Abstract
Long non-coding RNAs (lncRNAs) have been widely recognized as important regulators in myocardial infarction (MI) and other heart diseases. Our study aimed to investigate the mechanism and biological function of an unknown lncRNA zinc finger protein 561 antisense RNA 1 (ZNF561-AS1) in MI. After confirming the MI model was successful, we applied reverse transcription quantitative polymerase chain reaction and Western blot (WB) and found that the expression of NLR family pyrin domain containing 3 (NLRP3), interleukin (IL)-1β, and IL-18 was substantially increased in infarct and border zones of MI mice heart at 24 h and 72 h compared with that in sham-operated models. Moreover, we found that NLRP3 expression was promoted in hypoxia human cardiomyocytes (HCMs). Through cell function assays including CCK-8, 5-Ethynyl-2’-deoxyuridine (EdU), flow cytometry, and TdT-mediated dUTP Nick-End Labeling (TUNEL), supported by WB analysis, we verified that silencing of NLRP3 facilitated proliferation but impeded apoptosis of hypoxia-induced myocardial cell. Moreover, Ago2-RIP and RNA pull-down assays displayed that NLRP3 could combine with miR-223-3p. Luciferase reporter assays further confirmed that NLRP3 was directly targeted by miR-223-3p. Simultaneously, we found that miR-223-3p was the downstream gene of ZNF561-AS1. In addition, we conducted a series of rescue experiments to affirm that ZNF561-AS1 regulated cell proliferation and apoptosis in MI through miR-223-3p/NLRP3 axis.
Keywords: myocardial infarction, NLRP3, miR-223-3p, ZNF561-AS1
Introduction
Myocardial infarction (MI) is one of the most common ischemic heart diseases leading to death all over the world 1 . In recent years, the severity of MI has drawn increasing attention in humans. In acute MI, persistent and severe myocardial ischemia results in myocardial necrosis2,3. Moreover, many studies have explored that long non-coding RNAs (lncRNAs) are involved in the progression of MI4,5. Our study elucidated the molecular mechanism of a novel lncRNA zinc finger protein 561 antisense RNA 1 (ZNF561-AS1) in hypoxia-induced myocardial cells.
NLR family pyrin domain containing 3 (NLRP3) inflammasome has been reported to act as a modulator in MI6–8. NLRP3 inflammasome activation protects the heart during myocardial ischemia reperfusion 9 . Selective inhibition of the NLRP3 inflammasome could decline infarct size and preserve the cardiac function of MI 10 . Moreover, NLRP3 involvement in modulating cell malignant behaviors in different diseases has also been somewhat investigated. For instance, NLRP3 downregulation could markedly weaken proliferative, migratory, and invasive abilities of glioma cells while strengthening cell apoptosis 11 . In addition, NLRP3 knockdown has also been validated to significantly hamper oral squamous cell carcinoma (OSCC) cell viability and proliferation 12 . In our study, we examined the expression of NLRP3, as well as interleukin (IL)-1β and IL-18 expression, which were inflammatory markers in infarct and border zones of MI mice heart or in sham-operated models at 24 and 72 h to detect the differences. Afterward, we performed gain-of-function and loss-of function assays to confirm the influence of NLRP3 on proliferation and apoptosis of hypoxia-induced myocardial cells. Thereafter, we conducted further experiment to explore the underlying regulatory mechanism involving NLRP3 in hypoxia-induced myocardial cells.
Increasing evidence has implied that microRNAs (miRNAs) play crucial roles in the pathophysiological processes of MI13–15. LncRNA autophagy-promoting factor (APF) affects autophagy and MI by regulating miR-188-3p 16 . MiR-1231 aggravates arrhythmia by targeting CACNA2D2 in MI 17 . MiR-223 could promote cardiomyocyte apoptosis by inhibiting Foxo3a expression 18 . MiR-223 has also been proved to regulate cardiac fibrosis after MI by targeting RASA1 19 . In our study, we designed and implemented various experiments to ravel out the interaction between NLRP3 and miR-223-3p in hypoxia-induced myocardial cells. More importantly, we also applied bioinformatics programs to predict whether ZNF561-AS1 served as a potential lncRNA binding with miR-223-3p. In this respect, we further explored whether ZNF561-AS1/miR-223-3p/NLRP3 axis regulated cell proliferation and apoptosis of hypoxia-induced myocardial cells.
In a word, our study was to elucidate the role and relevant regulatory mechanism regarding ZNF561-AS1 in cardiomyocytes, which might provide a novel perspective to understand MI and offer a potential biomarker for MI.
Materials and Methods
Animals
The healthy adult Kunming male mice (25–30 g) were procured from Beijing Vital River Laboratory Animal Technology Co. Ltd. (Beijing, China) and used in this study. Mice were maintained under standard animal housing conditions and were free to feed and water for 7 days. All animal-related procedures were conducted in line with the Institutional Animal Care and Use Committee of the First Affiliated Hospital of Henan University and the Ethics Committee of the First Affiliated Hospital of Henan University.
Hematoxylin–Eosin (H&E) Staining
H&E staining was performed as previously described 20 . MI tissues were taken under aseptic conditions, subsequent to 24-h fixation with 4% formaldehyde. The processed tissues were stained with H&E, and then H&E-stained images were captured with a standard microscope.
Cell Culture and Treatment
Human cardiomyocytes (HCMs) used in this study were available from PromoCell GmbH (Heidelberg, Germany) and preserved in PromoCell cardiomyocyte culture medium with 10% fetal bovine serum (FBS; PromoCell). HCM was cultured in a humidified incubator supplying 5% CO2 at 37°C. The hypoxia HCMs were constructed under hypoxic conditions of 1% O2, 5% CO2, and 94% N2 for 48 h.
Total RNA Extraction and RT-qPCR
TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) was employed for extracting total RNA from cultured cells, as per the provided protocol. After that, complementary DNA (cDNA) synthesis was completed with the application of PrimeScript™ II Reverse Transcriptase (TaKaRa, Shiga, Japan), and reverse transcription quantitative polymerase chain reaction (RT-qPCR) procedure was achieved using SYBR Green PCR Kit (TaKaRa). Results of PCR were processed on the basis of 2−ΔΔCt method, standardized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) or U6. Primer sequences used in RT-qPCR are listed in Table 1. The assay included three bio-repeats.
Table 1.
