Abstract
Chemical exchange often serves as the first step in plant–microbe interactions and exchanges of various signals, nutrients, and metabolites continue throughout the interaction. Here, we highlight the role of metabolite exchanges and metabolic crosstalk in the microbiome–root–shoot–environment nexus. Roots secret a diverse set of metabolites; this assortment of root exudates, including secondary metabolites such as benzoxazinoids, coumarins, flavonoids, indolic compounds, and terpenes, shapes the rhizosphere microbiome. In turn, the rhizosphere microbiome affects plant growth and defense. These inter-kingdom chemical interactions are based on a metabolic circular economy, a seemingly wasteless system in which rhizosphere members exchange (i.e. consume, reuse, and redesign) metabolites. This review also describes the recently discovered phenomenon “Systemically Induced Root Exudation of Metabolites” in which the rhizosphere microbiome governs plant metabolism by inducing systemic responses that shift the metabolic profiles of root exudates. Metabolic exchange in the rhizosphere is based on chemical gradients that form specific microhabitats for microbial colonization and we describe recently developed high-resolution methods to study chemical interactions in the rhizosphere. Finally, we propose an action plan to advance the metabolic circular economy in the rhizosphere for sustainable solutions to the cumulative degradation of soil health in agricultural lands.
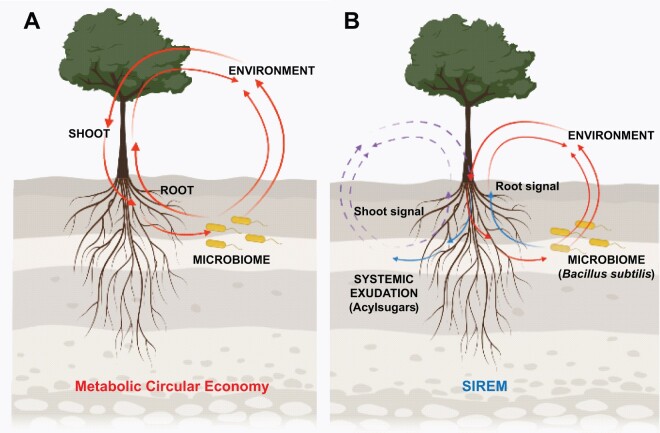
The microbiome–root–shoot–environment nexus is based on a metabolic circular economy, which influences rhizosphere interactions.
Introduction
Over millions of years, plants and microbes have developed various associations ranging from mutualistic to parasitic. Plants and their associated microbes can be considered holobionts, in which the host relies on its microbiome for specific functions and traits (Rosenberg et al., 2009). It is estimated that the number of microbial cells colonizing plants is higher than the sum of plant cells, particularly those colonizing the root (Mendes et al., 2013). The rhizosphere is considered the richest source of organic material in soil and therefore a hotspot for microbial growth and activity (Reinhold-Hurek et al., 2015). Moreover, the number of microbes in the rhizosphere (soil around the root zone) is 5–10 times higher than in nonrhizospheric soil (Groleau-Renaud et al., 2000). The rhizosphere microbiome provides diverse benefits to its plant host: microbes promote plant growth (Lugtenberg and Kamilova, 2009), support nutrient uptake (Weidner et al., 2015), improve tolerance to abiotic stress (Yang et al., 2009), defend the plant host against pathogens (Raaijmakers et al., 2009; Shi et al., 2017), and modulate the plant immune system to induce resistance (Bakker et al., 2013).
Roots select specific microbial populations and shape the microbiome composition in their vicinity (i.e. the rhizosphere) and internal tissues (i.e. the endosphere) (Bulgarelli et al., 2012; Bai et al., 2015; Uroz et al., 2019). To date, an exceptional number of reports have provided ample information regarding bacterial community structure in the rhizosphere of different plant species (mostly model and crop plants, but also some wild species) (Bai et al., 2015; Bulgarelli et al., 2015; Kawasaki et al., 2016). Although less well-studied than bacteria, fungal communities in the rhizosphere have also been systematically described (Berlanas et al., 2019). Archaea, oomycetes, protozoa, and viruses are also found in the rhizosphere (Mendes et al., 2013).
Chemical exchanges play an important role in the complex interactions among members of the root microbiome. Much of the research reporting the effect of root exudates on rhizosphere microbes was performed on dual relationships, such as plant interactions with nitrogen-fixing bacteria, mycorrhizal fungi, plant growth-promoting rhizobacteria, biocontrol microorganisms, and with pathogenic fungi and bacteria (Table 1). Understanding the chemodiversity and the chemical signaling affecting root activity and/or shaping rhizosphere microbial activity is hence pivotal to protect plants in nature and improve crop productivity.
Table 1.
Overview of studies reporting the effect of root exudates from different plant species on various rhizosphere microbes
| Plant species | Metabolite/whole exudate | Effect (±) | Rhizosphere microorganism | Reference |
|---|---|---|---|---|
| Alfalfa (M. sativa) | 7,4′-Dihydroxyflavone and naringenin | + | Acidobacteria | Szoboszlay et al. (2016) |
| Arabidopsis (A. thaliana) | Phytochemical extracts from root exudates | + | Microbiome | Badri et al. (2013) |
| Arabidopsis | Malic acid | + | B. subtilis FB17 | Rudrappa et al. (2008) |
| Arabidopsis | Scopoletin | − | F. oxysporum and V. dahliae | Stringlis et al. (2018) |
| Arabidopsis | Sideretin and fraxetin | − | Pseudomonas sp. Root329 | Voges et al. (2019) |
| Arabidopsis | Thalianin, thalianyl fatty acid esters, and arabidin | + | Proteobacteria | Huang et al. (2019) |
| − | Actinobacteria | |||
| Arabidopsis | Camalexin | + | Pseudomonas sp. CH267 | Koprivova et al. (2019) |
| Arabidopsis and alfalfa | Whole exudate effect | ± | Fungal community | Broeckling et al. (2008) |
| Arabidopsis abcg30 mutant | Whole exudate effect | + | Microbiome analysis (e.g. Bradyrhizobium) | Badri et al. (2009) |
| Arabidopsis myc2 and med25 mutants | Whole exudate effect | + | Microbiome analysis (Streptomyces, Bacillus, and Lysinibacillus) | Carvalhais et al. (2015) |
| Banana (Musa acuminata) | Malic and fumaric acids | + | B. amyloliquefaciens NJN-6 | Yuan et al. (2015) |
| Chinese tallow (Triadica sebifera) | Flavonoid | + | AM fungi | Tian et al. (2021) |
| Eucalyptus globulus ssp. Bicostata | Rutin | + | Pisolithus | Lagrange et al. (2001) |
| Common bean (Phaseolus vulgaris) | Flavonoid | + | Rhizobium leguminosarum | Aguilar et al. (1988) |
| Maize (Z. mays) | Benzoxazinoids | − | Flavobacteriaceae and Comamonadaceae | Cadot et al. (2021) |
| Maize | 2,4-Dihydroxy-7-methoxy-2H-1,4-benzoxazin-3(4H)-one | + | P. putida KT2440 | Neal et al. (2012) |
| Maize | (6R)-7,8-Dihydro- 3-oxo-ionone and (6R; 9R)-7,8-dihydro-3-oxo-ionol | − | F. oxysporum f. sp. melongenae | Park et al. (2004) |
| Maize | Flavones | + | Oxalobacteraceae | Yu et al. (2021) |
| Maize | Whole exudate effect | + | B. amyloliquefaciens SQR9 | Zhang et al. (2015) |
| Peanut (Arachis hypogaea) | Alanine and other amino acids | + | F. oxysporum and F. solani | Li et al. (2013) |
| Pine (Pinus radiata) | Quinic, lactic, maleic acids | + | Microbiome | Shi et al. (2011) |
| Potato (Solanum tuberosum) | Tyramine and other amino acids | + | S. subterranean | Balendres et al. (2016) |
| Sand Sedge (Carex arenaria) | Volatile | + | Soil bacteria | Schulz-Bohm et al. (2018) |
| Sugarbeet (Beta vulgaris) | Whole exudate effect | + | P. aeruginosa PA01 | Mark et al. (2005) |
| Tobacco | Whole exudate effect | + | Paenibacillus elgii | Das et al. (2010) |
| Tomato (S. lycopersicum) | α-Tomatine | + | Sphingomonadaceae | Nakayasu et al. (2021) |
| Tomato | Whole exudate effect | + | F. oxysporum f. sp. lycopersici | Scheffknecht et al. (2006) |
| Tomato | Whole exudate effect | + | Pseudomonas spp. | Kravchenko et al. (2003) |
| Tomato and cucumber (Cucumis sativus) | Citric acid | + | Pseudomonas fluorescens PCL1751, P. fluorescens PCL1753, Pantoea agglomerans PCA0067, and Aeromonas hydrophila PCA0081 | Kamilova et al. (2006) |
| Watermelon (Citrullus lanatus) | Chlorogenic acid | − | F. oxysporum f. sp. niveum | Ling et al. (2013) |
| Watermelon | Cinnamic acid | + | F. oxysporum f. sp. niveum | Ling et al. (2011) |
| Wild oat (Avena barbata) | Organic acids nicotinic, shikimic, salicylic, cinnamic and IAA | + | Microbacterium HA36, Flavobacterium HB58 and Cellulomonas HD24 | Zhalnina et al. (2018) |
Root growth, metabolism, and exudation are crucial for establishing interactions with rhizosphere microbiota. Root exudation is the main source of organic compounds released into the rhizosphere. Changes in soil parameters caused by root activity, known as soil conditioning, affect plant–soil feedback (Herrera Paredes and Lebeis, 2016). Soil conditioning has a major effect on microbial growth and activity in the rhizosphere. Conversely, rhizosphere microorganisms influence plant metabolism and performance (Korenblum and Aharoni, 2019). Microbial modulation of plant metabolism can be local or systemic; known systemic responses induced by microbial colonization of roots include: (1) nitrogen fixation (i.e. autoregulation of nodulation [AON]; [Reid et al., 2011]), (2) disease resistance (i.e. induced systemic resistance [Pieterse et al., 2014]) and, recently, (3) root exudation can be microbially modulated through a systemic response (i.e. SIREM for “Systemically Induced Root Exudation of Metabolites” [Korenblum et al., 2020]).
This review focuses on the chemical interaction between rhizosphere microbes and plant roots, including processes that modulate plant metabolism. We present a critical appraisal of plant root exudation and its effect on rhizosphere microbes, and compare the effect of conditioned and nonconditioned soils on the rhizosphere microbiome composition of the next generation of plant host (aka microbiome soil borne legacy) (Bakker et al., 2018). Root exudates shape the rhizosphere microbiome, which influences plant metabolism and exudation (as in SIREM). Finally, we bring a suit of evidence for host–microbiome and metabolome crosstalk, that is, plants eavesdrop on chemical communication between microbes, and vice-versa. Bringing these data together, it follows that the microbiome–root–shoot–environment nexus (Hou et al., 2021b, 2021a) is based on what can be delineated as “metabolic circular economy” (Figure 1) influencing rhizosphere interactions and plant health.
Figure 1.
