Highlights
-
•
We present a novel meta-approach, MLACP 2.0, and implement it as a user-friendly webserver for the accurate identification of ACPs.
-
•
MLACP 2.0 employed 11 different encoding schemes and eight different classifiers, including convolutional neural networks, to create a stable meta-model.
-
•
Benchmarking study has demonstrated that MLACP 2.0 achieves superior performance in ACP prediction compared to publicly available state-of-the-art predictors.
Keywords: Anticancer peptides, Convolutional neural network, Feature encodings, Conventional classifiers, Baseline models, Dataset construction
Abstract
Anticancer peptides are emerging anticancer drug that offers fewer side effects and is more effective than chemotherapy and targeted therapy. Predicting anticancer peptides from sequence information is one of the most challenging tasks in immunoinformatics. In the past ten years, machine learning-based approaches have been proposed for identifying ACP activity from peptide sequences. These methods include our previous method MLACP (developed in 2017) which made a significant impact on anticancer research. MLACP tool has been widely used by the research community, however, its robustness must be improved significantly for its continued practical application. In this study, the first large non-redundant training and independent datasets were constructed for ACP research. Using the training dataset, the study explored a wide range of feature encodings and developed their respective models using seven different conventional classifiers. Subsequently, a subset of encoding-based models was selected for each classifier based on their performance, whose predicted scores were concatenated and trained through a convolutional neural network (CNN), whose corresponding predictor is named MLACP 2.0. The evaluation of MLACP 2.0 with a very diverse independent dataset showed excellent performance and significantly outperformed the recent ACP prediction tools. Additionally, MLACP 2.0 exhibits superior performance during cross-validation and independent assessment when compared to CNN-based embedding models and conventional single models. Consequently, we anticipate that our proposed MLACP 2.0 will facilitate the design of hypothesis-driven experiments by making it easier to discover novel ACPs. The MLACP 2.0 is freely available at https://balalab-skku.org/mlacp2.
1. Introduction
Cancer is one of the prominent threats to human health, and it is often linked to a higher mortality rate as life expectancy increases, both in developed and developing countries [1]. In 2018, the World Health Organization (WHO) and the International Agency for Research on Cancer (IARC) reported that 18.1 million new cases and 9.6 million deaths were caused by cancer [2]. Cancer results from the uncontrolled proliferation of abnormal cells which invade normal tissues and organs and multiply in an uncontrolled manner [3]. The complexity and heterogeneity of cancer make its treatment difficult. Hence, cancer therapy must focus primarily on limiting the proliferation of cancer cells and inhibiting their spread [4].
With traditional surgery, precise removal of the cancerous part could not be guaranteed [5]. Radiotherapy, chemotherapy, and targeted therapy are the most common treatment options for cancer. In spite of this, these therapies are not very precise, as they fail to differentiate cancer cells from healthy cells and result in both damaging healthy cells and killing cancer cells. However, these therapies are expensive and have negative side effects on patients [6], [7]. Furthermore, cancer cells can develop resistance to chemotherapy drugs due to the fact that their genomes are changing dynamically [8]. Consequently, there is an urgent need to develop a novel cancer treatment that is free of adverse effects, reduces drug resistance, and specifically targets cancer cells.
Antimicrobial peptides (AMPs) are a diverse class of bioactive molecules that provide protection against bacteria, protozoa, fungi, and viruses [9]. A subset of AMPs exhibiting potential anticancer properties is referred to as anticancer peptides (ACPs), which are short peptides whose sequence length does not exceed 50 amino acid residues [10]. ACPs possess amphiphilic properties due to the presence of hydrophobic and positive residues that interact with the anionic membranes of cancer cells, selectively targeting and killing them [11]. ACPs can target cancer cells based on the membrane charge that forms an electrostatic interaction between the membrane and the ACPs, thus leaving normal cells untouched. This is the major advantage of using ACPs over currently available approaches in cancer treatment [12], [13]. Moreover, ACPs are naturally biological inhibitors, and they are also easy to synthesize, which makes them an ideal therapeutic agent to treat cancer [13]. In light of this, peptide-based therapeutics have emerged as a promising therapeutic agent for treating cancer compared to conventional therapies because they are less toxic, highly specific, capable of penetrating membranes, and easy to modify chemically [14], [15].
Several computational predictors have been proposed for the identification of ACPs. Many of these methods have been reviewed in recent literatures [16], [17], including our previous method, MLACP [18]. It was built using an imbalanced dataset, a linear integration of four different encodings, a support vector machine (SVM), and a random forest (RF). The MLACP is widely used among the research community, thus gaining popularity within the ACP research community. The number of experimentally verified ACPs is increasing exponentially; therefore, it is high time to update the previous version utilizing advanced computational techniques, thus increasing its accuracy and robustness.
The development of MLACP 2.0 involves the following steps: (i) Created a high-quality non-redundant training dataset and independent datasets based on extensive literature/database searches. (ii) Systematically evaluated 17 different feature encodings (including both conventional encodings and word embeddings) and built the corresponding model using seven different conventional classifiers (RF, gradient boosting (GB), SVM, extreme gradient boosting (XGB), AdaBoost (AB), light gradient boosting (LGB), and extremely randomized tree (ERT)). (iii) For each classifier, choose a subset of models based on certain criteria from 17 encoding-based models Matthews Correlation Coefficients (MCC) greater than the average MCC of 17 encoding-based models. Subsequently, the predicted probability of ACPs from the seven classifiers based on a subset of selected models was concatenated and trained using a convolutional neural network (CNN) for the final prediction, MLACP 2.0. Extensive benchmark experiments demonstrate the effectiveness of MLACP 2.0: it achieves a more accurate and stable performance compared with conventional single-encoding models and CNN-based one-hot encoding and word embedding models, on both the basis of cross-validation and independent assessment. On an independent test, MLACP 2.0 significantly outperformed the existing predictors. Using the proposed hybrid ensemble model, a user-friendly online predictor of MLACP (https://balalab-skku.org/mlacp2/) is implemented.
