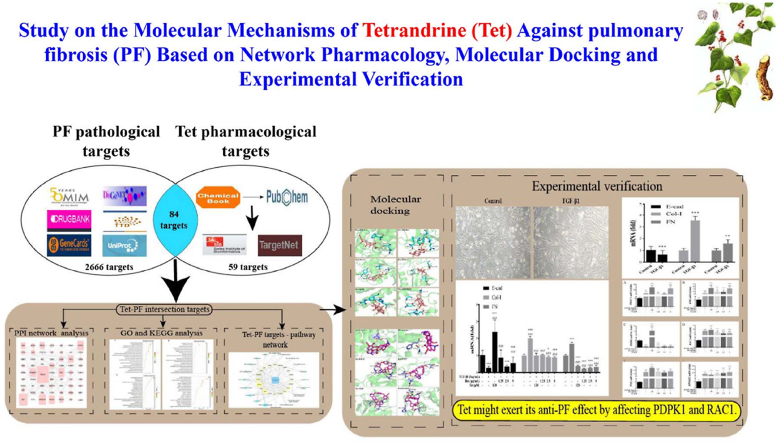
Abstract
Aims
This study aims to screen the potential targets of tetrandrine (Tet) against pulmonary fibrosis (PF) based on network pharmacological analysis, molecular docking and experimental verification.
Main methods
The network pharmacology methods were employed to predict targets, construct Tet-PF-intersection target-pathway networks, and screen the candidate targets. The molecular docking was performed using AutoDockTools1.5.6. TGF-β1-induced human lung adenocarcinoma A549 cells were used as an in vitro experimental verification model, taking dexamethasone (Dex) as the positive control, to verify the effects of Tet on the mRNA expression of the candidate targets.
Key findings
Six candidate targets were predicted based on network pharmacology and molecular docking, namely PIK3CA, PDPK1, RAC1, PTK2, KDR, and RPS6KB1. The experimental verification results showed that Dex and Tet presented quite different pharmacological effects. Specifically, compared with the model group, both Dex and Tet (5 μΜ) significantly increased the mRNA expression of PIK3CA and KDR (P < 0.001). Dex up-regulated the mRNA expression of PDPK1 and RAC1, while Tet (1.25 μΜ) down-regulated (P < 0.001). Dex up-regulated the mRNA expression of PTK2, but Tet had no effect. Dex down-regulated RPS6KB1 mRNA expression, while Tet (5 μΜ) up-regulated (P < 0.01).
Significance
Combined with the results of theoretical calculation and experimental verification, and considering the roles of these targets in the pathogenesis of PF, Tet might antagonize PF by acting on PDPK1 and RAC1. The results of this study will provide scientific reference for the prevention and clinical diagnosis and treatment of PF.
Keywords: Pulmonary fibrosis, Tetrandrine, Network pharmacology, Target prediction, Molecular docking, Experimental verification
Graphical abstract
Pulmonary fibrosis; Tetrandrine; Network pharmacology; Target prediction; Molecular docking; Experimental verification.
1. Introduction
Pulmonary fibrosis (PF) is a group of chronic progressive pulmonary interstitial diseases. It belongs to the category of fibroproliferative diseases and is in the final stage of interstitial lung diseases. In recent years, the incidence rate and prevalence rate of PF have been increasing year by year [1]. The prognosis of the disease is poor, and it is listed as a refractory disease by the World Health Organization (WHO). The main characteristics of the disease are the production of a large amount of collagen due to the proliferation of pulmonary stromal cells, excessive deposition of extracellular matrix components during the repair of lung injury, the significant adverse changes of alveolar epithelial cell phenotype, and the differentiation of fibroblasts into myofibroblasts [2]. At present, the clinically recognized treatment is the use of glucocorticoids, antioxidants, immunosuppressants, and anti-fibrosis drugs (pirfenidone, nidanib ethylsulfonate, etc.) [3, 4].
In addition to the above clinical drugs, we should also pay attention to the potential value of natural products in anti-PF. Natural products refer to the components or metabolites in animals and plants, marine organisms and microorganisms, as well as many endogenous components in humans and animals. Natural products are widely used in medicine, health science, biology, pharmacy, etc. [5, 6]. Natural products have been an important treasure house in new drug discovery. Recently, many natural products with unique structure and promising pharmacological potential for the treatment of PF have been reported [7, 8, 9].
Tetrandrine (Tet) is a bisbenzylisoquinoline-like alkaloid extracted from the root of Stephamia tetrandra S.Moore. Clinically, it is often used to prevent and treat arthritis, hypertension, tumor, silicosis (it has been used to treat silicosis for 50 years), liver fibrosis, lung cancer and other diseases because of its remarkable anti-inflammatory, analgesic, anti-hypertensive, anti-fibrosis, and anti-free-radical pharmacological effects [10, 11, 12, 13]. Besides, the latest research shows that natural compounds and their derivatives may be used for developing potent therapeutics with significant activity against SARS-COV-2, providing a promising frontline of fighting the coronaviruses pandemic [14].
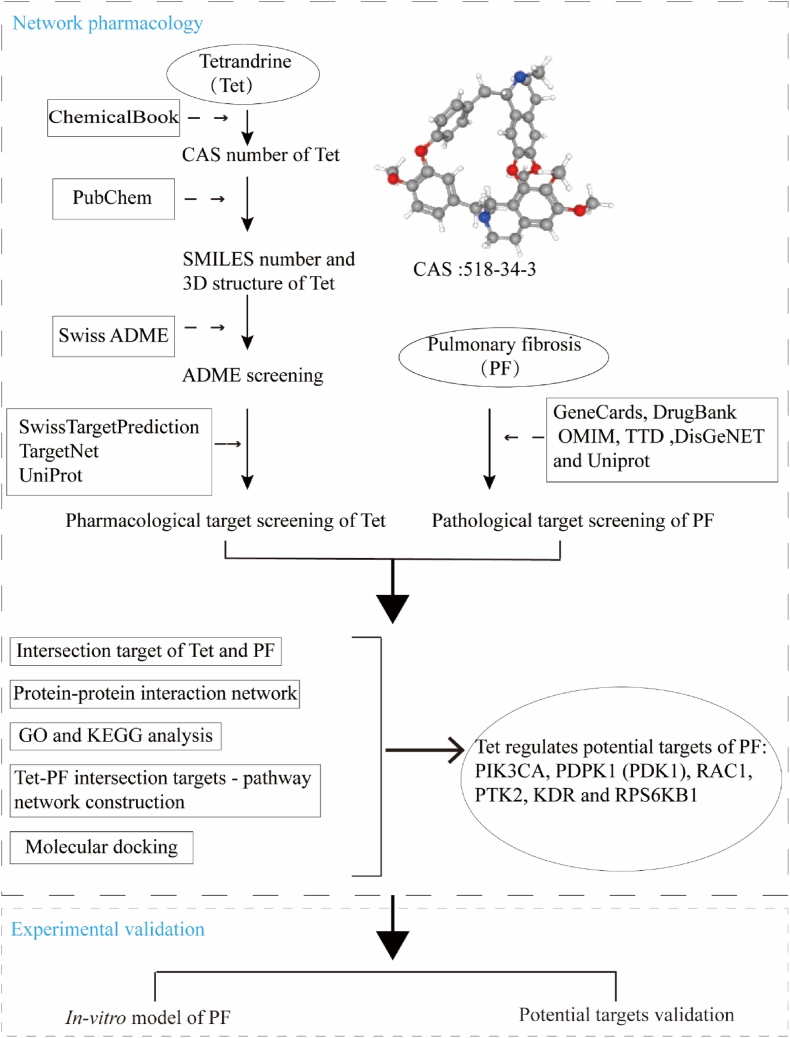
Although Tet has rich pharmacological effects, its mechanisms of action on PF has not been fully explained. Network pharmacology is an integrated approach that blends the disciplines of systems biology and bioinformatics and network science to investigate the underlying molecular mechanisms between drugs and therapeutic targets [15]. Molecular docking is a computer-aided drug simulation technique based on molecular structure that helps to predict the interactions that occur between molecules and biological targets by simulating molecular geometry and interaction forces to screen for the best way to bind small molecule compounds (drugs) to large molecule substances (proteins) [16, 17]. In this study, in order to further explore the underlying mechanisms of Tet in the treatment of PF, the targets and pathways of Tet in the treatment of PF based on network pharmacological analysis were predicted. Then, molecular docking simulation was performed between Tet and possible targets screened by network pharmacology. Finally, the above targets were verified by experiments, the workflow diagram for this study was shown in Figure 1. The results of this study will provide scientific reference for the basic research and clinical diagnosis and treatment of PF.
Figure 1.
Workflow chart of network pharmacological analysis and experimental verification in this study.
2. Materials and methods
2.1. Databases
The databases used in this study were listed in Table 1.
Table 1.
