Abstract
Objective
To evaluate pathological changes in cartilage and subchondral bone MRI biomarkers in a rabbit model of osteoarthritis (OA) and correlate these with histological variations.
Design
Transection of the anterior cruciate ligament was performed on the right knee of eighteen 12-week-old New Zealand white rabbits to induce OA. 3-Tesla MR images were obtained from 18 healthy control knees (left) and 18 knees with OA (right). Imaging biomarkers included volume, thickness, T1 and T2* cartilage parametric maps, and several subchondral bone features: bone volume to total volume ratio, trabecular thickness, trabecular spacing, trabecular number (TbN), 2D and 3D fractal dimensions, and quality of trabecular score (QTS). Microscopic analysis of the lateral femoral condyles was set as the ground truth.
Results
When healthy and osteoarthritic knees were compared, significant differences were seen in the T1 and T2* values of the femur and tibia cartilage and in the subchondral bone volume to total volume, TbN, and QTS of both the lateral and medial aspects of the femur and tibia. Histological findings revealed significant osteoarthritic changes between healthy and osteoarthritic knees in stain, structure, chondrocyte density, total score, and subchondral bone biomarker levels. A positive correlation was found between histological staining, structure, chondrocyte density, and total score variables in T1 and T2* cartilage biomarkers. A negative correlation was observed between histological subchondral bone variables and magnetic resonance D2D and QTS biomarkers.
Conclusion
Quantification of several cartilage and subchondral bone imaging biomarkers in a rabbit model of OA allows the detection of significant changes, which are correlated with histological findings.
Keywords: 3T MR, osteoarthritis, diagnosis, biomarkers, diagnostics, cartilage, subchondral bone
Introduction
Osteoarthritis (OA) is one of the most common diseases affecting the synovial joints. When chronic, this multifactorial degenerative disorder results in cartilage degeneration, subchondral bone exposure, and periarticular soft tissue changes. 1
Traditionally, radiography has been used in clinical practice to detect OA. However, plain films only allow visualization of bone structures and do not show a good correlation with the clinical signs. 2 MRI is increasingly being used in joint evaluation due to its high sensitivity to cartilage and periarticular soft tissue changes. 3
Different imaging biomarkers play an important role in the early detection of OA, especially in quantifying parameters. Namely, cartilage T1 relaxation time is used to assess proteoglycan content,4-6 and T2* relaxation time correlates with water content and collagen microstructure. 7 Increased cartilage T2* relaxation times have been shown to be a risk factor for OA development. 8 Furthermore, subchondral bone imaging biomarkers, including bone volume to total volume fraction (BV/TV), trabecular thickness (TbTh), and trabecular spacing (TbSp), provide relevant information about the histological trabecular bone structure. These biomarkers can be quantified using microcomputed tomography (microCT) 9 and MR. 10
Animal experiments were performed using either spontaneous or induced OA models. Induced small mammal models have been shown to provide significant advantages in terms of reproducibility, easy handling, and quick development of the pathology. 11 The rabbit anterior cruciate ligament transection (ACLT) model has been widely used to rapidly develop OA by causing joint destabilization. 12 Despite the biomechanical differences in joints and gait, 13 this model can predict the cartilage of the joint surface, subchondral bone, periarticular soft tissues, and osteophyte formation.
In experimental models, histological analysis is the standard method for evaluating cartilage and subchondral bone structures. A broad number of biomarkers have been registered, and overtime standardization of these biomarkers has been performed in different species.14-17 The Osteoarthritis Research Society International (OARSI) defines 5 basic principles when using grading scales for OA: simplicity, utility, scalability, extendibility, and comparability. 18
The challenge in recent years has been to establish a relationship between standardized histological biomarkers and new imaging biomarkers. This would allow OA evaluation of the cartilage and subchondral bone at different time periods using imaging biomarkers. Currently, few studies have taken this step, such as the use of computed tomography (CT) scans to evaluate subchondral bone 19 and MRI biomarkers to evaluate articular cartilage. 20
Our aim was to evaluate changes in cartilage and subchondral bone MR biomarkers of OA using an experimental rabbit model and to correlate these with histological variations.
Methods
Animal Model
Eighteen 12-week-old female New Zealand white rabbits (Oryctolagus cuniculus) underwent unilateral right ACLT under general anesthesia. The left knees (n = 18) were used as the healthy control group, while the operated right knees constituted the OA group (n = 18). MR examinations were performed 84 days after surgery. Immediately after MR, animals were euthanized, and the femoral condyles were harvested for histological study. All procedures were performed according to European legislation on the protection of animals and with the approval of the Local Government Animal Protection Ethics Committee (RD53/2013).
MRI
MR images were obtained using a 3T clinical scanner (Philips Achieva 3.0 TX, Amsterdam, The Netherlands) with a 16-channel coil (KNEE 16 COIL) ( Fig. 1A ).
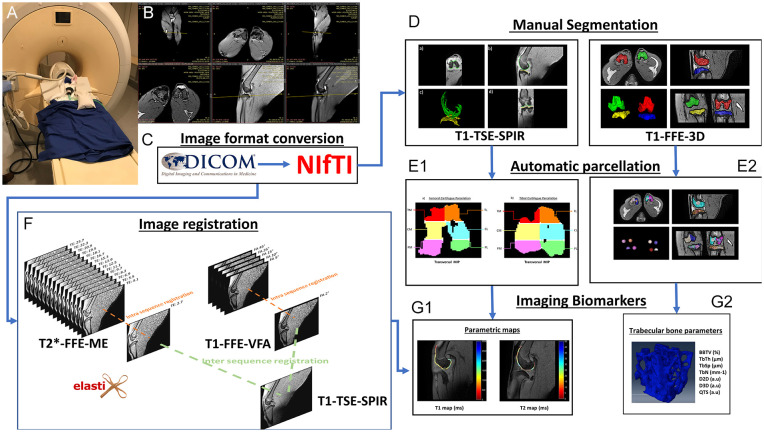
Figure 1.
The pipeline followed for the acquisition and analysis of MR images. (A) Placement of the rabbit in the knee coil and the MR equipment. (B) Acquisition planes used for different sequences. (C) Change from DICOM to NIfTI format. (D) Manual segmentation performed to delineate the cartilage and femoral and tibial condyles. (E1) Automatic cartilage parcellation on an MIP on the transverse plane. (E2) Automatic parcellation of the condyles and delimitation of the bone analysis volume. (F) Intra-sequence registration process for TEs and different flip angles, and their subsequent registration to the T1-TSE-SPIR sequence space. (G1) and (G2) Extracted image biomarkers for cartilage and trabecular bone. MR = magnetic resonance; TSE = turbo spin echo; FFE = fast field echo; VFA = variable flip angle; BV/TV = bone volume to total volume fraction; TbTh = trabecular thickness; TbSp = trabecular spacing; QTS = quality of trabecular score; NIfTI = Neuroimaging Informatics Technology Initiative; MIP = maximum intensity projection; TbN = trabecular number; D2D = two dimensional fractal dimension measurements; D3D = three dimensional fractal dimension measurements; DICOM = digital imaging and communication in medicine; ME = multi echo; SPIR = spectral presaturation with inversion recovery.
