A set of conserved effectors mediates multihost root compatibility in both pathogenic and endophytic strains of Fusarium oxysporum.
Abstract
Fungal interactions with plant roots, either beneficial or detrimental, have a crucial impact on agriculture and ecosystems. The cosmopolitan plant pathogen Fusarium oxysporum (Fo) provokes vascular wilts in more than a hundred different crops. Isolates of this fungus exhibit host-specific pathogenicity, which is conferred by lineage-specific Secreted In Xylem (SIX) effectors encoded on accessory genomic regions. However, such isolates also can colonize the roots of other plants asymptomatically as endophytes or even protect them against pathogenic strains. The molecular determinants of endophytic multihost compatibility are largely unknown. Here, we characterized a set of Fo candidate effectors from tomato (Solanum lycopersicum) root apoplastic fluid; these early root colonization (ERC) effectors are secreted during early biotrophic growth on main and alternative plant hosts. In contrast to SIX effectors, ERCs have homologs across the entire Fo species complex as well as in other plant-interacting fungi, suggesting a conserved role in fungus–plant associations. Targeted deletion of ERC genes in a pathogenic Fo isolate resulted in reduced virulence and rapid activation of plant immune responses, while ERC deletion in a nonpathogenic isolate led to impaired root colonization and biocontrol ability. Strikingly, some ERCs contribute to Fo infection on the nonvascular land plant Marchantia polymorpha, revealing an evolutionarily conserved mechanism for multihost colonization by root infecting fungi.
IN A NUTSHELL.
Background: Most interactions between plants and microbes occur belowground. The soil-inhabiting fungus Fusarium oxysporum (Fo) can enter the roots and colonize the water-transporting vessels of its host plant, causing wilt disease that provokes devastating losses in hundreds of different crops worldwide. Although any given strain of Fo only induces wilting on a single or a few plant species, such strains can still colonize the outer root layers of other plants without causing visible disease symptoms. Indeed, asymptomatic root colonization can even protect plants against infection by pathogenic strains. How Fo has acquired the capacity for multihost root colonization remains largely unknown.
Question: Initial colonization by Fo occurs predominantly through filaments called hyphae, which grow in the space between the plant cells known as the apoplast. We reasoned that the fungus must release molecules into the apoplastic space that promote its establishment in the root and used discovery proteomics to test this idea.
Findings: We found a set of previously uncharacterized proteins that we called early root colonization (ERC) effectors and showed that they contribute to fungal growth on diverse species of plants ranging from cultivated tomatoes to an evolutionarily distant nonvascular plant called Marchantia polymorpha. Intriguingly, the ERC proteins identified in our work are predicted to bind or modify the molecules that make up the plant cell wall and are found across a wide range of fungal lineages. This suggests that the cell wall is a major target for fungi establishing either detrimental or beneficial associations with plant roots.
Next steps: The discovery of a set of ERC proteins sheds new light on how root-colonizing fungi are able to infect a broad range of plant species. Future studies should aim to identify the host mechanisms targeted by these effectors and use this knowledge to protect crops from the harmful effects of fungal pathogens.
Introduction
Plant pathogenic microorganisms have evolved to infect aerial and below-ground organs resulting in devastating agricultural yield losses (Chaloner et al., 2021). Some pathogen species target the host vasculature resulting in systemic infections, while others remain restricted to nonvascular tissues, causing localized disease symptoms. These interactions are the result of distinct adaptations allowing pathogens to accommodate themselves inside the plant through secreted molecules termed effectors (Presti et al., 2015). Effectors are either secreted into the intercellular space (apoplast) or translocated inside plant cells, and facilitate pathogen infection by modulating diverse host functions including cell wall composition and intracellular signaling (Toruño et al., 2016). Plants in turn have evolved a multilayered immune system to detect and resist microbial invaders and thwart pathogen infection (Jones and Dangl, 2006; Ngou et al., 2021).
A particularly destructive group of soil-borne pathogens are vascular wilt fungi, which attack almost every crop except cereals and are extremely difficult to control (Berendsen et al., 2012; Gordon, 2017). The Fusarium oxysporum (Fo) species complex provokes devastating losses in global agriculture (Dean et al., 2012; Gordon, 2017), exemplified by the highly aggressive clone named tropical race 4 (TR4) that threatens to wipe out the world’s industrial Cavendish banana (Musa acuminata) production (Ordonez et al., 2015). Fo infection initiates in the soil, when the fungus senses chemical signals released by the roots that induce directed hyphal growth (Turrà et al., 2015). After entering the plant, Fo grows mainly between the cells of the root cortex. During this symptomless biotrophic stage, establishment of infection depends on conserved fungal compatibility mechanisms such as a mitogen-activated protein kinase cascade, controlling invasive growth (Di Pietro et al., 2001) or Rapid ALkalinization Factor (RALF), a small secreted protein mimicking plant regulatory peptides that trigger host alkalinization (Masachis et al., 2016). In a compatible pathogen–host interaction, Fo eventually enters and colonizes the xylem vessels causing characteristic vascular wilt symptoms and plant death (Gordon, 2017).
Although the Fo species complex collectively infects more than a hundred different crops, individual isolates only cause wilt disease on a single or a few related plant species and have accordingly been classified into formae speciales (ff. spp.; van Dam et al., 2016; Edel-Hermann and Lecomte, 2019). Such host-specific pathogenicity is conferred by accessory or lineage-specific (LS) genomic regions, which encode unique combinations of virulence determinants and can be horizontally transferred between Fo isolates (Ma et al., 2010; van Dam et al., 2016). These host-specific effector proteins were originally identified in the xylem sap of infected tomato (Solanum lycopersicum) plants and are thus known as Secreted In Xylem (SIX) effectors (Houterman et al., 2007; Gawehns et al., 2015). So far, 14 SIX proteins have been identified in the tomato pathogenic form Fo f. sp. lycopersici (Fol) and four of these, SIX1, SIX3, SIX5, and SIX6 are required for full virulence (Rep et al., 2004; Houterman et al., 2009; Gawehns et al., 2014; Ma et al., 2015). How SIX effectors determine host specificity is not fully understood, but SIX1, SIX3, and SIX4 were shown to function as avirulence (Avr) proteins in tomato effector-triggered immunity via recognition by the resistance (R) genes I-3, I-2, and I-1 (Takken and Rep, 2010). Recently, SIX1 and three other effectors (Foa1, Foa2, and Foa3) from Fo f. sp. conglutinans were found to suppress the pattern-triggered immunity (PTI)-associated oxidative burst in Arabidopsis thaliana (Tintor et al., 2020). Moreover, the SIX5–Avr2 effector pair alters plasmodesmatal exclusion to promote cell-to cell movement of Avr2 (Cao et al., 2018). Interestingly, sequence polymorphisms in six genes contributed to the emergence of different Fol races and avirulence genotypes. While the historically ancient race 1 contains all three Avr genes, race 2 (Fol4287) emerged by loss or mutation of Avr1 and race 3 subsequently emerged as a consequence of mutations in Avr2 (Takken and Rep, 2010; Inami et al., 2012).
Besides provoking vascular wilt disease on the main host, Fo isolates can also colonize the roots of alternative host plants where they grow asymptomatically as endophytes or even act as biocontrol agents to protect the plant against pathogenic Fo forms or other root pathogens (Katan, 1971; Fuchs et al., 1999; Alabouvette et al., 2009; Veloso and Díaz, 2012; de Lamo and Takken, 2020). The genetic determinants underlying multihost root compatibility in Fo remain unclear. In this study we investigated how Fo establishes either pathogenic or endophytic interactions with plant roots. By analyzing the apoplastic fluid (AF) of Fo-colonized tomato roots, we identified a set of fungal proteins termed early root colonization (ERC) effectors, which are secreted during early biotrophic growth on both main (pathogenic interaction) and alternative host plants (endophytic interaction). Interestingly, ERCs are found across the entire Fo species complex as well as in other phytopathogens and contribute to infection on evolutionarily distant vascular and nonvascular plant lineages, suggesting a broadly conserved role in fungus–plant interactions.
Results
Fo colonizes the roots of the main host as well as alternative host plants
Fo isolates were previously reported to asymptomatically colonize roots of other plant species in addition to the main host plant (Katan, 1971). The molecular basis of asymptomatic colonization of alternative hosts remains largely unknown. To address this question, we first tested the ability of a set of Fo reference isolates sequenced by the Broad Institute Fusarium Comparative Genome Initiative to colonize a single plant species, tomato (Supplemental Table S1). The set of isolates includes several plant pathogenic ff. spp. causing wilt on tomato (Solanum lycopersicum), melon (Cucumis melo), pea (Pisum sativum), banana (Musa acuminata), or crucifer species, as well as a nonpathogenic biocontrol strain (Alabouvette et al., 2009).
All tested isolates could be detected by quantitative polymerase chain reaction (qPCR) in roots and root crowns of tomato plants at 35-day post infection (dpi), but only the tomato pathogenic isolate (f. sp. lycopersici race 3) was found in the stems (Supplemental Figure S1A). Confocal microscopy of tomato roots inoculated with fluorescent mClover3-labeled strains of the tomato pathogenic isolate Fol4287 (f. sp. lycopersici race 2), the banana pathogenic isolate Foc54006 (f. sp. cubense), or the nonpathogenic biocontrol strain Fo47, confirmed the presence of fungal hyphae of all three strains which efficiently colonized the tomato root cortex (Figure 1A). Moreover, transmission electron microscopy (TEM) of Fol4287-infected plants at 3 dpi revealed that the fungus localized predominantly to the apoplast of root cortical cells, although occasional penetration events of cortical and endodermal cells were observed, accompanied by the loss of plant plasma membrane integrity (Figure 1B; Supplemental Figure S1B). Again, qPCR confirmed the presence of all three Fo isolates in tomato roots at 12 dpi, but only Fol4287 was detected in the stems (Figure 1C). Collectively, these results confirm that Fo isolates have a general capacity to colonize roots of both the main host and alternative host plants, in line with earlier reports (Katan, 1971; de Lamo and Takken, 2020). Our results further show that asymptomatic growth of Fo on alternative host plants is mostly restricted to roots and root crowns while the host-specific pathogenic forms of the fungus are able to invade the stems, eventually resulting in vascular wilt and plant death.
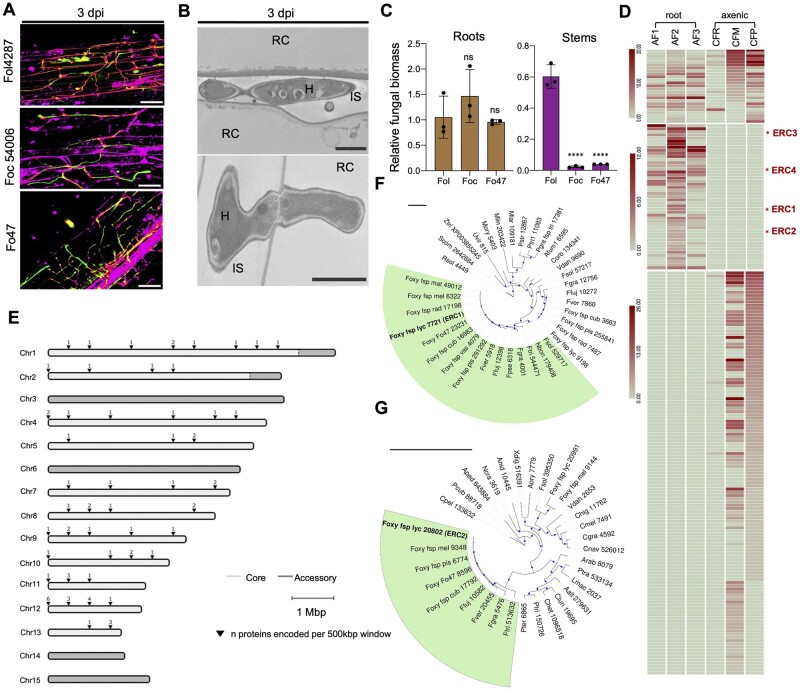
Figure 1.
