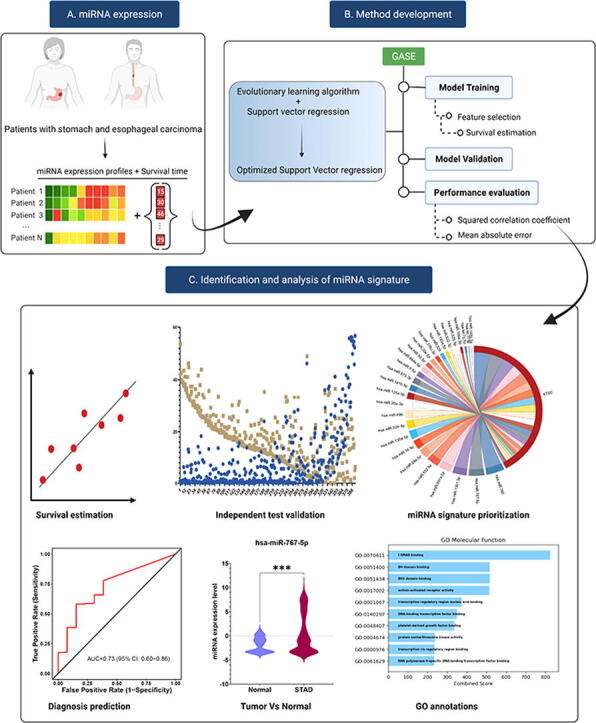
Graphical abstract
System overview of GASE. (A). MicroRNA expression profiles and survival time of patients with stomach and esophageal carcinoma are input of the system. (B) GASE method development and (C) microRNA discovery and analysis.
Keywords: miRNA signature, Machine learning, Survival estimation, Stomach and esophageal carcinoma
Abstract
Identifying a miRNA signature associated with survival will open a new window for developing miRNA-targeted treatment strategies in stomach and esophageal cancers (STEC). Here, using data from The Cancer Genome Atlas on 516 patients with STEC, we developed a Genetic Algorithm-based Survival Estimation method, GASE, to identify a miRNA signature that could estimate survival in patients with STEC. GASE identified 27 miRNAs as a survival miRNA signature and estimated the survival time with a mean squared correlation coefficient of 0.80 ± 0.01 and a mean absolute error of 0.44 ± 0.25 years between actual and estimated survival times, and showed a good estimation capability on an independent test cohort. The miRNAs of the signature were prioritized and analyzed to explore their roles in STEC. The diagnostic ability of the identified miRNA signature was analyzed, and identified some critical miRNAs in STEC. Further, miRNA-gene target enrichment analysis revealed the involvement of these miRNAs in various pathways, including the somatotrophic axis in mammals that involves the growth hormone and transforming growth factor beta signaling pathways, and gene ontology annotations. The identified miRNA signature provides evidence for survival-related miRNAs and their involvement in STEC, which would aid in developing miRNA-target based therapeutics.
1. Introduction
Stomach and esophageal carcinomas (STEC) are among the most prevalent malignant diseases causing thousands of deaths globally. Worldwide, stomach cancer ranks sixth in cancer incidence, with 1,089,103 new cases, and third in cancer morality, with 768,793 deaths, while esophageal cancer ranks tenth in cancer incidence, with 604,100 new cases, and sixth in cancer morality, with 544,076 deaths, based on estimates for the year 2020 [1]. STEC ranks higher in mortality than incidence because these cancers are often first diagnosed at an advanced stage. In the United States, diagnosis occurs at a localized, regional, and distant stage in 28 %, 32 %, and 40 %, respectively, of stomach cancer cases, and in 25 %, 29 %, and 31 %, respectively of esophageal cancer cases [2], [3]. For localized, regional, and metastatic disease, five-year survival is 64 %, 28.2 %, and 5.3 %, respectively, for stomach cancer, and 46.7 %, 25.1 %, and 4.8 %, respectively, for esophageal cancer [2], [3]. Treatment for STEC is selected based on disease stage [4], [5]. Surgery can be curative but is offered mainly in early disease stages. Chemotherapy and chemoradiotherapy provide an added survival benefit to surgery in early-stage disease and are offered without surgery in later disease stages. Targeted therapies (e.g., Trastuzumab, an inhibitor of human epidermal growth factor receptor 2) improve survival in STEC and are increasingly being used in STEC treatment [6], and immunotherapy and other emerging therapies continue to be evaluated for improvement in STEC survival [4], [5].
Biomarkers associated with STEC survival are potential targets for designing new STEC treatments to improve patient survival [7], [8]. MicroRNAs (miRNAs) function as oncogenes or tumor suppressor genes in STEC [6], [7] and have been investigated as biomarkers of STEC diagnosis and prognosis [9], [10]. Roles for miRNAs in STEC progression and survival have been described in several reports. For example, low levels of miR148a, a miRNA that suppresses cell invasion and migration, are associated with advanced clinical stage and poor prognosis in stomach cancer [11]. MiR-616-3p promotes angiogenesis and metastasis and is correlated with poor prognosis in stomach cancer [12]. Elevated miR-21 expression is linked to lymph node metastasis [13] and poor prognosis [14] in esophageal cancer. MiR-375 targets proteins involved in cancer cell proliferation and invasion [15], and its downregulation is associated with advanced cancer staging and poor prognosis in esophageal squamous cell carcinoma [16]. Aberrant miRNA expression has also been identified in STEC. Hwang et al. identified miRNAs, including miR-601, miR-107, miR-18a, miR-370, miR-300 and miR-96 that were significantly expressed in early gastric cancers when compared to normal samples [17]. A serum biomarker miRNA panel consisting of 12 miRNAs was developed for risk assessment in patients with gastric cancer [18]. Furthermore, several dysregulated miRNAs have been found in esophageal tumors that regulate carcinogenesis [19], [20]. A quantitative RT-qPCR study on patients with esophageal carcinoma revealed three miRNAs, including miR-34a-5p, miR-148a-3p and miR-181a-5p that were associated with the cancer progression [21].
In most studies, associations between miRNAs and STEC survival have been based on results from a single study sample assessed using the log-rank test to compare Kaplan-Meier survival curves or Cox proportional hazards regression analysis [22], [23], [24], [25]. A few other studies have employed discovery and validation stages in their design to increase the strength of the evidence supporting associations between miRNAs and STEC survival. These include studies that have identified differentially expressed miRNAs in STEC in the discovery stage and tested for association between the miRNAs and survival in an independent STEC study sample in the validation stage [26], [27], [28], [29], [30], [31]. Machine learning methods are also being applied to identify miRNAs associated with STEC survival. In a study of esophageal squamous cell carcinoma, a recursive feature elimination-support vector machine algorithm along with LASSO Cox proportional hazards regression was used to identify miRNAs associated with survival and build a prognostic model in a training sample, and the prognostic model was shown to correlate with survival in an independent, test sample [32]. While these previous reports indicate that miRNAs have potential clinical value as biomarkers of prognosis in STEC, they have not addressed whether miRNAs can predict STEC survival time in individual patients.
To design a personalized survival prediction model, it is necessary to identify biomarkers that show a robust association with survival in STEC patients. Accordingly, this study aimed to develop a genetic algorithm (GA)-based survival estimation method (GASE) to identify a survival-associated miRNA signature and estimate survival time in patients with STEC. A genetic algorithm (GA)-based survival estimation method (GASE) is proposed for estimating the survival time in STEC patients using miRNA expression profiles. GASE was developed using support vector regression (SVR) that incorporates an optimal feature selection algorithm inheritable bi-objective combinatorial genetic algorithm (IBCGA) [33]. The identified miRNA signature was analyzed further to explore miRNA association with STEC. The system overview of GASE is shown in the graphical abstract.
2. Material and methods
The miRNA expression profiles of patients with STEC were retrieved from The Cancer Genome Atlas (TCGA) database. These data were generated using an Illumina Hiseq 2000 sequencing platform. The number of patients with STEC in the initial dataset was 628. After excluding the patients without survival information and those whose survival time was less than 30 days, the final dataset consisted of 123 patients with miRNA expression profiles and clinical data, including days to death. Each miRNA expression profile consisting of 500 miRNAs was used for the survival estimation procedure. For the independent validation, we used a cohort of 393 patients who were alive with STEC at last follow-up in the TCGA.
