Abstract
The pH of the environment has been implicated in controlling the yeast-hypha transition and pathogenesis of Candida albicans. Several C. albicans genes, including PHR1 and PHR2, are pH dependent in their expression. To investigate the mechanism of pH-dependent expression, we have cloned and characterized PRR1 (for pH response regulator). PRR1 is homologous to palF, a component of the pH response pathway in Aspergillus nidulans. Expression of PRR1 was itself pH dependent, being maximal at acid pH but reduced severalfold at alkaline pH. In a prr1 null mutant the alkaline-induced expression of PHR1 was completely abolished. Conversely, expression of PHR2 was no longer repressed at alkaline pH. A prr1 null mutant exhibited no morphological abnormalities at either pH; however, it lost the ability to form hyphae on medium 199 and on 10% serum plates. The ability to filament on serum was not restored by forced expression of PHR1, indicating that additional PRR1-dependent genes are required for hyphal development. These developmental genes appear to be distinct from those controlled by the developmental regulator EFG1, since the EFG1-dependent gene HWP1 was expressed normally in the prr1 null mutant. We conclude that PRR1 encodes a component of the pH-dependent response pathway in C. albicans and that this pathway regulates the expression of multiple components of hyphal development.
Candida albicans is an opportunistic fungal pathogen of humans. It is the most frequent agent of superficial mucocutaneous infections, and, in immunocompromised hosts, it can cause life-threatening infections (38). The ability of this organism to cause infection depends, in part, on its ability to respond to changes in the pH of the environment (10). The pH response in C. albicans involves the differential expression of at least three genes, PHR1, PHR2, and PRA1 (34, 42, 44). PHR1 is an alkaline-expressed gene. It is highly expressed at between pH 7.0 and 8.0 but is not expressed at detectable levels at below pH 5.5 (42). PHR2 is an acid-expressed gene with an inverse pattern of expression (34). Mutations in PHR1 or PHR2 result in pH-conditional defects in growth, morphogenesis, and virulence (10, 34, 42). The significance of these genes and the pH response in the biology of C. albicans has prompted an investigation of the mechanisms controlling these responses.
The control of pH-dependent gene expression has been most extensively studied with Aspergillus nidulans. Seven genes critical to this regulation have been identified. These include six pal genes, palA, -B, -C, -F, -H, and -I, and pacC (2, 9, 11, 12, 31, 37, 48). pacC, the terminal component of this regulatory pathway, encodes a transcription factor containing a zinc finger motif that directly induces expression of alkaline-expressed genes and indirectly represses acid-expressed genes (43, 48). The pacC protein, PacC, is synthesized in an inactive form that is proteolytically activated at alkaline pH (39). Proteolysis of PacC is dependent upon the pal genes, although their role in this process is unknown (2, 11, 12, 37, 39).
The available data suggests that this response pathway is conserved, at least in part, in other fungi. In Yarrowia lipolytica a pacC homolog has been identified and shown to control alkaline-induced gene expression (27). In addition, four pal-like genes have been defined by mutation analysis (27). In Saccharomyces cerevisiae the pacC homolog, RIM101, was initially identified as controlling meiosis and haploid invasiveness (29, 46). More recently RIM101 and the yeast homology of palB, CPL1, have been implicated in a pH-dependent growth response of yeast (17). Genes homologous to palA and palI are also present in S. cerevisiae (11, 37). C. albicans sequences homologous to palA and pacC were recently reported (49). Null mutations in these genes affected morphological development, but it is not known if they affected the pH response (49).
To investigate the regulation of pH-dependent gene expression in C. albicans, we have isolated and characterized PRR1 (for pH response regulator), the C. albicans homolog of palF. Expression of PRR1 was pH dependent, and mutants lacking PRR1 were defective in pH-dependent regulation of gene expression. PHR1 was no longer induced at alkaline pH, and PHR2 was no longer repressed. Thus, PRR1 is a component of the pH response pathway in C. albicans. In addition, mutation of PRR1 resulted in a medium-conditional loss of hyphal development. This defect was not related to the altered expression of PHR1, suggesting that this pathway controls development-specific functions. This control was shown to be independent of the EFG1-dependent regulation of hyphal development.
MATERIALS AND METHODS
Strains and growth conditions.
The C. albicans strains utilized in this study and their genotypes are listed in Table 1. The strains were routinely cultured on either YPD (1% yeast extract, 2% Bacto Peptone, 2% glucose) or YNB (2% glucose, 0.67% Difco yeast nitrogen base) at 30°C. The effects of culture pH were assessed in medium 199 containing Earle's salts and glutamine but lacking sodium bicarbonate (Gibco-BRL) and containing 155 mM Tris or 150 mM HEPES adjusted to pH 7.5 or 4.0. To induce germ tube formation in liquid culture, yeast cells were inoculated at a density of 6 × 106 cells/ml into medium 199 (pH 7.5), Spider medium (30), the medium of Lee et al. (28), or 10% fetal calf serum (Difco) and incubated at 37°C with vigorous agitation on a rotary shaker. These same media were solidified with 2% agar to assay filamentation. Solidified media were spotted with 106 cells in 5 μl of water and incubated at 37°C for 3 to 6 days. To test for invasive hyphal growth, the surface of the plates was washed with sterile distilled water (20). In all assays, stationary-phase cells grown at 25°C in YPD were used as the inoculum. Media were supplemented with uridine (25 μg/ml) when necessary. Ura− auxotrophs were selected on medium containing 5′-fluoro-orotic acid (5′-FOA) as described previously (6, 16). Cell and colony morphologies were assessed by light microscopy.
TABLE 1.
