Abstract
Two ATP-binding cassette (ABC) exporters are present in Pseudomonas fluorescens no. 33; one is the recently reported AprDEF system and the other is HasDEF, which exports a heme acquisition protein, HasA. The hasDEF genes were cloned by DNA hybridization with a DNA probe coding for the LipB protein, one of the components of the Serratia marcescens ABC exporter Lip system. P. fluorescens HasA showed sequence identity of 40 to 49% with HasA proteins from Pseudomonas aeruginosa and Serratia marcescens. The P. fluorescens Has exporter secreted HasA proteins from P. fluorescens and P. aeruginosa but not S. marcescens HasA in Escherichia coli, whereas the Has exporter from S. marcescens allowed secretion of all three HasA proteins. The P. fluorescens HasDEF system also promoted the secretion of the lipase and alkaline protease of P. fluorescens. Hybrid exporter analysis demonstrated that the HasD proteins, which are ABC proteins, are involved in the discrimination of export substrates. Chimeric HasA proteins containing both P. fluorescens and S. marcescens sequences were produced and tested for secretion through the Has exporters. The C-terminal region of HasA was shown to be involved in the secretion specificity of the P. fluorescens Has exporter.
Many transport systems are involved in protein translocation across the cell membranes in gram-negative bacteria. One is the ATP-binding cassette (ABC) exporters, which are termed type I (6, 34). This system mediates a one-step secretion of proteins. The secretion differs from that through the sec gene-mediated pathway in that the secretory proteins lack an N-terminal signal sequence. The secretion system is composed of three specific components. The first component is situated in the inner membrane and belongs to the ABC transporters (17, 27). The second component is a member of the membrane fusion protein (MFP) family (8) and is a transport accessory protein that may associate with both the inner and outer membranes. The third component is an outer membrane protein (OMP).
ABC exporters translocate various polypeptides. One example is the Serratia marcescens HasSM system (23) composed of HasDSM (ABC protein), HasESM (MFP), and HasFSM (OMP) that promote secretion of the heme-binding protein HasA (HasASM) (25). S. marcescens also possesses another ABC exporter, the Lip system (LipB-LipC-LipD) (2), which mediates secretion of three unrelated proteins: the lipase LipASM (1), a metalloprotease, and the cell surface layer protein homologue SlaA. The structural gene of the secretory protein is usually linked to the genes encoding its specific ABC exporter. In S. marcescens, the genes for HasASM, HasDSM, and HasESM are encoded in the has operon (23) but the gene coding for HasFSM is located in another locus (5). Three components of the Lip system are encoded in the lipBCD operon, and the gene for SlaA is situated upstream of the operon (20).
Many proteins secreted through the ABC exporters possess a common C-terminal secretory signal. Some of them can be secreted by heterologous ABC exporters. For examples, the Erwinia chrysanthemi metalloprotease PrtC can be secreted via the HasSM and Lip systems (3, 4), the Pseudomonas aeruginosa AprPA system (AprDPA-AprEPA-AprFPA) (14) mediates secretion of the Pseudomonas fluorescens lipase LipAPF (9), and the E. chrysanthemi Prt system (PrtD-PrtE-PrtF) (22) promotes secretion of the P. aeruginosa alkaline protease AprAPA (10). However, efficient secretion of the proteins with the C-terminal signal through heterologous ABC exporters is not always possible. HasASM cannot be secreted via the Prt and Lip exporters (3, 4). Likewise, LipASM is secreted neither by the reconstituted HasSM system in Escherichia coli cells nor by the native HasSM system in S. marcescens (3). Analysis of hybrid exporters comprising components from ABC exporters revealed that one determinant of substrate specificity is the ABC protein (3, 4). Novel ABC exporters or secretory proteins showing unique secretion specificities would be useful tools for the analysis of the mechanism of substrate selectivity.
Among the secretory proteins exported through ABC exporters, HasA shows a unique secretion profile. Two HasA proteins have been identified from S. marcescens and P. aeruginosa (24, 26). Secretion of HasA is specific, and the HasSM system is the only one known. The P. aeruginosa Has exporter has been predicted but not identified yet. Since P. fluorescens belongs to the same rRNA homology group as P. aeruginosa (29), the presence of the Has system was expected in P. fluorescens. In this paper, we describe a Has system including a HasA protein and an ABC exporter from P. fluorescens. Specificity of HasA secretion through the Has exporter of this bacterium was studied by using chimeric HasA proteins.
MATERIALS AND METHODS
Strains, plasmids, and media.
E. coli K-12 DH5 (31) and P. fluorescens no. 33 (21) were used. The plasmids pSYC1000 (26), pUC/PFLipA13 (19), and pAK42 (19) encoding the P. aeruginosa HasA (HasAPA), P. fluorescens LipA (LipAPF), and P. fluorescens AprA (AprAPF) proteins, respectively, were used for secretion analysis. The aprDEFPF plasmid pACYC/AK60 was described previously (19). Luria-Bertani medium (31) was used for E. coli and P. fluorescens cells. Antibiotics were added at the following concentrations: ampicillin, 50 μg/ml; kanamycin, 50 μg/ml; and chloramphenicol, 20 μg/ml.
General methods.
DNA manipulations and hybridization analysis were carried out according to standard procedures (31). PCR was carried out through 30 cycles of denaturation at 95°C for 30 s, annealing at 54°C for 30 s, and extension at 72°C for 30 s with ExTaq purchased from Takara Shuzo (Kyoto, Japan). After being subcloned into pUC18 (31), pUC19 (31), pHSG299 (33), and pBluescript II SK(+) (31), the nucleotide sequence was determined with an automated DNA sequencer model 373A and a dideoxy chain termination procedure with fluorescence-labeled primers according to a protocol of the manufacturer (Perkin-Elmer Applied Biosystems). Nucleotide and amino acid sequence data were analyzed with the computer program GENETYX (Software Development, Tokyo, Japan).