2.1. Survival estimation method GASE
The GASE’s two primary objectives were to estimate the survival time and simultaneously identify the miRNA signature associated with survival in patients with STEC. GASE was developed using SVR and an optimal feature selection algorithm IBCGA. The optimization technique implemented in GASE was adopted from previous studies [34], [35], [36]. SVM is a supervised machine learning method, which has demonstrated good prediction capability in solving classification and regression problems in various biomedical fields, especially in cancer genomics [37]. SVR uses a nonlinear transformation to find the relation between input and output variables by generating a hyperplane that optimally fits in the high dimensional space and carries out the regression function [38]. The tuning of the parameters C, γ, and ν determine the performance of SVR; hence parameter tuning plays a vital role in the SVR modeling process. The minimization of the loss function can be optimized using the following objective function for the given input data points.
| (1) |
where ||w|| is the magnitude of the vector to the surface, C is a regularization parameter, ξi and ξi* are slack variables, ξi ≥ 0, ξi* ≥ 0, and i = 1,2,…N.
The optimal parameters of GASE were tuned based on an intelligent evolutionary algorithm (IEA) [39]. In the optimization process, IBCGA [33] was used to identify a small set of miRNAs while maximizing the fitness function in terms of squared correlation coefficient. GASE prediction performance was evaluated using two metrics, squared correlation coefficient and mean absolute error. IBGCA effectively solves bi-objective combinatorial problems where a small set of informative features will be selected from a large number of candidate features. The applications of IBCGA in identifying biomarkers in cancer research have been demonstrated in previous studies [34], [35], [36], [40], [41]. In the optimal feature selection process, all the candidate features were encoded into binary variables, including the parameters C, γ, and ν of the SVR. The detailed steps involved in IBCGA can be found in the supplementary methods. After identifying the miRNA signature, main effect difference (MED) [42] analysis was used to prioritize the miRNAs of the signature based on their contribution to the prediction performance.
2.2. Feature appearance score
To ensure robustness, we performed 50 independent runs of GASE and selected one feature set with the highest appearance score for the analysis. The feature appearance score (FAS) indicates the frequency of the features that appeared in the 50 independent runs. A feature set with a more significant appearance score suggests that the feature frequency in that particular set is higher when compared to other features across the independent runs. There are St features in the t-th signature. The frequency score for each feature m presented in the miRNA signatures can be calculated as follows.
| (2) |
where m is the miRNA of the t-th signature.
2.3. LASSO and elastic net
To evaluate the estimation ability of GASE, we compared the prediction performance with some standard regression methods, including ridge [43], Lasso [44] and elastic net [45]. We used the miRNA expression profiles and survival time of 123 patients with STEC as input. The minimum λ was selected after 100 independent runs of LASSO and elastic net using 10-CV.
2.4. Strong evidence on miRNA-gene target interaction
To identify the target genes of the selected miRNAs, we used the miRTarBase (9.0 beta) database [46] to extract the experimentally verified microRNA-target interactions (MTIs) with strong evidence, which are validated by reporter assay, Western blot, and qPCR.
2.5. Gene set enrichment test
Gene-set libraries are used to organize accumulated knowledge about the function of groups of genes. We used Enrichr [47], [48], which is a web-based application that includes the latest gene-set libraries, to perform gene-set enrichment analysis. We evaluated the ability of Enrichr to rank terms from gene-set libraries by combining the p-value computed using Fisher’s exact test with the z-score of the deviation from the expected rank by multiplying these two numbers as follows:
where z = z-score and p = p-value.
This study used six Gene-set libraries, including 1) WikiPathway Human 2021 [49], 2) Kyoto Encyclopedia of Genes and Genomes (KEGG), 3) MSigDB Hallmark [50], 4) Gene Ontology Molecular Function 2021 [51], 5) Gene Ontology Biological Process, and 6) Gene Ontology Cellular Component.
3. Results
3.1. GASE prediction performance
We used a survival estimation method, GASE, to identify a miRNA signature and estimate the survival time in patients with STEC. One hundred and twenty-three patients with miRNA expression profiles were retrieved from the TCGA database. GASE identified 27 miRNAs as a survival miRNA signature and estimated the survival time with a mean squared correlation coefficient (R2) of 0.80 ± 0.01 and a mean absolute error (MAE) of 0.44 ± 0.25 years between actual and estimated survival times.
A robust miRNA signature was selected by measuring the frequency appearance score (FAS) using 50 independent runs of GASE. A miRNA signature with the highest FAS indicates higher frequencies of miRNAs in the signature across the independent runs of GASE. The mean FAS obtained for the independent runs was 15.55 ± 1.45, while the highest FAS was 18.85 (shown in Supplementary Fig. S1 and Supplementary Table S1). The feature set with the highest FAS was selected for the analysis. This feature set obtained a R2 of 0.80 and a MAE of 0.43 years between actual and estimated survival times, and selected 27 miRNAs as a signature to estimate survival time in patients with STEC.
3.2. Prediction performance comparison and validation
Next, we compared GASE with some standard machine learning methods on their performance to predict survival times. The machine learning methods used in the comparison included ridge regression, least absolute shrinkage and selection operator (Lasso) and elastic net. Ridge regression obtained a R2 of 0.77 and a MAE of 0.54 years between actual and estimated survival times. Lasso obtained a R2 of 0.51 and a MAE of 0.69 years between actual and estimated survival times, and elastic net obtained a R2 of 0.50 and a MAE of 0.71 years between actual and estimated survival times, respectively. In comparison, GASE obtained a highest R2 of 0.83 and a MAE of 0.41 years between actual and estimated survival times (Table 1). The results indicated that the performance of GASE was better than that of the standard machine learning methods. The correlation plots of GASE and the other machine learning methods are shown in Supplementary Fig. S2A-D.
Table 1.
Prediction performance of GASE.
| Method | R2 | MAE (years) | Features selected |
|---|---|---|---|
| Ridge regression | 0.77 | 0.54 | 485 |
| LASSO | 0.51 | 0.69 | 28 |
| Elastic net | 0.50 | 0.71 | 30 |
| GASE-FAS | 0.80 | 0.43 | 27 |
| GASE-Best | 0.83 | 0.41 | 32 |
| GASE-Mean | 0.80 ± 0.01 | 0.44 ± 0.25 | 33.44 ± 3.59 |
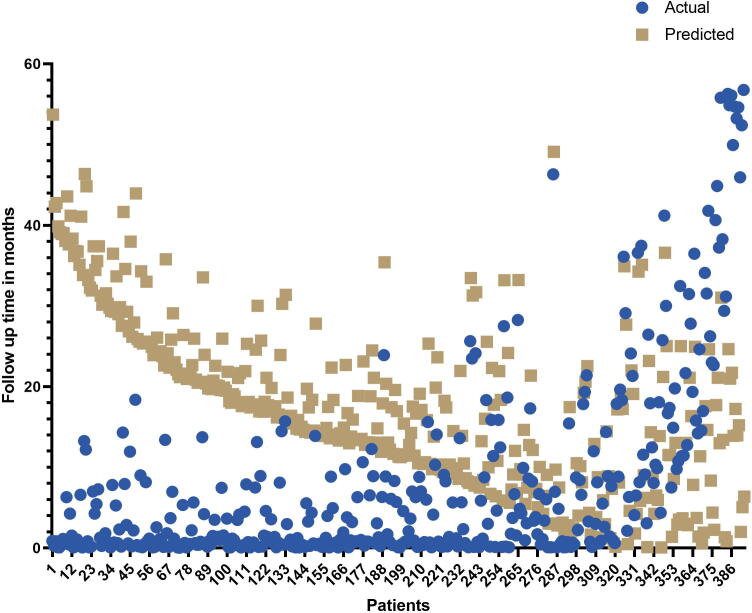
Next, the estimation ability of GASE was validated using a validation dataset consisting of 393 patients with STEC along with their follow-up times. The follow-up times of these patients were in the range of 0.3–56 months. We attempted to estimate the survival times of these patients using the GASE prediction model. The mean follow-up times observed in patients with STEC was 8.09 ± 12.09 months. The mean predicted survival time of these patients was 17.74 ± 10.50 months. GASE achieved an accuracy of 80.41 % for estimating the survival times of patients whose estimated survival times were higher than the follow-up times (mean follow-up time 4.0 ± 5.9 months). The mean estimated survival time of the 316 patients was 19.10 ± 10.28 months, and a mean prediction error of 12.15 months was obtained for the remaining patients. The results could be interpreted as follows: an estimated survival time that was higher than the patient’s follow-up time was considered as a correct prediction, whereas an estimated survival time that was lower than the follow-up time was a considered a prediction error. The follow-up and estimated survival times of these patients are shown in Fig. 1.