Primer Sequence.
| NLRP3 | F:GAGGCTGGCATCTGGATGAG R:TGTGTCCTGAGCCATGGAAG |
| IL-1β | F:CAGAAGTACCTGAGCTCGCC R:AGATTCGTAGCTGGATGCCG |
| IL-18 | F:CGCAGATGGCTCTTTGCTTT R:ATGGTCCGGGGTGCATTATC |
| miR-223-3p | F:GCCGAGtgtcagtttgtcaaat R:CTCAACTGGTGTCGTGGA |
| ZNF561-AS1 | F:GGGGGATGCTCCTCCAAAG R:TCACGTCACTGCATTCGAGC |
| GAPDH | F: GCTCTCTGCTCCTCCTGTTC R: TGGAATTTGCCATGGGTGGA |
| U6 | F: TCCCTTCGGGGACATCCG R: AATTTTGGACCATTTCTCGATTTGT |
GAPDH: glyceraldehyde 3-phosphate dehydrogenase; IL: interleukin; NLRP3: NLR family pyrin domain containing 3.
Western Blot (WB) Assay
The radioimmunoprecipitation assay (RIPA) buffer was applied to achieve the protein lysates. Total protein was extracted by PROTTOT-1KT. And then the concentration of protein was measured by Bradford Protein Assay Kit. Afterward, the proteins were treated with 12% sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to the poly(vinylidene fluoride) (PVDF) membranes. Subsequently, membranes blocked by 5% skimmed milk were co-cultivated with specific primary antibodies from Abcam (Cambridge, MA, USA), including anti-GAPDH (ab8245), anti-NLRP3 (ab263899), anti-caspase 1 (ab207802), anti-caspase 11 (ab246496), anti-Bcl-2 (ab32124), anti-Bax (ab32503), anti-total caspase-3 (ab32351), and anti-cleaved caspase-3 (ab32042) overnight at 4°C. After that, secondary antibodies were added for cultivation. Eventually, the proteins were measured via enhanced chemiluminescence (ECL) detection system. Bio-repeats were run in triplicate. GAPDH was regarded as the internal control.
Cell Transfection
The NLRP3-specifc and ZNF561-AS1-specific short hairpin RNAs (shRNAs) were designed by Genechem (Shanghai, China), along with their NC-shRNAs. The pcDNA3.1 vector targeting NLRP3 and empty vector were also obtained from Genechem. Moreover, miR-223-3p mimics and miR-223-3p inhibitor were synthesized by RiboBio (Guangzhou, China), together with NC mimics and NC inhibitor. Forty-eight-hour cell transfection was performed by means of Lipofectamine 3000 (Invitrogen). The assay included three bio-repeats.
Cell Counting Kit-8 (CCK-8) Assay
Cells with stable transfection during the logarithmic growth were seeded onto 96-well plates and cultivated with 5% CO2 at 37°C for 24 h. Then, CCK-8 solution was added to each well and the absorbance value at 450 nm was measured to determine the cell viability. The assay included three bio-repeats.
EdU Staining Assay
In all, 1 × 104 transfected cells were collected and plated into 96-well plates. 5-Ethynyl-2’-deoxyuridine (EdU) assay kit (RiboBio) was incubated with cells for 2 h at 37°C, and then DAPI staining solution was added for 5 min at room temperature. Fluorescence microscope (Olympus, Tokyo, Japan) was used for the fluorescent observation. The assay included three bio-repeats.
Flow Cytometry Apoptosis
In all, 1 × 106 transfected cells were prepared in six-well plates for flow cytometry assay, based on the standard method of Annexin V-FITC/PI double staining kit (Invitrogen). After treatment in the dark for 15 min, the samples were finally examined by flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA). The assay included three bio-repeats.
TUNEL Staining Assay
In all, 1 × 104 transfected cells in 96-well plates were prepared for TdT-mediated dUTP Nick-End Labeling (TUNEL) staining assay. Cells were treated with TUNEL assay kit (Merck KGaA, Darmstadt, Germany) and then fixed in 4% paraformaldehyde. Following culture with 0.1% Triton-X100, cells were stained by DAPI and observed using fluorescence microscope (Olympus). The assay included three bio-repeats.
RNA Immunoprecipitation
Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit was acquired for RIP assay, based on the provided protocol (Millipore, Bedford, MA, USA). Human Ago2 antibody and normal IgG antibody were used. After lysing, cell extracts were subjected to RIP buffer with the antibody-conjugated magnetic beads. The final precipitates were examined after RNA isolation. The assay included three bio-repeats.
RNA Pull-Down
RNA pull-down assay was monitored using Pierce Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher Scientific, Waltham, MA, USA). The wild-type (WT) and mutant (MUT) binding sites of ZNF561-AS1 or NLRP3 in miR-223-3p sequences were acquired and biotinylated. Then, the obtained probes were incubated with cell lysis and magnetic beads. The recovered RNA-RNA mixture was exposed to RT-qPCR. The assay included three bio-repeats.
Fluorescence In Situ Hybridization (FISH)
The specific FISH probe to ZNF561-AS1 was commercially synthesized by Ribobio and employed as instructed. After fixing, the collected cells were cultivated with the probe in the hybridization buffer, followed by nuclear staining with DAPI. Fluorescent detection was performed with the help of fluorescence microscope (Olympus). The assay included three bio-repeats.
Subcellular Fractionation Assay
The separation of cell nucleus and cytoplasm was achieved with the application of PARIS™ Kit (Invitrogen), in light of the user guide. Cell fractionation buffer was added for separating the cell cytoplasm. Expression levels of ZNF561-AS1, U6, and GAPDH were examined after isolating the total RNA. The assay included three bio-repeats.
Luciferase Reporter Gene Assay
ZNF561-AS1 fragment or NLRP3 3′-UTR (untranslated region), which contained the WT and MUT binding sites of miR-223-3p, was inserted into pmirGLO vector for luciferase assay. pmirGLO-NLRP3 3′-UTR WT/MUT or pmirGLO-ZNF561-AS1 WT/MUT was co-transfected with miR-223-3p mimics or miR-223-3p inhibitor in cells using Lipofectamine 3000. After transfection for 48 h, their relative luciferase activity was assayed using the dual-luciferase reporter assay system (Promega, Madison, WI, USA). The assay included three bio-repeats.
Immunohistochemistry (IHC)
NLRP3 expression in infarct zone, border zone, and remote zone after MI was analyzed via IHC staining as previously described 21 . Briefly, subsequent to, tissue specimens were embedded in paraffin and cut into 4-μm sections, followed by incubation with primary antibody against NLRP3 overnight at 4°C. After washing with phosphate-buffered saline (PBS), the sections were incubated with secondary antibody at room temperature. Finally, the tissue sections were stained with DAB for immunolabeling and the cell nucleus was stained with hematoxylin, followed by observation under microscope.
Statistical Analyses
All experiments included three bio-repeats, and the experimental data were exhibited as means ± standard deviation (SD). The data in group difference were estimated by analysis of variance (ANOVA) or Student’s t test, with P < 0.05 as the threshold of statistical significance. All graphs in this study were generated with the application of GraphPad PRISM 6 (GraphPad, San Diego, CA, USA).