Southern blot analysis.
The P. fluorescens chromosomal DNA was digested with one or a combination of the following restriction enzymes: EcoRI, HindIII, BamHI, SalI, and EcoRV. From each digested DNA, 10 μg was separated on a 0.7% agarose gel and then transferred onto a nylon membrane. The 0.9-kb PstI fragment of the S. marcescens lipB gene was prepared from pMWBCD10 (2). The blot was hybridized with the 32P-labeled DNA fragment at 55°C for 16 h and washed twice in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.5% sodium dodecyl sulfate (SDS) at room temperature, followed by a 5-min wash in 2× SSC–0.5% SDS at 45°C (low-stringency conditions). The filter was exposed to X-ray film at −70°C for 2 days. To confirm that the DNA fragment was related to the aprDPF gene, the 1.7-kb BglII-SacI fragment encoding the AprDPF N-terminal half was prepared from pAK42 (19). The blot was hybridized with the 32P-labeled probe at 65°C for 16 h and washed twice in 0.1× SSC–0.5% SDS at room temperature, followed by two 10-min washes in 0.1× SSC–0.5% SDS at 65°C (high-stringency conditions), and then exposed to X-ray film at −70°C overnight.
Cloning of the ABC exporter genes from P. fluorescens.
A genomic DNA library was constructed in E. coli with ligation of 1 μg of SalI-digested pUC19 and 10 μg of 9- to 23-kb SalI-digested P. fluorescens chromosomal DNA. The clones were isolated by colony hybridization with the above-described 32P-labeled lipB fragment under low-stringency conditions. To isolate all of the P. fluorescens ABC exporter genes, a genomic DNA library was constructed in E. coli with ligation of 1 μg of BamHI-EcoRV-digested pACYC184 (31) and 10 μg of BglII-EcoRV-digested P. fluorescens chromosomal DNA (6.4 to 9.7 kb). pOI78 was obtained from the library as the clone that hybridized with the 32P-labeled SalI-BglII (1.4-kb) fragment from pOIS90.
Plasmid construction.
The HasDEFPF plasmids pACYC/OI70 and pOI70R carrying the 6.4-kb BglII-StuI hasDEFPF fragment (blunt ended) were constructed in the EcoRV site of pACYC184 in the same and opposite orientations as the tet gene, respectively (Fig. 1). Deletion of the 1.4-kb SalI-StuI fragment from pACYC/OI70 produced pOI50. pOI701 and pOI702 were generated from pACYC/OI70 by destroying the SacI and EcoRI sites in the hasFPF and hasEPF genes, respectively. The hasEFPF plasmid pOI703 was produced by ligating the 5.4-kb NcoI fragment (blunt ended) with the EcoRV-digested pACYC184. Construction of the hasDPF plasmid pACYC/HasDPF was done by introducing the HindIII and blunt-ended EcoRI fragments of pACYC/OI70 into the HindIII-EcoRV-digested pACYC184. The 6.5-kb HindIII-ScaI hasDEFPF fragment of pACYC/OI70 was ligated with the HindIII-HincII-digested pMW219 (36), and then the 2.1-kb HindIII-NotI fragment was removed, resulting in the hasEFPF plasmid pMW/HasEFPF. The plasmids pACYC/HasDSM and pMW/HasESM containing the hasDSM and hasESM genes in pACYC184 and pMW218 (36), respectively, were produced from pMW/HasDE7 carrying the hasDESM genes of S. marcescens 8000 (unpublished data).
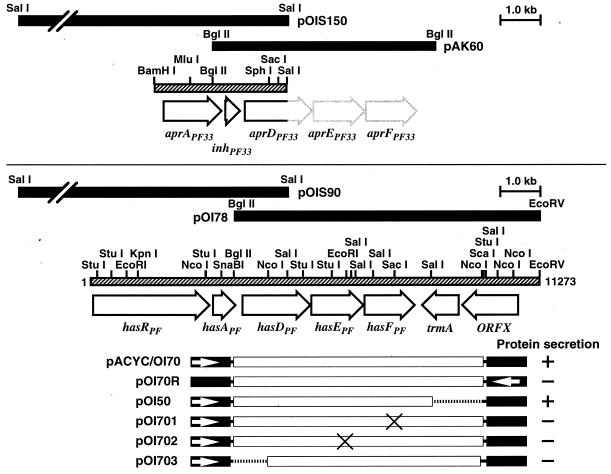
FIG. 1.
Physical maps of inserted DNA. The black bars and hatched bars represent the chromosomal DNA inserts and the regions for which the nucleotide sequences were determined, respectively. The plasmids pOIS150 and pOIS90 are shown in the upper and lower panels, respectively. The open arrows indicate ORFs. The plasmids encoding the HasDEFPF exporter mutants are shown below. The open and solid boxes represent the inserted DNA and the pACYC184 DNA, respectively. The open arrows show the tet promoter of pACYC184. The crosses and dotted lines indicate the positions of the mutations introduced and deletions, respectively. The ability to secrete HasAPF (+) is shown on the right.
Construction of HasA plasmids.