Fig. 1.
The GASE prediction performance on an independent test cohort of 393 patients with follow-up times.
3.3. Ranking of miRNA signature
The miRNAs of the identified miRNA signature were ranked based on their contribution towards estimating the survival time using main effect difference (MED) [42] analysis. A higher MED score represents greater contribution towards the prediction of survival time. A miRNA with a higher MED score indicates superior prediction ability towards the survival time estimation, whereas a lower-scoring miRNA indicates a smaller contribution to survival time estimation. The top 10 ranked miRNAs according to the MED analysis, include hsa-miR-760, hsa-miR-767-5p, hsa-miR-1301-3p, hsa-miR-891a-5p, hsa-miR-532-5p, hsa-miR-29a-5p, hsa-miR-16-5p, hsa-miR-130a-5p, hsa-miR-329-3p, and hsa-miR-496 (Table 2). The prioritization of miRNAs based on their contribution to the survival estimation is shown in Fig. 2.
Table 2.
Ranking of miRNA signature and corresponding MED scores.
| Rank | miRNA | MIMAT-ID | MED |
|---|---|---|---|
| 1 | hsa-miR-760 | MIMAT0004957 | 1.728135 |
| 2 | hsa-miR-767-5p | MIMAT0003882 | 1.480966 |
| 3 | hsa-miR-1301-3p | MIMAT0005797 | 1.344602 |
| 4 | hsa-miR-891a-5p | MIMAT0004902 | 1.14225 |
| 5 | hsa-miR-532-5p | MIMAT0002888 | 1.139153 |
| 6 | hsa-miR-29a-5p | MIMAT0004503 | 0.887408 |
| 7 | hsa-miR-16-5p | MIMAT0000069 | 0.88658 |
| 8 | hsa-miR-130a-5p | MIMAT0004593 | 0.863724 |
| 9 | hsa-miR-329-3p | MIMAT0001629 | 0.844311 |
| 10 | hsa-miR-496 | MIMAT0002818 | 0.818043 |
| 11 | hsa-miR-20a-3p | MIMAT0004493 | 0.724058 |
| 12 | hsa-miR-125a-5p | MIMAT0000443 | 0.63757 |
| 13 | hsa-miR-181b-5p | MIMAT0000257 | 0.590379 |
| 14 | hsa-miR-675-3p | MIMAT0006790 | 0.578151 |
| 15 | hsa-miR-9-5p | MIMAT0000441 | 0.484588 |
| 16 | hsa-miR-664a-5p | MIMAT0005948 | 0.425219 |
| 17 | hsa-miR-93-5p | MIMAT0000093 | 0.364274 |
| 18 | hsa-miR-30e-5p | MIMAT0000692 | 0.355408 |
| 19 | hsa-miR-376c-3p | MIMAT0000720 | 0.345478 |
| 20 | hsa-miR-326 | MIMAT0000756 | 0.312151 |
| 21 | hsa-miR-193a-5p | MIMAT0004614 | 0.275742 |
| 22 | hsa-miR-532-3p | MIMAT0004780 | 0.268942 |
| 23 | hsa-miR-625-3p | MIMAT0004808 | 0.259763 |
| 24 | hsa-miR-106a-5p | MIMAT0000103 | 0.213424 |
| 25 | hsa-let-7 g-5p | MIMAT0000414 | 0.152833 |
| 26 | hsa-let-7f-5p | MIMAT0000067 | 0.04358 |
| 27 | hsa-miR-193b-5p | MIMAT0004767 | 0.010963 |
Fig. 2.
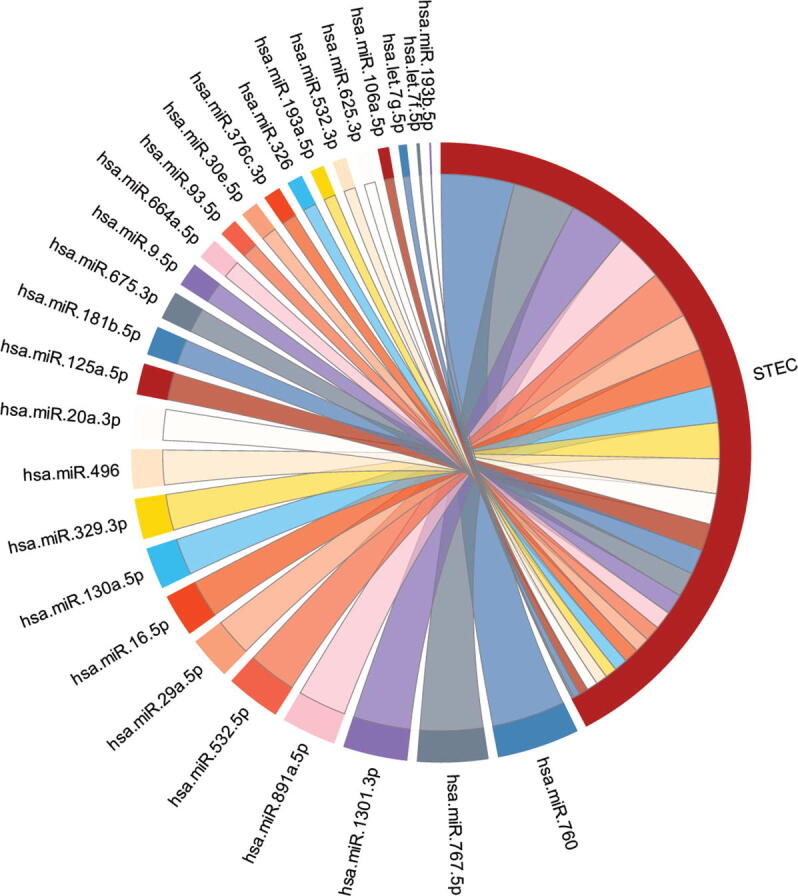
Chord diagram showing the prioritization of miRNAs of the signature based on their survival estimation ability in stomach and esophageal carcinoma. The size of the line is proportional to the percent contribution towards the survival estimation.