Metabolic circular economy in the rhizosphere. A, Schematic representation of chemical allocation in the phytobiome. The microbiome–root–shoot–environment nexus is based on a metabolic circular economy that influences rhizosphere interactions where metabolites are exchanged (i.e. consumed, reused and redecorated [redesigned]) by different rhizosphere microbiome members. B, SIREM leads to shifts in the chemical diversity in the rhizosphere and is microbiome-driven. SIREM (blue arrows) is a mechanism of the metabolic circular economy system (red and purple arrows). SIREM-related acylsugar exudation is induced by the soil bacterium B. subtilis.
Carbon sinks in the rhizosphere
Carbon allocation is vital for plants to adapt to environmental changes. The trade-off between carbon sink and source activities will finally govern the success of plant growth (Lemoine et al., 2013), but also has a major impact on plant interactions with its microbiota (Hennion et al., 2019). Consequently, belowground carbon allocation reflects a wide range of physiological and ecological strategies, such as nutrient mobilization (e.g. iron acquisition) and selecting the rhizosphere microbiome through root exudation. Plants exude large amounts of substances made of photosynthetically fixed carbon through the roots into the rhizosphere, the zone of soil under the immediate influence of plant roots (Hiltner, 1904). Root exudates contain a wide variety of small molecules including amino acids, carbohydrates, organic acids, hormones, vitamins, and different classes of specialized metabolites (Venturi and Keel, 2016; Sasse et al., 2018). Metabolite patterns and quantity of root exudates are dependent on plant species, age, and environment (Maurer et al., 2021).
Root exudation requires the transport of molecules to the root system from the shoot and/or through root cell layers and subsequent release to soil. We currently have little understanding of how metabolite secretion ensues and its associated regulatory mechanisms. To be secreted by cells of the epidermis and possibly by root cap cells, a molecule needs to traverse the plasma membrane and permeate the cell wall. Thus, membrane transporters are likely involved in root chemical secretion. Exudation of primary metabolites (e.g. sugars and amino acids) is facilitated by transporters (e.g. SWEET transporters) along the concentration gradient (Breia et al., 2021). As most secondary metabolites cannot simply diffuse through membranes, especially metabolites that are modified by glycosylation, acylation, or hydroxylation, the secretion of these molecules is likely an active process. In one of only a few examples, export of coumarins from roots in Arabidopsis thaliana is mediated by a Pleiotropic Drug Resistance (PDR)-Type ATP-Binding Cassette (ABC) transporter (ABCG37/PDR9; Ziegler et al., 2017). ABCG37/PDR9 was also associated with transport of the endogenous auxin precursor indole-3-butyric acid in Arabidopsis signifying a most likely promiscuous activity of such proteins (Růžička et al., 2010). Apart from the limited number of transporters associated with metabolite exudation (Weston et al., 2012; Sasse et al., 2018; Canarini et al., 2019) we also have a major gap of knowledge with respect to the precise location of root metabolite accumulation and the exact location of exudation. In recent work, we employed matrix-assisted Mass Spectrometry Imaging (MSI) and demonstrated the spatial localization of secondary metabolites in tomato (Solanum lycopersicum) roots (Korenblum et al., 2020). Some metabolites were specifically found on the tips of lateral roots (e.g. the acylsugar S1:5), while other metabolites were only detected on the hairs of the main root (e.g. acylsugar S4:19 and hydroxytomatine). MSI was also used in a different study to reveal the spatial distribution of metabolites involved in regulating biological nitrogen fixation within soybean root nodules (Veličković et al., 2018). An alternative to MSI techniques to track the precise location of metabolite accumulation in roots is the use of a biosensor. Pini et al. (2017), employed Rhizobium bio-reporter strains to map root secretion of sugars, polyols, amino acids, organic acids, or flavonoids in pea (Pisum sativum) roots and nodules. In pea nodules, dicarboxylates and sucrose are the main carbon sources. This evidence suggests that root exudation is likely variable along the root axis.
As early as 1904, Hiltner noted the selection of a unique population of microorganisms by the chemicals released from plant roots (Hiltner, 1904). Since then, a relatively small number of specific plant metabolites have been described to impact the root microbiome and several studies have tested the effect of total root exudates collections on soil microorganisms. Here, we provide a comprehensive list of studies that evaluated the effect of root exudates on the rhizosphere microbes, including plant volatiles (Table 1). Whole exudate collections are typically composed of a wide range of molecules, including high and low molecular weight compounds affecting both bacterial and fungi soil microorganisms. Arabidopsis and alfalfa (Medicago sativa) whole exudate extracts exhibit a plant-specific effect on soil fungal communities (Broeckling et al., 2008), while genetic modification of active metabolite transporters (Badri et al., 2009) or key regulators of pathogen defense genes (Carvalhais et al., 2015) resulted in changes of root exudates composition and microbial communities.
Metabolites exuded through the roots are either synthesized in the roots or supplied by the shoot. Roots are supplied with sugars that were synthesized in the leaves and are mostly used for maintenance of root growth during early growth stages (Hennion et al., 2019). Correspondingly, in Arabidopsis, sugars are exuded by roots in the greatest abundance early in the plant’s life cycle (7–10 days old) (Chaparro et al., 2014); in later stages (18–21 days old), sugars are still found in root exudates and have some contribution to the Arabidopsis rhizosphere microbiome structure (Badri et al., 2013). The rhizosphere effect of organic acids exuded from plant roots has been frequently studied (Kamilova et al., 2006; Rudrappa et al., 2008; Ling et al., 2011, 2013; Shi et al., 2011; Zhalnina et al., 2018). Apparently, rhizosphere bacteria grow preferentially on aromatic organic acids exuded by plants (i.e. malonic, malic, nicotinic, shikimic, salicylic, cinnamic, and indole-3-acetic acids) (Oburger et al., 2009; Zhalnina et al., 2018). Being one of the most labile carbon sources in the rhizosphere, when released by roots organic acids are quickly consumed (i.e. uptaken or biodegraded) by the rhizosphere microbiota (Oburger et al., 2009). These studies provided evidence that the chemical composition of root exudates, the substrate preference, and competition of soil microbes determine the structure and function of the rhizosphere microbiome. Soil conditioning through exudation of organic acids regulates the composition of the rhizosphere microbiome and apparently promotes the growth of microbes that assist plants fitness. For instance, malic and fumaric acids released by banana roots are crucial for Bacillus amyloliquefaciens NJN-6 colonization on the host roots. Bacillus amyloliquefaciens NJN-6, originally isolated from the rhizosphere of banana plants, was shown to protect these plants from Fusarium oxysporum f. sp. cubense and promote their growth (Yuan et al., 2015). Interestingly, organic acid treatment of soil was shown to improve soil physicochemical performance and affect the structure of the soil microbial community (by inducing the enrichment of plant growth-promoting bacteria). Following these findings, the authors suggested the use of organic acids as soil prebiotics (Macias-Benitez et al., 2020). Conversely, amino acids were associated with enhanced growth of pathogenic microorganisms. A typical example is the production of tyramine and other amino acids by potato roots, leading to Spongospora subterranea growth, a major crop-threatening pathogen (Balendres et al., 2016). Alanine and other amino acids secreted from peanuts were also found to promote the growth of F. oxysporum and Fusarium solani (Li et al., 2013). Amino acids in the rhizosphere may serve as sources of both carbon and nitrogen; while microbes seemingly prefer to uptake inorganic nitrogen, the ability to take up amino acids confers an advantage by some opportunistic soil pathogens (Moe, 2013).
The role of plant secondary metabolites in rhizosphere interactions
Besides organic acids and sugars that are essential carbon sources, secondary metabolites can also shape the rhizosphere microbiome. They function as semiochemicals mediating interactions or as toxic compounds deterring plant pathogens. For instance, flavonoids are hitherto one of the most studied chemical classes in root exudates. The various branches of the intricate flavonoid pathway exhibit diverse effects on soil microorganisms (Hassan and Mathesius, 2012; Weston and Mathesius, 2013). They are pivotal in attracting rhizobia to the root system of legume plants and induce nodule formation by activation of nod genes from the rhizobia. Legume-nodulating rhizobia use quorum-sensing (QS) N-acyl homoserine lactones (AHLs) to regulate this symbiotic interaction and flavonoids (e.g. genistein, apigenin, and daidzein) can also increase the production of autoinducers and consequently the expression of AHL synthesis genes in rhizobia (Pérez-Montaño et al., 2011). Conversely, certain flavonoids inhibit QS in Pseudomonas aeruginosa and Escherichia coli through allosteric inhibition of receptors (Paczkowski et al., 2017; Manner and Fallarero, 2018). Root excreted flavonoids are also well-known for inducing the establishment of arbuscular mycorrhizal (AM) symbiosis (Tian et al., 2021) and as defense molecules against soil-borne pathogens. Interestingly, the Nod Factors from Bradyrhizobium and Rhizobium were shown to induce exudation of flavonoids in higher amounts in soybean (Glycine max) plants (Schmidt et al., 1994). Maize (Zea mays) roots exude metabolites from different chemical classes, mostly benzoxazinoids (see below) and flavonoids are secreted into the rhizosphere. The flavone apigenin exuded by maize roots was shown to affect plant growth and nitrogen nutrition by a microbial-driven process, that is, oxalobacteraceae is enriched in apigenin-containing rhizosphere and promote plant growth and nitrogen acquisition (Yu et al., 2021).
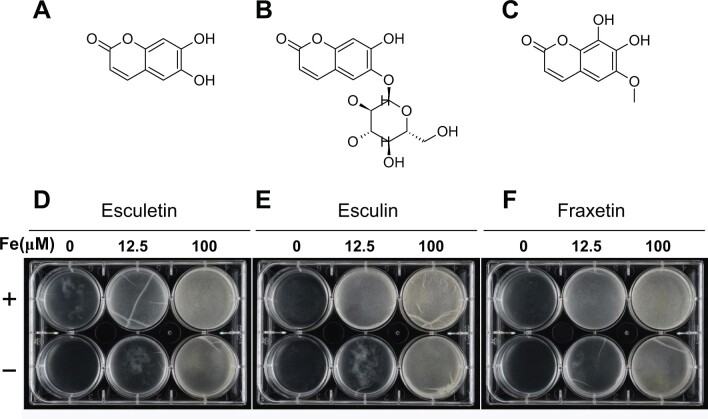
Another milestone in rhizosphere chemistry was the recent discovery that coumarins shape the Arabidopsis root microbiome when grown under low iron conditions (Stringlis et al., 2018; Voges, et al., 2019). Specifically, scopoletin inhibits the growth of two soil-borne fungal pathogens, F. oxysporum and Verticillium dahlia (Stringlis et al., 2018). While catecholic coumarins (e.g. fraxetin) inhibit the growth of the bacterial strain Pseudomonas sp. Root329, these molecules could improve Bacillus subtilis biofilm formation (Figure 2). Bacillus subtilis is widely used in agriculture as a biocontrol agent against various plant pathogens. Root colonization is dependent on its ability to form biofilm on roots and requires active iron acquisition from the soil milieu by producing catecholic siderophores (Chen et al., 2012; Rizzi et al., 2019). While B. subtilis likely uses the plant-derived catecholic coumarins as iron chelators; the proposed mode of action of these molecules against Pseudomonas involves the generation of reactive oxygen species (Voges et al., 2019). Besides flavonoids and coumarins, different phenylpropanoids can be secreted from watermelon roots and showed an opposite effect on the same soil microorganisms. Ling et al. (2013) showed that chlorogenic acid from watermelon has a negative effect on the pathogen F. oxysporum f. sp. niveum, while the upstream compound in the phenylpropanoid pathway, cinnamic acid, was found to support the growth of the same fungi (Ling et al., 2011).