2. Materials and methods
2.1. Construction of datasets
The objective of this study is to develop a prediction model using existing methods datasets and to evaluate the proposed model based on a newly constructed dataset. The training dataset was constructed by extracting the existing 37 methods' training datasets and separating them into ACPs and non-ACPs [16], [17]. Notably, several methods used the same datasets, and therefore some sequences are redundant. A CD-HIT [19] of 0.8 was applied among ACPs, which resulted in 1084 peptide sequences. The same cut-off was applied to non-ACP samples whose sequences overlapping with ACPs resulted in ∼ 7500 sequences. However, we randomly selected 1084 non-ACPs to avoid class bias during the model building and balanced the ACPs. This is the first time such a large non-redundant dataset has been used for training or model building in ACP prediction research.
Independent dataset: ACPs were extracted from the following 11 databases, CancerPPD [20], APD3 [21], PlantPepDB [22], DBAASP v3.0 [23], SATPdb [24], ADAM (https://bioinformatics.cs.ntou.edu.tw/ADAM), DRAMP 3.0 [25], [26], LAMP [27], Peptipedia [28], DbAMP [29], and AMPfun [30], resulting in 3725 ACPs. Secondly, a CD-HIT of 0.80 was applied to the collected ACPs that overlapped with the training ACP sequences, resulting in 769 sequences. Unlike previous studies, where random peptides were considered as non-ACPs, we considered other functional peptides (antihypertensive and antiviral, etc.), a small portion of random peptides, AMPs, and non-AMPs, and experimentally confirmed non-proinflammatory inducing peptides as non-ACPs, resulting 1287 non-redundant non-ACPs. This independent dataset can be used as a gold standard for evaluating future ACP prediction models. Furthermore, the supplementary information includes a brief description of the dataset length distribution and compositional analysis.
2.2. Feature encodings
The process of exploring different feature encodings on the same dataset is essential to understand and identify the appropriate encodings. Keeping this in mind, a wide range of features were used in this study, including 15 conventional encodings (dipeptide composition (DPC), dipeptide deviation from the expected mean (DDE), amino acid composition (AAC), composition transition and distribution (CTDC, CTDT, and CTDD), grouped DPC (GDPC), enhanced grouped AAC (EGAAC), grouped tripeptide composition (GTPC), BLOSUM62 (BLOS), enhanced AAC (EAAC), K-spaced conjoint triad (KSC), quasi sequence order (QSO) composition of k-spaced amino acid group pairs (CKSAAGP), and Z scale) and twoword embeddings are one-hot encoding (1OHE) and pretrained embedding from seq2vec. Among these 17 encodings, 11 encodings (AAC, CKSAAGP, CTDC, CTDD, CTDT, DDE, DPC, KSC, QSO, 1OHE, and seq2vec) are the most important and contributed significantly to the ACP prediction. Notably, nine of the conventional encodings contributed to the final prediction, whose encoding details are extensively described in our previous studies [31]. Using the same procedure AAC, CKSAAGP, CTDC, CTDD, CTDT, DDE, DPC, KSC, QSO encoded 20, 275, 39, 195, 39, 400, 400, 343, 100 D feature vectors, respectively. Notably, these features have been normalized as follows: . A brief description of these word embeddings is as follows:
2.3. 1OHE
The one-hot encoding method is quite popular among binary encoding techniques. The maximum length of peptides in our dataset is 50 amino acids. If the residues are<50 amino acids, a dummy residue X is added to the C-terminus. Therefore, each amino acid is represented by a 21-dimensional feature vector, where the standard amino acid is characterized by 1 at various positions and zero at the remaining 20 positions. Dummy residues, on the other hand, consist entirely of zeros. This resulted in a 1050-D feature vector.
2.4. Seq2vec
We utilized seq2vec’s pretrained embeddings to achieve the concept of transfer learning. Heinzinger et al. developed pretrained embeddings by training the ELMo model using millions of protein sequences extracted from UniRef50. For this study, we used the same pretrained embeddings that provide a 1024 D feature vector for a given peptide sequence.
2.5. MLACP 2.0 framework
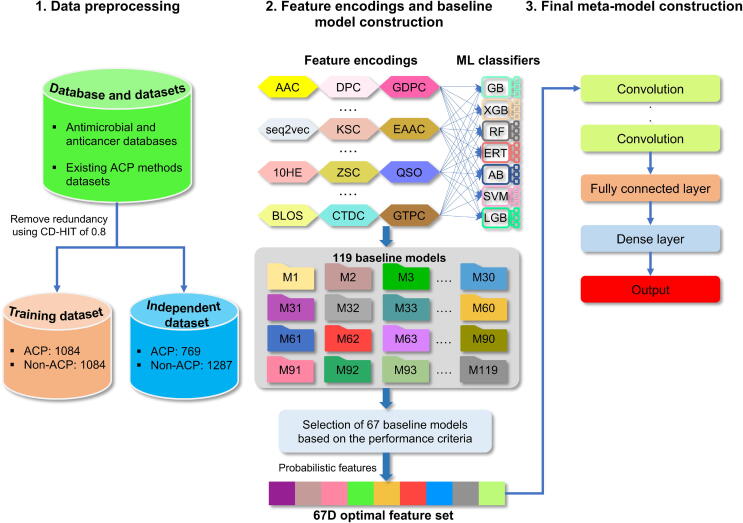
The MLACP 2.0 framework (Fig. 1) was developed using the training dataset and feature encodings mentioned above, and it consists of constructing baseline models and developing a meta-predictor.
Fig. 1.
Overview of MLACP 2.0. The process consists of three stages: preprocessing of the data, feature extraction and the construction of a baseline model using seven different classifiers, and the creation of the final meta-model.