Fo secretes a suite of conserved core effectors in the root intercellular space. A, Confocal microscopy of tomato root colonization of Fo isolates Fol4287 (tomato pathogen), Foc54006 (banana pathogen), or Fo47 (endophyte) expressing 3x-mClover3 at 3 days post infection (dpi). Fungal fluorescence (mClover3-green) is overlaid with propidium iodide staining of plant cell walls (magenta). PI also traces the non-cell-wall apoplastic space. Scale bars, 25 µm. B, Transmission electron micrographs showing hyphae of Fol4287 (H) growing intercellularly (top) or penetrating a root cortical cell (RC; bottom). IS, intercellular space. Scale bars, 2 µm (top); 1 µm (bottom). Note that the penetrating hypha displays an internal cell wall called septum which is related to its growth pattern. C, Fungal burden in roots and stems of tomato plants inoculated with the indicated Fo isolates were measured by qPCR of the Fo actin gene using total DNA extracted at 12 dpi. Fo DNA was calculated using the threshold cycle (ΔΔCt) method, normalized to the tomato gadph gene. Error bars indicate sd; n = 3 biological replicates. Asterisks indicate statistical significance versus Fol4287 [one-way analysis of variance (ANOVA), Bonferroni’s multiple comparison test, P < 0.05]. ns, nonsignificant. Experiments were performed three times with similar results. D, Heat map showing absolute counts of unique peptides of Fol4287 identified by LC‐MS/MS in three independent samples of tomato root AF at 3 dpi (AF 1, 2, 3) or in a single sample of filtrate from axenic cultures in minimal medium with tomato crushed roots (CFR) or sucrose as carbon source (CFM) or potato dextrose broth (CFP). Putative effectors ERC1-4 are highlighted with a red asterisk. Note differences in scale between sections of the heat map. E, Chromosomal distribution plot of the genes encoding Fo Fol4287 proteins identified in tomato AF, showing their localization exclusively in core genomic regions. Core and LS regions are shown in light and dark gray, respectively. F, G, Maximum likelihood phylogenetic trees based on the aligned amino acid sequences of the FOXG_11583 (ERC1) (F) and FOXG_04534 (ERC2) proteins (G), respectively. Number indicates MycoCosm protein ID. Size of blue dots represents bootstrap support for the branch with maximum 100 bootstraps (Tree scale = 1). Fungal species included in the analysis are listed in Supplemental Table S2.
Fo secretes a battery of core effector proteins in the root intercellular space
During plant infection, both pathogenic and mutualistic fungi deploy effector proteins to modulate host responses (Presti et al., 2015). Some effectors are highly specific for a given pathogen species, while others are conserved across a broad range of pathogens and endophytes (Irieda et al., 2019). Given the predominantly apoplastic growth of Fo during the early biotrophic stage (Figure 1, A and B), we performed discovery proteomics of AF collected from tomato roots infected with Fol4287 at 3 dpi to search for potential effectors. Liquid chromatography–mass spectrometry (LC–MS) analysis identified a total of 72 Fol4287 proteins present in AF, 32 of which were consistently detected across 3 biological replicates (Figure 1D; Supplemental Figure S1, C and D and Supplemental Data Set S1). Twenty‐five of the 72 apoplastic fungal proteins were also found in the filtrates of axenically grown fungal cultures, while 47 were exclusively detected in planta (Supplemental Data Set S1). The latter include different classes of cell wall degrading enzymes (CWDEs) such as polygalacturonase, pectate lyase, glucanase, or galactosidase, as well as other enzymes such as a choline dehydrogenase, a copper amine oxidase or a carbonic anhydrase (CA).
In an attempt to identify effector candidates we looked for proteins that (1) were specifically present in AF; (2) were consistently detected in all three biological AF replicates; (3) carry a predicted N-terminal secretion signal peptide for extracellular localization; (4) lack predicted transmembrane domains; and (5) contain multiple cysteines. Four of the identified Fo proteins fulfilled all five criteria and were selected for further analysis. We named these candidates ERC effectors (Figure 1D; Supplemental Figure S1, E and F and Supplemental Data Set S1). ERC1 (FOXG_11583), ERC2 (FOXG_04534), and ERC4 (FOXG_08211) are small secreted hypothetical proteins (SSPs) with a size range of 23–34 kDa, while ERC3 (FOXG_16902) is a 51-kDa protein with an α-L-arabinofuranosidase domain similar to those reported in effectors from the rice blast pathogen Pyricularia oryzae or the corn smut fungus Ustilago maydis (Lanver et al., 2014; Wu et al., 2016). α-L-arabinofuranosidases function in degradation of the plant cell wall polysaccharide arabinoxylan, thereby increasing its accessibility to xylanases (de Vries et al., 2000). A domain search in Pfam (Mistry et al., 2020) identified a putative cellulose-binding domain in the ERC1 sequence (Dumas et al., 2008) and ERC2 and ERC4 contain predicted lytic polysaccharide monooxygenase (LPMO) domains. LPMOs cleave cellulose, chitin and other polysaccharides through a novel oxidative mechanism and have been suggested to act on crystalline surface regions of the substrate to create attachment sites and enhance accessibility for canonical glycoside hydrolases (Vaaje-Kolstad et al., 2010).
We noted that almost all Fo proteins secreted in root AF, including ERCs, are encoded on core genomic regions that are shared across the entire Fo species complex (Figure 1E). This contrasts sharply with the previously reported SIX effector genes, which are all located on LS regions (Ma et al., 2010; van Dam et al., 2016). Moreover, unlike most SIX effectors, ERCs have predicted homologs outside the Fo species complex and are found in many asco- and basidiomycete species with distinct lifestyles including both phytopathogens and nonpathogens (Figure 1, F and G; Supplemental Figure S1G and Supplemental Data Set S2, A–D). Interestingly, ERC1 and ERC2 each have one paralog in Fol4287 (FOXG_12855 and FOXG_18882, respectively), which cluster separately and are also conserved across the entire Fo species complex, suggesting the presence of ERC1 and ERC2 candidate effector families (Figure 1, F and G; Supplemental Table S2). FOXG_18882 has 56% identity to ERC2 but is much shorter (97 amino acid (aa) versus 253 aa) and clusters with predicted homologs from F. solani and Fo f. sp. melonis (Figure 1G). ERC3 and ERC4 homologs were also found in some basidiomycetes such as the Agaricomycotina and Ustilaginomycotina, but not in the Pucciniomycotina (Supplemental Figure S1G; Supplemental Data Set S2, C and D). We conclude that Fo4287 secretes an array of putative core effectors during early stages of tomato root colonization that are conserved in diverse fungi including both pathogenic and nonpathogenic plant-associated species.
ERCs are upregulated during the early biotrophic infection stage
A hallmark of hemibiotrophic pathogens is a switch from biotrophy to necrotrophy coinciding with a marked shift in the gene expression pattern (O’Connell et al., 2012). In contrast to air-borne plant pathogens, the transcriptional dynamics of hemibiotrophic fungal root infection has not been explored in detail (Guo et al., 2021). We performed RNA sequencing (RNAseq) of tomato roots inoculated with Fol4287 during early stages (1, 2, and 3 dpi) of biotrophic growth in the root cortex as well as during a later stage (7 dpi) of invasion of the root vascular tissue, which likely marks the transition to necrotrophy. Importantly, 36 (77%) of the 47 genes encoding proteins previously identified in AF (Supplemental Data Set S1) showed a marked transcriptional upregulation during early infection stages as compared to axenic growth conditions (Supplemental Figure S2A).
Analysis of differentially expressed genes (DEGs; log2 fold change >2, P < 0.05) and principal component analysis revealed a major shift in the transcriptional profile between axenic and in planta conditions (Figure 2A; Supplemental Data Set S3, A–D) as well as between the early (1, 2, 3 dpi) and late (7 dpi) stages of infection (Supplemental Figure S2B). The total number of DEGs ranged from 1,500 to 1,800 upregulated and 600–800 downregulated genes depending on the time point (Figure 2A). Around 92% of the in planta-induced DEGs were located on core genomic regions. Gene ontology (GO) enrichment analysis of DEGs upregulated during the early colonization stages showed an abundance of fungal transcripts involved in amino acid biosynthesis and metabolic processes (Supplemental Figure S2C and Supplemental Data Set S4), which is indicative of biotrophic growth as previously reported for the interaction between Piriformospora indica and A. thaliana (Lahrmann et al., 2013).
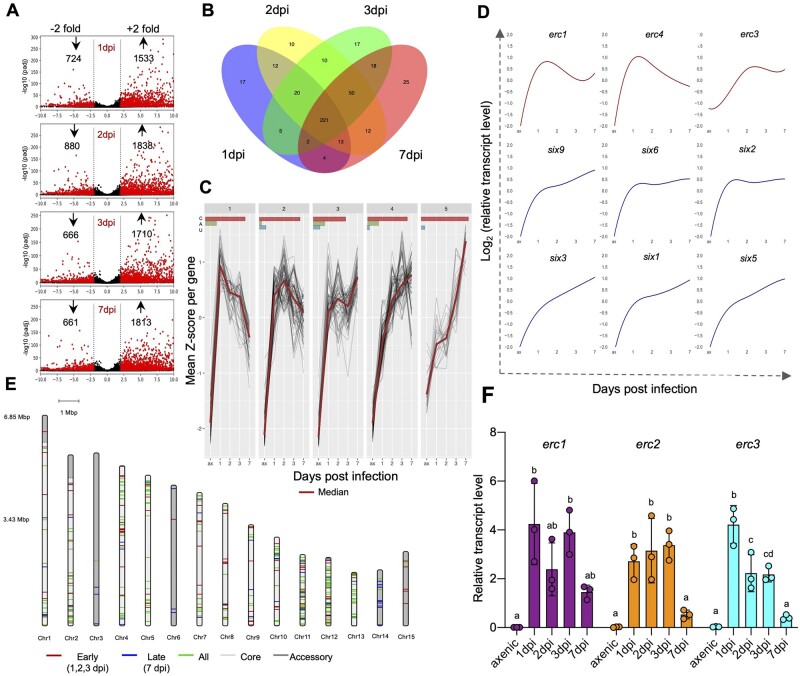
Figure 2.
ERCs are upregulated during early biotrophic stages of infection. A, Volcano plot showing pairwise differential expression analysis of Fol4287 genes at the indicated time points post infection in tomato roots versus axenic culture. Significantly DEGs are in red. B, Venn diagram of Fol4287 genes encoding predicted secreted proteins (SPs) upregulated in tomato roots at the indicated time points after inoculation versus axenic culture. A core set of 221 SPs are upregulated in planta across all the analyzed timepoints. C, Clustered expression profiles of genes encoding secreted proteins upregulated in tomato roots versus axenic culture (t test, P < 0.05). Red line, median expression per cluster. The bars at the top indicate the percentage of the genes located in core region (red), accessory region (green), or unmapped region (blue) of the genome represented in each cluster. D, Representative expression profiles of the indicated candidate effector genes plotted as Z-scores of log2 expression versus dpi. Profiles in red correspond to the indicated erc genes located on core genomic regions showing upregulation at early time points with a drop at 7 dpi. Profiles in blue correspond to known SIX effector genes encoded on LS chromosome 14 showing maximum upregulation at 7 dpi. E, Chromosomal distribution of in planta upregulated Fol4287 genes encoding predicted secreted proteins. Red bands are genes preferentially expressed at early infection stages (early to late expression ratio > 0.25). Blue bands are genes preferentially expressed at late infection stages (early to late expression ratio <0.25). Green bands are genes expressed at both early and late infection stages (differentially expressed in at least one early plus the 7 dpi data set versus axenic). Early refers to 1–3 dpi; late refers to 7 dpi. Core and LS genomic regions are shown in light and dark grey, respectively. F, Relative transcript levels of genes FOXG_11583 (erc1), FOXG_04534 (erc2), and FOXG_16902 (erc3) were measured by RT-qPCR of cDNA obtained from Fol4287 grown in minimal medium (axenic) or from roots of tomato plants inoculated with Fol4287 at 1, 2, 3, or 7 dpi. Transcript levels were calculated using the threshold cycle (ΔΔCt) method and normalized to the Fol4287 peptidyl prolyl isomerase (ppi) gene. Error bars indicate sd; n = 3 biological replicates. Different letters indicate statistically significant differences according to one way ANOVA, Bonferroni’s multiple comparison test (P < 0.05).
Around 6% (436) of the 6,894 Fo genes induced during the root colonization encode predicted secreted proteins. While many of these display differential expression profiles, we identified a set of 221 genes that were significantly upregulated at all four infection time points tested (Figure 2B). Importantly, the in planta-induced genes encoding predicted secreted proteins include previously characterized Fol4287 effectors belonging to the metalloprotease and serine protease families (Jashni et al., 2015; Supplemental Figure S2D).
We performed clustered expression profiling of in planta upregulated genes encoding predicted secreted proteins, based on the Z-scores for each gene expression value across different time points. This revealed five clusters, which collectively represent different variations of two fundamentally distinct expression patterns (Figure 2, C and D): (1) early expressed genes (clusters 1 and 2) which show highest transcript levels during early infection stages (1, 2, and 3 dpi) followed by a drop at 7 dpi; (2) late expressed genes (clusters 3–5), which show progressive upregulation with a higher expression at 7 dpi compared to the earlier timepoints. The first category includes the erc1 and erc4 genes, which are located in cluster 2, whereas the second category includes the six genes and erc3, which are located in cluster 4. Interestingly, most of the early upregulated secreted protein encoding genes are located on core genomic regions (except for some genes located on the less studied LS chromosome 15), while the late upregulated genes are predominantly located on accessory regions, particularly on the so-called pathogenicity chromosome 14 (Figure 2E). An enrichment Fischer’s exact statistical test of the late expressed genes versus all others (early expressed + expressed at all timepoints) shows an odds ratio of 26.881 (P = 4.812) while a test of the early expressed genes versus all others (late expressed + expressed at all timepoints) shows an odds-ratio of 2.0 (P = 1.0; Supplemental Table S3). Collectively, these results show that in planta-induced secreted protein encoding genes located on accessory genome regions are expressed preferentially at later time points of the infection process.