Results
NLRP3 Is Overexpressed in Mice MI Models and Hypoxia-Induced Myocardial Cells
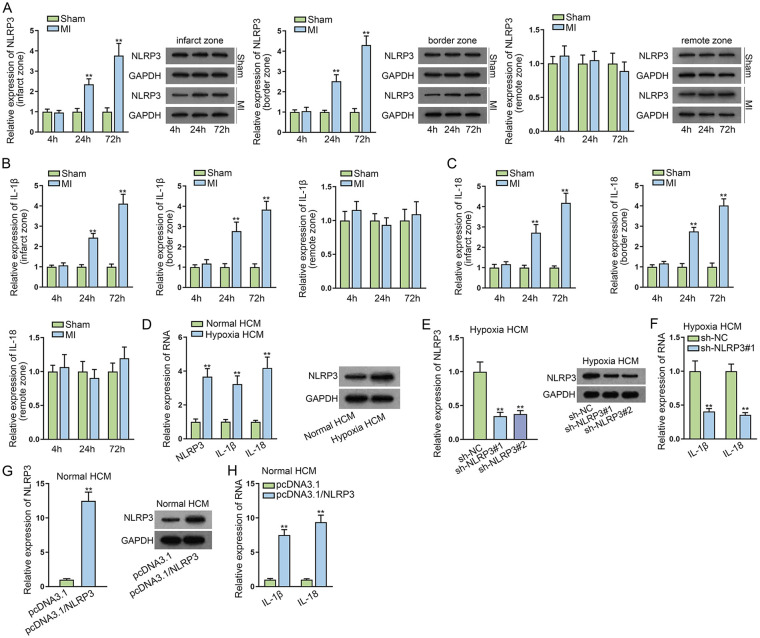
With the aim to confirm the successful establishment of MI model, H&E and 2,3,5-triphenyltetrazolium chloride (TTC) staining were first applied. As depicted in Supplementary Fig. 1A, the myocardial cells in the Sham group were well arranged and had normal nuclear structure, while the myocardial cells in the MI group were loosely connected and some were in disarray, with an overt increase in interstitial inflammatory exudation. Herein, it could be concluded that the MI model was successfully constructed. Subsequently, RT-qPCR, WB, and IHC were applied to detect the expression of NLRP3 in infarct zone, border zone, and remote zone after MI for 4, 24, and 72 h, respectively. In infarct zone and border zone, NLRP3 expression was significantly elevated in MI mice hearts at 24 and 72 h compared with that in sham-operated animals. Inversely, no change in NLRP3 expression was found in the remote zone of MI mice (Fig. 1A and Supplementary Fig. 2A). Consistently, the expression of IL-1β and IL-18 was much more upregulated in infarct zone and border zone of MI mice hearts at 24 and 72 h than that in sham-operated animals. The expression of IL-1β and IL-18 had no significant difference in the remote zone of MI mice (Fig. 1B, C). Furthermore, the expression of NLRP3, IL-1β, and IL-18 was apparently higher in hypoxia HCMs compared with the normal group, and NLRP3 protein level was also higher in hypoxia HCMs relative to normal HCMs (Fig. 1D). Then we knocked down NLRP3 in hypoxia HCMs, and the knockdown efficiency was determined by RT-qPCR and WB (Fig. 1E). Then, RT-qPCR assays evidenced that the expression of IL-1β and IL-18 was markedly lessened by NLRP3 silencing in hypoxia HCMs (Fig. 1F). On the contrary, we overexpressed NLRP3 in normal HCMs and found that the expression of IL-1β and IL-18 was obviously promoted in normal HCMs transfected with pcDNA3.1/NLRP3 (Fig. 1G, H). At the same time, the expression of caspase-1 (canonical NLRP3/caspase-1/IL-1β pathway) and caspase-11 (noncanonical capase-11 pathway) was also tested through WB in different groups. According to the result, the expression of caspase-1 notably increased in MI groups at 24 and 72 h in infarct zone and border zone compared with sham-operated groups, while no obvious change was observed in the expression of caspase-11 (Supplementary Fig. 2B). Also, the expression of caspase-1 was much more in hypoxia HCMs than in normal HCMs, and the expression of caspase-11 showed no variations (Supplementary Fig. 2C). Meanwhile, caspase-1 expression was decreased in hypoxia HCMs after NLRP3 knockdown and was increased in normal HCMs after NLRP3 augment, while the expression of caspase-11 showed no significant difference in different groups (Supplementary Fig. 2D, E). Together, NLRP3 was overexpressed in mice MI models and hypoxia-induced myocardial cells.
Figure 1.
NLRP3 is overexpressed in mice MI models and hypoxia-induced myocardial cells. (A) RT-qPCR and WB detected the NLRP3 level in infarct zone, border zone, and remote zone after MI for 4, 24, and 72 h, respectively. (B, C) RT-qPCR and WB measured IL-1β and IL-18 expression in infarct zone, border zone, and remote zone after MI for 4, 24, and 72 h, respectively. (D) The expression of NLRP3, IL-1β, and IL-18 was assessed in hypoxia HCMs compared with the normal group through RT-qPCR and WB. (E) The expression of NLRP3 was detected in hypoxia HCMs transfected with shRNAs targeting NLRP3 via RT-qPCR and WB. (F) RT-qPCR examined the expression of IL-1β and IL-18 after NLRP3 silencing in hypoxia HCMs. (G) RT-qPCR and WB evaluated NLRP3 level in normal HCMs transfected with pcDNA3.1/NLRP3. (H) The expression of IL-1β and IL-18 was examined in normal HCMs transfected with pcDNA3.1/NLRP3 using RT-qPCR. HCM: human cardiomyocytes; MI: myocardial infarction; NLRP3: NLR family pyrin domain containing 3; NLRP3: NLR family pyrin domain containing 3; RT-qPCR: reverse transcription quantitative polymerase chain rection; WB: western blot. **P < 0.01.