C. albicans strains used in this study
| Strain | Parental strain | Genotype | Source or reference |
|---|---|---|---|
| SC5314 | Clinical isolate | 19 | |
| CAI4 | Δura3::imm434/Δura3::imm434 | 16 | |
| CAI12 | CAI4 | Δura3::imm434/URA3 | This work |
| CAPM1 | CAI4 | Δprr1::hisG-URA3-hisG/PRR1 Δura3::imm434/Δura3::imm434 | This work |
| CAPM2 | CAPM1 | Δprr1::hisG/PRR1 Δura3::imm434/Δura3::imm434 | This work |
| CAPM3 | CAPM2 | Δprr1::hisG/Δprr1::hisG-URA3-hisG Δura3::imm434/Δura3::imm434 | This work |
| CAPM4 | CAPM3 | Δprr1::hisG/Δprr1::hisG Δura3::imm434/Δura3::imm434 | This work |
| CAPM5 | CAPM4 | PRR1-pUC18-URA3-Δprr1::hisG/Δprr1::hisG Δura3::imm434/Δura3::imm434 | This work |
| CAPM6 | CAPM4 | Δprr1::hisG/Δprr1::hisG TEF1pr::PHR1::URA3 Δura3::imm434/Δura3::imm434 | This work |
| CAR4 | CAR26 | Δprr2::hisG/Δprr2::hisG TEF1pr::PHR1::URA3 Δura3::imm434/Δura3::imm434 | This work |
Identification and cloning of PRR1.
Sequence data for C. albicans was obtained from the Stanford DNA Sequencing and Technology Center website (45a). A BLASTN search of the C. albicans genome sequences was conducted by using the sequence of A. nidulans palF as a query (1, 32). This identified a partial sequence related to palF, and the corresponding gene was named PRR1 (for pH response regulator). This region was amplified by PCR with genomic DNA from SC5314 as a template and the primers 5′-AGTGATGATTTGGTTGCTTG-3′ and 5′-CGGATTTGGGATAGGTTC-3′. After 33 cycles consisting of incubations at 95°C for 60 s, 48°C for 45 s, and 72°C for 120 s, the amplified product was gel purified with the Geneclean II Kit (Bio 101 Inc.) and used as a probe for hybridization screening of a C. albicans λGEM-12 genomic library (5). About 25,000 phage plaques were blotted onto nylon filters (Hybond N+; Amersham) and hybridized with digoxigenin-labeled probe. The probe was labeled and hybridization was detected with the DIG DNA Labeling and Detection Kit (Boehringer Mannheim Biochemicals). Positive plaques were purified and characterized by restriction endonuclease mapping. A 6-kb BamHI DNA fragment, which hybridized to the PCR product, was gel purified and subcloned into pUC18 to generate plasmid pAP1.
DNA sequence analysis.
Nucleotide sequencing was performed by PCR with the dRhodamine terminator cycle sequencing Ready Reaction kit with AmpliTaq DNA polymerase, FS (Perkin-Elmer), and custom-synthesized oligonucleotide primers. Nucleotide and protein sequence analyses were conducted with DNA Strider (33) and LaserGene (DNASTAR). Homology searches of the nonredundant GenBank database were done by using the BLAST algorithm (1). Multiple-sequence alignments were constructed by using Clustal W 1.6 (47).
Strain constructions.
To construct a prr1 null mutant, a 2,475-bp EcoRI fragment was isolated from pAP1 and subcloned into the EcoRI site of pUC18 to generate plasmid pAP2. This fragment contains 1,069 bp of sequence upstream of the coding region and all but the last 456 bp of the coding region. A 1,074-bp region flanked by NsiI-BglII sites was deleted from pAP2 and replaced with a 3.8-kb BglII-PstI fragment from plasmid pMB-7 containing hisG-URA3-hisG (16). The resulting plasmid, pAP3, was digested with BanII, releasing a 4,428-bp DNA fragment containing the hisG-URA3-hisG fragment flanked by 478 bp of PRR1 sequence on the 5′ end and 94 bp on the 3′ end. Approximately 5 μg of transforming DNA was used to sequentially disrupt both PRR1 alleles in strain CAI4 by previously published methods (16) except that transformation was performed by the lithium acetate method (18).
To obtain a reconstituted strain, a 2.5-kb SstI-KpnI fragment from plasmid pAP1 was purified and ligated into the SstI-KpnI sites of plasmid pSMS-44 (42), which contains the EcoRV-XbaI fragment of URA3. The resulting plasmid, designated pAP4, was linearized at the unique AflII site 832 bp upstream of PRR1 and used to transform a Ura− Δprr1 strain.
To restore PHR1 expression in the prr1 null mutant, plasmid pSMS-43 (42), containing the C. albicans EF-1α promoter driving expression of the PHR1 gene, was linearized at the unique HpaI site within the URA3 sequences and used to transform strain CAPM4 to Ura+, generating strain CAPM6. An analogous construct was obtained in a prr2 null mutant, CAR26 (41). Constitutive expression of PHR1 was verified for both strains by Northern blot analysis. The integration events in all transformants were characterized by Southern blot analysis.
The control strain CAI12 was constructed by transformation of strain CAI4 with a PstI-BglII fragment containing URA3 (26) to restore one allele at the URA3 locus.
Southern blot analysis.
Genomic DNA for Southern analysis was prepared as previously described (25). About 5 μg of genomic DNA was digested to completion with BamHI or double digested with NsiI and BglII and electrophoresed through a 0.8% agarose gel. The fractionated DNA was transferred to positively charged nylon membranes (Hybond N+; Amersham), and the membranes were fixed by UV irradiation. The blots were prehybridized and hybridized in 1× phosphate buffer solution (0.5 M sodium phosphate [pH 7.2], 5% sodium dodecyl sulfate, 10 mM EDTA) at 65°C. A 1.7-kb NsiI fragment of plasmid pAP1 was labeled by random priming with the DNA Ready-to-Go Labeling Beads (−dCTP) Kit and [α-32P]dCTP (Amersham Pharmacia Biotech Inc.) and used as a probe. Following hybridization, the filters were washed three times for 15 min each in 0.1× phosphate buffer and exposed to X-ray film, using intensifying screens, at −70°C overnight.
