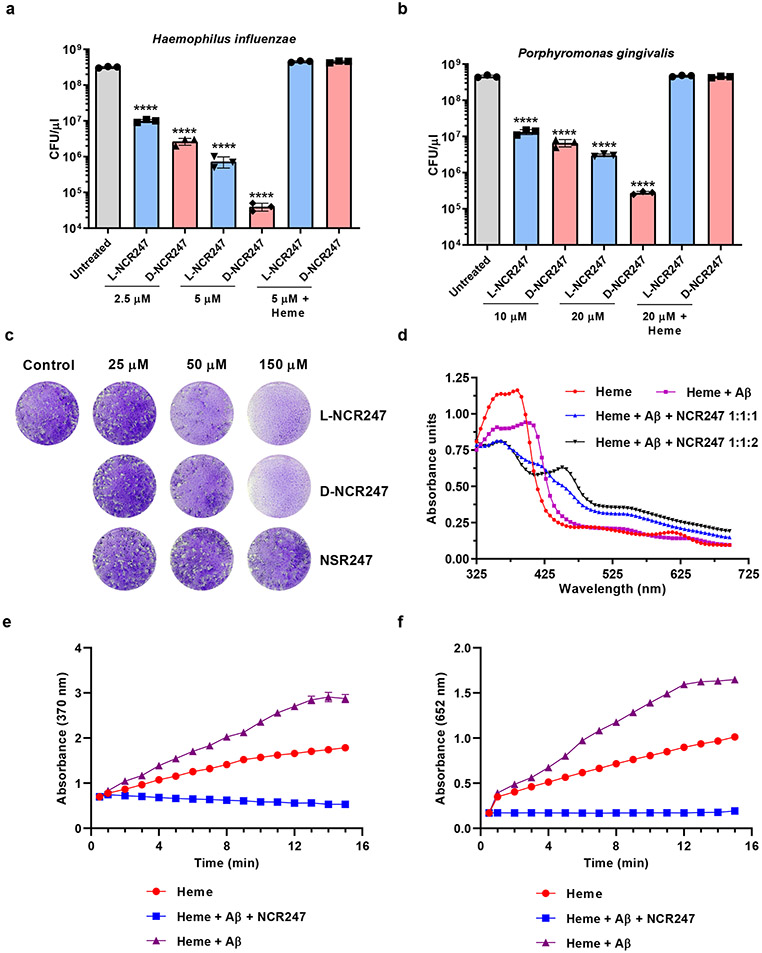
L- and D-NCR247’s special combination of haem-binding and sequestering characteristics suggest that these peptides and their derivatives might be exploited in a variety of clinical applications, including the following examples. First, NCR247’s ability to sequester extracellular haem could block the growth of pathogens that are haem auxotrophs or require extra haem for some stage of their life cycle, such as egg-laying. Important pathogens that require exogenous haem include not only bacterial pathogens51, but also a striking number of eukaryotic parasites52 and worms53, some of which infect significant fractions of the world’s population but lack an effective treatment. First, in Fig. 4a and b we tested this idea with naturally occurring bacterial haem auxotrophs Hemophilus influenzae (infections) and Porphyromonas gingivalis (periodontal disease), and found that NCR247 can prevent the growth of both of these pathogens. The D-enantiomer is more effective in killing than the L-enantiomer because of its protease resistance. Further, we showed that killing of these bacterial cells with NCR247 can be overcome by addition of haem supporting our hypothesis that sequestration of haem by NCR247 makes the haem unavailable to the pathogen. We also tested whether NCR247 could prevent the infectivity of apicomplexan parasite Toxoplasma gondii (toxoplasmosis). In Fig. 4c we show that 6 hr. treatment of T. gondii with L- or D-NCR247 leads to a massive reduction in the capacity of the parasite to form plaques on monolayers of human foreskin cell cultures54. The NSR247 variant, which does not bind haem, does not cause this phenotype even when used at high concentrations. Second, D- or L-NCR247 peptide attached to a solid support could be used to remove the free haem that accumulates in stored blood and is associated with the deleterious effects of large volume transfusions used in trauma-hemorrhage patients 55,56,57 or from the blood that is lysed during hemodialysis procedures for kidney disease, hemolytic diseases, etc58. Extended data Fig 9c and 9d show that NCR247 attached to magnetic beads can effectively pull down most of the free haem reported to be present in 42-day old blood 59. Third, since haem sequestration by NCR247 would prevent the free haem from exerting its pro-oxidant, pro-inflammatory, and cytotoxic effects, it could potentially be used to relieve the severity of a wide range of human diseases and conditions that result in the release of a large amount of toxic free haem into the plasma, such as sepsis or malaria, hemolytic human diseases such as Sickle Cell Disease and β-thalassemia, or by Ischemia Reperfusion Injury60,61. Interestingly, haem binding by human Aβ(1-42) amyloid peptides (KD=140 nM) triggers a peroxidase activity and that has been postulated to be an important contributor to neurodegeneration associated with human Alzheimer’s Disease, because of its ability to oxidize neurotransmitters 62,63,64. Fig. 4d shows the appearance of NCR247 specific peaks upon equimolar addition of NCR247 to the Aβ (1-42) amyloid peptide-haem complex and Fig. 4e and 4f show that equimolar addition of NCR247 to the Aβ (1-42)-haem complex quenches its peroxidase activity. We note that these extremely limited experiments were designed only to demonstrate simple proofs of principle. Of course, future investigations of L and D-NCR247s possible clinical potential will require a great deal of additional research including the use of whole animal infection models, pharmacokinetics studies and evaluation of delivery mechanisms.
Fig.4∣. Potential therapeutic applications of NCR247-haem binding.
a and b, Reduction of viable cell number of Haemophilus influenzae (a) and Porphyromonas gingivalis (b) upon treatment with increasing concentrations of L and D-NCR247. c, Images of crystal-violet stained fibroblast monolayers with plaques formed by the parasite Toxoplasma gondii pre-treated with L-NCR247, D-NCR247, or L-NSR247. d, UV-Vis spectrum of Amyloid Beta(Aβ) peptide- heme complex (1:1), Aβ:heme complex with equimolar NCR247(1:1:1) (* indicates an intermediary peak formed at 420 nm) and Aβ -heme complex with excess NCR247 (1:1:2), ** represents the appearance of typical NCR247-heme complex peaks at 366 nm and 450 nm. e and f, Peroxidase activity of Heme, Aβ-heme complex and NCR247 + Aβ-heme complex on a chromogenic substrate TMB as measured by absorption at 370 nm (e) and 652nm (f). c and d, Representative image from two independent replicates. In a, b and e, data are presented as mean of three independent replicates ± s.d. In a and b ****P<0.0001 vs untreated sample; two-way analysis of variance (ANOVA) with multiple comparisons.
