Summary
We developed a multi-unit microscope for all-optical inter-layers circuits interrogation. The system performs two-photon (2P) functional imaging and 2P multiplexed holographic optogenetics at axially distinct planes. We demonstrated the capability of the system to map, in the mouse retina, the functional connectivity between rod bipolar cells (RBCs) and ganglion cells (GCs) by activating single or defined groups of RBCs while recording the evoked response in the GC layer with cell-type specificity and single-cell resolution. We then used a logistic model to probe the functional connectivity between cell types by deriving the “cellular receptive field” describing how RBCs impact each GC type. With the capability to simultaneously image and control neuronal activity at axially distinct planes, the system enables a precise interrogation of multi-layered circuits. Understanding this information transfer is a promising avenue to dissect complex neural circuits and understand the neural basis of computations.
Keywords: computer-generated holography, temporal focusing, retina, optogenetics, two-photon excitation, calcium imaging
Graphical abstract
Highlights
-
•
An optical system is developed for the study of inter-layer retinal circuits
-
•
The system achieves holographic stimulation and imaging on axially distinct planes
-
•
Functional connectivity is mapped between rod bipolar and ganglion cells
-
•
Potential to map functional inter-layers connectivity in different brain regions
Motivation
Understanding how signals propagate through the multiple retina cell layers is of key importance to optimize the illumination schemes for high-resolution visual restoration through optogenetic therapy.
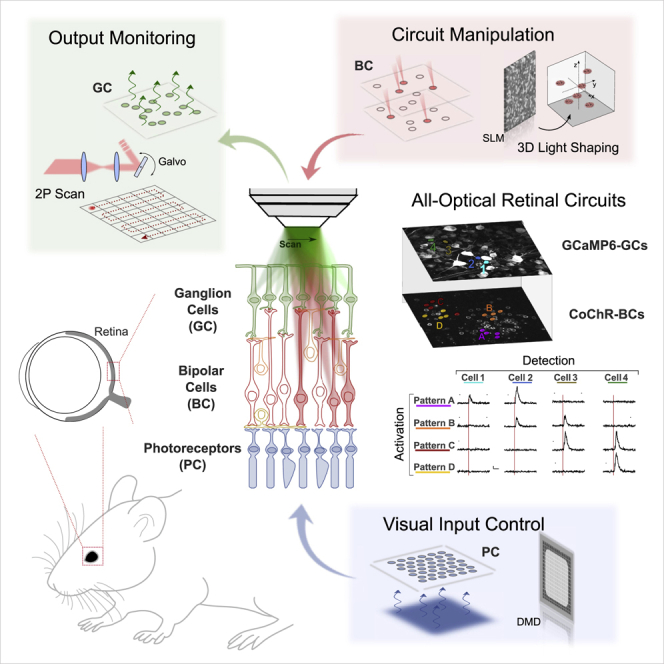
To enable these types of studies, we demonstrate the use of a multi-unit microscope combining holographic light pattering and functional imaging. The system enables manipulating the retinal input at the photoreceptors or at the inner retina layers and analyzing the evoked responses at the ganglion cell layer with cell-type specificity and single-cell resolution. The same approach can enable all-optical inter-layers circuit interrogation in different brain regions.
Spampinato et al. demonstrate an optical system for two-photon functional imaging and holographic optogenetic photostimulation at axially distinct planes. The system is used to map the functional connectivity between bipolar cells and ganglion cells in the mouse retina. This approach can enable all-optical inter-layers circuit interrogation in different brain regions.
Introduction
The optogenetics revolution started with the discovery of the genes Channelrhodopsin-2 (Nagel et al., 2003) and Halorhodopsin (Zhang et al., 2007) and of their light sensitivity, and continued with the demonstration of their expression in neuronal cells (Boyden et al., 2005). Here, light-induced conformational changes of the two opsins permit the direct transduction of photons into electrical currents of opposite polarities, thus activating or inhibiting neuronal signals non-invasively. So far, there have been two main applications for optogenetics: the manipulation of neuronal circuits to establish the causal role of specific patterns of neural activity and cell types in the control of behaviors or pathologies (Boyden, 2015), and the expression of optogenetic actuators or inhibitors in the retina for vision restoration (Roska and Sahel, 2018).
