Highlights
-
•
A novel CNN model was applied to multi-parameter MRI sequences.
-
•
A learnable subtraction layer and hierarchical classification are proposed.
-
•
LSTM enables efficient patient-level classification.
-
•
Using APTw data as additional input can significantly boost diagnostic performance.
-
•
Class activation maps visually highlight the evidence for classification.
Keywords: Amide proton transfer-weighted MRI, Convolutional neural network, Glioma, Treatment effect
Abstract
The purpose of this study was to develop and verify a convolutional neural network (CNN)-based deep-learning algorithm to identify tumor progression versus response by adding amide proton transfer-weighted (APTw) MRI data to structural MR images as the proposed model input. 145 scans with 2175 MR instances from 98 patients with malignant glioma (acquired between April 2010 and February 2018) were re-analyzed. An end-to-end classification framework based on a ResNet backbone was developed. The architecture includes a learnable subtraction layer and a hierarchical classification paradigm, and synthesizes information over multiple MR slices using a long short-term memory. Areas under the receiver-operating-characteristic curves (AUCs) were used to assess the impact of adding APTw MRI to structural MRI (, , FLAIR, and GdT1w) on classification of tumor response vs. progression, both on the slice- and scan-level. With both APTw and structural MRI data, adding a learnable subtraction layer and a hierarchical classification paradigm to the backbone ResNet model improved the slice-level classification performance from an AUC of 0.85 to 0.90. Adding APTw data to structural MR images as input to our proposed CNN classification framework led to an increase in AUCs from 0.88 to 0.90 for the slice-level classification (P < 0.001), and from 0.85 to 0.90 for the scan-level classification (P < 0.05). Generated saliency maps highlighted the vast majority of lesions. Complementing structural MRI sequences with protein-based APTw MRI enhanced CNN-based classification of recurrent glioma at the slice and scan levels. Addition of APTw MRI to structural MRI sequences enhanced CNN-based classification of recurrent glioma at the slice and scan levels.
1. Introduction
Despite maximum feasible surgical resection followed by radiotherapy with concurrent chemotherapy, malignant gliomas eventually progress, with a median survival of 12–15 months for glioblastoma (Stupp et al., 2005). Radiographic evaluation plays a critical role in the management of post-treatment malignant gliomas, in which magnetic resonance imaging (MRI) following the response-assessment-in-neuro-oncology (RANO) criteria remains the standard (Wen et al., 2010). However, structural MR images used in the clinical setting, including -, -, fluid-attenuated inversion recovery (FLAIR), and gadolinium-enhanced T1w ( MR images, are not sufficiently tissue-specific to guide treatment decisions (Wen et al., 2010). These limitations have immediate clinical consequences confounding post-treatment diagnostics and treatment planning and complicating the procedures for new therapy development. Therefore, reliable, automated imaging diagnostic tools to assess malignant glioma response to therapies are urgently needed.
Amide proton transfer-weighted (APTw) imaging, based on chemical exchange saturation transfer (CEST) MRI contrast mechanism (Ward et al., 2000), is an emerging molecular MRI technique that was designed to detect endogenous cellular proteins and peptides in tissue (Zhou et al., 2019). The APTw hyperintensity observed in malignant glioma is most likely associated with increased cytosolic protein content due to higher cellularity and slightly increased intracellular pH (Lee et al., 2017, Ray et al., 2019, Yan et al., 2015). Numerous research groups worldwide have confirmed that the hyperintensity on APTw images is a reliable imaging marker of malignant glioma before (Jiang et al., 2017, Jiang et al., 2022, Sotirios et al., 2020) and after (Liu et al., 2020, Park et al., 2020a, Park et al., 2020b) treatment. Notably, consensus recommendations on clinical APTw imaging approaches at 3 T for brain tumors have recently been published (Zhou et al., 2022).
On the other hand, advances in artificial intelligence and computer vision have achieved powerful solutions for improving medical imaging techniques and automatic diagnosis (Eijgelaar et al., 2020, Hu et al., 2021, Verma et al., 2020), including CEST MRI (Cohen et al., 2018, Glang et al., 2020, Goldenberg et al., 2019). Convolutional neural networks (CNNs) have recently been applied successfully to neuro-oncological imaging (Buda et al., 2020, Chang et al., 2018, Choi et al., 2019, Havaei et al., 2015). However, the number of studies on post-treatment image analysis to predict true progression for patients with malignant gliomas (Bacchi et al., 2019, Li et al., 2020) is still limited. This study aims to develop and verify a CNN-based deep-learning algorithm to identify tumor recurrence through a cross-sectional, multi-modal MRI exam. Our CNN analysis results demonstrate that adding protein-based APTw MRI to traditional structural MR images can significantly increase the accuracy of treatment response assessment compared to using traditional MR images only.
2. Materials and methods
2.1. Patient Enrollment and annotation
This study is a secondary analysis of previously collected data, and part of the data used here have been published (Guo et al., 2021, Jiang et al., 2019, Ma et al., 2016). This study, based on the de-identified data, was approved by the Institutional Review Board (IRB) and the need for consent for this de-identified data reanalysis was waived. Patient inclusion criteria were as follows: ≥20 years old; diagnosis of WHO grade III or IV glioma; status-post initial surgery and chemoradiation or radiotherapy alone; suspected tumor recurrence and completed APTw imaging (in addition to structural MRI sequences) study after the completion of therapy; and an integrated clinical diagnosis of tumor recurrence or treatment effect.
Each lesion was annotated as “response to treatment” (including complete response, partial response, stable disease, radiation necrosis, and pseudoprogression) or “progressive disease“ (including progression and pseudoresponse), according to the RANO criteria for two-dimensional (2D) images for each instance (Vogelbaum et al., 2012, Wen et al., 2010). Here, one instance indicates a set of APTw, T1w, T2w, FLAIR, and GdT1w MR images acquired at the same slice level. For the scan-level classification, scans with one or more slices of “progressive disease” were assigned as “progression”, and all other scans assigned as “response”. Notably, if patients underwent surgery within four weeks after the observed APTw-MRI scan, histopathologic diagnosis took priority over the longitudinal MRI analysis.
