Strigolactones (SLs) are a group of phytohormones that control many aspects of plant development, including the suppression of branching. In rice, SLs repress the formation of tillers, an important trait linked to biomass and grain yield (Yan et al., 1998). SLs bind to the receptor DWARF14 (D14), triggering downstream events that eventually lead to the degradation of the transcriptional repressor DWARF53 (D53) and subsequent activation of SL target genes. While an extensive body of literature is available about SL perception and signaling (Aquino et al., 2021), less is known about how other processes regulate the SL pathway.
In a new publication, Tianzhen Liu, Xin Zhang, Huan Zhang, Zhijun Cheng,and colleagues (Liu et al., 2022) present evidence that tillering in rice is repressed by splicing of the D14 pre-mRNA. The authors identified a rice mutant that displayed retarded growth and a higher number of tillers, which they named dwarf and high tillering1 (dht1). DHT1 is a predicted heterogeneous nuclear ribonucleoprotein (hnRNP)-like protein with two putative RNA-binding motifs (RRMs). During intron splicing, a large number of proteins including hnRNPs and serine–arginine-rich (SR) proteins ensure the accurate recognition of the splice site by the precatalytic spliceosome (Wahl et al., 2009). By sequencing the genomic region in the mutant, Liu and colleagues detected a single-nucleotide substitution in DHT1 causing an isoleucine-to-phenylalanine substitution. When they reintroduced the gene into the mutant, they restored the phenotype, and vice versa, knocking out DHT1 with CRISPR exacerbated the phenotype, producing even more tillers. The authors then used DHT1-GFP fusion proteins to show that DHT1 is localized in nuclear speckles. Using domain deletions, they were able to demonstrate that DHT1’s C-terminal domain and RRM1 are required for speckle localization.
They then set out to identify possible interaction partners of DHT1. After tagging a series of SR proteins with mCherry, they showed that a number of these proteins colocalize with DHT1, and their interactions with DHT1 were confirmed using in vitro pull downs, Co-IP assays, and firefly luciferase complementation imaging. Next, Liu and colleagues investigated the events at the gene regulation level. They used RNA immunoprecipitation sequencing (RIP-seq) to identify DHT1 target genes and RNA-seq to look for potential splicing defects. An overlap of these two results obtained a list of 116 genes with potential alternative splicing caused by DHT1. One of these genes was the SL receptor D14, whose splicing efficiency and transcript level decreased in the dht1 mutant.
The authors explored further how DHT1 influences SL signaling. They noticed that dht1 mutant plants not only exuded more SL than wild-type plants, but that they were also less sensitive to the synthetic SL analog GR24. Quantitative reverse transcription PCR (RT-qPCR) revealed that DHT1 expression was reduced in several SL biosynthesis and signaling mutants but remained unaltered in SL-insensitive dwarf53 (d53) mutant plants. These results strongly suggested that DHT1 participates in SL signaling. To test this hypothesis, the authors used immunoblots to compare protein levels of D14 and D53 between the dht1 mutant and wild-type. While D14 turned out to be decreased in the dht1 mutant, D53 levels were significantly higher and the D53 protein was no longer degraded after application of GR24. This nicely demonstrated that DHT1 acts as a stabilizing factor in SL signaling. Finally, when either D14 or D53 was overexpressed in the dht1 mutant, the tiller number phenotype was restored, suggesting that these two proteins act downstream of DHT1 in the regulation of rice tillering.
By elegantly combining in planta and in vitro methods, Liu and colleagues have identified and characterized an hnRNP-like protein that is required for the splicing of many introns, including the single intron of D14, thus establishing a connection between posttranscriptional splicing and SL signaling (see Figure). Due to the pleiotropic phenotypes of the dht1 mutant, it is hard to judge to what extent DHT1 evolved to specifically regulate SL signaling. Since DHT1 also binds to many other genes that are involved in plant growth and development, it will be exciting to see future reports about this protein.
Figure.
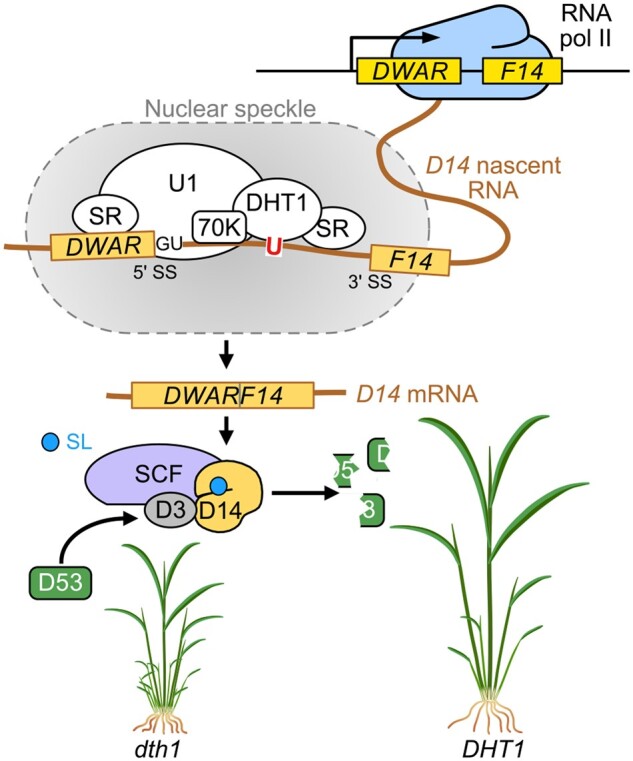
DHT1 regulates rice tillering through D14 splicing. DHT1 is part of a spliceosome complex localized in nuclear speckles. Production of the correct D14 mRNA produces a functional SL receptor at the beginning of the SL perception cascade. Adapted from Liu et al. (2022), Figure 7.
References
- Aquino B, Bradley JM, Lumba S (2021) On the outside looking in: roles of endogenous and exogenous strigolactones. Plant J 105: 322–334 [DOI] [PubMed] [Google Scholar]
- Liu T, Zhang X, Zhang H, Cheng Z, Lin Q, Wan J (2022) Dwarf and high tillering1 represses rice tillering through mediating the splicing of D14 pre-mRNA. Plant Cell 34: 3301–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R (2009) The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Yan JQ, Zhu J, He CX, Benmoussa M, Wu P (1998) Quantitative trait loci analysis for the developmental behavior of tiller number in rice (Oryza sativa L.). Theor Appl Genet 97: 267–274 [Google Scholar]