In the latter application, the transgenic expression of photosensitive opsins in specific retinal cell types is used to restore the conversion of light into an electric signal, which is otherwise compromised in the case of retinal dystrophies. Specifically, a pioneering study on vision restoration has shown that halorhodopsin expression in non-functional but surviving “dormant” cones preserves the propagation of visual inputs in all layers of the retina and restore visual responses in mice (Busskamp et al., 2010). Alternatively, in pathological states that leave an insufficient concentration (or absence) of remaining cones, a photosensitive opsin can be expressed in the inner or ganglion retinal cell layers (Bi et al., 2006; Gauvain et al., 2021). In this last case, it is preferable to target neurons that lie upstream in the circuit, as this enables restoration of vision as close as possible to natural vision (Busskamp and Roska, 2011). This makes targeting of bipolar cells (BCs), when possible, the preferred strategy. Recently, the design of potent AAV capsids, combined with specific enhancing sequences (Chaffiol et al., 2017; Lu et al., 2016; Macé et al., 2015), enables improved gene delivery to a large number of ON-BC types. Pioneering experiments in mice retina have shown that optogenetic targeting of BCs (Chaffiol et al., 2017; Lu et al., 2016; Macé et al., 2015) enables reactivation of ON and OFF visual pathways (Chaffiol et al., 2017; Lu et al., 2016; Macé et al., 2015). In these first investigations, full-field flashes of visible light were used to excite all of the retinal bipolar layer. This proved to be sufficient to induce light-evoked locomotory behavior (Macé et al., 2015) but lacked the spatial resolution to reproduce physiological activity pattern both in the BCs and ganglion cells (GCs). This will be of key importance to understand retinal processing and how to optimize light stimulation to restore high-resolution vision, but until now has remained technologically out of reach.
To refine these types of studies, we have designed a multi-unit microscope capable of manipulating the retinal input at the photoreceptors or at the inner retina layers and to analyze the evoked responses at the GC layer with cell type specificity and single-cell resolution. Precise control of the activation of genetically identified cells in the inner layers of the retina is achieved through the use of two-photon (2P) excitation with temporally focused holographic beams combined with the use of highly specific AAV capsids for opsin expression (Lu et al., 2016). Spatiotemporal recording of light-evoked signals on the GC layer is done using 2P Ca2+ imaging. Patterned excitation of the photoreceptors using visible light modulation through a digital micromirror device (DMD) is used to classify the different GC types.
We demonstrate the capabilities of the system to investigate the rod bipolar cell (RBC) pathway, one of the main circuits that processes the signal coming from the rod photoreceptors, and with known anatomical connectivity. Specifically, we demonstrated the use of the system for high-throughout functional connectivity mapping among RBCs and multiple GC types. To derive, at each excitation of the BCs, the response of all GCs contained in the field of view (FOV), we used 2P GCaMP imaging. Since GCaMP does not have the necessary sensitivity to reveal subthreshold responses generated by the excitation of one cell at a time, we sequentially activated groups of 2 to 10 cells. Using a logistic model, we could estimate from the responses to many different patterns of stimulation the “cellular receptive field,” describing how the activation of a defined RBC impacts each GC type.
The suggested experimental strategy can be extended to manipulate the input/output signals of other retina pathways and dissect their functioning and be extended to the investigation of functional inter-layer connectivity in different brain regions.