2.2. MRI data collection
APTw images were obtained on a 3 T human MRI scanner (Achieva; Philips Medical Systems) using body coil excitation and a 32-channel phased-array coil for reception. Three-dimensional (3D) APTw imaging was based on a previously published sequence (Jiang et al., 2019, Ma et al., 2016), with the following image parameters: radiofrequency saturation duration, 830 ms; saturation power, 2 µT; field of view (FOV), 212×186×66 mm3; resolution, 0.82×0.82×4.4 mm3 (reconstructed); and matrix, 256×256×15 (reconstructed). was acquired with imaging parameters: TR, 4 sec; echo time (TE), 80 ms; 60 slices; thickness, 2.2 mm, and FLAIR was acquired with imaging parameters: TR, 11 sec; TE, 120 ms; inversion recovery time, 2.8 s; 60 slices; thickness, 2.2 mm. and images were acquired with the following parameters: 3D magnetization-prepared-rapid-gradient-echo sequence; TR, 3 s; TE, 3.7 ms; inversion recovery time, 843 ms; flip angle, 8; 150 slices; isotropic voxel, 1.1 ), and the dose of Gd contrast agents was 0.2 mL/kg body weight. The anatomic MRI sequences (T1w, T2w, FLAIR, and GdT1w) had the image parameters: FOV, 212×172×165 (or 212×189×132) mm3; resolution, 0.41×0.41×1.1 mm3 (reconstructed); and matrix, 512×512×150 (reconstructed). For each scan, due to the fact that the 3D APTw MRI protocol provided 15 slices, volumetric MR images used 15 instances. Each instance included T1w, T2w, FLAIR, GdT1w, and APTw images with the matrix shape of 5 (sequences) × 256 (pixels) × 256 (pixels). Instances were the input of proposed slice-level feature extractor CNN.
2.3. Data preprocessing
Data preprocessing steps, including co-registration (Lowekamp et al., 2013), skull-stripping (Lipkova et al., 2019), N4-bias field correction (Tustison et al., 2010), and MRI standardization (Nyúl et al., 2000), were performed sequentially. Notably, based on our experience during the image preprocessing, in order to preserve the distinguishing radiographic patterns on APTw-MRI, MRI scale standardization was not performed on APTw-MRI. We used a rigid-body registration for the co-registration across the APTw images and the anatomical MR images (T1w, T2w, FLAIR, and GdT1w) (Zhang et al., 2016). This was performed through the saturated images at 3.5 ppm to reconstruct the APTw images, which share the same spatial information with the APTw images. Preprocessing was performed by a medical imaging engineer and supervised by a radiologist who also verified the image preprocessing outputs. Lesions of post-treatment malignant glioma that cover the regions of abnormal intensities on multiparameter MR images were segmented on the co-registered FLAIR MR images. Then, the pre-surgery and all clinical follow-up MR images, together with all clinical reports in the electronic medical record system, were reviewed serially for annotation.
2.4. Deep-learning classification pipeline
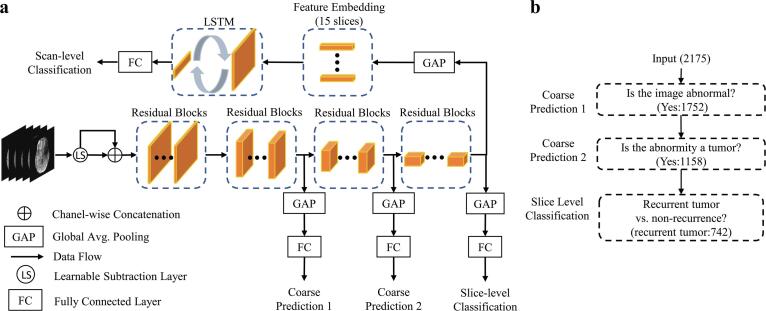
The classification framework consisted of two main stages: slice-level classification and scan-level classification (Fig. 1a). During the slice-level classification, a CNN using three concatenated residual learning blocks (ResNet-18) (He et al., 2016) served as a feature extractor and the backbone model in a previous study. This backbone architecture without any modification was denoted as the standard model. As explained below, we introduced a learnable subtraction module (LS) and a hierarchical classification (HC) paradigm as modifications to the standard ResNet-18 architecture. To aggregate predictions across all slices and obtain a scan-level prediction, a long short-term memory (LSTM) module was added that sequentially processed all embedded feature representations of all slices. The detailed descriptions of each module are presented in the following sub-sections.
Fig. 1.
(a) Overview of the proposed deep-learning pipeline, which consists of a ResNet-18 backbone, a learnable subtraction module, three classification branches for the hierarchical training paradigm, and an LSTM module for scan-level prediction. (b) The schematics of the proposed hierarchical training. The tumor progression classification is decomposed into three binary classification sub-tasks. The number in parentheses is the number of instances reported for the entire dataset.
2.5. Slice-level feature extractor CNN
The classification consists of three hierarchical binary-classification sub-tasks (Fig. 1b). The ResNet-18 architecture was modified according to this hierarchy by inserting classification branches for three binary classification tasks above at increasing depths into the network. This procedure increases gradient flow as additional gradients are injected during back-propagation, and promotes generalizable learning since the extracted features must inform several related tasks rather than only one. Binary cross entropy loss was adopted for each branch and is defined as follows:
where × is the predicted probability and y is the binary indicator (0 or 1) for the target label. The loss function of a CNN is a weighted summation of all branches, , and is defined as follows:
where D is the number of branches, is the binary cross entropy loss of the corresponding branch, and the weight is used to control the relative importance of each branch.
The most accurate slice-level CNN classification framework was used to perform the sanity check.