3.4. Diagnosis prediction
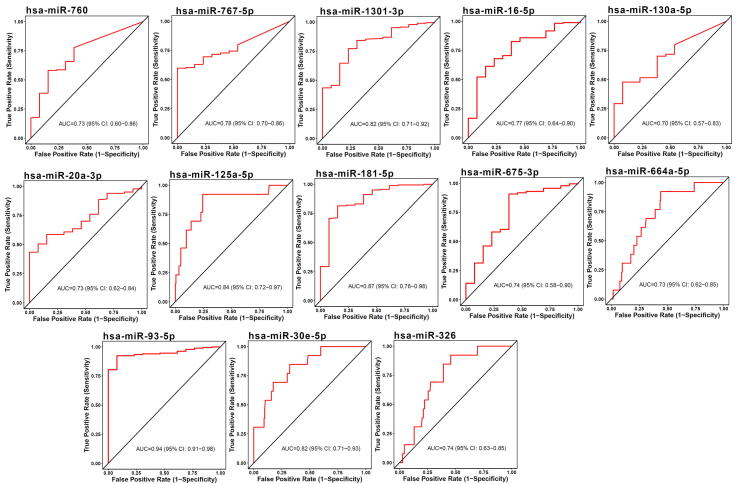
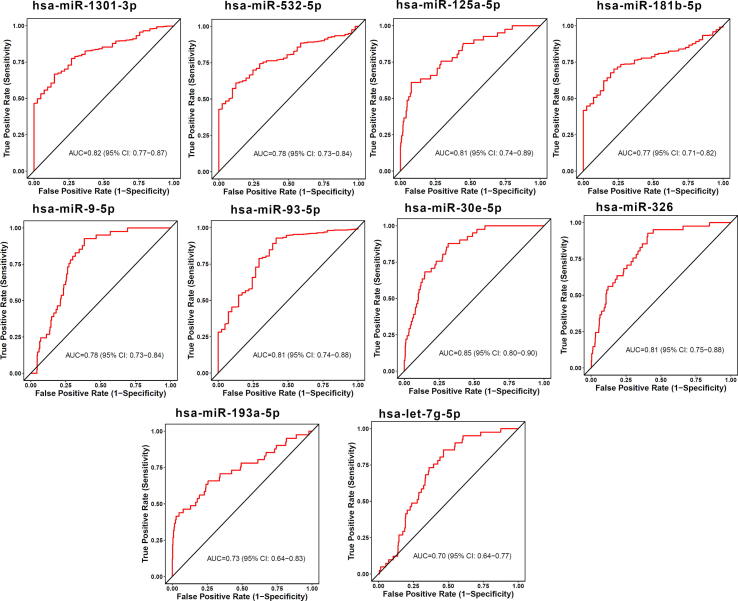
The diagnostic ability of the identified miRNA signature was measured by distinguishing healthy and STEC patients using CancerMiRNome database [52]. The individual miRNAs that compose the miRNA signature had AUCs in a range of 0.49–0.94 for distinguishing healthy from STEC patients, as shown in Table 3. Among the signature miRNAs, 13 miRNAs, including hsa-miR-93-5p, hsa-miR-1 81b-5p, hsa-miR-125a-5p, hsa-miR-1301-3p, hsa-miR-30e-5p, hsa-miR-767-5p, hsa-miR-16-5p, hsa-miR-675-3p, hsa-miR-326, hsa-miR-760, hsa-miR-20a-3p, hsa-miR-664a-5p, and hsa-miR-130a-5p were good diagnostic predictors of esophageal carcinoma (ESCA) (AUC ≥ 0.70), as shown in Fig. 3. Ten miRNAs, including hsa-miR-30e-5p, hsa-miR-1301-3p, hsa-miR-125a-5p, hsa-miR-93-5p, hsa-miR-326, hsa-miR-532-5p, hsa-miR-9-5p, hsa-miR-181b-5p, hsa-miR-193a-5p, and hsa-let-7 g-5p were good diagnostic predictors of stomach adenocarcinoma (STAD) (AUC ≥ 0.7), as shown in Fig. 4.
Table 3.
Diagnosis prediction of patients with STEC using the miRNA signature.
| miRNAs | ESCA-AUC | STAD-AUC |
|---|---|---|
| hsa-miR-760 | 0.73 | 0.60 |
| hsa-miR-767-5p | 0.78 | 0.63 |
| hsa-miR-1301-3p | 0.82 | 0.82 |
| hsa-miR-891a-5p | 0.52 | 0.53 |
| hsa-miR-532-5p | 0.59 | 0.78 |
| hsa-miR-29a-5p | 0.57 | 0.64 |
| hsa-miR-16-5p | 0.77 | 0.49 |
| hsa-miR-130a-5p | 0.70 | 0.56 |
| hsa-miR-329-3p | 0.55 | 0.53 |
| hsa-miR-496 | 0.60 | 0.59 |
| hsa-miR-20a-3p | 0.73 | 0.62 |
| hsa-miR-125a-5p | 0.84 | 0.81 |
| hsa-miR-181b-5p | 0.87 | 0.77 |
| hsa-miR-675-3p | 0.74 | 0.54 |
| hsa-miR-9-5p | 0.49 | 0.78 |
| hsa-miR-664a-5p | 0.73 | 0.69 |
| hsa-miR-93-5p | 0.94 | 0.81 |
| hsa-miR-30e-5p | 0.82 | 0.85 |
| hsa-miR-376c-3p | 0.57 | 0.66 |
| hsa-miR-326 | 0.74 | 0.81 |
| hsa-miR-193a-5p | 0.59 | 0.73 |
| hsa-miR-532-3p | 0.52 | 0.68 |
| hsa-miR-625-3p | 0.68 | 0.5 |
| hsa-miR-106a-5p | 0.62 | 0.51 |
| hsa-let-7 g-5p | 0.64 | 0.7 |
| hsa-let-7f-5p | 0.50 | 0.62 |
| hsa-miR-193b-5p | 0.58 | 0.58 |
Abbreviation: ESCA-Esophageal carcinoma, STAD-Stomach adenocarcinoma, AUC-Area under the receiver operating curve.
Fig. 3.
Diagnosis prediction ability of miRNAs was evaluated in ESCA using ROC curves.
Fig. 4.
Diagnosis prediction ability of miRNAs was evaluated in STAD using ROC curves.
3.5. Expression differences of the miRNA signature
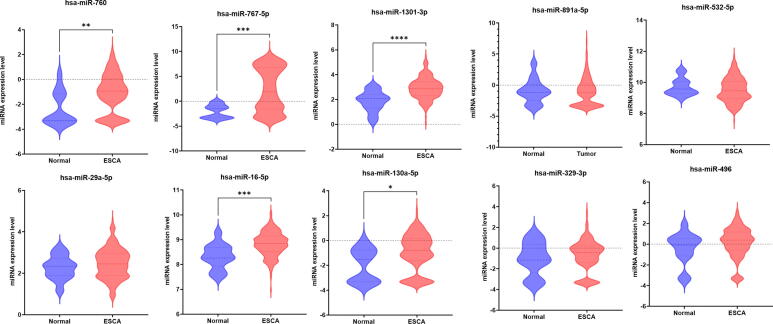
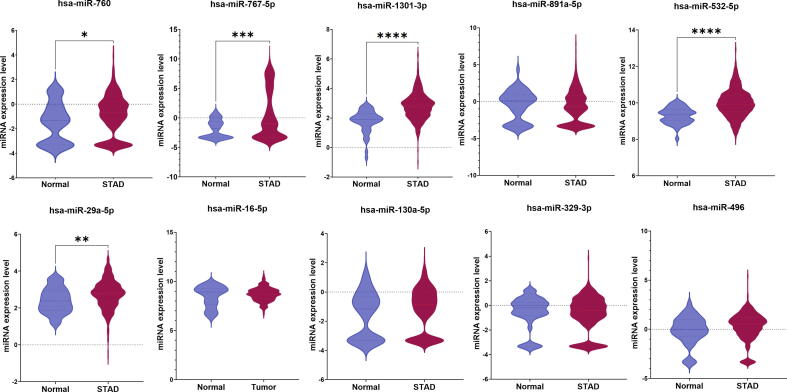
Expression difference analysis was performed to measure the significance in the expression levels of the identified miRNA signature between normal and tumor tissues of ESCA and STAD patients using the CancerMiRNome database [52]. There were 14 miRNAs, including hsa-miR-625-3p, hsa-miR-664a-5p, hsa-miR-326, hsa-miR-130a-5p, hsa-miR-20a-3p, hsa-miR-675-3p, hsa-miR-760, hsa-miR-16-5p, hsa-miR-767-5p, hsa-miR-1301-3p, hsa-miR-125a-5p, hsa-miR-181b-5p, hsa-miR-93-5p, and hsa-miR-30e-5p which showed a significant difference (p < 0.05) between normal and ESCA samples (Table 4). There were 19 miRNAs, including hsa-miR-664a-5p, hsa-miR-767-5p, hsa-let-7 g-5p, hsa-let-7f-5p, hsa-miR-376c-3p, hsa-miR-29a-5p, hsa-miR-760, hsa-miR-1301-3p, hsa-miR-532-5p, hsa-miR-20a-3p, hsa-miR-125a-5p, hsa-miR-181b-5p, hsa-miR-9-5p, hsa-miR-93-5p, hsa-miR-30e-5p, hsa-miR-326, hsa-miR-193a-5p, hsa-miR-532-3p, and hsa-miR-193b-5p, that had significantly different expression between normal and STAD patients (Table 4). The top 10 ranked miRNAs and their expression differences between healthy and ESCA and STAD patients are shown in Fig. 5, Fig. 6, respectively.