Results
The multi-unit optical system
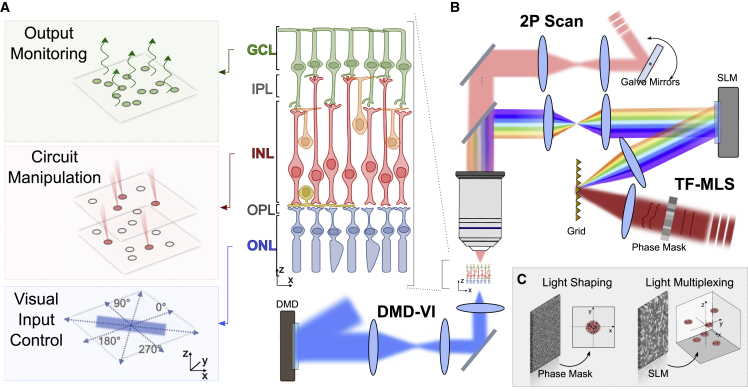
The optical microscope (Figure 1 and S1A) comprises a first module for patterned photoreceptor illumination where visible light from a light-emitting diode is projected on a DMD conjugated to the sample plane through a condenser. This enables the projection of spatiotemporally controlled full-field stimuli at variable spatial and temporal frequencies on the retina oriented with the photoreceptor on the bottom side of the recording chamber. A second module includes a compact multiplexed temporally focused light-shaping (MTF-LS) module (Accanto et al., 2018) composed of a fused silica phase mask coupled to a temporally focused multiplexing unit for the generation of arbitrarily defined and axially confined 3D excitation patterns through the upper objective. Illumination was provided by a fiber amplifier laser source, emitting at 1,030 nm and operating at 500 kHz repetition rate (Satsuma HP, Amplitude) for efficient 2P optogenetic activation (Chaigneau et al., 2016; Ronzitti et al., 2017a). Finally, a 2P scanning galvo-based imaging system, also coupled through the upper objective, is used to perform imaging and functional imaging at the GC and inner layers. The microscope is coupled to a patch-clamp platform for electrophysiological recording of retinal cells to characterize opsin expression, and optimize illumination protocols and achievable resolution. This required using a long working distance objective to enable simultaneous electrophysiological recording and imaging.
Figure 1.
System design
(A) The conceptual scheme to dissect retinal circuits needs to include spatiotemporal control of visual inputs, flexible manipulation of retinal information flux at single-cell resolution, and monitoring of the retinal output signal.
(B) The all-optical architecture combined a multi-light-path architecture allowing bottom-side spatiotemporal control of photoreceptors by means of digital micromirror device visual inputs (DMD-VI), top-side manipulation of retinal processing via optogenetic light targeting by means of multiplexed temporally focusing light shaping (MTF-LS), and monitoring of activity of GCs via 2PE scan functional imaging.
(C) The MTF-LS system is based on a two-step phase modulation, including a first modulation based on a fixed custom-made transmissive phase mask to shape the illumination in the form of a circular holographic spot and a second modulation based on an SLM to multiplex the holographic spot on multiple arbitrarily defined xyz locations.
MTF-LS unit
The MTF-LS (Accanto et al., 2018) comprises a beam-shaping unit, a temporal focusing (TF) unit, and a spatial multiplexing unit. The beam-shaping unit in our realization is a static phase mask, which spatially modulates the phase of the incoming illumination beam to produce a holographic 2D shape (here a circular 10-μm-diameter spot that matches the size of BCs) (Figures 1B and 1C). The generated intensity pattern is successively focused on a dispersive grating to enable TF, which provides enhanced axial confinement of the illumination pattern at the sample and robust propagating through scattering tissues (Bègue et al., 2013; Papagiakoumou et al., 2013). A further holographic phase modulation provided by a reconfigurable liquid-crystal spatial light modulator (SLM) allows to dynamically replicate the 2D temporally focused shape in multiple positions in the sample volume (Figure 1C).
The transmissive fixed fused silica phase mask for beam shaping, as originally proposed in (Accanto et al., 2018), allows the generation of static intensity patterns without a zero-order component and with high diffraction efficiency.
The static dielectric hologram phase mask was fabricated as a diffractive optical element (DOE) where the phase change is related to the depth profile of the diffractive optic by the following equation (Herzig, 1997).
where λ0 is the wavelength of operation, n(λ0) is the refractive index of the substrate element, and ψ(x,y) is the desired phase variation. Since, phase differences can be wrapped between 0 and 2π, the maximum height required for a DOE is on the order 2λ0 − 3λ0 depending on the refractive index of the material. This makes it possible to implement DOEs in extremely thin substrates.