2.6. Learnable subtraction module and long short-term memory
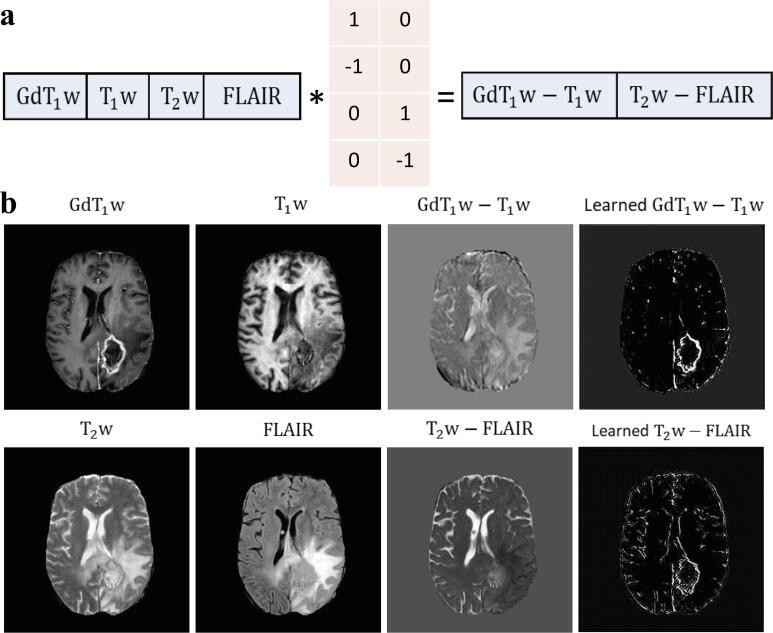
Informed by the radiologic reading workflow for such images, we proposed an LS module on top of the CNN to perform a learnable-parameter-adjusted image subtraction (Fig. 2a). The LS module was calculated between GdT1w and T1w, as well as between T2w and FLAIR images for image comparison. An LSTM was proposed to obtain all extracted, slice-based features from the same scan as the sequential input for scan-level classification.
Fig. 2.
Visual illustration of the concept of the learnable subtraction module. The schematics of the learnable subtraction in a pixel-wise dot product (a) and an example of visualization of image subtraction (b). * denotes the dot product. The learnable parameters for this pixel-wise operation were implemented by a 1×1 convolutional layer with a kernel size of 1 using predefined initializing parameters. Notably, in order to avoid trivial solutions during training, instead of initializing with zero, we set those zero values to a small number (i.e., 1e-3).
2.7. Long short-term memory
The proposed LSTM takes all extracted slice-level features from a scan as a sequential input to perform scan-level diagnosis. For each element of the input sequence, LSTM was computed as follows:
where , , and are the input, cell state, and hidden state for slice t, respectively. is the sigmoid function and is the hyperbolic tangent function. is the Hadamard product. , , , and are the input, forget, cell, and output gates, respectively. We denoted the output features of the last layer of the LSTM as Then, H was passed through a fully connected layer to produce the final scan-level prediction based on all extracted slice-level features of a single scan from the slice-level CNN. Thus, the dependence between input instances and scan-level prediction in our task was modeled.
2.8. Training and implementation details
We adopted the binary cross entropy loss and the Adam (Kingma and Ba, 2015) optimizer to minimize the loss function, with an exponential decay rate β = (0.9, 0.999) for both CNN and LSTM. The initial learning rate was set to 10e−4 and 10e−2 for training CNN and LSTM, respectively. The learning rate of first five epochs was an initial learning rate × 0.1 and constant in the first 50 epochs, and then linearly decayed to zero in the last 50 epochs, with a warm-start, learning-rate scheduler. The batch size was set to 15 and 8 for training CNN and LSTM, respectively. The importance parameter was set to 1 for all branches. The inference time for the proposed CNN and LSTM was 0.018 s per instance and 0.001 s per scan, respectively. We implemented the proposed approach on an Ubuntu 18.04 computer using an NVIDIA 2080Ti GPU and PyTorch. We utilized the class activation map (CAM) to expose the attention of the CNN on the input slices, which highlights the most informative image regions relevant to the predicted class (Zhou et al., 2016). High-response regions with a salience score of higher than the 95th percentile value were further generated, which were denoted by CAM*.
2.9. Statistical analysis and model evaluation
Scan-based data was split into 70% training, 10% validation, and 20% testing according to the chronological order of the MRI scan date (Table 1). The rationale of chronological order data split is to simulate the real-world clinical practice. Our data split reflects the attempt of attaining unbiased performance estimates in prospective deployment. Notably, for the scan-level analysis, all instances from the same scan were treated as an individual sample to prevent data sharing across the split datasets, and the scans from the same patient were not shared between training and testing datasets. The diagnostic performances of the proposed methods, including sensitivity, specificity, and the area under the receiver-operating-characteristic (ROC) curve (AUC), were measured on the testing dataset. Notably, to explicitly evaluate the incremental diagnostic impact of APTw MRI, the ROC curves were compared between inputs with and without APTw (DeLong et al., 1988). We ran all experiments three times and reported the mean metrics. Data analysis was performed with the Python scikit-learn package (Pedregosa et al., 2011).
Table 1.
Participant Demographic Information, and Basic Characteristics of the Datasets.
| Parameter | Dataset |
||
|---|---|---|---|
| Training | Validation | Testing | |
| No. of instancesa | 1530 | 210 | 435 |
| No. of scans | 102 | 14 | 29 |
| No. of patients | 72 | 10 | 16 |
| No. of females (scan based, %) | 31 (30.4%) | 2 (14.3%) | 16 (55.2%) |
| Age (scan based, year) | 52.7 | 46.1 | 50.4 |
| No. of grade IV (%) | 63 (61.8%) | 9 (64.3%) | 22 (75.9%) |
| Time interval after radiation (day) b | 257 (95–448) | 183 (89–375) | 287 (190–542) |
| No. of first progression (%) | 66 (64.7%) | 5 (35.7%) | 11 (37.9%) |
| No. of progression at the slice level (%) | 547 (35.8%) | 59 (28.1%) | 136 (31.3%) |
| No. of progression at the scan level (%) | 61 (59.8%) | 7 (50.0%) | 18 (62.1%) |
| Enrollment time of first scan (YYYY/MM) c | 2010/04 | 2014/10 | 2015/07 |
| Enrollment time of last scan (YYYY/MM) c | 2014/10 | 2015/07 | 2018/02 |
Note. a One instance indicates a set of APTw, T1w, T2w, FLAIR, and GdT1w MR images acquired at the same slice level. b Time interval after radiation indicates the timing of research MRI acquisition with regard to the end of radiation, described as median with 25th percentile and 75th percentile.