Silencing of NLRP3 Promotes Proliferation and Suppresses Apoptosis of Myocardial Cells
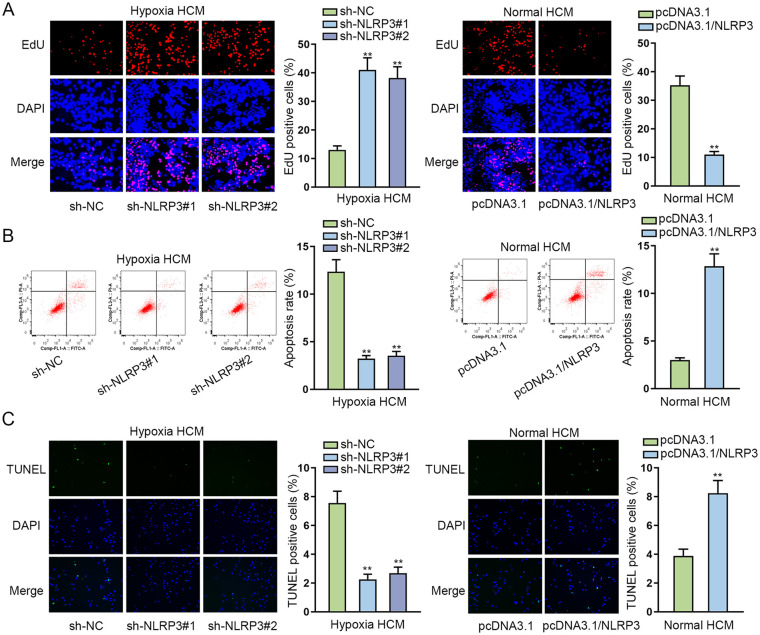
To investigate the role of NLRP3 in cardiomyocytes, cell function assays were conducted. CCK-8 assay was implemented for investigating how NLRP3 impacts cell viability, and it was suggested from the results that the decline in NLRP3 expression resulted in enhanced viability of HCMs under hypoxia condition, while an increment in NLRP3 level overtly inhibited the viability of HCMs under normal condition (Supplementary Fig. 3A). The results of EdU assays disclosed that silencing of NLRP3 increased the proportion of EdU-positive cells in hypoxia HCMs, while the overexpression of NLRP3 decreased the proportion of EdU-positive cells in normal HCMs (Fig. 2A). Besides, it was revealed in flow cytometry and TUNEL assays that the knockdown of NLRP3 reduced the apoptosis rate of hypoxia HCMs and upregulation of NLRP3 enhanced the apoptosis rate of normal HCMs (Fig. 2B, C). Furthermore, WB analysis was applied for the measurement of apoptosis-related genes, including Bcl-2, Bax, and cleaved caspase-3. From the results, NLRP3 silence led to augmentation of Bcl-2 and reduction of Bax and cleaved caspase-3 in hypoxia HCM, while NLRP3 overexpression led to the opposite consequences in HCM under normal condition, reflecting that NLRP3 downregulation weakened the apoptotic ability of hypoxia HCMs, while NLRP3 upregulation strengthened the apoptosis of HCMs under normal condition (Supplementary Fig. 3B). Totally, silencing of NLRP3 promoted the proliferation and suppressed apoptosis of myocardial cells.
Figure 2.
Silencing of NLRP3 promotes proliferation and suppresses apoptosis of hypoxia-induced myocardial cells. (A) EdU assays testified the proliferation ability of hypoxia HCMs transfected with shRNAs targeting NLRP3 or of normal HCMs transfected with pcDNA3.1/NLRP3. (B, C) Flow cytometry and TUNEL assays examined the apoptosis rate of hypoxia HCMs transfected with shRNAs targeting NLRP3 or of normal HCMs transfected with pcDNA3.1/NLRP3. HCM: human cardiomyocytes; NLRP3: NLR family pyrin domain containing 3; shRNA: short hairpin RNA; DAPI: 4’,6-diamidino-2-phenylindole. **P < 0.01.
NLRP3 Is Directly Targeted by miR-223-3p
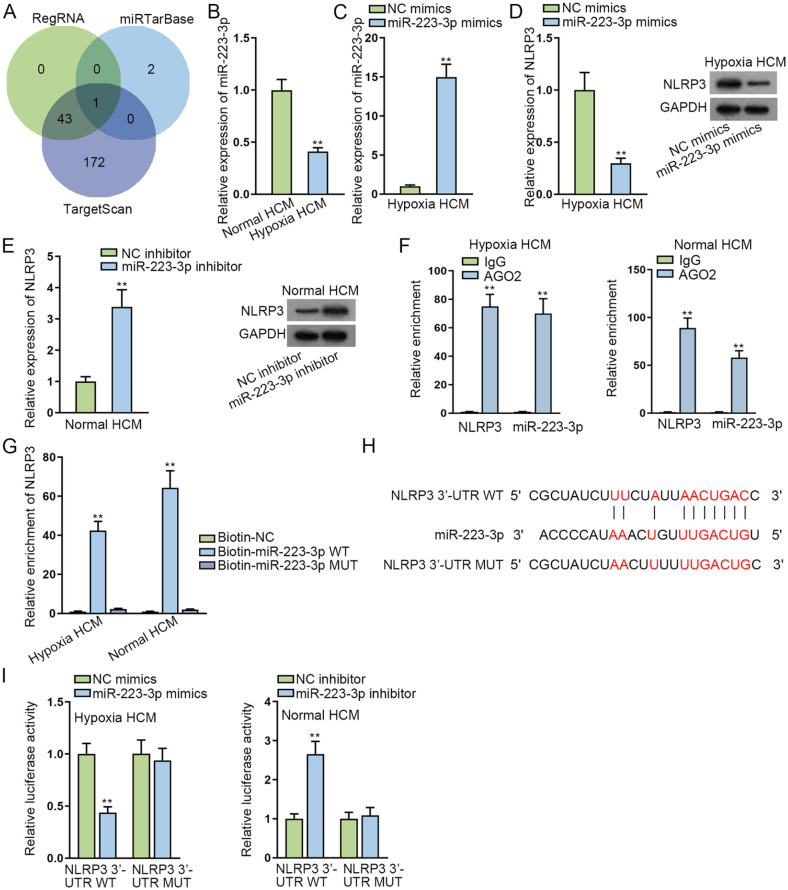
Through the analysis of bioinformatics programs including RegRNA, miRTarBase, and TargetScan, we discovered that miR-223-3p was the potential miRNA combining with NLRP3 (Fig. 3A). It was indicated from RT-qPCR analysis that in infarct zone and border zone, miR-223-3p expression significantly declined in MI mice hearts at 24 and 72 h compared with that in sham-operated animals. Inversely, miR-223-3p expression hardly changed with time in the remote zone of MI mice (Supplementary Fig. 3C). Subsequently, RT-qPCR examined that miR-223-3p had low expression in hypoxia HCMs (Fig. 3B). Then we elevated miR-223-3p expression in hypoxia HCMs while reducing miR-223-3p expression in normal HCMs. The overexpression efficiency was tested via RT-qPCR (Fig. 3C). The results from Fig. 3D, E demonstrated that the expression of NLRP3 was lessened by upregulated miR-223-3p in hypoxia HCMs and was promoted by miR-223-3p inhibitor in normal HCMs. Furthermore, in Ago2-RIP assay, large enrichment of NLRP3 and miR-223-3p was discovered in Ago2 groups (Fig. 3F). RNA pull-down experiments showed that NLRP3 was only largely enriched in Biotin-miR-223-3p WT groups (Fig. 3G). In addition, we predicted a binding site between NLRP3 and miR-223-3p using starBase (http://starbase.sysu.edu.cn) website (Fig. 3H). Luciferase reporter experiments further confirmed that the luciferase activity remarkably declined in hypoxia HCMs co-transfected with miR-223-3p mimics and NLRP3 3′-UTR WT, while it increased in normal HCMs co-transfected with miR-223-3p inhibitor and NLRP3 3′-UTR WT (Fig. 3I). Totally, NLRP3 was directly targeted by miR-223-3p.