Figure 2.
Coumarins from Arabidopsis root exudate improve biofilm formation of the soil bacterium B. subtilis. Structures of the coumarins Esculetin (A), Esculin (B), and Fraxetin (C) are depicted. The effect of these molecules (D, E, and F, respectively) in a B. subtilis pellicle assay conducted with a final concentration of 5 μM of each coumarin is shown. The sign (+) on the left side indicates coumarin treatment and the (−) sign indicates control (without coumarin). Bacterial cells were inoculated into the growth medium with different iron concentrations, and incubated at 23ºC for 2 days in dark before imaging. Biofilm formation appears as a bright layer in the positive wells. In low iron condition, root-derived coumarins improve B. subtilis biofilm formation.
Arabidopsis root-derived triterpenes also exert a dual effect on the root microbiome. A blend of specific root-derived triterpenes (namely thalianin, thalianyl medium-chain fatty acid esters, and arabidin) in Arabidopsis, may increase the proliferation of bacterial strains belonging to Proteobacteria strains while it inhibits the growth of Actinobacteria strains. Terpenes are the largest group of plant secondary metabolites showing a vast diversity of chemistries (Chen et al., 2004; Huang and Osbourn, 2019; Muchlinski et al., 2019). This class of metabolites likely confers a broad spectrum of biological activities in the rhizosphere; yet no molecular mechanism of action of root-derived terpenes on rhizosphere bacteria was disclosed. Nevertheless, monoterpenes are known to exhibit antibacterial activity, for example, carvacrol and thymol disturb membrane integrity of nonplant associated bacteria (Solórzano-Santos and Miranda-Novales, 2012). The sesquiterpene lactone strigolactone, which was first identified in root exudates of cotton plants a few decades ago (Cook et al., 1966), influences AM symbiosis and rhizobial root interactions (López-Ráez et al., 2017). In rhizobial interactions, strigolactones affect swarming motility of the rhizobacteria. In AM symbiosis, strigolactones are exuded into the rhizosphere and attract AM fungi; in return, the fungal counterpart provides phosphate and facilitates plant water uptake. Several strigolactones were isolated and associated to the induction of hyphal branching in AM fungi (AMF), which is a critical step in host recognition; for instance 5-deoxystrigol in Lotus japonicus (Akiyama et al., 2005), orobanchol and solanacol in tomato plants (López-Ráez et al., 2008). For additional details on the role of strigolactones in plant interactions with beneficial and detrimental organisms, see for example the review by López-Ráez et al. (2017).
The localization of biosynthesis of indole-derived specialized metabolites, such as camalexin and benzoxazinoids, is critical to root exudation and interactions. Arabidopsis and other Brassicaceae plants produce and secrete camalexin in the shoot and in the root as a phytoalexin against pathogens. Interestingly, root-specific biosynthesis of camalexin that is dependent on the activity of the cytochrome P450 CYP71A27 in Arabidopsis, is pivotal to the plant interactions with multiple microbial strains (Koprivova et al., 2019). A remarkable association of the activity of CYP71A27 in roots and the sulfatase activity in bacteria appeared crucial to plant growth promotion by Pseudomonas sp. CH267 and MPI9 strains. Pharmacologically effective doses of camalexin complemented both effects of the CYP71A27 gene knockout, the sulfatase activity, and the plant growth promotion by Pseudomonas sp. CH267. The plant defense benzoxazinoids (BX), specifically the aglycones 2,4-dihydroxy-1,4-benzoxazin-3-one and 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one glucoside, are predominantly secreted from crown roots (roots originating from the stem) as compared to primary roots (roots developing from the radicle). The rhizosphere microbial community structure from BX-producing wild-type maize differs from that of a BX-deficient bx1 mutant of maize (Hu et al., 2018), and crown root-associated communities of both wild-type and BX-deficient mutants show reduced diversity indices when compared with primary root-associated communities (Cotton et al., 2019). Mutant lines have been used to identify the role of BX and other plant metabolites in rhizosphere interactions, but those specific metabolic changes in mutant lines may affect overall plant metabolome and consequently change the metabolic pattern of the root exudates. For instance, Cotton et al. (2019) showed that benzoxazinoids deficient bx1 and bx2 mutants of maize influence the rhizosphere microbiome by an endogenous regulatory activity on a wider spectrum of plant-derived rhizosphere signals (e.g. flavonoids). This key observation highlights a challenge in deciphering the role of secondary metabolites in the rhizosphere; metabolic engineering primarily targets one metabolite or gene but it usually generates off-target metabolic changes (Lv et al., 2014). Plant metabolism is highly intertwined and perturbation of a single gene leads to multiple consequences on metabolic flux. Thus, besides complementation assays, as used in the case of camalexin by Koprivova et al. (2019), a system-wide analysis might be assisting in evaluating the whole exudate effect on the root microbiome.
Plant–soil feedback and root–shoot–root systemic responses
By changing soil properties, root exudation has a fundamental role in plant–soil feedbacks also in further generation of plants, the so-called microbiome driven “soil-borne legacy” (Bakker et al., 2018). The best-studied case of the benefits of soil conditioning on new generations growing in the same soil is the agricultural phenomenon of disease-suppressive soils. Improved suppression of fungal and bacterial plant pathogens is associated with the enrichment of antagonistic microbial members in the soil microbiome. Examples include the take-all disease of wheat (Triticum aestivum) (Berendsen et al., 2012), scab disease of potato (Sagova-Mareckova et al., 2015) and black rot disease of tobacco (Nicotiana tabacum) (Almario et al., 2014). In maize, benzoxazinoids recruit rhizosphere bacteria (e.g. the plant beneficial bacterium Pseudomonas putida) that enhance jasmonate signaling and defense responses in the next generation of plants (Hu et al., 2018). While soil conditioning functions in assembling a more health-promoting root microbiota, benzoxazinoids secretion also enriches various potential plant pathogenic fungi (Cadot et al., 2021). Some soils are naturally suppressive, but disease suppression can also be determined by the plant host selection of antibiotic producing bacteria from the native soil microbiota. For instance, a specific tomato variety (Hawaii 7996) recruits a rhizospheric flavobacterium against the soil-borne pathogen Ralstonia solanacearum and rhizosphere transplantation of this resistant plant transfers the ability to control disease symptoms in a susceptible tomato variety (Kwak et al., 2018). Specific root exudation pattern likely results in the recruitment of this flavobacterium by resistant plants. In Arabidopsis, alteration of root exudation patterns and increased resistance to Pseudomonas syringae were observed in plants grown in conditioned soils (i.e. soils used to grow multiple plant generations that were infected by P. syringae) (Yuan et al., 2018). Infected plants exuded through the roots increased amounts of amino acids and long-chain organic acids. The addition of these molecules into the rhizosphere was found to elicit disease suppression.
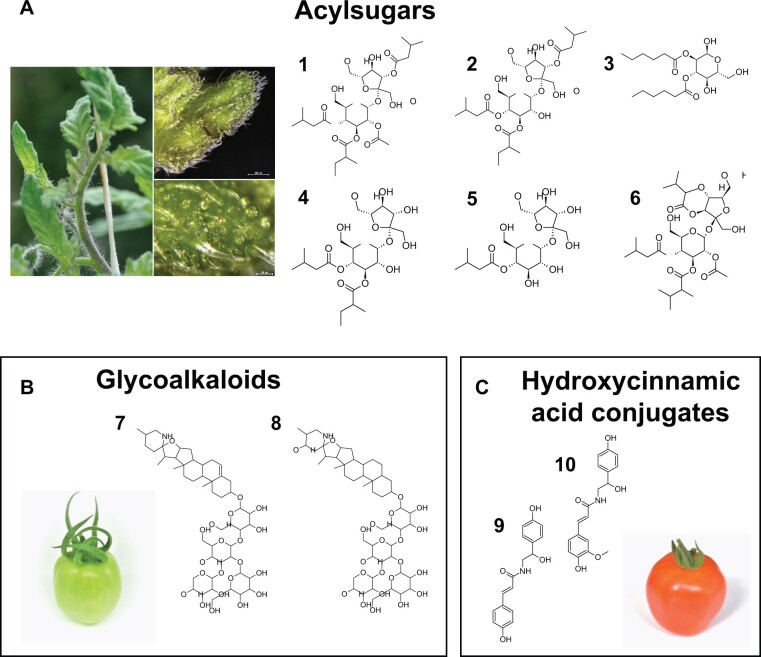
Microbial-driven changes in root metabolite profiles, gene expression and developmental patterns were reported numerous times. SIREM (Korenblum et al., 2020) is a root–root long-distance signaling mediated by the root microbiome. In SIREM, tomato rhizosphere microbiome modulates the chemical diversity secreted to the rhizosphere by changing the patterns of root exudation through a systemic root–shoot-root signaling mechanism. Using a split-root hydroponic system, different soil microbiomes introduced to one root side (“local side”) were used to evaluate the effect of the root microbiomes on the metabolites secreted in the second root side (“systemic side”; kept under axenic conditions). Three root microbiome treatments were prepared with high, medium, and low microbial diversity levels (termed HD, MD, and LD, respectively). Following 7 days of root microbiome treatment, metabolic profiling of root exudates in the “systemic side” revealed that 53.3%–75.4% of the total mass features (detected in electrospray negative or positive ionization mode, respectively) were significantly modulated by one of the three microbiome treatments. We detected a total of 115 metabolites that were significantly enriched or depleted in the systemic side that was modulated by the “local side” root microbiome. SIREM-induced metabolites represented different chemical classes including aliphatic and aromatic alcohol glycosides, fatty acids, hydroxycinnamic acid conjugates, organic acid derivatives, sulfur-containing compounds, steroidal glycoalkaloids (SGAs), steroidal saponins, and acylsugars. The latter class of metabolites is known to be produced by foliar glandular trichomes of tomato and other members of the Solanaceae family (Figure 3). These molecules consist of either glucose or sucrose backbones esterified with three to four acyl chains, each containing 2–12 carbons. In SIREM, acylsucroses (26 metabolites) and acylglucoses (G2:12 [6, 6]; seven isomers) were exuded by the “systemic side” of tomato roots; the sugar moiety and the length of the acyl chains differed according to the root microbiome composition. HD-treated plants mostly exuded acylsugars that are uniquely secreted in the course of SIREM in tomato; acylsucroses with C5 acyl chains (S1:5, S2:10, and S3:15). As in the case of acylsugars, hydroxycinnamic acid amides conjugated to tyramine or octopamine, were reported for the first time to be secreted by roots. While acylsugars and hydroxycinnamic acid conjugates were induced in HD-treated plants, ferulic acid glycosides were suppressed in LD-treated plant exudates. Tomato SGAs commonly found in tomato fruits were also SIREM-modulated in roots, such as hydroxytomatine, dehydrotomatine, and α-tomatine (Korenblum et al., 2020). Interestingly, α-tomatine was recently associated with enrichment of Sphingomonadaceae in the tomato rhizosphere, suggesting its role in belowground chemical communication (Nakayasu et al., 2021).