Cartilage imaging was performed with 3 different sagittal sequences: a high-resolution turbo spin echo T1-weighted sequence with fat suppression (T1-TSE-SPIR) (TE = 9 ms and TR = 1,105 ms, SENSE factor = 1.9, acquisition matrix = 432 × 432 × 50, voxel size = 0.27 × 0.27 × 0.5 and 5 min 54 s duration); a fast field echo (FFE) T2*-weighted sequence with 16 echoes (T2*-FFE-ME) (TE1 = 2.7, ΔTE = 1.4, TR = 39 ms, SENSE factor = 1.4, flip angle = 25°, acquisition matrix = 512 × 512 × 50, voxel size = 0.23 × 0.23 × 0.5 and 5 min 51 s duration); and an FFE T1-weighted variable flip angle sequence (T1-FFE-VFA) (TE = 4.6 ms, TR = 14 ms, SENSE factor = 2, 6 flip angles = 2°-5°-10°-15°-25°-45°, acquisition matrix = 512 × 512 × 50, voxel size = 0.23 × 0.23 × 0.5 and 2 min 6 s duration).
Subchondral bone imaging was performed with a 3D high-resolution T1-weighted balanced fast field echo (T1-FFE-3D) sequence acquired on the transversal plane (TE = 3.5 ms, TR = 16 ms, flip angle = 25°, SENSE factor = 1.5, acquisition matrix = 480 × 480, 120 slices, voxel size = 0.25 × 0.25 × 0.25 mm, 3 signal averages, and 22 min 58 s duration) ( Fig. 1B ).
Image preparation
Imaging biomarkers were extracted using the pipeline described in Figure 1 . Prior to analysis, MR images were converted to NIfTI (Neuroimaging Informatics Technology Initiative) format ( Fig. 1C ) for cartilage and subchondral bone segmentation using open-access ITK-SNAP software ( Fig. 1D ).21,22 Automatic femoral and tibial 6-segment cartilage parcellation included the medial anterior region (TM), lateral anterior region (TL), medial central region (CM), lateral central region (CL), medial posterior region (PM), and lateral posterior region (PL) ( Fig. 1E1 ).9,23
Automatic subchondral tibia and femur bone parcellation labeled both epiphyses as medial and lateral, respectively. On each parcel, the centroid was calculated by defining a 5-mm diameter sphere as the region of interest to measure trabeculae metrics ( Fig. 1E2 ). 9
The Elastix toolbox was used for spatial intrasequence (different TEs and flip angles) and intersequence registration into a common geometric space corresponding to T1-TSE-SPIR. Registration was performed with non-rigid registration using B-splines and a parametric approach where different levels of resolution allowed macroscopic approximation to be the basis for adjusting each iteration ( Fig. 1F ). 10
Image processing
Imaging biomarkers were extracted using an ad hoc program written in MATLAB (R2016b, MathWorks, Natick, MA) for both cartilage and subchondral bone.
Whole cartilage from 6 segments of the femur and tibia samples was analyzed. Cartilage volume and thickness analyses were performed for each segment. The thickness analysis used 2D skeletonization and contour detection algorithms, where a transform distance was applied, providing the minimum distance of each voxel to the contour. Finally, the resulting image was multiplied by the skeletonization image, providing half the value of cartilage thickness ( Fig. 1G1 ). 10
Cartilage T1 relaxation time analysis was computed with all flip angles in a voxel-wise approach using the method described by Alberich-Bayarri et al. 10 and Fram et al. 24 The transversal T2* relaxation time analysis used all TEs and the method described elsewhere. 25
Trabecular bone volume analysis used an algorithm based on the local Laplacian to reduce heterogenicity and partial volume effects to obtain bone volume fraction. 26 Thresholding and super-resolution resizing were performed after heterogenicity and partial volume corrections, following Manjón et al.’s 27 and Otsu’s 28 algorithms. BV/TV, considering the trabecular bone volume percentage included in the volume of interest (VOI), was calculated using the ratio between the number of voxels in the trabeculae and the total number of voxels in the VOI. TbTh and trabecular separation were calculated based on the distance transformation of the skeleton on the contour, as previously described for the cartilage thickness analysis. Trabecular number (TbN) was calculated as the ratio between BV/TV and TbTh. Spatial distribution of the trabeculae was also evaluated by calculating the D2D and D3D fractal dimensions, which provide information on how trabeculae are dispersed in space. 29 In addition, a novel image biomarker quality of trabecular score (QTS) was calculated. This biomarker provides a single score that reflects the quality of the bone trabecula (patent filing ID: 201931050) ( Fig. 1G2 ).
Histological Study
Following MRI scan, sacrificed animals and stifle joints were dissected carefully and femoral condyles were isolated and preserved at −80 °C for further analysis. A total of 36 femoral condyles (18 left and 18 right) were fixed in 4% formaldehyde and decalcified to acquire cuts from the lateral condyle of each condyle. After paraffin inclusion, 4-µm lateral condylar sagittal cuts were made using a microtome and prepared on slides for staining. Samples were stained with 2 different staining techniques: hematoxylin-eosin using Dako Cover Stainer® Hematoxylin-Eosin (Agilent, Santa Clara, CA) and Masson trichrome using Dako Artisan Link Pro® Masson trichrome (Agilent). All samples were scanned using a digital scanner (Pannoramic 250 Flash®; 3DHISTECH Ltd, Budapest, Hungary) and evaluated using specific slide viewer software (CaseViewer 2.2®; 3DHISTECH Ltd) ( Fig. 2 ). Cartilage analysis consisted of the evaluation of staining, structure, chondrocyte density, and cluster formation following the scale described by Laverty et al. 14 Similarly, subchondral bone analysis consisted of the evaluation of trabeculae, grading the basophilia and fragmentation of the tidemark, mesenchymal changes in the marrow, and thickening of the subchondral bone. For this purpose, calcified cartilage and subchondral bone damage scores described by Gerwin et al. 17 were used.