Figure 3.
NLRP3 is directly targeted by miR-223-3p. (A) Bioinformatics programs including RegRNA, miRTarBase, and TargetScan predicted miR-223-3p was the potential miRNA combining with NLRP3. (B) RT-qPCR examined miR-223-3p expression in normal and hypoxia HCMs. (C) MiR-223-3p expression was detected in hypoxia HCMs transfected with miR-223-3p mimics. (D, E) RT-qPCR and WB examined NLRP3 expression after miR-223-3p upregulation in hypoxia HCMs or after miR-223-3p inhibition in normal HCMs. (F) Ago2-RIP assay indicated the enrichment of NLRP3 and miR-223-3p in Ago2 groups. (G) RNA pull-down experiments showed the enrichment of NLRP3 in Biotin-miR-223-3p WT groups. (H) StarBase predicted the binding site between NLRP3 and miR-223-3p. (I) The affinity between NLRP3 and miR-223-3p was further investigated via luciferase reporter experiments. HCM: human cardiomyocytes; miRNA: microRNA; NLRP3: NLR family pyrin domain containing 3; RT-qPCR: reverse transcription polymerase chain reaction; WT: wild type. **P < 0.01.
MiR-223-3p is the Downstream Gene of ZNF561-AS1
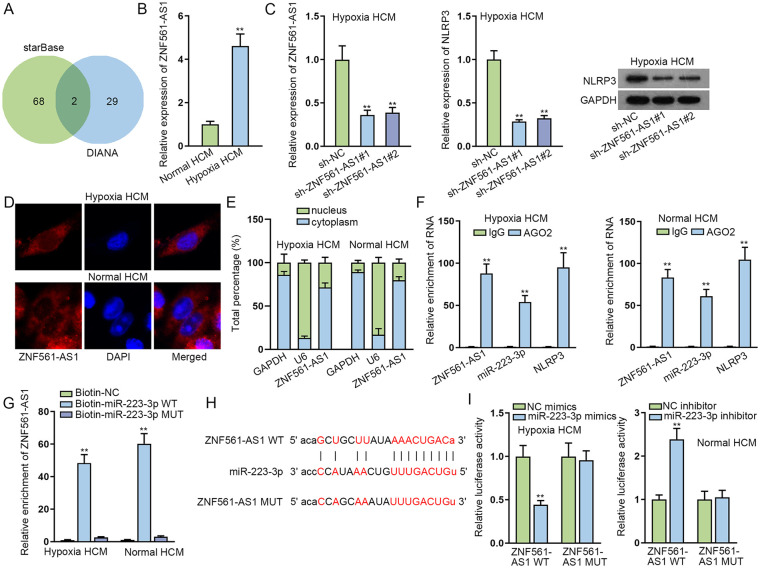
We applied starBase and DIANA (http://diana.imis.athena-innovation.gr/DianaTools/index.php) to screen out two lncRNAs (KCNQ1OT1 and ZNF561-AS1) that combined with miR-223-3p (Fig. 4A). As KCNQ1OT1 has been reported to act as a regulator in MI 22 , we selected ZNF561-AS1 for subsequent experiments. Based on RT-qPCR analysis, ZNF561-AS1 expression in infarct zone and border zone was significantly elevated in MI mice hearts at 24 and 72 h compared with that in sham-operated animals, while no obvious changes were discovered in ZNF561-AS1 expression in the remote zone (Supplementary Fig. 3D). RT-qPCR results also indicated that ZNF561-AS1 was overexpressed in hypoxia HCMs relative to normal HCMs (Fig. 4B). Then we silenced the expression of ZNF561-AS1 in hypoxia HCMs and found that NLRP3 expression also declined by ZNF561-AS1 reduction in hypoxia HCMs (Fig. 4C). Furthermore, nuclear separation and FISH assays showed that ZNF561-AS1 mostly existed in the cytoplasm (Fig. 4D, E). The results of Ago2-RIP assays manifested that ZNF561-AS1, miR-223-3p, and NLRP3 were definitely enriched in Ago2 groups, suggesting these three RNAs co-existed in the RNA-induced silencing complex (RISC) (Fig. 4F). RNA pull-down experiments displayed large enrichment of ZNF561-AS1 only in Bio-miR-223-3p WT groups (Fig. 4G). In addition, we predicted a binding site between ZNF561-AS1 and miR-223-3p using starBase (Fig. 4H). The luciferase activity was lessened in hypoxia HCMs co-transfected with miR-223-3p mimics and ZNF561-AS1 WT, while it increased in normal HCMs co-transfected with miR-223-3p inhibitor and ZNF561-AS1 WT (Fig. 4I). All these results evidenced that miR-223-3p was the downstream gene of ZNF561-AS1.
Figure 4.
MiR-223-3p is the downstream gene of ZNF561-AS1. (A) StarBase and DIANA screened out two lncRNAs (KCNQ1OT1 and ZNF561-AS1) that combined with miR-223-3p. (B) RT-qPCR analysis indicated ZNF561-AS1 expression was overtly higher in hypoxia HCMs. (C) RT-qPCR detected the expression of ZNF561-AS1 in hypoxia HCMs transfected with shRNAs targeting ZNF561-AS1 and NLRP3 expression was tested via RT-qPCR along with WB in hypoxia HCMs after ZNF561-AS1 was inhibited. (D, E) FISH and nuclear separation assays showed the location of ZNF561-AS1. (F) Ago2-RIP assays manifested the enrichment of ZNF561-AS1, miR-223-3p, and NLRP3 in Ago2 groups. (G) RNA pull-down experiments displayed the enrichment of ZNF561-AS1 in Bio-miR-223-3p WT groups. (H) The binding site between ZNF561-AS1 and miR-223-3p was observed using starBase. (I) Luciferase reporter assays were implemented for further validation of the affinity between miR-223-3p and ZNF561-AS1. FISH: fluorescence in situ hybridization; HCM: human cardiomyocytes; lncRNA: long non-coding RNAs; NLRP3: NLR family pyrin domain containing 3; RNA: ribonucleic acid; WB: Western blot; WT: wild type. **P < 0.01.