Regarding the expression of erc genes, the RNAseq data are mostly in line with those from proteomics of AF at 3 dpi (Figure 1, D and E). Indeed, reverse transcription-quantitative polymerase chain reaction (RT–qPCR) analysis confirmed the early in planta upregulation and a subsequent downregulation of three of the erc genes at 7 dpi (Figure 2F; Supplemental Figure S2E). Taken together, our results suggest that Fol4287 undergoes a major transcriptional shift between the early stages of biotrophic root colonization (1, 2, and 3 dpi) and the later stage at 7 dpi, most likely coinciding with the transition from the root cortex to the xylem. The finding that core effector candidates such as ERCs are specifically upregulated during the initial stage suggests a potential role in the establishment of early fungus–plant compatibility.
ERCs contribute to infection and to modulation of root immunity on the main plant host
To test the role of ERCs in colonization and virulence of Fol4287 on the main plant host tomato, we generated isogenic deletion mutants lacking either the FOXG_11583, FOXG_04534, FOXG_16902, or FOXG_08211 gene, which were named Δerc1, Δerc2, Δerc3, and Δerc4 respectively (Supplemental Figure S3, A–E). Phenotypic analysis of these mutants revealed no detectable changes in vegetative growth, colony morphology, or resistance to different types of stresses (Supplemental Figure S3F). Interestingly, tomato plants inoculated with the Δerc1, Δerc2, and Δerc3 mutants, but not those inoculated with the Δerc4 mutant, showed significantly reduced wilt symptoms, fresh weight loss, and mortality and accumulated less in planta fungal biomass than those inoculated with the Fol4287 wild-type strain (Figure 3, A–C; Supplemental Figure S3, G–I; Supplemental Figure S4, A and B). Trypan blue staining confirmed that the Δerc1, Δerc2, and Δerc3 mutants were less efficient in colonizing tomato roots than the wild-type (Supplemental Figure S3J). Importantly, complemented strains of these mutants obtained by reintroduction of the wild-type allele (Supplemental Figure S3K; Δerc1-C#19, Δerc2-C#32, and Δerc3-C#11) showed fully restored virulence and root colonization (Figure 3, A–C; Supplemental Figure S3, H and I).
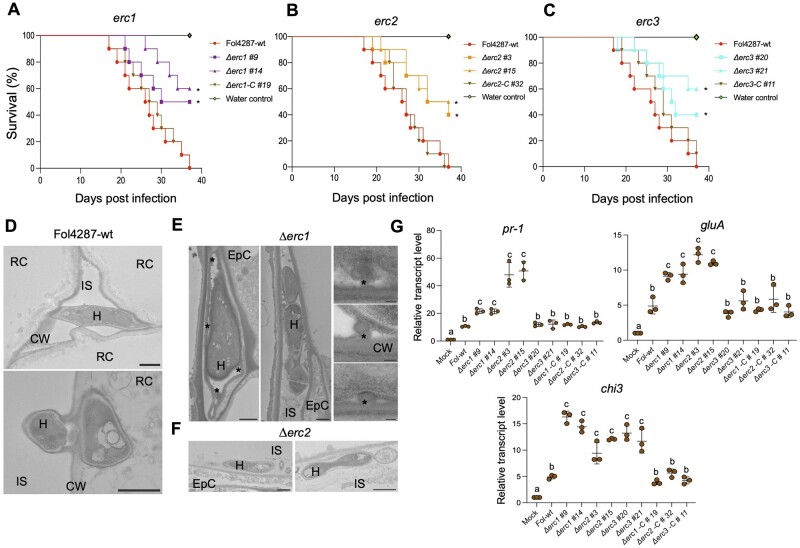
Figure 3.
ERCs contribute to root colonization, virulence and suppression of the plant immune response. A–C, ERCs have a role in virulence of Fo. Kaplan–Meier plot showing the survival of tomato plants inoculated with the Fol4287-wt strain or the indicated single gene knockout mutants. Number of independent experiments = 3; 10 plants/treatment. Data shown are from one representative experiment. *P <0.05, versus the wild-type according to log-rank test. Note that mortality caused by the Δerc1, Δerc2, and Δerc3 single mutants is significantly lower than that caused by the wild-type or the Δerc1-C#19, Δerc2-C#32, and Δerc3-C#11 complemented strains. D, TEM micrographs showing hyphae (H) of the Fol4287 wild-type (wt) growing between (top) or penetrating into tomato root cortical cells (RC; bottom). CW, plant cell wall; IS, intercellular space. Scale bars, 1 µm. E, F, TEM micrographs showing hyphae (H) of the Fol4287 Δerc1 (e) or Δerc2 (f) mutants growing between tomato root epidermal cells (EpC). Note in (E) that hyphae are encapsulated by protrusions of amorphous granular material (asterisks) and in (F) hyphae are located close to the root periphery. Scale bars in (E), 1 µm in left and center images; 0.1 µm in right image; in (F) 1 µm. G, Transcript levels of tomato defense genes pr-1, gluA, and chi3 were measured by RT-qPCR of cDNA obtained from tomato roots at 2 days post inoculation with the indicated fungal strains. Transcript levels were calculated using the ΔΔCt method, normalized to the tomato gadph gene and expressed relative to those of the wild-type infected control. Error bars indicate sd; n = 3 biological replicates. Different letters indicate statistically significant differences according to one way ANOVA, Bonferroni’s multiple comparison test (P < 0.05).
TEM analysis of tomato roots inoculated with Δerc1 or Δerc2 at 3 dpi revealed the absence of penetration of root cortical cells (Figure 3, E and F; Supplemental Figure S4C) as compared to those infected by the wild-type strain where successful penetration events were frequently observed (Figure 3D; Supplemental Figure S4C). In the Δerc1–tomato interaction we detected the presence of characteristic protrusions from the walls of root cortex cells encapsulating the fungal hyphae as well as the secretion of an amorphous granular material (asterisks in Figure 3E, arrowheads in Supplemental Figure S4C). Similar structures have been reported previously in plants infected with vascular fungal pathogens including Fusarium, Verticillium, or Ceratocystis and were suggested to contribute to plant resistance by preventing plant cell wall degradation and inhibiting hyphal spread (Pegg, 1976; Bishop and Cooper, 1983; Araujo et al., 2014). In line with this idea, the growth of the Δerc1 and Δerc2 mutants remained largely restricted to the intercellular space in the root cortex, while the wild-type showed both inter- and intracellular growth in the cortical root cells (Figure 3, D–F).
The diversity of the ultrastructural phenotypes of the Δerc single mutants suggest that these effectors may have different, nonredundant roles in promoting host root colonization. To test this idea, we generated Δerc1Δerc2 and Δerc2Δerc3 double knockout mutants. The double mutants displayed similar levels of virulence and fungal burden as the single mutants, and although minor differences were observed during plant colonization in the Δerc1Δerc2 strain as determined by trypan blue staining and qPCR, they were not significant compared to the single mutants (Supplemental Figure S5, A–E).
To test whether ERCs contribute to suppression of host defense or prevent its activation, we measured the transcript levels of known plant defense genes at 2 dpi. We found a two- to four-fold higher induction of the pr-1, gluA, and chi3 genes encoding pathogenesis-related protein 1, basic β-1,3-glucanase, and acidic chitinase, respectively (Masachis et al., 2016), in tomato roots infected with the Δerc1 and Δerc2 mutants as compared to those infected with the wild-type or the complemented strains, while for Δerc3 only an increased induction of chi3 gene was observed (Figure 3G). We also measured the expression of the previously reported tomato PTI marker genes GRAS2, PTI5, and Lrr22 (Kim et al., 2009) encoding, respectively, a transcription factor related to abiotic and biotic stress, an ethylene response family transcription factor, and a homolog of the Arabidopsis Lrr22 receptor-like kinase (Nguyen et al., 2010). Interestingly, the transcript levels of GRAS2 and PTI5 were significantly higher in roots inoculated with Δerc1 mutant, while Lrr22 transcript levels were higher in plants infected with the Δerc2 and Δerc3 mutants respectively, as compared to roots inoculated with the wild-type (Supplemental Figure S6). Taken together, these results suggest that ERCs contribute to infection and virulence of Fol4287 on the main plant host and may have a role in preventing the plant immune responses.
ERCs contribute to endophytic colonization and biocontrol activity of a nonpathogenic Fo strain
Fo has been known for decades to grow as an endophyte on roots of alternative host plants without inducing detectable disease symptoms (Katan, 1971; Fuchs et al., 1999; de Lamo and Takken, 2020). Moreover, certain Fo isolates appear to be nonpathogenic, such as Fo47, a well-characterized biocontrol strain that was originally isolated from a soil naturally suppressive to Fusarium wilt (Alabouvette et al., 2009). Fo47 was recently reported to trigger endophyte-mediated resistance against plant pathogenic forms of Fo (Constantin et al., 2020). Here, we confirmed that co-inoculation of tomato plants with Fo47 resulted in a marked reduction of mortality caused by Fol4287, as well as in a decrease of in planta biomass of Fol4287, when compared to single inoculation with Fol4287 (Supplemental Figure S7, A and B).
Interestingly, Fol4287 and Fo47 were both able to penetrate the root endodermis cells and reach the xylem vessels, but Fol4287 consistently showed more profuse spread in the xylem than Fo47 (Figure 4A). TEM analysis revealed the presence of fungal membrane tubules in penetration hyphae of Fol4287, but not in Fo47 (Supplemental Figure S7, C and D). Interestingly, similar structures were previously reported during invasive growth of plant symbionts and pathogens (Roth et al., 2019). We also observed aborted penetration events into tomato endodermis cells by Fo47 hyphae that were sometimes devoid of cytosol (Figure 4A, arrows; Supplemental Figure S7, E to G). In rare occasions where Fo47 hyphae were observed inside xylem vessels, they were often encapsulated by an amorphous granular material which also encrusted the plant cell walls and blocked the pits between the xylem vessel and adjacent cells (Figure 4A, asterisks; Supplemental Figure S7H, arrowheads). In contrast, such material was absent from the xylem pits of uninoculated plants and only rarely observed in xylem vessels colonized by Fol4287 where hyphae successfully spread across xylem vessels (Figure 4A, arrowheads; Supplemental Figure S7, I–K). This observation is reminiscent of the deposition of phenolic compounds described previously in the interaction between tomato and the vascular wilt fungus Verticillium alboatrum (Pegg, 1976). The deposition of amorphous granular material, as well as phenolic infusions, lignification, or incorporation of calcium into pit membranes have been associated with plant defense against vascular wilt fungi by inhibiting hyphal spread and plant cell wall degradation (Pegg, 1976; Bishop and Cooper, 1983; Araujo et al., 2014). Overall, our findings indicate that the commitment of different Fo isolates toward pathogenic versus endophytic lifestyle occurs mainly at the level of endodermis penetration and xylem colonization, which are successfully completed by the pathogenic strain Fol4287 but reduced or blocked in the endophytic isolate Fo47. These results also confirm that Fo47 is able to colonize tomato roots to a certain extent and to protect the plant against wilt disease caused by the pathogenic form Fol4287.
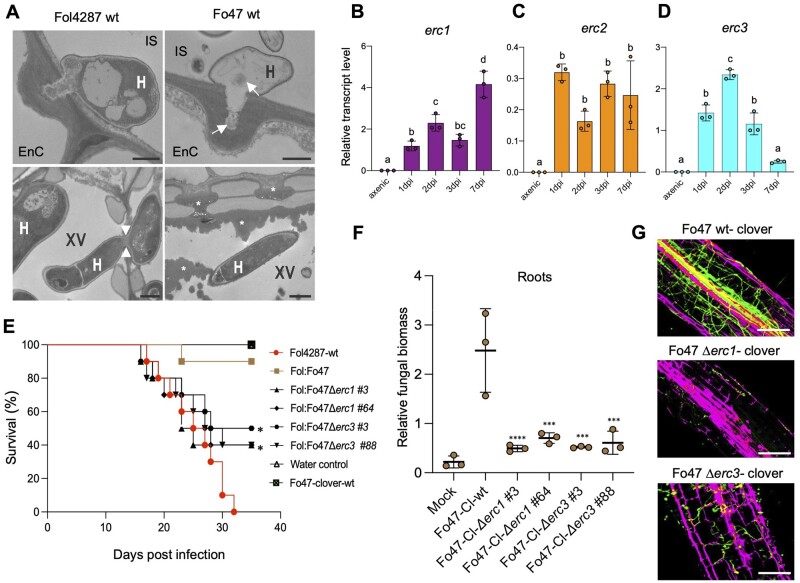
Figure 4.