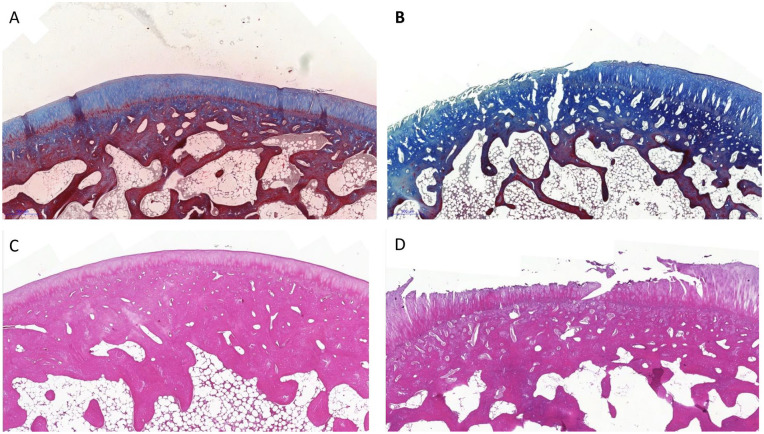
Figure 2.
Masson trichrome stain in (A) a control group case and (B) an osteoarthritis group case. H&E stain in (C) a control group case and (D) an osteoarthritis group case. H&E = hematoxylin and eosin.
Statistical Analysis
Statistical analysis was performed using R statistical software version 4.0.4. 30 Control and OA were considered independent groups. Normality of the variables was verified using the Shapiro-Wilk test. Homoscedasticity was assessed using Levene’s test. Comparisons between histological (stain, structure, chondrocyte density, cluster formation, total score, and subchondral bone) and imaging (cartilage volume, thickness, T1, T2*, and BV/TV, TbTh, TbSp, TbN, D2D, D3D, and QTS) variables, and groups (OA and control) were performed using the btwrim() function, which is included in the WRS2 package. 31 This function computes a 2-way between-within-subjects analysis of variance on the trimmed means. Finally, a general linear model was used to study the relationship between histological variables () and protocols. This was performed using the yuend() and yuend() functions for dependent and dependent sample t tests on robust location measures, including effect sizes. Differences were considered significant at a confidence interval of 95% and P value of <0.05.
Finally, the correlation matrix was obtained for the histological and imaging variables by means of Spearman correlation and P values on the upper triangle. Only the correlations between pairwise variables with P-value <0.05 were considered.
Results
MRI Results
Cartilage MRI results
Cartilage volume and thickness analyses did not reveal any statistically significant differences between the groups. The levels of all cartilage T1 and T2* imaging biomarkers were significantly higher in the OA group than in the control group, except for femoral T1_CM, tibial T2*_TM, and T2*_TL. T1 and T2* cartilage results are presented in Table 1 .
Table 1.
Cartilage T1 and T2* Biomarkers Compared Between OA and Control Groups, With Statistical Significance Labeled.
| Cartilage MRI | OA |
Control |
OA vs. Control |
||||
|---|---|---|---|---|---|---|---|
| Maximum | Minimum | Median | Maximum | Minimum | Median | P Value | |
| T1 whole (ms) | |||||||
| Femur | 1,530.00 | 987.00 | 1,220.00 | 1,090.00 | 708.00 | 887.00 | <0.01 |
| Tibia | 1,190.00 | 901.00 | 1,000.00 | 1,090.00 | 682.00 | 789.00 | <0.01 |
| T1_TM (ms) | |||||||
| Femur | 1,770.00 | 883.00 | 1,200.00 | 1,180.00 | 712.00 | 893.00 | <0.01 |
| Tibia | 1,210.00 | 472.00 | 919.00 | 1,250.00 | 347.00 | 613.00 | <0.01 |
| T1_TL (ms) | |||||||
| Femur | 1,530.00 | 873.00 | 1,140.00 | 1,050.00 | 652.00 | 867.00 | <0.01 |
| Tibia | 1,180.00 | 579.00 | 875.00 | 738.00 | 348.00 | 515.00 | <0.01 |
| T1_CM (ms) | |||||||
| Femur | 1,450.00 | 886.00 | 1,070.00 | 1,220.00 | 836.00 | 1,010.00 | 0.16 |
| Tibia | 1,340.00 | 855.00 | 1,080.00 | 1,550.00 | 820.00 | 973.00 | <0.01 |
| T1_CL (ms) | |||||||
| Femur | 1,340.00 | 981.00 | 1,070.00 | 1,230.00 | 639.00 | 913.00 | <0.01 |
| Tibia | 1,140.00 | 929.00 | 1,060.00 | 1,070.00 | 597.00 | 822.00 | <0.01 |
| T1_PM (ms) | |||||||
| Femur | 1,520.00 | 942.00 | 1,320.00 | 1,040.00 | 592.00 | 839.00 | <0.01 |
| Tibia | 1,440.00 | 831.00 | 1,110.00 | 1,210.00 | 697.00 | 856.00 | <0.01 |
| T1_PL (ms) | |||||||
| Femur | 1,440.00 | 1,030.00 | 1,230.00 | 1,060.00 | 705.00 | 853.00 | <0.01 |
| Tibia | 1,330.00 | 840.00 | 1,070.00 | 1,080.00 | 721.00 | 880.00 | <0.01 |
| T2* whole (ms) | |||||||
| Femur | 30.10 | 15.60 | 22.20 | 18.00 | 12.30 | 14.80 | <0.01 |
| Tibia | 21.70 | 10.50 | 13.50 | 12.70 | 8.61 | 10.50 | <0.01 |
| T2*_TM (ms) | |||||||
| Femur | 41.20 | 11.90 | 23.30 | 23.60 | 11.80 | 15.10 | <0.01 |
| Tibia | 14.90 | 7.84 | 11.00 | 14.80 | 7.27 | 9.68 | 0.14 |
| T2*_TL (ms) | |||||||
| Femur | 35.10 | 13.00 | 22.40 | 22.50 | 12.80 | 17.90 | <0.05 |
| Tibia | 25.80 | 6.88 | 9.85 | 15.20 | 7.44 | 10.60 | 0.27 |
| T2*_CM (ms) | |||||||
| Femur | 22.70 | 9.37 | 14.60 | 14.10 | 7.74 | 10.50 | <0.01 |
| Tibia | 19.40 | 7.37 | 10.40 | 12.90 | 7.70 | 9.49 | <0.05 |
| T2*_CL (ms) | |||||||
| Femur | 20.80 | 9.85 | 14.50 | 13.40 | 7.41 | 9.29 | <0.01 |
| Tibia | 17.90 | 7.95 | 10.50 | 10.30 | 5.95 | 7.97 | <0.05 |
| T2*_PM (ms) | |||||||
| Femur | 42.00 | 14.80 | 27.50 | 19.90 | 10.40 | 13.50 | <0.01 |
| Tibia | 28.20 | 11.40 | 18.00 | 15.50 | 8.06 | 11.30 | <0.005 |
| T2*_PL (ms) | |||||||
| Femur | 34.60 | 18.20 | 24.70 | 19.10 | 12.10 | 14.90 | <0.01 |
| Tibia | 27.00 | 9.45 | 14.50 | 15.40 | 8.99 | 11.20 | <0.01 |
MRI = magnetic resonance imaging; OA = osteoarthritis; TM = medial anterior region; TL = lateral anterior region; CM = medial central region; CL = lateral central region; PM = medial posterior region; PL = lateral posterior region.