Northern blot analysis.
To obtain RNA samples, cells were grown to stationary phase in YPD at 25°C and used to inoculate 300 ml of medium 199, pH 7.5 or 4.0, at a density of 6 × 106 cells/ml. The cultures were incubated with vigorous agitation at 25°C for 2 h. Yeast cells were collected by centrifugation at 4,000 × g for 10 min and washed with ice-cold sterile distilled water. Total RNA was prepared by selective precipitation with LiCl as previously described (40). Samples containing 10 μg of RNA were fractionated by electrophoresis in agarose gels containing MOPS (morpholinepropanesulfonic acid) and formaldehyde essentially as described previously (7) except that 2.2% formaldehyde was included in the running buffer. The gels were blotted and hybridized as described for Southern blot analysis. The blots were hybridized with the 867-bp PCR fragment containing part of the PRR1 open reading frame, a 1,063-bp AatII-NdeI fragment of PHR1 (42), a 1,257-bp BamHI-NheI fragment of PHR2 (34), or a 4.3-kb EcoRI fragment of HWP1 (45). The blots were subsequently stripped in boiling 0.1% sodium dodecyl sulfate and hybridized with a 1.9-kb SalI fragment of ACT1 DNA. Hybridization intensity was quantified by using a model 445SI PhosphorImager (Molecular Dynamics) and ImageQuant software and normalized to ACT1 mRNA. All Northern data was confirmed with at least two independent RNA preparations. Transcript sizes were approximated by comparison with rRNA.
RESULTS
Identification and cloning of the palF ortholog.
Several C. albicans genes are differentially expressed in relation to the ambient pH of the culture medium (34, 42, 44). The response to pH affects in vitro dimorphism and is critical to virulence (3, 8, 10, 14). To investigate the mechanism(s) that controls the pH response in C. albicans, we searched for homologs of the genes that control pH responsiveness in A. nidulans (48). By using the nucleotide sequence of palF (32) in a BLASTN search of the C. albicans genome sequences, a short homologous sequence of 867 nucleotides was identified. This fragment encoded a putative peptide 27% identical and 44% similar to PalF, the palF protein. A genomic clone was isolated, and the nucleotide sequence of the corresponding region was determined. Within the 3,336 bp of sequence obtained, a single extended open reading frame of 1,863 bp was identified. The predicted 621-amino-acid protein was 24% identical to PalF (Fig. 1), and the identity was distributed along the entire lengths of the proteins. On the basis of this homology and the potential role of the gene in the pH response, the gene was designated PRR1 (for pH response regulator). A BLASTP search of the nonredundant GenBank database revealed no other related genes except for two adjacent open reading frames in the S. cerevisiae genome, YGLO46w (accession no. Z72568) and YGLO45w (accession no. Z72567), as previously noted for palF (32) (Fig. 1). No functional motifs were identified in a search of the BLOCKS (21, 22) and PROSITE (4) databases. However, a potential nuclear localization signal, PRPKPKR, located between positions 205 and 211 (23), was identified by PSORT II (35).
FIG. 1.
Alignment of the amino acid sequences of Prr1p, PalF, YGL045w, and YGL046w. Identical residues are boxed.
Transcription of PRR1.
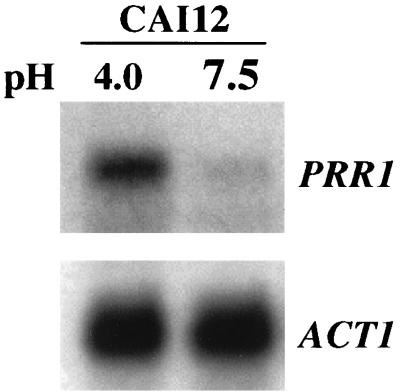
Northern analysis was performed to define the conditions under which PRR1 is expressed. Because of the potential role of PRR1 in the pH response, the effect of ambient pH on PRR1 expression was examined. The probe hybridized with a single transcript of approximately 2 kb, consistent with the size of the predicted open reading frame (Fig. 2). The intensity of this band was maximal at pH 4.0 and was reduced about threefold at pH 7.5 when normalized against actin mRNA. Thus, expression of PRR1 was pH dependent and elevated at acidic pH. The pH dependence of PRR1 expression was further confirmed by the effect of PRR2 mutations on PRR1 expression, as discussed in the accompanying paper (41).
FIG. 2.
Expression of PRR1 as a function of ambient pH. RNA was prepared from strain CAI12 cultured at pH 4.0 or 7.5. A Northern blot of these samples was hybridized with either PRR1 or ACT1 DNA as indicated.
PRR1 is required for pH-dependent gene expression.
Recessive mutations in palF mimic the effect of growth at acid pH by blocking the induction of alkaline-expressed genes and preventing the repression of acid-expressed genes under alkaline growth conditions (9, 48). If PRR1 is a functional homolog of palF, then a prr1 null mutant should have a similar phenotype. To test this possibility, PRR1 mutants were constructed (16), and expression of PHR1, an alkaline-expressed gene, and PHR2, an acid-expressed gene, was examined in the mutants.