ERCs are required for endophytic colonization and biocontrol activity of a non-pathogenic Fo strain. A, TEM micrographs showing hyphae (H) of Fol4287 and Fo47 attempting penetration of tomato root endodermis cells (EnC; top) or growing inside xylem vessels (XV; bottom). Note that the penetration hypha of Fo47 is devoid of cytosol and contains only remnants of cell components (arrows). Arrowheads indicate a Fol4287 hypha penetrating an adjacent xylem vessel. Asterisks indicate the deposition of amorphous granular material encapsulating a Fo47 hypha in a xylem vessel or blocking the pits between vessels to inhibit cell-to-cell movement of the fungus. CW, cell wall; IS, intercellular space. Scale bars = 1 µm in top images; 2 µm in bottom images. B–D, Relative transcript levels of the genes FOZG_11686 (erc1), FOZG_02496 (erc2), and FOZG_12886 (erc3) in isolate Fo47 (biocontrol strain) were measured by RT-qPCR of cDNA obtained from fungal mycelium grown in minimal medium (axenic) or from roots of tomato plants inoculated with Fo47 at 1, 2, 3, or 7 dpi. Transcript levels were calculated using the threshold cycle (ΔΔCt) method and normalized to the Fo peptidyl prolyl isomerase (ppi) gene. Error bars indicate sd; n = 3 biological replicates. Different letters indicate statistically significant differences according to one-way ANOVA, Bonferroni’s multiple comparison test (P < 0.05). E, Kaplan–Meier plot showing the survival of tomato plants inoculated with the Fol4287 or Fo47-3x-mClover3 wild-type strains alone or co-inoculated with Fol4287 plus Fo47-3x-mClover3 wild-type or the indicated Fo47-3x-mClover3 Δerc mutants. Number of independent experiments = 3; 10 plants/treatment. Data shown are from one representative experiment. *P <0.05, versus Fol4287 alone according to log-rank test. Note that protection provided by the Fo47-3x-mClover3 Δerc1 and Δerc3 mutants is significantly lower than that provided by the Fo47-3x-mClover3 wild-type strain. F, Fungal burden in roots of tomato plants inoculated with the indicated Fo isolates was measured by qPCR of the Fo47-specific gene (FOBG_10856) using total DNA extracted at 10 dpi. Fo DNA was calculated using the threshold cycle (ΔΔCt) method, normalized to the tomato gadph gene and expressed relative to that in roots inoculated with Fo47-3x-mClover3-wt. Error bars indicate sd; n = 3 biological replicates. Asterisks indicate statistical significance versus Fo47-mClover wild-type (one-way ANOVA, Bonferroni’s multiple comparison test, P < 0.05). The experiment was performed three times with similar results. G, Confocal microscopy images showing tomato roots inoculated with the Fo47-mClover3 wild-type strain or the indicated Fo47-3x-mClover3 Δerc mutants at 3 dpi. Fungal fluorescence (green) is overlaid with propidium iodide staining of plant cells (magenta). Scale bars, 25 µm.
Because ERCs are encoded on core genomic regions present both in pathogenic and nonpathogenic Fo isolates, we asked whether they might also contribute to endophytic root colonization and possibly, biocontrol activity. In support of this idea, erc1, erc2, and erc3 genes were transcriptionally upregulated during early stages of tomato root colonization in the nonpathogenic isolate Fo47 (biocontrol interaction) and in the banana pathogenic isolate Foc54006 (alternative host interaction), as previously observed in the pathogenic interaction with Fol4287 (Figure 4, B–D; Supplemental Figure S8, A–C). Moreover, we found that the erc1 and erc2 orthologs of the vascular wilt pathogen Verticillium dahliae VdLS17 (Klosterman et al., 2011), VDAG_04446, and VDAG_07135, were also transcriptionally induced during early stages of tomato root infection (Supplemental Figure S8, D–F).
We next asked whether ERCs are required for efficient root colonization and biocontrol activity of Fo47. Indeed, Δerc1 and Δerc3 deletion mutants obtained in a Fo47-mClover3 background showed reduced ability to colonize tomato roots compared to the wild-type Fo47 mClover3 strain, as observed in an overview of lateral roots including the vasculature (Figure 4, E–G). Importantly, these two mutants were significantly less efficient in protecting tomato plants against mortality caused by Fol4287 (Figure 4, E–F). Overall, these results suggest that ERCs are used by vascular fungal pathogens with contrasting lifestyles––pathogenic versus endophytic––to establish associations with plant roots.
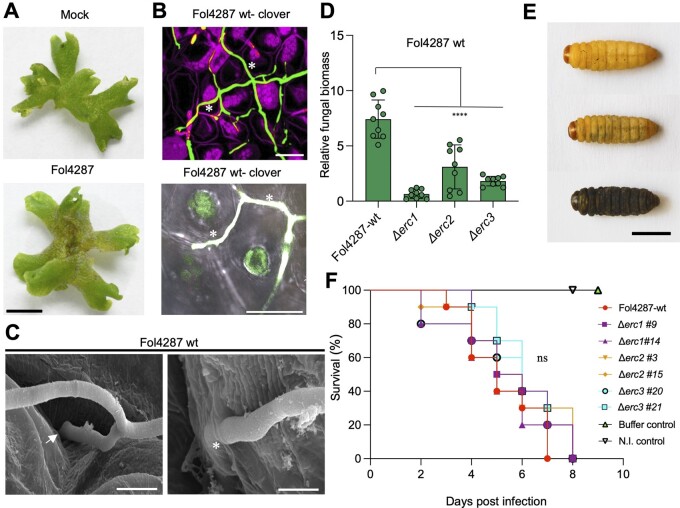
ERCs contribute to Fo infection on the nonvascular land plant Marchantia polymorpha
The broad distribution of ERC homologs across the fungal kingdom suggests a conserved role of these core effectors in plant colonization. To experimentally test this idea, we took advantage of a recently established Fo infection model in the liverwort Marchantia polymorpha (Redkar et al., 2022a), which has emerged as a nonvascular model for molecular plant–microbe interactions (Upson et al., 2018). As previously reported (Redkar et al., 2022a), Fol4287 caused visible disease syptoms on Marchantia thalli and displayed mostly intercellular hyphal growth similar to that observed in the tomato root cortex (Figure 5, A and B). Scanning electron microscopy (SEM) showed Fol4287 hyphae entering the thalli intercellularly, either by growing between cells or through air pores (Figure 5C; Supplemental Figure S9A).
Figure 5.
ERCs are conserved compatibility factors on both vascular and nonvascular plants but not on animal hosts. A, Macroscopic disease symptoms of M. polymorpha Tak-1 thalli 5 dpi with 105 microconidia mL−1 of Fol4287 or water (mock). Images are representative of three independent experiments. Scale bar, 1 cm. B, Confocal microscopy showing intercellular growth of Fol4287-3x-mClover3 on a Tak-1 thallus at 3 dpi. Fungal fluorescence (green) is overlaid with propidium iodide staining of plant cells (magenta). Asterisks indicate intercellular hyphal growth. Scale bar, 25 µm. C, SEM micrographs showing hyphae (H) entering thalli of Marchantia, either intercellularly (arrow) or by direct penetration (asterisks). Scale bars = 5 µm in top image; 2 µm bottom image. D, Fungal burden in 3-week-old Marchantia thalli inoculated with the Fol4287 wild type strain or the indicated Δerc mutants was measured by qPCR of the Fo actin gene using total DNA extracted at 6 dpi. The relative amount of fungal DNA was normalized to the Mp EF1a gene. Statistical significance versus wild-type (P < 0.05, Unpaired t test) is indicated by an asterisk. Error bars indicate sd (n = 3). E, Progressive melanization and killing of G. mellonella larvae after inoculation with 1.6 × 104 microconidia of Fol4287 and incubation at 30°C. Images are taken at 3 dpi. Scale bar, 1 cm. F, Kaplan–Meier plots showing the survival of G. mellonella larvae after injection of 1.6 × 104 microconidia of Fol4287, the Δerc1, Δerc2, and Δerc3, deletion mutants or buffer control into the hemocoel and incubation at 30°C. Number of independent experiments = 3; 10 larvae/treatment. Data shown are from one representative experiment. ns, nonsignificant versus Fol4287 according to log-rank test.
In addition, we observed occasional events of direct penetration of cells (Figure 5C). Similar to infection of tomato plants, transcript levels of the erc1, erc2, and erc3 genes were markedly upregulated during growth of Fol4287 in Marchantia thalli compared to the axenic control (Supplemental Figure S9, B–D) although the relative transcript levels of erc2 were lower than those of erc1 and erc3. Thalli inoculated with the Fol4287 Δerc1, Δerc2, or Δerc3 mutants showed reduced severity of macroscopic disease symptoms such as progressive maceration of the thallus tissue (Supplemental Figure S9, E and F) and contained less fungal biomass than those inoculated with the Fol4287 wild-type strain (Figure 5D). We conclude that ERCs contribute to fungal infection on evolutionarily distant plant host lineages independent of the presence of a true vasculature.
Besides causing disease in plants, Fo has also been reported as an opportunistic pathogen of humans (Nucci and Anaissie, 2007). Fol4287 was previously shown to cause mortality on immunodepressed mice and larvae of the invertebrate insect model host Galleria mellonella (Ortoneda et al., 2004; Navarro-Velasco et al., 2011). Here, we found that mortality caused by the Δerc1, Δerc2, or Δerc3 mutants on G. mellonella larvae did not differ significantly from that caused by the Fol4287 wild-type strain (Figure 5, E and F; Supplemental Figure S9G), suggesting that ERCs are dispensable for infection on the animal host G. mellonella.
Discussion
The soil-borne vascular pathogen Fo causes systemic infections and wilting on a broad range of crops, with individual strains exhibiting exquisite host specificity determined by LS effector proteins. However, pathogenic isolates can also colonize roots of alternative host plants without causing wilt and hence behave as true endophytes (Gordon, 2017). The ability of Fo to grow on alternative host plants is both of ecological and agronomical relevance, as it increases the persistence of the fungus in the soil in the absence of the main host (Smith and Snyder, 1975; Pereira et al., 2019). Colonization of multiple plant hosts is thought to involve compatibility factors that are conserved across a wide range of Fo isolates, but the nature of these molecules has so far remained elusive. By integrating discovery proteomics with early-stage RNA sequencing (RNA-seq) and targeted gene knockout analysis, we show here that colonization of both main and alternative host plants by Fo is mediated by a set of conserved ERC effectors, which are encoded by core genomic regions and secreted into the root apoplast during the initial stages of infection.
Our transcriptomic analysis of early infection stages (1, 2, 3, and 7 dpi) identified a set of candidate effector genes encoding predicted secreted fungal proteins that are specifically upregulated in planta. Most of these are encoded on core genomic regions and upregulated during all four infection time points. Interestingly, the ∼10% that show maximum induction during the earlier time points (1–3 dpi) are largely encoded on core genomic regions, whereas the remaining ∼5% which show maximum induction at the late time point (7 dpi) are mainly encoded on the LS pathogenicity chromosome 14 and include the SIX effector set. This finding suggests that the lifestyle transition from endophytic to pathogenic growth is associated with a transcriptional shift involving different waves of effector genes, in line with a previous study in the hemibiotrophic pathogen Colletotrichum (O’Connell et al., 2012).
Among the in planta-induced secreted proteins detected both in the proteomic and transcriptomic data sets, we identified four early effector candidates which are conserved across the entire Fo species complex as well as in other fungi. These ERCs are upregulated during colonization of both the main host and alternative host plants, indicating that they have a broadly conserved role in promoting fungus–root associations rather than a host-specific function in vascular colonization and wilting, as previously described for the LS-encoded SIX effectors.
In a recent study, the number of LS genes encoding candidate effectors were found to be higher in pathogenic than in endophytic Fo isolates (Constantin et al., 2021). Moreover, it was noted that the promoter regions of SIX effector genes are frequently associated with a miniature impala (mimp) transposable element (Schmidt et al., 2013; van Dam et al., 2016). Similar to SIX effectors, genes encoding ERCs were specifically upregulated in planta, but not in axenic minimal medium with crushed tomato roots. We did not detect an association of erc genes with known transposable elements, possibly due to the fact that ercs are located in core genomic regions that are less rich in transposons (Ma et al. 2010). Whether erc promoters contain conserved cis-acting elements for in planta transcriptional upregulation remains a topic for future investigation and may provide clues for the prediction of additional root compatibility factor candidates.
Importantly, targeted deletion of three of the four identified erc genes led to significant reduction in root colonization in the tomato pathogenic isolate Fol4287 and this effect was confirmed in two erc homologs in the non-pathogenic isolate Fo47. Moreover, the loss of ERCs in Fol4287 resulted in lower virulence and rapid activation of plant immune responses, while in Fo47 it led to reduced biocontrol activity against the pathogenic Fo isolate. While the molecular function of ERCs is currently unknown, it is worth noting that all four proteins contain predicted domains associated with binding or modification of plant cell walls. This suggests that they may contribute to the mobilization of carbohydrates in the apoplast or protect the fungus from the plant immune response, in line with the vital role of primary cell wall cellulose in early defense responses to vascular wilt pathogens (Menna et al., 2021). Both hypotheses are supported by TEM analysis of the Δerc1 and Δerc2 mutants, which were unable to progress from inter- to intracellular growth and remained confined to the intercellular space by distinctive plant cell wall protrusions. Moreover, the increased induction of immunity marker genes in plants infected with the Δerc mutants compared to those infected with the wild-type strain suggests that these mutants are impaired in their ability to prevent activation of host immune responses, possibly at the level of cell wall remodeling. Indeed, a recent study found that the co-incidence of physical damage and microbe associated molecular pattern recognition strongly induces local immune responses in Arabidopsis roots during bacterial infection (Zhou et al., 2020).