Discussion
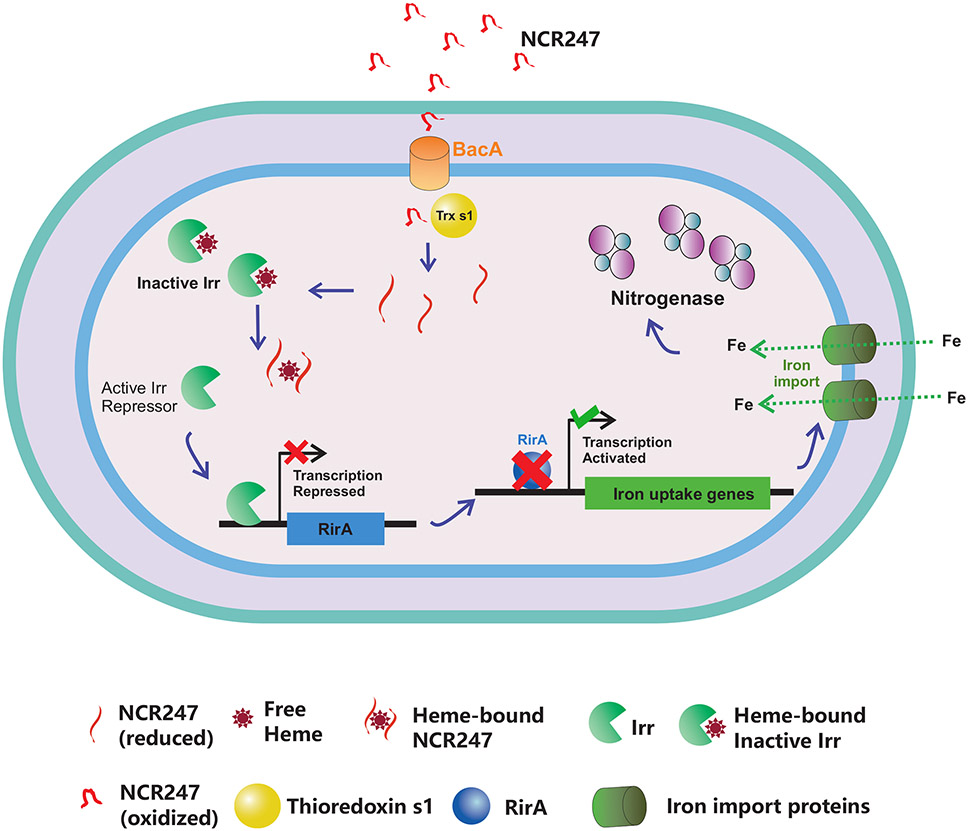
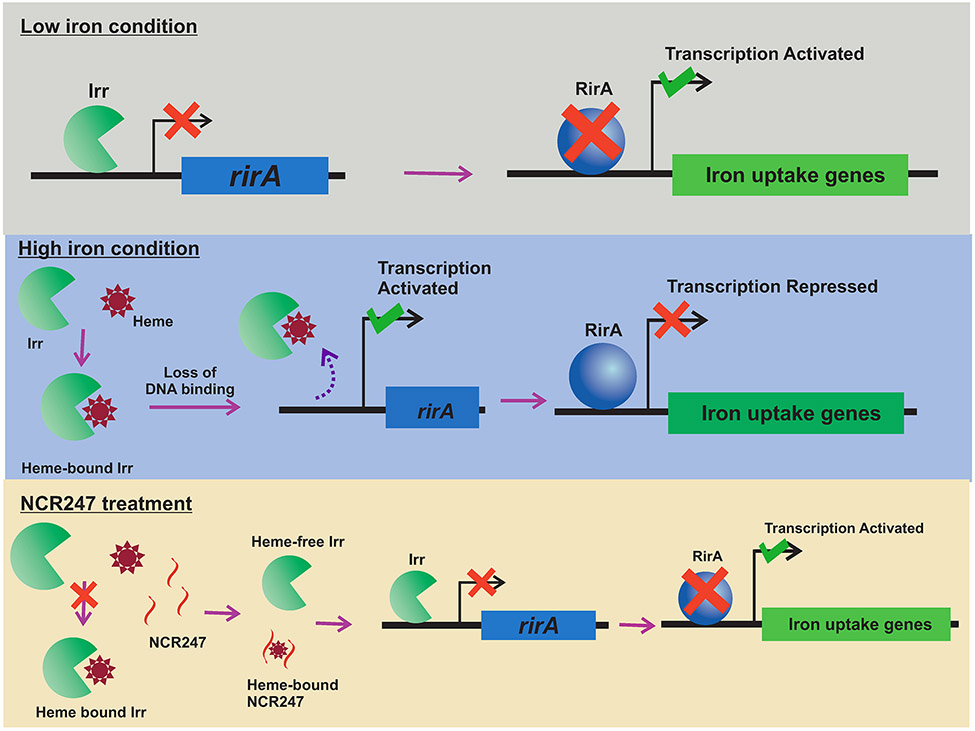
We report that plant-produced peptide NCR247 can bind to and sequester haem, which in turn stimulates rhizobia to import the iron that is needed for nitrogenase functioning. This finding is the first report, to our knowledge, of a plant peptide modulating the metal homeostasis of symbiotic bacteria in order to benefit the plant. We suggest a model mechanism, which is consistent with our present data, for how a haem-sequestering NCR peptide like NCR247 can affect nitrogen fixation in specific legume-rhizobium symbioses65 (Fig. 5). Our data demonstrate that NCR247 plays a critical active role in bacterial physiology by being imported into the bacterial cytoplasm where it directly interacts with the metabolite haem in a 1:1 manner that makes the haem and its iron biologically inaccessible. Cells then respond to the resulting physiological condition of haem deprivation by inducing all their RirA-regulated iron import genes even if they already have enough iron to satisfy their normal metabolic needs. We also show that this causes an increase in iron import that results in an elevated iron content of the cells. We present data indicating that NCR247 is essential for establishing an effective nitrogen fixing symbiosis. Although import of excess iron can result in oxidative stress in bacteria, that problem is avoided in the legume-rhizobia symbiosis by the expression of NCR247 in the microaerophilic nitrogen-fixing zone of the nodule. We suggest that when haem sequestration by NCR247 induces increased iron import into the rhizobia, the newly imported iron is made available for nitrogenase and other symbiotic purposes instead of being used for the synthesis of replacement haem because of the simultaneous down-regulation of the haem biosynthetic pathway.
Fig.5∣. NCR247 sequesters haem and overrides bacterial iron regulation to aid in symbiosis.
Model for the proposed role of NCR247 in iron regulation of bacteria and symbiosis. NCR247 secreted by the Medicago plant enters the cytoplasm of S. meliloti through the inner membrane protein BacA. NCR247 could be then reduced by plant produced Thioredoxin s1. Reduced NCR247 sequesters heme tightly. This leads to unavailability of heme and stabilization of heme regulated transcriptional repressor Irr even under iron sufficient conditions. Active Irr represses rirA. RirA is a transcriptional repressor of iron uptake genes. This leads to an increase in transcription of iron uptake genes and ultimately results in an increase in iron import into the cell. The need for iron in nodule increases during nitrogen fixation and nitrogenase (the key nitrogen fixing enzyme) requires numerous iron atoms structurally and functionally. Thus, NCR247 mediated boost in iron import could improve nitrogen fixation to ultimately benefit the plant.
Iron import into bacteroids during symbiosis is essential for nitrogenase activity, yet the identities of the S. meliloti transporters that function in planta remain unknown. Unlike Bradyrhizobium japonicum, where FeoB is the primary iron importer66, S. meliloti lacks FeoB and so future studies are needed to identify the iron importer in its bacteroids. The gene expression patterns within various zones of nodules, where multiple NCR peptides are present, is more complex and does not fully overlap with what we observe in vitro upon treatment with NCR247 alone (Supplementary Table 1 derived from our previous publication11) highlighting the need for more detailed studies of genes involved in iron homeostasis in various zones of nodules. Even though CRISPR knockdown of NCR247 in Medicago truncatula indicated its importance in symbiosis, a full transgenic knockout line will be required to further analyse its role in symbiosis.
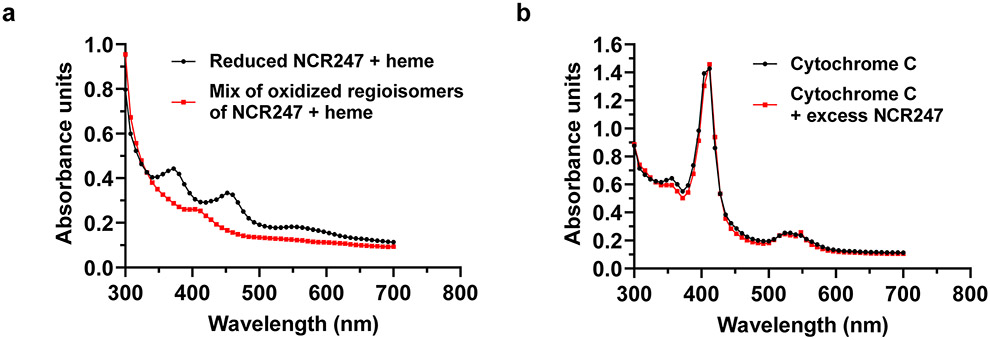
NCR247 is a secreted peptide, so it is likely to be converted to an oxidized form in the endoplasmic reticulum of the host cell during its transport to the bacteroids67. The reducing environment of the bacteroid cytoplasm, as well as the symbiotically important glutaredoxins (SmGRX1)68 and secreted plant thioredoxin (Trx1)69 would reduce oxidized NCR247 and facilitate haem binding in the cytoplasm of bacteroids. Since oxidized NCR247 is not capable of binding haem (Extended data Fig. 10a), it seems unlikely that NCR247 would interact with haem in the peri-bacteroid space. Cytochromes are known to have a role in symbiotic respiration70 but NCR247 cannot sequester cytochrome haem because of the covalent attachment (Extended data Fig. 10b). On the other hand, the Kd of the high affinity binding site of Irr from R. leguminosorum is ~1.0 ± 0.1 × 10−7 m (about 100-fold lesser than NCR247) which might enable NCR247 to sequester haem away from Irr.
Further investigations of the sequence dependence of haem binding and multimerization and the structure of the NCR247-haem complex will reveal new insights into the molecular details of the haem sequestration process. Furthermore, since NCR247 elicits multiple responses including inhibition of cell division, as well as transcriptional and translational responses that affect different cellular processes7,10, future studies will be needed to differentiate the responses that are solely due to NCR247’s ability to bind haem from its other functions. Since-NCR247 is known to interact with proteins71 (chiral interactions), haem, and membrane (non-chiral interactions), a more complete understanding of the mechanism of action of NCR247 can be gained by using the L and D-NCR247 enantiomers to distinguish the physiological effects caused by chiral and non -chiral interactions. Also, a ΔbacA mutant can be used to distinguish NCR247-dependent physiological effects caused outside inside cytoplasm from those inside the cytoplasm.