ZNF561-AS1 Regulates MI through miR-223-3p/NLRP3 Axis
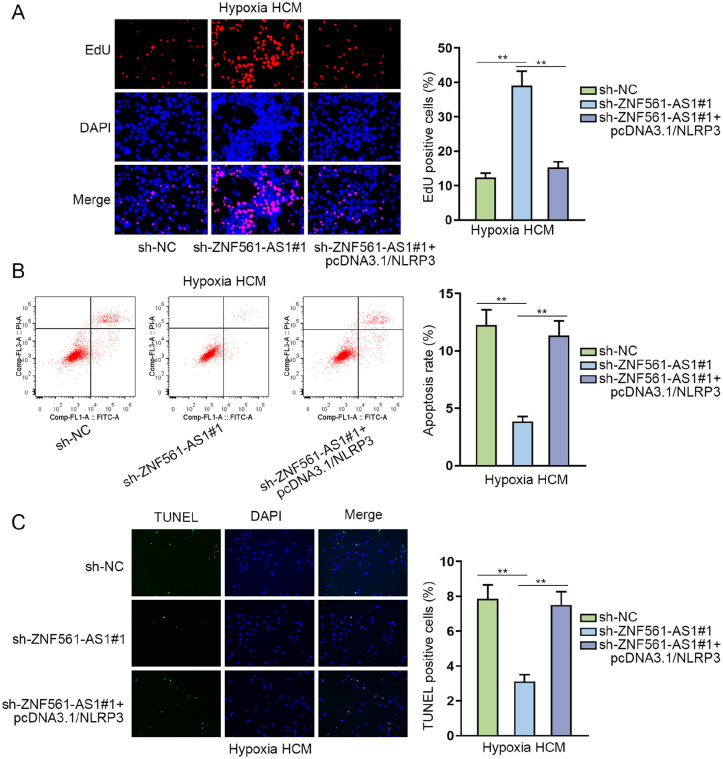
To further verify the interaction between ZNF561-AS1 and NLRP3 in cardiomyocytes, we performed rescue experiments. According to the results of CCK-8 assays, the enhanced viability of hypoxia HCMs, resulting from ZNF561-AS1 depletion, was recovered in response to NLRP3 upregulation (Supplementary Fig. 3E). As shown in Fig. 5A, it was revealed in EdU assay that silenced ZNF561-AS1 promoted cell proliferation of hypoxia HCMs, while NLRP3 upregulation reversed this status. Moreover, the results from flow cytometry and TUNEL assays showed that the apoptosis rate was inhibited by ZNF561-AS1 knockdown and then enhanced by NLRP3 overexpression (Fig. 5B, C). Meanwhile, the expression of cleaved caspase-3, Bcl-2, and Bax in hypoxia HCMs was also examined through WB. The result indicated that the transfection of sh-ZNF561-AS1 led to a decline in Bax and cleaved caspase-3 expression, which was rescued as a result of pcDNA3.1/NLRP3 transfection, while the augment in Bcl-2 level caused by ZNF561-AS1 silence was reversed in response to NLRP3 upregulation (Supplementary Fig. 3F). Totally, ZNF561-AS1 regulated cell proliferation and apoptosis in MI through miR-223-3p/NLRP3 axis.
Figure 5.
ZNF561-AS1 regulates hypoxia HCM proliferation and apoptosis through miR-223-3p/NLRP3 axis. (A) EdU assay examined cell proliferation of hypoxia HCMs transfected with sh-ZNF561-AS1#1 or sh-ZNF561-AS1#1+pcDNA3.1/NLRP3. (B, C) Flow cytometry and TUNEL assays detected the apoptosis rate of hypoxia HCMs transfected with sh-ZNF561-AS1#1 or sh-ZNF561-AS1#1+pcDNA3.1/NLRP3. HCM: human cardiomyocytes; NLRP3: NLR family pyrin domain containing 3; DAPI: 4’,6-diamidino-2-phenylindole. **P < 0.01.
Discussion
Accumulating studies have shown the important roles of lncRNAs in the progression of cardiovascular diseases. LncRNA CAIF-p53-myocardin axis reduces autophagy and MI 23 . Knockdown of lncRNA KCNQ1OT1 protects against myocardial ischemia/reperfusion injury following acute MI through regulating AdipoR1 and p38 MAPK/NF-κB signal pathway 24 . Downregulation of lncRNA MALAT1 negatively modulates acute MI via miR-320/Pten axis 25 . In our study, we focused on investigating the role of a novel lncRNA ZNF561-AS1 in regulating the activities of hypoxia-induced myocardial cells.
It has been claimed that IL-1β and IL-18 are inflammatory cytokines implicated in the activation of NLRP3 inflammasome in MI26,27. Accordingly, our research examined that the expression of NLRP3, IL-1β, and IL-18 was significantly increased in infarct and border zones of MI mice heart at 24 and 72 h compared with that in sham-operated models. Then we performed cell function assays and confirmed that the knockdown of NLRP3 facilitated cell proliferation and inhibited apoptosis of hypoxia-induced myocardial cells. Thus, we speculated whether NLRP3 was regulated by lncRNA in hypoxia-induced myocardial cells.
It is well known that lncRNAs could modulate mRNA expression through functioning as competing endogenous RNAs (ceRNAs) by sequestering miRNA. As reported previously, miRNAs serve as biomarkers which are implicated in the cardiovascular diseases. FGF21 protects myocardial ischemia-reperfusion injury through regulating miR-145 to inhibit Angpt2 expression 28 . Upregulated miRNA-221 negatively affects hypoxia/reoxygenation in association with autophagy via the DDIT4/mTORC1 and Tp53inp1/p62 pathways 29 . MiR-128/SOX7 regulates IL-33/sST2 signaling pathway to alleviate myocardial ischemia injury in acute MI 30 . In our study, we applied several bioinformatics programs and found that miR-223-3p combined with NLRP3. Consistently, miR-223-3p has once been reported to participate in the development of cardiovascular diseases. Specifically, miR-223 inhibition affects cardiac functional deterioration and cardiac fibrosis through regulating RASA1 19 . MiR-223-3p/KLF15 improves hypoxia-induced injury through inhibiting cardiomyocyte apoptosis and oxidative stress 31 . In addition, we further performed luciferase reporter assays to confirm that NLRP3 was the target gene of miR-223-3p. Therefore, we continued to investigate the upstream gene of miR-223-3p.