An interesting finding is that ERC2 and ERC4 carry predicted LPMO domains. LPMOs are copper-dependent enzymes that oxidatively cleave polysaccharide chains in their crystalline multichain regions and act in synergy with other plant cell wall-degrading enzymes (PCWDEs) such as hydrolases to enhance plant cell wall deconstruction (Vaaje-Kolstad et al., 2010). A recent study in P. infestans reported that a secreted pectin monooxygenase with a LPMO domain contributes to plant infection by promoting pectin cleavage (Jagadeeswaran et al., 2021; Sabbadin et al., 2021). We speculate that LPMO effectors such as ERC2 or ERC4 are used by Fo for initial cleavage of the polysaccharide backbone of the plant cell walls to aid pathogen accommodation in the root cortex apoplast. Interestingly, LPMOs have also been associated with mutualistic and commensal symbioses (Vandhana et al., 2021). The ectomycorrhizal fungus Laccaria bicolor expresses three AA9 LPMOs during formation of the hartig net in Poplus trichocarpa (Veneault-Fourrey et al., 2014) which together with other PCWDEs may contribute to the loosening of the plant cell wall (Zhang et al., 2018).
In vascular wilt fungi, secretion of PCWDEs has so far been mainly associated with colonization of the xylem and induction of wilt symptoms (Cooper and Wood, 1973; Cooper et al., 1978). However, knowledge of the role of cell wall modifying proteins during the early biotrophic infection stages and their requirement for plant compatibility is limited (Redkar et al., 2022b). Progressive loss of ligninolytic and cellulolytic enzymes has been associated with the evolution of symbiotic from saprotrophic lifestyle during fungus–plant associations (Zuccaro et al., 2014; Kohler et al., 2015; Miyauchi et al., 2020). However, the precise role of PCWDEs across the parasitism/mutualism continuum of the endophytic fungal lifestyle is largely unknown. Based on the large fraction of predicted fungal cell wall modifying enzymes detected in the proteome of tomato root AF, we propose that this group of enzymes plays an essential role during early root establishment of Fo. A similar enrichment of CWDEs was also observed in the AF during symbiotic root interaction with the endophytic fungus Piriformospora indica (Nizam et al., 2019). Moreover, a recent large-scale genomic analysis in A. thaliana roots identified a set of cell wall modifying enzymes as genetic determinants of fungal endophytism (Mesny et al., 2021). Taken together, these findings suggest that fungal modification of root cell walls plays a key role during early biotrophic establishment, both in endophytic and pathogenic interactions.
Currently, there is limited understanding on how plants can engage with beneficial microbes and at the same time restrict invasion by pathogens (Thoms et al., 2021). Recent evidence suggests that plant roots employ cell-layer-specific programs to differentially respond to beneficial and pathogenic microbes (Fröschel et al., 2021). Comparative TEM analysis of the pathogenic isolate Fol4287 and the biocontrol isolate Fo47 revealed significant differences in the ultrastructural development of the two isolates in the inner root cell layers, where growth of the Fo47 hyphae was largely abrogated in contrast to Fol4287, whereas both strains exhibited similar growth in root cortical cells. This is in line with the finding that erc genes are expressed predominantly during the initial infection stage and thus are likely to play a role in early root colonization, a process that is shared between endophytic and pathogenic Fo strains. By contrast, host-specific LS effectors were preferentially expressed at the later infection stage, suggesting that they function mainly during entry and colonization of the xylem coinciding with the switch to pathogenicity. Importantly, our results in the nonvascular bryophyte model M. polymorpha establish that ERCs are induced during plant intercellular growth of Fo independent of vasculature signatures. The finding that some ERCs also contribute to Fo infection on this evolutionarily distant land plant lineage further supports a broadly conserved role of ERCs in fungus–plant interactions.
Taking these results together, our study uncovers a suite of previously uncharacterized early root compatibility effectors, which are secreted by a vascular wilt fungus during the initial asymptomatic infection stages. ERCs contribute to host colonization by both pathogenic and non-pathogenic isolates on evolutionarily distant plant species such as tomato and the liverwort M. polymorpha that lacks a differentiated vasculature. While the precise modes of action of ERCs remain to be elucidated, our results suggest that they may target evolutionarily ancient plant processes and thus have broadly conserved roles in fungus–plant interactions.
Materials and methods
Fungal strains and transformants
Fungal strains used in this study are listed in Supplemental Table S4. All the generated knockouts are derivatives of Fo f. sp. lycopersici isolate 4287 (NRRL34936) or Fo Fo47 (NRRL54002). Strain cultures and storage were performed as described (Di Pietro et al., 2001). Phenotypic analysis of colony growth was done as previously reported (López-Berges et al., 2010). Targeted gene replacement with the hygromycin resistance cassette and complementation of the mutants by co-transformation with the phleomycin resistance cassette were performed as previously reported (López-Berges et al., 2010). Oligonucleotides used to generate PCR fragments for knockout generation by gene replacement, and complementation of mutants are listed in Supplemental Table S5.
Plant growth conditions and infection assays
Tomato seeds (Solanum lycopersicum cv. Monika F1 hybrid from Syngenta Seeds, Almería; susceptible to Fol4287 race 2) were surface sterilized in 1% sodium hypochlorite for 30 mins and potted in vermiculite supplemented with one-fourth (v/v) of universal peat substrate with all nutrients (Projar, Barcelona, Spain). Seedlings were grown in a growth chamber maintained at following conditions (15-/9-h light/dark cycle with white light at 6,000 lux intensity and 28°C). Marchantia polymorpha accession Takaragaike-1 (Tak-1; male) was used for generating Marchantia thalli. Marchantia polymorpha gemmae were grown on plates of half Gamborg’s B5 medium as described before (Gimenez-Ibanez et al., 2019).
Tomato root infection assays with Fo were performed as previously described on 2-week old seedlings (Di Pietro and Roncero, 1998) using a dipping protocol with 5 × 106 Fo microconidia mL−1. Survival was recorded daily and mortality curves were plotted by the Kaplan–Meier method and compared among groups using the log-rank test. Death of the infected plant was diagnosed as a complete collapse of the stem, without any green parts left accompanied by visible proliferation of the fungal mycelium on the dead tissue. The fresh weight of whole infected seedlings (including shoot and root tissue) was determined at 25 dpi. Marchantia polymorpha inoculation with Fo microconidia was performed as described (Redkar et al., 2022a). Severity of disease symptoms in M. polymorpha was quantified based on the macerated area of the infected thalli as previously described (Redkar et al., 2022a).
Galleria mellonella pathogenicity assays
Infection assays in G. mellonella larvae were performed as described previously (Navarro-Velasco et al., 2011). Briefly, a Burkard Auto Applicator (Burkard Manufacturing, UK) with a 1-mL syringe was used to inject 8 μL of the microconidial suspension (2×107 microconidia mL−1 in 1× PBS) into the hemocoel. Injected larvae were incubated in ventilated glass bottles at 30°C and survival was recorded daily. Larvae were considered dead when they displayed no movement and were melanized (Figure 5E). Mortality curves were plotted by the Kaplan–Meier method and compared among groups using the log-rank test. The experiment was performed three times with similar results. Data presented are from one representative experiment.
Generation of Fol-mClover3 or Fo47-mClover3 or Foc-mClover3-tagged Fo transformants
Plasmid pUC57 backbone carrying three copies of a Fo codon-optimized mClover3 (Bajar et al., 2016) gene (Fo-mClover3), followed by three copies of the FLAG octapeptide tag coding region (3xFLAG) and driven by the Aspergillus nidulans gpdA promoter and the SV40 late polyadenylation signal was synthesized by ProteoGenix (Schiltigheim, France). Codon-optimization of mClover3 was performed in accordance with Fo f. sp. lycopersici codon usage and GC content data retrieved from the Codon Usage Database (http://www.kazusa.or.jp/codon/).
Fo-mClover3-labeled strains of Fo f. sp. lycopersici (NRRL 34936), Fo f. sp. cubense (NRRL54006), and the Fo biocontrol isolate Fo47 (NRRL54002) were obtained by co-transforming fungal protoplasts with a Fo-mClover3 expression cassette (amplified from pUC57-Fo-mClover3 plasmid with primers gpdA-15b + SV40 Rev) and a hygromycin resistance cassette amplified from plasmid pAN7-1 (Punt et al., 1987) with primers gpdA-15b + Trpc8B, as previously described (Masachis et al., 2016). Cytoplasmic Fo-mClover3 expression was observed and quantified in at least 20 independent transformants using a Zeiss Axio Imager M2 microscope (Zeiss, Barcelona, Spain) equipped with an Evolve Photometrics EM512 digital camera (Photometrics Technology, Tucson, AZ, USA) and a GFP filter set (BP 450/490, FT 510, LP 515). Fungal transformants showing brightest fluorescence were used in subsequent microscopy analysis.
Laser scanning confocal microscopy
Laser scanning confocal microscopy was performed using a Zeiss 880 Confocal microscope with Airyscan. Solanum lycopersicum roots or M. polymorpha thalli inoculated with fluorescent transformants of wild-type Fol4287, Foc54006 (TR4), Fo47 or different knockout derivatives thereof expressing cytoplasmic Fo-mClover3 were visualized at an excitation of 488 nm and emission detected at 495–540 nm. The maturation zone of the secondary lateral roots or the adaxial surface of infected M. polymorpha thalli were sampled routinely for microscopy analysis. To visualize plant cell walls, samples were co-stained by 15 min incubation in 2 mg mL−1 propidium iodide (PI) in water in the dark for 15 mins before imaging. PI fluorescence was visualized at an excitation of 561 nm, and emission detected at 570–640 nm.
Sample preparation for TEM/SEM analysis
Sample preparation for TEM was carried out according to a protocol previously reported with slight modifications (Simon et al., 2013). Briefly, all secondary roots were fixed for 90 min with 2.5% glutaraldehyde in 0.06-M Sorensen phosphate buffer at pH 7.2. After four washes of 10 min each in the same buffer, the samples were post-fixed in 1% osmium tetroxide for 90 min. Samples were then rinsed four times 10 min each in buffer and dehydrated in a graded series of increasing concentrations of acetone (50%, 70%, 90%, and 100%) for 20 min per concentration. After dehydration samples were gradually infiltrated with increasing concentrations of EMBed812 resin (30%, 60%, and 100%) mixed with acetone for a minimum of 3 h per step. Finally, samples were embedded in pure, fresh EMBed812 resin and polymerized for 48 h at 60°C. Ultrathin sections (80 nm) were cut with a Leica EM Ultracut UC7 ultramicrotome (Leica Microsystems, Vienna, Austria), post-stained 5 min with 1% (w/v) lead citrate dissolved in 0.6-M NaOH and subsequently 15 min with 2% (w/v) uranyl-acetate dissolved in distilled water. Sections were analyzed using a JEM1010 transmission electron microscope (JEOL, Tokyo, Japan). All TEM images of the root samples are derived from the maturation zone of secondary lateral roots.
For SEM, sample preparation was carried out as described (Matthaeus et al., 2020) with slight modifications. Small pieces (1 mm2) were cut from Marchantia thalli and fixed for 90 min with 2.5% glutaraldehyde in 0.06-M Sorensen phosphate buffer at pH 7.2. After four washes of 10 min each in the same buffer, the samples were dehydrated in a graded series of increasing concentrations of ethanol (50%, 70%, 90%, and 100%) for 20 min per concentration. Dehydrated samples were critical point dried (Leica EM CPD 300; Leica Microsystems) using a customized program for plant leaves of about 80 min duration (settings for CO2 inlet: speed = medium and delay = 120 s; settings for exchange: speed = 5 and cycles = 18; settings for gas release: heat = medium and speed = medium). Samples were then mounted on aluminum stubs with carbon tape and sputter coated with 10 nm iridium (Leica EM ACE 600, Leica Microsystems) and imaged using a FEI Versa 3D scanning electron microscope (FEI, Hillsboro, OR, USA) under high vacuum condition.