Although these insights were gained through fundamental research into a plant-bacterial symbiosis, several characteristics of L- and D-NCR247’s haem binding and sequestering abilities suggest the potential for similar chemically-synthesizable small peptides to have multiple applications as potential therapeutics to improve human health. Over the past decade, free haem has been implicated in the pathology of an extremely diverse variety of genetic and non-genetic human diseases and conditions and so haem has begun to attract attention as a target for therapeutic interventions60. In addition to the potential clinical applications briefly discussed above, haem is also involved in diseases caused by infectious agents that result in the release of free haem (e.g., malaria72 and sepsis73), cancer74, kidney disease75, immune-mediated inflammatory diseases76, cardiovascular disease77, atherosclerosis78, and neurodegeneration79. However, the challenge is that there has not been a “small molecule” (i.e., obtainable by chemical synthesis) that has the necessary characteristics for controlling or removing free haem. It will be interesting to investigate whether L- or D-NCR247 or their derivatives could serve as useful drugs in this capacity. Also, a striking number of important bacterial pathogens51 and eukaryotic parasites52 and worms53, - some of which infect significant fractions of the world’s population but lack an effective treatment - critically need to import haem from their environment to live. In principle, these pathogens could be controlled if there was a way to limit the free haem in their extracellular environments, but there has been no way to do that. The striking preliminary data presented in this paper demonstrating such a function in in vitro models, leads us to suggest that L- and D-NCR247 and their derivatives should be further investigated with respect to this class of clinical applications as well.
Finally, about ~700 NCR peptides have been identified in M. truncatula yet their molecular mechanism of action remains unexplored. Studying their detailed mechanism will not only deepen the understanding about symbiosis but also could help in developing various new diagnostic and therapeutic applications.
Online Methods
Primers, plasmids and strains used in this study are provided in Supplementary Table 2.
Growth conditions
Sinorhizhobium meliloti wild type strain RM1021 and irr deletion mutant were routinely grown in LB medium supplemented with 2.5 mM CaCl2 and 2.5 mM MgSO4 (LBMC) in the presence of 200 μg/ml Streptomycin at 30°C for 48 hours. When mentioned, S. meliloti were grown in minimal media (MM) with composition as described previously81. For making iron-free MM, FeCl3 was omitted from the MM. Metal free water (VWR ARISTAR® ULTRA) was used to make MM. All flasks and tubes were washed with 6 M HCl and then with metal free water before autoclaving. Escherichia coli strains were routinely grown in LB medium at 37°C. When required 100 μg/ml of neomycin, 50 μg/ml of kanamycin, and 25 μg/ml of chloramphenicol were used.
Irr mutant generation
Irr (SMc00329) deletion mutant Δirr and RirA (SMc00785) deletion mutant (ΔrirA) was created as described previously82. In short, 500 bp flanking regions of irr or rirA (omitting the gene) were combined using overlap extension PCR and cloned into pK18MobSacB83. This was then transformed into S. meliloti 1021 using triparental mating and resulting colonies were selected in 5% Sucrose medium. The colonies that grew were then screened for loss of the pK18MobSacB plasmid by their inability to grow on Neomycin.
Growth curve
All growth curve experiments were performed in a Tecan SPARK 10M microplate reader using polystyrene flat bottomed, non-treated, sterile 96 well plates. Overnight cultures grown in LBMC were washed and were subcultured (1:100 dilution) in minimal medium supplemented with respective iron concentrations. The plates were programed to continuously shake at 150 rpm and temperature maintained at 30° C. Optical density was measured at 600 nm every 60 minutes.
ICP-MS
200 μL of protein sample was mixed with 2 ml of 2% HNO3 and ICP-MS was performed as described previously 84. For bacterial samples, 1 ml of sample was spun down and the pellet was resuspended in 40 μL of 100% HNO3 and heated at 98°C for 1 hour. The supernatant of the solution was mixed with metal free water to make up to 2 mL and ICP-MS analysis was performed as described previously84. Same number of cells were spun down for protein analysis through BSA method and data were normalized to the amount of protein in each sample. For ICP-MS analysis of nodules same procedure was followed, except nodules were first crushed in PBS, portion of sample kept aside for protein quantification and then remaining was treated with HNO3. Agilent ICP-MS instrumentation with MassHunter 4.4 was used to collect data.
Haem preparation
Hemin solutions were always prepared in 0.1 M NaOH and used within 30 min of preparation. 1:1000 and 1:500 dilution of the stock solution was made and stock concentration was measured by pyridine hemochrome assay as described previously 85 by measuring the absorption at 557 nm using the extinction coefficient of pyridine hemochromogen (34.7 mM−1 cm−1).
Mass spectrometry
LC-MS analyses were performed on an LC/MS quadrupole time-of-flight (Q-TOF) mass spectrometer from Agilent (Santa Clara, CA) with an electrospray ionization (ESI) source. The mass spectrometer was coupled with a High-Performance Liquid Chromatography system from Agilent (Santa Clara, CA). Agilent MassHunter Workstation Software -Data Acquisition Version B.05.01 was used to collect and Quantitative analysis Version B.07.00 was used to analyse the data. Haem samples were analyzed in positive mode using a COSMOSIL 5C18-AR-II Packed Column, 4.6 mm I.D. x 150 mm C18 reverse phase column from Nacalai USA (San Diego, CA). The mobile phases were water (A) or acetonitrile (B). A linear gradient was run from 10% to 50% B over 30 min, at 100 μL/min. The ESI source parameters were: spray voltage, 4 kV; gas temperature, 340 °C; drying gas, 8 L/min; nebulizer, 20 psig; fragmentor, 175 V.
Biotinylation of haem
Hemin was biotinylated using the method exactly as described previously16. Biotin hydrazide, DCC was purchased from Sigma. COSMOSIL 5C18-AR-II Packed Column, 4.6 mm I.D. x 150 mm from Nacalai USA (San Diego, CA) was used to separate the products. LC-MS was used to verify the correct molecular weight (969.4 Da) of biotinylated haem (in which only one of the two propionate groups of protoheme was conjugated with biotin hydrazide) as described above.
Iron uptake assay
Radioactive 55Fe uptake assays were performed as described previously66 with some modifications. Cells were grown in LB to an O.D. 600 nm of 0.2. 20 ml cultures were then spun down, and suspended in 20 ml minimal media with 5 μM FeSO4 for 1 hour. 2 μM NCR247 and then 100 μM sodium ascorbate was added, and cells were incubated for additional 30 mins. At time 0, 1 μM 55FeCl3 mixed with ascorbate was added. At given time points, 1 ml aliquots were taken and quenched in 3 ml of ice-cold quench buffer (0.1M Tris, 1 mM ascorbate, and 100 μM FeSO4, pH 6.0). The cells were collected immediately after quenching on 0.45-μm filters, presoaked in quench buffer using a Millipore sigma 1225 vacuum filtration unit. The radioactive 55Fe content of the filters with cells was counted using a scintillation counter. Internalized 55Fe levels were normalized to the protein levels in the cell.
RNA isolation and qRT-PCR analysis
Cells were grown in LB until they reached an O.D. 600 nm of 0.2. Then cells were spun down and suspended in minimal media with or without appropriate FeSO4 concentrations for 1 hour. NCR247 was then added and 5 ml of appropriate cultures were spun down at given time intervals. Total RNA was extracted using Trizol (Thermo Fisher Scientific) method. Qiagen RNeasy kit was used to purify the RNA. On-column DNA removal was carried out using DNase I from NEB. 500ng of each RNA sample was used to make cDNA using the iScript cDNA synthesis kit (Biorad). qRT-PCRs were performed as described previously86. The standard curve method was used for relative quantification. In short, a standard curve was generated for each gene of interest (including SMc00128) by setting up qpCR reactions to amplify increasing amounts of S. meliloti Rm 1021 genomic DNA. All the primer sets used resulted in a proportional dose response curve with R2>0.99 confirming their efficiency. This curve was then used for extrapolating relative expression level of each gene of interest in a particular sample to obtain the starting quantities (SQ). This value is then normalized to the SQ values of SMc00128 obtained for respective sample. SMc00128 was used as a control gene since the expression levels did not change with iron and were used as a control in previous NCR247 studies7 and shown in Supplementary Fig. 1. These normalized values are then expressed as an average of triplicates, with standard deviation (s.d.) represented by the error bars.
Peptides
All chemically synthesized peptides were purchased from Genscript. The purity of all peptides was > 99% and verified by HPLC. The mass of each peptide was verified by MS analysis.
MBP-NCR247 protein purification
The coding sequence of NCR247 was ordered from gene synthesis (IDT). The amplified NCR247 sequence was cloned into plasmid pET28A downstream of the T7 promoter for expression of NCR247 with an N-terminal maltose-binding-protein (MBP) tag (Addgene)87. This plasmid was then transformed into an E. coli BL21(DE3) strain harboring a pRARE plasmid (carries genes for co-expression of various rare tRNAs in E. coli to compensate for unfavorable codon usage88). Cells were grown to mid-exponential phase at 37°C in LB media and expression of MBP-NCR247 was induced with 1mM IPTG. Cells were then shifted to 16°C and were grown overnight. Cells were collected using centrifugation at 8000g for 30 min and re-suspended in a lysis buffer (20 mM Tris-HCl, 200 mM NaCl, 1 mM EDTA, cOmplete Mini EDTA free protease inhibitor (Roche), pH 7.4). Cells were then lysed mechanically using a French press. Cell lysates were separated by centrifugation at 10000g for 15 min and passed through a 0.45 μm filter. Proteins were purified using the MBPTrap HP column (GE) according to the manufacturer's instructions. Eluted protein was then loaded onto a size exclusion column (GE Hiload 16/60 Superdex 75 pg) equilibrated with Buffer B (20 mM Tris-HCl, 200 mM NaCl, 1 mM EDTA). Gel Filtration Standard (Biorad) was used to estimate the molecular weight of peaks. Unicorn 7 software was used to collect all FPLC data.