According to the above analyses, based on bioinformatics prediction, ZNF561-AS1 was screened out as the underlying lncRNA that combined with miR-223-3p. And the expression of ZNF561-AS1 was notably upregulated in hypoxia-induced myocardial cell. Furthermore, we also found a binding site between ZNF561-AS1 and miR-223-3p. More importantly, we conducted a series of rescue experiments to further validate ZNF561-AS1 inhibited cell proliferation and promoted apoptosis of hypoxia-induced myocardial cells through regulating NLRP3 expression.
In conclusion, our study investigated the mechanism about ZNF561-AS1/miR-223-3p/NLRP3 axis in the research of cell biological behaviors in MI. Our research might provide a novel molecular target to prevent myocardial cells from hypoxia-induced apoptosis.
Supplemental Material
Supplemental material, sj-tif-1-cll-10.1177_09636897221077928 for ZNF561-AS1 Regulates Cell Proliferation and Apoptosis in Myocardial Infarction Through miR-223-3p/NLRP3 Axis by Xiaoyu Li, Jun Long, Ligeng Zong, Chengcheng Zhang, Zhongxin Yang and Shengnan Guo in Cell Transplantation
Supplemental material, sj-tif-2-cll-10.1177_09636897221077928 for ZNF561-AS1 Regulates Cell Proliferation and Apoptosis in Myocardial Infarction Through miR-223-3p/NLRP3 Axis by Xiaoyu Li, Jun Long, Ligeng Zong, Chengcheng Zhang, Zhongxin Yang and Shengnan Guo in Cell Transplantation
Supplemental material, sj-tif-3-cll-10.1177_09636897221077928 for ZNF561-AS1 Regulates Cell Proliferation and Apoptosis in Myocardial Infarction Through miR-223-3p/NLRP3 Axis by Xiaoyu Li, Jun Long, Ligeng Zong, Chengcheng Zhang, Zhongxin Yang and Shengnan Guo in Cell Transplantation
Acknowledgments
Thanks a lot for all supports from our laboratory.
Footnotes
Ethical Approval and Consent to Participate: This study was approved by the ethic committee of the First Affiliated Hospital of Henan University.
Statement of Human and Animal Rights: All animal-related procedures were conducted in line with the Institutional Animal Care and Use Committee of the First Affiliated Hospital of Henan University and the Ethics Committee of the First Affiliated Hospital of Henan University.
Statement of Informed Consent: There are no human subjects in this study, and informed consent is not applicable.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by The role and mechanism of microRNA-15a in myocardial angiogenesis after ischemia (202102310376); Study on the value of microRNA-15a in the prevention and treatment of ischemic cardiomyopathy (2018020305); Interaction mechanism of IRT1/NF-κB in ginsenoside RB1 against vascular endothelial senescence (2018020307); Protective effect of Irisin/PPARA on NF-κB-mediated cell pyrotosis in atherosclerosis (LHGJ20190501); Study on the value of resveratrol and Notch-PINL signaling pathway in the prevention and treatment of atherosclerosis (1903044); Study on the effect and mechanism of siRT1 mercapto modification mediated autophagy in the anti-aging effect of hydrogen sulfide on endothelial cells (2003025).
ORCID iD: Jun Long  https://orcid.org/0000-0002-7930-7085
https://orcid.org/0000-0002-7930-7085
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Wang Y, Zhang H, Chai F, Liu X, Berk M. The effects of escitalopram on myocardial apoptosis and the expression of Bax and Bcl-2 during myocardial ischemia/reperfusion in a model of rats with depression. BMC Psychiatry. 2014;14:349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kajstura J, Cheng W, Reiss K, Clark WA, Sonnenblick EH, Krajewski S, Reed JC, Olivetti G, Anversa P. Apoptotic and necrotic myocyte cell deaths are independent contributing variables of infarct size in rats. Lab Invest. 1996;74(1):86–107. [PubMed] [Google Scholar]
- 3. Thygesen K, Alpert JS, Jaffe AS, Chaitman BR, Bax JJ, Morrow DA, White HD. Fourth universal definition of myocardial infarction (2018). Eur Heart J. 2019;40(3):237–69. [DOI] [PubMed] [Google Scholar]
- 4. Busch A, Eken SM, Maegdefessel L. Prospective and therapeutic screening value of non-coding RNA as biomarkers in cardiovascular disease. Ann Transl Med. 2016;4(12):236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yan Y, Zhang B, Liu N, Qi C, Xiao Y, Tian X, Li T, Liu B. Circulating long noncoding RNA UCA1 as a novel biomarker of acute myocardial infarction. Biomed Res Int. 2016;2016: 8079372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Toldo S, Abbate A. The NLRP3 inflammasome in acute myocardial infarction. Nat Rev Cardiol. 2018;15(4):203–14. [DOI] [PubMed] [Google Scholar]
- 7. Takahashi M. NLRP3 in myocardial ischaemia-reperfusion injury: inflammasome-dependent or -independent role in different cell types. Cardiovasc Res. 2013;99(1):4–5. [DOI] [PubMed] [Google Scholar]
- 8. Takahashi M. NLRP3 inflammasome as a novel player in myocardial infarction. Int Heart J. 2014;55(2):101–5. [DOI] [PubMed] [Google Scholar]
- 9. Sandanger O, Gao E, Ranheim T, Bliksoen M, Kaasboll OJ, Alfsnes K, Nymo SH, Rashidi A, Ohm IK, Attramadal H, Aukrust P, et al. NLRP3 inflammasome activation during myocardial ischemia reperfusion is cardioprotective. Biochem Biophys Res Commun. 2016;469(4):1012–20. [DOI] [PubMed] [Google Scholar]
- 10. van Hout GP, Bosch L, Ellenbroek GH, de Haan JJ, van Solinge WW, Cooper MA, Arslan F, de Jager SC, Robertson AA, Pasterkamp G, Hoefer IE. The selective NLRP3-inflammasome inhibitor MCC950 reduces infarct size and preserves cardiac function in a pig model of myocardial infarction. Eur Heart J. 2017;38(11):828–36. [DOI] [PubMed] [Google Scholar]