To simulate the practical use of this algorithm in a simulated prospective study, the whole dataset was split according to the chronological order of the MRI scan date.
3. Results
3.1. Patient demographic information
A total of 145 scans obtained from 98 patients between April 2010 and February 2018 were included in this study. Patient demographic information and basic lesion characteristics are shown in Table 1. Based on the integrated clinical pathologic results, 86 scans were classified as “progression“, while the remaining 59 scans were classified as “response”. Based on the slice-level features, this led to 2175 instances in total, 742 of which were grouped as “progressive disease“.
3.2. Slice-level diagnostic performance
The slice-level classification performance was first evaluated in the standard CNN model, where the contribution of using GdT1w and APTw as a part of the input was investigated (Table 2). In the slice-level backbone CNN model using , , and FLAIR MRI data as the baseline input, the AUC for distinguishing progressive disease from non-progression was 0.77 (CI, 0.70–0.81). Adding or APTw MRI alone to the baseline input, the AUCs were increased to 0.82 (CI, 0.79–0.87) or 0.84 (CI, 0.81–0.89), respectively. Adding GdT1w and APTw MRI jointly achieved the highest AUC, with 0.85 (CI, 0.82–0.89). This input ablation study indicates that the use of and APTw MRI data, either individually or jointly, significantly improved the progressive disease vs. non-progression classification. Moreover, APTw MRI yielded a marginally higher value compared to .
Table 2.
Comparisons of performances with different MRI sequence data as input for the slice-level classification in the backbone model.
| Data Input |
Diagnostic Performances |
|||||||
|---|---|---|---|---|---|---|---|---|
| APTw | GdT1w | T1w | T2w | FLAIR | AUC (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) | P value# |
| √ | √ | √ | 0.77 (0.70, 0.81) | 0.61 (0.53, 0.69) | 0.81 (0.77, 0.85) | – | ||
| √ | √ | √ | √ | 0.82 (0.79, 0.87) | 0.82 (0.75, 0.88) | 0.75 (0.70, 0.80) | P < 0.001 | |
| √ | √ | √ | √ | 0.84 (0.81, 0.89) | 0.87 (0.81, 0.92) | 0.68 (0.62, 0.73) | P < 0.001 | |
| √ | √ | √ | √ | √ | 0.85 (0.82, 0.89) | 0.92 (0.87, 0.96) | 0.63 (0.58, 0.69) | P < 0.001 |
Note. √ indicates that the input instance contains the corresponding MR sequence.
# P value for the ROC curve comparison with respect to the input of T1w, T2w and FLAIR data.
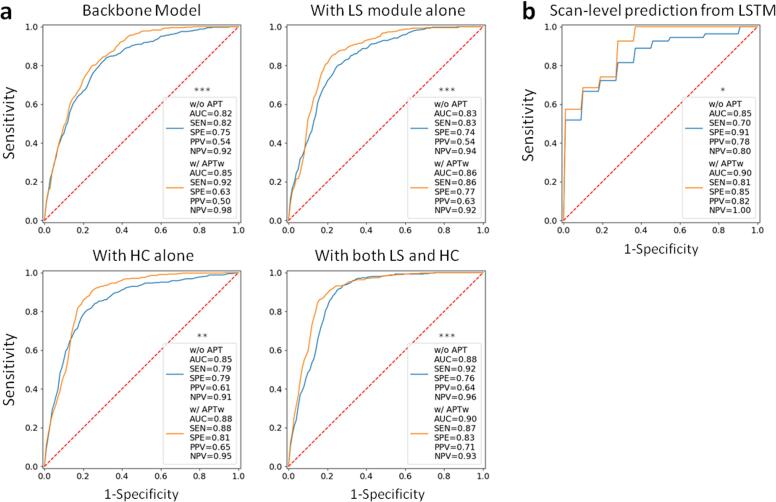
A thorough ablation study to analyze the separate or joint effects of the LS module and a HC paradigm was further performed (Fig. 3a). For input without APTw data (, , FLAIR, and ), the AUCs were increased from 0.82 to 0.83 (adding the LS module), to 0.85 (adding the HC paradigm), and to 0.88 (adding them jointly), respectively. For input with APTw images (APTw, , , FLAIR, and ), the corresponding AUCs were increased from 0.85 to 0.86, to 0.88, and to 0.90. Adding APTw images is capable of significantly improving the classification performance compared to the structural images, with the LS module and the HC paradigm, either jointly (P < 0.01) or separately (p < 0.001 and p < 0.01 respectively), Notably, the best performance was achieved by the model that employed the LS module and the HC paradigm jointly, using APTw MRI data (AUC, 0.90; sensitivity, 0.87; specificity, 0.83). In addition, images of learned GdT1w – T1w and T2w – FLAIR subtractions, as the output of LS, show the distinguishable features that the module learned from the comparisons between GdT1w and T1w images, as well as between T2w and FLAIR images (Fig. 2b).
Fig. 3.
(a) ROC curves showing the slice-level diagnostic performance of the models evaluated (the standard model and with the LS module and HC paradigm, alone and jointly; two inputs, structural MR images, including , , FLAIR, and GdT1w, without or with APTw MRI data). (b) ROC curves showing the scan-level prediction from the LSTM. *, **, and *** denote the ROC comparison (with APTw vs. without APTw data) results with P < 0.05, < 0.01 and < 0.001, respectively.
Two examples that applied CAM to generate saliency maps that highlighted the regions on which the model focused during inference are shown in Fig. 4. The derived CAMs, and especially the CAM*s, covered the vast majority of lesions, including the Gd-enhancing regions, surgical cavity, and edema areas for both lesions with progressive disease and response.
Fig. 4.