Negative staining and TEM
Freshly ionized carbon-coated grids were floated on a 10 μl drop of sample for 1 minute. The grid was washed with 5 drops of 2% acidic UA. Excess UA was drawn off with grade 50 Whatman filter paper. Grids were allowed to air dry and imaged with a Hitachi 7800 at 100KV.
Fluorescence measurements
All fluorescence measurements were performed in 96-well black plates and measurements were taken in a Tecan Spark plate reader. For the peptide haem quenching experiment: 100 nM of FITC-NCR247 or FITC-NSR247 was added to the wells and increasing concentrations of molar equivalents of haem was added and fluorescence was noted. Fluorescent measurements from equal amount of FITC-NCR247 or FITC-NSR247in the same buffer without any added heme was considered as 100%.
For fluorescence quenching experiment from extracts: S. meliloti were grown in LBMC medium supplemented with 100 μM EDDHA or 20 μM ALA+100 μM FeSO4. Cells were collected at saturation and resuspended in a lysis buffer (20 mM Tris-HCl, 200 mM NaCl, cOmplete Mini EDTA free protease inhibitor (Roche), pH 7.4). Cells were then lysed mechanically using a French press. Cell lysates were separated by centrifugation at 10000g for 15 min. The supernatant was then normalized for total protein using BCA method. 100 μg of extracts were loaded in 96-well black plates in triplicates and made up to final volume of 195 ul. 50 nM in 5μL of either FITC-NCR247 or FITC-NSR247 were mixed with the extracts and fluorescent measurements were taken. Fluorescent measurements from equal amount of FITC-NCR247 or FITC-NSR247in the same buffer was considered as 100%.
Peroxidase assay
Pierce TMB Substrate Kit was used to measure the peroxidase activity of the haem and equimolar NCR247 was added to haem according to the manufacturer’s instructions. In short, 100 μL of TMB substrate solution (1:1 of TMB substrate and Peroxide solution) was added to 96 well black, polystyrene flat bottomed, non-treated, sterile plates. 5 μM heme or 5 μM NCR247 + 5 μM heme was added to the wells and the progression of the reaction was measured in a Tecan Spark plate reader. UV-Vis absorption values at 370 nm and 652 nm are noted every minute over a time period of 16 mins.
Mass photometry
All solutions were twice filtered with 0.22 μm syringe filters immediately prior to mass photometry measurements. Microscope coverslips (No. 1.5, 24 x 50 mm, Marienfeld) were cleaned by sequential submersion in Milli-Q water and 100% ethanol twice each followed by drying with optical lens paper. The final ethanol wash was dried with an air stream. Silicon gaskets were placed on clean microscope coverslips. Each measurement was acquired by adding 18 μL of storage buffer (20 mM Tris-HCl, 200 mM NaCl, 1 mM EDTA) to a gasket well. Following an autofocus stabilization, 2 μL of NCR247-MBP was added to the well to reach a final concentration of 1 μg/ml. Movies were recorded with a duration of 60 s at 1 kHz. A Contrast-to-mass calibration was performed using NativeMark Unstained Protein Standard. Data were gathered using the OneMP (Refeyn) with AcquireMP (Refeyn) 2.3.0. Movies were processed and analyzed using DiscoverMP (Refeyn) 2.3.0. Statistics were calculated after removing negative mass values from the data.
UV-VIS spectroscopy
All absorption spectra were collected in a Tecan SPARK 10M microplate reader using black polystyrene 96-well plates. Absorbance from 300 nm to 700 nm was recorded with a 5 nm interval after an initial shaking at 100 rpm for 15 secs. Heme binding to synthesized peptide: Water (pH 7.4) was used in all reactions and was first added to the wells. Peptides solubilized in water were added and then heme solubilized in 0.1 M NaOH was added, mixed thoroughly and measurements were taken immediately. Peptide and heme volume were kept to a maximum of 5 ul in volume. For all the absorption curves (except Extended Fig. 3a) 5 μM heme and 5μM of respective peptide was used. The same was also repeated with 0.1 M Tris buffer (pH 7.4) as a base buffer instead of water.
EPR spectroscopy
EPR spectra were collected in a Bruker EMX-Plus spectrometer at 10K with a Bruker/ColdEdge 4K waveguide cryogen-free cryostat. Xenon 1.1b.155 software was used to collect and process spectra. Spectra were recorded at 9.37 GHz with a modulation amplitude of 8G, microwave power of 0.2518 mW, and a 100 kHz modulation frequency. A center field of 3850G, a sweep time of 60s, and a sweep width of 7300G were used. Each spectrum shown is an average of 10 scans.
Raman Spectroscopy
The Raman data was collected on a Horiba XploRa confocal Raman microscope using a 405 nm diode laser at 5.4 mW of power. The system is based on an Olympus BX41 upright microscope. The 2400 grooves/mm blaze grating was used. An Olympus 50X long working distance objective with a NA of 0.5 was used. A 180-second exposure was used with an entrance slit of 200 μm and a confocal aperture of 500 μm. Two accumulations were averaged together for automatic cosmic ray removal. The denoise filter in Labspec 6 was used to smooth the data. A thermoelectric cooled (−70°C), Syncerity camera was used to collect the spectra. A uEye camera by IDS Imaging was used for the optical image of the sample.
Biolayer interferometry
Biolayer interferometry was carried out using a ForteBio Octet RED96 biolayer interferometer, following the manufacturer's instructions for a standard kinetic assay. Streptavidin-coated biosensor tips were incubated in 200 μl assay buffer (Water [pH 7.4]), each for 60s. Then biotinylated heme (or NCR247 in the case of Fe(III) PPIX dimethyl ester chloride) was loaded onto each biosensor tip at the defined concentration until the binding signal reached a value of >1.4. Biosensor tip loading was followed by incubation in assay buffer for 60s. Association between the ligand-heme and the analyte-various variants of NCR247 (Increasing concentrations in assay buffer) was observed over a time frame of ~116 s, in assay buffer. To stop binding kinetics for dissociation, the biosensor tips were placed back into assay buffer not containing any analyte, for 120 s. Curves were fit with Global fit analysis and Data analysis and KD calculation was performed using Fortebio Octet data analyses 8.2 software as described previously89.
Oxalic acid assay for measurement of haem
Total amount of heme in protein and plasma samples was measured by Oxalic acid method as described previously 90,91. To 50 μL of protein sample, 450 μL of 20 mM oxalic acid was added and stored at 4°C overnight. Then 500 μL of 2 M Oxalic acid was added and the sample was split into two. One of the samples was heated to 98°C for 30 mins. An unheated sample was used as a blank. 96-well black plates and measurements were taken in a Tecan Spark plate reader. The porphyrin fluorescence (excitation 400 nm, emission 620 nm) was measured for each sample. The standard curve was determined using the same method for various concentrations of hemin chloride.
Cell viability assays for H. influenza and P. gingivalis
Haemophilus influenzae Rd [KW20] was obtained from ATCC (51907). Standard growth and culturing techniques were followed as described previously92. Cultures were grown in Brain Heart Infusion broth (BHI) supplemented with 7.5 μM of hemin and 2 μg/ml NAD with or without the addition of peptide for 24 hours. The number of viable cells for every reaction mixture was then determined by serially diluting and spotting 10-μl aliquots in triplicates on BHI agar plates supplemented with 15 μM hemin and 3 μM NAD.
Porphyromonas gingivalis 2561 was obtained from ATCC (33277). Pre–reduced, anaerobically sterilized Brucella Broth and BRU - Brucella Blood Agar – were purchased from Anaerobe systems (CA, USA). They were opened just before use. Static cultures and plates were incubated at 37°C in an incubation chamber from BD GasPak™ EZ Container Systems. Anaerobic conditions were maintained by using BD BBL CO2 gas generators and BD BB GasPak CO2 indicators. Cultures were grown anaerobically in Brucella Broth with or without addition of peptides for 48 hours and viable cells for every reaction mixture were then determined by serially diluting and spotting 10-μl aliquots in triplicates on Brucella Blood Agar.
Pull-down assay of haem from stored blood
Expired units of whole blood were obtained from the American Red Cross through a local hospital, and plasma was separated from whole blood by centrifugation for 10 minutes at 1500 x g in 10 ml BD Vacutainer Plastic Blood Collection Tubes with K2EDTA. The supernatant was collected and divided into several 500 μL aliquots. Washed Streptavidin T1 MyOne Dynabeads (Invitrogen) were incubated with excess N-terminal Biotin-labeled NCR247 for 30 mins. After being washed according to the manufacturer’s protocol, 300 μL of plasma was added and further incubated at 4°C with rotation for 2 hours. Beads were then collected by using a magnetic stand (Dynamag-2 Life technologies) and washed with 300 μL PBS 3 times. The beads were then resuspended in 50 μL of 20 mM Oxalic acid and porphyrin content was measured as mentioned above. Beads not incubated with Biotin-NCR247 were used as a control and were subjected to the same procedure. Unheated sample from the Oxalic acid method was used as blank for the respective heated samples. Original plasma was serially diluted in PBS and the porphyrin content was measured by the same oxalic acid method.