Databases used for pharmacological target screening of Tet and pathological target screening of PF.
| Databases | Websites |
|---|---|
| PubChem | https://pubchem.ncbi.nlm.nih.gov/ |
| ChemicalBook | https://www.chemicalbook.com/ProductIndex.aspx |
| Swiss ADME | http://targetnet.scbdd.com/ |
| SwissTargetPrediction | http://www.swisstargetprediction.ch/ |
| TargetNet | http://targetnet.scbdd.com/ |
| UniProt | https://www.uniprot.org/ |
| GeneCards | https://www.genecards.org/ |
| DrugBank | https://go.drugbank.com/ |
| OMIM | https://www.omim.org/ |
| TTD | http://db.idrblab.net/ttd/ |
| DisGeNET | https://www.disgenet.org/ |
| STRING | https://string-db.org/cgi/about |
| DAVID 6.8 | https://david.ncifcrf.gov/ |
| RCSB PDB | https://www.rcsb.org/ |
| Venny | https://bioinfogp.cnb.csic.es/tools/venny/ |
| Bioinformatics | http://www.bioinformatics.com.cn/ |
2.2. Pharmacological target screening of Tet
The CAS number of Tet was retrieved from the ChemicalBook database. The canonical SMILES string and 3D structure were downloaded from the PubChem database and saved in "SDF" format. The 3D structure was uploaded to the SwissTargetPrediction platform (Download Apr 20, 2021). Target prediction was carried out on TargetNet platform (Download Apr 14, 2021) by using the canonical SMILES string. After standardized processing by UniProt database [18], the targets predicted by the two databases were merged and the duplicate targets were deleted.
2.3. Pathological target screening of PF
With "pulmonary fibrosis" as the keyword, GeneCards (Download Apr 30, 2021), DrugBank (Download Apr 15, 2021), OMIM (Download Apr 14, 2021), TTD (Download Apr 23, 2021) and DisGeNET (Download May 12, 2021) databases were used to predict the pathological targets of PF. The targets obtained from the above five databases were normalized by the UniProt database and duplicate values were removed.
2.4. Intersection target screening and protein-protein interaction network construction
The Venny 2.1.0 online analysis tool was employed to analyze the intersection of Tet pharmacological targets and PF pathological targets, and draw the Wayne diagram. The above intersection targets were submitted to the STRING 11.0 database to construct the PPI network. In the column of "multiple proteins", the organism was set to "Homo sapiens", the lowest interaction score was set to "highest confidence (0.900)", nodes without connection in the network were set to hide, and other parameters were set to the default values. The obtained data were downloaded in TSV format and imported into Cytoscape 3.8.2 software for visualization and analysis.
2.5. GO and KEGG analysis
The intersection targets of Tet pharmacological targets and PF pathological targets were imported into DAVID 6.8 database. The identifier was set to "Official Gene Symbol", the list type was set to "Gene List", and the species type was set to "Homo sapiens" for GO and KEGG analysis. The top 20 entries were filtered according to the number of target genes and P value. The "Bioinformatics" online platform was used for visualization analysis.
2.6. Tet-PF intersection targets - pathway network construction
The network file (network.xlsx) and the attribute file (type.xlsx) were created in an Excel spreadsheet. The Tet-PF intersection targets - pathway network was constructed by using Cytoscape3.8.2 software. The network topology analysis and centrality properties (including degree, closeness centrality, and betweenness centrality) were employed by "Analyze Network" in Cytoscape3.8.2 software.
2.7. Molecular docking
The targets with node degrees greater than the median were selected to dock with dexamethasone (Dex) and Tet. The target protein structures were downloaded from the RCSB PDB database, and the screening principles were as follows. Firstly, the species was selected as "Homo sapiens". Secondly, the source of the protein structure was obtained by X-crystal diffraction. Thirdly, the resolution should be as high as possible (Resolution: Best to Worst (Resolution <3Å)). Fourthly, the type of polymeric entity should be protein. Fifthly, a composite crystal structure containing the original ligand was required. The SDF format of Tet was converted to Mol2 format using Open Babel. The PyMOL software was used to remove the water molecules and small molecule ligands from the target proteins downloaded from the PDB database. The AutoDockTools1.5.6 software was used to dock the target proteins with Dex and Tet. The binding energy was calculated using AutoGrid. To improve the accuracy of the calculation, the parameter "Number of GA Runs" was set to 50, the Lamarckian Genetic Algorithm was employed as the output method, and the rest were set to the default values. After setting the above parameters, the molecular docking operation was executed.
Considering the accuracy of the docking method, it was processed with a sampling algorithm, and its quantification index was the root-mean-square deviation (RMSD) of the atomic positions. The ligands in the crystal structures downloaded from the PDB database were extracted using PyMOL software, after which the predicted ligand conformations were extracted by re-docking the ligands to the protein molecules using AutoDockTools1.5.6 software. The "align" tool of PyMOL was used to analyze the difference between the predicted conformation and the conformation in the original crystal and calculate the RMSD value. In order to screen out the best conformation of molecular docking, the screening threshold values of the binding energy and the RMSD value were -5 kcal/mol and 2Å, respectively [19, 20]. Finally, PyMOL software was used for visualization.
3. In vitro experimental verification
3.1. Materials
Dex and Tet were purchased from MedChemExpress Co., Ltd. Transforming growth factor-β1 (TGF-β1) was purchased from PeproTech Inc. RNAsimple Total RNA Kit, FastKing gDNA Dispelling RT SuperMix, and RealUniversal Color PreMix (SYBR Green) were purchased from Tiangen Biotech (Beijing) Co., Ltd. The PCR primers were synthesized by Sangon Biotech (Shanghai) Co., Ltd. The primer sequences were shown in Table 2.
Table 2.
Primers used in RT-qPCR.
| Gene | GenBank accession number | Primer Sequence (5′-3′) | Product Size | |
|---|---|---|---|---|
| GAPDH | NM_01357943 | Forward | CAGGAGGCATTGCTGATGAT | 138 |
| Reverse | GAAGGCTGGGGCTCATTT | |||
| E-cad | NM_001317185 | Forward | CTGATTCTGCTGCTCTCTTGCTGTTTC | 127 |
| Reverse | GGTCCTCTTCTCCGCCTCCTTC | |||
| Col-Ⅰ | NM_001200 | Forward | TCCCGACAGAACTCAGTGCTATCTC | 102 |
| Reverse | GACACACCCACAACCCTCCACAAC | |||
| FN | NM_001365522 | Forward | AGAGGCATAAGGTTCGGGAAGAGG | 80 |
| Reverse | CGAGTCATCCGTAGGTTGGTTCAAG | |||
| PIK3CA | NM_006218 | Forward | CGGTGACTGTGTGGGACTTATTGAG | 111 |
| Reverse | TGTAGTGTGTGGCTGTTGAACTGC | |||
| PDPK1 | NM_001261816 | Forward | TGTCCCAACACTCCCATCATCCCCTAG | 80 |
| Reverse | CTTCGTCCTCCTCCTCACACTCCGTACTGCTCTATGTTGCTGCCTGAC | |||
| RAC1 | NM_0018890 | Forward | TGTCCCAACACTCCCATCATCCCCTAG | 143 |
| Reverse | ACAGCACCAATCTCCTTAGCCATG | |||
| PTK2 | NM_001387644 | Forward | AGGCAGTATTGACAGGGAGGATGG | 111 |
| Reverse | CGAGGCGGTTTCTTTGGTGGAG | |||
| KDR | NM_002253 | Forward | AGGGAGTCTGTGGCATCTGAAGG | 80 |
| Reverse | GTGGTGTCTGTGTCATCGGAGTG | |||
| RPS6KB1 | NM_001272044 | Forward | TGCTGTGGATTGGTGGAGTTTGG | 148 |
| Reverse | TCTGGCTTCTTGTGTGAGGTAGGG |
3.2. Cell culture
Human lung adenocarcinoma A549 cells were cultured in DMEM culture medium containing 10% fetal bovine serum and 1% penicillin-streptomycin (complete medium) and incubated at 37 °C in a humidified 5% CO2 atmosphere.
3.3. Construction and characterization of TGF-β1-induced A549 fibrosis model
The TGF-β1-induced A549 fibrosis model was established by referring to the relevant literature [21]. Briefly, A549 cells at the logarithmic growth stage were inoculated in 24-well-plates and incubated for 24 h, followed by starvation treatment for 24 h. The culture medium was discarded, and DMEM culture medium containing 3% fetal bovine serum was added to the culture wells of the control group, while DMEM culture medium containing 3% fetal bovine serum and TGF (5 ng/mL) was added to the culture wells of the model group. After 48 h, the morphology of cells was observed by inverted microscope. The cells of each group were collected and the total RNA was extracted. The mRNA expression levels of epithelial marker - E-cadherin (E-cad), interstitial markers - type I collagen (CoL-I) and fibronectin (FN) were measured by RT-qPCR, combined with the changes of cell morphology, to comprehensively judge whether the cell fibrosis model was successful [22, 23].