Figure 3.
Root exudate chemical diversity modulated by rhizosphere microbiome. SIREM-related metabolites that are exuded from “systemic side” roots of tomato plants, which were previously detected in other tomato tissues, depicted in (A), tomato trichomes; B, unripe fruit and; C, ripe fruit. Structures of SIREM-related metabolites: A, acylsugars; B, glycoalkaloids; and C, hydroxycinnamic acid conjugates are presented. (1) S4:17 (2,5,5,5); (2) S3:15 (5,5,5); (3) G2:12 (6,6); (4) S2:10 (5,5); (5) S1:5; (6) S4:19 (2,5,6,6); (7) dehydrotomatine; (8) hydroxytomatine; (9) coumaroyloctopamine; and (10) feruloyloctopamine. The acyl chains positions were not unambiguously confirmed in the acylsugar structures and were assigned arbitrarily.
Another SIREM-related molecule that was detected in exudates of microbiome-treated plants was azelaic acid (AzA). It accumulated in the “systemic side” root tissue in a glycosylated form. The AzA aglycone is commonly found in leaves of various plants after pathogen attack (Lim et al., 2017); neither the aglycone nor the glycosylated form were reported previously to occur in roots nor in exudates. Following the challenge of split-root plants with AzA only, AzA was detected in systemic parts of the split-root plants, both shoot and root, in its glycosylated form (i.e. AzA-hexose). The AzA aglycone is exuded through the “systemic side” root. Additionally, AzA treatment induces systemic exudation of other metabolites, for example, the SGA α-tomatine was higher in exudates of AzA-treated plants as compared to untreated plants (Korenblum et al., 2020).
SIREM is dependent on the colonization of roots by specific bacteria and possibly on the interaction of plant roots with other soil microbes. For instance, B. subtilis induced the exudation of specific tetraacylsucroses and bacteria belonging to the order Pseudomonadales were correlated to the systemic exudation of ferulic acid glycosides. Therefore, SIREM represents a microbial-driven systemic root exudation mechanism that likely promotes soil conditioning. We also suggested that AzA or AzA-hexose are SIREM-inducing molecules that might be reprograming plant metabolism. The detection of specific chemistries (with large structural diversity) in root exudates modulated by root microbiome suggests exclusive rhizosphere functions, likely important to interactions belowground. Future research is required to elucidate acylsugars’ biosynthesis in roots of Solanaceae plants and unravel the specific role of these metabolites exudation in belowground interactions. Moreover, AzA biosynthesis and transport in planta also requires further research.
Plant systemic signaling mechanisms that regulate soil microbiome–root–shoot–root interactions remain little investigated to date. One challenge in such experiments is the isolation of the “local side” root from the “systemic side” root. The “split-root” experimental system is an excellent tool to reveal systemic signaling controlling root interactions with the rhizosphere microbiome (Kassaw and Frugoli, 2012; Larrainzar et al., 2014). The AON pathway is part of legume root–rhizobium symbiosis, in which nodule number is controlled by a systemic mechanism (Pervent et al., 2021). When soil N is limiting, legume plants are triggered to exude flavonoids (e.g. luteolin and apigenin), which recruit rhizobia to the roots and induce nodule formation (Weston and Mathesius, 2013). In the nodules, the enzyme nitrogenase catalyzes the reduction of N2 to NH4+. Bacterial infection and nodule formation are controlled by nitrate availability to roots. Notably, the number of nodules is systemically regulated through a signal that is produced in the nodule/root tissue (small peptides of the CLAVATA3 /EMBRYO SURROUNDING REGION-RELATED (CLE) family [Mortier et al., 2012]). The peptides are transported to the shoot, and then a shoot-derived secondary signal is transmitted back to roots to inhibit further nodulation in distal parts of the root system. This intricate long-distance relay of small peptides (including microRNAs and hormones; Okuma et al., 2020) that is crucial for balancing between susceptibility to rhizobia colonization (i.e. nodule formation and influx of nitrogen) and the efflux of carbon compounds derived from photosynthesis to maintain the nodule active. Interestingly, autoregulation of symbiosis has also been reported for the AMF root colonization, a “common symbiosis pathway” that controls the establishment of both root nodulation and the AMF–plant symbiosis (Ikeda et al., 2010). Nodulation systemically influences AMF root colonization and the other way around (Catford et al., 2003). In AMF–plant symbiosis, CLE peptides are also produced by the fungal counterpart, which positively regulates symbiosis (Le Marquer et al., 2019).
Metabolic crosstalk in the rhizosphere
While there is no evidence of a universal language in the rhizosphere, it is clear that extensive chemical communication occurs between plants and its microbiome. The plant host and its microbiome coexisted and coevolved for millions of years. During this period of time both counterparts have been exposed to numerous chemicals, among them signaling molecules, produced and released by the other. Therefore, plants enhance and interfere with bacterial communication systems and similarly bacterial signal molecules can influence plant metabolism. Aside from modulation of metabolism, crosstalk (or “hijacking”) of inter-kingdom signals (such as bacterial auto-inducers and host plant hormones) has broad implications for bacterial colonization on roots and plant fitness.
The inoculation of crops with beneficial microbes has been long explored to improve plant yield (Sessitsch et al., 2018). Several studies showed the effect of plant growth-promoting rhizobacteria on plant growth is based on their ability to produce phytohormones such as gibberellins (GAs), auxins, cytokinins (CKs), ethylene, and abscisic acid (ABA) (Freebairn and Buddenhagen, 1964; Morrone et al., 2009; Kudoyarova et al., 2014; Shahzad et al., 2017; Keswani et al., 2020). GAs were first discovered in the fungal rice pathogen Gibberella fujikuroi, but they are also produced by other fungi and bacteria (Salazar-Cerezo et al., 2018). Active GAs (i.e. GA1, GA3, GA4, and GA7) are pivotal for plant growth and its interaction with microbes. GA biosynthesis was unraveled recently in rhizobia, which independently evolved a biosynthetic pathway divergent from the plant and fungal ones (Nett et al., 2017). Bradyrhizobium diazoefficiens and other rhizobia contain an operon encoding the enzymes to produce GA. While two rhizobial diterpene cyclases (CPS and KS) share some homology with the plant and fungal cyclases, the other enzymes involved in rhizobial GA biosynthesis share little or no homology with the plant and fungi proteins (Nett et al., 2017). Similarly, plants and microbes are able to produce auxin. The most common auxin indole-3-acetic acid (IAA) evolved independently in fungi (first detected in the spent media of a yeast culture), bacteria, and plants (Duca et al., 2014). In plants, auxin plays a role amongst others in cell division, tissue differentiation, and plant growth while in fungi, IAA affects cell expansion, disturbs cell division, and in some species induces spore germination (Fu et al., 2015). It is estimated that over 80% of the rhizospheric bacteria are capable of synthesizing IAA (Spaepen and Vanderleyden, 2011). For instance, both rhizobacterial strains, Bacillus megaterium UMCV1 and Azospirillum brasilense Sp245, produce auxins and induce root-architectural alterations such as increased number of lateral roots and longer root hairs (López-Bucio et al., 2007; Spaepen and Vanderleyden, 2011). In both plants and microbes, IAA is synthesized either by a tryptophan-dependent pathway or by a tryptophan-free way. The production of auxins in bacteria seems to depend on the availability of precursors in root secretions. l-tryptophan has been identified in root exudates and is suggested as the main precursor of the synthesis of bacterial auxins (Kamilova et al., 2006; Fu et al., 2015). Therefore, auxin concentration in the rhizosphere is highly dependent on plant–microbe interactions. The effect of auxin on bacteria is diverse; it may function as a signaling molecule affecting gene expression, regulate antibiotic synthesis, and pathogenesis antagonizing plant defense responses (Fu et al., 2015; Kunkel and Harper, 2018; Matilla et al., 2018). CKs are involved in the interactions between roots and soil microorganisms and have been reported to play an important role in defense against biotrophic pathogens. Arabidopsis plants treated with trans-zeatin before P. syringae pv. tomato DC3000 inoculation, led to decreased susceptibility to the bacterial pathogen (Choi et al., 2010). The convergent evolution of GA, IAA, and CK biosynthesis suggests that these molecules were favored as a widespread physiological code in plants and microbes.
The gaseous hormone ethylene is also produced by microbes, however, the main microbial modulation in the rhizosphere impacting ethylene balance in plants is the reduction of plant ethylene levels via degradation of its immediate precursor 1-aminocyclopropane-1-carboxylate (ACC) (Gamalero and Glick, 2015). The catabolic activity of the microbial enzyme ACC deaminase lowers local levels of the hormone in plants, and then the low ethylene concentration allows plant growth under stressed conditions. In return, ACC is a nitrogen source to the rhizosphere microbiota. This mutualistic relationship between plants and ACC deaminase-producing bacteria has a great potential to promote plant stress tolerance as ethylene displays a wide range of biological effects in plants (Liu et al., 2019). As ethylene, ACC acts as a signaling molecule in several plant processes such as root–shoot signaling (Yoon and Kieber, 2013; Van de Poel, 2020), thus the interaction of plants with ACC deaminase-producing bacteria might decrease the degree of ACC signaling of specific plant functions. For instance, plant roots typically respond to flooding by synthesizing a high level of ACC. Due to lack of oxygen, ACC is translocated from roots to shoots, where it becomes a substrate for ACC oxidase and is converted to ethylene (Gamalero and Glick, 2015). Experiments with the “split-root” system demonstrated that a positive message (i.e. ACC) produced in roots was transmitted through the xylem and stimulated shoot ethylene production (Jackson, 2002). The abiotic stress plant hormone ABA is also produced by rhizosphere microbes and can be perceived by the plant hosts thereby improving drought resistance (e.g. A. brasilense sp 245; Cohen et al., 2008). ABA accumulation in soil can negatively affect seed germination, inhibit root growth and increase plant disease susceptibility (Yuzikhin et al., 2021). ABA root concentration balance interplays with other phytohormones in disease resistance, such as salicylic acid and ethylene. Interestingly, many rhizobacteria are capable of affecting the balance of one or more plant hormones. For example, Burkholderia phytofirmans PsJN affects both ethylene and auxin levels in plants. This same strain produces the autoinducer AHL that mediates QS in the rhizosphere and is crucial for root colonization (Zúñiga et al., 2013).