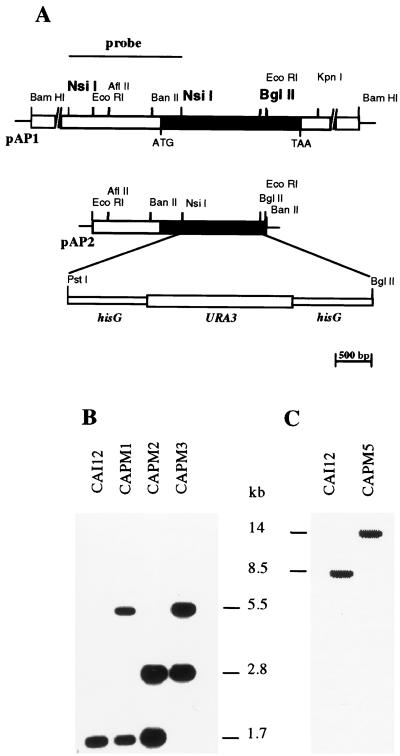
The mutants were constructed by targeted gene replacement. The cells were transformed with a deletion-and-replacement construct in which a 1,074-bp segment of PRR1 was removed and replaced with a hisG-URA3-hisG cassette (Fig. 3). Southern blot analysis of a representative Ura+ transformant, strain CAPM1, demonstrated the presence of a 1.7-kb NsiI-BglII band characteristic of the wild-type PRR1 and of the predicted 5.5-kb band representing the disrupted allele (Fig. 3). After 5′-FOA selection, the latter band was reduced to 2.8 kb, as shown in the representative heterozygous mutant CAPM2 (Fig. 3). Transformation of CAPM2 yielded a homozygous null mutant, CAPM3, in which the 1.7-kb fragment was absent and replaced by the 5.5-kb fragment, indicative of the replacement event (Fig. 3). Northern analysis confirmed the absence of a PRR1 transcript in the null mutant CAPM3 (data not shown).
FIG. 3.
Construction of PRR1 mutants. (A) Partial restriction map of the 6-kb BamHI genomic fragment containing the PRR1 gene in plasmid pAP1 and the subcloned EcoRI fragment in plasmid pAP2. The black bar indicates the coding region of PRR1. The region replaced by the URA3 cassette is also indicated. The overlined region of pAP1 was used as a hybridization probe in the Southern blot show in panel B. The BglII and NsiI recognition sites relevant to the Southern blot analysis are indicated in boldface. (B) Genomic DNAs from the parental strain CAI12 and representative PRR1 mutants CAPM1, CAPM2, and CAPM3 were double digested with NsiI and BglII and used in Southern blot analysis. (C) Genomic DNAs from CAI12 and one representative revertant strain, CAPM5, were digested with BamHI.
A wild-type PRR1 allele was introduced into a prr1 null mutant by targeted integration of plasmid pAP4 in strain CAPM4. Strain CAPM4 is a Ura− strain derived from CAPM3 by 5′-FOA selection. Southern blot analysis of BamHI-digested genomic DNA revealed one hybridizing band of about 8.5 kb in the control strain, CAI12. DNA from the transformed strain, CAPM5, showed this band as well as an approximately 14-kb band corresponding to the reconstructed allele (Fig. 3). Expression of PRR1 in the revertant was confirmed by Northern analysis (data not shown).
The effect of PRR1 mutation on pH-dependent gene expression was examined by Northern blot analysis. The various mutants were cultured at either acidic pH or alkaline pH, and RNA from these cells was hybridized with either PHR1 (42) or PHR2 (34). In the control strain CAI12, the expected expression pattern of PHR1 was observed (Fig. 4). PHR1 was abundant in cells grown at pH 7.5 but undetectable in cells grown at pH 4.0. The same pattern was exhibited by strain CAPM1, which lacks one allele of PRR1. In contrast, PHR1 was not expressed in the homozygous null mutant, CAPM3, cultured at alkaline pH (Fig. 4). Alkaline-induced expression was restored upon introduction of a wild-type allele of PRR1 (Fig. 4). Hybridization with PHR2 revealed the expected expression pattern in CAI12 and an identical pattern in the heterozygous mutant CAPM1 (Fig. 4). In the homozygous null mutant, CAPM3, however, PHR2 was highly expressed at both acid and alkaline pH (Fig. 4). Repression at alkaline pH was restored upon reintroduction of PRR1 (Fig. 4). These results demonstrate that PRR1, like its palF homolog, regulates the pH response and is required for induction of alkaline-expressed genes and repression of acid-expressed genes.
FIG. 4.
Effect of PRR1 mutations on PHR1 and PHR2 expression. Total RNAs were isolated from the indicated strains cultured at either pH 4.0 or 7.5. A Northern blot of the samples was prepared and sequentially hybridized with of PHR1, PHR2, and ACT1 DNAs.
Effect of PRR1 mutations on growth and hyphal development.
Ambient pH has a dramatic effect on hyphal development in vitro; the process is inhibited at acid pH and induced at neutral to alkaline pH (3, 8, 14). Having established the role of PRR1 in the pH response, its role in determining cell morphology and hyphal development was examined. The null mutant exhibited a typical yeast morphology when grown in a variety of liquid media at 25°C, under acidic or alkaline conditions. However, a high percentage of multibudded cells was observed when the Δprr1 mutant was grown until stationary phase in YPD at 25°C.