3.4. Exposure experiment and measurement of relative gene expression levels
A549 cells in logarithmic growth stage were inoculated into 24-well-plates. The exposure groups were set as follows: the control group (complete medium), the model group (5 ng/mL TGF-β1), Dex positive control group (5 ng/mL TGF-β1 and 120 μg/mL Dex), Tet low dose group (5 ng/mL TGF-β1 and 1.25 μΜ Tet), Tet medium dose group (5 ng/mL TGF-β1 and 2.5 μΜ Tet), Tet high dose group (5 ng/mL TGF-β1 and 5 μΜ Tet). Cells were treated for 48 h. The treated cells were collected and the total RNA was extracted. The mRNA expression levels of E-cad, Col-Ⅰ, FN, PIK3CA, PDPK1, RAC1, PTK2, KDR, RPS6KB1 were detected by RT-qPCR.
3.5. Statistical analysis
The verification experiment was repeated three times. The relative mRNA expression levels were calculated using the 2-△△CT method. The data were expressed as mean ± standard deviation (mean ± SD). The data were statistically analyzed using SPSS 25.0 and plotted using GraphPad Prism8.0 software. The data were analyzed with one-way analysis of variance (ANOVA) followed by the Post-Hoc LSD test. P < 0.05 was considered statistically significant difference.
4. Results
4.1. Screening results of Tet pharmacological targets
After screened by SwissTargetPrediction database, 105 targets were obtained, and 59 targets were obtained after screened by TargetNet database (with values greater than zero). After merging and deleting duplicate values, 143 Tet pharmacological targets were finally obtained (Supplementary Excel Table 1).
4.2. PF pathological target screening
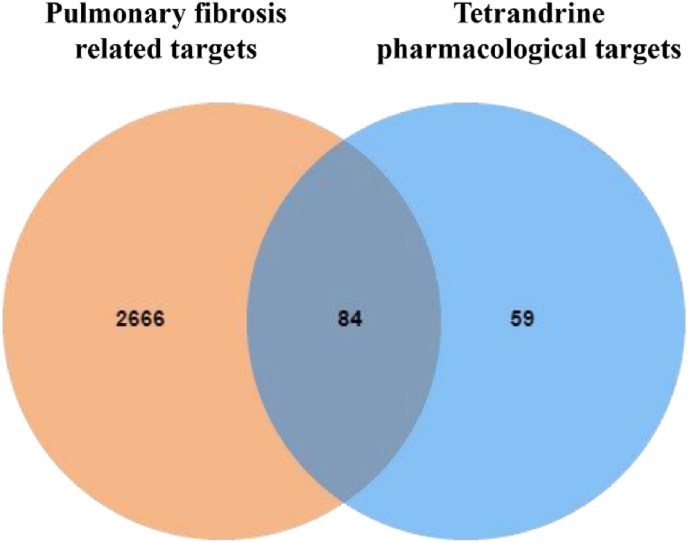
After the prediction and analysis of GeneCards and DisGeNET databases, 5440 and 924 PF pathological targets were obtained respectively. Then, the targets corresponding to the values greater than or equal to the median were further screened, and 2722 (score ≥3.68) and 199 (score ≥0.064) PF pathological targets were obtained respectively. After the prediction and analysis of DrugBank, OMIM, and TTD databases, 19475 and 5 PF pathological targets were obtained respectively. After merging and deleting duplicate values, 2750 PF pathological targets were finally obtained. The analysis results produced by Venny2.1.0 showed that there were 84 Tet-PF intersection targets (Figure 2, Supplementary Excel Table 1).
Figure 2.
Venn diagram of Tet pharmacological targets and PF pathological targets.
4.3. PPI network construction and analysis
There were 62 nodes (proteins) and 165 edges (indicating the relationship between proteins) in PPI network. The node area represented the degree value of the node. The larger the node area, the greater the corresponding degree value, indicating that the node was more important, such as PIK3CA, SRC, HSP90AA1, PDPK1, RAC1, PRKCA, etc (Figure 3, Table 3).
Figure 3.
PPI network of the potential targets of Tet against PF.
Table 3.
The detailed information on the screened targets of PPI network.
| Gene name | Degree | Gene name | Degree |
|---|---|---|---|
| PIK3CA | 24 | PTK2 | 11 |
| SRC | 17 | PRKCD | 10 |
| HSP90AA1 | 15 | KDR | 9 |
| PDPK1 | 13 | TBXA2R | 9 |
| RAC1 | 13 | CHRM1 | 8 |
| PRKCA | 12 | PRKCB | 8 |
| RPS6KB1 | 12 | JAK2 | 7 |
Note: Only the top 14 targets in terms of degree value are listed here.
4.4. GO and KEGG analysis
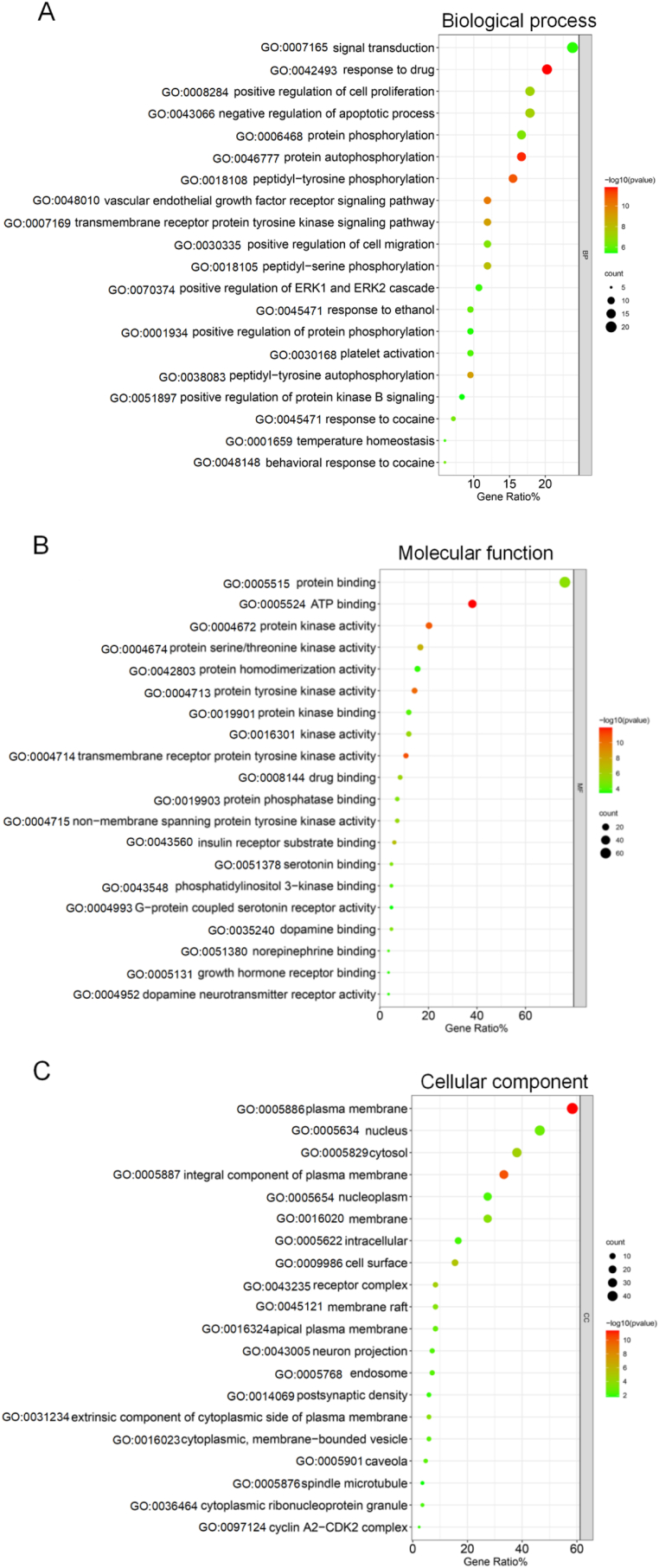
Go analysis included biological process (BP), molecular function (MF) and cellular component (CC). The results of BP analysis showed that the above intersection targets involved 244 entries, including response to drug, protein autophosphorylation, peptidyl-tyrosine phosphorylation, vascular endothelial growth factor receptor signaling pathway, peptidyl-tyrosine autophosphorylation, etc. There were 60 entries related to MF, which were mainly involved in ATP binding, transmembrane receptor protein tyrosine kinase activity, protein kinase activity, protein tyrosine kinase activity, protein serine/threonine kinase activity, etc. There were 35 entries related to CC, which were mainly located in plasma membrane, integral component of plasma membrane, cell surface, receptor complex, cytosol, etc. The top 20 entries of BP, MF, and CC were sorted according to P value (P < 0.05) and count value, and bubble diagrams were drawn with the "Bioinformatics" online platform (Figure 4).
Figure 4.
Bubble diagram of GO analysis results of the potential targets of Tet against PF. Notes: Only the top 20 entries sorted according to P value (P < 0.05) and the corresponding count value were listed. The size of the bubble indicated the number of targets enriched on this entry, and the color indicates P value.