- 11. Yin XF, Zhang Q, Chen ZY, Wang HF, Li X, Wang HX, Li HX, Kang CM, Chu S, Li KF, Li Y, et al. NLRP3 in human glioma is correlated with increased WHO grade, and regulates cellular proliferation, apoptosis and metastasis via epithelial-mesenchymal transition and the PTEN/AKT signaling pathway. Int J Oncol. 2018;53(3):973–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang H, Luo Q, Feng X, Zhang R, Li J, Chen F. NLRP3 promotes tumor growth and metastasis in human oral squamous cell carcinoma. BMC Cancer. 2018;18(1):500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Boon RA, Dimmeler S. MicroRNAs in myocardial infarction. Nat Rev Cardiol. 2015;12(3):135–42. [DOI] [PubMed] [Google Scholar]
- 14. Zhou SS, Jin JP, Wang JQ, Zhang ZG, Freedman JH, Zheng Y, Cai L. miRNAS in cardiovascular diseases: potential biomarkers, therapeutic targets and challenges. Acta Pharmacol Sin. 2018;39(7):1073–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang C, Jing Q. Non-coding RNAs as biomarkers for acute myocardial infarction. Acta Pharmacol Sin. 2018;39(7):1110–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang K, Liu CY, Zhou LY, Wang JX, Wang M, Zhao B, Zhao WK, Xu SJ, Fan LH, Zhang XJ, Feng C, et al. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat Commun. 2015;6:6779. [DOI] [PubMed] [Google Scholar]
- 17. Zhang J, Wu L, Li Z, Fu G. miR-1231 exacerbates arrhythmia by targeting calciumchannel gene CACNA2D2 in myocardial infarction. Am J Transl Res. 2017;9(4):1822–33. [PMC free article] [PubMed] [Google Scholar]
- 18. Wang PP, Zhang YJ, Xie T, Sun J, Wang XD. MiR-223 promotes cardiomyocyte apoptosis by inhibiting Foxo3a expression. Eur Rev Med Pharmacol Sci. 2018;22(18):6119–26. [DOI] [PubMed] [Google Scholar]
- 19. Liu X, Xu Y, Deng Y, Li H. MicroRNA-223 Regulates Cardiac Fibrosis After Myocardial Infarction by Targeting RASA1. Cell Physiol Biochem. 2018;46(4):1439–54. [DOI] [PubMed] [Google Scholar]
- 20. Dong FF, Dong SH, Liang Y, Wang K, Qin YW, Zhao XX. MiR-34a promotes myocardial infarction in rats by inhibiting the activity of SIRT1. Eur Rev Med Pharmacol Sci. 2019;23(16):7059–65. [DOI] [PubMed] [Google Scholar]
- 21. Luo Q, Zhang S, Wei H, Pang X, Zhang H. Roles of Foxp3 in the occurrence and development of cervical cancer. Int J Clin Exp Pathol. 2015;8(8):8717–30. [PMC free article] [PubMed] [Google Scholar]
- 22. Smillie CL, Sirey T, Ponting CP. Complexities of post-transcriptional regulation and the modeling of ceRNA crosstalk. Crit Rev Biochem Mol Biol. 2018;53(3):231–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu CY, Zhang YH, Li RB, Zhou LY, An T, Zhang RC, Zhai M, Huang Y, Yan KW, Dong YH, Ponnusamy M, et al. LncRNA CAIF inhibits autophagy and attenuates myocardial infarction by blocking p53-mediated myocardin transcription. Nat Commun. 2018;9(1):29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li X, Dai Y, Yan S, Shi Y, Han B, Li J, Cha L, Mu J. Down-regulation of lncRNA KCNQ1OT1 protects against myocardial ischemia/reperfusion injury following acute myocardial infarction. Biochem Biophys Res Commun. 2017;491(4):1026–33. [DOI] [PubMed] [Google Scholar]
- 25. Hu H, Wu J, Li D, Zhou J, Yu H, Ma L. Knockdown of lncRNA MALAT1 attenuates acute myocardial infarction through miR-320-Pten axis. Biomed Pharmacother. 2018;106:738–46. [DOI] [PubMed] [Google Scholar]
- 26. Yang TC, Chang PY, Lu SC. L5-LDL from ST-elevation myocardial infarction patients induces IL-1beta production via LOX-1 and NLRP3 inflammasome activation in macrophages. Am J Physiol Heart Circ Physiol. 2017;312(2):H265–74. [DOI] [PubMed] [Google Scholar]
- 27. Sandanger O, Ranheim T, Vinge LE, Bliksoen M, Alfsnes K, Finsen AV, Dahl CP, Askevold ET, Florholmen G, Christensen G, Fitzgerald KA, et al. The NLRP3 inflammasome is up-regulated in cardiac fibroblasts and mediates myocardial ischaemia-reperfusion injury. Cardiovasc Res. 2013;99(1):164–74. [DOI] [PubMed] [Google Scholar]
- 28. Yuan Y, Sun S, Jiao N, Shu Y, Zhang Y. Upregulation of HOXA10 protein expression predicts poor prognosis for colorectal cancer. Genet Test Mol Biomarkers. 2018;22(6):390–97. [DOI] [PubMed] [Google Scholar]
- 29. Chen Q, Zhou Y, Richards AM, Wang P. Up-regulation of miRNA-221 inhibits hypoxia/reoxygenation-induced autophagy through the DDIT4/mTORC1 and Tp53inp1/p62 pathways. Biochem Biophys Res Commun. 2016;474(1):168–74. [DOI] [PubMed] [Google Scholar]
- 30. Yang J, Hu F, Fu X, Jiang Z, Zhang W, Chen K. MiR-128/SOX7 alleviates myocardial ischemia injury by regulating IL-33/sST2 in acute myocardial infarction. Biol Chem. 2019;400(4):533–44. [DOI] [PubMed] [Google Scholar]
- 31. Tang Q, Li MY, Su YF, Fu J, Zou ZY, Wang Y, Li SN. Absence of miR-223-3p ameliorates hypoxia-induced injury through repressing cardiomyocyte apoptosis and oxidative stress by targeting KLF15. Eur J Pharmacol. 2018;841:67–74. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-tif-1-cll-10.1177_09636897221077928 for ZNF561-AS1 Regulates Cell Proliferation and Apoptosis in Myocardial Infarction Through miR-223-3p/NLRP3 Axis by Xiaoyu Li, Jun Long, Ligeng Zong, Chengcheng Zhang, Zhongxin Yang and Shengnan Guo in Cell Transplantation
Supplemental material, sj-tif-2-cll-10.1177_09636897221077928 for ZNF561-AS1 Regulates Cell Proliferation and Apoptosis in Myocardial Infarction Through miR-223-3p/NLRP3 Axis by Xiaoyu Li, Jun Long, Ligeng Zong, Chengcheng Zhang, Zhongxin Yang and Shengnan Guo in Cell Transplantation
Supplemental material, sj-tif-3-cll-10.1177_09636897221077928 for ZNF561-AS1 Regulates Cell Proliferation and Apoptosis in Myocardial Infarction Through miR-223-3p/NLRP3 Axis by Xiaoyu Li, Jun Long, Ligeng Zong, Chengcheng Zhang, Zhongxin Yang and Shengnan Guo in Cell Transplantation