Construction of baseline models: We utilized seven different classifiers (RF, ERT, SVM GB, AB, LGB, and XGB) that have been extensively applied in Bioinformatics and computational biology [32], [33], [34], [35], [36], [37]. For each classifier, there are a set of hyperparameters that determine the performance of the model during cross-validation. We optimized the hyperparameters using a grid search approach and 10-fold cross-validation. To construct each baseline model, 10-fold cross-validation was repeated five times with random portioning of the training samples, and the median parameters were taken as the final optimal values. These values were then used to construct the final baseline model. The hyperparameters search range for each classifier is as follows: (i) LGB seven hyperparameters are: num_leaves 50 to 1000] with an interval of 20, max_bins [200 to 400] with an interval of 10, n_estimators [100 to 2000] with an interval of 10, min_child_samples [30 to 400] with an interval of 10, max_depth [5 to 12] with an interval of 1, learning_rate [10-6 to 10-1], and bagging fraction is 0.8. (ii) RF and ERT have the same hyperparameters with different model construction procedures, whose search ranges are: n_estimators [50, 75, …, 3000], max_features [1, 2, …,20], and min_samples_split [2, 3, …,10]. (iii) SVM two hyperparameters are with a step size of 2 and with a step size of 2-1. (iv) AB three hyperparameters are: n_estimators [10, 20, …, 500], max_depth [1, 2, …, 11], and learning_rate [1, 0.5, 0.25, 0.1, 0.05, 0.01]. (v) GB hyperparameters search ranges are n_estimators [10, 20, …, 500], learning_rate [1, 0.5, 0.25, 0.1, 0.05, 0.01], and max_features [1, 2, …,10]. (vi) four XGB hyperparameters are: n_estimators [10, 20, …, 2000], max_depth [1, 2, …, 11]; learning_rate [1.0, 0.5, 0.25, 0.1, 0.05, 0.01, 0.001, 0.0001, 0.2, 0.3], and eta [0.0001, 0.001, 0.002, 0.01, 0.02, 0.05, 1.0]. Notably, the names of hyperparameters are derived from those given in their corresponding package. This work implemented the scikit-learn version 0.24.2 [38] library for five classifiers (SVM, RF, GB, AB, and ERT) lightGBM version 3.3.0 [39], and XGBoost version 0.82 python package to carry out the classification task.
Finally, a total of 119 baseline models were generated, and a set of models was selected based on the following criteria: (i) computed cumulative score (CS) of Mathews correlation coefficient, accuracy (ACC), and area under the ROC curve (AUC) for each baseline model. (ii) selected a set of baseline models for each classifier, whose CS is greater than the mean CS. Consequently, this filtering resulted in 67 baseline models. The predicted probability values of the selected models (67D) are used for the development of the meta predictor.
Construction of meta-predictor: A similar CNN network architecture was adopted based on recent studies [40], [41]. However, we optimized four filters, epochs, and batch size using 10-fold cross-validation. Specifically, the predicted probability of 67D feature vector input to four 1D convolutional layers. The kernel sizes associated with these layers are 4, 5, 6, and 7, with corresponding filters of 20, 8, 32, and 8, respectively. After the convolutional operation, the activation function of the rectified linear unit (ReLU) was applied, which can be described as follows:
| (1) |
For each of the four 1D convolutional layers, 20, 8, 32, and 8 feature maps were generated. Following each 1D convolutional layer, a 1D Global Max Pooling layer was applied, whose purpose was to find the maximum values from the feature maps that were converted to univariate feature vectors and concatenated subsequently. Additionally, a dropout rate of 0.5 is used for the independent component layer. Then, three dense layers of 32, 16, and 8 neurons, respectively, were applied with the ReLU activation function. Finally, a dense layer composed of a single neuron with a sigmoid function is applied, which produces a value between 0 and 1. If the value is greater than 0.5, the peptide belongs to ACP, otherwise non-ACP. Adam optimization was used to update the network weights. Notably, CNN was implemented using Keras deep learning library [42].
| (2) |
2.6. Model evaluation
Furthermore, we considered the commonly used six evaluation metrics to evaluate the model performance [16], [43], including MCC, Sensitivity (Sn), Specificity (Sp), ACC, and AUC. The definition of the metrics is as follows:
| (3) |
where TP, TN, FP, FN, respectively denote true positives, true negatives, false positives, and false negatives. Furthermore, ROC curves and AUC values were used to assess overall performance.
3. Results and discussion
3.1. Construction of baseline models
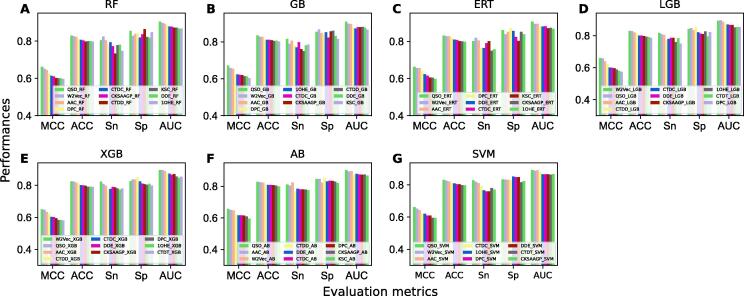
This study utilized 17 different encodings (QSO, seq2vec, AAC, DPC, CTDC, CTDD, CTDT, CKSAAGP, KSC, DDE, 1OHE, EAAC, GTPC, BLOS, ZSC, GPDC, and EGAAC) that mostly covered composition, physicochemical properties, evolutionary information, word embedding, and position-specific information. The discriminative ability of each of these encodings was evaluated by seven different conventional classifiers (RF, ERT, GB, AB, SVM, LGB, and XGB). Table S1-S7 shows the performances of these encodings to each classifier. Result shows that RF, ERT, GB, XGB, LGB, AB, and SVM have ACC ranges of 72.0–83.0 %, 72.0–83.1 %, 71.3–83.5 %, 70.6–82.5 %, 70.0–83.0 %, 71.7–82.8 %, 71.1–83.1 %, respectively. We observed that roughly 10 % ACC gap between 17 baseline models produced by each classifier. The QSO encoding achieved the best performance based on four classifiers and the seq2vec encoding achieved the best performance based on two classifiers (XGB and LGB). Generally, most ACP predictors demonstrated high accuracy of >90.0 % in training datasets that contained homologous sequences with high similarity [44], [45]. While the best baseline model (QSO-GB) achieved the maximum ACC of 83.5 %, it indicates a drop in performance due to the highly non-redundant training dataset, which may limit the overestimation of model performance.