Parasite and host cell culture
T. gondii parasites (strain RH, ATCC 50838) were grown in human foreskin fibroblasts (HFFs) maintained in DMEM (GIBCO) supplemented with 3% inactivated fetal calf serum (IFS) and 10 μg/mL gentamicin (Thermo Fisher Scientific), referred to as D3. Where noted, DMEM supplemented with 10% IFS and 10 μg/mL gentamicin was used, referred to as D10.
Plaque Assays
Freshly lysed parasites were filtered through 5 μm filters and spun down at 1000 x g and 18-for 10 min. Parasites were resuspended to 6E6 parasites/mL in Fluorobrite media supplemented with 3% IFS. Parasites were incubated with either peptide or vehicle for 6 hours at 37- and 5% CO2. Parasites were then spun down, washed once in 2 mL of Fluorobrite supplemented with 3% IFS, and resuspended in 1 mL of media. 2000 parasites were inoculated into each well of 6-well plates of HFFs maintained in D10 and allowed to grow undisturbed for 9 days. Plates were washed with PBS and fixed for 10 min at room temperature with 100% ethanol. Monolayers were visualized by staining for 5 min at room temperature with crystal violet solution, followed by two washes with PBS, one wash with water, and drying overnight.
Plant growth and inoculation
Three-day-old alfalfa or M. truncatula seedlings were inoculated with S. meliloti strains 1 ml OD600 of 0.05 in sterile water on Jensen’s agar exactly as described previously 93. S. meliloti strains were grown in minimal medium supplemented with 5 μM FeSO4 prior to inoculation. Plants were grown at 25°C with a light/dark cycle of 16/8 h, respectively.
Bacteroid isolation
Bacteroids were isolated from 28-day old nodules using the Percoll gradient (Sigma) method described previously 94,95. 50 nodules were removed from plants inoculated with Wild type or Δirr and immediately washed in wash buffer (0.35 M mannitol, 3 mM MgSO4, and 25 mM MES-KOH pH 7.0). After surface sterilization with 95% ethanol, nodules were crushed and filtered by miracloth (Millipore) assay. The filtrate was layered on [1 ml 80% (v/v):3 ml 60% (v/v):1 ml 30% (v/v)] prepared in wash buffer and centrifuged for 4000 g at 4°C for 15 mins. The bacteroids were then diluted in wash buffer and vortexed for 1 min for release from the peribacteroid units. After the release of bacteroids, they were immediately used for iron uptake assay.
Cytotoxic activity assay
Human embryonic kidney (HEK-293-ATCC-CRL-1573) cell lines were cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% Fetal Bovine Serum (FBS). When cells reached 104-105 cells per well, cells were supplemented with fresh DMEM without FBS, and then treated with 100 μM of NCR247 (L and D). After treatment for 24 h, MTT (Abcam, United Kingdom) stock solution was added to each well at a final concentration of 500 μg/mL and incubated in the dark for 4 h at 37°C. The absorbance at 570 nm was measured in the Tecan plate reader.
Haemolysis assay
Standard methods of hemolysis assay as previously published was followed96. Human red blood cells (hRBC) were washed three times with phosphate-buffered saline (PBS: 10 mM Na2HPO4, 1.76 mM K2HPO4, pH 7.4, containing 173 mM NaCl, and 2.7 mM KCl). Two-fold serial dilutions of the peptide solutions were then added to 50 μl aliquots of hRBC in PBS to adjust a final volume to 100 μl and hRBC concentration to 4% (v/v) in each well of a 96-well plate. The suspension was incubated for 1.5 h at 37°C under stirring at 1000 rpm. The plates were centrifuged at 2000 g for 5 min. Supernatant aliquots of 50 μl were transferred into flat-bottomed 96-well microplates, and the release of hemoglobin was monitored by measuring the absorbance at 405 nm in a microplate reader. hRBC in PBS (0% lysis control) and 0.1% Triton X-100 (100% lysis control) were used as negative and positive controls, respectively. Hemolytic activity was expressed as a percentage of hemolysis calculated according to the following equation: Hemolysis (%) = (OD405nm sample - OD405nm 0% lysis control)/ (OD405nm 100% lysis control - OD405nm 0% lysis control) * 100
CRISPR vector generation:
gRNAs were designed manually and BLAST search was done through phytozome to avoid overlap with other exons. ATUM program was run to confirm the validity of gRNAs. Primers for cloning to pDirect 22c vector were designed from (http://crispr-multiplex.cbs.umn.edu/assembly.php) - the Voytas lab website tools. They were then cloned into pDirect22c97 by golden gate cloning using NEBridge golden gate assembly kit. The sequenced vectors were then electroporated to Agrobacterium rhizogenes for hairy root transformation.
Hairy root transformation:
Hairy root transformation was performed as described previously98. Standard CTAB method was used for DNA extraction (OPS diagnostics- CTAB protocol for isolating DNA from plant tissues). The sequences were verified using PCR amplification of ~1000bp region around the gene, followed by Sanger sequencing.
Statistical Analysis
Details of statistical analyses are presented in the figure legends. Statistical analysis was performed on Prism software (GraphPad 6.01) using two-way ANOVA with multiple comparisons for repeated measurements.
Extended Data
Extended Data Fig. 1. NCR247 treatment induces an increase in expression of iron uptake genes.
a-d, qRT PCR analysis shows an increase in expression of genes involved in iron uptake (foxA (a), fhuP (b), rhrA (c), and fbpA (d)) upon treatment with 2 μM NCR247 for 30 mins. Cells were grown in iron sufficient medium (5 μM). e-i, qRT PCR analysis show increase in transcript levels of genes involved in iron uptake (hmuP(e), foxA (f), fhup (g), rhrA (h) and fbpA (i)) when grown in iron-replete medium (30 μM) upon NCR247 treatment for 30 mins. In a-i, the data are expressed as starting quantities (SQ) of respective mRNAs normalized to the control gene SMc00128 and are presented as average of three technical replicates ± s.d. j, Growth pattern of 2 μM NCR247 treated cells when grown in minimal medium lacking Fe, Mn, and Zn or medium supplemented with 30 μM of either FeSO4, MnCl2 or ZnSO4. Data are presented as mean of three biological replicates ± s.d. k, Change in Mn, Co and Zn content of 2 μM NCR247 treated S. meliloti when compared to untreated cells as measured by ICP-MS analysis. Data are presented as mean of two biological replicates ± s.d. In a **P<0.004, b *** P=0.0005, d***P=0.0006**, e*P=0.0463, f*P=0.0478, h*P=.0452, k** P=0.001, *** P=0.0006 and in c,g,i****P<0.0001 vs untreated sample; two-way analysis of variance (ANOVA) with multiple comparisons.
Extended Data Fig. 2. NCR247 binds haem and NSR247 lacks haem binding ability.
a, Purified MBP-NCR247 is a reddish colored protein. b, UV-Vis spectra of MBP-NCR247 showing peaks at 362 nm, 418 nm, and 540 nm with a slight shoulder at 580 nm. c, LC-MS spectrum from MBP-NCR247 (top) when compared to heme standard (Bottom). d, Metal content of purified MBP and MBP-NCR247 (measured using ICP-MS analysis) showing the presence of iron and absence of any other metal. Data are presented as mean of three biological replicates ± s.d. e, EPR spectrum of NSR247 with heme (g values =5.56) indicating a high spin ferric heme (spectrum similar to free heme). f, Resonance –Raman spectrum of NSR247 with heme shows prominent ν peaks indicative of a Fe3+, five-coordinate, high spin (5cHS) b-type heme. g and h, Lack of change in expression of iron uptake gene (hmuP) upon treatment with 2 μM (g) or 15 μM (h) NSR247. Data are expressed as starting quantities (SQ) of respective mRNAs normalized to the control gene SMc00128 and are presented as average of three technical replicates ± s.d. ****P<0.0001 NCR247 treated WT vs Untreated; two-way analysis of variance (ANOVA) with multiple comparisons. i, UV-Vis spectra of NCR247- Ferrous heme complex, after reduction by excess Sodium dithionite in an anaerobic chamber, indicating peaks at 420 nm and 550 nm. j, Representative raw image of association and dissociation steps (before and after diving red line) in an Octet bio-layer interferometry experiment (detailed in methods). Biotinylated heme was used as ligand and NCR247 was used as analyte. In a, b, c, e, f, and i, representative data from three independent experiments is shown.
Extended Data Fig. 3. Haem-induced multimerization of NCR247.
a, UV-Vis spectrum of NCR247-heme complex upon addition of increasing concentrations of heme (0.2 to 20 molar equivalents). Peak at 366 nm visibly increases in height even after peaks at 450 nm and 580 nm are saturated. b, Size-exclusion chromatogram of native MBP-NSR247 from E. coli grown with ALA (predominant monomer) c, Mass photometry analysis indicating the average molecular weight of the species that existed after addition of half molar equivalent, equimolar, and excess heme to the monomer fraction of purified MBP-NCR247. Data are presented as mean of three independent replicates ± s.d. d, Whole view of the grid used for negative staining made from the hexameric fraction of purified MBP-NCR247 showing multiple daisy like species marked in red. e, Tris tricine SDS gel showing multimerization of NCR247 peptide upon addition of heme (FePPIX) and CoPPIX and crosslinking with formaldehyde. In a, b, d and e representative data from three independent experiments is shown.