Subchondral bone MRI results
Significant differences were observed between BV/TV, TbN, QTS, medial femoral TbTh, medial and lateral tibial TbSp, lateral femoral D2D and D3D, and medial tibial D3D groups. No other significant differences were noted ( Table 2 ).
Table 2.
Subchondral Bone MRI Biomarkers Compared Between OA and Control Groups, With Statistical Significance Labeled.
| Subchondral Bone MRI | OA |
Control |
OA vs. Control |
||||
|---|---|---|---|---|---|---|---|
| Maximum | Minimum | Median | Maximum | Minimum | Median | P Value | |
| BV/TV (%) | |||||||
| Femur | |||||||
| Medial | 44.30 | 26.60 | 35.10 | 46.30 | 29.00 | 39.10 | <0.01 |
| Lateral | 47.80 | 31.10 | 36.20 | 46.20 | 32.80 | 39.00 | <0.05 |
| Tibia | |||||||
| Medial | 56.20 | 37.40 | 43.20 | 49.70 | 30.20 | 37.40 | <0.01 |
| Lateral | 41.20 | 31.00 | 36.00 | 56.10 | 42.20 | 46.70 | <0.01 |
| TbTh_mean (µm) | |||||||
| Femur | |||||||
| Medial | 340.00 | 265.00 | 274.00 | 369.00 | 271.00 | 286.00 | <0.01 |
| Lateral | 306.00 | 266.00 | 279.00 | 340.00 | 269.00 | 278.00 | 0.63 |
| Tibia | |||||||
| Medial | 362.00 | 256.00 | 269.00 | 360.00 | 264.00 | 274.00 | 0.69 |
| Lateral | 301.00 | 262.00 | 275.00 | 356.00 | 262.00 | 278.00 | 0.32 |
| TbSp_mean (µm) | |||||||
| Femur | |||||||
| Medial | 427.00 | 316.00 | 372.00 | 454.00 | 313.00 | 353.00 | 0.43 |
| Lateral | 435.00 | 299.00 | 356.00 | 428.00 | 294.00 | 341.00 | 0.14 |
| Tibia | |||||||
| Medial | 353.00 | 266.00 | 296.00 | 456.00 | 312.00 | 368.00 | <0.01 |
| Lateral | 471.00 | 320.00 | 368.00 | 314.00 | 258.00 | 293.00 | <0.01 |
| TbN (mm−1) | |||||||
| Femur | |||||||
| Medial | 1.46 | 0.95 | 1.26 | 1.55 | 1.04 | 1.38 | <0.05 |
| Lateral | 1.70 | 1.12 | 1.28 | 1.68 | 1.17 | 1.40 | <0.05 |
| Tibia | |||||||
| Medial | 2.16 | 1.30 | 1.58 | 1.51 | 1.11 | 1.37 | <0.01 |
| Lateral | 1.44 | 1.12 | 1.29 | 2.02 | 1.51 | 1.67 | <0.01 |
| D2D (a.u) | |||||||
| Femur | |||||||
| Medial | 1.59 | 1.40 | 1.53 | 1.63 | 1.37 | 1.53 | 0.73 |
| Lateral | 1.56 | 1.33 | 1.50 | 1.62 | 1.45 | 1.55 | <0.01 |
| Tibia | |||||||
| Medial | 1.61 | 1.10 | 1.49 | 1.57 | 1.37 | 1.49 | 0.9 |
| Lateral | 1.57 | 1.32 | 1.49 | 1.59 | 1.43 | 1.48 | 0.41 |
| D3D (a.u) | |||||||
| Femur | |||||||
| Medial | 2.13 | 2.02 | 2.07 | 2.14 | 1.99 | 2.09 | 0.19 |
| Lateral | 2.13 | 2.00 | 2.09 | 2.15 | 2.06 | 2.12 | <0.05 |
| Tibia | |||||||
| Medial | 2.16 | 1.74 | 2.03 | 2.11 | 1.94 | 2.07 | <0.05 |
| Lateral | 2.10 | 1.97 | 2.06 | 2.16 | 1.91 | 2.06 | 0.99 |
| QTS (a.u) | |||||||
| Femur | |||||||
| Medial | 3.47 | 0.95 | 1.81 | 3.35 | 1.26 | 2.41 | <0.01 |
| Lateral | 3.03 | 1.35 | 2.03 | 3.31 | 1.68 | 2.35 | <0.05 |
| Tibia | |||||||
| Medial | 4.46 | 1.69 | 2.68 | 4.13 | 1.15 | 2.05 | <0.05 |
| Lateral | 2.65 | 1.27 | 1.88 | 4.48 | 2.40 | 3.00 | <0.01 |
MRI = magnetic resonance imaging; OA = osteoarthritis; BV/TV = bone volume to total volume fraction; QTS = quality of trabecular score; TbTh = trabecular thickness; TbSp = trabecular spacing; TbN = trabecular number; D2D = two dimensional fractal dimension measurements; D3D = three dimensional fractal dimension measurements.
Histological Results
Microscopic analysis
The control knees had a normal appearance with no degenerative changes. Microscopic findings associated with OA in this group were absent in all variables.
In contrast, the OA group exhibited significant changes in all biomarkers with a P value <0.001 except for cluster formation, which was insignificant ( Table 3 ). The main changes observed in the histological sections were loss of superficial and intermediate layers, loss of stain intensity from the cartilage matrix, increased cellular density, and irregular distribution along the affected areas ( Fig. 2 ).
Table 3.