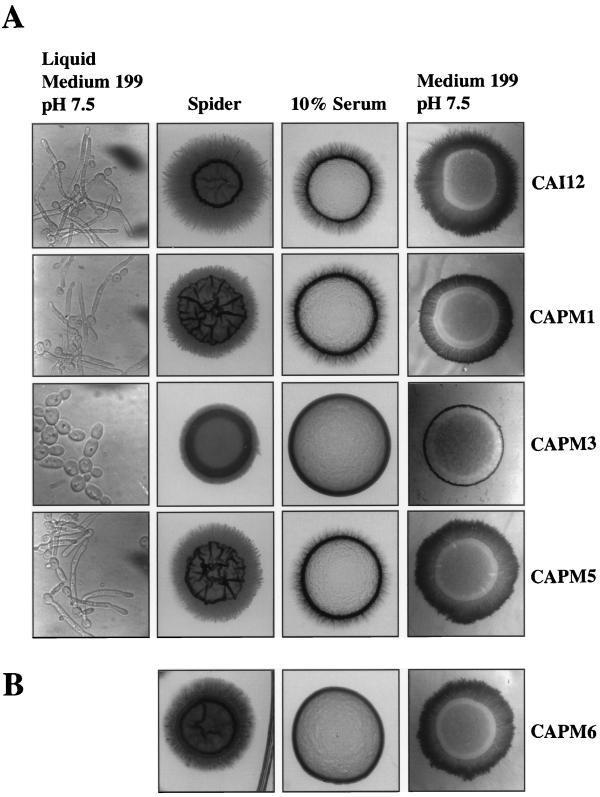
No differences were observed between CAI12 and the prr1 null mutant in the frequency or rate of germ tube formation when inoculated into Spider medium, the medium of Lee et al. (28), or 10% serum and incubated at 37°C (data not shown). However, the null mutant formed no germ tubes in medium 199 buffered at pH 7.5 (Fig. 5A). Instead, chains of typical yeast cells were formed. This defect was not evident in either heterozygous mutant, CAPM1 or CAPM5, and was verified with three independent null mutants.
FIG. 5.
Effect of PRR1 mutations on hyphal development. (A) Photomicrographs of CAI12 and the PRR1 mutants induced to form germ tubes in medium 199, pH 7.5, and to filament on agar-solidified medium. (B) Filamentation of a prr1 null mutant containing a constitutively expressed PHR1 allele.
Mutation of PRR1 had a more significant consequence for hyphal development on agar-solidified media (Fig. 5A). On 10% serum plates the heterozygous mutant CAPM1 exhibited a modest reduction in filamentation in comparison with the control strain CAI12. However, no hyphae were observed emanating from colonies of the null mutant CAPM3. Upon reintroduction of the gene, as in strain CAPM5, filamentation was restored to the same extent as observed for the heterozygous disruptant. Filamentation was similarly absent from the null mutant cultured on medium 199, pH 7.5, plates. In contrast, filamentation was greatly reduced, but not eliminated, when the null mutant was cultured on either Spider medium or the medium of Lee et al. (28). Again, the null phenotype was largely reversed in the revertant strain. The same null phenotype was observed in three independent mutants.
The null mutant also exhibited medium-conditional defects in invasion of the agar below the colony dome. Invasiveness was observed on Spider medium, the medium of Lee et al. (28), and even medium 199, despite the absence of filamentation. However, the mutant was not invasive on 10% serum (data not shown).
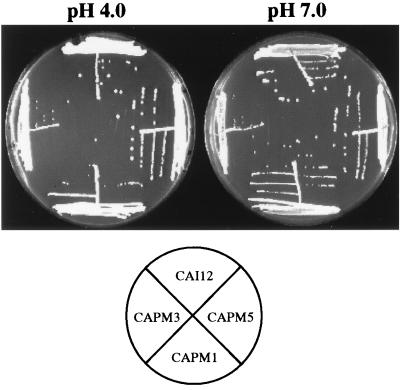
Mutation of palF imparts a pH-conditional growth defect. The mutants are unable to grow at alkaline pH (9, 13, 48). The prr1 null mutant grew equally well at either pH 4.0 or 7.0, although the colonies appeared to be slightly smaller than those of the control strain (Fig. 6). This was observed for all three independent mutants.
FIG. 6.
Influence of pH on growth of prr1 null mutants. Cells of all indicated strains were streaked on YNB agar adjusted to pH 4.0 or 7.0 and incubated for 3 days at 30°C.
PRR1 controls development-specific functions.
The absence of hyphal development in the prr1 null mutant may reflect its role in controlling development-specific functions or may simply reflect the absence or reduction of PHR1 expression. The cellular morphology of the mutant in medium 199 at pH 7.5 (Fig. 5A) is similar to that of a phr1 null mutant at pH 7.0 (42). To distinguish between these possibilities, expression of PHR1 was restored in the Δprr1 mutant by placing PHR1 under the control of a constitutive, non-pH-regulated promoter. As shown in Fig. 5B, the constitutive expression of PHR1 in this strain, CAPM6, did not restore the ability to form hyphae on 10% serum plates. However, on Spider medium and medium 199, filamentation was enhanced compared to that of the null mutant but did not match that of the control strain CAI12. Thus, the absence of PHR1 partially limits hyphal development under some conditions, but PRR1 apparently controls additional functions required for filamentation.
Because the function of PRR1 is unknown, it is conceivable that it has a dual function, one within the pH response pathway and the other within a development-specific pathway. To test this possibility, PHR1 was constitutively expressed in a Prr1+ Prr2− background. PRR2 is a second component of the pH response pathway that is homologous to Aspergillus pacC (41). pacC encodes the terminal component of the pH response pathway, a zinc finger transcription factor (48). A prr2 null mutant is phenotypically similar to a prr1 null mutant in altered expression of pH-dependent genes and in its failure to filament on serum and medium 199 (41). As seen with the prr1 null mutant, constitutive expression of PHR1 partially restored filamentation on medium 199 but failed to restore filamentation on 10% serum (data not shown). This suggests that PRR1 acts via the pH response pathway in its control of hyphal development.
PRR1 control of development is independent of EFG1.