3.2. Construction of MLACP 2.0
It is likely that considering the 119 baseline models (17 baseline models 7 classifiers) for the final model construction is not appropriate due to the performance gap of (∼10 %) for each classifier among 17 baseline models. Therefore, we calculated the average MCC from 17 baseline models for each classifier and then considered only models whose performance was above average MCC, resulting in 10 baseline models respectively from RF, GB, LGB, and XGB, and 9 respectively from ERT, SVM, and AB. In total, we obtained 67 baseline models, whose performance is shown in Fig. 2, whose ACC is in the range of 0.800–0.835. Next, we examined how many unique encodings contributed to the 67 baseline models. Out of 17 encodings, only 11 encodings (AAC, CKSAAGP, CTDC, CTDD, CTDT, DDE, DPC, KSC, QSO, seq2vec, and 1OHE) contributed to the selected baseline models, which are mostly composition-based and word embeddings. The remaining six encodings (EAAC, EGAAC, GTPC, GDPC, ZSC, and BLOS) based models were excluded because of their relatively lower performance during the training. Based upon the selected baseline models, the prediction probability of ACPs is concatenated and treated as a novel feature vector, which is then trained with CNN to develop the final prediction model. Notably, we also tested with the other seven classifiers employed for the baseline model construction, but CNN has the edge in terms of robustness (Figure S8). Hence, we selected CNN for the meta-model construction, named MLACP 2.0. It achieved MCC, ACC, Sn, Sp, and AUC of 0.694, 0.846, 0.815, 0.876, and 0.915, respectively.
Fig. 2.
Performance comparison of the baseline models selected for each classifier. (A) random forest (RF), (B) gradient boosting (GB), (C) extremely randomized tree (ERT), (D) light gradient boosting (LGB), (E) extreme gradient boosting (XGB), (F) AdaBoost (AB), and (G) support vector machine (SVM).
3.3. Comparison of MLACP 2.0 with different approaches to training dataset
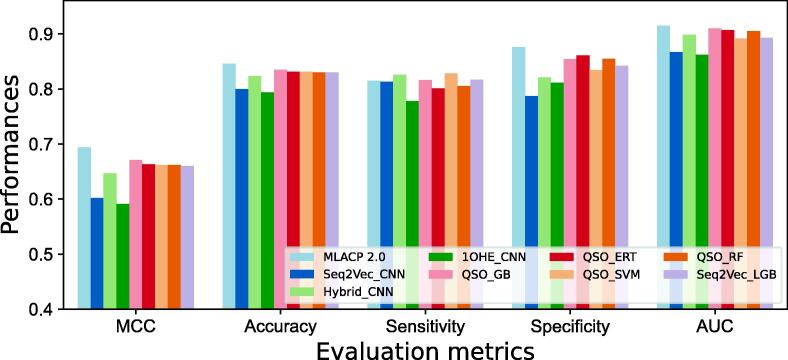
For the purpose of illustrating the advantages of using probabilistic features in MLACP 2.0, we have also developed CNN-based word embedding models, namely seq2vec-CNN and 1OHE-CNN, as well as a CNN model based on hybrid features (a linear integration of eleven encodings). Fig. 3 compares the performance of MLACP 2.0 with the top five baseline models, CNN-based word embeddings, and hybrid feature models. Compared to the best five baseline models, the CNN-hybrid model performs similarly to the best five baseline models, and significantly better than the CNN-word embedding model, indicating that automated word embedding features are not as effective as feature engineering in ACP prediction. MLACP 2.0 outperforms the best five baseline models as well as CNN-based models. More specifically, the improvements of MLACP 2.0 are 2.3–10.3 % in MCC, 1.1–5.2 % in ACC, and 0.5–4.8 % in AUC, demonstrating that a systematic approach to evaluating multiple encodings in tandem with the selection of a set of baseline models utilized for meta-model construction led to improved performance.
Fig. 3.
Performance comparison of MLACP 2.0 with the top five baseline models and with other approaches based on the training dataset.
3.4. Evaluation of MLACP 2.0 and the state-of-the-art methods on an independent dataset
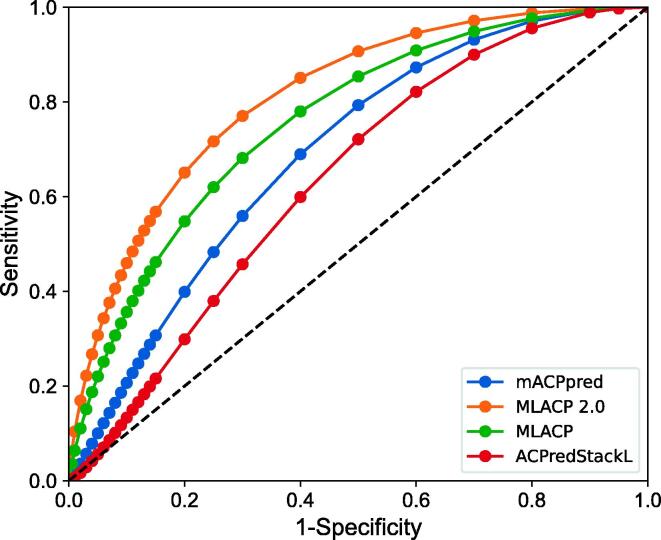
The independent dataset was used to evaluate MLACP 2.0 along with the previous version and the two best ACP predictors (mACPpred and ACPpredStackL) [16], [46]. Notably, two of the methods were reported to be the best predictors in previous studies by unbiased evaluations. In contrast to the routine independent datasets used in previous studies, a challenging dataset was created. Independent datasets have the following important characteristics: (i) none of the ACPs share >80 % sequence identity with the training dataset; and (ii) the non-ACPs were constructed considering several practical scenarios, including other functional peptides and experimentally characterized negative examples. According to Table 1, MLACP 2.0 achieves MCC, ACC, Sn, Sp, and AUC values of 0.513, 0.765, 0.750, and 0.773, and 0.817, respectively. In particular, MLACP 2.0 improved the MCC of 16.2–28.0 %, the ACC of 0.8–13.7 %, and the AUC of 7.3–17.7 % compared to the existing predictors. Furthermore, MLACP 2.0 predictor performance is more balanced (low difference between Sn and Sp) when compared to the existing predictors, demonstrating that MLACP 2.0 performs well on unseen data and is better suited for practical applications.