Histological Results of OA Group Compared to the Control Group, With Statistical Significance Labeled.
| Histological Biomarkers | OA | OA vs. Control | ||
|---|---|---|---|---|
| Maximum | Minimum | Median | P Value | |
| Stain | 5 | 1 | 2.61 | <0.01 |
| Structure | 8 | 1 | 3.61 | <0.01 |
| Chondrocyte density | 3 | 0 | 1.5 | <0.01 |
| Cluster formation | 3 | 0 | 0.38 | 0.3 |
| Subchondral bone | 4 | 1 | 1.61 | <0.01 |
| Total | 19 | 2 | 8.11 | <0.01 |
OA = osteoarthritis
Correlations between histological and MRI biomarkers
Cartilage volume and thickness were not considered in this analysis, as they were not significantly different between the groups. Histological cartilage biomarkers, including stain, structure, chondrocyte density, and total score histological biomarkers, were positively correlated with all lateral cartilage T1 and T2* MR biomarkers. Subchondral bone histological biomarkers also showed a positive correlation with T1 and T2* MR biomarkers. Cluster formation alone did not reveal any significant differences or correlations with cartilage biomarkers ( Table 4 ).
Table 4.
Correlation Between T1 and T2* MRI Biomarkers With the Different Histological Biomarkers.
| T1 and T2 vs. Histological Biomarkers | T1 Biomarker |
T2* Biomarker |
||||||
|---|---|---|---|---|---|---|---|---|
| T1 Whole (ms) | T1_TL (ms) | T1_CL (ms) | T1_PL (ms) | T2* Whole (ms) | T2*_TL (ms) | T2*_CL (ms) | T2*_PL (ms) | |
| Stain | ||||||||
| Correlation | 0.77 | 0.78 | 0.65 | 0.81 | 0.78 | 0.54 | 0.77 | 0.83 |
| P value | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| Structure | ||||||||
| Correlation | 0.78 | 0.77 | 0.67 | 0.81 | 0.74 | 0.42 | 0.73 | 0.80 |
| P value | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.05 | <0.01 | <0.01 |
| Chondrocyte density | ||||||||
| Correlation | 0.70 | 0.75 | 0.56 | 0.75 | 0.70 | 0.41 | 0.68 | 0.80 |
| P value | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.05 | <0.01 | <0.01 |
| Cluster formation | ||||||||
| Correlation | 0.25 | 0.29 | 0.14 | 0.31 | 0.17 | −0.01 | 0.29 | 0.30 |
| P value | NA | NA | NA | NA | NA | NA | NA | NA |
| TOTAL_SCORE | ||||||||
| Correlation | 0.76 | 0.78 | 0.65 | 0.82 | 0.75 | 0.42 | 0.75 | 0.83 |
| P value | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| Subchondral bone | ||||||||
| Correlation | 0.79 | 0.76 | 0.70 | 0.82 | 0.76 | 0.41 | 0.76 | 0.83 |
| P value | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.05 | <0.01 | <0.01 |
TL = lateral anterior region; CL = lateral central region; PL = lateral posterior region; NA = non-applicable; MRI = magnetic resonance imagin.
The subchondral bone MR biomarkers D2D and QTS were significantly correlated with histological subchondral bone biomarkers. In addition, D2D and QTS also presented a significant correlation with the remaining histological variables, except for cluster formation. BV/TV was significantly correlated with structure, chondrocyte density, and total score, while TbN was only correlated with structure. TbTh, TbSp, and D3D were not correlated with any of the histological biomarkers (Table 5; Fig. 3).
Table 5.
Correlation Between Subchondral Bone MRI Biomarkers With the Different Histological Hallmarks a .
| Subchondral Bone vs. Histological Biomarkers | BV/TV (%) | TbTh_Mean (µm) | TbSp_Mean (µm) | TbN (mm−1) | D2D (a.u) | D3D (a.u) | QTS (a.u) |
|---|---|---|---|---|---|---|---|
| Stain | |||||||
| Correlation | −0.33 | −0.18 | 0.17 | −0.27 | −0.40 | −0.23 | −0.37 |
| P value | NA | NA | NA | NA | <0.05 | NA | <0.05 |
| Structure | |||||||
| Correlation | −0.38 | −0.17 | 0.29 | −0.33 | −0.36 | −0.29 | −0.41 |
| P value | <0.05 | NA | NA | <0.05 | <0.05 | NA | <0.05 |
| Chondrocyte density | |||||||
| Correlation | −0.38 | −0.23 | 0.23 | −0.30 | −0.35 | −0.20 | −0.43 |
| P-value | <0.05 | NA | NA | NA | 0.05 | NA | <0.01 |
| Cluster formation | |||||||
| Correlation | −0.13 | −0.11 | 0.03 | −0.10 | −0.21 | −0.03 | −0.17 |
| P value | NA | NA | NA | NA | NA | NA | NA |
| TOTAL_SCORE | |||||||
| Correlation | −0.37 | −0.20 | −0.23 | −0.31 | −0.39 | −0.25 | −0.41 |
| P-value | <0.05 | NA | NA | NA | <0.05 | NA | <0.05 |
| Subchondral bone | |||||||
| Correlation | −0.37 | −0.20 | 0.23 | −0.31 | −0.39 | −0.25 | −0.41 |
| P value | NA | NA | NA | NA | <0.01 | NA | <0.05 |
D2D and QTS demonstrate a negative correlation; the remaining significant results also show a negative correlation between MRI and histologic biomarkers.
BV/TV = bone volume to total volume fraction; QTS = quality of trabecular score; TbTh = trabecular thickness; TbSp = trabecular spacing; TbN = trabecular number; D2D = two dimensional fractal dimension measurements; D3D = three dimensional fractal dimension measurements; NA = non-applicable; MRI = magnetic resonance imaging.
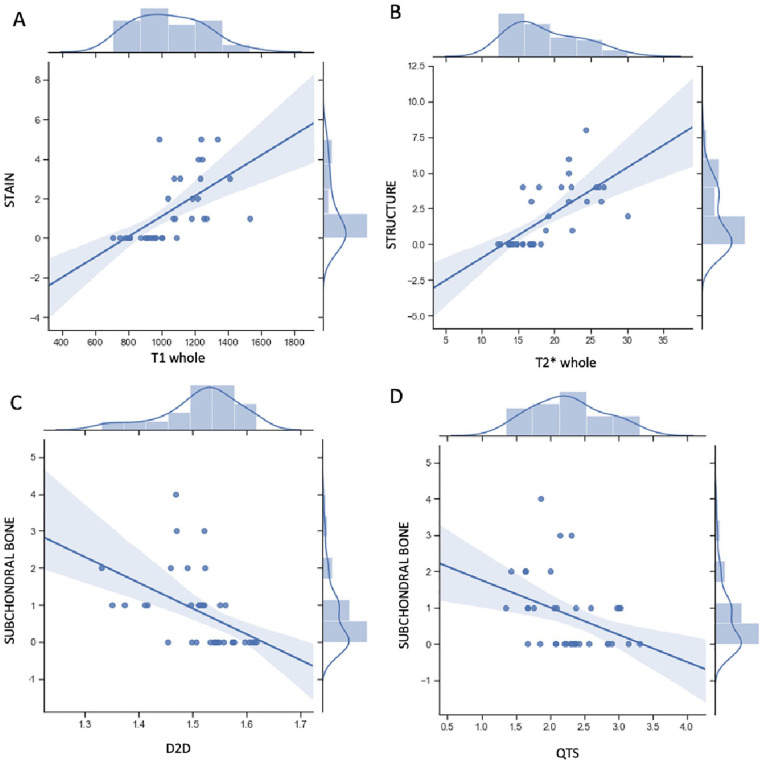
Figure 3.