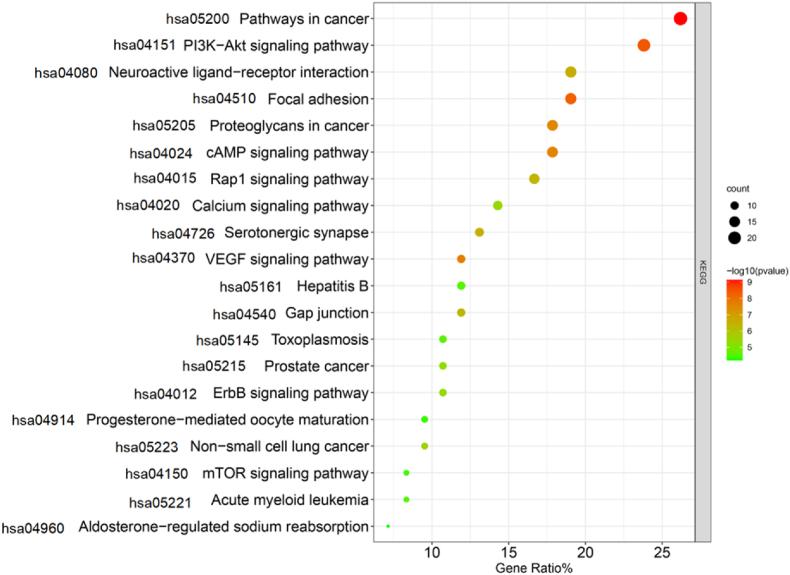
KEGG analysis results showed that 84 Tet-PF intersection targets involved 83 pathways, mainly including PI3K-AKT signaling pathway, pathways in cancer, focal adhesion, proteoglycans in cancer, and cAMP signaling pathway, etc (Figure 5).
Figure 5.
Bubble diagram of KEGG analysis results of the potential targets of Tet against PF. Notes: Only the top 20 entries sorted according to P value (P < 0.05) and the corresponding count value were listed. The size of the bubble indicated the number of targets enriched on this entry, and the color indicates P value.
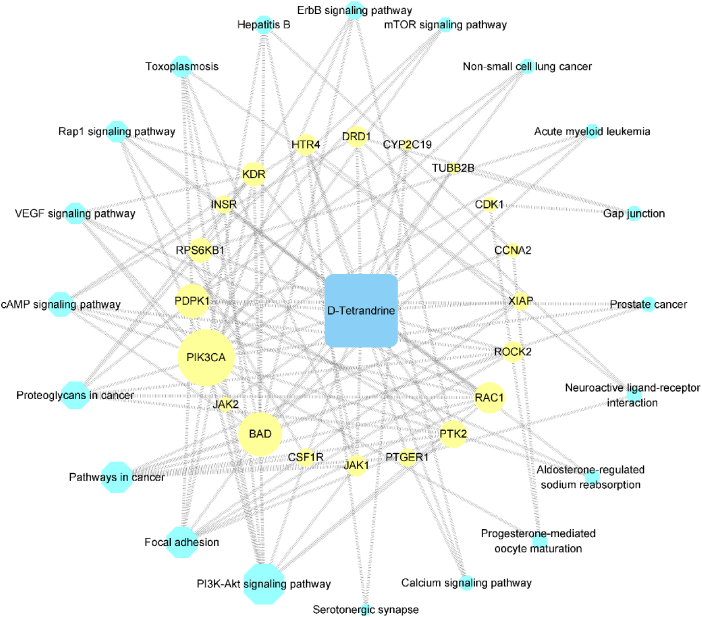
4.5. Tet-PF intersection targets - pathway network
Figure 6 showed Tet-PF intersection targets - pathway network built by Cytoscape3.8.2. The results of KEGG pathway analysis showed that a total of 59 targets were involved in the top 20 pathways. Further analysis showed that a total of 20 intersection targets were involved. The larger the node area, the greater the degree value, which means that the more nodes connected to this node, the more important regulation role of this node in the network. There were 41 nodes (20 targets, 20 pathways, 1 compound) and 113 edges in this network. Among these 20 targets, the more important targets for network regulation included PIK3CA, BAD, PDPK1, RAC1, PTK2, RPS6KB1, etc (Table 4).
Figure 6.
Tet-PF intersection targets - pathway network. Notes: The text box with yellow shading represented Tet-PF intersection targets. The text box with blue shading represented the pathway. The area of the node represented the size of the degree value.
Table 4.
Basic information on Tet-PF intersection targets.
| Targets | Full Name | Degree |
|---|---|---|
| PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | 16 |
| BAD | Bcl2-associated agonist of cell death | 12 |
| PDPK1 (PDK1) | 3-phosphoinositide-dependent protein kinase 1 | 9 |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 | 8 |
| PTK2 | Focal adhesion kinase 1 | 7 |
| KDR | Vascular endothelial growth factor receptor 2 | 6 |
| RPS6KB1 | Ribosomal protein S6 kinase beta-1 | 6 |
| DRD1 | D(1A) dopamine receptor | 5 |
| HTR4 | 5-hydroxytryptamine receptor 4 | 5 |
| JAK1 | Tyrosine-protein kinase JAK1 | 5 |
| ROCK2 | Rho-associated protein kinase 2 | 5 |
| INSR | Insulin receptor | 4 |
| CSF1R | Macrophage colony-stimulating factor 1 receptor | 4 |
| PTGER1 | Prostaglandin E2 receptor EP1 subtype | 4 |
| XIAP | E3 ubiquitin-protein ligase XIAP | 4 |
| CDK1 | Cyclin-dependent kinase 1 | 3 |
| JAK2 | Tyrosine-protein kinase JAK2 | 3 |
| CCNA2 | Cyclin-A | 3 |
| TUBB2B | Tubulin beta-2B chain | 2 |
| CYP2C19 | Cytochrome P450 2C19 | 2 |
4.6. Molecular docking
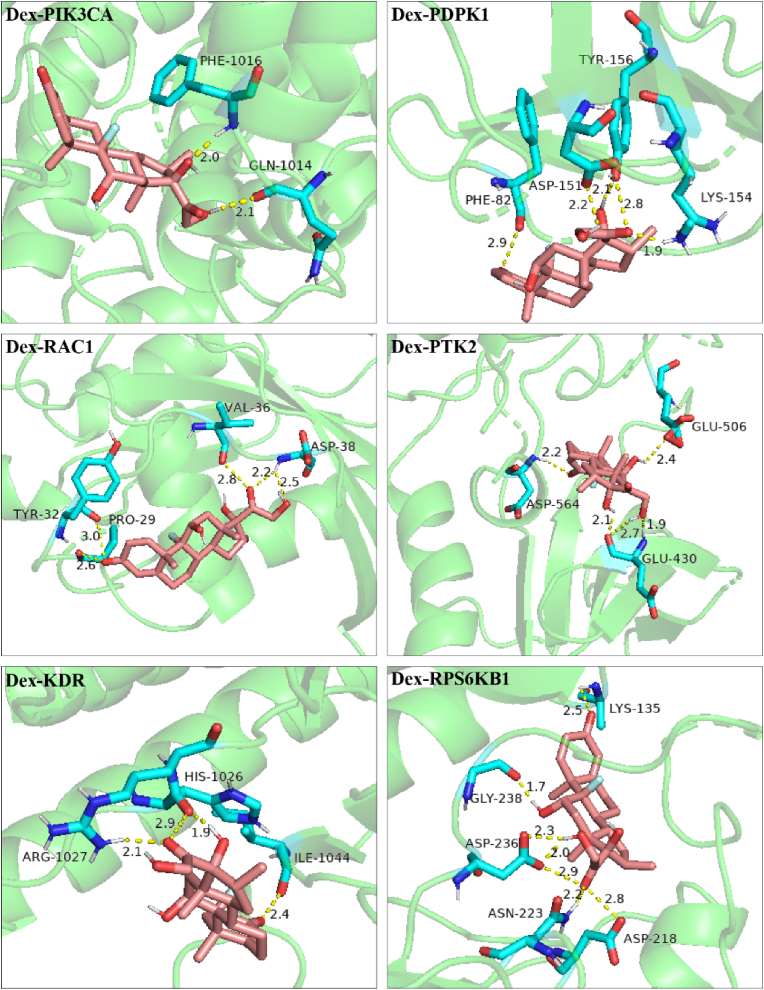
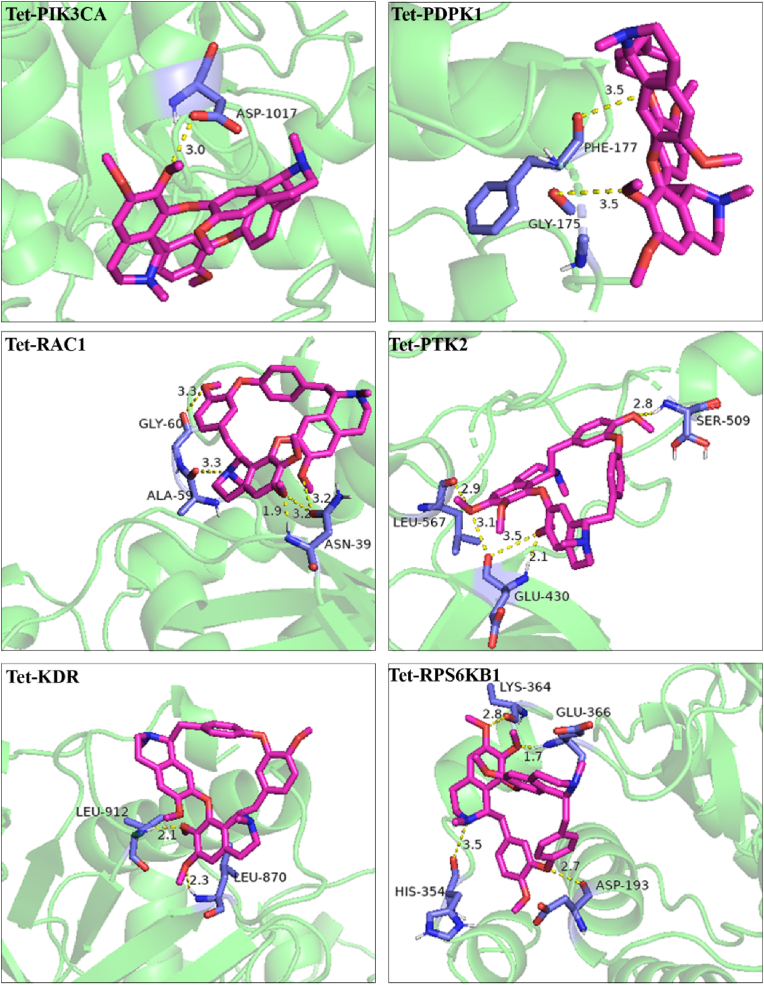
According to the network topology analysis data, the median of the potential targets was 5. Considering both the degree value (greater than 5) and the source of protein structure (from X-ray diffraction), 6 key target proteins (PIK3CA, PDPK1, RAC1, PTK2, KDR, RPS6KB1) were screened. The molecular docking results showed that the calculated RMSD values of small molecules (Dex and Tet) docking with the active sites of these 6 proteins were less than 2 Å, suggesting that the above docking results had good reproducibility. The binding energies of the above docking were all less than -5.0 kcal mol−1, indicating that the ligand molecule binds well to the receptor protein (Table 5). The visual analysis results showed that Dex and Tet might bind to the amino acids near the active sites through hydrogen bond (Figures 7 and 8).