While AHL production by rhizosphere microbes is recognized to modulate plant gene expression and metabolism, host metabolites can cross-signal with microbial QS signals to modulate bacterial gene expression and root colonization (Joshi et al., 2021). The rhizosphere harbors a high amount of bacteria that employ QS mechanisms (Elasri et al., 2001). Among 129 bacterial isolates from cottonwood tree rhizosphere, 40% were tested positive for AHL production; all positive isolates belonged to the Proteobacteria phylum (Schaefer et al., 2013). A classical AHL QS system consists of a LuxI-type protein (AHL synthase) that interacts with the cognate LuxR-type protein (a transcription factor) (Steindler et al., 2008). Various luxR homologs were detected in the genomes of the cottonwood proteobacterial isolates, some of these homologs were suggested to be members of a subfamily of LuxRs that respond to plant signals rather than to bacterial AHLs (Schaefer et al., 2013). Bacterial AHLs can function as inter-kingdom signals on a widespread signaling network between plants and bacteria. As many plant beneficial and pathogenic bacteria require QS to successfully colonize the host plant, these bacteria can use their QS molecules to regulate plant growth (Schenk et al., 2012). Several plant species have been shown to respond to AHLs influencing and reprogramming plant gene expression (González and Venturi, 2013). Recently, four AHL molecules (N-(3-oxohexanoyl)-l-homoserine lactone [oxo-C6-HSL], N-(3-oxooctanoyl)-l-homoserine lactone [oxo-C8-HSL], N-(3-oxododecanoyl)-l-homoserine lactone [oxo-C12-HSL], and N-(3-oxotetradecanoyl)-l-homoserine lactone [oxo-C14-HSL]) and combinations of these molecules were tested for their effect on Arabidopsis growth and resistance against P. syringae pathovar tomato. Some of these AHL molecules, when treated independently, positively influenced plant growth, while others induced resistance by AHL-driven priming (Shrestha and Schikora, 2020). Many plant-associated bacteria (e.g. rhizobia, xanthomonads, and pseudomonads) have a LuxR-like protein that lacks an AHL synthase and these proteins are regarded as LuxR solo or orphan (Patankar and González, 2009; González and Venturi, 2013). LuxR solo proteins bind and respond to plant compounds. For instance, the plant phenylpropanoid p-coumaric acid accumulates in the rhizosphere. This metabolite activates the 4-coumaroyl-homoserine lactone synthase of Bradyrhizobium sp., which uses the plant-derived p-coumaric acid and endogenous S-adenosylmethionine to generate the hybrid signal molecule p-coumaroyl homoserine lactone (i.e. p-coumaroyl-HSL), which finally induces genes related to chemotaxis (Schaefer et al., 2008). Metabolism crosstalk is thus an emerging field in the rhizosphere inter-kingdom signaling, where plant host metabolites can be used as alternative substrates in bacteria (e.g. p-coumaroyl-HSL).
Future perspectives
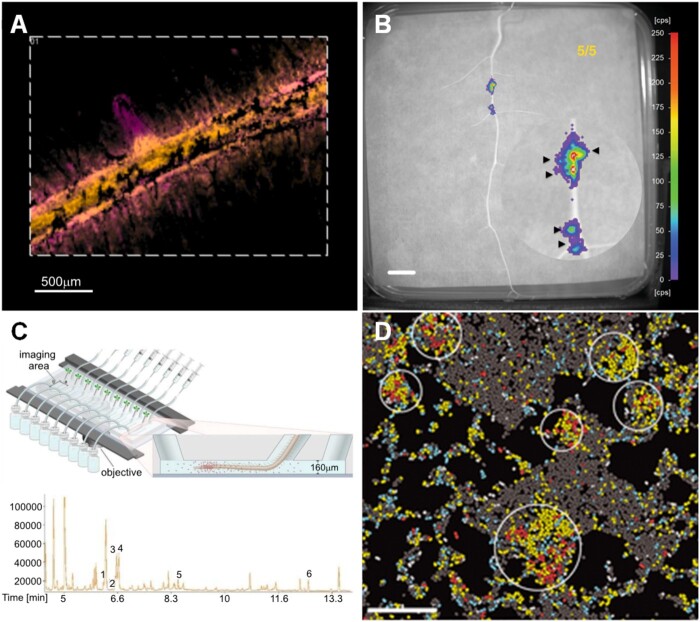
The fusion of plant and microbial small molecules as a concerted effort (Wang and Seyedsayamdost, 2017), and the biosynthesis of metabolites induced by external signals or depending on the microbiome context (Korenblum and Aharoni, 2019) are unquestionably pertinent in complex ecosystems such as the rhizosphere. However, the effect of these phenomena is not restricted to the rhizosphere and likely influences the plant host at the “phytobiome scale” (i.e. the network of the whole plant with their microbiome, other organisms, and the environment; Leach et al., 2017). Systemic processes are particularly important in the microbiome–root–shoot–environment nexus; the rhizosphere microbiota can induce various physiological changes in plants, including promotion of growth, improved health, and modulation of root exudation (e.g. in SIREM). The understanding of the internal and external chemical signals induced by the rhizosphere microbiome in systemic responses will provide tools for better bioinoculants technology and exudate-oriented plant breeding. Moreover, in this “metabolic circular economy” in the rhizosphere, the chemical spectrum is extensive and there is likely no metabolic waste. It is estimated that one plant species produces a few thousand metabolites (Fernie et al., 2004), but only a few tens of root-derived metabolites are known to have a role on the rhizosphere microbiome (Table 1). Besides reducing this knowledge gap and systematic evaluation of the metabolic connectivity among root exudates and the rhizosphere microbiome in model and nonmodel plants further research on the spatial distribution of root-secreted metabolites along the root axis using high-resolution methods will reveal detailed localization of metabolites in roots (Figure 4). These technologies include the combination of biosensors with microfluidic systems for in vivo spatiotemporal mapping of root secretion and microbial colonization (Massalha et al., 2017; Pini et al., 2017; Geddes et al., 2019) and MSI at the root cell-type resolution (Veličković et al., 2018; Korenblum et al., 2020). The spatial distribution of metabolites using these methods can be advanced by comparing mutants and wild-type plants. Additionally, spatially resolved metabolite localization at the single cell (Taylor et al., 2021) or the effect of metabolites on the microbiome transcriptomics at the single-cell resolution (Dar et al., 2021) are cutting-edge techniques that will further allow super-resolution of the metabolic coupling in the rhizosphere. Moreover, better understanding of the modulation of phytobiome metabolism will certainly advance the discovery of novel chemistries and the fundamental evolutionary trajectories of the metabolic crosstalk in the rhizosphere, that is, the chemically driven ecological rules in the rhizosphere. Gaining this knowledge is fundamental for the development of innovative strategies for sustainable agriculture and environmental protection.
Figure 4.
High-resolution methods to study chemical interactions in the rhizosphere. A, MSI-Imaging of tomato roots, metabolites are distributed in specific root regions as indicated by the overlap of false color images, hydroxytomatine (m/z 1050.54 ± 0.01 Da) in yellow and dehydrotomatine (m/z 1,032.54 ± 0.01 Da) in magenta (color hue was adjusted from Korenblum et al., 2020). B, NightOwl images of bioluminescence of R. leguminosarum Rlv3841/pOPS0046 (a rhizopine biosensor) growing on the surface of M. sativa roots allowed the detection of rhizopine exudation (Geddes et al., 2019). C, The microfluidic device TRIS consists of a series of 6.4 µL chambers, which allowed single-root exudate metabolomics. Ion chromatogram of a metabolic profile of single-root exudates analyzed by gas chromatography–mass spectrometry. Metabolites identified: 1, phosphoric acid; 2, isoleucine; 3, glycine; 4, succinic acid; 5, pyroglutamic acid; and 6, myoinositol (Massalha et al., 2017). D, Transcriptome imaging using par-seq FISH allowed the visualization of the spatial organization of transcriptional programs in P. aeruginosa at single-cell resolution. Using par-seq FISH, transcriptional states of individual cells can be identified and mapped onto the biofilm image (with permission from Dar et al., 2021).
Acknowledgments
We thank A. Goldshmidt (Plant Science Institute, ARO-Volcani Center) for providing M82 plants and H. Zemach (Plant Science Institute, ARO-Volcani Center) for technical help with imaging tomato trichrome. We also thank the Adelis Foundation, Leona M. and Harry B. Helmsley Charitable Trust, Jeanne and Joseph Nissim Foundation for Life Sciences, Tom and Sondra Rykoff Family Foundation Research, and the Raymond Burton Plant Genome Research Fund for supporting the A.A. lab activity.
Funding
The rhizosphere work in the A.A. lab is supported by the European Research Council Advanced Grant (ERC-2019-ADG; #884316; SIREM).
Conflict of interest statement. None declared.
Contributor Information
Elisa Korenblum, Institute of Plant Science, Agricultural Research Organization, The Volcani Center, Rishon LeTsiyon 7528809, Israel.
Hassan Massalha, Theory of Condensed Matter Group, Cavendish Laboratory, Wellcome Sanger Institute, University of Cambridge, Cambridge CB2 1TN, UK.
Asaph Aharoni, Department of Plant and Environmental Sciences, Weizmann Institute of Science, Rehovot 7610001, Israel.
A.A. is the incumbent of the Peter J. Cohn Professorial Chair. E.K. and A.A. wrote the paper. E.K generated Figures 1, 3, and 4. E.K. and H.M. contributed the data and generated Figure 2.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) are: Elisa Korenblum (elisak@volcani.agri.gov.il) and Asaph Aharoni (asaph.aharoni@weizmann.ac.il).