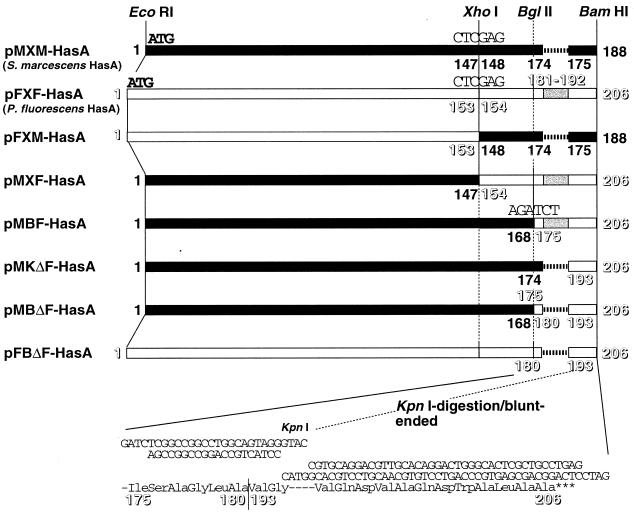
Inserting the 4.1-kb EcoRI-SalI fragment of pOIS90 into the corresponding sites of pUC18 produced the hasAPF plasmid pUC/HasAPF. The 0.6-kb DNA fragment containing the hasASM gene was amplified by PCR with a primer set (5′-GGGAATTCTAATTCATCAATGGAGATAGAGAAATG-3′ and 5′-GGGGATCCGGCGGGCAAACGGCCGCGATCAGG-3′) and pSYC134 (25) as a template. After digestion with EcoRI and BamHI, the fragment was inserted into the corresponding sites of pUC18, generating pUC/HasASM. The plasmids encoding chimeras between HasASM and HasAPF were produced by creating the XhoI sites at Leu-Glu (amino acid residues 153 to 154 in HasAPF and amino acid residues 147 to 148 in HasASM) by PCR with pUC/HasASM and pUC/HasAPF as templates (Fig. 2). The fragments encoding the N-terminal regions of HasASM and HasAPF were generated with the primer sets RV primer (5′-CAGGAAACAGCTATGAC-3′) plus 5′-CCGGATCCCTCGAGCGCGCCGGTATCGCCGGACATCAGG-3′) and 5′-GCGAATTCGAGGTTAAGTGATGACTATTTCTG-3′ plus 5′-CCGGATCCCTCGAGTGCCGAGGTGTTGCCTTGCATCAG-3′, respectively. The EcoRI-BamHI-digested PCR products (ca. 0.45 kb) were introduced into the corresponding sites of pUC18, resulting in pMX and pFX encoding the HasASM and HasAPF N-terminal regions with the XhoI site, respectively. The DNA fragments coding for the HasASM and HasAPF C-terminal regions were obtained by the primer sets 5′-TTCCTCGAGACCGCGCTGAACGGCATCCTCGACGACTA-3′ plus 5′-GTTTTCCCAGTCACGAC-3′ and 5′-GGGAATTCCTCGAGACCGTGCTCAACAACCTGCTGGACG-3′ plus 5′-CCGGATCCTCAGGCAGCGAGTGCCCAGTCCTG-3′, respectively. The amplified fragments were cloned into pMX and pFX by using the XhoI and BamHI sites, producing four HasA plasmids, pMXM-HasA, pMXF-HasA, pFXM-HasA, and pFXF-HasA (Fig. 2).
FIG. 2.
Construction of the HasA chimera plasmids encoding HasA chimeras between HasASM and HasAPF. The construction of the plasmids is illustrated schematically. The solid and open boxes represent amino acid sequences of HasASM and HasAPF, respectively. The HasAPF C-terminal inserts are shaded. The amino acid residue numbers of the HasAPF sequence are shown in outline. The XhoI, BglII, and KpnI sites introduced are shown. The synthetic oligonucleotides encoding the C-terminal sequences of HasAPF (amino acid residues 175 to 180 and 193 to 206), which are introduced in pMKΔF-HasA, pMBΔF-HasA, and pFBΔF-HasA, are shown, with a deduced amino acid sequence below. The dashed lines indicate gaps to maximize homology at the C terminus.
The DNA fragment encoding HasASM (amino acid residues 1 to 168) was produced by PCR with the RV primer, the primer 5′-GGAGATCTGATCGAAGGTGGAGTTGACGC-3′, and the template DNA pUC/HasASM. The amplified fragment (0.5 kb) was digested by EcoRI and BglII and then introduced into the corresponding sites of pFXF-HasA, resulting in pMBF-HasA. The synthetic oligonucleotides 5′-CGTGCAGGACGTTGCACAGGACTGGGCACTCGCTGCCTGAG-3′ and 5′-GATCCTCAGGCAGCGAGTGCCCAGTCCTGTGCAACGTCCTGCACGGTAC-3′, encoding the HasAPF C terminus (amino acid residues 195 to 206) were inserted into the KpnI and BamHI sites of pUC18, generating pUC/PFlinker12. The DNA encoding HasASM (amino acid residues 1 to 175) was produced by PCR with the RV primer and the primer 5′-CCGGTACCCCACCGCCGTCGCCGCCGCCAC-3′ and the template DNA pUC/HasASM. The PCR product was digested with EcoRI and KpnI and introduced into the corresponding sites of pUC/PFlinker12. The KpnI site of the resultant plasmid was destroyed to produce pUC/MKΔF-HasA.
The pMBΔF-HasA plasmid was created as follows: the 0.95-kb ScaI-KpnI fragment of pUC/PFlinker12 was ligated with the 2.2-kb ScaI-BglII fragment of pMBF-HasA and the BglII-KpnI linker 5′-GATCTCGGCCGGCCTGGCAGTAGGGTAC-3′ plus 5′-CCTACTGCCAGGCCGGCCGA-3′. The resultant ampicillin-resistant plasmid was digested with KpnI and then blunt ended to produce pMBΔF-HasA. The plasmid encoding HasAPF lacking the C-terminal insert (pFBΔF-HasA) was produced by inserting the 0.5-kb EcoRI-BglII fragment of pFXF-HasA into the corresponding sites of pMBΔF-HasA.