Table 5.
Basic information of docking of Dex and Tet with the screened targets.
| Targets | PBD ID | Degree (Dex/Tet) | RMSD (Å) (Dex/Tet) | Binding Energy (kcal/mol) (Dex/Tet) |
|---|---|---|---|---|
| PIK3CA | 6pys | 16/16 | 1.444/0.001 | -7.51/-8.35 |
| PDPK1(PDK1) | 5lvo | 9/9 | 0.789/0.001 | -7.86/-9.51 |
| RAC1 | 5o33 | 8/8 | 1.530/0.000 | -7.36/-8.54 |
| PTK2 | 6yoj | 7/7 | 0.841/0.001 | -7.55/-8.51 |
| KDR | 2xir | 6/6 | 1.038/0.001 | -7.26/-7.77 |
| RPS6KB1 | 3wf7 | 6/6 | 0.849/0.001 | -8.48/-7.52 |
Notes: The determination of the above protein list was based on the degree value (greater than 5) and the source of protein structure (from X-ray diffraction).
Figure 7.
Molecular docking diagram of Dex to the screened targets.
Figure 8.
Molecular docking diagram of Tet to the screened targets.
4.7. Verification experiment
4.7.1. Identification of in-vitro model of PF
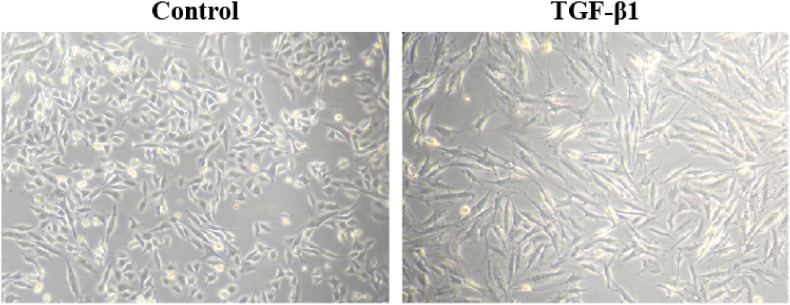
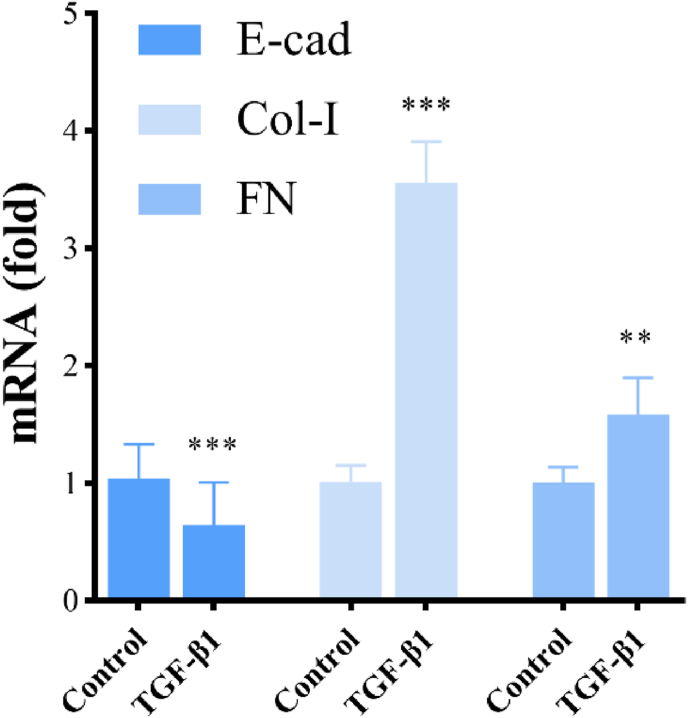
Microscopic examination showed that A549 cells in the control group appeared cobblestone-like while the cells became elongated and shuttle-shaped in the TGF-β1 treatment group (Figure 9). The results of RT-qPCR showed that the mRNA expression of E-cad in the TGF-β1 treatment group was decreased to 62% (P < 0.05), while the mRNA expression of Col-Ⅰ and FN was increased to 3.52-fold and 1.57-fold (P < 0.01) compared with the control group. These results indicated that the exposure of TGF-β1 (5 ng/mL) for 48 h could induce EMT in A549 cells (Figure 10).
Figure 9.
TGF-β1 (5 ng/mL) exposure for 48 h induced distinct morphological changes in A549 cells (200×).
Figure 10.
The mRNA expression of E-cad, Col-I and FN in A549 cells after TGF-β1 (5 ng/mL) exposure for 48 h. Notes: The asterisk indicated that there was a significant difference compared with the control group (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001).
4.7.2. Effect of Dex and Tet on the relative mRNA expression of E-cad, Col-Ⅰ and FN in TGF-β1-induced A549 cells
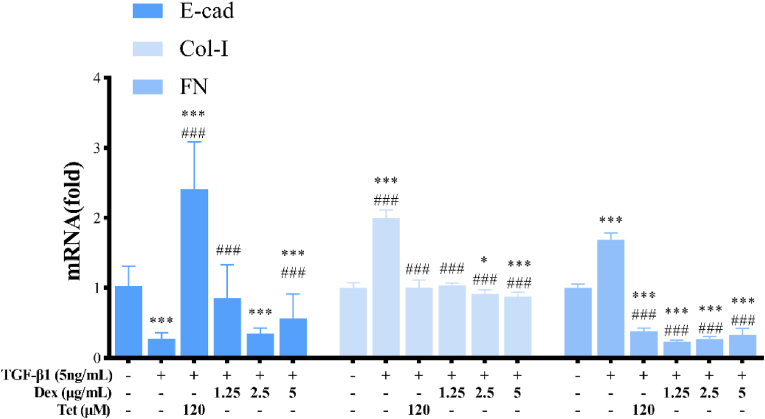
Compared with the TGF-β1 treatment group, the mRNA expression of E-cad in Dex group was increased to 8.64 times (P < 0.001), while the mRNA expression levels of Col-Ⅰ and FN was decreased to 50% and 23% respectively (P < 0.001). The results in Tet group were similar. Taking Tet low dose group as an example, the mRNA expression of E-cad was up-regulated to 3.06 times (P < 0.001), while the mRNA expression of CoL-I and FN were down-regulated to 52% and 14%, respectively (P < 0.001) (Figure 11).
Figure 11.
The mRNA expression of E-cad, Col-I, and FN in TGF-β1-induced A549 cells by Dex and Tet. Notes: The asterisk indicated that there was a significant difference compared with the control group (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001), and the asterisk indicated that there was a significant difference compared with the TGF-β1 treatment group (#P < 0.05, ##P < 0.01, ###P < 0.001).