Extended Data Fig. 4. NCR247 drives iron uptake by controlling Irr mediated iron regulation.
a, Fluorescence of FITC-NCR247 quenched by increasing concentrations of heme. Fluorescence of FITC-NSR247 remains unquenched even after addition of excess heme. b-f, Decrease in expression of genes involved in iron uptake (fhuP (b), fbpA (c), rhrA (d), hmuP (e), foxA (f)) in a 2 μM NCR247 treated Δirr when compared to NCR247 treated wild type S. meliloti, when grown in iron-replete medium (30 μM). NCR247 was treated for 30 mins g and h, Growth pattern of untreated(g) and NCR247 treated (h) wildtype and Δirr cells in iron-depleted (-Fe) and iron-replete media (30 μM). i, Derepressed expression of hmuP in an untreated ΔrirA when compared to wildtype S. meliloti as measured by qRT-PCR analysis. j, Increased uptake of 55Fe in untreated ΔrirA when compared to untreated and NCR247 treated wildtype S. meliloti. k and l, Growth pattern of untreated (k) and NCR247 treated (l) wildtype and ΔrirA cells in iron-depleted (-Fe), iron sufficient (5 μM) and iron-replete media (30 μM). In a, g, h, j, k and l data are presented as mean of three independent replicates ± s.d. In b-f and i, The data are expressed as starting quantities (SQ) of respective mRNAs normalized to the control gene SMc00128 and are presented as average of three technical replicates ± s.d. In b-f, ****P<0.0001 NCR247 treated WT vs Δirr - samples; In i, ***P=0.0003 WT untreated vs WT NCR247 treated; two-way analysis of variance (ANOVA) with multiple comparisons.
Extended Data Fig. 5. Model of regulation of iron metabolism by Irr and RirA in S. meliloti and proposed mechanism of action of NCR247.
In S. meliloti regulation of iron status is controlled by two anti-parallel regulators Irr and RirA. Both Irr and RirA bind to DNA elements upstream of genes and repress gene expression. However, Irr senses iron status through heme97 (Irr loses DNA binding ability upon heme binding28) and RirA through Fe-S cluster formation (functional Fe-S cluster binding on RirA is needed for RirA to bind DNA29). During low intracellular iron concentrations, due to low intracellular heme availability, Irr remains stable and represses genes involved in iron storage, iron export, and also rirA25. Hence, in this condition, the expression of rirA is lowered and the availability of Fe-S cluster is scarce. This lack of RirA leads to an increase in expression of iron uptake genes. At high iron concentration, heme is available to bind Irr and this leads to the inability of Irr to bind DNA for repression. This leads to an increase in transcription of rirA and repression by RirA leads to a decrease in expression of iron uptake genes to prevent further iron uptake. When NCR247 is present during these conditions, it sequesters heme and hence heme is not available to inactivate Irr mediated repression. This leads to unusual availability of active Irr and repression of rirA. This leads to activation of iron uptake genes. Thus, NCR247 treatment leads to an iron starvation response and increase in import of iron even during iron sufficient and replete conditions.
Extended Data Fig. 6. Nodules of M. sativa and M. truncatula inoculated with Δirr S. meliloti are pale and deformed.
a and b, Representative image of 21-day nodules of M. sativa inoculated with wild type (Rm1021) (a) or Δirr S. meliloti (b). c and d, Representative image of 21-day nodules of M. truncatula (A17) inoculated with wild type (Rm1021) (c) or Δirr S. meliloti (d). In a-d, representative image from 7 sets of plants with each pair is shown. e and f, Table representing shoot height of the plant and number of nodules elicited for each pair. A total of 7 plants from each pair were considered for measurements.
Extended Data Fig. 7. CRISPR construct and genetic localization of the mutants.
a, Sequence of genetic locus of NCR247. Pale green is the 5’UTR region, dark yellow is the NCR247 coding region (signal sequence and coding region separated by an intron), Grey is part of the 3’UTR region. Location of gRNAs are marked in bold. The homozygous deleted region common in all the mutants is designated in red. b, Sequences of the gRNAs used in two different CRISPR constructs used. c, Location of deletions and substitution mutations in the 8 homozygous hairy roots obtained when compared to the wildtype sequence. Mutant 1-6 was obtained when the CRISPR construct 1 was used and Mutant 7-8 was obtained using CRISPR construct 2. d, Total iron content of the whole nodules isolated from wildtype and NCR247 knockdown roots as measured by ICP-MS analysis. 28 nodules from each type were collected and analysed in two technical replicates. In d, **P=0.007 WT vs NCR247 knockdown; two tailed unpaired t test.
Extended Data Fig. 8. Sequences similar to NCR247.
a, UV-Vis spectrum of NCR peptides (NCR169, NCR035, NCR211, and NCR247) with heme indicating that only NCR247 shows a spectrum characteristic of heme binding proteins. b, Sequence alignment of NCR247 from the plants M. sativa and M. truncatula. c and d, Sequence alignment of NCR247 from M. truncatula and C-terminal region of DppD (protein involved in heme transport) of Hemophilus influenzae (c) and E. coli (d). In b, c, and d, alignments were performed using CLUSTAL Omega. NCR247 from M. sativa and C-terminal end of DppD were significantly similar sequences with a e-value less than 5 obtained in a BLAST search. e and f, Sequence similar to NCR247 from C-terminal end of DppD of E. coli, tagged to MBP purifies as a reddish colored protein (e) and shows EPR spectrum (mixture of high and low spin heme) similar to other haem binding proteins98 g and h, Chemically synthesized peptide with sequence similar to NCR247 from C-terminal end of DppD of H. influenzae shows reddish color upon binding heme (g) and exhibits a UV-Vis spectrum (366 nm, 427 nm, and 540 nm) characteristic of haem binding proteins(h). In a and e-h representative data from three independent experiments is shown.
Extended Data Fig. 9. NCR247 suitability for potential therapeutic applications.
a, Standard cytotoxicity assay (Methods) on HEK293 cell line indicates negligible hemolysis by L and D-NCR247. b, Standard hemolysis assay (Methods) on hRBC indicates negligible hemolysis by L and D-NCR247. In a and b, Triton X-100 was used as a positive lysis control and data was normalized to PBS blank. Data are presented as mean of three independent replicates ± s.d. c and d, Pull down of heme by biotinylated NCR247 from 0-day old (c) and 42-day old plasma (d). Oxalic acid assay was used to measure the total heme content of the pull-down. Data are presented as mean of three independent replicates (with three technical replicates for each) ± s.d. In a, b, **P=0.0016, ***P=0.0009 vs TritonX-100 treated sample and in d, and ****P<0.0001 vs beads only; two-way analysis of variance (ANOVA) with multiple comparisons.
Extended Data Fig. 10. NCR247 sequestering haeme from haemoproteins in planta is less probable.
a, UV-Vis spectrum showing poor interaction of oxidized regioisomers of NCR247 and a presence of minor Soret band with heme when compared to reduced NCR247. b, UV-Vis spectrum showing inability of NCR247 to sequester heme from Cytochrome c. The absorption spectrum of Cytochrome c remains unaltered even after addition of excess NCR247.
Supplementary Material
Acknowledgements
This work was supported by NIH Grants (R01 GM031030 to G.C.W., R01 AI158501 to S.L., ES028374 and HL120877 to M.B.Y., F32 GM129882 to M.C.A, R35 GM126982 to C.L.D.). D.M.A is supported by a grant to the MIT from the Howard Hughes Medical Institute through the James H. Gilliam Fellowships for Advanced Study program. Areej Alhhazmi was supported by the KACST-MIT Ibn Khaldun Fellowship for Saudi Arabian Women. G.C.W. is an American Cancer Society Professor. C.L.D. is a Howard Hughes Medical Institute Investigator. This work was completed in part with resources at the MIT Department of Chemistry Instrumentation Facility with the help of John Grimes and Walt Massefski. Bio-Instrumentation Facility at the Department of Biology, MIT, and the Octet biolayer interferometry system (NIH S10 OD016326) is greatly acknowledged. We acknowledge the MIT Center for Environmental Health Sciences (NIH P30 ES002109). This work was performed in part at the Harvard University Center for Nanoscale Systems (CNS); a member of the National Nanotechnology Coordinated Infrastructure Network (NNCI), which is supported by the National Science Foundation under NSF award no. ECCS-2025158. We thank Dr. Arthur McClelland for his assistance with the Raman spectroscopy and Nicki Watson for her assistance with the negative staining and TEM analysis. We thank Dr. Dong Wang from University of Massachusetts, Amherst for providing with materials and advice on the CRISPR knockout experiment. We thank Kelsey Miller for her assistance with EPR interpretations.
Footnotes
Competing interests
A provisional patent application directed to heme-binding small peptides has been filed by MIT with inventors G.C.W., S.S., and V.M.P.B. All the other authors do not have any competing interests.
Data availability
All data in this manuscript is available. Source data for all the graphs and charts for all the figures and raw images are provided with the manuscript.