Table 4.
Expression differences of the miRNA signature between normal and tumor tissues.
| miRNA signature | Normal vs ESCA | Normal vs STAD |
|---|---|---|
| p-value | p-value | |
| hsa-miR-760 | 0.003 | 0.0297 |
| hsa-miR-767-5p | 0.0004 | 0.0003 |
| hsa-miR-1301-3p | 0.0001 | <0.0001 |
| hsa-miR-891a-5p | 0.8733 | 0.6737 |
| hsa-miR-532-5p | 0.3987 | <0.0001 |
| hsa-miR-29a-5p | 0.3481 | 0.008 |
| hsa-miR-16-5p | 0.0005 | 0.0911 |
| hsa-miR-130a-5p | 0.0115 | 0.1496 |
| hsa-miR-329-3p | 0.4576 | 0.6845 |
| hsa-miR-496 | 0.1857 | 0.1031 |
| hsa-miR-20a-3p | 0.005 | <0.0001 |
| hsa-miR-125a-5p | <0.0001 | <0.0001 |
| hsa-miR-181b-5p | <0.0001 | <0.0001 |
| hsa-miR-675-3p | 0.0033 | 0.1292 |
| hsa-miR-9-5p | 0.9717 | <0.0001 |
| hsa-miR-664a-5p | 0.0177 | 0.0002 |
| hsa-miR-93-5p | <0.0001 | <0.0001 |
| hsa-miR-30e-5p | <0.0001 | <0.0001 |
| hsa-miR-376c-3p | 0.5389 | 0.0033 |
| hsa-miR-326 | 0.0117 | <0.0001 |
| hsa-miR-193a-5p | 0.3494 | <0.0001 |
| hsa-miR-532-3p | 0.8464 | <0.0001 |
| hsa-miR-625-3p | 0.023 | 0.9569 |
| hsa-miR-106a-5p | 0.1783 | 0.9017 |
| hsa-let-7 g-5p | 0.3872 | 0.0003 |
| hsa-let-7f-5p | 0.7968 | 0.0026 |
| hsa-miR-193b-5p | 0.343 | <0.0001 |
Abbreviation: ESCA-Esophageal carcinoma, STAD-Stomach adenocarcinoma.
Fig. 5.
Comparison of expression of the top 10 ranked miRNAs between normal and ESCA samples using boxplot representation (* indicates p < 0.05).
Fig. 6.
Comparison of expression of the top 10 ranked miRNAs between normal and STAD samples using boxplot representation. (* indicates p < 0.05).
3.6. MiRNA-gene target enrichment analysis
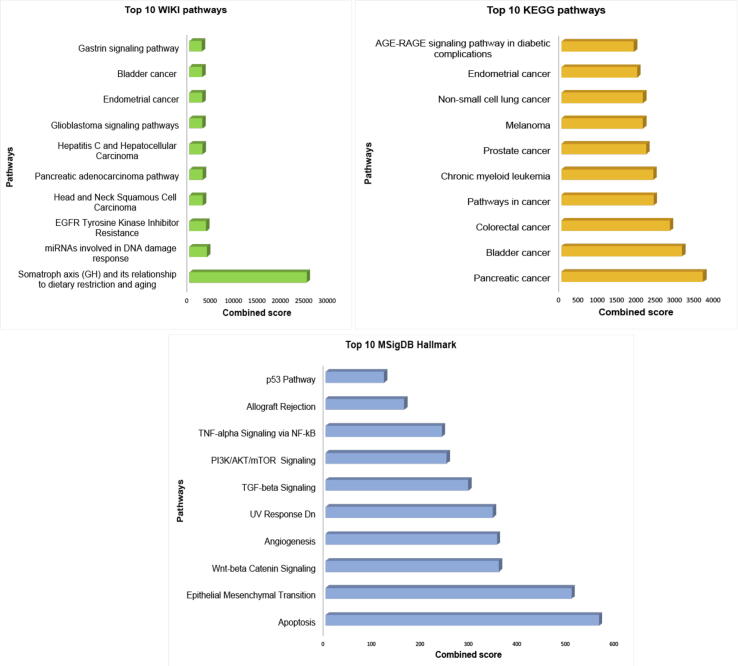
There were 558 miRNA target interactions (MTI) with strong evidence, which included 32 miRNAs and 352 target genes from miRTarBase (Supplementary Table S2). We performed gene-set enrichment analysis using three pathway libraries: WikiPathway, KEGG, and MSigDB Hallmark, shown in Fig. 7. The highly enriched pathways in WikiPathway, KEGG, and MSigDB Hallmark were the somatotrophic axis and its relationship to dietary restriction and aging (WP4186) (adjusted p-value: 1.34E-10, Odds ratio: 117888, combined score: 2,862,498), pancreatic cancer (adjusted p-value:1.54E-34, Odds ratio:44.55, combined score: 3655.72), and apoptosis (adjusted p-value:5.17E-20, Odds ratio: 12.68, combined score: 563.12), respectively, shown in Supplementary Tables S3-S5. Additionally, the miRNA signature-gene interaction network was built using miRTarBase [46], TarBase V8. [53] and miRecords [54]. There were 28,057 edges associated with 10,525 genes. We reduced the low priority edges using the shortest path network measures [55]. The final network, consisting of 832 edges 93 targeted genes, is shown in Supplementary Fig. S3.
Fig. 7.
The pathways enrichment analysis of miRNA signature targeted genes in three categories, (A) wiki pathways, (B) KEGG pathways, and (C) MSigDB hallmark.
The Gene Ontology (GO) annotations of the target genes were in three categories: biological process, molecular function, and cellular component. The highly enriched pathways for biological process, molecular function, and cellular component were positive regulation of smooth muscle cell apoptosis process (GO:0034393), I-SMAD binding (GO:0070411), and serine/threonine protein kinase complex (GO:1902554), respectively, as shown in Supplementary Figs. S4-S6 and Supplementary Table S6.
3.7. MiRNAs in cancers
The roles of the top 10 ranked miRNAs in various diseases and cancers were examined using the Human microRNA Disease Database (HMDD v3.2) [56], miRTarbase, and by reviewing the scientific literature. The information from these resources indicate that the top 10 ranked miRNAs are involved in STEC. A quantitative real-time PCR analysis reported that hsa-miR-760 is significantly downregulated in ESCA tissues and cell lines, suggesting that this miRNA could be used as a prognostic indicator [57]. Significant differential expression of hsa-miR-769-5p was observed in ESCA tissue when compared to the normal tissues [58]. Over-expression of hsa-miR-1301-3p induces cell proliferation and tumorigenesis in gastric cancer tissues [59]. Wu et al. reported the differential expression of hsa-miR-1301-3p in ESCA, suggesting that this miRNA could be used as a prognostic biomarker for ESCA [60]. Zhang and colleagues reported the downregulation of hsa-miR-532-5p in gastric cancer cells, and its expression is associated with poorer survival in patients with gastric cancer [61]. Tokumaru and colleagues demonstrated the association of hsa-miR-29a with overall survival in patients with gastric cancer, and lower expression of hsa-miR-29a worsens the overall survival in patients with gastric cancer [62]. Hsa-miR-16-5p has been utilized as a prospective biomarker for prognosis prediction in patients with gastric cancer and ESCA [63], [64]. Hsa-miR-130a-5p affects cell growth, migration and invasion by targeting cannabinoid receptor 1 in gastric cancer cells [65]; it also deregulates PTEN and controls malignant cell survival and tumor growth in multiple cancers [66]. Hsa-miR-329-3p acts as a tumor suppressor by targeting T lymphoma invasion and metastasis in gastric cancer cells and could be utilized as potential therapeutic target [67]. Hsa-miR-496 is downregulated in gastric cancer cell lines, and it inhibits cell proliferation via targeting Lyn kinase in gastric cancer cell lines [68]. Among the top 10 ranked miRNAs, the roles of two miRNAs, hsa-miR-769-5p and hsa-miR-891a-5p, have not been reported previously in either STAD or ESCA.