(A) Positive correlation between histological stain marker with MRI T1 lateral femur biomarker. (B) Positive correlation between histological structure marker with MRI T2* lateral femur biomarker. (C) and (D) Negative correlation between histological subchondral bone biomarker with MRI D2D and QTS biomarkers, respectively. MRI = magnetic resonance imaging; QTS = quality of trabecular score; D2D = two dimensional fractal dimension measurements.
Discussion
This study reveals how quantification of MR biomarkers allows for detection of OA changes in a rabbit model and demonstrates a strong correlation between different MRI cartilage and subchondral bone MRI biomarkers and histological hallmarks of OA.
Histology is the reference standard used to evaluate cartilage 14 and subchondral bone 17 and to quantify degeneration in OA. As reported in previous studies, ACLT rabbit model has displayed excellent outcomes regarding disease progression. 11 These histological results demonstrate degenerative osteoarthritic changes in cartilage and subchondral knee bone ( Fig. 3 ), with a large difference between healthy and OA groups ( Table 1 ). The literature describes a decrease in the quantity and quality of proteoglycans in the extracellular matrix, structural changes due to the loss of superficial and deep layers of the articular cartilage, usually seen after mechanical alterations, and an increase, in early stages, of extracellular matrix cellularity due to compensatory mechanisms following cartilage insult. 15 Rabbits are a suitable animal model of OA that also allow visualization of correlation between histology and MRI biomarkers.
Cluster formation has been studied in patients with end-stage disease and aids in chondrocyte proliferation and joint repair. 32 The present experimental model has exhibited a mild-moderate OA degree. This might explain why cluster formation was limited in our study and why no significant differences were observed in the control group.
Most cartilage and subchondral bone imaging studies have been performed in humans. Traditionally, subchondral bone has been evaluated using CT because of its high accuracy in analyzing bone structures. MRI studies have been conducted focusing on T1 and T2* relaxation times, 5 some of which have implemented the use of delayed contrast-enhanced MRI to determine the quantity of glycosaminoglycans. 33 Human cartilage volume and thickness are commonly analyzed using MR images. Frisbie et al. 34 measured the average thickness of cartilage in rabbits as 0.3 mm, compared with 2.2-2.5 mm in humans. Therefore, the voxel size of our study (0.27 × 0.27 × 0.4) represents a limitation in quantifying cartilage thickness and volume. This limitation leads to overestimation of cartilage thickness and volume in rabbits and explains why no significant differences were observed between the experimental groups.
In the ACLT rabbit model, the induction of OA leads to a modified gait. 35 The lateral compartment of the rabbit knee is most affected when analyzing the cartilage surface. 36 Although cartilage changes are seen predominantly in the lateral compartment, 37 some researchers have described similar subchondral CT bone changes in the medial compartment. 9 This model results in alteration of weightbearing forces and mechanical erosion of the articular cartilage, leading to overload on the subchondral bone. Kajabi et al. 37 suggested that this erosion might be due to instability of the joint, combined with the rotational and translational abnormal movements of the tibia relative to the femur. Joint overload generates an initial increase in density of the underlying subchondral bone, thereby altering the trabecular microstructure. 9
Subchondral bone changes were observed predominantly on the medial aspect of the condyles. 9 In our study, both the femoral condyles and tibial plateaus displayed significant differences between groups in the medial and lateral compartments. Florea et al. hypothesized that the medial aspect of the femoral condyles undergoes remodeling and resorption due to minor weightbearing forces passing through the medial compartment. Another hypothesis is that due to inflammation, osteoclasts reach the area and contribute to remodeling, and which both stifle compartments are subjected to, but the lateral aspect presents higher loading and osteoblast activity compared with the medial aspect.
Our results are consistent with previous findings in the lateral cartilage and medial subchondral bone and could be explained by rotational and translational instability movements that wear out the lateral cartilage surface while the medial compartment experiences redistribution of the weight load on the joint.9,37 The authors suggested that menisci could also protect cartilage erosion against shear forces in the medial compartment. The medial meniscus attaches completely to the tibia, whereas the lateral meniscus attaches to the tibia and femur.
MR evaluation revealed significant changes in the subchondral bone in both compartments of the stifle joint at 12 weeks. BV/TV, TbN, and QTS displayed differences in both the medial and lateral compartments of the tibia and femur. These results can be explained by the time length of our study, which was considerably longer than the 4 weeks of Florea et al.’s study. This postulates that the subchondral bone changes occur at the medial aspect of the femoral condyles. No statistical difference was identified in the medial compartment, with a possible explanation being that compensatory mechanisms may be acting at this stage. 9 Our study generated different results under similar conditions, but at 12 weeks. In this case, compensatory mechanisms may start to fail, and significant changes can be observed.
Prior to this study, correlations between histological and MR biomarkers of OA had not been previously established in a rabbit model. Cartilage analysis revealed a positive correlation between T1 and T2* relaxation times and histological biomarker staining, structure, and chondrocyte density. T1 relaxation time was used to quantify proteoglycans in the extracellular matrix. 5 Histological biomarker staining has been used for the same proteoglycan measurement purposes in other studies. 15 Similarly, T2* is used to quantify the water content and collagen network 5 such as histological biomarker structure. 15 Chondrocytes produce proteoglycans and contribute to the quality of the matrix structure. 38 Therefore, chondrocyte density histological biomarkers can be related to both T1 and T2* MR biomarkers.
For subchondral bone analysis, MRI D2D and QTS biomarkers were significantly correlated with histological subchondral bone biomarkers. This can be explained by the association of the 2 MRI biomarkers with degenerative changes in the femoral condyle and adjacent cartilage areas. 29
Some study limitations are that only the lateral aspect of the femoral condyles was evaluated histologically, and the sensitivity of the technique did not allow evaluation of cartilage volume and thickness due to the acquisition voxel size. Last, further studies are required to assess how histological and MR biomarkers behave at different time points as the disease progresses.