The nucleotide sequences of all of these hasA genes were confirmed by sequencing. These hasA genes were expressed under control of the lacZ promoter of pUC18.
Analysis of protein secretion.
The E. coli cells carrying the plasmids coding for the exporter and secretory proteins were cultured in Luria-Bertani medium at 30°C for 40 h with vigorous shaking. The polypeptides in the supernatants were precipitated with trichloroacetic acid at a final concentration of 10% and were subjected to SDS-polyacrylamide gel electrophoresis (PAGE). Protein analysis was carried out independently three times, and similar results were confirmed. β-Galactosidase activity was measured as described by Miller (28) to assess cell lysis. The levels of extracellular β-galactosidase activity were <0.5% of that of whole cells, indicating no significant cell lysis.
SDS-PAGE and immunoblot analysis.
The precast gel PAGEL (12.5%) (ATTO, Tokyo, Japan) was used for SDS-PAGE. The proteins in gels were stained by Coomassie brilliant blue G-250 or electrophoretically transferred to an Immobilon P filter (Millipore) for immunodetection. Peptide corresponding to amino acid residues 41 to 60 of HasAPF was synthesized, and the antiserum was obtained by injecting rabbits with the peptide in Freund's complete adjuvant (15). The antiserum to LipASM (1) was used for detection of LipAPF. The antibody against HasASM was a generous gift of Cécile Wandersman. The antiserum raised against P. fluorescens AprA was kindly provided by Tamotsu Hoshino. The blots were blocked by soaking them in Block Ace (Dainippon Pharmaceutical, Osaka, Japan) overnight at 4°C and incubated with antiserum at room temperature for 2 h (diluted 1:1,000 to 1:4,000 in phosphate-buffered saline containing 0.1% Tween 20). They were washed and incubated with horseradish peroxidase-conjugated anti-rabbit immunoglobulin G, and the bound antibody was detected with the enhanced chemiluminescence system (Amersham).
Nucleotide sequence accession number.
The hasRADEFPF sequence has been submitted to the GenBank, EMBL, and DDBJ databases with accession no. AB023289.
RESULTS
Cloning of the ABC exporter homologue genes from P. fluorescens.
The ABC proteins of the Prt, Lip, HasSM, and AprPA systems have 54 to 60% identity. Therefore, Southern analysis of the isolated P. fluorescens chromosomal DNA was performed with the 32P-labeled lipB fragment encoding the N-terminal portion well conserved among ABC proteins under low-stringency conditions. Two positive signals were consistently detected, irrespective of the enzyme used (data not shown). Hybridization analysis revealed that one of the signals was found to come from the aprDPF DNA fragment. To identify the origin of the other signal, a genomic DNA library of P. fluorescens was constructed. Two clones, pOIS90 and pOIS150, which contained SalI fragments of 9 and 15 kb, respectively, with different restriction patterns, were isolated by colony hybridization with the lipB probe under low-stringency conditions (Fig. 1).
Nucleotide sequence analysis revealed that pOIS150 and pOIS90 encode a part of the aprPF operon (19) and the N-terminal-to-central region of a novel ABC protein, respectively. The entire complex of novel ABC exporter genes was isolated as the 7.8-kb BglII-EcoRV fragment cloned into pACYC184 (pOI78 [Fig. 1]). Southern hybridization revealed that the cloned fragments came from the P. fluorescens chromosomal DNA without rearrangement (data not shown).
Nucleotide sequence analysis of the new ABC exporter genes.
The inserted DNA fragment of pOI78 contained five open reading frames (ORFs) predicted to encode proteins of 580 (Mr, 61,803), 443 (Mr, 48,491), 439 (Mr, 48,658), 363 (Mr, 41,240), and 431 (Mr, 44,894) amino acid residues. The first three ORFs are contiguous, and the last two ORFs are located on the opposite strand (Fig. 1). A putative ribosome binding site (32) is present upstream of each ORF. The ORF1 product possessed features of ABC proteins, which are ATP-binding and ABC motifs, and was 67, 60, 62, 59, and 60% identical to HasDSM, AprDPF, AprDPA, PrtD, and LipB proteins, respectively. The ORF2 product showed 50 to 52% identity to the MFPs of the exporters. The ORF3 product, a putative OMP, was 57, 58, 52, 54, 22, and 26% identical to AprFPF, AprFPA, PrtF, LipD, TolC (35), and HasFSM, respectively.
Upstream of the exporter genes, two ORFs encoding proteins of 206 (Mr, 21,243) and 916 (Mr, 101,186) amino acid residues were found (Fig. 1). The product of the former ORF (hasAPF) was a homologue of the heme-binding proteins, showing 41 and 40% identity with HasASM and HasAPA, respectively. The latter (the hasRPF gene product) was 43% identical to S. marcescens HasRSM (12), which is an OMP capable of binding HasA. On the basis of this gene organization and the secretion function described below, the exporter genes downstream of the hasRAPF genes were designated hasDPF, hasEPF, and hasFPF, respectively. No obvious promoter sequence (16) was found upstream of or within the hasRADEFPF complex. Thus, P. fluorescens appears to possess a heme acquisition system and at least two kinds of ABC exporter, AprDEFPF and HasDEFPF.
The hasRADEFPF genes were not followed by a typical rho-independent terminator (30), and the downstream ORF was in the opposite orientation. The product of this ORF was 48% identical to tRNA (uracil-5-)-methyltransferases (EC 2.1.1.35) from E. coli (13), and the ORF was designated trmA. ORFX, predicted to encode a protein that was 52% identical to hypothetical proteins reported in E. coli (7), was found upstream of the trmA gene in the same orientation as trmA.