Table 1.
Performance of different methods on independent datasets.
| Methods | MCC | Accuracy | Sensitivity | Specificity | AUC | P-value |
|---|---|---|---|---|---|---|
| MLACP 2.0 | 0.513 | 0.765 | 0.750 | 0.773 | 0.817 | |
| MLACP | 0.256 | 0.677 | 0.283 | 0.911 | 0.744 | 0.000003 |
| mACPpred | 0.351 | 0.677 | 0.700 | 0.663 | 0.704 | <0.000001 |
| ACPredStackL | 0.233 | 0.628 | 0.588 | 0.651 | 0.640 | <0.000001 |
It is difficult to obtain statistical estimates from the threshold-based comparison described above. Consequently, we compared two AUC values of different methods using ROC and calculated the P value for observed differences based on the results of a two-tailed test [47]. According to Fig. 4 and Table 1, the MLACP 2.0 outperformed the existing predictors on the independent dataset by a significant amount. One limitation of the proposed method is that it cannot predict peptides with more than 50 amino acid residues.
Fig. 4.
Comparison of binormal receiver operating characteristics (ROC) curves for ACPs prediction using different methods on an independent dataset.
3.5. Model interpretation
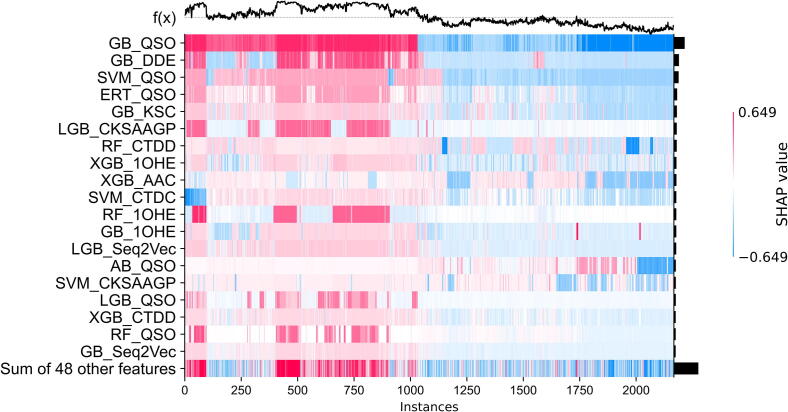
MLACP 2.0 was trained using the optimal probabilistic feature vector. As a result, it performed better and more competitively than the previous predictors. The contribution and directionality of the probabilistic features contributed to the meta-model are unknown. Based on a series of recent studies, we have conducted a model interpretation analysis using SHapley Additive exPlanation (SHAP) [48], in order to illustrate the most significant features and their relationship with the outcomes of MLACP 2.0. Fig. 4 shows that MLACP 2.0 generates predictions in the form of line charts above the heatmap matrix (f(x)), each feature's global importance is illustrated in the form of bar graphs on the right-hand side of the heatmap, and the top 20 most important features are listed in order of their global importance. As shown in Fig. 5, we observed that six baseline models based on QSO encoding, and three baseline models based on 1OHE, CTDD, seq2vec, and CKSAAGP encoding respectively contributed two baseline models, four encodings (DDE, CTDC, KSC, and AAC) based their respective model contributed the most in the final MLACP 2.0 prediction. The importance of physicochemical properties has been highlighted in previous studies [49], [50]. Our analysis also indicates that CKSAAGP is one of the influential features in MLACP 2.0 performance. Among the baseline model comparisons, QSOs had the best performances compared to other encodings (Fig. 2). Therefore, it is not surprising that these models contributed the most to MLACP 2.0. It is interesting to note that the SHAP analysis accurately identifies this phenomenon.
Fig. 5.
A heatmap plot of the SHAP values for the top 20 probabilistic features based on the training dataset for identifying ACPs.
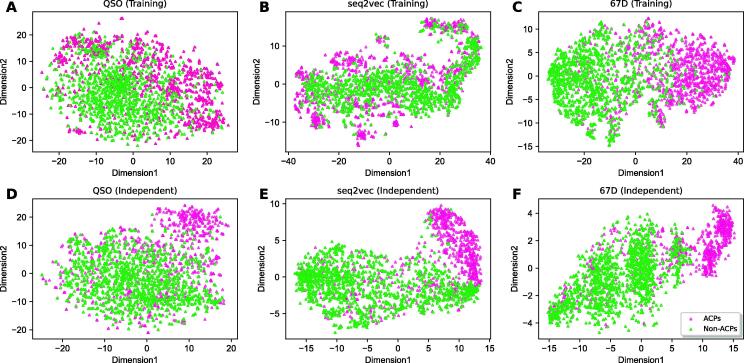
Moreover, t-Distributed Stochastic Neighbor Embedding (t-SNE) was applied to reduce the multidimensional features to two-dimensional plots to understand the relationship between two classes for different encodings. As shown in Fig. 6, QSO and seq2vec encodings (achieved superior performance among baseline models), where ACPs and non-ACPs overlap substantially. It is interesting to note that such overlaps were significantly reduced in the probabilistic features (Fig. 6). The pattern observed in the training was the same on the independent dataset, although non-ACPs are extremely diverse, demonstrating the robustness of our approach.
Fig. 6.