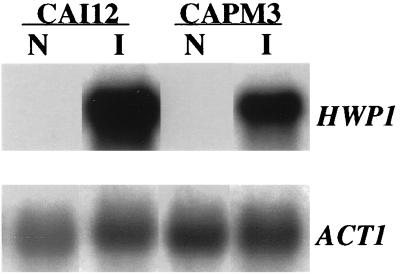
Multiple regulatory genes influence the process of hyphal development. We recently identified HWP1 as a downstream target of one of these regulators, EFG1 (45). EFG1 is required to induce HWP1 expression, and HWP1 is required for normal filamentation. To determine if the pH response pathway, as defined by PRR1, controls filamentation via EFG1, the effect of PRR1 mutations on HWP1 expression was examined. The control strain CAI12 and the prr1 null mutant CAPM3 were cultured in medium 199, either at pH 4.0 and 25°C or at pH 7.5 and 37°C. These conditions repress and strongly induce expression of HWP1, respectively (45). RNA from these cells was examined by Northern blot analysis with HWP1 as a probe. As shown in Fig. 7, expression of HWP1 was induced in the absence of PRR1, indicating that the pH response pathway is not required for the expression of EFG1-dependent genes.
FIG. 7.
Effect of PRR1 mutation on HWP1 expression. Total RNA was extracted from either CAI12 or the prr1 null mutant CAPM3 cultured under noninducing (lanes N) or hypha-inducing (lanes I) conditions. A Northern blot was prepared and probed with HWP1 and ACT1 (as a control).
DISCUSSION
Previous studies established that several genes in C. albicans are differentially expressed in relation to the ambient pH of the culture medium (34, 42, 44). Furthermore, this pH response appears to be essential for virulence (10). The regulation of pH-dependent gene expression has been extensively studied with A. nidulans and entails a set of six pal genes which are required for the alkaline-induced, proteolytic activation of the pacC-encoded transcription factor (2, 9, 11, 12, 31, 37). This mechanism of the pH response is conserved, at least in part, in Y. lipolytica and S. cerevisiae (17, 27). To determine if pH-dependent gene expression was similarly regulated in C. albicans, we cloned and characterized PRR1, the C. albicans homolog of palF.
The C. albicans homolog, PRR1, was homologous to palF along its entire length. No motifs that would provide clues to the biochemical function of this protein were identified in Prr1p. However, a potential nuclear localization site is present in Prr1p. Although this sequence was not conserved in PalF, PalF does contain other potential nuclear localization signals, suggesting that these proteins may function within the nucleus. Unlike palA, palB, and palI, for which homologs have been identified in other species, no additional full-length homologs of palF were identified. As previously reported (32), two adjacent open reading frames related to the amino and carboxy termini of PalF were identified in the S. cerevisiae genome sequences. The available expression data suggests that these are independently expressed (24) and not simply the result of a sequencing error. This may indicate that PalF and Prr1p are bifunctional proteins and that these functions have became separated in yeast.
Although the biochemical function of Prr1p is unclear, Prr1p was clearly demonstrated to play a role in the pH response of C. albicans. PHR1 and PHR2 are alkaline-expressed and acid-expressed genes, respectively (34, 42). The balanced, reciprocal expression pattern of these genes suggested a coordinated control via a common regulatory mechanism (34). Indeed, expression of both genes was affected in a homozygous prr1 null mutant. Expression of PHR1 was no longer induced at alkaline pH, and expression of PHR2 was no longer repressed. This parallels the effect of palF mutations on pH-dependent gene expression in A. nidulans and indicates that the pH response pathway has been conserved between these species.
PRR1 not only is a component of the pH response pathway but is itself regulated by this response. Expression was substantially reduced at alkaline pH. This observation was somewhat surprising, assuming that Prr1p, like PalF, is required at alkaline pH to activate the terminal transcription factor. However, as shown in related studies (41), the alkaline repression of PRR1 likely reflects a feedback loop to prevent runaway expression of PRR2, the homolog of pacC. It is not known if this is a conserved regulatory feature, since the expression of palF in relation to ambient pH has not been reported. However, pH does not affect the expression of palA, -B, -C, -H, or -I (11, 12, 36, 37).
In addition to controlling expression of PHR1 and PHR2, which encode proteins involved in cell wall biosynthesis (15), PRR1 also controls development-specific functions. prr1 null mutants exhibited medium-conditional defects in germ tube induction and filamentation on agar-solidified media. The mutants lost the ability to form germ tubes in medium 199, but not in other media. Filamentation was blocked on 10% serum and on medium 199 and was partially reduced on Spider medium or the medium of Lee et al. (28). These defects were not due to differential acidification of the media by the mutant, since the media were strongly buffered and the pH did not change over the course of the experiments. Nor were they due to the absence of PHR1 expression in the mutant. This was directly demonstrated by forced expression of PHR1 in a prr1 null mutant, which failed to restore filamentation on 10% serum. This suggests that the mutant fails to express functions required for hyphal development.
Constitutive expression of PHR1 did restore some measure of filamentation on medium 199. This was not expected, because PHR2 is constitutively expressed in the PRR1 mutant and Phr2p is functionally homologous to Phr1p (34). In fact, unlike a phr1 null mutant (42), the PRR1 mutants exhibited a normal yeast morphology at alkaline pH despite the absence of PHR1 expression. This presumably is due to the expressed Phr2p substituting for Phr1p. However, the ability of the constitutive PHR1 allele to partially restore filamentation suggests that there may be important functional distinctions between Phr1p and Phr2p. Alternatively, the level of PHR2 expression may simply be inadequate for hyphal development under these conditions. This presupposes that different levels of these proteins are needed for yeast versus hyphal wall synthesis.
It remains to be determined how the pH response pathway is integrated with other signal pathways that control differentiation in C. albicans. However, in the absence of PRR1, the EFG1-dependent expression of HWP1 was still noted under the conditions tested. This suggests that the pH response pathway is distinct from the EFG1-dependent signaling pathway and probably acts on a separate set of morphogenic functions. Finally, it should be noted that the role of PRR1 in development is consistent with, and provides a partial mechanistic explanation for, the longstanding observation that ambient pH strongly influences hyphal development in vitro (3, 8, 14).