Protein secretion through the P. fluorescens ABC exporter.
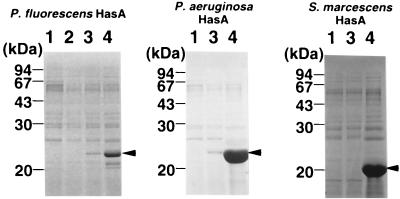
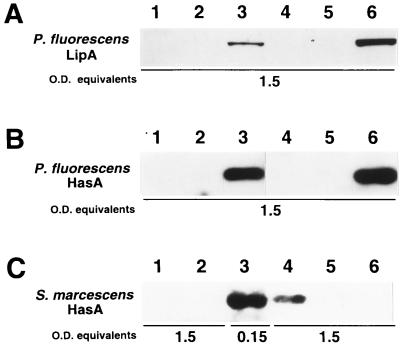
The abilities of AprPF and HasPF to secrete protein were examined in recombinant E. coli (Fig. 3). First, we investigated secretion of HasAPF. HasAPF secretion was promoted by the HasPF exporter, but secretion of HasAPF through AprPF encoded by pACYC/AK60 (19) was not detectable even on immunoblot analysis with the antiserum against HasAPF (data not shown). The HasPF exporter plasmids lacking one of the three components (pOI701, pOI702, or pOI703) were unable to secrete HasAPF (Fig. 1). Since pOI701 deficient in the gene for the OMP HasFPF did not allow HasAPF secretion in E. coli DH5 (carrying the tolC gene), HasFPF function cannot be replaced by the E. coli TolC. pOI70R did not direct the E. coli cells to secrete HasAPF, showing that expression of hasDEFPF is driven by an exogenous promoter, Ptet in pACYC184. Interestingly, the HasPF exporter also secreted HasAPA but not HasASM. On the other hand, the HasSM system encoded by pK150 (4) secreted HasAPF, HasAPA, and HasASM (Fig. 3). The levels of HasA secretion through the HasPF exporter were low compared with those through the HasSM exporter.
FIG. 3.
HasA secretion through the Has exporters. The plasmids pUC/HasAPF (left), pSYC1000 (middle), and pUC/HasASM (right) encoding the HasA proteins from P. fluorescens, P. aeruginosa, and S. marcescens, respectively, were introduced into the E. coli cells carrying the AprPF (pACYC/AK60), HasPF (pACYC/OI70), and HasSM (pK150) exporters. The polypeptides in the supernatants of the cultured media (1.5 optical density equivalents) were concentrated and then subjected to SDS-PAGE (12.5% acrylamide). The gels were stained with Coomassie brilliant blue G-250. The arrowheads indicate the positions of the HasA proteins, and the positions of molecular mass markers are shown on the left of each gel. Lanes 1, pACYC184; lane 2, pACYC/AK60; lanes 3, pACYC/OI70; lanes 4, pK150.
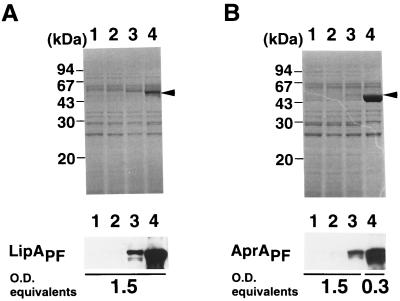
In addition to HasA secretion, the HasPF exporter also promoted secretion of the P. fluorescens lipase LipAPF and alkaline protease AprAPF in E. coli cultured at 30°C (Fig. 4). Under these conditions, the AprPF exporter, which is temperature sensitive (19), failed to secrete these proteins. Interestingly, LipAPF was secreted through the HasSM exporter although the exporter did not promote secretion of LipASM, which is 66% identical to LipAPF.
FIG. 4.
Secretion of LipAPF (A) and AprAPF (B) through the Has exporters. E. coli cells carrying LipAPF and AprAPF plasmids (pUC/PFLipA13 and pAK42, respectively) with the various exporters were cultured and tested for protein secretion as described in the legend to Fig. 3. The proteins were visualized by Coomassie brilliant blue G-250 (upper gels) and analyzed by immunoblotting with antisera against LipASM and AprAPF (lower gels). The arrowheads indicate the positions of the LipAPF and AprAPF proteins, and the positions of molecular mass markers are shown on the left of each gel. The positions of molecular mass markers are shown on the left of the upper gels. Optical density (O.D.) equivalents are indicated below. Lanes 1, pACYC184; lanes 2, pACYC/AK60; lanes 3, pACYC/OI70; lanes 4, pK150.
Secretion analysis of the hybrid Has exporters.
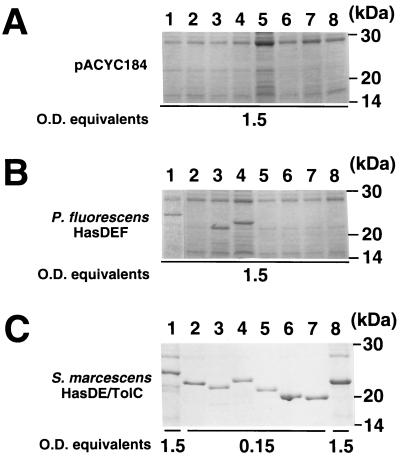
To investigate the functional overlap of two Has exporters, HasPF and HasSM, secretion analysis was carried out with hybrid exporters having components from each system (Fig. 5). The hasDSM or hasDPF plasmid and the hasESM or hasEFPF plasmid were introduced into the E. coli cells carrying the plasmids encoding LipAPF, HasAPF, and HasASM. Only exporters composed of components from the same system (HasDSM-HasESM-TolC or HasDPF-HasEPF-HasFPF) were functional for LipAPF and HasAPF secretion (Fig. 5A and B, lanes 3 and 6). The hybrid exporter HasDSM-HasEPF-HasFPF allowed HasASM secretion at a very low level (3%) compared with that through HasDSM-HasESM-TolC (Fig. 5C, lanes 3 and 4); all other hybrid systems were nonfunctional. The finding that HasEPF and HasFPF operated as an MFP and an OMP for HasASM secretion, respectively, albeit very inefficiently, suggested that HasDPF plays a key role for substrate specificity in the HasPF system.