References
- 1.Gibson KE, Kobayashi H & Walker GC Molecular determinants of a symbiotic chronic infection. Annu. Rev. Genet 42, 413–441 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van De Velde W et al. Plant peptides govern terminal differentiation of bacteria in symbiosis. Science (80-. ) 327, 1122–1126 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Kim M et al. An antimicrobial peptide essential for bacterial survival in the nitrogen-fixing symbiosis. Proc. Natl. Acad. Sci. U. S. A 112, 15238–15243 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horváth B et al. Loss of the nodule-specific cysteine rich peptide, NCR169, abolishes symbiotic nitrogen fixation in the Medicago truncatula dnf7 mutant. doi: 10.1073/pnas.1500777112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mikuláss KR et al. Antimicrobial nodule-specific cysteine-rich peptides disturb the integrity of bacterial outer and inner membranes and cause loss of membrane potential. Ann. Clin. Microbiol. Antimicrob 15, 43 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farkas A et al. Medicago truncatula symbiotic peptide NCR247 contributes to bacteroid differentiation through multiple mechanisms. Proc. Natl. Acad. Sci. U. S. A 111, 5183–5188 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Penterman J et al. Host plant peptides elicit a transcriptional response to control the Sinorhizobium meliloti cell cycle during symbiosis. doi: 10.1073/pnas.1400450111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shabab M et al. Disulfide cross-linking influences symbiotic activities of nodule peptide NCR247. doi: 10.1073/pnas.1610724113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chao TC, Buhrmester J, Hansmeier N, Pühler A & Weidner S Role of the regulatory gene rirA in the transcriptional response of Sinorhizobium meliloti to iron limitation. Appl. Environ. Microbiol 71, 5969–5982 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shabab M et al. Disulfide cross-linking influences symbiotic activities of nodule peptide NCR247. Proc. Natl. Acad. Sci. U. S. A 113, 10157–10162 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Penterman J et al. Host plant peptides elicit a transcriptional response to control the Sinorhizobium meliloti cell cycle during symbiosis. Proc. Natl. Acad. Sci. U. S. A 111, 3561–3566 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barr I et al. DiGeorge Critical Region 8 (DGCR8) is a double-cysteine-ligated heme protein. J. Biol. Chem 286, 16716–16725 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kupke T, Klare JP & Brügger B Heme binding of transmembrane signaling proteins undergoing regulated intramembrane proteolysis. Commun. Biol 3, 1–16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barr I et al. Ferric, not ferrous, heme activates RNA-binding protein DGCR8 for primary microRNA processing. Proc. Natl. Acad. Sci. U. S. A 109, 1919–1924 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Girvan HM et al. Analysis of Heme Iron Coordination in DGCR8: The Heme-Binding Component of the Microprocessor Complex. Biochemistry 55, 5073–5083 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Ishida M, Dohmae N, Shiro Y & Isogai Y Synthesis of biotinylated heme and its application to panning heme-binding proteins. Anal. Biochem 321, 138–141 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Kühl T et al. Analysis of Fe(III) heme binding to cysteine-containing heme-regulatory motifs in proteins. ACS Chem. Biol 8, 1785–1793 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Shimizu T Binding of cysteine thiolate to the Fe(III) heme complex is critical for the function of heme sensor proteins. J. Inorg. Biochem 108, 171–177 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Li T, Bonkovsky HL & Guo JT Structural analysis of heme proteins: Implications for design and prediction. BMC Struct. Biol 11, 1–13 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brewitz HH et al. Heme interacts with histidine- and tyrosine-based protein motifs and inhibits enzymatic activity of chloramphenicol acetyltransferase from Escherichia coli. Biochim. Biophys. Acta - Gen. Subj 1860, 1343–1353 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Juhász T et al. Interplay between membrane active host defense peptides and heme modulates their assemblies and in vitro activity. Sci. Reports ∣ 11, 18328 (123AD). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferguson GP, Roop RM & Walker GC Deficiency of a Sinorhizobium meliloti bacA mutant in alfalfa symbiosis correlates with alteration of the cell envelope. J. Bacteriol 184, 5625–5632 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guefrachi I et al. Bradyrhizobium BclA Is a Peptide Transporter Required for Bacterial Differentiation in Symbiosis with Aeschynomene Legumes. 28, (2015). [DOI] [PubMed] [Google Scholar]
- 24.Marlow VL et al. Essential role for the BacA protein in the uptake of a truncated eukaryotic peptide in Sinorhizobium meliloti. J. Bacteriol 191, 1519–1527 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeda S, Kamiya N & Nagamune T A novel protein-based heme sensor consisting of green fluorescent protein and apocytochrome b562. Anal. Biochem 317, 116–119 (2003). [DOI] [PubMed] [Google Scholar]
- 26.O’Brian MR Perception and Homeostatic Control of Iron in the Rhizobia and Related Bacteria. Annu. Rev. Microbiol 69, 229–245 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Hibbing ME & Fuqua C Antiparallel and interlinked control of cellular iron levels by the Irr and RirA regulators of Agrobacterium tumefaciens. J. Bacteriol 193, 3461–3472 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H et al. Insights into irr and rira gene regulation on the virulence of brucella melitensis m5-90. Can. J. Microbiol 66, 351–358 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Costa D, Amarelle V, Valverde C, O’Brian MR & Fabiano E The Irr and RirA proteins participate in a complex regulatory circuit and act in concert to modulate bacterioferritin expression in Ensifer meliloti 1021. Appl. Environ. Microbiol 83, 895–912 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singleton C et al. Heme-responsive DNA binding by the global iron regulator Irr from rhizobium leguminosarum. J. Biol. Chem 285, 16023–16031 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pellicer Martinez MT et al. Sensing iron availability via the fragile [4Fe-4S] cluster of the bacterial transcriptional repressor RirA †. (2017) doi: 10.1039/c7sc02801f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brear EM, Day DA & Smith PMC Iron: An essential micronutrient for the legume-rhizobium symbiosis. Front. Plant Sci 4, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.González-Guerrero M, Matthiadis A, Sáez Á & Long TA Fixating on metals: New insights into the role of metals in nodulation and symbiotic nitrogen fixation. Front. Plant Sci 5, 45 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seibert M, Lien S, Weaver PF & Janzen AF Photobiological Production of Hydrogen and Electricity. 273–229 (1981) doi: 10.1016/b978-0-08-025388-6.50039-8. [DOI] [Google Scholar]
- 35.Einsle O et al. Nitrogenase MoFe-protein at 1.16 Å resolution: A central ligand in the FeMo-cofactor. Science (80-. ) 297, 1696–1700 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Terry RE, Soerensen KU, Von Jolley D & Brown JC The role of active Bradyrhizobium japonicum in iron stress response of soybeans. Plant Soil 130, 225–230 (1991). [Google Scholar]
- 37.Roux B et al. An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J. 77, 817–837 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Hamza I, Chauhan S, Hassett R & O’Brian MR The bacterial irr protein is required for coordination of heme biosynthesis with iron availability. J. Biol. Chem 273, 21669–21674 (1998). [DOI] [PubMed] [Google Scholar]
- 39.Montiel J et al. Morphotype of bacteroids in different legumes correlates with the number and type of symbiotic NCR peptides. doi: 10.1073/pnas.1704217114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Létoffé S, Delepelaire P & Wandersman C The housekeeping dipeptide permease is the Escherichia coli heme transporter and functions with two optional peptide binding proteins. Proc. Natl. Acad. Sci. U. S. A 103, 12891–12896 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morton DJ, Seale TW, Vanwagoner TM, Whitby PW & Stull TL The dppBCDF gene cluster of Haemophilus influenzae: Role in heme utilization. BMC Res. Notes 2, 166 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitra A, Ko YH, Cingolani G & Niederweis M Heme and hemoglobin utilization by Mycobacterium tuberculosis. Nat. Commun 10, 1–14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamal JKA & Behere DV Binding of heme to human serum albumin: Steady-state fluorescence, circular dichroism and optical difference spectroscopic studies. Indian J. Biochem. Biophys 42, 7–12 (2005). [PubMed] [Google Scholar]
- 44.Hrkal Z, Vodrážka Z & Kalousek I Transfer of Heme from Ferrihemoglobin and Ferrihemoglobin Isolated Chains to Hemopexin. Eur. J. Biochem 43, 73–78 (1974). [DOI] [PubMed] [Google Scholar]
- 45.Wang T et al. Heme Sequestration as an Effective Strategy for the Suppression of Tumor Growth and Progression. Mol. Cancer Ther 20, 2506–2518 (2021). [DOI] [PubMed] [Google Scholar]
- 46.Lima RM, Kylarová S, Mergaert P & Kondorosi É Unexplored Arsenals of Legume Peptides With Potential for Their Applications in Medicine and Agriculture. Front. Microbiol 11, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Srivastava S et al. Cysteine-rich antimicrobial peptides from plants: The future of antimicrobial therapy. Phytotherapy Research vol. 35 256–277 (2021). [DOI] [PubMed] [Google Scholar]
- 48.Lehrer RI & Ganz T Endogenous vertebrate antibiotics. Defensins, protegrins, and other cysteine-rich antimicrobial peptides. Ann. N. Y. Acad. Sci 797, 228–239 (1996). [DOI] [PubMed] [Google Scholar]