Examples of applying CAM to generate saliency maps for an instance with progressive disease (a) and an instance with non-progressive disease (b). An artifact from the adjacent nasal sinus (white arrow) is visible on the APTw image of case (a). For the annotation, the cavity (including surgical cavity and cavity with liquefactive necrosis) within the lesion was labeled for each instance. In addition to the tumor portion (annotated as either progression or response) and cavity, the remaining portion of the lesion was defined as edema. On the lesion masks, gray, blue, yellow, and green represent the annotated portions with edema, cavity, tumor progression, and tumor response, respectively. High-response regions with a salience score of higher than the 95th percentile value were denoted by CAM*. On CAM and CAM*, red regions indicate a stronger contribution and blue regions have little to no contribution toward the classification. They, especially CAM*, cover the majority of lesions, including the gadolinium-enhancing regions, surgical cavity, and edema areas for instances of both progressive disease and non-progression. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. Scan-level diagnostic performance
The AUCs for the classification of true progression versus response were 0.85 (sensitivity, 0.70; specificity, 0.91) and 0.90 (sensitivity, 0.81; specificity, 0.85) for input without and with APTw images, respectively (Fig. 3b). A significant incremental value of APTw images over structural MRIs was also observed in the scan-level classifier (P < 0.05).
4. Discussion
Determining whether a person with malignant glioma has tumor progression versus response after chemoradiation remains pivotal in the postoperative management of people with malignant gliomas. We present a multiparameter MRI processing pipeline for the classification of post-treatment patients with malignant gliomas, which provides an end-to-end model for both slice-level and scan-level assessment. For 145 scans from 98 patients, we made the following observations: (i) multiparameter MRIs, including structural MRIs and APTw MRIs, are capable of building a compelling deep-learning classification model; (ii) APTw MRI provides incremental diagnostic performance compared to structural MRIs as the input for both slice-level and scan-level classifications; (iii) LS and HC are able to improve the classification performance for the backbone model; and (iv) generated CAMs, especially CAM*s, visually highlighting the evidence for classification, localize the majority of abnormalities on structural MRIs and APTw MRI.
Our proposed approach has the following three advantages. First, we developed the LS module. When assessing MR images, besides other imaging modalities, patient history and lab tests, radiologists usually observe the Gd-enhancement pattern and the water-signal suppression pattern (Eisele et al., 2016, Wen et al., 2010, Wolf, 2019) by comprehensively comparing the GdT1w and T1w images, as well as the T2w and FLAIR images. However, direct image-subtraction might not be optimal for the subsequent feature extraction due to the variety of the original intensities on the structural MRIs. The learnable-parameter-adjusted image subtraction derived from the proposed LS demonstrated more discriminative features (Fig. 2), thus improving the classification performance both with and without APTw data. Second, we introduced the HC paradigm to the CNN by decomposing the final task into three hierarchic subtasks (Kowsari et al., 2017, Zhu and Bain, 2017). The shallower layers in the CNN can provide hierarchical priors for the subsequent deeper layers, thus serving as an extra supervised guidance on the intermediate layers. As a result, without increasing the depth of the network, these two coarse prediction branches boosted the performance in this study. Last, because of the extreme intratumoral heterogeneity, it is not uncommon that a progressive lesion partly contains some regions that either are not tumor (i.e., cavity or edema) or non-progressive aspects of the tumor (details in Methods). In this scenario, simply averaging the slice-level logits from the CNN will result in impractical high specificity and low sensitivity for scan-level classification. In this study, slice-level pipeline is more specific for regional analysis, which is for essential for diagnostic surgery procedure, while the scan-level pipeline corresponds to patient/case diagnosis. Thus, the slice-level and scan-level diagnostic performances are slightly different. They might be less comparable due to distinguishing clinical scenarios. An LSTM (Hochreiter and Schmidhuber, 1997) was recently introduced to multiparametric MRI data with favorable results (Lee et al., 2020). Inspired by this, we proposed a set of data from the five MRI sequences as a spatial sequence input for the scan-level classification framework. This LSTM model could thus incorporate the entire information via 75 MR slices (15 instances × 5 MR sequences) affiliated with a scan for the scan-level classification. Furthermore, using chronological order data split in this study reflects the attempt of attaining unbiased performance estimates in prospective deployment. However, with this data split method, the clinical and demographic characteristics could barely be controlled to evenly distribute among data splits, such as gender ratio or first progression v.s. in subsequent therapy this study.
In this study, pairs of models using data with and without APTw MRI were compared to explore the incremental value of APTw MRI to structural MRIs. Our data show that APTw MRI improves the diagnostic performance for both slice-level and scan-level classifications. This is consistent with previous studies that demonstrated the value of APTw MRI for post-treatment malignant gliomas (Zhou et al., 2019). Furthermore, CAMs were calculated from the slice-level CNN. As a visualization and attribution method with which to elucidate CNNs, CAM explanations correspond to the gradient of the class score (logit) with respect to the feature map of the last convolutional unit of a CNN (Selvaraju et al., 2016). This mapping uses the gradients of any target concept flowing into the final convolutional layer to produce a coarse localization map, that highlighs the important regions in the image for predicting the concept. CAM is capable of generating images that highlight salient regions, arguably acting as a localizer for important regions that contain highly discriminative information, with great promise for clinical translation (Fong and Vedaldi, 2017, Sundararajan et al., 2017). The CAMs, especially the CAM*s of our study, substantially localized the majority of regions of abnormal MRI signals (Fig. 4). The results indicate that the proposed methods can successfully extract truly and biologically relevant distinguishable information from multi-modality MRIs.