References
- Aguilar JMM, Ashby AM, Richards AJM, Loake GJ, Watson MD, Shaw CH (1988) Chemotaxis of Rhizobium leguminosarum biovar phaseoli towards flavonoid inducers of the symbiotic nodulation genes. Microbiology 134: 2741–2746 [Google Scholar]
- Akiyama K, Matsuzaki K, Hayashi H (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435: 824–827 [DOI] [PubMed] [Google Scholar]
- Almario J, Muller D, Défago G, Moënne-Loccoz Y (2014) Rhizosphere ecology and phytoprotection in soils naturally suppressive to Thielaviopsis black root rot of tobacco. Environ Microbiol 16: 1949–1960 [DOI] [PubMed] [Google Scholar]
- Badri DV, Quintana N, El Kassis EG, Kim HK, Choi YH, Sugiyama A, Verpoorte R, Martinoia E, Manter DK, Vivanco JM (2009) An ABC transporter mutation alters root exudation of phytochemicals that provoke an overhaul of natural soil microbiota. Plant Physiol 151: 2006–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badri DV, Chaparro JM, Zhang R, Shen Q, Vivanco JM (2013) Application of natural blends of phytochemicals derived from the root exudates of Arabidopsis to the soil reveal that phenolic-related compounds predominantly modulate the soil microbiome. J Biol Chem 288: 4502–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Müller DB, Srinivas G, Garrido-Oter R, Potthoff E, Rott M, Dombrowski N, Münch PC, Spaepen S, Remus-Emsermann M, et al. (2015) Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528: 364–369 [DOI] [PubMed] [Google Scholar]
- Bakker PAHM, Pieterse CMJ, de Jonge R, Berendsen RL (2018) The soil-borne legacy. Cell 172: 1178–1180 [DOI] [PubMed] [Google Scholar]
- Bakker PAHMHM, Doornbos RF, Zamioudis C, Berendsen RL, Pieterse CMJCMJ (2013) Induced systemic resistance and the rhizosphere microbiome. Plant Pathol J 29: 136–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balendres MA, Nichols DS, Tegg RS, Wilson CR (2016) Metabolomes of potato root exudates: compounds that stimulate resting spore germination of the soil-borne pathogen Spongospora subterranea. J Agric Food Chem 64: 7466–7474 [DOI] [PubMed] [Google Scholar]
- Berendsen RL, Pieterse CMJ, Bakker PAHM (2012) The rhizosphere microbiome and plant health. Trends Plant Sci 17: 478–486 [DOI] [PubMed] [Google Scholar]
- Berlanas C, Berbegal M, Elena G, Laidani M, Cibriain JF, Sagües A, Gramaje D (2019) The fungal and bacterial rhizosphere microbiome associated with grapevine rootstock genotypes in mature and young vineyards. Front Microbiol 10: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breia R, Conde A, Badim H, Fortes AM, Gerós H, Granell A (2021) Plant SWEETs: from sugar transport to plant–pathogen interaction and more unexpected physiological roles. Plant Physiol 186: 836–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeckling CD, Broz AK, Bergelson J, Manter DK, Vivanco JM (2008) Root exudates regulate soil fungal community composition and diversity. Appl Environ Microbiol 74: 738–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgarelli D, Rott M, Schlaeppi K, van Themaat EVL, Ahmadinejad N, Assenza F, Rauf P, Huettel B, Reinhardt R, Schmelzer E, et al. (2012) Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488: 91–95 [DOI] [PubMed] [Google Scholar]
- Bulgarelli D, Garrido-Oter R, Münch PC, Weiman A, Dröge J, Pan Y, McHardy AC, Schulze-Lefert P (2015) Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 17: 392–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadot S, Guan H, Bigalke M, Walser JC, Jander G, Erb M, van der Heijden MGA, Schlaeppi K (2021) Specific and conserved patterns of microbiota-structuring by maize benzoxazinoids in the field. Microbiome 9: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canarini A, Kaiser C, Merchant A, Richter A, Wanek W (2019) Root exudation of primary metabolites: mechanisms and their roles in plant responses to environmental stimuli. Front Plant Sci 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalhais LC, Dennis PG, Badri DV, Kidd BN, Vivanco JM, Schenk PM (2015) Linking jasmonic acid signaling, root exudates, and rhizosphere microbiomes. Mol Plant Microbe Interact 28: 1049–1058 [DOI] [PubMed] [Google Scholar]
- Catford JG, Staehelin C, Lerat S, Piché Y, Vierheilig H (2003) Suppression of arbuscular mycorrhizal colonization and nodulation in split-root systems of alfalfa after pre-inoculation and treatment with Nod factors. J Exp Bot 54: 1481–1487 [DOI] [PubMed] [Google Scholar]
- Chaparro JM, Badri DV, Vivanco JM (2014) Rhizosphere microbiome assemblage is affected by plant development. ISME J 8: 790–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Ro DK, Petri J, Gershenzon J, Bohlmann J, Pichersky E, Tholl D (2004) Characterization of a root-specific Arabidopsis terpene synthase responsible for the formation of the volatile monoterpene 1,8-cineole. Plant Physiol 135: 1956–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Cao S, Chai Y, Clardy J, Kolter R, Guo JH, Losick R (2012) A Bacillus subtilis sensor kinase involved in triggering biofilm formation on the roots of tomato plants. Mol Microbiol 85: 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Huh SU, Kojima M, Sakakibara H, Paek KH, Hwang I (2010) Developmental cell the cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis. Dev Cell 19: 284–295 [DOI] [PubMed] [Google Scholar]
- Cohen AC, Bottini R, Piccoli PN (2008) Azospirillum brasilense Sp 245 produces ABA in chemically-defined culture medium and increases ABA content in Arabidopsis plants. Plant Growth Regul 54: 97–103 [Google Scholar]
- Cotton TEA, Pétriacq P, Cameron DD, Meselmani MAl, Schwarzenbacher R, Rolfe SA, Ton J (2019) Metabolic regulation of the maize rhizobiome by benzoxazinoids. ISME J 13: 1647–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook CE, Whichard LP, Turner B, Wall ME, Egley GH (1966) Germination of witchweed (Striga lutea Lour.): isolation and properties of a potent stimulant. Science 154: 1189–1190 [DOI] [PubMed] [Google Scholar]
- Dar D, Dar N, Cai L, Newman DK (2021) Spatial transcriptomics of planktonic and sessile bacterial populations at single-cell resolution. Science 373: eabi4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das SN, Dutta S, Kondreddy A, Chilukoti N, Pullabhotla SVSRN, Vadlamudi S, Podile AR (2010) Plant growth-promoting chitinolytic Paenibacillus elgii responds positively to tobacco root exudates. J Plant Growth Regul 29: 409–418 [Google Scholar]
- Duca D, Lorv J, Patten CL, Rose D, Glick BR (2014) Indole-3-acetic acid in plant–microbe interactions. Anton Leeuw 106: 85–125 [DOI] [PubMed] [Google Scholar]
- Elasri M, Delorme S, Lemanceau P, Stewart G, Laue B, Glickmann E, Oger PM, Dessaux Y (2001) Acyl-homoserine lactone production is more common among plant-associated Pseudomonas spp. than among soilborne Pseudomonas spp. Appl Environ Microbiol 67: 1198–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Trethewey RN, Krotzky AJ, Willmitzer L (2004) Metabolite profiling: from diagnostics to systems biology. Nat Rev Mol Cell Biol 5: 763–769 [DOI] [PubMed] [Google Scholar]
- Freebairn HT, Buddenhagen IW (1964) Ethylene production by Pseudomonas solanacearum . Nature 202: 313–314 [DOI] [PubMed] [Google Scholar]
- Fu SF, Wei JY, Chen HW, Liu YY, Lu HY, Chou JY (2015) Indole-3-acetic acid: a widespread physiological code in interactions of fungi with other organisms. Plant Signal Behav 10: e1048052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamalero E, Glick BR (2015) Bacterial modulation of plant ethylene levels. Plant Physiol 169: 13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geddes BA, Paramasivan P, Joffrin A, Thompson AL, Christensen K, Jorrin B, Brett P, Conway SJ, Oldroyd GED, Poole PS (2019) Engineering transkingdom signalling in plants to control gene expression in rhizosphere bacteria. Nat Commun 10: 3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine R, La Camera S, Atanassova R, Dédaldéchamp F, Allario T, Pourtau N, Bonnemain JL, Laloi M, Coutos-Thévenot P, Maurousset L et al. (2013) Source-to-sink transport of sugar and regulation by environmental factors. Front Plant Sci 4: 272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González JF, Venturi V (2013) A novel widespread interkingdom signaling circuit. Trends Plant Sci 18: 167–174 [DOI] [PubMed] [Google Scholar]
- Groleau-Renaud V, Plantureux S, Tubeileh A, Guckert A (2000) Influence of microflora and composition of root bathing solution on root exudation of maize plants. J Plant Nutr 23: 1283–1301 [Google Scholar]
- Hassan S, Mathesius U (2012) The role of flavonoids in root–rhizosphere signalling: opportunities and challenges for improving plant-microbe interactions. J Exp Bot 63: 3429–3444 [DOI] [PubMed] [Google Scholar]
- Hennion N, Durand M, Vriet C, Doidy J, Maurousset L, Lemoine R, Pourtau N (2019) Sugars en route to the roots. Transport, metabolism and storage within plant roots and towards microorganisms of the rhizosphere. Physiol Plant 165: 44–57 [DOI] [PubMed] [Google Scholar]
- Herrera Paredes S, Lebeis SL (2016) Giving back to the community: microbial mechanisms of plant–soil interactions. Funct Ecol 30: 1043–1052 [Google Scholar]
- Hiltner L (1904) Über neuere erfahrungen und probleme auf dem gebiete der bodenbakteriologie unter besonderer berücksichtigung der gründüngung und brache. Arab Dtsch Landwirtsch Gesellschaft 98: 59–78 [Google Scholar]
- Hou S, Thiergart T, Vannier N, Mesny F, Ziegler J, Pickel B, Hacquard S (2021a) A microbiota–root–shoot circuit favours Arabidopsis growth over defence under suboptimal light. Nat Plants 7: 1078–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou S, Wolinska KW, Hacquard S (2021b) Microbiota–root–shoot–environment axis and stress tolerance in plants. Curr Opin Plant Biol 62: 102028. [DOI] [PubMed] [Google Scholar]
- Hu L, Robert CAM, Cadot S, Zhang X, Ye M, Li B, Manzo D, Chervet N, Steinger T, Van Der Heijden MGA, et al. (2018) Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat Commun 9: 2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AC, Osbourn A (2019) Plant terpenes that mediate below-ground interactions: prospects for bioengineering terpenoids for plant protection. Pest Manag Sci 75: 2368–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AC, Jiang T, Liu YX, Bai YC, Reed J, Qu B, Goossens A, Nützmann HW, Bai Y, Osbourn A (2019) A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science 364: eaau6389. [DOI] [PubMed] [Google Scholar]
- Ikeda S, Okubo T, Anda M, Nakashita H, Yasuda M, Sato S, Kaneko T, Tabata S, Eda S, Momiyama A, et al. (2010) Community- and genome-based views of plant-associated bacteria: plant-bacterial interactions in soybean and rice. Plant Cell Physiol 51: 1398–1410 [DOI] [PubMed] [Google Scholar]
- Jackson MB (2002) Long-distance signaling from roots to shoots assessed: the flooding story. J Exp Bot 53: 175–181 [DOI] [PubMed] [Google Scholar]
- Joshi JR, Khazanov N, Charkowski A, Faigenboim A, Senderowitz H, Yedidia I (2021) Interkingdom signaling interference: the effect of plant-derived small molecules on quorum sensing in plant-pathogenic bacteria. Annu Rev Phytopathol 59: 153–190 [DOI] [PubMed] [Google Scholar]