Quantification of fungal burden by qPCR and gene expression by RT–qPCR
qPCR for quantification of gene expression or of fungal biomass in S. lycopersicum or M. polymorpha plants was performed as described previously (Redkar et al., 2022a). Briefly, RNA or DNA was extracted from roots of five tomato plants or three entire M. polymorpha thalli at the desired time points and considered as one biological replicate. RNA isolation was performed using the Tripure RNA isolation reagent (Roche, Spain) with DNase treatment (Roche, Spain). Reverse transcription was carried out using Transcriptor Universal cDNA Master (Roche, Spain). Genomic DNA was extracted using a modified chloroform: octanol extraction protocol (Torres et al., 1993) and used for quantification of fungal biomass. qPCR was performed using CFX96 Touch Real-Time PCR (Bio-Rad). Cycling conditions were 10 min at 95°C followed by 40 cycles of 10 s at 95°C, 10 s at 62°C, and 20 s at 72°C. Data were analyzed using the ΔΔCt method (Livak and Schmittgen, 2001) by calculating the ratio of the plant housekeeping genes SlGapdh (tomato) or MpEF1a (M. polymorpha; Carella et al., 2018) versus the Fol4287-specific six1 gene (FOXG_16418) or the Fo47-specific gene on Chr7 (FOBG_10856) to calculate the fungal burden. Expression profiling of the erc genes was carried out by using Fol4287-peptidyl prolyl isomerase gene (FOXG_08379; Redkar et al., 2022a). Moreover, the S. lycopersicum defense-related genes were tested by using the primers from previous report (Aimé et al., 2013). Expression analysis of the PTI marker genes from tomato were selected from Nguyen et al. (2010). For expression profiling of V. dahliae erc genes, VdGAPDH was used as a reference (Kombrink et al., 2017). All primers used for the qPCR analysis are listed in Supplemental Table S5.
Isolation of tomato AF and fungal culture filtrate
For isolation of AF, roots of 120 2-week old tomato plants were used at 72 h after inoculation. AF isolation was carried out as previously described (Wawra et al., 2016) with minor modification. Briefly, whole roots were thoroughly washed in running water and cut into pieces of 2 cm in length. Samples were vacuum infiltrated with deionized water three times for 15 min at 100 mbar with 5-min atmospheric pressure breaks. Roots were then thoroughly dried by gentle tapping on a tissue paper. Bundled infiltrated roots were centrifuged in 10-mL syringe barrels at 2,200 rpm for 15 min at 4°C. Pooled AF was flash frozen and stored at −80°C.
To process the protein samples for LC–MS, the isolated AF (∼500–700 μL) was concentrated on an Amicon Ultra 0.5-mL −3 kDa cutoff (Merck Millipore, Germany) and the concentrate was further resolved in 20 μL of 1×sodium dodecyl sulphate (SDS) sample buffer and 25 μL were used. Proteins were analyzed by SDS–polyacrylamide (10%) gel electrophoresis with a short run, followed by Coomassie blue staining. Protein bands of 1-cm block on the resolving gel were eluted and tested to identify proteins in AF. An In-gel Trypsin digestion was carried out and samples were analyzed by LC–MS. Identification of tomato and Fo proteins was carried out at the RTP, Proteomics Facility, University of Warwick, UK.
Filtrates from 50 mL of axenic cultures of Fo-secreted proteins were obtained as previously described (Nizam et al., 2019). Briefly, 3-day-old cultures of Fol4287 grown in potato dextrose broth (PDB); or in liquid minimal medium (MM) containing 25-mM NaNO3 as the sole nitrogen source (Puhalla, 1985; López-Berges et al., 2010;) and supplemented either with crushed roots or sucrose as carbon source, were harvested by filteration through cheesecloth and subsequently through a 0.45-μm syringe filter (Merck Millipore, Germany). Proteins were precipitated by adding 10% (v/v, final concentration) trichloroacetic acid, incubating at −20°C overnight and subsequent centrifugation at 12,000 rpm. Pelleted secreted proteins were resolved in 50 μL of 1× SDS sample buffer and 25 μL were used for SDS–PAGE analysis.
Immunoblot analysis
To exclude cytoplasmic contamination an immunoblot was performed on the isolated AF along with the total root extract as positive control with tubulin antibody. For immunoblotting, proteins from SDS–PAGE gels were transferred onto nitrocellulose membranes using the Transblot Turbo RTA Midi Transfer kit (BioRad, USA). Mouse anti-α-tubulin antibody (Sigma-Aldrich; T9026) was used at a 1:5,000 dilution to determine leakage of cytoplasmic contamination into the apoplastic fluid. The membrane was visualized using the ECL Select Western blotting detection reagent (GE Healthcare, Chicago, IL, USA) in a LAS-3000 detection system (Fujifilm, Barcelona, Spain).
Phylogenetic analysis and tree construction
Protein sequences of putative effectors were obtained from the Department of Energy-Joint Genome Institute MycoCosm database (https://mycocosm.jgi.doe.gov/mycocosm/home). A blastp search was performed using ERC1, ERC2, and ERC3 against the MycoCosm database. For phylogenetic tree construction, protein sequences were selected based on sequence homology. Multiple sequence alignment was performed using MUSCLE (v3.8.425) with default parameters over 100 iterations (Edgar, 2004). The phylogenetic tree was constructed with RAxML (v8.2.12) using the PROTGAMMAWAG model for protein sequence alignment (Stamatakis et al., 2004). Bootstrap support was determined for all the phylogenetic trees with a convergence test to confirm sufficient sampling.
RNA-seq and data analysis
Roots of five 2-week-old tomato plants were collected at 1, 2, 3, or 7 dpi with Fol4287 and considered as one biological replicate. The axenic growth control was generated as follows; fresh microconidia of Fol4287 obtained from a 3-day culture in PDB as described (López-Berges et al., 2010) were inoculated in 200-mL fresh PDB at a concentration of 2 × 106 microconidia mL−1. After 13-h incubation at 28°C, the germlings were harvested by filtration through a cheesecloth membrane, resuspended in 200-mL MM containing NaNO3 as nitrogen source and incubated an additional 5 h at 28°C. Germlings were then harvested and flash frozen for RNA isolation. Total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen, Germany) and treated with DNAse I using the Turbo DNA Free Kit (Invitrogen, Germany) according to the manufacturer’s instructions. RNA sequencing was performed by Novogene, UK. For library preparation mRNA was captured through poly-A enrichment on the total RNA, and a TruSeq RNA Library Preparation Kit (Illumina, USA) was used to build the libraries according to the manufacturer’s protocol. Libraries were sequenced on a NovaSeq6000 sequencing platform (Illumina). Paired-end 150-bp reads were obtained for each RNAseq library.
Read mapping and differential expression analysis
Transcript quantification was performed with Salmon (Patro et al., 2017). RNA-seq paired-end read data sets were quasi-mapped against the reference transcriptome of Fo (GCF_000149955.1_ASM14995v2_rna.fna, obtained from NCBI RefSeq). Adaptor-trimmed reads have been uploaded to the ArrayExpress database. Differential gene expression analysis on gene and transcript level was analyzed using DESeq2 (1.28.1; Love et al., 2014), following a pairwise comparison between Fo samples in planta as compared to the axenic growth control. GO-terms for Fo were obtained by processing the GCF_000149955.1 proteins with annotF (https://github.com/gemygk/AnnotF), wrapping Blast (Altschul et al., 1990), Blast2GO (Conesa et al., 2005), and InterProScan (Quevillon et al., 2005). GO-term analysis was done in R using the topGO package (Alexa et al., 2006; 2.40.0), by testing for enrichment in each infection timepoint as compared to the axenic growth control. DEGs (absolute LFC [log2 fold change] > 2 and adjusted P <0.01) were used to create volcano plots and hierarchical clustering of samples. Secreted proteins from the DEGs were predicted with SignalP5.0 (Almagro Armenteros et al., 2019), using a minimal signal peptide threshold of 0.75. Rlog-transformed expression counts of differentially expressed secreted genes were transformed to Z-scores and grouped into five clusters using the R-functions hclust and cutree. Scripts used to analyze RNA-seq data sets and visualize DEGs are available at https://github.com/cschu/redkar_et_al_erc_tpc_2022. Expression plots for the effectors of interest and for the amino acid transporters enriched in GO processes were created in Python by plotting the Z-scores of log2 expression values of the candidate gene on the y-axis versus, time in dpi on the x-axis.
Chromosomal locations of DEGs were visualized using a custom Python script https://github.com/cschu/chrom_plot. To this aim, DEGs were assigned to three different groups: early expressed genes (average of all three expression values for 7 dpi/average of all nine expression values for 1–3 dpi > 0.25) and represented in red; late expressed genes (average of all nine expression values for 1–3 dpi/average of all three expression values for 7 dpi < 0.25) and represented in blue; or expressed at all timepoints (DE at 7 dpi and at least one additional timepoint) and represented in green.
Targeted knockouts of the Fo erc genes
Targeted gene replacement with the hygromycin resistance cassette and complementation of the mutants by co-transformation with the phleomycin resistance cassette were performed as reported previously (López-Berges et al., 2010). Oligonucleotides used to generate PCR fragments for gene replacement, complementation or identification of mutants are listed (Supplemental Table S5). PCR was performed with a High Fidelity Phusion Polymerase (NEB, Germany), using a Mini personal thermal cycler (BioRad).
Statistical analysis
No statistical methods were used to predetermine sample size. Statistical analysis was carried out using the GraphPad Prism 9.0 software (San Diego, USA) and the data on biomass quantification and expression profiling were plotted using the same tool. For statistical analysis, all data were tested with a nonparametric or mixed one-way ANOVA followed by Bonferroni’s multiple comparison test or an unpaired t test for statistical significance. Data points with different letters indicate significant differences of P < 0.05 for Bonferroni’s test results. Data points are plotted onto the graphs, and the number of samples or the description of error bars are indicated in the corresponding figure legends. (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001; otherwise, not significant) between samples. For the Kaplan–Meier plots showing comparison between percent survival of plants, log-rank (Mantel–Cox) test was performed to calculate the statistical significance (P < 0.05) of survival compared to wild-type infected plants. Summary of the statistical analyses of data sets from all figures is given in Supplemental Data Set S5. Fisher’s exact test was performed to calculate the enrichment of the secreted proteins during different infection stages on either the core or accessory genome regions.
Accession numbers
Sequence data for the genes described in this study can be found at the Fungal and oomycete Informatics Resources (FungiDB) under the following accession numbers: erc1, FOXG_11583; erc2, FOXG_04534; erc3, FOXG_16902; erc4, FOXG_08211; ppi, FOXG_08379. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD026400. The RNA-seq data sets generated and analyzed in this study have been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-10597.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. F. oxysporum colonizes the roots of main and alternative host plants and secretes conserved core effectors.
Supplemental Figure S2. Transcriptional dynamics in F. oxysporumduring early stages of root infection.
Supplemental Figure S3. Generation and phenotyping of erc knockout mutants in F. oxysporum.
Supplemental Figure S4. ERCs have a role in root colonization and virulence on the main plant host.
Supplemental Figure S5. ERCs do not have an additive role in root colonization and virulence on the main host plant.
Supplemental Figure S6. ERCs contribute to suppression of PAMP-triggered defense in tomato roots.
Supplemental Figure S7. ERCs contribute to root colonization and biocontrol ability of Fo47.
Supplemental Figure S8. ERCs are upregulated during interaction of different vascular wilt fungi with the main and alternative plant hosts.
Supplemental Figure S9. ERCs promote fungal compatibility on a broad range of plant hosts.
Supplemental Table S1. Fo reference isolates from the Broad Institute Fusarium Comparative Genome Initiative used to test colonization on Solanum lycopersicum (Tomato).
Supplemental Table S2. Protein sequences of all the ERC homologs from all fungal phyla which are included in the phylogenetic tree.
Supplemental Table S3. Fischer’s exact test for distribution of secreted proteins across core and accessory regions of the F. oxysporum (Fol4287) genome.
Supplemental Table S4. F. oxysporum strains used in the study.
Supplemental Table S5. Oligonucleotides used in the study.
Supplemental Data Set S1. Fol4287 proteins consistently identified in discovery proteomics through LC–MS.
Supplemental Data Set S2. Blast analysis of ERC1, ERC2, ERC3, and ERC4 homologs across all fungal phyla.
Supplemental Data Set S3. F. oxysporum differentially expressed genes (DEGs) upregulated (UP) at 1, 2, 3, and 7 dpi.
Supplemental Data Set S4 . Overrepresentation of the Gene Ontology terms in Category Biological Processes during F. oxysporum early colonization timepoints.
Supplemental Data Set S5. Summary of the statistical analyses.
Supplementary Material
Acknowledgments
We thank M.I.G. Roncero (Universidad de Córdoba, Spain) for critical reading of the manuscript. We acknowledge the Central Service for Research Support (SCAI) of the University of Córdoba for use of the confocal microscopy facility. The high-performance computing resources and services in this work were supported by the Earlham Institute Scientific Computing group alongside the Norwich BioScience Institutes Partnership Computing infrastructure for Science (CiS) group.