There are several limitations to this study. First, the proposed models were trained and tested on single-center data. Our next work will incorporate data from multiple external institutions to create a generalizable algorithm. Second, for the scan-level prediction, the proposed method was developed to use the embedded 2D-CNN-features. An alternative is to directly use volumetric data to build 3D CNN algorithms. However, there was a discrepancy in resolution along the z-axis between APTw images and the structural MR sequences (4.4 mm vs. 1.1 mm). Consequently, a dramatically compromised fidelity for APTw images due to resampling for an isotropic 3D volume, impedes a 3D CNN. Third, several critical genetic markers, such as the status of isocitrate dehydrogenase (IDH) mutation, 1p19q codeletion and (O-6-methylguanine-DNA methyltransferase (MGMT) methylation, were not included in the CNN due to limited, assessable genetic data. Only with these additional data, the added value of APTw imaging in these definite glioma patient subsets can be established. We will collect and analyze genetic profiles in our ongoing prospective study. Forth, perfusion MRI, an advanced MR imaging with great diagnostic value for post-treatment glioma patients, was not included in the analysis of this study. The lack of any analysis with perfusion and APTw images left an unexplored question for the further study. Last but not least, malignant gliomas infiltrated throughout the whole brain. However, APTw MRI only covered up to 66 mm in the z direction (4.4 mm × 15 slices) due to the technique limitation. This led to the fact that a whole brain comparison was not performed in this study.
5. Conclusion
We propose a deep learning-based pipeline to identify true tumor progression versus treatment effects after radiation for patients with malignant glioma utilizing multiparameter MRIs. The proposed learnable subtraction layer shows promise, which indicates the improvement on the data usage plays an important role in increasing automated analysis outcome. The boosted performance achieved by supervising the natural hierarchy of general-to-specific order under targets class demonstrates that the proposed hierarchical classification paradigm can provide prior for deeper layers as a good guide during the training. The AUCs of our best-performing models (0.90 for both slice-level and scan-level models) verifies our motivation that complementing structural with functional APTw MRI can further improve the diagnostic performance. Based on this performance, the proposed method could be a highly efficient solution that could help clinical experts to make precise diagnoses for patients with post-treatment malignant gliomas.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported in part by grants from the National Institutes of Health (R37CA248077, R01CA228188, and P41031771). The authors thank Ms. Lindsay Blair for patient coordination and Ms. Mary McAllister for editorial assistance.
Data availability
The authors do not have permission to share data.
References
- Bacchi S., Zerner T., Dongas J., Asahina A.T., Abou-Hamden A., Otto S., Oakden-Rayner L., Patel S. Deep learning in the detection of high-grade glioma recurrence using multiple MRI sequences: A pilot study. J Clin Neurosci. 2019;70:11–13. doi: 10.1016/j.jocn.2019.10.003. [DOI] [PubMed] [Google Scholar]
- Buda M., AlBadawy E.A., Saha A., Mazurowski M.A. Deep Radiogenomics of Lower-Grade Gliomas: Convolutional Neural Networks Predict Tumor Genomic Subtypes Using MR Images. Radiol Artif Intell. 2020;2(1):e180050. doi: 10.1148/ryai.2019180050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- K. Chang, K., Bai, H.X., Zhou, H., Su, C., Bi, W.L., Agbodza, E., Kavouridis, V.K., Senders, J.T., Boaro, A., Beers, A. 2018. Residual convolutional neural network for the determination of IDH status in low-and high-grade gliomas from MR imaging. Clinical Cancer Research. 24. 1073-1081. [DOI] [PMC free article] [PubMed]
- Choi K.S., Choi S.H., Jeong B. Prediction of IDH genotype in gliomas with dynamic susceptibility contrast perfusion MR imaging using an explainable recurrent neural network. Neuro-oncology. 2019;21:1197–1209. doi: 10.1093/neuonc/noz095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen O., Huang S., McMahon M.T., Rosen M.S., Farrar C.T. Rapid and quantitative chemical exchange saturation transfer (CEST) imaging with magnetic resonance fingerprinting (MRF) Magn Reson Med. 2018;80(6):2449–2463. doi: 10.1002/mrm.27221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong E.R., DeLong D.M., Clarke-Pearson D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- Eijgelaar R.S., Visser M., Müller D.M.J., Barkhof F., Vrenken H., van Herk M., Bello L., Conti Nibali M., Rossi M., Sciortino T., Berger M.S., Hervey-Jumper S., Kiesel B., Widhalm G., Furtner J., Robe P.A.J.T., Mandonnet E., De Witt Hamer P.C., de Munck J.C., Witte M.G. Robust Deep Learning-based Segmentation of Glioblastoma on Routine Clinical MRI Scans Using Sparsified Training. Radiol Artif Intell. 2020;2(5):e190103. doi: 10.1148/ryai.2020190103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisele S.C., Wen P.Y., Lee E.Q. Assessment of brain tumor response: RANO and its offspring. Curr. Treat. Options Oncol. 2016;17:35. doi: 10.1007/s11864-016-0413-5. [DOI] [PubMed] [Google Scholar]
- Fong, R.C., Vedaldi, A., 2017. Interpretable explanations of black boxes by meaningful perturbation. Proceedings of the IEEE International Conference on Computer Vision, pp. 3429-3437.
- Glang F., Deshmane A., Prokudin S., Martin F., Herz K., Lindig T., Bender B., Scheffler K., Zaiss M. DeepCEST 3T: Robust MRI parameter determination and uncertainty quantification with neural networks-application to CEST imaging of the human brain at 3T. Magn Reson Med. 2020;84(1):450–466. doi: 10.1002/mrm.28117. [DOI] [PubMed] [Google Scholar]
- Goldenberg J.M., Cárdenas-Rodríguez J., Pagel M.D. Machine learning improves classification of preclinical models of pancreatic cancer with chemical exchange saturation transfer MRI. Magn Reson Med. 2019;81(1):594–601. doi: 10.1002/mrm.27439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P., Wang P., Yasarla R., Zhou J., Patel V.M., Jiang S. Anatomic and molecular MR image synthesis using confidence guided CNNs. IEEE Trans. Med. Imaging. 2021;40(10):2832–2844. doi: 10.1109/TMI.2020.3046460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaei, M., Dutil, F., Pal, C., Larochelle, H., Jodoin, P.-M. 2015. A convolutional neural network approach to brain tumor segmentation. BrainLes. 2015. Springer. 195-208.
- He, K., Zhang, X., Ren, S., Sun, J. 2016. Deep residual learning for image recognition. Proceedings of the IEEE conference on computer vision and pattern recognition. 770-778.