- Kamilova F, Kravchenko LV, Shaposhnikov AI, Azarova T, Makarova N, Lugtenberg B (2006) Organic acids, sugars, and L-tryptophan in exudates of vegetables growing on stonewool and their effects on activities of rhizosphere bacteria. Mol Plant Microbe Interact 19: 250–256 [DOI] [PubMed] [Google Scholar]
- Kassaw TK, Frugoli JA (2012) Simple and efficient methods to generate split roots and grafted plants useful for long-distance signaling studies in Medicago truncatula and other small plants. Plant Methods 8: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki A, Donn S, Ryan PR, Mathesius U, Devilla R, Jones A, Watt M (2016) Microbiome and exudates of the root and rhizosphere of Brachypodium distachyon, a model for wheat. PLoS ONE 11: 1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keswani C, Singh SP, Cueto L, García-Estrada C, Mezaache-Aichour S, Glare TR, Borriss R, Singh SP, Blázquez MA, Sansinenea E (2020) Auxins of microbial origin and their use in agriculture. Appl Microbiol Biotechnol 104: 8549–8565 [DOI] [PubMed] [Google Scholar]
- Koprivova A, Schuck S, Jacoby RP, Klinkhammer I, Welter B, Leson L, Martyn A, Nauen J, Grabenhorst N, Mandelkow JF, et al (2019) Root-specific camalexin biosynthesis controls the plant growth-promoting effects of multiple bacterial strains. Proc Natl Acad Sci USA 116: 15735–15744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenblum E, Aharoni A (2019) Phytobiome metabolism: beneficial soil microbes steer crop plants’ secondary metabolism. Pest Manag Sci 75: 2378–2384 [DOI] [PubMed] [Google Scholar]
- Korenblum E, Dong Y, Szymanski J, Panda S, Jozwiak A, Massalha H, Meir S, Rogachev I, Aharoni A (2020) Rhizosphere microbiome mediates systemic root metabolite exudation by root-to-root signaling. Proc Natl Acad Sci USA 117: 3874–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravchenko LV, Azarova TS, Leonova-Erko EI, Shaposhnikov AI, Makarova NM, Tikhonovich IA (2003) Root exudates of tomato plants and their effect on the growth and antifungal activity of Pseudomonas strains. Microbiology 72: 37–41 [PubMed] [Google Scholar]
- Kudoyarova GR, Melentiev AI, Martynenko EV, Timergalina LN, Arkhipova TN, Shendel GV, Kuz’mina LY, Dodd IC, Veselov SY (2014) Cytokinin producing bacteria stimulate amino acid deposition by wheat roots. Plant Physiol Biochem 83: 285–291 [DOI] [PubMed] [Google Scholar]
- Kunkel BN, Harper CP (2018) The roles of auxin during interactions between bacterial plant pathogens and their hosts. J Exp Bot 69: 245–254 [DOI] [PubMed] [Google Scholar]
- Kwak MJ, Kong HG, Choi K, Kwon SK, Song JY, Lee J, Lee PA, Choi SY, Seo M, Lee HJ, et al. (2018) Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat Biotechnol 36: 1100–1109 [DOI] [PubMed] [Google Scholar]
- Lagrange H, Jay-Allgmand C, Lapeyrie F (2001) Rutin, the phenolglycoside from eucalyptus root exudates, stimulates Pisolithus hyphal growth at picomolar concentrations. New Phytol 149: 349–355 [DOI] [PubMed] [Google Scholar]
- Larrainzar E, Gil-Quintana E, Arrese-Igor C, Gonzlez EM, Marino D (2014) Split-root systems applied to the study of the legume-rhizobial symbiosis: what have we learned? J Integr Plant Biol 56: 1118–1124 [DOI] [PubMed] [Google Scholar]
- Le Marquer M, Bécard G, Frei dit Frey N (2019) Arbuscular mycorrhizal fungi possess a CLAVATA3/embryo surrounding region-related gene that positively regulates symbiosis. New Phytol 222: 1030–1042 [DOI] [PubMed] [Google Scholar]
- Leach JE, Triplett LR, Argueso CT, Trivedi P (2017) Communication in the phytobiome. Cell 169: 587–596 [DOI] [PubMed] [Google Scholar]
- Li X, Zhang TL, Wang XX, Hua K, Zhao L, Han ZM (2013) The composition of root exudates from two different resistant peanut cultivars and their effects on the growth of soil-borne pathogen. Int J Biol Sci 9: 164–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim GH, Singhal R, Kachroo A, Kachroo P (2017) Fatty acid- and lipid-mediated signaling in plant defense. Annu Rev Phytopathol 55: 505–536 [DOI] [PubMed] [Google Scholar]
- Ling N, Huang Q, Guo S, Shen Q (2011) Paenibacillus polymyxa SQR-21 systemically affects root exudates of watermelon to decrease the conidial germination of Fusarium oxysporum f. sp. niveum. Plant Soil 341: 485–493 [Google Scholar]
- Ling N, Zhang W, Wang D, Mao J, Huang Q, Guo S, Shen Q (2013) Root exudates from grafted-root watermelon showed a certain contribution in inhibiting Fusarium oxysporum f. sp. niveum. PLoS ONE 8: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Khan MY, Carvalhais LC, Delgado-Baquerizo M, Yan L, Crawford M, Dennis PG, Singh B, Schenk PM (2019) Soil amendments with ethylene precursor alleviate negative impacts of salinity on soil microbial properties and productivity. Sci Rep 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bucio J, Campos-Cuevas JC, Hernández-Calderón E, Velásquez-Becerra C, Farías-Rodríguez R, Macías-Rodríguez LI, Valencia-Cantero E (2007) Bacillus megaterium rhizobacteria promote growth and alter root-system architecture through an auxin- and ethylene-independent signaling mechanism in Arabidopsis thaliana. Mol Plant Microbe Interact 20: 207–217 [DOI] [PubMed] [Google Scholar]
- López-Ráez JA, Charnikhova T, Gómez-Roldán V, Matusova R, Kohlen W, De Vos R, Verstappen F, Puech-Pages V, Bécard G, Mulder P, et al. (2008) Tomato strigolactones are derived from carotenoids and their biosynthesis is promoted by phosphate starvation. New Phytol 178: 863–874 [DOI] [PubMed] [Google Scholar]
- López-Ráez JA, Shirasu K, Foo E (2017) Strigolactones in plant interactions with beneficial and detrimental organisms: the Yin and Yang. Trends Plant Sci 22: 527–537 [DOI] [PubMed] [Google Scholar]
- Lugtenberg B, Kamilova F (2009) Plant-growth-promoting rhizobacteria. Annu Rev Microbiol 63: 541–556 [DOI] [PubMed] [Google Scholar]
- Lv Q, Cheng R, Shi T (2014) Regulatory network rewiring for secondary metabolism in Arabidopsis thaliana under various conditions. BMC Plant Biol 14: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias-Benitez S, Garcia-Martinez AM, Caballero Jimenez P, Gonzalez JM, Tejada Moral M, Parrado Rubio J (2020) Rhizospheric organic acids as biostimulants: monitoring feedbacks on soil microorganisms and biochemical properties. Front Plant Sci 11: 633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manner S, Fallarero A (2018) Screening of natural product derivatives identifies two structurally related flavonoids as potent quorum sensing inhibitors against gram-negative bacteria. Int J Mol Sci 19: 1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark GL, Dow JM, Kiely PD, Higgins H, Haynes J, Baysse C, Abbas A, Foley T, Franks A, Morrissey J, et al. (2005) Transcriptome profiling of bacterial responses to root exudates identifies genes involved in microbe–plant interactions. Proc Natl Acad Sci USA 102: 17454–17459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massalha H, Korenblum E, Malitsky S, Shapiro OH, Aharoni A (2017) Live imaging of root–bacteria interactions in a microfluidics setup. Proc Natl Acad Sci USA 114: 4549–4554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilla MA, Daddaoua A, Chini A, Morel B, Krell T (2018) An auxin controls bacterial antibiotics production. Nucleic Acids Res 46: 11229–11238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer D, Malique F, Alfarraj S, Albasher G, Horn MA, Butterbach-Bahl K, Dannenmann M, Rennenberg H (2021) Interactive regulation of root exudation and rhizosphere denitrification by plant metabolite content and soil properties. Plant Soil 467: 107–127 [Google Scholar]
- Mendes R, Garbeva P, Raaijmakers JM (2013) The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev 37: 634–663 [DOI] [PubMed] [Google Scholar]
- Moe LA (2013) Amino acids in the rhizosphere: from plants to microbes. Am J Bot 100: 1692–1705 [DOI] [PubMed] [Google Scholar]
- Morrone D, Chambers J, Lowry L, Kim G, Anterola A, Bender K, Peters RJ (2009) Gibberellin biosynthesis in bacteria: separate ent-copalyl diphosphate and ent-kaurene synthases in Bradyrhizobium japonicum. FEBS Lett 583: 475–480 [DOI] [PubMed] [Google Scholar]
- Mortier V, De Wever E, Vuylsteke M, Holsters M, Goormachtig S (2012) Nodule numbers are governed by interaction between CLE peptides and cytokinin signaling. Plant J 70: 367–376 [DOI] [PubMed] [Google Scholar]
- Muchlinski A, Chen X, Lovell JT, Köllner TG, Pelot KA, Zerbe P, Ruggiero M, Callaway LM, Laliberte S, Chen F, et al. (2019) Biosynthesis and emission of stress-induced volatile terpenes in roots and leaves of switchgrass (Panicum virgatum L.). Front Plant Sci 10: 1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayasu M, Ohno K, Takamatsu K, Aoki Y, Yamazaki S, Takase H, Shoji T, Yazaki K, Sugiyama A (2021) Tomato roots secrete tomatine to modulate the bacterial assemblage of the rhizosphere. Plant Physiol 186: 270–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal AL, Ahmad S, Gordon-Weeks R, Ton J (2012) Benzoxazinoids in root exudates of maize attract Pseudomonas putida to the rhizosphere. PLoS ONE 7: e35498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett RS, Montanares M, Marcassa A, Lu X, Nagel R, Charles TC, Hedden P, Rojas MC, Peters RJ (2017) Elucidation of gibberellin biosynthesis in bacteria reveals convergent evolution. Nat Chem Biol 13: 69–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oburger E, Kirk GJD, Wenzel WW, Puschenreiter M, Jones DL (2009) Interactive effects of organic acids in the rhizosphere. Soil Biol Biochem 41: 449–457 [Google Scholar]
- Okuma N, Soyano T, Suzaki T, Kawaguchi M (2020) MIR2111-5 locus and shoot-accumulated mature miR2111 systemically enhance nodulation depending on HAR1 in Lotus japonicus. Nat Commun 11: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paczkowski JE, Mukherjee S, McCready AR, Cong JP, Aquino CJ, Kim H, Henke BR, Smith CD, Bassler BL (2017) Flavonoids suppress Pseudomonas aeruginosa virulence through allosteric inhibition of quorum-sensing receptors. J Biol Chem 292: 4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Takano Y, Matsuura H, Yoshihara T (2004) Antifungal compounds from the root and root exudate of Zea mays. Biosci Biotechnol Biochem 68: 1366–1368 [DOI] [PubMed] [Google Scholar]
- Patankar AV, González JE (2009) Orphan LuxR regulators of quorum sensing. FEMS Microbiol Rev 33: 739–756 [DOI] [PubMed] [Google Scholar]
- Pérez-Montaño F, Guasch-Vidal B, González-Barroso S, López-Baena FJ, Cubo T, Ollero FJ, Gil-Serrano AM, Rodríguez-Carvajal MÁ, Bellogín RA, Espuny MR (2011) Nodulation-gene-inducing flavonoids increase overall production of autoinducers and expression of N-acyl homoserine lactone synthesis genes in rhizobia. Res Microbiol 162: 715–723 [DOI] [PubMed] [Google Scholar]