The t-SNE distribution of the ACPs and non-ACPs in two-dimensional space. The pink and limegreen represent ACPs and non-ACPs, respectively. A–C represents the QSO, seq2vec, and probabilistic features (67D) based on the training dataset. (D-F) show the corresponding distribution for the independent dataset. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.6. Webserver implementation
In order to make the MLACP 2.0 algorithm widely accessible to users, a webserver has been developed, which can be found at https://balalab-skku.org/mlacp2. The web server was built using Django, Python, CSS, HTML, and JavaScript programming languages, as well as a PostgreSQL database for storage and retrieval of job results. Users can find instructions on how to use MLACP 2.0 on the home page and it also includes links to the curated datasets used in the study. The user may upload a file containing multiple FASTA sequences or paste one or more query sequences in FASTA format for prediction. The results of a successful job are displayed in a separate interface, where they can also be downloaded in CSV format for later use. On the submission page, users can view the results of previously completed jobs by entering the job ID into the 'find job' option.
4. Conclusion
In this study, we developed an improved version of our previous ACP predictor, named MLACP 2.0, based on peptide sequence information. For the development of the second-generation tool, we first constructed non-redundant training and independent datasets based on extensive database and literature searches. It is noteworthy that this is the first study to use such a large non-redundant dataset for modeling or training. Second, 17 different feature encodings and seven different classifiers were employed to develop a pool of baseline models. In the next step, a set of baseline models was manually identified whose predicted ACP values were integrated and trained with CNN to yield the final model.
Several factors contribute to the improved performance of MLACP 2.0, including (i) a reduced training dataset coupled with a meta-model approach; (ii) the predicted probabilistic features have a high intrinsic discriminatory ability on both datasets, resulting in improved performance. Interestingly, this approach can be extended to predict other peptide therapeutic functions [51], [52], [53], [54]. Despite its promising performance, MLACP 2.0 also has room for improvement. (i) A novel sequence-based encoding system that is independent of composition and physicochemical properties are expected to be developed and applied in the future. (ii) The use of feature selection techniques [55], [56], [57], [58], [59] might help quantify the contribution of each encoding to distinguishing ACPs and non-ACPs. (iii) It may also be possible to develop ensemble deep learning models or hybrid models (conventional and deep learning models) [60] to improve the performance of ACPs when additional datasets become available in the future.
Funding
This work is supported by the National Research Foundation of Korea (NRF) funded by the Korean government (MSIT) (2021R1A2C1014338 and 2021R1C1C1007833), the SungKyunKwan University and the BK21 FOUR (Graduate School Innovation) funded by the Ministry of Education (MOE, Korea) and NRF.
Author Contributions
B.M., and J.Y.J, conceived the project and designed the experiments. L.T.P., H.W.P., B.M., T.M., and T.P., performed the experiments and analyzed the data, and B.M, T.P., L.T.P., and H.W.P., wrote the manuscript. All authors read and approved the final manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.07.043.
Contributor Information
Young-Jun Jeon, Email: jeon2020@skku.edu.
Balachandran Manavalan, Email: bala2022@skku.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Ortega-Garcia M.B., Mesa A., Moya E.L.J., Rueda B., Lopez-Ordono G., Garcia J.A., et al. Uncovering Tumour Heterogeneity through PKR and nc886 Analysis in Metastatic Colon Cancer Patients Treated with 5-FU-Based Chemotherapy. Cancers (Basel) 2020;12 doi: 10.3390/cancers12020379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Palumbo M.O., Kavan P., Miller W.H., Jr., Panasci L., Assouline S., Johnson N., et al. Systemic cancer therapy: achievements and challenges that lie ahead. Front Pharmacol. 2013;4:57. doi: 10.3389/fphar.2013.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holohan C., Van Schaeybroeck S., Longley D.B., Johnston P.G. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. 2013;13:714–726. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 5.An Z., Flores-Borja F., Irshad S., Deng J., Ng T. Pleiotropic role and bidirectional immunomodulation of innate lymphoid cells in cancer. Front Immunol. 2019;10:3111. doi: 10.3389/fimmu.2019.03111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaspar D., Veiga A.S., Castanho M.A. From antimicrobial to anticancer peptides. A review. Front Microbiol. 2013;4:294. doi: 10.3389/fmicb.2013.00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morel D., Jeffery D., Aspeslagh S., Almouzni G., Postel-Vinay S. Combining epigenetic drugs with other therapies for solid tumours - past lessons and future promise. Nat Rev Clin Oncol. 2020;17:91–107. doi: 10.1038/s41571-019-0267-4. [DOI] [PubMed] [Google Scholar]
- 8.Zahreddine H., Borden K.L. Mechanisms and insights into drug resistance in cancer. Front Pharmacol. 2013;14:4–28. doi: 10.3389/fphar.2013.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raffatellu M. Learning from bacterial competition in the host to develop antimicrobials. Nat Med. 2018;24:1097–1103. doi: 10.1038/s41591-018-0145-0. [DOI] [PubMed] [Google Scholar]
- 10.Xie M., Liu D., Yang Y. Anti-cancer peptides: classification, mechanism of action, reconstruction and modification. Open Biol. 2020;10 doi: 10.1098/rsob.200004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shoombuatong W., Schaduangrat N., Nantasenamat C. Unraveling the bioactivity of anticancer peptides as deduced from machine learning. EXCLI J. 2018;17:734. doi: 10.17179/excli2018-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schweizer F. Cationic amphiphilic peptides with cancer-selective toxicity. Eur J Pharmacol. 2009;625:190–194. doi: 10.1016/j.ejphar.2009.08.043. [DOI] [PubMed] [Google Scholar]