Funding
This work was supported by grants from the Spanish Ministry of Science and Innovation (MICINN, grant PID2019-108045RB-I00) and Junta de Andalucía (P20_00179) to A.D.P. A.R. and M.S. were supported by the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska‐Curie grant agreements No. 750669 and 797256. A.R. also acknowledges funding from Juan de la Cierva Incorporación grant from the Spanish Research Agency (IJC2018-038468-I) as well as financial support from Ramalingaswami Re-entry Fellowship (BT/RLF/Re-entry/72/2020), Department of Biotechnology, Government of India. C.S. was supported by BBSRC strategic funding, Core Capability Grant BB/CCG1720/1, BBS/E/T/000PR9816. S.G.I is funded by the Spanish Ministry for Science and Innovation RTI2018-094526-J-I00 and the CSIC 2021AT006 grants. Research in the R.S. lab was funded by the Spanish Ministry for Science and Innovation grant PID2019-107012RB-100 (MICINN/FEDER).
Conflict of interest statement. None declared.
Contributor Information
Amey Redkar, Departamento de Genética, Universidad de Córdoba, 14071 Córdoba, Spain.
Mugdha Sabale, Departamento de Genética, Universidad de Córdoba, 14071 Córdoba, Spain.
Christian Schudoma, Earlham Institute, Norwich Research Park, Norwich NR4 7UZ, UK.
Bernd Zechmann, Baylor University, Center for Microscopy and Imaging, Waco, Texas 76798, USA.
Yogesh K Gupta, The Sainsbury Laboratory, Norwich Research Park, Norwich NR4 7UH, UK.
Manuel S López-Berges, Departamento de Genética, Universidad de Córdoba, 14071 Córdoba, Spain.
Giovanni Venturini, Departamento de Genética, Universidad de Córdoba, 14071 Córdoba, Spain.
Selena Gimenez-Ibanez, Plant Molecular Genetics Department, Centro Nacional de Biotecnologıa-CSIC (CNB-CSIC), 28049 Madrid, Spain.
David Turrà, Department of Agriculture and Center for Studies on Bioinspired Agro-enviromental Technology, Università di Napoli Federico II, 80055 Portici, Italy.
Roberto Solano, Plant Molecular Genetics Department, Centro Nacional de Biotecnologıa-CSIC (CNB-CSIC), 28049 Madrid, Spain.
Antonio Di Pietro, Departamento de Genética, Universidad de Córdoba, 14071 Córdoba, Spain.
A.R. and A.D.P. conceptualized the work, designed the experiments and supervised the conducted research. A.R., M.S., B.Z., Y.K.G., M.S.L.B., S.G.I., G.V., and D.T. carried out the experiments and analyzed the data. C.S. performed all the bioinformatic analysis. R.S. and S.G.I contributed in sample material and gave intellectual input for the Marchantia-related experiments. A.R. and A.D.P. wrote the article. All authors participated in reviewing and editing the final version of the article.
The author(s) responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) are: Amey Redkar (aredkar@unipune.ac.in) and Antonio Di Pietro (ge2dipia@uco.es).
References
- Alabouvette C, Olivain C, Migheli Q, Steinberg C (2009). Microbiological control of soil-borne phytopathogenic fungi with special emphasis on wilt-inducing Fusarium oxysporum. New Phytol 184: 529–544 [DOI] [PubMed] [Google Scholar]
- Aimé S, Alabouvette C, Steinberg C, Olivain C (2013) The endophytic strain Fusarium oxysporum Fo47: a good candidate for priming the defense responses in tomato roots. Mol Plant-Microbe Interact 26: 918–926 [DOI] [PubMed] [Google Scholar]
- Alexa A, Rahnenführer J, Lengauer T (2006) Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22: 1600–1607 [DOI] [PubMed] [Google Scholar]
- Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H (2019) SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37: 420–423 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990). Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Araujo L, Bispo WMS, Cacique IS, Moreira WR, Rodrigues FÁ (2014) Resistance in mango against infection by Ceratocystis fimbriata. Phytopathology 104: 820–833 [DOI] [PubMed] [Google Scholar]
- Bajar BT, Wang ES, Lam AJ, Kim BB, Jacobs CL, Howe ES, Davidson MW, Lin MZ, Chu J (2016) Improving brightness and photostability of green and red fluorescent proteins for live cell imaging and FRET reporting. Sci Rep 6: 20889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen RL, Pieterse CMJ, Bakker PAHM (2012) The rhizosphere microbiome and plant health. Trends Plant Sci 17: 478–486 [DOI] [PubMed] [Google Scholar]
- Bishop GD, Cooper RM (1983) An ultrastructural study of vascular colonization in three vascular wilt diseases I. Colonization of susceptible cultivars. Physiol Plant Pathol 23: 323–343 [Google Scholar]
- Carella P, Gogleva A, Tomaselli M, Alfs C, Schornack S (2018) Phytophthora palmivora establishes tissue-specific intracellular infection structures in the earliest divergent land plant lineage. Proc Natl Acad Sci USA 115: E3846–E3855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Blekemolen MC, Tintor N, Cornelissen BJC, Takken FLW (2018) The Fusarium oxysporum Avr2-Six5 effector pair alters plasmodesmatal exclusion selectivity to facilitate cell-to-cell movement of Avr2. Mol Plant 11: 691–705 [DOI] [PubMed] [Google Scholar]
- Chaloner TM, Gurr SJ, Bebber DP (2021) Plant pathogen infection risk tracks global crop yields under climate change. Nat Climate Change 11: 710–715 [Google Scholar]
- Cooper RM, Wood RKS (1973) Induction of synthesis of extracellular cell-wall degrading enzymes in vascular wilt fungi. Nature 246: 309–311 [Google Scholar]
- Cooper RM, Rankin B, Wood RKS (1978) Cell-wall degrading enzymes of vascular wilt fungi. II. Properties and modes of action of polysaccharidase of Verticillium albo-atrum and Fusarium oxysporum f.sp. lycopersici. Physiol Plant Pathol 13: 101–134 [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676 [DOI] [PubMed] [Google Scholar]
- Constantin ME, Vlieger BV, Takken FLW, Rep M (2020) Diminished pathogen and enhanced endophyte colonization upon coinoculation of endophytic and pathogenic Fusarium strains. Microorganisms 8: 544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantin ME, Fokkens L, de Sain M, Takken FLW, Rep M (2021) Number of candidate effector genes in accessory genomes differentiates pathogenic from endophytic Fusarium oxysporum strains. Front Plant Sci 12: 761740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean R, Van Kan JAL, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD, Rudd JJ, Dickman M, Kahmann R, Ellis J, et al. (2012). The Top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol 13: 414–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lamo FJ, Takken FLW (2020) Biocontrol by Fusarium oxysporum using endophyte-mediated resistance. Front Plant Sci 11: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries RP, Kester HCM, Poulsen CH, Benen JAE, Visser J (2000) Synergy between enzymes from Aspergillus involved in the degradation of plant cell wall polysaccharides. Carbohydr Res 327: 401–410 [DOI] [PubMed] [Google Scholar]
- Di Pietro A, García-Maceira FI, Méglecz E, Roncero MIG (2001) A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol Microbiol 39: 1140–1152 [PubMed] [Google Scholar]
- Di Pietro A, Roncero MIG (1998) Cloning, expression, and role in pathogenicity of pg1 encoding the major extracellular endopolygalacturonase of the vascular wilt pathogen Fusarium oxysporum. Mol Plant-Microbe Interact 11: 91–98 [DOI] [PubMed] [Google Scholar]
- Dumas B, Bottin A, Gaulin E, Esquerré-Tugayé M-T (2008) Cellulose-binding domains: cellulose associated-defensive sensing partners? Trends Plant Sci 13: 160–164 [DOI] [PubMed] [Google Scholar]
- Edel-Hermann V, Lecomte C (2019) Current status of Fusarium oxysporum formae speciales and races. Phytopathology 109: 512–530 [DOI] [PubMed] [Google Scholar]
- Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröschel C, Komorek J, Attard A, Marsell A, Lopez-Arboleda WA, Le Berre J, Wolf E, Geldner N, Waller F, Korte A, et al. (2021). Plant roots employ cell-layer-specific programs to respond to pathogenic and beneficial microbes. Cell Host Microbe 29: 299–310.e7 [DOI] [PubMed] [Google Scholar]
- Fuchs JG, Moënne-Loccoz Y, Défago G (1999) Ability of nonpathogenic Fusarium oxysporum Fo47 to protect tomato against Fusarium wilt. Biol Control 14: 105–110 [Google Scholar]
- Gawehns F, Houterman PM, Ichou FA, Michielse CB, Hijdra M, Cornelissen BJC, Rep M, Takken FLW (2014) The Fusarium oxysporum effector Six6 contributes to virulence and suppresses I-2-mediated cell death. Mol Plant-Microbe Interact 27: 336–348 [DOI] [PubMed] [Google Scholar]
- Gawehns F, Ma L, Bruning O, Houterman PM, Boeren S, Cornelissen BJC, Rep M, Takken FLW (2015) The effector repertoire of Fusarium oxysporum determines the tomato xylem proteome composition following infection. Front Plant Sci 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Zamarreño AM, García-Mina JM, Solano R (2019) An evolutionarily ancient immune system governs the interactions between Pseudomonas syringae and an early-diverging land plant lineage. Curr Biol 29: 2270–2281.e74 [DOI] [PubMed] [Google Scholar]
- Gordon TR (2017) Fusarium oxysporum and the Fusarium Wilt Syndrome. Annu Rev Phytopathol 55: 23–39 [DOI] [PubMed] [Google Scholar]
- Guo L, Yu H, Wang B, Vescio K, DeIulio GA, Yang H, Berg A, Zhang L, Steinberg C, Edel-Hermann V, et al. (2021) Metatranscriptomic comparison of endophytic and pathogenic Fusarium–Arabidopsis interactions reveals plant transcriptional plasticity. Mol Plant-Microbe Interact 34: 1071–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houterman PM, Speijer D, Dekker HL, De Koster CG, Cornelissen BJC, Rep M (2007) The mixed xylem sap proteome of Fusarium oxysporum-infected tomato plants. Mol Plant Pathol 8: 215–221 [DOI] [PubMed] [Google Scholar]
- Houterman PM, Ma L, Van Ooijen G, De Vroomen MJ, Cornelissen BJC, Takken FLW, Rep M (2009) The effector protein Avr2 of the xylem-colonizing fungus Fusarium oxysporum activates the tomato resistance protein I-2 intracellularly. Plant J 58: 970–978 [DOI] [PubMed] [Google Scholar]
- Inami K, Yoshioka-Akiyama C, Morita Y, Yamasaki M, Teraoka T, Arie T (2012). A genetic mechanism for emergence of races in Fusarium oxysporum f. sp. lycopersici: inactivation of avirulence gene AVR1 by transposon insertion. PLoS One 7: e44101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadeeswaran G, Veale L, Mort AJ (2021) Do lytic polysaccharide monooxygenases aid in plant pathogenesis and herbivory? Trends Plant Sci 26: 142–155 [DOI] [PubMed] [Google Scholar]
- Jashni MK, Dols IHM, Iida Y, Boeren S, Beenen HG, Mehrabi R, Collemare J, de Wit PJGM (2015) Synergistic action of a metalloprotease and a serine protease from Fusarium oxysporum f. sp. lycopersici cleaves chitin-binding tomato chitinases, reduces their antifungal activity, and enhances fungal virulence. Mol Plant-Microbe Interact 28: 996–1008 [DOI] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Irieda H, Inoue Y, Mori M, Yamada K, Oshikawa Y, Saitoh H, Uemura A, Terauchi R, Kitakura S, Kosaka A, et al. (2019) Conserved fungal effector suppresses PAMP-triggered immunity by targeting plant immune kinases. Proc Natl Acad Sci USA 116: 496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katan J (1971) Symptomless carriers of the tomato Fusarium wilt pathogen. Phytopathology 61: 1213–1217 [Google Scholar]
- Kim J-G, Li X, Roden JA, Taylor KW, Aakre CD, Su B, Lalonde S, Kirik A, Chen Y, Baranage G, et al. (2009). Xanthomonas T3S effector XopN suppresses PAMP-triggered immunity and interacts with a tomato atypical receptor-like kinase and TFT1. Plant Cell 21: 1305–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klosterman SJ, Subbarao KV, Kang S, Veronese P, Gold SE, Thomma BP, Chen Z, Henrissat B, Lee YH, Park J, et al. (2011) Comparative genomics yields insights into niche adaptation of plant vascular wilt pathogens. PLoS Pathogens 7: e1002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Kuo A, Nagy LG, Morin E, Barry KW, Buscot F, Canbäck B, Choi C, Cichocki N, Clum A, et al. (2015) Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nat Genet 47: 410–415 [DOI] [PubMed] [Google Scholar]