- 49.Halai R & Craik DJ Conotoxins: Natural product drug leads. Natural Product Reports vol. 26 526–536 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Layer RT & McIntosh JM Conotoxins: Therapeutic potential and application. Marine Drugs vol. 4 119–142 (2006). [Google Scholar]
- 51.Richard KL, Kelley BR & Johnson JG Heme uptake and utilization by gram-negative bacterial pathogens. Front. Cell. Infect. Microbiol 9, 81 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kořený L, Oborník M & Lukeš J Make It, Take It, or Leave It: Heme Metabolism of Parasites. PLoS Pathog. 9, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perner J, Gasser RB, Oliveira PL & Kopáček P Haem Biology in Metazoan Parasites – ‘The Bright Side of Haem’. Trends Parasitol. 35, 213–225 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Bergmann A et al. Toxoplasma gondii requires its plant-like heme biosynthesis pathway for infection. PLoS Pathog. 16, e1008499 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wagener BM et al. Role of heme in lung bacterial infection after trauma hemorrhage and stored red blood cell transfusion: A preclinical experimental study. PLoS Med. 15, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee JS & Kim-Shapiro DB Stored blood: How old is too old? J. Clin. Invest 127, 100–102 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Graw JA et al. Haptoglobin or hemopexin therapy prevents acute adverse effects of resuscitation after prolonged storage of red cells. Circulation 134, 945–960 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ofori-Acquah SF et al. Hemopexin deficiency promotes acute kidney injury in sickle cell disease. Blood 135, 1044–1048 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gouveia Z et al. Characterization of plasma labile heme in hemolytic conditions. FEBS J. 284, 3278–3301 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Immenschuh S, Vijayan V, Janciauskiene S & Gueler F Heme as a target for therapeutic interventions. Front. Pharmacol 8, 146 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith A & McCulloh RJ Mechanisms of haem toxicity in haemolysis and protection by the haem-binding protein, haemopexin. ISBT Sci. Ser 12, 119–133 (2017). [Google Scholar]
- 62.Seal M, Ghosh C, Basu O & Dey SG Cytochrome c peroxidase activity of heme bound amyloid β peptides. J. Biol. Inorg. Chem 21, 683–690 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Ghosh C, Seal M, Mukherjee S & Ghosh Dey S Alzheimer’s Disease: A Heme-Aβ Perspective. Accounts of Chemical Research vol. 48 2556–2564 (2015). [DOI] [PubMed] [Google Scholar]
- 64.Atamna H & Boyle K Amyloid-β peptide binds with heme to form a peroxidase: Relationship to the cytopathologies of Alzheimer’s disease. Proc. Natl. Acad. Sci. U. S. A 103, 3381–3386 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Downie JA & Kondorosi E Why Should Nodule Cysteine-Rich (NCR) Peptides Be Absent From Nodules of Some Groups of Legumes but Essential for Symbiotic N-Fixation in Others? Front. Agron 3, 42 (2021). [Google Scholar]
- 66.Sankari S & O’Brian MR The Bradyrhizobium japonicum ferrous iron transporter FeoAB is required for ferric iron utilization in free living aerobic cells and for symbiosis. J. Biol. Chem 291, 15653–15662 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sevier CS & Kaiser CA Formation and transfer of disulphide bonds in living cells. Nat. Rev. Mol. Cell Biol 3, 836–847 (2002). [DOI] [PubMed] [Google Scholar]
- 68.Benyamina SM et al. Two Sinorhizobium meliloti glutaredoxins regulate iron metabolism and symbiotic bacteroid differentiation. Environ. Microbiol 15, 795–810 (2013). [DOI] [PubMed] [Google Scholar]
- 69.Ribeiro CW et al. Regulation of Differentiation of Nitrogen-Fixing Bacteria by Microsymbiont Targeting of Plant Thioredoxin s1. Curr. Biol 27, 250–256 (2017). [DOI] [PubMed] [Google Scholar]
- 70.Delgado MJ, Bedmar EJ & Downie JA Genes involved in the formation and assembly of rhizobial cytochromes and their role in symbiotic nitrogen fixation. Adv. Microb. Physiol 40, 191–231 (1998). [DOI] [PubMed] [Google Scholar]
- 71.Farkas A et al. Medicago truncatula symbiotic peptide NCR247 contributes to bacteroid differentiation through multiple mechanisms. Proc. Natl. Acad. Sci. U. S. A 111, 5183–5188 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Seixas E et al. Heme oxygenase-1 affords protection against noncerebral forms of severe malaria. Proc. Natl. Acad. Sci. U. S. A 106, 15837–15842 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Larsen R et al. A central role for free heme in the pathogenesis of severe sepsis. Sci. Transl. Med 2, 51ra71–51ra71 (2010). [DOI] [PubMed] [Google Scholar]
- 74.Fiorito V, Chiabrando D, Petrillo S, Bertino F & Tolosano E The Multifaceted Role of Heme in Cancer. Front. Oncol 9, 1540 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ofori-Acquah SF et al. Hemopexin deficiency promotes acute kidney injury in sickle cell disease. Blood 135, 1044–1048 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Larsen R, Gouveia Z, Soares MP & Gozzelino R Heme cytotoxicity and the pathogenesis of immune-mediated inflammatory diseases. Front. Pharmacol 3 MAY, 77 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vinchi F et al. Hemopexin therapy improves cardiovascular function by preventing heme-induced endothelial toxicity in mouse models of hemolytic diseases. Circulation 127, 1317–1329 (2013). [DOI] [PubMed] [Google Scholar]
- 78.Kishimoto Y, Kondo K & Momiyama Y The Protective Role of Heme Oxygenase-1 in Atherosclerotic Diseases. Int. J. Mol. Sci 20, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chiabrando D, Fiorito V, Petrillo S & Tolosano E Unraveling the role of heme in neurodegeneration. Frontiers in Neuroscience vol. 12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Roux B et al. An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J. 77, 817–837 (2014). [DOI] [PubMed] [Google Scholar]
- 81.Robertsen BK, Åman P, Darvill AG, McNeil M & Albersheim P Host-Symbiont Interactions. Plant Physiol. 67, 389–400 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Arnold MFF et al. Genome-Wide Sensitivity Analysis of the Microsymbiont Sinorhizobium meliloti to Symbiotically Important, Defensin-Like Host Peptides. (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schäfer A et al. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145, 69–73 (1994). [DOI] [PubMed] [Google Scholar]
- 84.Babu VMP, Sankari S, Budnick JA, Caswell CC & Walker GC Sinorhizobium meliloti YbeY is a zinc-dependent single-strand specific endoribonuclease that plays an important role in 16S ribosomal RNA processing. Nucleic Acids Res. 48, 332–348 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Barr I & Guo F Pyridine Hemochromagen Assay for Determining the Concentration of Heme in Purified Protein Solutions. Bio-Protocol 5, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang J et al. Bradyrhizobium japonicum senses iron through the status of haem to regulate iron homeostasis and metabolism. Mol. Microbiol 60, 427–437 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ghosal A et al. C21orf57 is a human homologue of bacterial YbeY proteins. Biochem. Biophys. Res. Commun 484, 612–617 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Guo Y, Wallace SS & Bandaru V A novel bicistronic vector for overexpressing Mycobacterium tuberculosis proteins in Escherichia coli. Protein Expr. Purif 65, 230–237 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shah NB & Duncan TM Bio-layer interferometry for measuring kinetics of protein-protein interactions and allosteric ligand effects. J. Vis. Exp 84, 51383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sassa S Sequential induction of heme pathway enzymes during erythroid differentiation of mouse friend leukemia virus-infected cells*. J. Exp. Med 143, 305–315 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Michener JK, Nielsen J & Smolke CD Identification and treatment of heme depletion attributed to overexpression of a lineage of evolved P450 monooxygenases. Proc. Natl. Acad. Sci. U. S. A 109, 19504–19509 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Poje G & Redfield RJ General methods for culturing Haemophilus influenzae. Methods Mol. Med 71, 51–56 (2003). [DOI] [PubMed] [Google Scholar]
- 93.Leigh JA, Signer ER & Walker GC Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc. Natl. Acad. Sci. U. S. A 82, 6231–6235 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ferguson AP et al. Importance of unusually modified lipid A in Sinorhizobium stress resistance and legume symbiosis. Mol. Microbiol 56, 68–80 (2005). [DOI] [PubMed] [Google Scholar]
- 95.Natera SHA, Guerreiro N & Djordjevic MA Proteome analysis of differentially displayed proteins as a tool for the investigation of symbiosis. Mol. Plant-Microbe Interact 13, 995–1009 (2000). [DOI] [PubMed] [Google Scholar]
- 96.Tucker AT et al. Discovery of Next-Generation Antimicrobials through Bacterial Self-Screening of Surface-Displayed Peptide Libraries. Cell 172, 618–628.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Čermák T et al. A Multipurpose Toolkit to Enable Advanced Genome Engineering in Plants. Plant Cell 29, 1196–1217 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Haney CH & Long SR Plant flotillins are required for infection by nitrogen-fixing bacteria. Proc. Natl. Acad. Sci. U. S. A 107, 478–483 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Qi Z, Hamza I & O’Brian MR Heme is an effector molecule for iron-dependent degradation of the bacterial iron response regulator (Irr) protein. Proc. Natl. Acad. Sci. U. S. A 96, 13056–13061 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Aldag C et al. Probing the role of the proximal heme ligand in cytochrome P450cam by recombinant incorporation of selenocysteine. Proc. Natl. Acad. Sci. U. S. A 106, 5481–5486 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data in this manuscript is available. Source data for all the graphs and charts for all the figures and raw images are provided with the manuscript.