Although DOEs can be manufactured using a number of techniques, we chose to fabricate them using a multi-level binary photolithography approach. In this approach, the lithography process is repeated multiple times to approximate a continuous surface by 2M staircase like steps, M being the number of binary amplitude masks used. We used an eight-level staircase pattern that exhibits ∼95% diffraction efficiency (Herzig, 1997). The phase masks were made entirely out of UV-grade fused silica (Corning 7980) because of its high transmissivity (>90%) in the visible-NIR wavelength region (0.25–2 μm) and its high laser-induced damage threshold for femtosecond pulses (Chimier et al., 2011).
The temporally focused shape generated by the phase mask at the grating plane is then multiplexed to multiple sample locations by the SLM. This is done by addressing the SLM with holographic phase masks, which generate 2D or 3D distributions of diffraction-limited spots located at the target positions. The achievable FOV depends on the wavelength, λ, the SLM pixel size at the objective back aperture, a, and the objective focal length, fobj (Ronzitti et al., 2017b). This configuration, when combined with 2P scan imaging, enables decoupling the imaging from the photostimulation plane.
Multi-plane light targeting
Here, we use the capability of MTF-LS to decouple the imaging and photostimulation planes to manipulate BC activity while recording GC light-evoked responses through the same objective.
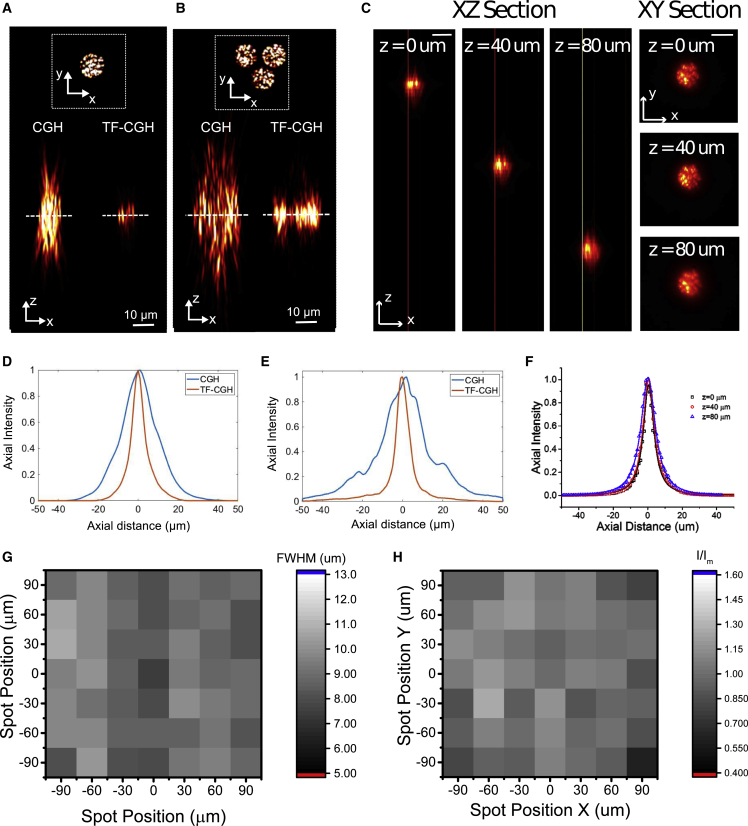
To characterize the accessible axial range and axial resolution we used a configuration featuring two opposite-facing objectives (Lutz et al., 2008). An illumination objective was used to excite a thin fluorescent layer with holographic patterns. Placed at the opposite side, a detection objective imaged the emitted fluorescence on a camera. By keeping the detection objective in a fixed position and moving the illumination objective along the axial direction, we measured the axial displacement and resolution of the holographic patterns.
We demonstrated that the illumination spot could be displaced axially in a range comparable with the distance between GCs and BCs (i.e., 0–80 μm), while maintaining an axial confinement of the illumination spot below 10 μm (full width at half maximum [FWHM] of the axial intensity distribution) (Figures 2A–2F). This value represents a 2- to 3-fold improvement with respect to a non-temporally focused holographic spot of the same size and is maintained independently of the number and distribution of spots (Papagiakoumou et al., 2008). We observed nearly uniform FWHMs by positioning 10-μm-diameter holographic spots in a FOV of 200 × 200 μm2 at 70 μm below the focal plane (FWHM 8.9 ± 0.75 μm, mean ± standard deviation [SD]) (Figures 2G and S1). The illumination intensity variability was around ±12% (±SD of intensities) across the FOV (Figures 2H and S1). This indicates that the system can provide nearly homogenous photostimulation of opsin-expressing cells across multiple layers while maintaining the imaging focus on the GC layer.