- Kombrink A, Rovenich H, Shi-Kunne X, Rojas-Padilla E, van den Berg GCM, Domazakis E, de Jonge R, Valkenburg D-J, Sánchez-Vallet A, Seidl MF, et al. (2017) Verticillium dahliae LysM effectors differentially contribute to virulence on plant hosts. Mol Plant Pathol 18: 596–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahrmann U, Ding Y, Banhara A, Rath M, Hajirezaei MR, Döhlemann S, von Wirén N, Parniske M, Zuccaro A (2013) Host-related metabolic cues affect colonization strategies of a root endophyte. Proc Natl Acad Sci USA 110: 13965–13970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanver D, Berndt P, Tollot M, Naik V, Vranes M, Warmann T, Münch K, Rössel N, Kahmann R (2014) Plant surface cues prime Ustilago maydis for biotrophic development. PLoS Pathogens 10: e1004272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- López-Berges MS, Rispail N, Prados-Rosales RC, Di Pietro A (2010) A nitrogen response pathway regulates virulence functions in Fusarium oxysporum via the protein kinase TOR and the bZIP protein MeaB. Plant Cell 22: 2459–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L-J, van der Does HC, Borkovich KA, Coleman JJ, Daboussi MJ, Di Pietro A, Dufresne M, Freitag M, Grabherr M, Henrissat B, et al. (2010). Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464: 367–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Houterman PM, Gawehns F, Cao L, Sillo F, Richter H, Clavijo-Ortiz MJ, Schmidt SM, Boeren S, Vervoort J, et al. (2015). The AVR2–SIX5 gene pair is required to activate I-2-mediated immunity in tomato. New Phytol 208: 507–518 [DOI] [PubMed] [Google Scholar]
- Masachis S, Segorbe D, Turrà D, Leon-Ruiz M, Fürst U, El Ghalid M, Leonard G, López-Berges MS, Richards TA, Felix G, et al. (2016) A fungal pathogen secretes plant alkalinizing peptides to increase infection. Nat Microbiol 1: 16043. [DOI] [PubMed] [Google Scholar]
- Matthaeus WJ, Schmidt J, White JD, Zechmann B (2020) Novel perspectives on stomatal impressions: rapid and non-invasive surface characterization of plant leaves by scanning electron microscopy. PLoS One 15: e0238589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menna A, Dora S, Sancho-Andrés G, Kashyap A, Meena MK, Sklodowski K, Gasperini D, Coll NS, Sánchez-Rodríguez C (2021) A primary cell wall cellulose-dependent defense mechanism against vascular pathogens revealed by time-resolved dual transcriptomics. BMC Biol 19: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesny F, Miyauchi S, Thiergart T, Pickel B, Atanasova L, Karlsson M, Hüttel B, Barry KW, Haridas S, Chen C, et al. (2021). Genetic determinants of endophytism in the Arabidopsis root mycobiome. bioRxiv, 2021.2004.2028.441743 [DOI] [PMC free article] [PubMed]
- Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, Tosatto SCE, Paladin L, Raj S, Richardson LJ, et al. (2020) Pfam: the protein families database in 2021. Nucleic Acids Res 49: D412–D419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyauchi S, Kiss E, Kuo A, Drula E, Kohler A, Sánchez-García M, Morin E, Andreopoulos B, Barry KW, Bonito G, et al. (2020). Large-scale genome sequencing of mycorrhizal fungi provides insights into the early evolution of symbiotic traits. Nat Commun 11: 5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Velasco GY, Prados-Rosales RC, Ortíz-Urquiza A, Quesada-Moraga E, Di Pietro A (2011) Galleria mellonella as model host for the trans-kingdom pathogen Fusarium oxysporum. Fungal Genet Biol 48: 1124–1129 [DOI] [PubMed] [Google Scholar]
- Ngou BPM, Ahn H-K, Ding P, Jones JDG (2021) Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592: 110–115 [DOI] [PubMed] [Google Scholar]
- Nguyen HP, Chakravarthy S, Velásquez AC, McLane HL, Zeng L, Nakayashiki H, Park D-H, Collmer A, Martin GB (2010) Methods to study PAMP-triggered immunity using tomato and Nicotiana benthamiana. Mol Plant-Microbe Interact 23: 991–999 [DOI] [PubMed] [Google Scholar]
- Nizam S, Qiang X, Wawra S, Nostadt R, Getzke F, Schwanke F, Dreyer I, Langen G, Zuccaro A (2019) Serendipita indica E5′NT modulates extracellular nucleotide levels in the plant apoplast and affects fungal colonization. EMBO Rep 20: e47430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nucci M, Anaissie E (2007) Fusarium infections in immunocompromised patients. Clin Microbiol Rev 20: 695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell RJ, Thon MR, Hacquard S, Amyotte SG, Kleemann J, Torres MF, Damm U, Buiate EA, Epstein L, Alkan N, et al. (2012). Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat Genet 44: 1060–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordonez N, Seidl MF, Waalwijk C, Drenth A, Kilian A, Thomma BPHJ, Ploetz RC, Kema GHJ (2015) Worse comes to worst: bananas and panama disease—when plant and pathogen clones meet. PLoS Pathogens 11: e1005197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortoneda M, Guarro J, Madrid MP, Caracuel Z, Roncero MIG, Mayayo E, Di Pietro A (2004) Fusarium oxysporum as a multihost model for the genetic dissection of fungal virulence in plants and mammals. Infect Immunity 72: 1760–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C (2017) Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14: 417–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg GF (1976) Transmission electron microscopy of Verticillium alboatrum hyphae in xylem vessels of tomato plants. Physiol Plant Pathol 8: 22l–224 [Google Scholar]
- Pereira E, Vázquez de Aldana BR, San Emeterio L, Zabalgogeazcoa I (2019) A survey of culturable fungal endophytes from Festuca rubra subsp. pruinosa, a grass from marine cliffs, reveals a core microbiome. Front Microbiol 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presti LL, Lanver D, Schweizer G, Tanaka S, Liang L, Tollot M, Zuccaro A, Reissmann S, Kahmann R (2015) Fungal effectors and plant susceptibility. Annu Rev Plant Biol 66: 513–545 [DOI] [PubMed] [Google Scholar]
- Puhalla JE (1985) Classification of strains of Fusarium oxysporum on the basis of vegetative incompatibility. Can J Bot 63: 179–183 [Google Scholar]
- Punt PJ, Oliver RP, Dingemanse MA, Pouwels PH, van den Hondel CAMJJ (1987) Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene 56: 117–124 [DOI] [PubMed] [Google Scholar]
- Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R (2005) InterProScan: protein domains identifier. Nucleic Acids Res 33: W116–W120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redkar A, Gimenez Ibanez S, Sabale M, Zechmann B, Solano R, Di Pietro A (2022a) Marchantia polymorpha model reveals conserved infection mechanisms in the vascular wilt fungal pathogen Fusarium oxysporum. New Phytol 234: 227–241 [DOI] [PubMed] [Google Scholar]
- Redkar A, Sabale M, Zuccaro A, Di Pietro A (2022b) Determinants of endophytic and pathogenic lifestyle in root colonizing fungi. Curr Opin Plant Biol 67: 102226. [DOI] [PubMed] [Google Scholar]
- Rep M, Van Der Does HC, Meijer M, Van Wijk R, Houterman PM, Dekker HL, De Koster CG, Cornelissen BJC (2004) A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato. Mol Microbiol 53: 1373–1383 [DOI] [PubMed] [Google Scholar]
- Roth R, Hillmer S, Funaya C, Chiapello M, Schumacher K, Lo Presti L, Kahmann R, Paszkowski U (2019) Arbuscular cell invasion coincides with extracellular vesicles and membrane tubules. Nat Plants 5: 204–211 [DOI] [PubMed] [Google Scholar]
- Sabbadin F, Urresti S, Henrissat B, Avrova AO, Welsh LRJ, Lindley PJ, Csukai M, Squires JN, Walton PH, Davies GJ, et al. (2021) Secreted pectin monooxygenases drive plant infection by pathogenic oomycetes. Science 373: 774–779 [DOI] [PubMed] [Google Scholar]
- Schmidt SM, Houterman PM, Schreiver I, Ma L, Amyotte S, Chellappan B, Boeren S, Takken FLW, Rep M (2013) MITEs in the promoters of effector genes allow prediction of novel virulence genes in Fusarium oxysporum. BMC Genomics 14: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon UK, Polanschütz LM, Koffler BE, Zechmann B (2013) High resolution imaging of temporal and spatial changes of subcellular ascorbate, glutathione and H2O2 distribution during botrytis cinerea infection in Arabidopsis. PLoS One 8: e65811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SN, Snyder WC (1975) Persistence of Fusarium oxysporum f. sp. vasinfectum in fields in absence of cotton. Phytopathology 65: 190–196 [Google Scholar]
- Stamatakis A, Ludwig T, Meier H (2004) RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21: 456–463 [DOI] [PubMed] [Google Scholar]
- Takken F, Rep M (2010) The arms race between tomato and Fusarium oxysporum. Mol Plant Pathol 11: 309–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms D, Liang Y, Haney CH (2021) Maintaining symbiotic homeostasis: how do plants engage with beneficial microorganisms while at the same time restricting pathogens? Mol Plant-Microbe Interact 34: 462–469 [DOI] [PubMed] [Google Scholar]
- Tintor N, Paauw M, Rep M, Takken FLW (2020) The root-invading pathogen Fusarium oxysporum targets pattern-triggered immunity using both cytoplasmic and apoplastic effectors. New Phytol 227: 1479–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres AM, Weeden NF, Martín A (1993) Linkage among isozyme, RFLP and RAPD markers in Vicia faba. Theoret Appl Genet 85: 937–945 [DOI] [PubMed] [Google Scholar]
- Toruño TY, Stergiopoulos I, Coaker G (2016) Plant-pathogen effectors: cellular probes interfering with plant defenses in spatial and temporal manners. Annu Rev Phytopathol 54: 419–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrà D, El Ghalid M, Rossi F, Di Pietro A (2015) Fungal pathogen uses sex pheromone receptor for chemotropic sensing of host plant signals. Nature 527: 521–524 [DOI] [PubMed] [Google Scholar]
- Upson JL, Zess EK, Białas A, Wu C-h, Kamoun S (2018) The coming of age of EvoMPMI: evolutionary molecular plant–microbe interactions across multiple timescales. Curr Opin Plant Biol 44: 108–116 [DOI] [PubMed] [Google Scholar]
- Vaaje-Kolstad G, Westereng B, Horn SJ, Liu Z, Zhai H, Sørlie M, Eijsink VGH (2010) An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 330: 219–222 [DOI] [PubMed] [Google Scholar]
- van Dam P, Fokkens L, Schmidt SM, Linmans JHJ, Kistler HC, Ma L-J, Rep M (2016) Effector profiles distinguish formae speciales of Fusarium oxysporum. Environ Microbiol 18: 4087–4102 [DOI] [PubMed] [Google Scholar]
- van Dam P, Rep M (2017) The distribution of miniature impala elements and SIX genes in the fusarium genus is suggestive of horizontal gene transfer. J Mol Evol 85: 14–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandhana TM, Reyre J-L, Sushmaa D, Berrin J-G, Bissaro B, Madhuprakash J (2022) On the expansion of biological functions of lytic polysaccharide monooxygenases. New Phytol. doi: 10.1111/nph.17921 [DOI] [PubMed] [Google Scholar]
- Veloso J, Díaz J (2012) Fusarium oxysporum Fo47 confers protection to pepper plants against Verticillium dahliae and Phytophthora capsici, and induces the expression of defence genes. Plant Pathol 61: 281–288 [Google Scholar]
- Veneault-Fourrey C, Commun C, Kohler A, Morin E, Balestrini R, Plett J, Danchin E, Coutinho P, Wiebenga A, de Vries RP, et al. (2014) Genomic and transcriptomic analysis of Laccaria bicolor CAZome reveals insights into polysaccharides remodelling during symbiosis establishment. Fungal Genet Biol 72: 168–181 [DOI] [PubMed] [Google Scholar]
- Wawra S, Fesel P, Widmer H, Timm M, Seibel J, Leson L, Kesseler L, Nostadt R, Hilbert M, Langen G, et al. (2016). The fungal-specific β-glucan-binding lectin FGB1 alters cell-wall composition and suppresses glucan-triggered immunity in plants. Nat Commun 7: 13188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Wang Y, Park S-Y, Kim SG, Yoo JS, Park S, Gupta R, Kang KY, Kim ST (2016) Secreted alpha-N-arabinofuranosidase B protein is required for the full virulence of Magnaporthe oryzae and triggers host defences. PLoS One 11: e0165149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Anasontzis GE, Labourel A, Champion C, Haon M, Kemppainen M, Commun C, Deveau A, Pardo A, Veneault-Fourrey C, et al. (2018) The ectomycorrhizal basidiomycete Laccaria bicolor releases a secreted β-1,4 endoglucanase that plays a key role in symbiosis development. New Phytol 220: 1309–1321 [DOI] [PubMed] [Google Scholar]
- Zhou F, Emonet A, Dénervaud Tendon V, Marhavy P, Wu D, Lahaye T, Geldner N (2020) Co-incidence of damage and microbial patterns controls localized immune responses in roots. Cell 180: 440–453.e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccaro A, Lahrmann U, Langen G (2014) Broad compatibility in fungal root symbioses. Curr Opin Plant Biol 20: 135–145 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.