- Hochreiter S., Schmidhuber J. Long short-term memory. Neural Comput. 1997;9(8):1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- Hu Q., Whitney H.M., Li H., Ji Y.u., Liu P., Giger M.L. Improved Classification of Benign and Malignant Breast Lesions Using Deep Feature Maximum Intensity Projection MRI in Breast Cancer Diagnosis Using Dynamic Contrast-enhanced MRI. Radiol Artif Intell. 2021;3(3):e200159. doi: 10.1148/ryai.2021200159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, S., Eberhart, C.G., Lim, M., Heo, H.-Y., Zhang, Y., Blair, L., Wen, Z., Holdhoff, M., Lin, D., Huang, P., Qin, H., Quinones-Hinojosa, A., Weingart, J.D., Barker, P.B., Pomper, M.G., Laterra, J., van Zijl, P.C.M., Blakeley, J.O., Zhou, J. 2019. Identifying recurrent malignant glioma after treatment using amide proton transfer-weighted MR imaging: A validation study with image-guided stereotactic biopsy. Clin. Cancer Res. 25. 552-561. [DOI] [PMC free article] [PubMed]
- Jiang S., Eberhart C.G., Zhang Y., Heo H.-Y., Wen Z., Blair L., Qin H., Lim M., Quinones-Hinojosa A., Weingart J.D., Barker P.B., Pomper M.G., Laterra J., van Zijl P.C.M., Blakeley J.O., Zhou J. Amide proton transfer-weighted magnetic resonance image-guided stereotactic biopsy in patients with newly diagnosed gliomas. Eur. J. Cancer. 2017;83:9–18. doi: 10.1016/j.ejca.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Wen Z., Ahn S.S., Cai K., Paech D., Eberhart C.G., Zhou J. Applications of chemical exchange saturation transfer magnetic resonance imaging in identifying genetic markers in gliomas. NMR Biomed. 2022;e4731 doi: 10.1002/nbm.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingma, D.P., Ba, J. 2015. Adam: A method for stochastic optimization. 3rd International Conference for Learning Representations. San Diego.
- Kowsari K., Brown D.E., Heidarysafa M., Meimandi K.J., Gerber M.S., Barnes L.E. 2017 16th IEEE international conference on machine learning and applications (ICMLA). IEEE. 2017. Hdltex: Hierarchical deep learning for text classification; pp. 364–371. [Google Scholar]
- Lee D.H., Heo H.Y., Zhang K., Zhang Y., Jiang S., Zhao X., Zhou J. Quantitative assessment of the effects of water proton concentration and water T1 changes on amide proton transfer (APT) and nuclear overhauser enhancement (NOE) MRI: The origin of the APT imaging signal in brain tumor. Magn. Reson. Med. 2017;77:855–863. doi: 10.1002/mrm.26131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Wang N., Turk S., Mohammed S., Lobo R., Kim J., Liao E., Camelo-Piragua S., Kim M., Junck L., Bapuraj J., Srinivasan A., Rao A. Discriminating pseudoprogression and true progression in diffuse infiltrating glioma using multi-parametric MRI data through deep learning. Sci Rep. 2020;10:20331. doi: 10.1038/s41598-020-77389-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Tang H., Chan M.D., Zhou X., Qian X. DC-AL GAN: Pseudoprogression and true tumor progression of glioblastoma multiform image classification based on DCGAN and AlexNet. Med Phys. 2020;47(3):1139–1150. doi: 10.1002/mp.14003. [DOI] [PubMed] [Google Scholar]
- Lipkova J., Angelikopoulos P., Wu S., Alberts E., Wiestler B., Diehl C., Preibisch C., Pyka T., Combs S.E., Hadjidoukas P., Van Leemput K., Koumoutsakos P., Lowengrub J., Menze B. Personalized Radiotherapy Design for Glioblastoma: Integrating Mathematical Tumor Models, Multimodal Scans, and Bayesian Inference. IEEE transactions on medical imaging. 2019;38(8):1875–1884. doi: 10.1109/TMI.2019.2902044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Li C., Chen Y., Lv X., Lv Y., Zhou J., Xi S., Dou W., Qian L., Zheng H., Wu Y., Chen Z. Diagnostic performance of multiparametric MRI in the evaluation of treatment response in glioma patients at 3T. Journal of Magnetic Resonance Imaging. 2020;51(4):1154–1161. doi: 10.1002/jmri.26900. [DOI] [PubMed] [Google Scholar]
- Lowekamp B.C., Chen D.T., Ibáñez L., Blezek D. The design of SimpleITK. Frontiers in neuroinformatics. 2013;7:45. doi: 10.3389/fninf.2013.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B.o., Blakeley J.O., Hong X., Zhang H., Jiang S., Blair L., Zhang Y.i., Heo H.-Y., Zhang M., van Zijl P.C.M., Zhou J. Applying amide proton transfer-weighted MRI to distinguish pseudoprogression from true progression in malignant gliomas. J. Magn. Reson. Imaging. 2016;44(2):456–462. doi: 10.1002/jmri.25159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyúl L.G., Udupa J.K., Zhang X. New variants of a method of MRI scale standardization. IEEE transactions on medical imaging. 2000;19:143–150. doi: 10.1109/42.836373. [DOI] [PubMed] [Google Scholar]
- Park J.E., Kim H.S., Park S.Y., Jung S.C., Kim J.H., Heo H.-Y. Identification of Early Response to Anti-Angiogenic Therapy in Recurrent Glioblastoma: Amide Proton Transfer–weighted and Perfusion-weighted MRI compared with Diffusion-weighted MRI. Radiology. 2020;295(2):397–406. doi: 10.1148/radiol.2020191376. [DOI] [PubMed] [Google Scholar]
- Park Y.W., Ahn S.S., Kim E.H., Kang S.-G., Chang J.H., Kim S.H., Zhou J., Lee S.-K. Differentiation of recurrent diffuse glioma from treatment-induced change using amide proton transfer imaging: incremental value to diffusion and perfusion parameters. Neuroradiology. 2021;63(3):363–372. doi: 10.1007/s00234-020-02542-5. [DOI] [PubMed] [Google Scholar]
- Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V. Scikit-learn: Machine learning in Python. the Journal of machine Learning research. 2011;12:2825–2830. [Google Scholar]
- Ray, K.J., Simard, M.A., Larkin, J.R., Coates, J., Kinchesh, P., Smart, S.C., Higgins, G.S., Chappell, M.A., Sibson, N.R. 2019. Tumor pH and protein concentration contribute to the signal of amide proton transfer magnetic resonance imaging. Cancer Research 79. 1343-1352. [DOI] [PMC free article] [PubMed]
- Selvaraju, R.R., Das, A., Vedantam, R., Cogswell, M., Parikh, D., Batra, D., 2016. Grad-CAM: Why did you say that? arXiv preprint arXiv:1611.07450.