FIG. 5.
HasA and LipA secretion by hybrid Has exporters. The plasmids pACYC/HasDPF and pACYC/HasDSM were used as the plasmids encoding the ABC proteins HasDPF and HasDSM8000, respectively. MFP and OMP were provided by pMW/HasEFPF (hasEFPF) and pMW/HasESM. These plasmids were introduced into the E. coli cells harboring the LipAPF plasmid pUC/PFLipA13 (A), the HasAPF plasmid pUC/HasAPF (B), and the HasASM plasmid pUC/HasASM (C). The cells were cultured and tested for protein secretion as described in the legend to Fig. 3. The gels were analyzed by immunoblotting with antisera against LipASM, HasAPF, and HasASM. Secretory proteins are indicated on the left and optical density (O.D.) equivalents are shown below. Lanes 1, pACYC184 plus pMW/HasESM; lanes 2, pACYC184 plus pMW/HasEFPF; lanes 3, pACYC/HasDSM plus pMW/HasESM; lanes 4, pACYC/HasDSM plus pMW/HasEFPF; lanes 5, pACYC/HasDPF plus pMW/HasESM; lanes 6, pACYC/HasDPF plus pMW/HasEFPF.
Chimeric analysis of HasA secretion through the Has exporter.
The most notable difference among the HasA sequences in the three species is the apparent deletion, in HasASM, of a segment close to the C terminus corresponding to amino acid residues 181 to 192 of HasAPF (Fig. 2). To investigate whether the substrate specificity of the HasPF exporters on HasA secretion depends on this sequence diversity, we constructed chimeras containing the HasASM and HasAPF sequences and examined their secretion by Has exporters, HasPF and HasSM (Fig. 2) (see Materials and Methods). First, C-terminal HasA chimeras (pFXM-HasA and pMXF-HasA) were created and then tested for secretion from the E. coli cells carrying the Has exporters (Fig. 6). The HasSM system allowed secretion of all these chimeras (Fig. 6C). The HasPF exporter secreted HasAPF and a chimera produced by pMXF-HasA (Fig. 6B, lanes 1 and 3) but failed to secrete HasASM and a chimera from pFXM-HasA (Fig. 6B, lanes 2 and 7), indicating that the C-terminal region of HasAPF is involved in HasAPF secretion via the HasPF exporter. To define the sequence related to the substrate specificity, we created further C-terminal chimeras (Fig. 2). A pMBF-HasA-encoded chimera that is HasASM containing the HasAPF C-terminal sequence of 32 amino acid residues (amino acid residues 175 to 206) was secreted by both Has exporters (Fig. 6B and C, lanes 4). Chimeras encoded in pMKΔF-HasA and pMBΔF-HasA, which carry a C terminus of HasAPF but lack the HasAPF insert (Ala181 to Leu192), were exported by HasSM (Fig. 6C, lanes 5 and 6) but not by HasPF (Fig. 6B, lanes 5 and 6). Deletion of the insert from HasAPF (pFBΔF-HasA) did not affect secretion of the chimera through the HasSM system (Fig. 6C, lane 8), whereas the HasPF exporter aborted secretion (Fig. 6B, lane 8). These findings showed that the inserted segment close to the HasAPF C terminus may play an important role in HasAPF recognition by the HasPF exporter.
FIG. 6.
Secretion of chimeras between HasASM and HasAPF. Eight hasA plasmids encoding HasASM and HasAPF and six HasA chimeras were introduced into the E. coli cells carrying pACYC184 (A), pK150 (B), and pACYC/OI70 (C). The cells were cultured and tested for protein secretion as described in the legend to Fig. 3. The gels were stained with Coomassie brilliant blue G-250. Optical density (O.D.) equivalents are indicated below each gel. Lanes 1, pFXF-HasA; lanes 2, pFXM-HasA; lanes 3, pMXF-HasA; lanes 4, pMBF-HasA; lanes 5, pMBΔF-HasA; lanes 6, pMKΔF-HasA; lanes 7, pMXM-HasA; lanes 8, pFBΔF-HasA.
DISCUSSION
The P. fluorescens genes encoding an unidentified ABC exporter were isolated together with the secretory protein gene. The secretory protein HasAPF, which is similar to HasASM and also lacks an N-terminal signal peptide, was shown to possess heme-binding activity, a typical feature of HasA (unpublished data). The gene organization of the hasRADEFPF operon showed a typical structure of the genes coding for the ABC exporter and its secretory protein. The HasPF exporter secreted HasAPF and also mediated secretion of LipAPF and AprAPF more efficiently than AprPF did in the recombinant E. coli system. The presence of two distinct ABC exporters, AprPF and HasPF, in P. fluorescens demonstrated that S. marcescens is not unique in possessing two ABC exporters, the Has and Lip systems (2, 23).
Several differences between the HasPF and HasSM systems were observed. The OMP HasFPF was essential for secretion, and its function could not be replaced with TolC, in spite of HasFSM being replaceable with TolC. The hasDEFPF operon coded for all three secretory components, whereas the HasSM system is encoded by the hasDESM operon and the unlinked hasFSM gene. Of interest was the substrate specificity of the HasPF exporter. This system promoted secretion of HasAPF and HasAPA but not HasASM, whereas the HasSM exporter allowed secretion of all HasA proteins tested. Hybrid exporter analysis indicated that an ABC protein (HasDPF) was responsible for the substrate specificity, as in other ABC exporters (3, 4).