Additionally, a miRNA-disease network was constructed for the miRNA signature using miRNet 2.0 [55]. The miRNAs of the signature were observed to be involved in several diseases. In the miRNA-disease association network, there were 12 nodes (miRNAs) with 132 edges associated with 85 diseases, shown in Supplementary Fig. S7.
4. Discussion
MiRNAs provide a way to explore disease mechanisms in various cancers, including STEC. The clinical applications of miRNAs in cancer rely on identifying miRNA signatures as potential biomarkers and developing miRNA-target based therapeutics. Accordingly, we developed a survival time estimation method, GASE, to identify a miRNA signature that was correlated with STEC patient survival. Computational methods for feature selection often suffer from issues related to data quality and high dimensionality, especially when dealing with biomedical data. To address the challenges to identifying the right biomarker, we used an optimal feature selection algorithm, IBCGA, which is good at identifying s small number of important features from a large number of candidate features. The optimization method was previously utilized to estimate the survival time in various cancers [34], [35], [36]. In this study, we exclusively focused on identifying a miRNA signature in patients with STEC. The proposed method, GASE, identified 27 miRNAs as a survival miRNA signature and performed better than standard machine learning methods in estimating survival time. Our evaluation of the diagnostic ability of the identified miRNAs revealed that 13 miRNAs were good diagnostic predictors (AUC ≥ 0.7) in ESCA and 10 miRNAs in STAD. The differential expression analysis between tumor and normal samples from patients with STEC revealed that several miRNAs had significantly different expression between tumor and normal samples. Further, previous reports provide evidence supporting the importance of the top 10 ranked miRNAs of the signature in STEC.
The miRNA-gene target interaction analysis showed that the target genes were highly enriched in the somatotrophic axis and its relationship to dietary restriction and the aging (WP4186) pathway. The somatotrophic axis in mammals involves signaling by growth hormone (GH), which is produced by the anterior pituitary, and its secondary mediator, insulin-like growth factor 1 (IGF-1). In a previous study, growth hormone–releasing hormone and its receptor (GHRH-R) were found primarily in the anterior pituitary gland, gastric cancers, other solid tumors, and lymphomas. Increased levels of GHRH-R in tumor samples from patients with gastric cancer are associated with poor outcomes [69]. Another important enriched pathway of the miRNA signature was transforming growth factor beta (TGF-β) signaling pathway. TGF-β is a cytokine that participates in both physiological and pathological processes including tumorigenesis [70]. During tumor progression, TGF-β signaling regulates the immune/inflammatory response and the tumor microenvironment. It also regulates tumor growth, epithelial-mesenchymal transition (EMT), and cancer cell stemness depending on tumor stage and cellular context [71]. EMT is also an enriched pathway from MSigDB Hallmark (adjusted p-value: 8.24E-19, Odds ratio: 11.13, combined score: 506.81), which is consistent with this biological mechanism. Abnormal TGF-β signaling has been associated with progression of gastrointestinal cancer [72], which includes esophageal, gastric, liver, colorectal, and pancreatic carcinomas that, collectively, are major causes of cancer-related deaths worldwide [73]. Several TGF-β-based therapeutics have been developed for the treatment of gastrointestinal cancers and have displayed efficacy in clinical trials [74], [75]. Additional support for the role of TGF-β signaling in STEC was obtained from the GO annotation analysis, which showed that I-SMAD binding (GO:0070411) was enriched in the GO molecular function category (adjusted p-value: 1.03E-06). Nuclear accumulation of active SMAD complexes is crucial for the transduction of TGF-β superfamily signals from transmembrane receptors to the nucleus.
The top hits for gene target enrichment analysis also indicated that the miRNA signature was related to miRNAs involved in DNA damage response, epidermal growth factor receptor tyrosine kinase inhibitor resistance, apoptosis, Wnt/beta-catenin signaling, and angiogenesis. DNA damage response pathways are known to be related to therapy resistance in STEC [76], [77], and resistance to epidermal growth factor receptor tyrosine kinase inhibitors are relevant to survival in STEC, consistent with the use of epidermal growth factor receptor tyrosine kinase inhibitors as targeted therapy in STEC [78], [79]. The Wnt/beta-catenin signaling pathway has been implicated in cancer progression in STEC [80], and the dysregulation of apoptosis and angiogenesis are known to promote tumor growth [81], [82]. This suggests the miRNAs in the signature and the putative gene targets of these miRNAs are possible molecular targets for exploitation in the pursuit to create new therapies for STEC.
In addition to being associated with survival, the miRNAs in the signature could discriminate between healthy and STEC patients, and were differentially expressed between the healthy and tumor tissues of patients with STEC. This suggests that the capability of these miRNAs to function as prognostic or diagnostic biomarkers. Further investigation is needed to determine the utility of the miRNA signature as a prognostic biomarker for monitoring response to therapy or predicting survival after therapy in STEC patients and as a biomarker for early STEC diagnosis. Other questions for study are whether the miRNA signature can perform as a biomarker in STEC of different types and stages, and whether the miRNA signature can be detected in blood at a level of accuracy comparable to that in tumor tissue (to allow for the possibility of performing liquid biopsies for biomarker detection).
In conclusion, a better understanding of the miRNA signature in survival predictions will aid in developing treatment strategies for STEC. We anticipate that the miRNA signature identified here could help in understanding the roles of miRNAs in STEC and developing miRNA-based cancer therapeutics.
Funding
This work was supported in part by the Marshfield Clinic Research Institute, Marshfield, WI. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
S.Y.S. designed the system, carried out the detail study and supervised the study. S.Y.S, M.T, T.C, P.A, S.K.S, A.B and S.Y.H, participated in data analysis, manuscript preparation and discussed the results. All authors have read and approved the final manuscript.
Availability of data and materials
All the data used in this analysis can be found on the TCGA data portal [https://portal.gdc.cancer.gov/].
Ethics approval and consent to participate
Not applicable.
Consent to publish
Not applicable.