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant GM47727 from the National Institutes of Health and the Burroughs Wellcome Fund Scholar Award in Molecular Pathogenic Mycology. Sequencing of C. albicans was accomplished with the support of the NIDR and the Burroughs Wellcome Fund.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Arst H N J, Bignell E, Tilburn J. Two new genes involved in signalling ambient pH in Aspergillus nidulans. Mol Gen Genet. 1994;245:787–790. doi: 10.1007/BF00297286. [DOI] [PubMed] [Google Scholar]
- 3.Auger P, Joly J. Factors influencing germ tube production in Candida albicans. Mycopathologia. 1977;61:183–186. doi: 10.1007/BF00468014. [DOI] [PubMed] [Google Scholar]
- 4.Bairoch A. PROSITE: a dictionary of sites and patterns in proteins. Nucleic Acids Res. 1992;20:2013–2022. doi: 10.1093/nar/20.suppl.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birse C E, Irwin M Y, Fonzi W A, Sypherd P S. Cloning and characterization of ECE1, a gene expressed in association with cell elongation of the dimorphic pathogen Candida albicans. Infect Immun. 1993;61:3648–3655. doi: 10.1128/iai.61.9.3648-3655.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boeke J D, LaCroute F, Fink G R. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 7.Brown T, Mackey K. Analysis of RNA by Northern and slot blot hybridization. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: Greene Publishing Associates; 1997. pp. 4.9.1–4.9.16. [Google Scholar]
- 8.Buffo J, Herman M A, Soll D R. A characterization of pH-regulated dimorphism in Candida albicans. Mycopathology. 1984;85:21–30. doi: 10.1007/BF00436698. [DOI] [PubMed] [Google Scholar]
- 9.Caddick M X, Brownlee A G, Arst H N J. Regulation of gene expression by pH of the growth medium in Aspergillus nidulans. Mol Gen Genet. 1986;203:346–353. doi: 10.1007/BF00333978. [DOI] [PubMed] [Google Scholar]
- 10.De Bernardis F, Mühlschlegel F A, Cassone A, Fonzi W A. The pH of the host niche controls gene expression in and virulence of Candida albicans. Infect Immun. 1998;66:3317–3325. doi: 10.1128/iai.66.7.3317-3325.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denison S H, Negrete-Urtasun S, Mingot J M, Tilburn J, Mayer W A, Goel A, Espeso E A, Peñalva M A, Arst H N., Jr Putative membrane components of signal transduction pathways for ambient pH regulation in Aspergillus and meiosis in Saccharomyces are homologous. Mol Microbiol. 1998;30:259–264. doi: 10.1046/j.1365-2958.1998.01058.x. [DOI] [PubMed] [Google Scholar]
- 12.Denison S H, Orejas M, Arst H N., Jr Signaling of ambient pH in Aspergillus involves a cysteine protease. J Biol Chem. 1995;270:28519–28522. doi: 10.1074/jbc.270.48.28519. [DOI] [PubMed] [Google Scholar]
- 13.Espeso E A, Tilburn J, Arst H N, Peñalva M A. pH regulation is a major determinant in expression of a fungal penicillin biosynthetic gene. EMBO J. 1993;12:3947–3956. doi: 10.1002/j.1460-2075.1993.tb06072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans E G V, Odds F C, Richardson M D, Holland K T. Optimum conditions for initiation of filamentation in Candida albicans. Can J Microbiol. 1974;21:338–342. doi: 10.1139/m75-048. [DOI] [PubMed] [Google Scholar]
- 15.Fonzi W A. PHR1 and PHR2 of Candida albicans encode glycosidases required for the proper cross-linking of β-1,3-glucans and β-1,6-glucans. J Bacteriol. 1999;181:7070–7079. doi: 10.1128/jb.181.22.7070-7079.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fonzi W A, Irwin M Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Futai E, Maeda T, Sorimachi H, Kitamoto K, Ishiura S, Suzuki K. The protease activity of a calpain-like cysteine protease in Saccharomyces cerevisiae is required for alkaline adaptation and sporulation. Mol Gen Genet. 1999;260:559–568. doi: 10.1007/s004380050929. [DOI] [PubMed] [Google Scholar]
- 18.Gietz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gillum A M, Tsay E Y H, Kirsch D R. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet. 1984;198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- 20.Gimeno C J, Ljungdahl P O, Styles C A, Fink G R. Unipolar cell division in the yeast S. cerevisiae leads to filamentous growth: regulation by starvation. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- 21.Henikoff J G, Henikoff S, Pietrokovski S. New features of the Blocks Database servers. Nucleic Acids Res. 1999;27:226–228. doi: 10.1093/nar/27.1.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henikoff S, Henikoff J G, Pietrokovski S. Blocks+: a non-redundant database of protein alignment blocks derived from multiple compilations. Bioinformatics. 1999;15:471–479. doi: 10.1093/bioinformatics/15.6.471. [DOI] [PubMed] [Google Scholar]
- 23.Hicks G R, Raikhel N V. Protein import into the nucleus: an integrated view. Annu Rev Cell Dev Biol. 1995;11:155–188. doi: 10.1146/annurev.cb.11.110195.001103. [DOI] [PubMed] [Google Scholar]
- 24.Hodges P E, McKee A H Z, Davis B P, Payne W E, Garrels J I. The yeast proteome database (YPD): a model for the organization and presentation of genome-wide functional data. Nucleic Acids Res. 1999;27:69–73. doi: 10.1093/nar/27.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffman C S. Preparation of yeast DNA, RNA, and proteins. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: Greene Publishing Associates; 1997. pp. 13.11.1–13.11.4. [Google Scholar]