4.7.3. Effect of Dex and Tet on the mRNA expression of the selected targets in A549 cells
4.7.3.1. The mRNA expression of PIK3CA and KDR
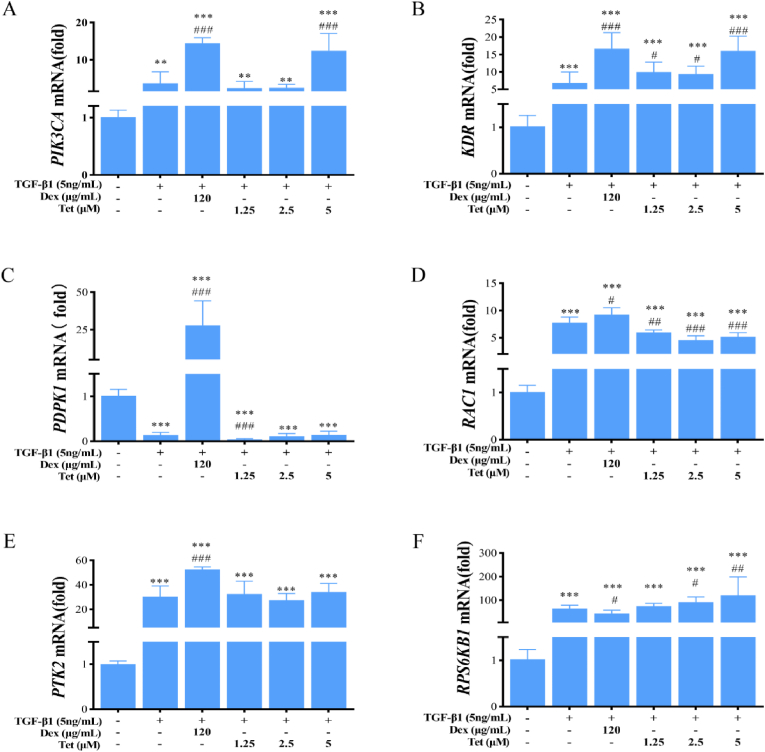
Compared with the control group, the mRNA expression of PIK3CA and KDR in the model group were up-regulated (P < 0.01). Compared with the model group, the mRNA expression of PIK3CA and KDR in Dex group were up-regulated to 3.91-fold and 2.45-fold, respectively (P < 0.001). The results of Tet group were similar. Taking Tet high dose group as an example, the mRNA expression of PIK3CA and KDR were up-regulated to 3.38-fold and 2.36-fold, respectively (P < 0.001) (Figure 12A,B).
Figure 12.
The mRNA expression levels of the screened targets in TGF-β1-induced A549 cells by Dex and Tet. Notes: The asterisk indicated that there was a significant difference compared with the control group (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001), and the asterisk indicated that there was a significant difference compared with the TGF-β1 treatment group (#P < 0.05, ##P < 0.01, ###P < 0.001).
4.7.3.2. The mRNA expression of PDPK1
Compared with the control group, the mRNA expression of PDPK1 in the model group were down-regulated (P < 0.001). Compared with the model group, the mRNA expression of PDPK1 in Dex group was up-regulated to 197.54-fold (P < 0.001), while the expression in Tet low dose group was down-regulated to 0.31-fold (P < 0.001), and there was no significant difference in other Tet groups (Figure 12C).
4.7.3.3. The mRNA expression of RAC1
Compared with the control group, the mRNA expression of RAC1 in the model group was increased (P < 0.001). Compared with the model group, the mRNA expression of RAC1 in Dex group was up-regulated to 1.19-fold (P < 0.05), while the expression in Tet group was down-regulated (P < 0.01) (Figure 12D).
4.7.3.4. The mRNA expression of PTK2
Compared with the control group, the mRNA expression of PTK2 in the model group was increased (P < 0.001). Compared with the model group, the mRNA expression of PTK2 in Dex group was up-regulated to 1.73-fold (P < 0.001), while the expression in Tet group was not statistically significant (Figure 12E).
4.7.3.5. The mRNA expression of RPS6KB1
Compared with the control group, the mRNA expression of RPS6KB1 in the model group was increased (P < 0.001). Compared with the model group, the mRNA expression of RPS6KB1 in Dex group was down-regulated to 0.67-fold (P < 0.05), while the expression in Tet medium and high dose groups were up-regulated 1.42-fold and 1.88-fold, respectively (P < 0.05) (Figure 12F).
5. Discussion
5.1. Natural products as powerful tools against PF
PF refers to a group of interstitial lung diseases. Pathologically, persistent inflammatory injury of lung tissue causes repeated destruction, repair and excessive deposition of extracellular matrix, and thus leads to the destruction of normal lung tissue structure and loss of function. At present, the prevalence and mortality of PF is increasing. Taking idiopathic pulmonary fibrosis as an example, the estimated the adjusted global incidence and prevalence of IPF to be in the range of 0.09–1.30 and 0.33–4.51 per 10000 persons, respectively [24]. The exact mechanism is not completely clear, and there is a lack of specific and effective treatment in clinic.
As a huge resource treasure-house, natural products have become an important source of modern drug research and development. In recent years, many natural products, such as Tet, have been reported to have anti-PF effects [25]. Finding active monomers with clear pharmacological effects from natural products, clarifying the mechanism of action and treating the multi-level characteristics of PF can give full play to the multi-target treatment advantages of natural products. Therefore, in this study, the underlying mechanism of Tet against PF was studied based on network pharmacology, molecular docking and experimental verification.
Network pharmacology analysis can provide as many possibilities as possible. Molecular docking can theoretically verify the clues obtained in network pharmacology analysis. The experimental verification can further confirm the candidate molecules obtained in network pharmacology and molecular docking research. Using the progressive and funnel-shaped research paradigm of the combination of network pharmacology, molecular docking and verification experiment can not only avoid the "castle in the air" -like conclusion of pure theoretical research, but also avoid "The Blind Men and the Elephant" -like conclusion of pure experimental research.
5.2. The results of network pharmacology and molecular docking suggest the candidate targets of downstream experimental verification
In this study, based on the network pharmacology method, the intersection targets of Tet pharmacological targets and PF pathological network targets were screened, and 6 candidate molecules were obtained, namely PIK3CA, PDPK1 (PDK1), RAC1, PTK2, KDR and RPS6KB1. The molecular docking method was used to theoretically verify the possibility of Tet acting on the protein molecules encoded by these targets. The results showed that Tet could stably bind to these 6 molecules, suggesting that Tet may produce the anti-PF effect by acting on these targets. Finally, TGF-β1-induced A549 cells were used as the in vitro experimental verification model, the effects of Tet exposure on the above targets were studied from mRNA expression level.
5.3. Selection of positive control in validation experiment, and the effects of Dex and Tet on the relative mRNA expression levels of EMT markers
A large number of studies have confirmed that TGF-β1 can down-regulate E-cad (epithelial marker) expression, and up-regulate Col-Ⅰ and FN (interstitial markers) expression, induce extracellular matrix deposition, and thus affect the progression of PF [26, 27, 28]. A549 cells are basal epithelial cells of human alveolar adenocarcinoma, belonging to type II alveolar epithelium. It is a commonly used cell model to study the molecular mechanism of EMT. Therefore, in this study, TGF-β1-induced A549 cells were used as a model for in vitro experimental verification.
Dex is a synthetic glucocorticoid, which has the effects of anti-inflammatory, improving vascular permeability and immunosuppression. It is widely used in clinic and can prevent the occurrence and development of PF. Some studies have shown that Dex can reduce bleomycin-induced PF in mice via TGF- β, Smad3 and JAK-STAT signaling pathways [29]. Therefore, Dex was selected as the positive control in this study. Compared with the model group, Dex and Tet up-regulated the mRNA expression level of E-cad and down-regulated the mRNA expression levels of Col-Ⅰ and FN, suggesting that both Dex and Tet could suppress EMT in epithelial cells by up-regulating the epithelial markers and down-regulating the mesenchymal markers, so as to play an anti-PF role [30, 31].
5.4. Different effects of Dex and Tet on the relative mRNA expression levels of key regulatory genes in TGF-β1-induced A549 cells
Phosphatidylinositol-3 kinase catalytic subunit α (PIK3CA) gene encodes class I PI3K catalytic subunit P110α protein. PIK3CA has been identified as an oncogene. PIK3CA mutation will increase kinase activity, then continue to stimulate downstream AKT, activate PI3K/AKT signaling pathway, and eventually increase cell invasion and metastasis. In this study, TGF-β1 stimulation up-regulated the expression level of PIK3CA in A549 cells. Similarly, Song et al. found that TGF-β1 increased the expression of PIK3CA and PIK3CB in mouse primary lung telocytes, while decreased the expression of PIK3CD and PIK3CG [32]. KDR/Flk-1 tyrosine kinase, one of the two vascular endothelial growth factor (VEGF) receptors, induces mitogenesis and differentiation of vascular endothelial cells [33]. The up-regulated expression level of VEGF/VEGFR (KDR/Flk-1) was related to the increased number of pulmonary microvessels and degree of PF [34]. Chen et al. found that Tet could effectively suppressed the growth of and induced apoptosis in A549 cells. ELISA and Western blotting results showed that Tet significantly up-regulated the protein expression of PARP, Bax, intercellular adhesion molecule-1 (ICAM-1) and VEGF, while significantly suppressed the phosphorylation of Akt and protein expression of HIF-1α, suggesting that Tet might suppress A549 cell viability and induce apoptosis via the VEGF/HIF-1α/ICAM-1 signaling pathway [35]. In this study, both Dex and Tet (5 μΜ) up-regulated the expression levels of PIK3CA and KDR. Taking the roles of the above two molecules in the pathogenesis of PF, these effects are considered to be secondary effects.