CRediT authorship contribution statement
Srinivasulu Yerukala Sathipati: Conceptualization, Data curation, Writing – original draft, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Supervision. Ming-Ju Tsai: Validation, Visualization, Formal analysis. Tonia Carter: Formal analysis, Data curation, Writing - review & editing. Patrick Allaire: Formal analysis. Sanjay K Shukla: Formal analysis. Afshin Beheshti: Formal analysis, Writing - review & editing. Shinn-Ying Ho: Formal analysis.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.08.025.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Sung H., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Jim M.A., et al. Stomach cancer survival in the United States by race and stage (2001–2009): Findings from the CONCORD-2 study. Cancer. 2017;123(Suppl 24):4994–5013. doi: 10.1002/cncr.30881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uhlenhopp D.J., Then E.O., Sunkara T., Gaduputi V. Epidemiology of esophageal cancer: update in global trends, etiology and risk factors. Clin J Gastroenterol. 2020;13:1010–1021. doi: 10.1007/s12328-020-01237-x. [DOI] [PubMed] [Google Scholar]
- 4.Sexton R.E., Al Hallak M.N., Diab M., Azmi A.S. Gastric cancer: a comprehensive review of current and future treatment strategies. Cancer Metastasis Rev. 2020;39:1179–1203. doi: 10.1007/s10555-020-09925-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Short M.W., Burgers K.G., Fry V.T. Esophageal cancer. Am Fam Physician. 2017;95:22–28. [PubMed] [Google Scholar]
- 6.Bang Y.J., et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–697. doi: 10.1016/s0140-6736(10)61121-x. [DOI] [PubMed] [Google Scholar]
- 7.Tsai M.M., et al. Potential diagnostic, prognostic and therapeutic targets of MicroRNAs in human gastric cancer. Int J Mol Sci. 2016;17 doi: 10.3390/ijms17060945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma P., Sharma R. miRNA-mRNA crosstalk in esophageal cancer: From diagnosis to therapy. Crit Rev Oncol Hematol. 2015;96:449–462. doi: 10.1016/j.critrevonc.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Jiang C., Chen X., Alattar M., Wei J., Liu H. MicroRNAs in tumorigenesis, metastasis, diagnosis and prognosis of gastric cancer. Cancer Gene Ther. 2015;22:291–301. doi: 10.1038/cgt.2015.19. [DOI] [PubMed] [Google Scholar]
- 10.Zarrilli G., et al. miRNAs involved in esophageal carcinogenesis and miRNA-related therapeutic perspectives in esophageal carcinoma. Int J Mol Sci. 2021;22:3640. doi: 10.3390/ijms22073640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang S.H., et al. microRNA-148a suppresses human gastric cancer cell metastasis by reversing epithelial-to-mesenchymal transition. Tumour Biol. 2013;34:3705–3712. doi: 10.1007/s13277-013-0954-1. [DOI] [PubMed] [Google Scholar]
- 12.Wu Z.H., et al. MiR-616-3p promotes angiogenesis and EMT in gastric cancer via the PTEN/AKT/mTOR pathway. Biochem Biophys Res Commun. 2018;501:1068–1073. doi: 10.1016/j.bbrc.2018.05.109. [DOI] [PubMed] [Google Scholar]
- 13.Wen S.W., et al. Characterization and effects of miR-21 expression in esophageal cancer. Genet Mol Res. 2015;14:8810–8818. doi: 10.4238/2015.August.3.4. [DOI] [PubMed] [Google Scholar]
- 14.Wen S.W., et al. Association of miR-21 with esophageal cancer prognosis: a meta-analysis. Genet Mol Res. 2015;14:6578–6582. doi: 10.4238/2015.June.12.12. [DOI] [PubMed] [Google Scholar]
- 15.Hu C., et al. MicroRNA-375 suppresses esophageal cancer cell growth and invasion by repressing metadherin expression. Oncol Lett. 2017;13:4769–4775. doi: 10.3892/ol.2017.6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu H., et al. MicroRNA-375 inhibits esophageal squamous cell carcinoma proliferation through direct targeting of SP1. Exp Ther Med. 2019;17:1509–1516. doi: 10.3892/etm.2018.7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang J., et al. MicroRNA expression profiles in gastric carcinogenesis. Sci Rep. 2018;8:14393. doi: 10.1038/s41598-018-32782-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.So J.B.Y., et al. Development and validation of a serum microRNA biomarker panel for detecting gastric cancer in a high-risk population. Gut. 2021;70:829. doi: 10.1136/gutjnl-2020-322065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu J., Wang Y., Wu X. MicroRNA in the pathogenesis and prognosis of esophageal cancer. Curr Pharm Des. 2013;19:1292–1300. doi: 10.2174/138161213804805775. [DOI] [PubMed] [Google Scholar]
- 20.Zarrilli G., et al. miRNAs involved in esophageal carcinogenesis and miRNA-related therapeutic perspectives in esophageal carcinoma. Int J Mol Sci. 2021;22 doi: 10.3390/ijms22073640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin Z., et al. Potential miRNA biomarkers for the diagnosis and prognosis of esophageal cancer detected by a novel absolute quantitative RT-qPCR method. Sci Rep. 2020;10:20065. doi: 10.1038/s41598-020-77119-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y.N., et al. MicroRNA-575 regulates development of gastric cancer by targeting PTEN. Biomed Pharmacother. 2019;113:108716. doi: 10.1016/j.biopha.2019.108716. [DOI] [PubMed] [Google Scholar]
- 23.Hezova R., et al. MiR-205 functions as a tumor suppressor in adenocarcinoma and an oncogene in squamous cell carcinoma of esophagus. Tumour Biol. 2016;37:8007–8018. doi: 10.1007/s13277-015-4656-8. [DOI] [PubMed] [Google Scholar]
- 24.Zhang C., et al. Downregulation of microRNA-376a in gastric cancer and association with poor prognosis. Cell Physiol Biochem. 2018;51:2010–2018. doi: 10.1159/000495820. [DOI] [PubMed] [Google Scholar]
- 25.Winther M., et al. Evaluation of miR-21 and miR-375 as prognostic biomarkers in esophageal cancer. Acta Oncol. 2015;54:1582–1591. doi: 10.3109/0284186x.2015.1064161. [DOI] [PubMed] [Google Scholar]
- 26.Zhang C., Zhang C.D., Ma M.H., Dai D.Q. Three-microRNA signature identified by bioinformatics analysis predicts prognosis of gastric cancer patients. World J Gastroenterol. 2018;24:1206–1215. doi: 10.3748/wjg.v24.i11.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He R., et al. Reduced miR-203 predicts metastasis and poor survival in esophageal carcinoma. Aging. 2019;11:12114–12130. doi: 10.18632/aging.102543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X., et al. Construction of a nine-MicroRNA-based signature to predict the overall survival of esophageal cancer patients. Front Genet. 2021;12 doi: 10.3389/fgene.2021.670405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X., et al. Survival prediction of gastric cancer by a seven-microRNA signature. Gut. 2010;59:579–585. doi: 10.1136/gut.2008.175497. [DOI] [PubMed] [Google Scholar]
- 30.Matsui D., et al. Primary tumor microRNA signature predicts recurrence and survival in patients with locally advanced esophageal adenocarcinoma. Oncotarget. 2016;7:81281–81291. doi: 10.18632/oncotarget.12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu J., et al. Immune-Related Nine-MicroRNA Signature for Predicting the Prognosis of Gastric Cancer. Front Genet. 2021;12:690598. doi: 10.3389/fgene.2021.690598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu J., et al. Characterization of a five-microRNA signature as a prognostic biomarker for esophageal squamous cell carcinoma. Sci Rep. 2019;9:19847. doi: 10.1038/s41598-019-56367-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ho S.-Y., Chen J.-H., Huang M.-H. Inheritable genetic algorithm for biobjective 0/1 combinatorial optimization problems and its applications. IEEE Trans Syst Man Cybern Part B (Cybernetics) 2004;34:609–620. doi: 10.1109/tsmcb.2003.817090. [DOI] [PubMed] [Google Scholar]
- 34.Yerukala Sathipati S, Ho S-Y. Identifying the miRNA signature associated with survival time in patients with lung adenocarcinoma using miRNA expression profiles. Sci Rep 2017;7:7507, doi:10.1038/s41598-017-07739-y. [DOI] [PMC free article] [PubMed]
- 35.Sathipati S.Y., Sahu D., Huang H.-C., Lin Y., Ho S.-Y. Identification and characterization of the lncRNA signature associated with overall survival in patients with neuroblastoma. Sci Rep. 2019;9:1–13. doi: 10.1038/s41598-019-41553-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sathipati S.Y., Ho S.-Y. Identification of the miRNA signature associated with survival in patients with ovarian cancer. Aging (Albany NY) 2021;13:12660. doi: 10.18632/aging.202940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang S., et al. Applications of support vector machine (SVM) LEARNING IN CANCER GENOMIcs. Cancer Genomics Proteomics. 2018;15:41–51. doi: 10.21873/cgp.20063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang C.-C., Lin C.-J. Training v-support vector regression: theory and algorithms. Neural Comput. 2002;14:1959–1977. doi: 10.1162/089976602760128081. [DOI] [PubMed] [Google Scholar]