- Sotirios B., Demetriou E., Topriceanu C.C., Zakrzewska Z. The role of APT imaging in gliomas grading: A systematic review and meta-analysis. Eur. J. Radiol. 2020;133:109353. doi: 10.1016/j.ejrad.2020.109353. [DOI] [PubMed] [Google Scholar]
- Stupp R., Mason W.P., van den Bent M.J., Weller M., Fisher B., Taphoorn M.J.B., Belanger K., Brandes A.A., Marosi C., Bogdahn U., Curschmann J., Janzer R.C., Ludwin S.K., Gorlia T., Allgeier A., Lacombe D., Cairncross J.G., Eisenhauer E., Mirimanoff R.O. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- Sundararajan, M., Taly, A., Yan, Q., 2017. Axiomatic attribution for deep networks. Proceedings of the 34th International Conference on Machine Learning-Volume 70. JMLR. org, pp. 3319-3328.
- Tustison N.J., Avants B.B., Cook P.A., Yuanjie Zheng, Egan A., Yushkevich P.A., Gee J.C. N4ITK: improved N3 bias correction. IEEE transactions on medical imaging. 2010;29(6):1310–1320. doi: 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R., Correa R., Hill V.B., Statsevych V., Bera K., Beig N., Mahammedi A., Madabhushi A., Ahluwalia M., Tiwari P. Tumor Habitat-derived Radiomic Features at Pretreatment MRI That Are Prognostic for Progression-free Survival in Glioblastoma Are Associated with Key Morphologic Attributes at Histopathologic Examination: A Feasibility Study. Radiol Artif Intell. 2020;2(6):e190168. doi: 10.1148/ryai.2020190168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelbaum M.A., Jost S., Aghi M.K., Heimberger A.B., Sampson J.H., Wen P.Y., Macdonald D.R., Van den Bent M.J., Chang S.M. Application of novel response/progression measures for surgically delivered therapies for gliomas: Response assessment in neuro-oncology (RANO) working group. Neurosurgery. 2012;70:234–243. doi: 10.1227/NEU.0b013e318223f5a7. [DOI] [PubMed] [Google Scholar]
- Ward K.M., Aletras A.H., Balaban R.S. A new class of contrast agents for MRI based on proton chemical exchange dependent saturation transfer (CEST) J. Magn. Reson. 2000;143(1):79–87. doi: 10.1006/jmre.1999.1956. [DOI] [PubMed] [Google Scholar]
- Wen P.Y., Macdonald D.R., Reardon D.A., Cloughesy T.F., Sorensen A.G., Galanis E., DeGroot J., Wick W., Gilbert M.R., Lassman A.B., Tsien C., Mikkelsen T., Wong E.T., Chamberlain M.C., Stupp R., Lamborn K.R., Vogelbaum M.A., van den Bent M.J., Chang S.M. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. Journal of Clinical Oncology. 2010;28(11):1963–1972. doi: 10.1200/JCO.2009.26.3541. [DOI] [PubMed] [Google Scholar]
- Wolf R.L. MRI of Recurrent Glioblastoma: Reliability and Reality. Radiology. 2019;290(2):477–478. doi: 10.1148/radiol.2018182530. [DOI] [PubMed] [Google Scholar]
- Yan K., Fu Z., Yang C., Zhang K., Jiang S., Lee D.H., Heo H.Y., Zhang Y., Cole R.N., Van Eyk J.E., Zhou J. Assessing amide proton transfer (APT) MRI contrast origins in 9L gliosarcoma in the rat brain using proteomic analysis. Mol. Imaging Biol. 2015;17:479–487. doi: 10.1007/s11307-015-0828-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.i., Heo H.-Y., Lee D.-H., Zhao X., Jiang S., Zhang K., Li H., Zhou J. Selecting the reference image for registration of CEST series. Journal of Magnetic Resonance Imaging. 2016;43(3):756–761. doi: 10.1002/jmri.25027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, B., Khosla, A., Lapedriza, A., Oliva, A., Torralba, A. 2016. Learning deep features for discriminative localization. Proceedings of the IEEE conference on computer vision and pattern recognition. 2921-2929.
- Zhou J., Heo H.-Y., Knutsson L., van Zijl P.C.M., Jiang S. APT-weighted MRI: Techniques, current neuro applications, and challenging issues. J. Magn. Reson. Imaging. 2019;50(2):347–364. doi: 10.1002/jmri.26645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Zaiss M., Knutsson L., Sun P.Z., Ahn S.S., Aime S., Bachert P., Blakeley J.O., Cai K., Chappell M.A., Chen M., Gochberg D.F., Goerke S., Heo H.-Y., Jiang S., Jin T., Kim S.-G., Laterra J., Paech D., Pagel M.D., Park J.E., Reddy R., Sakata A., Sartoretti-Schefer S., Sherry A.D., Smith S.A., Stanisz G.J., Sundgren P.C., Togao O., Vandsburger M., Wen Z., Wu Y., Zhang Y., Zhu W., Zu Z., van Zijl P.C.M. Review and consensus recommendations on clinical APT-weighted imaging approaches at 3T: Application to brain tumors. Magn. Reson. Med. 2022 doi: 10.1002/mrm.29241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X., Bain, M., 2017. B-CNN: branch convolutional neural network for hierarchical classification. arXiv preprint arXiv:1709.09890.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors do not have permission to share data.