Figure 2.
Optical characterization of the system
(A and B) Simulation of the axial propagation of one single 10-μm-diameter holographic spot (A) and of three 10-μm-diameter holographic spots placed at a distance of 12.5 μm one from the other (center to center) arranged in a triangle configuration (B) without (left; CGH) and with temporal focusing (right; TF-CGH). The simulation spans in ±50 μm around the focal plane (dashed line). The images show the xy (top) and the xz (bottom) orthogonal maximum simulated fluorescence intensity projection of the spots.
(C) Axial displacement of a 10-μm-diameter temporally focused holographic spot. (Left) xz orthogonal maximum fluorescence intensity projection of the spot for different axial displacements. (Right) Corresponding xy 2P fluorescence intensity cross-section. Scale bar, 10 μm.
(D and E) Axial profile of the simulated fluorescence intensity of (D) the spot shown in (A) and of (E) the three spots shown in (B) for the two cases of CGH (blue line) and TF-CGH (red line). The FWHM of the axial profile is 17 μm for the CGH spot and 7 μm for the TF-CGH spot.
(F) Axial profile of the fluorescence intensity of axially displaced spots shown in (C). Solid lines indicate Lorentzian fit.
(G) 2D map of the FWHM of the axial profiles of the fluorescence intensity given by a matrix of spots (30 μm inter-spot distance) displayed 70 μm below the focal plane (each pixel represents one spot).
(H) Normalized illumination intensity of a matrix of spots displayed 70 μm below the focal plane (each pixel represents a spot).
Targeted holographic stimulation of RBCs
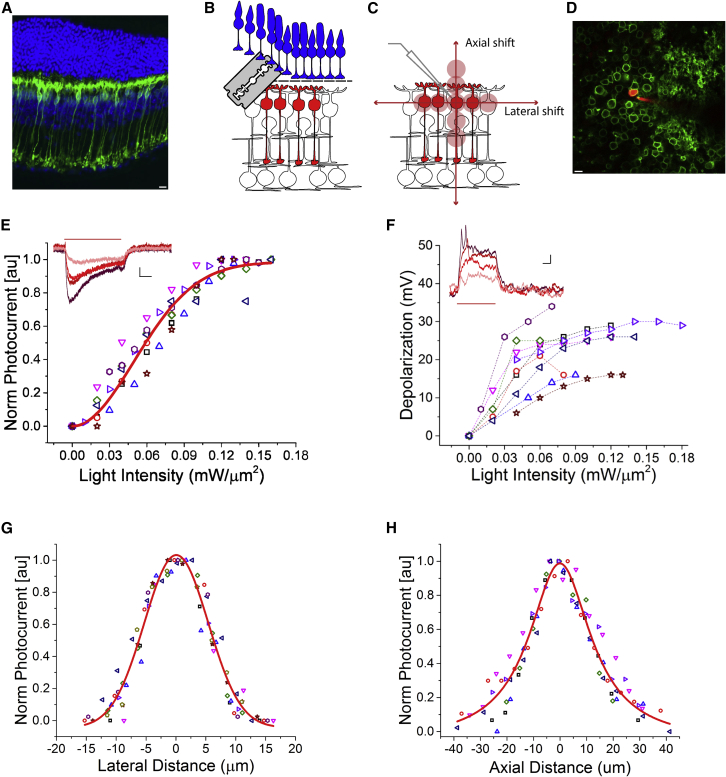
To demonstrate the capability of the system to generate physiological activation and reach cellular resolution, we performed electrophysiological recording of RBCs expressing the opsin CoChR while stimulating them with targeted 2P excitation.