- 26.Kelly R, Miller S M, Kurtz M B, Kirsch D R. Directed mutagenesis in Candida albicans: one-step gene disruption to isolate ura3 mutants. Mol Cell Biol. 1987;7:199–207. doi: 10.1128/mcb.7.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lambert M, Blanchin-Roland S, Le Louedec F, Lepingle A, Gaillardin C. Genetic analysis of regulatory mutants affecting synthesis of extracellular proteinases in the yeast Yarrowia lipolytica: identification of a RIM101/pacC homolog. Mol Cell Biol. 1997;17:3966–3976. doi: 10.1128/mcb.17.7.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee K L, Buckley H R, Campbell C C. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia. 1975;13:148–153. doi: 10.1080/00362177585190271. [DOI] [PubMed] [Google Scholar]
- 29.Li W, Mitchell A P. Proteolytic activation of Rim1p, a positive regulator of yeast sporulation and invasive growth. Genetics. 1997;145:63–73. doi: 10.1093/genetics/145.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu H, Kohler J, Fink G R. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- 31.MacCabe A P, Van den Hombergh J P, Tilburn J, Arst H N, Jr, Visser J. Identification, cloning and analysis of the Aspergillus niger gene pacC, a wide domain regulatory gene responsive to ambient pH. Mol Gen Genet. 1996;250:367–374. doi: 10.1007/BF02174395. [DOI] [PubMed] [Google Scholar]
- 32.Maccheroni W, Jr, May G S, Martinez-Rossi N M, Rossi A. The sequence of palF, an environmental pH response gene in Aspergillus nidulans. Gene. 1997;194:163–167. doi: 10.1016/s0378-1119(97)00095-4. [DOI] [PubMed] [Google Scholar]
- 33.Marck C. “DNA Strider”: a C program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 1988;16:1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mühlschlegel F A, Fonzi W A. PHR2 of Candida albicans encodes a functional homolog of the pH-regulated gene PHR1 with an inverted pattern of expression. Mol Cell Biol. 1997;17:5960–5967. doi: 10.1128/mcb.17.10.5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Negrete-Urtasun S, Reiter E, Diez E, Tilburn J, Espeso E A, Peñalva M A, Arst H N., Jr Ambient pH signal transduction in Aspergillus: completion of gene characterization. Mol Microbiol. 1999;33:994–1003. doi: 10.1046/j.1365-2958.1999.01540.x. [DOI] [PubMed] [Google Scholar]
- 37.Negrete-Urtasun S, Denison S H, Arst H N., Jr Characterization of the pH signal transduction pathway gene palA of Aspergillus nidulans and identification of possible homologs. J Bacteriol. 1997;179:1832–1835. doi: 10.1128/jb.179.5.1832-1835.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Odds F C. Candida and candidosis. A review and bibliography. 2nd ed. London, United Kingdom: Bailliere Tindal; 1988. [Google Scholar]
- 39.Orejas M, Espeso E A, Tilburn J, Sarkar S, Arst H N, Jr, Peñalva M A. Activation of the Aspergillus PacC transcription factor in response to alkaline ambient pH requires proteolysis of the carboxy-terminal moiety. Genes Dev. 1995;9:1622–1632. doi: 10.1101/gad.9.13.1622. [DOI] [PubMed] [Google Scholar]
- 40.Ramon A M, Gil R, Burgal M, Sentandreu R, Valentin E. A novel cell wall protein specific to the mycelial form of Yarrowia lipolytica. Yeast. 1996;12:1535–1548. doi: 10.1002/(SICI)1097-0061(199612)12:15%3C1535::AID-YEA59%3E3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 41.Ramon A M, Porta A, Fonzi W A. Effect of environmental pH on morphological development of Candida albicans is mediated via the PacC-related transcription factor encoded by PRR2. J Bacteriol. 1999;181:1061–99. doi: 10.1128/jb.181.24.7524-7530.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saporito-Irwin S M, Birse C E, Sypherd P S, Fonzi W A. PHR1, a pH-regulated gene of Candida albicans, is required for morphogenesis. Mol Cell Biol. 1995;15:601–613. doi: 10.1128/mcb.15.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarkar S, Caddick M X, Bignell E, Tilburn J, Arst H N., Jr Regulation of gene expression by ambient pH in Aspergillus: genes expressed at acid pH. Biochem Soc Trans. 1996;24:360–363. doi: 10.1042/bst0240360. [DOI] [PubMed] [Google Scholar]
- 44.Sentandreu M, Elorza M V, Sentandreu R, Fonzi W A. Cloning and characterization of PRA1, a gene encoding a novel pH-regulated antigen of Candida albicans. J Bacteriol. 1998;180:282–289. doi: 10.1128/jb.180.2.282-289.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharkey L L, McNemar M D, Saporito-Irwin S M, Sypherd P S, Fonzi W A. HWP1 functions in the morphological development of Candida albicans downstream of EFG1, TUP1, and RBF1. J Bacteriol. 1999;181:5273–5279. doi: 10.1128/jb.181.17.5273-5279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45a.Stanford DNA Sequencing and Technology Center. July 1999, revision date. [Online.] http://www-sequence.stanford.edu/group/candida. [June 1999, last date accessed.]
- 46.Su S S Y, Mitchell A P. Molecular characterization of the yeast meiotic regulatory gene RIM1. Nucleic Acids Res. 1993;21:3789–3797. doi: 10.1093/nar/21.16.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tilburn J, Sarkar E A, Orejas M, Mungroo J, Peñalva M A, Arst H N J. The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 1995;14:779–790. doi: 10.1002/j.1460-2075.1995.tb07056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson R B, Davis D, Mitchell A P. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol. 1999;181:1868–1874. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]