Phosphatidylinositol 3-kinase/phosphatidylinositide-dependent protein kinase 1 (PDPK1)/Akt signaling plays a critical role in activating proliferation and survival pathways in PF. Chen et al. reported that the protein levels of matrix metalloproteinases 9 (MMP-9), phosphorylated PI3K, PDK1, Akt and NF-κB in human renal cell carcinoma 786-O and 769-P cells were markedly reduced after Tet treatment [36]. A more powerful evidence was that targeting inhibition of HIF-1α/PDK1 axis could significantly reduce bleomycin induced PF [37], which was basically consistent with the results of low-dose-Tet intervention in this study.
Rac1, a member of Rho GTPase superfamily, plays an important role in the pathogenesis of PF. The mitochondrial import and direct electron transfer from cytochrome c to Rac1 modulates mitochondrial H2O2 production in alveolar macrophages, and Rac1 null mice fail to develop PF [38, 39]. Osborn-Heaford et al. employed alveolar macrophages from normal volunteers and patients with IPF and asbestosis, bleomycin (1.3–2.0 U/kg) or chrysotile (100 μg) intratracheally administered Rac1 null and Rac2 knockout mice, and human THP-1 macrophages as experimental materials, and found that targeting the isoprenoid pathway to alter Rac1 geranylgeranylation could halt the progression of PF after lung injury [40]. Studies showed that cigarette smoke extract could up-regulate the mRNA and protein expression of Rac1, and thus induce EMT in A549 cells [41]. Similarly, in this study, TGF-β1 exposure up-regulated the mRNA expression of Rac1 in A549 cells, while Tet exposure down-regulated, suggesting that this molecule might be one of the target for Tet to play an anti-PF role.
P70S6K1 is a serine/threonine kinase encoded by RPS6KB1. It is the downstream target of mTOR. The phosphorylation of mTOR can cause its activation, so as to further mediate downstream molecular biological events, including but not limited to promoting the pathogenesis of PF. The research showed that mTOR overactivation in alveolar epithelial cells and compromised autophagy in the lungs are involved in the pathogenesis of PF [42], and targeting the S6K pathway selectively modified the progression of PF in the subpleural compartment of the lung [43]. Similarly, in this study, TGF-β1 exposure up-regulated the mRNA expression of RPS6KB1 in A549 cells. Dex exposure down-regulated the mRNA expression of RPS6KB1, while Tet up-regulated, suggesting that this molecule might be one of the target for Dex (but not for Tet) to play an anti-PF role.
Taken together, even though Dex and Tet have similar efficacy, they show similar but different effects on the candidate targets of anti-PF screened by network pharmacology and molecular docking. Considering the roles of these molecules in the pathogenesis of PF, Tet might exert the anti-PF effect by acting on PDPK1 and RAC1.
6. Conclusion and perspectives
In this study, network pharmacology, molecular docking and verification experiment were used to reveal the underlying mechanisms of Tet in the pathogenesis of PF. The results showed that Tet might exert its anti-PF effect by affecting PDPK1 and RAC1. The results of this study will provide scientific reference for the basic research and clinical treatment of PF.
Before carrying out this research, we designed a broad-brush outline on the basis of consulting relevant literature and classic bibliographies. Further, on the basis of the results of the pre-research, we made necessary adjustments, such as the selection of the network pharmacology database, the determination standard of differential expression indicators, the basis for the selection of indicators in the experimental verification, etc. The adjusted road map was taken as the final road map.
To sum up, this study did not fully comply with the published technical roadmap of a certain study. But according to the research needs, on the basis of following the basic network pharmacological analysis and experimental verification research strategy as a whole, some adjustments in line with the specifications have been made.
In the experimental verification, this study uses the human lung adenocarcinoma A549 cell model, which is widely used in the study of the injury effects and molecular mechanisms of respiratory system caused by numerous exogenous factors. In terms of the research results, we can answer from a profile how Tet antagonizes the EMT promoting effect caused by TGF-β1 to a certain extent.
In order to further understand the molecular mechanism of Tet reversing EMT, we plan to use mouse modeling to explore the in vivo pharmacological effect of Tet.
Declarations
Author contribution statement
Jie Li: Performed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Yi Wang: Contributed reagents, materials, analysis tools or data.
Rui Wang; Ying-Chi Zhang: Analyzed and interpreted the data.
Meng-Yu Wu; Jing Shan: Performed the experiments; Contributed reagents, materials, analysis tools or data.
Hai-Ming Xu: Conceived and designed the experiments; Wrote the paper.
Funding statement
Professor Hai-Ming Xu was supported by National Natural Science Foundation of China [81660527], Ningxia Natural Science Foundation [2022AAC05027].
Data availability statement
Data will be made available on request.
Declaration of interest's statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
Not applicable.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Hutchinson J., Fogarty A., Hubbard R., McKeever T. Global incidence and mortality of idiopathic pulmonary fibrosis: a systematic review. Eur. Respir. J. 2015;46(3):795–806. doi: 10.1183/09031936.00185114. [DOI] [PubMed] [Google Scholar]
- 2.Karampitsakos T., Woolard T., Bouros D., Tzouvelekis A. Toll-like receptors in the pathogenesis of pulmonary fibrosis. Eur. J. Pharmacol. 2017;808:35–43. doi: 10.1016/j.ejphar.2016.06.045. [DOI] [PubMed] [Google Scholar]
- 3.Glass D.S., Grossfeld D., Renna H.A., Agarwala P., Spiegler P., Kasselman L.J., et al. Idiopathic pulmonary fibrosis: molecular mechanisms and potential treatment approaches. Respir Investig. 2020;58(5):320–335. doi: 10.1016/j.resinv.2020.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Torrisi S.E., Kahn N., Vancheri C., Kreuter M. Evolution and treatment of idiopathic pulmonary fibrosis. Presse Med. 2020;49(2) doi: 10.1016/j.lpm.2020.104025. [DOI] [PubMed] [Google Scholar]
- 5.Yao H., Liu J., Xu S., Zhu Z., Xu J. The structural modification of natural products for novel drug discovery. Expet Opin. Drug Discov. 2017;12(2):121–140. doi: 10.1080/17460441.2016.1272757. [DOI] [PubMed] [Google Scholar]
- 6.Ahmed H.M., Nabavi S., Behzad S. Herbal drugs and natural products in the light of nanotechnology and nanomedicine for developing drug formulations. Mini Rev. Med. Chem. 2021;21(3):302–313. doi: 10.2174/1389557520666200916143240. [DOI] [PubMed] [Google Scholar]
- 7.Wang L., Li S., Yao Y., Yin W., Ye T. The role of natural products in the prevention and treatment of pulmonary fibrosis: a review. Food Funct. 2021;12(3):990–1007. doi: 10.1039/d0fo03001e. [DOI] [PubMed] [Google Scholar]
- 8.Hosseini S.A., Zahedipour F., Sathyapalan T., Jamialahmadi T., Sahebkar A. Pulmonary fibrosis: therapeutic and mechanistic insights into the role of phytochemicals. Biofactors. 2021;47(3):250–269. doi: 10.1002/biof.1713. [DOI] [PubMed] [Google Scholar]
- 9.Huang Y.Y., Deng J.H., Tian Y.J., Liang J.H., Xie X., Huang Y., et al. Mangostanin derivatives as novel and orally active phosphodiesterase 4 inhibitors for the treatment of idiopathic pulmonary fibrosis with improved safety. J. Med. Chem. 2021;64(18):13736–13751. doi: 10.1021/acs.jmedchem.1c01085. [DOI] [PubMed] [Google Scholar]
- 10.Wang R., Ma T.M., Liu F., Gao H.Q. [Research progress on pharmacological action and clinical application of Stephania Tetrandrae Radix] Zhongguo Zhongyao Zazhi. 2017;42(4):634–639. doi: 10.19540/j.cnki.cjcmm.20170121.024. [DOI] [PubMed] [Google Scholar]
- 11.Chan E.W.C., Wong S.K., Chan H.T. An overview on the chemistry, pharmacology and anticancer properties of tetrandrine and fangchinoline (alkaloids) from Stephania tetrandra roots. J. Integr. Med. 2021;19(4):311–316. doi: 10.1016/j.joim.2021.01.001. [DOI] [PubMed] [Google Scholar]
- 12.Wang S., Fu J.L., Hao H.F., Jiao Y.N., Li P.P., Han S.Y. Metabolic reprogramming by traditional Chinese medicine and its role in effective cancer therapy. Pharmacol. Res. 2021;170 doi: 10.1016/j.phrs.2021.105728. [DOI] [PubMed] [Google Scholar]
- 13.Mir R.H., Shah A.J., Mohi-Ud-Din R., Pottoo F.H., Dar M.A., Jachak S.M., et al. Natural anti-inflammatory compounds as drug candidates in Alzheimer's disease. Curr. Med. Chem. 2021;28(23):4799–4825. doi: 10.2174/0929867327666200730213215. [DOI] [PubMed] [Google Scholar]
- 14.Khalifa S.A.M., Yosri N., El-Mallah M.F., Ghonaim R., Guo Z., Musharraf S.G., et al. Screening for natural and derived bio-active compounds in preclinical and clinical studies: one of the frontlines of fighting the coronaviruses pandemic. Phytomedicine. 2021;85 doi: 10.1016/j.phymed.2020.153311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S. Network pharmacology evaluation method guidance - draft. World J. Traditional Chin. Med. 2021;7(1):148. doi: 10.19540/j.cnki.cjcmm.20210914.702. [DOI] [PubMed] [Google Scholar]
- 16.Liu K., Tao X., Su J., Li F., Mu F., Zhao S., et al. Network pharmacology and molecular docking reveal the effective substances and active mechanisms of Dalbergia Odoriferain protecting against ischemic stroke. PLoS One. 2021;16(9) doi: 10.1371/journal.pone.0255736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinzi L., Rastelli G. Molecular docking: shifting paradigms in drug discovery. Int. J. Mol. Sci. 2019;20(18) doi: 10.3390/ijms20184331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.UniProt C. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49(D1):D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huey W, Morris GM, Forli S. Using AutoDock 4 and AutoDock Vina with AutoDockTools: A Tutorial.