HasA proteins were 40 to 49% identical to each other, but several insertions and deletions were observed. The secretion signal of the proteins secreted by the ABC exporter is known to be situated at the C terminus (11), often containing a motif consisting of negatively charged amino acid residues followed by several hydrophobic residues. The sequences D-W-A-L-A-A, D-L-A-L-A-A, and E-L-L-A-A are identified in the C-termini of HasAPF, HasAPA, and HasASM, respectively (26). These motifs are expected to be essential but not sufficient for secretion. In some cases, the signal was located in the last 15 to 30 amino acids but at a low secretion level (10, 11).
We postulated that the inserts close to the C terminus, which are found in HasAPF and HasAPA but not HasASM and contain several conserved amino acid residues, may be involved in the specific secretion via the HasPF exporter. Interestingly, a HasAPF inserted segment (Ala181 to Leu192) close to the C terminus was shown to be responsible for the substrate specificity on HasA secretion via the HasPF exporter. Lack of the insert caused failure of secretion. Very recently, conformation of the C terminus of HasASM, which is the last 15 residues, containing the motif essential for secretion, has been studied by 1H nuclear magnetic resonance (18). This peptide was shown to be highly flexible and unstructured. It is of interest that the C-terminal insert, which we showed to be responsible for the specificity of secretion, is situated just upstream of the last 15 amino acid residues. Since HasSM allows the secretion of all three HasA proteins and HasA chimeras, further secretion analysis of the HasA chimeras and mutants with amino acid substitutions through HasPF and other ABC exporters may be of interest.
ACKNOWLEDGMENTS
We are grateful to Yumiko Kawashima for DNA sequencing. The strain P. fluorescens no. 33 was kindly provided by Haruto Kumura. We gratefully acknowledge Sylvie Létoffé, Philippe Delepelaire, and Cécile Wandersman for generous gifts of the anti-HasASM antibody and the plasmids containing hasASM, hasAPA, and hasDESM. The antiserum against the P. fluorescens AprA is a kind gift from Tamotsu Hoshino.
REFERENCES
- 1.Akatsuka H, Kawai E, Omori K, Komatsubara S, Shibatani T, Tosa T. The lipA gene of Serratia marcescens which encodes an extracellular lipase having no N-terminal signal peptide. J Bacteriol. 1994;176:1949–1956. doi: 10.1128/jb.176.7.1949-1956.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akatsuka H, Kawai E, Omori K, Shibatani T. The three genes lipB, lipC, and lipD involved in the extracellular secretion of the Serratia marcescens lipase which lacks an N-terminal signal peptide. J Bacteriol. 1995;177:6381–6389. doi: 10.1128/jb.177.22.6381-6389.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akatsuka H, Binet R, Kawai E, Wandersman C, Omori K. Lipase secretion by bacterial hybrid ATP-binding cassette exporters: molecular recognition of the LipBCD, PrtDEF, and HasDEF exporters. J Bacteriol. 1997;176:4754–4760. doi: 10.1128/jb.179.15.4754-4760.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Binet R, Wandersman C. Protein secretion by bacterial hybrid ABC-transporters: specific functions of the membrane ATPase and membrane fusion protein. EMBO J. 1995;14:2298–2306. doi: 10.1002/j.1460-2075.1995.tb07224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Binet R, Wandersman C. Cloning of the Serratia marcescens hasF gene encoding the Has ABC exporter outer membrane component: a TolC analogue. Mol Microbiol. 1996;22:265–273. doi: 10.1046/j.1365-2958.1996.00103.x. [DOI] [PubMed] [Google Scholar]
- 6.Binet R, Létoffé S, Ghigo J-M, Delepelaire P, Wandersman C. Protein secretion by Gram-negative bacterial ABC exporters—a review. Gene. 1997;192:7–11. doi: 10.1016/s0378-1119(96)00829-3. [DOI] [PubMed] [Google Scholar]
- 7.Burland V, Plunkett G, Daniels D L, Blattner F R. DNA sequence and analysis of 136 kilobases of the Escherichia coli genome: organizational symmetry around the origin of replication. Genomics. 1993;16:551–561. doi: 10.1006/geno.1993.1230. [DOI] [PubMed] [Google Scholar]
- 8.Dinh T, Paulsen I T, Saier M H., Jr A family of extracytoplasmic proteins that allow transport of large molecules across the outer membranes of gram-negative bacteria. J Bacteriol. 1994;176:3825–3831. doi: 10.1128/jb.176.13.3825-3831.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duong F, Soscia C, Lazdunski A, Murgier M. The Pseudomonas fluorescens lipase has C-terminal secretion signal and is secreted by a three-component bacterial ABC-exporter system. Mol Microbiol. 1994;11:1117–1126. doi: 10.1111/j.1365-2958.1994.tb00388.x. [DOI] [PubMed] [Google Scholar]
- 10.Duong F, Lazdunski A, Murgier M. Protein secretion by heterologous bacterial ABC-transporters: the C-terminus secretion signal of the secreted protein conferred high recognition specificity. Mol Microbiol. 1996;21:459–470. doi: 10.1111/j.1365-2958.1996.tb02555.x. [DOI] [PubMed] [Google Scholar]
- 11.Ghigo J-M, Wandersman C. A carboxyl-terminal four amino acid motif is required for secretion of the metalloprotease PrtG through the Erwinia chrysanthemi protease secretion pathway. J Biol Chem. 1994;269:8979–8985. [PubMed] [Google Scholar]