- 39.Ho S.-Y., Shu L.-S., Chen J.-H. Intelligent evolutionary algorithms for large parameter optimization problems. IEEE Trans Evol Comput. 2004;8:522–541. [Google Scholar]
- 40.Sathipati S.Y., Ho S.-Y. Identifying a miRNA signature for predicting the stage of breast cancer. Sci Rep. 2018;8:1–11. doi: 10.1038/s41598-018-34604-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sathipati S.Y., Ho S.-Y. Novel miRNA signature for predicting the stage of hepatocellular carcinoma. Sci Rep. 2020;10:1–12. doi: 10.1038/s41598-020-71324-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tung C.-W., Ho S.-Y. Computational identification of ubiquitylation sites from protein sequences. BMC Bioinf. 2008;9:1–15. doi: 10.1186/1471-2105-9-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoerl A.E., Kennard R.W. Ridge regression: Biased estimation for nonorthogonal problems. Technometrics. 1970;12:55–67. [Google Scholar]
- 44.Tibshirani R. Regression Shrinkage and Selection Via the Lasso. J Roy Stat Soc: Ser B (Methodol) 1996;58:267–288. doi: 10.1111/j.2517-6161.1996.tb02080.x. [DOI] [Google Scholar]
- 45.Zou H., Hastie T. Regularization and variable selection via the elastic net. J R Stat Soc Series B (Stat Methodol) 2005;67:301–320. [Google Scholar]
- 46.Huang H.Y., et al. miRTarBase 2020: updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res. 2020;48:D148–D154. doi: 10.1093/nar/gkz896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen E.Y., et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuleshov M.V., et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kutmon M., et al. WikiPathways: capturing the full diversity of pathway knowledge. Nucleic Acids Res. 2016;44:D488–D494. doi: 10.1093/nar/gkv1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liberzon A., et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res 2021:49;D325-34, doi:10.1093/nar/gkaa1113. [DOI] [PMC free article] [PubMed]
- 52.Li R., et al. CancerMIRNome: an interactive analysis and visualization database for miRNome profiles of human cancer. Nucleic Acids Res. 2022;50:D1139–D1146. doi: 10.1093/nar/gkab784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karagkouni D., et al. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 2018;46:D239–D245. doi: 10.1093/nar/gkx1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiao F., et al. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37:D105–D110. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang L., Zhou G., Soufan O., Xia J. miRNet 2.0: network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020;48:W244–W251. doi: 10.1093/nar/gkaa467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang Z., et al. HMDD v3.0: a database for experimentally supported human microRNA-disease associations. Nucleic Acids Res. 2019;47:D1013–D1017. doi: 10.1093/nar/gky1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang X., et al. miR-760 exerts an antioncogenic effect in esophageal squamous cell carcinoma by negatively driving fat metabolism via targeting c-Myc. J Cell Biochem. 2020;121:2950–2961. doi: 10.1002/jcb.29540. [DOI] [PubMed] [Google Scholar]
- 58.Cheng L., et al. Identifying the differentially expressed microRNAs in esophagus squamous cell carcinoma of Kazakh patients in Xinjiang. Oncol Lett. 2019;17:2657–2668. doi: 10.3892/ol.2019.9904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luo D., et al. miR-1301-3p promotes cell proliferation and facilitates cell cycle progression via targeting SIRT1 in gastric cancer. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.664242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu K., Zhang C., Zhang C., Dai D. A novel three-miRNA signature identified using bioinformatics predicts survival in esophageal carcinoma. Biomed Res Int. 2020;2020:5973082. doi: 10.1155/2020/5973082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang J.X., et al. LINC01410-miR-532-NCF2-NF-kB feedback loop promotes gastric cancer angiogenesis and metastasis. Oncogene. 2018;37:2660–2675. doi: 10.1038/s41388-018-0162-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 62.Tokumaru Y., et al. Low expression of miR-29a is associated with aggressive biology and worse survival in gastric cancer. Sci Rep. 2021;11:14134. doi: 10.1038/s41598-021-93681-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang J., et al. Circulating MiR-16-5p and MiR-19b-3p as two novel potential biomarkers to indicate progression of gastric cancer. Theranostics. 2015;5:733–745. doi: 10.7150/thno.10305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Butz F., et al. MicroRNA profiling in oesophageal adenocarcinoma cell lines and patient serum samples reveals a role for miR-451a in radiation resistance. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21238898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xian X., Tang L., Wu C., Huang L. miR-23b-3p and miR-130a-5p affect cell growth, migration and invasion by targeting CB1R via the Wnt/β-catenin signaling pathway in gastric carcinoma. Onco Targets Ther. 2018;11:7503–7512. doi: 10.2147/ott.S181706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wei H., et al. miR-130a deregulates PTEN and stimulates tumor growth. Cancer Res. 2017;77:6168–6178. doi: 10.1158/0008-5472.Can-17-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li Z., et al. By downregulating TIAM1 expression, microRNA-329 suppresses gastric cancer invasion and growth. Oncotarget. 2015;6:17559–17569. doi: 10.18632/oncotarget.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Su R., Zhao E., Zhang J. miR-496 inhibits proliferation via LYN and AKT pathway in gastric cancer. Open Med (Wars) 2021;16:1206–1214. doi: 10.1515/med-2021-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gan J., et al. Growth hormone-releasing hormone receptor antagonists inhibit human gastric cancer through downregulation of PAK1-STAT3/NF-κB signaling. Proc Natl Acad Sci U S A. 2016;113:14745–14750. doi: 10.1073/pnas.1618582114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Massagué J. TGFβ signalling in context. Nat Rev Mol Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.David C.J., Massagué J. Contextual determinants of TGFβ action in development, immunity and cancer. Nat Rev Mol Cell Biol. 2018;19:419–435. doi: 10.1038/s41580-018-0007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Katz L.H., et al. TGF-β signaling in liver and gastrointestinal cancers. Cancer Lett. 2016;379:166–172. doi: 10.1016/j.canlet.2016.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Toomey P.G., Vohra N.A., Ghansah T., Sarnaik A.A., Pilon-Thomas S.A. Immunotherapy for gastrointestinal malignancies. Cancer Control. 2013;20:32–42. doi: 10.1177/107327481302000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Neuzillet C., et al. Targeting the TGFβ pathway for cancer therapy. Pharmacol Ther. 2015;147:22–31. doi: 10.1016/j.pharmthera.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 75.Liu S., Ren J., Ten Dijke P. Targeting TGFβ signal transduction for cancer therapy. Signal Transduct Target Ther. 2021;6:8. doi: 10.1038/s41392-020-00436-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu J., et al. Bcl-2-associated transcription factor 1 Ser290 phosphorylation mediates DNA damage response and regulates radiosensitivity in gastric cancer. J Transl Med. 2021;19:339. doi: 10.1186/s12967-021-03004-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang H., et al. Cancer-associated fibroblast–promoted LncRNA <em>DNM3OS</em> confers radioresistance by regulating DNA damage response in esophageal squamous cell carcinoma. Clin Cancer Res. 2019;25:1989. doi: 10.1158/1078-0432.CCR-18-0773. [DOI] [PubMed] [Google Scholar]
- 78.Zhu Y., Zhu X., Wei X., Tang C., Zhang W. HER2-targeted therapies in gastric cancer. Biochim Biophys Acta Rev Cancer. 2021;1876:188549. doi: 10.1016/j.bbcan.2021.188549. [DOI] [PubMed] [Google Scholar]
- 79.Ayyappan S., Prabhakar D., Sharma N. Epidermal growth factor receptor (EGFR)-targeted therapies in esophagogastric cancer. Anticancer Res. 2013;33:4139–4155. [PubMed] [Google Scholar]
- 80.Luan F., et al. TNFRSF11B activates Wnt/β-catenin signaling and promotes gastric cancer progression. Int J Biol Sci. 2020;16:1956–1971. doi: 10.7150/ijbs.43630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nienhüser H., Schmidt T. Angiogenesis and anti-angiogenic therapy in gastric cancer. Int J Mol Sci. 2017;19 doi: 10.3390/ijms19010043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lin E.W., Karakasheva T.A., Hicks P.D., Bass A.J., Rustgi A.K. The tumor microenvironment in esophageal cancer. Oncogene. 2016;35:5337–5349. doi: 10.1038/onc.2016.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data used in this analysis can be found on the TCGA data portal [https://portal.gdc.cancer.gov/].