- 21.Kasai H., Allen J.T., Mason R.M., Kamimura T., Zhang Z. TGF-beta1 induces human alveolar epithelial to mesenchymal cell transition (EMT) Respir. Res. 2005;6:56. doi: 10.1186/1465-9921-6-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang C., Zhu X., Hua Y., Zhao Q., Wang K., Zhen L., et al. YY1 mediates TGF-beta1-induced EMT and pro-fibrogenesis in alveolar epithelial cells. Respir. Res. 2019;20(1):249. doi: 10.1186/s12931-019-1223-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanemaru R., Takahashi F., Kato M., Mitsuishi Y., Tajima K., Ihara H., et al. Dasatinib suppresses TGFbeta-mediated epithelial-mesenchymal transition in alveolar epithelial cells and inhibits pulmonary fibrosis. Lung. 2018;196(5):531–541. doi: 10.1007/s00408-018-0134-6. [DOI] [PubMed] [Google Scholar]
- 24.Maher T.M., Bendstrup E., Dron L., Langley J., Smith G., Khalid J.M., et al. Global incidence and prevalence of idiopathic pulmonary fibrosis. Respir. Res. 2021;22(1):197. doi: 10.1186/s12931-021-01791-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luan F., He X., Zeng N. Tetrandrine: a review of its anticancer potentials, clinical settings, pharmacokinetics and drug delivery systems. J. Pharm. Pharmacol. 2020;72(11):1491–1512. doi: 10.1111/jphp.13339. [DOI] [PubMed] [Google Scholar]
- 26.Petrini I., Barachini S., Carnicelli V., Galimberti S., Modeo L., Boni R., et al. ED-B fibronectin expression is a marker of epithelial-mesenchymal transition in translational oncology. Oncotarget. 2017;8(3):4914–4921. doi: 10.18632/oncotarget.13615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi L., Dong N., Fang X., Wang X. Regulatory mechanisms of TGF-beta1-induced fibrogenesis of human alveolar epithelial cells. J. Cell Mol. Med. 2016;20(11):2183–2193. doi: 10.1111/jcmm.12918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morbini P., Inghilleri S., Campo I., Oggionni T., Zorzetto M., Luisetti M. Incomplete expression of epithelial-mesenchymal transition markers in idiopathic pulmonary fibrosis. Pathol. Res. Pract. 2011;207(9):559–567. doi: 10.1016/j.prp.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 29.Shi K.Y., Jiang J.Z., Ma T.L., Xie J., Duan L.R., Chen R.H., et al. Dexamethasone attenuates bleomycin-induced lung fibrosis in mice through TGF-beta, Smad3 and JAK-STAT pathway. Int. J. Clin. Exp. Med. 2014;7(9):2645–2650. [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang F., Chen L., Zhou Y., Ding D., Hu Q., Liu Y., et al. Dexamethasone prevents the Epstein-Barr virus induced epithelial-mesenchymal transition in A549 cells. J. Med. Virol. 2020 doi: 10.1002/jmv.25999. [DOI] [PubMed] [Google Scholar]
- 31.Liu Y., Zhong W., Zhang J., Chen W., Lu Y., Qiao Y., et al. Tetrandrine modulates rheb-mTOR signaling-mediated selective autophagy and protects pulmonary fibrosis. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.739220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song D., Tang L., Wang L., Huang J., Zeng T., Fang H., et al. Roles of TGFbeta1 in the expression of phosphoinositide 3-kinase isoform genes and sensitivity and response of lung telocytes to PI3K inhibitors. Cell Biol. Toxicol. 2020;36(1):51–64. doi: 10.1007/s10565-019-09487-3. [DOI] [PubMed] [Google Scholar]
- 33.Guo H., Zhou H., Lu J., Qu Y., Yu D., Tong Y. Vascular endothelial growth factor: an attractive target in the treatment of hypoxic/ischemic brain injury. Neural Regen. Res. 2016;11(1):174–179. doi: 10.4103/1673-5374.175067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ou X.M., Li W.C., Liu D.S., Li Y.P., Wen F.Q., Feng Y.L., et al. VEGFR-2 antagonist SU5416 attenuates bleomycin-induced pulmonary fibrosis in mice. Int. Immunopharm. 2009;9(1):70–79. doi: 10.1016/j.intimp.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 35.Chen Z., Zhao L., Zhao F., Yang G., Wang J.J. Tetrandrine suppresses lung cancer growth and induces apoptosis, potentially via the VEGF/HIF-1alpha/ICAM-1 signaling pathway. Oncol. Lett. 2018;15(5):7433–7437. doi: 10.3892/ol.2018.8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen S., Liu W., Wang K., Fan Y., Chen J., Ma J., et al. Tetrandrine inhibits migration and invasion of human renal cell carcinoma by regulating Akt/NF-kappaB/MMP-9 signaling. PLoS One. 2017;12(3) doi: 10.1371/journal.pone.0173725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goodwin J., Choi H., Hsieh M.H., Neugent M.L., Ahn J.M., Hayenga H.N., et al. Targeting hypoxia-inducible factor-1alpha/pyruvate dehydrogenase kinase 1 Axis by dichloroacetate suppresses bleomycin-induced pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 2018;58(2):216–231. doi: 10.1165/rcmb.2016-0186OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osborn-Heaford H.L., Ryan A.J., Murthy S., Racila A.M., He C., Sieren J.C., et al. Mitochondrial Rac1 GTPase import and electron transfer from cytochrome c are required for pulmonary fibrosis. J. Biol. Chem. 2012;287(5):3301–3312. doi: 10.1074/jbc.M111.308387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murthy S., Ryan A., He C., Mallampalli R.K., Carter A.B. Rac1-mediated mitochondrial H2O2 generation regulates MMP-9 gene expression in macrophages via inhibition of SP-1 and AP-1. J. Biol. Chem. 2010;285(32):25062–25073. doi: 10.1074/jbc.M109.099655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osborn-Heaford H.L., Murthy S., Gu L., Larson-Casey J.L., Ryan A.J., Shi L., et al. Targeting the isoprenoid pathway to abrogate progression of pulmonary fibrosis. Free Radic. Biol. Med. 2015;86:47–56. doi: 10.1016/j.freeradbiomed.2015.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen H.J., Sun Y.H., Zhang S.J., Jiang J.X., Dong X.W., Jia Y.L., et al. Cigarette smoke-induced alveolar epithelial–mesenchymal transition is mediated by Rac1 activation. BBA - General Subjects. 2014;1840(6):1838–1849. doi: 10.1016/j.bbagen.2014.01.033. [DOI] [PubMed] [Google Scholar]
- 42.Yao-Song G., Lianmei W., Xinlun T., Xue L., Aiping M., Weixun Z., et al. mTOR overactivation and compromised autophagy in the pathogenesis of pulmonary fibrosis. PLoS One. 2015;10(9) doi: 10.1371/journal.pone.0138625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Madala S.K., Thomas G., Edukulla R., Davidson C., Schmidt S., Schehr A., et al. p70 ribosomal S6 kinase regulates subpleural fibrosis following transforming growth factor-α expression in the lung. Am. J. Physiol. Lung Cell Mol. Physiol. 2016 doi: 10.1152/ajplung.00063.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.