- 13.Soon T.N., Chia A.Y.Y., Yap W.H., Tang Y.Q. Anticancer mechanisms of bioactive peptides. Protein Pept Lett. 2020;27:823–830. doi: 10.2174/0929866527666200409102747. [DOI] [PubMed] [Google Scholar]
- 14.Fosgerau K., Hoffmann T. Peptide therapeutics: current status and future directions. Drug Discov Today. 2015;20:122–128. doi: 10.1016/j.drudis.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 15.Lau J.L., Dunn M.K. Therapeutic peptides: Historical perspectives, current development trends, and future directions. Bioorg Med Chem. 2018;26:2700–2707. doi: 10.1016/j.bmc.2017.06.052. [DOI] [PubMed] [Google Scholar]
- 16.Basith S., Manavalan B., Hwan Shin T., Lee G. Machine intelligence in peptide therapeutics: A next-generation tool for rapid disease screening. Med Res Rev. 2020;40:1276–1314. doi: 10.1002/med.21658. [DOI] [PubMed] [Google Scholar]
- 17.Basith S., Manavalan B., Shin T.H., Lee D.Y., Lee G. Evolution of machine learning algorithms in the prediction and design of anticancer peptides. Curr Protein Pept Sci. 2020;21:1242–1250. doi: 10.2174/1389203721666200117171403. [DOI] [PubMed] [Google Scholar]
- 18.Manavalan B., Basith S., Shin T.H., Choi S., Kim M.O., Lee G. MLACP: machine-learning-based prediction of anticancer peptides. Oncotarget. 2017;8:77121–77136. doi: 10.18632/oncotarget.20365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu L., Niu B., Zhu Z., Wu S., Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tyagi A., Tuknait A., Anand P., Gupta S., Sharma M., Mathur D., et al. CancerPPD: a database of anticancer peptides and proteins. Nucleic Acids Res. 2015;43:D837–D843. doi: 10.1093/nar/gku892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang G., Li X., Wang Z. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016;44:D1087–D1093. doi: 10.1093/nar/gkv1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Das D., Jaiswal M., Khan F.N., Ahamad S., Kumar S. PlantPepDB: A manually curated plant peptide database. Sci Rep. 2020;10:2194. doi: 10.1038/s41598-020-59165-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pirtskhalava M., Amstrong A.A., Grigolava M., Chubinidze M., Alimbarashvili E., Vishnepolsky B., et al. DBAASP v3: database of antimicrobial/cytotoxic activity and structure of peptides as a resource for development of new therapeutics. Nucleic Acids Res. 2021;49:D288–D297. doi: 10.1093/nar/gkaa991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh S., Chaudhary K., Dhanda S.K., Bhalla S., Usmani S.S., Gautam A., et al. SATPdb: a database of structurally annotated therapeutic peptides. Nucleic Acids Res. 2016;44:D1119–D1126. doi: 10.1093/nar/gkv1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan L., Sun J., Zhou M., Zhou J., Lao X., Zheng H., et al. DRAMP: a comprehensive data repository of antimicrobial peptides. Sci Rep. 2016;6:24482. doi: 10.1038/srep24482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi G., Kang X., Dong F., Liu Y., Zhu N., Hu Y., et al. DRAMP 3.0: an enhanced comprehensive data repository of antimicrobial peptides. Nucleic Acids Res. 2022;50:D488–D496. doi: 10.1093/nar/gkab651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao X., Wu H., Lu H., Li G., Huang Q. LAMP: A Database Linking Antimicrobial Peptides. PLoS ONE. 2013;8:e66557. doi: 10.1371/journal.pone.0066557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quiroz C., Saavedra Y.B., Armijo-Galdames B., Amado-Hinojosa J., Olivera-Nappa A., Sanchez-Daza A., et al. Peptipedia: a user-friendly web application and a comprehensive database for peptide research supported by Machine Learning approach. Database (Oxford) 2021;2021 doi: 10.1093/database/baab055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jhong J.H., Chi Y.H., Li W.C., Lin T.H., Huang K.Y., Lee T.Y. dbAMP: an integrated resource for exploring antimicrobial peptides with functional activities and physicochemical properties on transcriptome and proteome data. Nucleic Acids Res. 2019;47:D285–D297. doi: 10.1093/nar/gky1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung C.R., Kuo T.R., Wu L.C., Lee T.Y., Horng J.T. Characterization and identification of antimicrobial peptides with different functional activities. Brief Bioinform. 2019 doi: 10.1093/bib/bbz043. [DOI] [PubMed] [Google Scholar]
- 31.Manavalan B., Patra M.C. MLCPP 2.0: an updated cell-penetrating peptides and their uptake efficiency predictor. J Mol Biol. 2022;434:167604. doi: 10.1016/j.jmb.2022.167604. [DOI] [PubMed] [Google Scholar]
- 32.Wang X., Li F., Xu J., Rong J., Webb G.I., Ge Z., et al. ASPIRER: a new computational approach for identifying non-classical secreted proteins based on deep learning. Brief Bioinform. 2022;23 doi: 10.1093/bib/bbac031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malik A., Subramaniyam S., Kim C.B., Manavalan B. SortPred: The first machine learning based predictor to identify bacterial sortases and their classes using sequence-derived information. Comput Struct Biotechnol J. 2022;20:165–174. doi: 10.1016/j.csbj.2021.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li F., Guo X., Xiang D., Pitt M.E., Bainomugisa A., Coin L.J.M. Computational analysis and prediction of PE_PGRS proteins using machine learning. Comput Struct Biotechnol J. 2022;20:662–674. doi: 10.1016/j.csbj.2022.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chai D., Jia C., Zheng J., Zou Q., Li F. Staem5: A novel computational approachfor accurate prediction of m5C site. Mol Ther Nucleic Acids. 2021;26:1027–1034. doi: 10.1016/j.omtn.2021.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei L., He W., Malik A., Su R., Cui L., Manavalan B. Computational prediction and interpretation of cell-specific replication origin sites from multiple eukaryotes by exploiting stacking framework. Brief Bioinform. 2021;22 doi: 10.1093/bib/bbaa275. [DOI] [PubMed] [Google Scholar]