Conclusions
Quantification of changes in cartilage and subchondral bone MRI biomarkers is feasible in a 12-week rabbit model of OA and provides an excellent correlation with histopathological changes.
Footnotes
Acknowledgments and Funding: The authors would like to acknowledge Bioiberica SAU for supporting this study and Professor Jose Ignacio Redondo Garcia for the statistical analysis. The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: A.T.-E. is the recipient of a PFIS grant (FI20/00239), Instituto de Salud Carlos III. This project was funded by Bioiberica S.A.U., Esplugues de Llobregat, Spain.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical Approval: The ethical and institutional boards of the Fundación Instituto Inv. Sanitaria La Fe, Valencia, Spain approved this study (ID: 2017/VSC/PEA/00177).
ORCID iDs: Vicente Sifre  https://orcid.org/0000-0003-4874-1812
https://orcid.org/0000-0003-4874-1812
Amadeo Ten-Esteve  https://orcid.org/0000-0002-3339-7767
https://orcid.org/0000-0002-3339-7767
Ángel Alberich-Bayarri  https://orcid.org/0000-0002-5932-2392
https://orcid.org/0000-0002-5932-2392
Sergi Segarra  https://orcid.org/0000-0002-6680-4146
https://orcid.org/0000-0002-6680-4146
Luis Martí-Bonmatí  https://orcid.org/0000-0002-8234-010X
https://orcid.org/0000-0002-8234-010X
References
- 1. Joseph GB, McCulloch CE, Nevitt MC, Neumann J, Gersing AS, Kretzschmar M, et al. Tool for osteoarthritis risk prediction (TOARP) over 8 years using baseline clinical data, X-ray, and MRI: data from the osteoarthritis initiative. J Magn Reson Imaging. 2018;47(6):1517-26. doi: 10.1002/jmri.25892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wu PT, Shao CJ, Wu KC, Wu TT, Chern TC, Kuo LC, et al. Pain in patients with equal radiographic grades of osteoarthritis in both knees: the value of gray scale ultrasound. Osteoarthritis Cartilage. 2012;20(12):1507-13. doi: 10.1016/j.joca.2012.08.021. [DOI] [PubMed] [Google Scholar]
- 3. Alizai H, Walter W, Khodarahmi I, Burke CJ. Cartilage imaging in osteoarthritis. Semin Musculoskelet Radiol. 2019;23(5):569-77. doi: 10.1055/s-0039-1695720. [DOI] [PubMed] [Google Scholar]
- 4. Duvvuri U, Kudchodkar S, Reddy R, Leigh JS. T(1rho) relaxation can assess longitudinal proteoglycan loss from articular cartilage in vitro. Osteoarthritis Cartilage. 2002;10(11):838-44. doi: 10.1053/joca.2002.0826. [DOI] [PubMed] [Google Scholar]
- 5. Mittal S, Pradhan G, Singh S, Batra R. T1 and T2 mapping of articular cartilage and menisci in early osteoarthritis of the knee using 3-Tesla magnetic resonance imaging. Pol J Radiol. 2019;84:e549-64. doi: 10.5114/pjr.2019.91375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sanz-Requena R, Martí-Bonmatí L, Hervás V, Vega M, Alberich-Bayarri A, García-Martí G, et al. [Modification of longitudinal relaxation time (T1) as a biomarker of patellar cartilage degeneration]. Radiologia. 2010;52(3):221-7. doi: 10.1016/j.rx.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 7. Nieminen MT, Rieppo J, Töyräs J, Hakumäki JM, Silvennoinen J, Hyttinen MM, et al. T2 relaxation reveals spatial collagen architecture in articular cartilage: a comparative quantitative MRI and polarized light microscopic study. Magn Reson Med. 2001;46(3):487-93. doi: 10.1002/mrm.1218. [DOI] [PubMed] [Google Scholar]
- 8. Liess C, Lüsse S, Karger N, Heller M, Glüer CC. Detection of changes in cartilage water content using MRI T2-mapping in vivo. Osteoarthritis Cartilage. 2002;10(12):907-13. doi: 10.1053/joca.2002.0847. [DOI] [PubMed] [Google Scholar]
- 9. Florea C, Malo MK, Rautiainen J, Mäkelä JT, Fick JM, Nieminen MT, et al. Alterations in subchondral bone plate, trabecular bone and articular cartilage properties of rabbit femoral condyles at 4 weeks after anterior cruciate ligament transection. Osteoarthritis Cartilage. 2015;23(3):414-22. doi: 10.1016/j.joca.2014.11.023. [DOI] [PubMed] [Google Scholar]
- 10. Alberich-Bayarri A, Marti-Bonmati L, Sanz-Requena R, Belloch E, Moratal D. In vivo trabecular bone morphologic and mechanical relationship using high-resolution 3-T MRI. AJR Am J Roentgenol. 2008;191(3):721-6. doi: 10.2214/AJR.07.3528. [DOI] [PubMed] [Google Scholar]
- 11. Serra CI, Soler C. Animal models of osteoarthritis in small mammals. Vet Clin North Am Exot Anim Pract. 2019;22(2):211-21. doi: 10.1016/j.cvex.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 12. Kuyinu EL, Narayanan G, Nair LS, Laurencin CT. Animal models of osteoarthritis: classification, update, and measurement of outcomes. J Orthop Surg Res. 2016;11(1):19. doi: 10.1186/s13018-016-0346-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McCoy AM. Animal models of osteoarthritis: comparisons and key considerations. Vet Pathol. 2015;52(5):803-18. doi: 10.1177/0300985815588611. [DOI] [PubMed] [Google Scholar]
- 14. Laverty S, Girard CA, Williams JM, Hunziker EB, Pritzker KP. The OARSI histopathology initiative—recommendations for histological assessments of osteoarthritis in the rabbit. Osteoarthritis Cartilage. 2010;18(Suppl 3):S53-65. doi: 10.1016/j.joca.2010.05.029. [DOI] [PubMed] [Google Scholar]
- 15. Cook JL, Kuroki K, Visco D, Pelletier JP, Schulz L, Lafeber FP. The OARSI histopathology initiative—recommendations for histological assessments of osteoarthritis in the dog. Osteoarthritis Cartilage. 2010;18(Suppl 3):S66-79. doi: 10.1016/j.joca.2010.04.017. [DOI] [PubMed] [Google Scholar]
- 16. Glasson SS, Chambers MG, Van Den Berg WB, Little CB. The OARSI histopathology initiative—recommendations for histological assessments of osteoarthritis in the mouse. Osteoarthritis Cartilage. 2010;18(Suppl 3):S17-23. doi: 10.1016/j.joca.2010.05.025. [DOI] [PubMed] [Google Scholar]