- Pervent M, Lambert I, Tauzin M, Karouani A, Nigg M, Jardinaud MF, Severac D, Colella S, Martin-Magniette ML, Lepetit M (2021) Systemic control of nodule formation by plant nitrogen demand requires autoregulation-dependent and independent mechanisms . J Exp Bot 72: 7942–7956 [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, Zamioudis C, Berendsen RL, Weller DM, Van Wees SCM, Bakker PAHM (2014) Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol 52: 347–375 [DOI] [PubMed] [Google Scholar]
- Pini F, East AK, Appia-Ayme C, Tomek J, Karunakaran R, Mendoza-Suárez M, Edwards A, Terpolilli JJ, Roworth J, Downie JA, et al. (2017) Bacterial biosensors for in vivo spatiotemporal mapping of root secretion. Plant Physiol 174: 1289–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers JM, Paulitz TC, Steinberg C, Alabouvette C, Moënne-Loccoz Y (2009) The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 321: 341–361 [Google Scholar]
- Reid DE, Ferguson BJ, Hayashi S, Lin YH, Gresshoff PM (2011) Molecular mechanisms controlling legume autoregulation of nodulation. Ann Bot 108: 789–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold-Hurek B, Bünger W, Burbano CS, Sabale M, Hurek T (2015) Roots shaping their microbiome: global hotspots for microbial activity. Annu Rev Phytopathol 53: 403–424 [DOI] [PubMed] [Google Scholar]
- Rizzi A, Roy S, Bellenger JP, Beauregard PB (2019) Iron homeostasis in Bacillus subtilis requires siderophore production and biofilm formation. Appl Environ Microbiol 85: e02439–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg E, Sharon G, Zilber-Rosenberg I (2009) The hologenome theory of evolution contains Lamarckian aspects within a Darwinian framework. Environ Microbiol 11: 2959–2962 [DOI] [PubMed] [Google Scholar]
- Rudrappa T, Czymmek KJ, Paré PW, Bais HP (2008) Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol 148: 1547–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Růžička K, Strader LC, Bailly A, Yang H, Blakeslee J, Łangowski L, Nejedlá E, Fujita H, Itoh H, Syōno K, et al. (2010) Arabidopsis PIS1 encodes the ABCG37 transporter of auxinic compounds including the auxin precursor indole-3-butyric acid. Proc Natl Acad Sci USA 107: 10749–10753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagova-Mareckova M, Daniel O, Omelka M, Kristufek V, Divis J, Kopecky J (2015) Determination of factors associated with natural soil suppressivity to potato common scab. PLoS ONE 10: e0116291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar-Cerezo S, Martínez-Montiel N, García-Sánchez J, Pérez-y-Terrón R, Martínez-Contreras RD (2018) Gibberellin biosynthesis and metabolism: a convergent route for plants, fungi and bacteria. Microbiol Res 208: 85–98 [DOI] [PubMed] [Google Scholar]
- Sasse J, Martinoia E, Northen T (2018) Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci 23: 25–41 [DOI] [PubMed] [Google Scholar]
- Schaefer AL, Greenberg EP, Oliver CM, Oda Y, Huang JJ, Bittan-Banin G, Peres CM, Schmidt S, Juhaszova K, Sufrin JR, et al. (2008) A new class of homoserine lactone quorum-sensing signals. Nature 454: 595–599. [DOI] [PubMed] [Google Scholar]
- Schaefer AL, Lappala CR, Morlen RP, Pelletier DA, Lu TYS, Lankford PK, Harwood CS, Greenberg EP (2013) LuxR- and luxI-type quorum-sensing circuits are prevalent in members of the Populus deltoides microbiome. Appl Environ Microbiol 79: 5745–5752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffknecht S, Mammerler R, Steinkellner S, Vierheilig H (2006) Root exudates of mycorrhizal tomato plants exhibit a different effect on microconidia germination of Fusarium oxysporum f. sp. lycopersici than root exudates from non-mycorrhizal tomato plants. Mycorrhiza 16: 365–370 [DOI] [PubMed] [Google Scholar]
- Schenk ST, Stein E, Kogel KH, Schikora A (2012) Arabidopsis growth and defense are modulated by bacterial quorum sensing molecules. Plant Signal Behav 7: 178–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt PE, Broughton WJ, Werner D (1994) Nod factors of Bradyrhizobium japonicum and Rhizobium sp. NGR234 induce flavonoid accumulation in soybean root exudate. Mol Plant Microbe Interact 7: 384–390 [Google Scholar]
- Schulz-Bohm K, Gerards S, Hundscheid M, Melenhorst J, De Boer W, Garbeva P (2018) Calling from distance: Attraction of soil bacteria by plant root volatiles. ISME J 12: 1252–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessitsch A, Brader G, Pfaffenbichler N, Gusenbauer D, Mitter B (2018) The contribution of plant microbiota to economy growth. Microb Biotechnol 11: 801–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahzad R, Khan AL, Bilal S, Waqas M, Kang SM, Lee IJ (2017) Inoculation of abscisic acid-producing endophytic bacteria enhances salinity stress tolerance in Oryza sativa. Environ Exp Bot 136: 68–77 [Google Scholar]
- Shi L, Du N, Shu S, Sun J, Li S, Guo S (2017) Paenibacillus polymyxa NSY50 suppresses Fusarium wilt in cucumbers by regulating the rhizospheric microbial community. Sci Rep 7: 41234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S, Richardson AE, O’Callaghan M, Deangelis KM, Jones EE, Stewart A, Firestone MK, Condron LM (2011) Effects of selected root exudate components on soil bacterial communities. FEMS Microbiol Ecol 77: 600–610 [DOI] [PubMed] [Google Scholar]
- Shrestha A, Schikora A (2020) AHL-priming for enhanced resistance as a tool in sustainable agriculture. FEMS Microbiol Ecol 96: fiaa226. [DOI] [PubMed] [Google Scholar]
- Solórzano-Santos F, Miranda-Novales MG (2012) Essential oils from aromatic herbs as antimicrobial agents. Curr Opin Biotechnol 23: 136–141 [DOI] [PubMed] [Google Scholar]
- Spaepen S, Vanderleyden J (2011) Auxin and plant-microbe interactions. Cold Spring Harb Perspect Biol 3: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steindler L, Bertani I, De Sordi L, Bigirimana J, Venturi V (2008) The presence, type and role of N-acyl homoserine lactone quorum sensing in fluorescent Pseudomonas originally isolated from rice rhizospheres are unpredictable. FEMS Microbiol Lett 288: 102–111 [DOI] [PubMed] [Google Scholar]
- Stringlis IA, Yu K, Feussner K, De Jonge R, Van Bentum S, Van Verk MC, Berendsen RL, Bakker PAHM, Feussner I, Pieterse CMJ (2018) MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc Natl Acad Sci USA 115: E5213–E5222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szoboszlay M, White-Monsant A, Moe LA (2016) The effect of root exudate 7,4′-dihydroxyflavone and naringenin on soil bacterial community structure. PLoS ONE 11: e0146555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MJ, Lukowski JK, Anderton CR (2021) Spatially resolved mass spectrometry at the single cell: recent innovations in proteomics and metabolomics. J Am Soc Mass Spectrom 32: 872–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Pei Y, Huang W, Ding J, Siemann E (2021) Increasing flavonoid concentrations in root exudates enhance associations between arbuscular mycorrhizal fungi and an invasive plant. ISME J 15: 1919–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uroz S, Courty PE, Oger P (2019) Plant symbionts are engineers of the plant-associated microbiome. Trends Plant Sci 24: 905–916 [DOI] [PubMed] [Google Scholar]
- Van de Poel B (2020) Ethylene’s fraternal twin steals the spotlight. Nat Plants 6: 1309–1310 [DOI] [PubMed] [Google Scholar]
- Veličković D, Agtuca BJ, Stopka SA, Vertes A, Koppenaal DW, Paša-Tolić L, Stacey G, Anderton CR (2018) Observed metabolic asymmetry within soybean root nodules reflects unexpected complexity in rhizobacteria–legume metabolite exchange. ISME J 12: 2335–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturi V, Keel C (2016) Signaling in the rhizosphere. Trends Plant Sci 21: 187–198 [DOI] [PubMed] [Google Scholar]
- Voges MJEEE, Bai Y, Schulze-Lefert P, Sattely ES (2019) Plant-derived coumarins shape the composition of an Arabidopsis synthetic root microbiome. Proc Natl Acad Sci USA 116: 12558–12565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Seyedsayamdost MR (2017) Opinion: hijacking exogenous signals to generate new secondary metabolites during symbiotic interactions. Nat Rev Chem 1: 1–8 [Google Scholar]
- Weidner S, Koller R, Latz E, Kowalchuk G, Bonkowski M, Scheu S, Jousset A (2015) Bacterial diversity amplifies nutrient-based plant-soil feedbacks. Funct Ecol 29: 1341–1349 [Google Scholar]
- Weston LA, Mathesius U (2013) Flavonoids: their structure, biosynthesis and role in the rhizosphere, including allelopathy. J Chem Ecol 39: 283–297 [DOI] [PubMed] [Google Scholar]
- Weston LA, Ryan PR, Watt M (2012) Mechanisms for cellular transport and release of allelochemicals from plant roots into the rhizosphere. J Exp Bot 63: 3445–3454 [DOI] [PubMed] [Google Scholar]
- Yang J, Kloepper JW, Ryu CM (2009) Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci 14: 1–4 [DOI] [PubMed] [Google Scholar]
- Yoon GM, Kieber JJ (2013) 1-Aminocyclopropane-1-carboxylic acid as a signalling molecule in plants. AoB Plants 5: plt017 [Google Scholar]
- Yu P, He X, Baer M, Beirinckx S, Tian T, Moya YAT, Zhang X, Deichmann M, Frey FP, Bresgen V, et al. (2021) Plant flavones enrich rhizosphere Oxalobacteraceae to improve maize performance under nitrogen deprivation. Nat Plants 7: 481–499 [DOI] [PubMed] [Google Scholar]
- Yuan J, Zhang N, Huang Q, Raza W, Li R, Vivanco JM, Shen Q (2015) Organic acids from root exudates of banana help root colonization of PGPR strain Bacillus amyloliquefaciens NJN-6. Sci Rep 5: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Zhao J, Wen T, Zhao M, Li R, Goossens P, Huang Q, Bai Y, Vivanco JM, Kowalchuk GA, et al. (2018) Root exudates drive the soil-borne legacy of aboveground pathogen infection. Microbiome 6: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzikhin OS, Gogoleva NE, Shaposhnikov AI, Konnova TA, Osipova EV, Syrova DS, Ermakova EA, Shevchenko VP, Nagaev IY, Shevchenko KV, et al. (2021) Rhizosphere bacterium Rhodococcus sp. P1Y metabolizes abscisic acid to form dehydrovomifoliol. Biomolecules 11: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhalnina K, Louie K, Hao Z, Mansoori N, da Rocha UN, Shi S, Cho H, Karaoz U, Loqué D, Bowen BP, et al. (2018) Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat Microbiol 3: 1–11 [DOI] [PubMed] [Google Scholar]
- Zhang N, Yang D, Wang D, Miao Y, Shao J, Zhou X, Xu Z, Li Q, Feng H, Li S, et al. (2015) Whole transcriptomic analysis of the plant-beneficial rhizobacterium Bacillus amyloliquefaciens SQR9 during enhanced biofilm formation regulated by maize root exudates. BMC Genomics 16: 685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler J, Schmidt S, Strehmel N, Scheel D, Abel S (2017) Arabidopsis transporter ABCG37/PDR9 contributes primarily highly oxygenated coumarins to root exudation. Sci Rep 7: 3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zúñiga A, Poupin MJ, Donoso R, Ledger T, Guiliani N, Gutiérrez RA, González B (2013) Quorum sensing and indole-3-acetic acid degradation play a role in colonization and plant growth promotion of Arabidopsis thaliana by Burkholderia phytofirmans PsJN. Mol Plant Microbe Interact 26: 546–553 [DOI] [PubMed] [Google Scholar]