- 37.Basith S., Hasan M.M., Lee G., Wei L., Manavalan B. Integrative machine learning framework for the identification of cell-specific enhancers from the human genome. Brief Bioinform. 2021;22 doi: 10.1093/bib/bbab252. [DOI] [PubMed] [Google Scholar]
- 38.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., et al. Scikit-learn: Machine learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 39.Ke G., Meng Q., Finley T., Wang T., Chen W., Ma W., et al. Lightgbm: A highly efficient gradient boosting decision tree. Adv Neural Inform Process Syst. 2017;30:3146–3154. [Google Scholar]
- 40.Sharma R., Shrivastava S., Singh S.K., Kumar A., Singh A.K., Deep-AVPpred S.S. Artificial intelligence driven discovery of peptide drugs for viral infections. IEEE J Biomed Health Inform. 2021 doi: 10.1109/JBHI.2021.3130825. [DOI] [PubMed] [Google Scholar]
- 41.Sharma R., Shrivastava S., Kumar Singh S., Kumar A., Saxena S., Kumar S.R. Deep-AFPpred: identifying novel antifungal peptides using pretrained embeddings from seq2vec with 1DCNN-BiLSTM. Brief Bioinform. 2022;23 doi: 10.1093/bib/bbab422. [DOI] [PubMed] [Google Scholar]
- 42.Chollet F. Simon and Schuster; 2021. Deep learning with Python. [Google Scholar]
- 43.Hasan M.M., Shoombuatong W., Kurata H., Manavalan B. Critical evaluation of web-based DNA N6-methyladenine site prediction tools. Brief Funct Genomics. 2021;20:258–272. doi: 10.1093/bfgp/elaa028. [DOI] [PubMed] [Google Scholar]
- 44.Agrawal P., Bhagat D., Mahalwal M., Sharma N., Raghava G.P.S. AntiCP 2.0: an updated model for predicting anticancer peptides. Brief Bioinform. 2021;22 doi: 10.1093/bib/bbaa153. [DOI] [PubMed] [Google Scholar]
- 45.Charoenkwan P., Chiangjong W., Lee V.S., Nantasenamat C., Hasan M.M., Shoombuatong W. Improved prediction and characterization of anticancer activities of peptides using a novel flexible scoring card method. Sci Rep. 2021;11:3017. doi: 10.1038/s41598-021-82513-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang X., Li F., Chen J., Li J., Wu H., Li S., et al. Large-scale comparative review and assessment of computational methods for anti-cancer peptide identification. Brief Bioinform. 2021;22 doi: 10.1093/bib/bbaa312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hanley J.A., McNeil B.J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 48.Štrumbelj E., Kononenko I. Explaining prediction models and individual predictions with feature contributions. Knowl Inf Syst. 2014;41:647–665. [Google Scholar]
- 49.Chen J., Cheong H.H., Siu S.W.I. xDeep-AcPEP: deep learning method for anticancer peptide activity prediction based on convolutional neural network and multitask learning. J Chem Inf Model. 2021;61:3789–3803. doi: 10.1021/acs.jcim.1c00181. [DOI] [PubMed] [Google Scholar]
- 50.Yan J., Zhang B., Zhou M., Kwok H.F., Siu S.W.I. Multi-Branch-CNN: Classification of ion channel interacting peptides using multi-branch convolutional neural network. Comput Biol Med. 2022;147 doi: 10.1016/j.compbiomed.2022.105717. [DOI] [PubMed] [Google Scholar]
- 51.Hasan M.M., Schaduangrat N., Basith S., Lee G., Shoombuatong W., Manavalan B. HLPpred-Fuse: improved and robust prediction of hemolytic peptide and its activity by fusing multiple feature representation. Bioinformatics. 2020;36:3350–3356. doi: 10.1093/bioinformatics/btaa160. [DOI] [PubMed] [Google Scholar]
- 52.Kalyan G., Junghare V., Khan M.F., Pal S., Bhattacharya S., Guha S., et al. Anti-hypertensive peptide predictor: a machine learning-empowered web server for prediction of food-derived peptides with potential angiotensin-converting enzyme-i inhibitory activity. J Agric Food Chem. 2021;69:14995–15004. doi: 10.1021/acs.jafc.1c04555. [DOI] [PubMed] [Google Scholar]
- 53.Manavalan B., Basith S., Shin T.H., Wei L., Lee G. mAHTPred: a sequence-based meta-predictor for improving the prediction of anti-hypertensive peptides using effective feature representation. Bioinformatics. 2019;35:2757–2765. doi: 10.1093/bioinformatics/bty1047. [DOI] [PubMed] [Google Scholar]
- 54.Timmons P.B., Hewage C.M. HAPPENN is a novel tool for hemolytic activity prediction for therapeutic peptides which employs neural networks. Sci Rep. 2020;10:10869. doi: 10.1038/s41598-020-67701-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen Z., Zhao P., Li C., Li F., Xiang D., Chen Y.Z., et al. iLearnPlus: a comprehensive and automated machine-learning platform for nucleic acid and protein sequence analysis, prediction and visualization. Nucleic Acids Res. 2021;49:e60. doi: 10.1093/nar/gkab122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen Z., Zhao P., Li F., Marquez-Lago T.T., Leier A., Revote J., et al. iLearn: an integrated platform and meta-learner for feature engineering, machine-learning analysis and modeling of DNA, RNA and protein sequence data. Brief Bioinform. 2020;21:1047–1057. doi: 10.1093/bib/bbz041. [DOI] [PubMed] [Google Scholar]
- 57.Li H.L., Pang Y.H., Liu B. BioSeq-BLM: a platform for analyzing DNA, RNA and protein sequences based on biological language models. Nucleic Acids Res. 2021;49:e129. doi: 10.1093/nar/gkab829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu B. BioSeq-Analysis: a platform for DNA, RNA and protein sequence analysis based on machine learning approaches. Brief Bioinform. 2019;20:1280–1294. doi: 10.1093/bib/bbx165. [DOI] [PubMed] [Google Scholar]
- 59.Liu B., Gao X., Zhang H. BioSeq-Analysis2.0: an updated platform for analyzing DNA, RNA and protein sequences at sequence level and residue level based on machine learning approaches. Nucleic Acids Res. 2019;47:e127. doi: 10.1093/nar/gkz740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hasan M.M., Tsukiyama S., Cho J.Y., Kurata H., Alam M.A., Liu X., et al. Deepm5C: A deep-learning-based hybrid framework for identifying human RNA N5-methylcytosine sites using a stacking strategy. Mol Ther. 2022 doi: 10.1016/j.ymthe.2022.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.