- 17. Gerwin N, Bendele AM, Glasson S, Carlson CS. The OARSI histopathology initiative—recommendations for histological assessments of osteoarthritis in the rat. Osteoarthritis Cartilage. 2010;18(Suppl 3):S24-34. doi: 10.1016/j.joca.2010.05.030. [DOI] [PubMed] [Google Scholar]
- 18. Pritzker KPH, Gay S, Jimenez SA, Ostergaard K, Pelletier JP, Revell PA, et al. Osteoarthritis cartilage histopathology: grading and staging. Osteoarthr Cartilage. 2006;14(1):13-29. doi: 10.1016/j.joca.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 19. Sulaiman SZS, Tan WM, Radzi R, Shafie IN, Ajat M, Mansor R, et al. Comparison of bone and articular cartilage changes in osteoarthritis: a micro-computed tomography and histological study of surgically and chemically induced osteoarthritic rabbit models. J Orthop Surg Res. 2021;16(1):663. doi: 10.1186/s13018-021-02781-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martig S, Boisclair J, Konar M, Spreng D, Lang J. MRI characteristics and histology of bone marrow lesions in dogs with experimentally induced osteoarthritis. Vet Radiol Ultrasound. 2007;48(2):105-12. doi: 10.1111/j.1740-8261.2007.00213.x. [DOI] [PubMed] [Google Scholar]
- 21. Matsui H, Shimizu M, Tsuji H. Cartilage and subchondral bone interaction in osteoarthrosis of human knee joint: a histological and histomorphometric study. Microsc Res Techniq. 1997;37(4):333-42. doi: [DOI] [PubMed] [Google Scholar]
- 22. Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, et al. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31(3):1116-28. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 23. Bobinac D, Spanjol J, Zoricic S, Maric I. Changes in articular cartilage and subchondral bone histomorphometry in osteoarthritic knee joints in humans. Bone. 2003;32(3):284-90. doi: 10.1016/s8756-3282(02)00982-1. [DOI] [PubMed] [Google Scholar]
- 24. Fram EK, Herfkens RJ, Johnson GA, Glover GH, Karis JP, Shimakawa A, et al. Rapid calculation of T1 using variable flip angle gradient refocused imaging. Magn Reson Imaging. 1987;5(3):201-8. doi: 10.1016/0730-725x(87)90021-x. [DOI] [PubMed] [Google Scholar]
- 25. Li D, Dhawale P, Rubin PJ, Haacke EM, Gropler RJ. Myocardial signal response to dipyridamole and dobutamine: demonstration of the BOLD effect using a double-echo gradient-echo sequence. Magn Reson Med. 1996;36(1):16-20. doi: 10.1002/mrm.1910360105. [DOI] [PubMed] [Google Scholar]
- 26. Vasilic B, Wehrli FW. A novel local thresholding algorithm for trabecular bone volume fraction mapping in the limited spatial resolution regime of in vivo MRI. IEEE Trans Med Imaging. 2005;24(12):1574-85. doi: 10.1109/tmi.2005.859192. [DOI] [PubMed] [Google Scholar]
- 27. Manjón JV, Coupé P, Buades A, Collins DL, Robles M. MRI superresolution using self-similarity and image priors. Int J Biomed Imaging. 2010;2010:425891. doi: 10.1155/2010/425891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Otsu N. A Threshold selection method from gray-level histograms. Ieee Transactions Syst Man Cybern. 1979;9(1):62-6. doi: 10.1109/tsmc.1979.4310076. [DOI] [Google Scholar]
- 29. Alberich-Bayarri Á, Martí-Bonmatí L, Pérez MA, Sanz-Requena R, Lerma-Garrido JJ, García-Martí G, et al. Assessment of 2D and 3D fractal dimension measurements of trabecular bone from high-spatial resolution magnetic resonance images at 3 T. Med Phys. 2010;37(9):4930-7. doi: 10.1118/1.3481509. [DOI] [PubMed] [Google Scholar]
- 30. Champely S. pwr: basic functions for power analysis (R Package Version 1.3-0). n.d. [cited 2021 Mar 23]. Available from: https://CRAN.R-project.org/package=pwr.
- 31. Mair P, Wilcox R. Robust statistical methods in R using the WRS2 package. Behav Res Methods. 2020;52(2):464-88. doi: 10.3758/s13428-019-01246-w. [DOI] [PubMed] [Google Scholar]
- 32. Hoshiyama Y, Otsuki S, Oda S, Kurokawa Y, Nakajima M, Jotoku T, et al. Chondrocyte clusters adjacent to sites of cartilage degeneration have characteristics of progenitor cells. J Orthop Res. 2015;33(4):548-55. doi: 10.1002/jor.22782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Naughton JF, Stewart MC, Ciobanu L, Constable PD. Contrast magnetic resonance imaging for measurement of cartilage glycosaminoglycan content in dogs: a pilot study. Vet Comp Orthop Traumatol. 2013;26(2):100-4. doi: 10.3415/VCOT-12-03-0033. [DOI] [PubMed] [Google Scholar]
- 34. Frisbie DD, Cross MW, McIlwraith CW. A comparative study of articular cartilage thickness in the stifle of animal species used in human pre-clinical studies compared to articular cartilage thickness in the human knee. Vet Comp Orthop Traumatol. 2006;19(3):142-6. doi: 10.1055/s-0038-1632990. [DOI] [PubMed] [Google Scholar]
- 35. Bray RC, Shrive NG, Frank CB, Chimich DD. The early effects of joint immobilization on medial collateral ligament healing in an ACL-deficient knee: a gross anatomic and biomechanical investigation in the adult rabbit model. J Orthop Res. 1992;10(2):157-66. doi: 10.1002/jor.1100100202. [DOI] [PubMed] [Google Scholar]
- 36. Gushue DL, Houck J, Lerner AL. Rabbit knee joint biomechanics: motion analysis and modeling of forces during hopping. J Orthop Res. 2005;23(4):735-42. doi: 10.1016/j.orthres.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 37. Kajabi AW, Casula V, Ojanen S, Finnilä MA, Herzog W, Saarakkala S, et al. Multiparametric MR imaging reveals early cartilage degeneration at 2 and 8 weeks after ACL transection in a rabbit model. J Orthop Res. 2020;38(9):1974-86. doi: 10.1002/jor.24644. [DOI] [PubMed] [Google Scholar]
- 38. Gigante A, Callegari L. The role of intra-articular hyaluronan (Sinovial) in the treatment of osteoarthritis. Rheumatol Int. 2011;31(4):427-44. doi: 10.1007/s00296-010-1660-6. [DOI] [PubMed] [Google Scholar]