- 12.Ghigo J-M, Létoffé S, Wandersman C. A new type of hemophore-dependent heme acquisition system of Serratia marcescens reconstituted in Escherichia coli. J Bacteriol. 1997;179:3572–3579. doi: 10.1128/jb.179.11.3572-3579.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gustafsson C, Lindstroem P H, Hagervall T G, Esberg K B, Bjoerk G R. The trmA promoter has regulatory features and sequence elements in common with the rRNA P1 promoter family of Escherichia coli. J Bacteriol. 1991;173:1757–1764. doi: 10.1128/jb.173.5.1757-1764.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guzzo J, Duong F, Wandersman C, Murgier M, Lazdunski A. The secretion genes of Pseudomonas aeruginosa alkaline protease are functionally related to those of Erwinia chrysanthemi proteases and Escherichia coli α-haemolysin. Mol Microbiol. 1991;5:447–453. doi: 10.1111/j.1365-2958.1991.tb02128.x. [DOI] [PubMed] [Google Scholar]
- 15.Harlow E, Lane D. Antibody: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 16.Hawley D K, McClure W R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higgins C F. ABC-transporters from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 18.Izadi-Pruneyre N, Wolff N, Redeker V, Wandersman C, Delepierre M, Lecroisey A. NMR studies of the C-terminal secretion signal of the haem-binding protein, HasA. Eur J Biochem. 1999;261:562–568. doi: 10.1046/j.1432-1327.1999.00305.x. [DOI] [PubMed] [Google Scholar]
- 19.Kawai E, Idei A, Kumura H, Shimazaki K, Akatsuka H, Omori K. The ABC-exporter genes involved in the lipase secretion are clustered with lipase, alkaline protease, and serine protease homologues in Pseudomonas fluorescens No. 33. Biochim Biophys Acta. 1999;1446:377–382. doi: 10.1016/s0167-4781(99)00094-9. [DOI] [PubMed] [Google Scholar]
- 20.Kawai E, Akatsuka H, Idei A, Shibatani T, Omori K. Serratia marcescens S-layer protein is secreted extracellularly via an ATP-binding cassette exporter, the Lip system. Mol Microbiol. 1998;27:941–952. doi: 10.1046/j.1365-2958.1998.00739.x. [DOI] [PubMed] [Google Scholar]
- 21.Kumura H, Mikawa K, Saito Z. Purification and some properties of proteinase from Pseudomonas fluorescens No. 33. J Dairy Res. 1993;60:229–237. doi: 10.1017/s0022029900027540. [DOI] [PubMed] [Google Scholar]
- 22.Létoffé S, Delepelaire P, Wandersman C. Protease secretion by Erwinia chrysanthemi: the specific secretion functions are analogous to those of Escherichia coli α-haemolysin. EMBO J. 1990;9:1375–1382. doi: 10.1002/j.1460-2075.1990.tb08252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Létoffé S, Ghigo J-M, Wandersman C. Identification of two components of the Serratia marcescens metalloprotease transporter: protease SM secretion in Escherichia coli is TolC dependent. J Bacteriol. 1993;175:7321–7328. doi: 10.1128/jb.175.22.7321-7328.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Létoffé S, Ghigo J-M, Wandersman C. Iron acquisition from heme and hemoglobin by a Serratia marcescens extracellular protein. Proc Natl Acad Sci USA. 1994;91:9876–9880. doi: 10.1073/pnas.91.21.9876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Létoffé S, Ghigo J-M, Wandersman C. Secretion of the Serratia marcescens HasA protein by an ABC transporter. J Bacteriol. 1994;176:5372–5377. doi: 10.1128/jb.176.17.5372-5377.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Létoffé S, Redeker V, Wandersman C. Isolation and characterization of an extracellular haem-binding protein from Pseudomonas aeruginosa that shares function and sequence similarities with Serratia marcescens HasA haemphore. Mol Microbiol. 1998;28:1223–1234. doi: 10.1046/j.1365-2958.1998.00885.x. [DOI] [PubMed] [Google Scholar]
- 27.Linton K J, Higgins C F. The Escherichia coli ATP-binding cassette (ABC) proteins. Mol Microbiol. 1998;28:5–13. doi: 10.1046/j.1365-2958.1998.00764.x. [DOI] [PubMed] [Google Scholar]
- 28.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. [Google Scholar]
- 29.Palleroni N J. Pseudomonas classification. Antonie Leeuwenhoek. 1993;64:231–251. doi: 10.1007/BF00873084. [DOI] [PubMed] [Google Scholar]
- 30.Rosenberg M, Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 32.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takeshita S, Sato M, Toba M, Masahashi W, Hashimoto-Gotoh T. High-copy-number plasmid vectors for lacZ α-complementation and chloramphenicol- or kanamycin-resistant selection. Gene. 1987;61:63–74. doi: 10.1016/0378-1119(87)90365-9. [DOI] [PubMed] [Google Scholar]
- 34.Wandersman C. Secretion across the bacterial outer membrane. Trends Genet. 1992;8:317–322. doi: 10.1016/0168-9525(92)90264-5. [DOI] [PubMed] [Google Scholar]
- 35.Wandersman C, Delepelaire P. TolC, and Escherichia coli outer membrane protein required for hemolysin secretion. Proc Natl Acad Sci USA. 1990;87:4776–4780. doi: 10.1073/pnas.87.12.4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamaguchi K, Masamune Y. Autogenous regulation of synthesis of the replication protein in plasmid pSC101. Mol Gen Genet. 1985;200:362–367. doi: 10.1007/BF00425718. [DOI] [PubMed] [Google Scholar]