To selectively target RBCs we injected a variant of AAV2, 7m8, in the vitreous of the mouse eye, to express CoChR fused with the GFP protein under the control of In4sIn3-200En-mGluR500P, a promoter specific to RBCs (Lu et al., 2016) (Figure 3A; see also STAR Methods and Data S1). To patch RBCs in a whole-mount configuration, we removed the photoreceptor layer and turned the retina upside down to access the RBCs directly from the top (Figures 3B and 3C, STAR Methods). This is practically equivalent to stimulating with 2P illumination from the GC side since the retina is an almost transparent tissue (except for the photoreceptor layer [Chen, 1993]).
Figure 3.
2P holographic stimulation enables physiological responses in RBCs with high spatial selectivity
(A) Retinal slice showing the expression of CoChR-GFP in RBCs. Green, GFP; blue, DAPI. Scale bar, 10 μm.
(B and C) Schematic of the experiment. We first removed the photoreceptor layer by cutting the retina with a vibratome (see STAR Methods), and then patched single RBCs expressing CoChR-GFP. We stimulated the cell with a 10-μm-diameter holographic spot, which we successively moved laterally and axially to estimate the photostimulation selectivity. Power was set to depolarize the RBC in its physiological range.
(D) Representative two-photon image of a patched RBC. Green, CoChR-GFP-expressing cells; red, Alexa 594 dye, inside the pipette and filling the recorded cell.
(E) Peak photocurrent versus light intensity, normalized to the maximum for each recorded cell. Each symbol corresponds to a different cell. Red line, saturation curve. Inset: representative light-evoked photocurrents. Different traces correspond to different illumination intensities (from 0.02 mW/μm2 [light red] to 0.08 mW/μm2 [dark red] with 0.02 mW/μm2 steps). Vertical scale bar, 10 pA. Horizontal scale bar, 100 ms. Red horizontal bar indicates the photostimulation time (500 ms).
(F) Peak voltage depolarization in response to different stimulations versus light intensity. Each symbol and dashed curve correspond to one cell. Inset: representative light-evoked depolarizations. Different traces correspond to different illumination intensities (trace color codes) as in (E). Vertical scale bar, 3 mV. Horizontal scale bar, 100 ms.
(G) Peak photocurrent as a function of lateral displacement, normalized to the maximum for each recorded cell. Each symbol corresponds to a different cell. Red curve, Gaussian fit.
(H) Same as (G) for axial displacements. Red curve, Lorentzian fit.
We patched fluorescent RBCs under 2P guidance (Figure 3D) and checked the morphology of the patched cell by filling it with Alexa 594. By using various light intensities, in the voltage clamp we measured the induced photocurrents that saturate at around 0.14 mW/μm2 (Figures 3E, S2A, and S2D). To characterize the homogeneity of opsin expression in RBCs from different preparations, we imaged the patched cell and correlated the cell membrane fluorescence with the peak photocurrent (stimulation power: 1.2 ± 0.02 mW/μm2). We found that higher photocurrents corresponded to a higher cell membrane fluorescence (Figure S2C).
In the current clamp, light-evoked depolarizations ranging from 10 to 27 mV were obtained in most of the cells between 0.07 and 0.12 mW/μm2 illumination (n = 8) (Figures 3F and S2B). Higher depolarizations (V = 37 mV; I = 0.07 mW/μm2, n = 1) were measured in one cell, which exhibited particularly high photocurrents (445 pA; I = 0.08 mW/μm2, n = 1) (Figures 3F and S2D). Depolarization variability was likely due to differences in expression of the opsin and the variable presence of intrinsic voltage-gated ion channels. Previous studies have shown that, under physiological conditions, visual stimulation could lead to a depolarization between 4 and 26 mV (15 ± 7 mV [mean ± SD]) (Euler and Masland, 2000). These results suggested that using excitation powers in the range of 0.03 mW/μm2 < I < 0.09 mW/μm2, yields physiological activations, i.e., comparable with the ones that would be evoked by visual stimulation.
Next, we checked the photostimulation spatial resolution achievable using our system. To this end, we measured the photo-induced current while the holographic spot was laterally and axially displaced around the RBC soma (Figure 3G), keeping the illumination intensity, I, in the physiologically tolerated range (0.03 mW/μm2 < I < 0.09 mW/μm2). We observed a 50% drop of photocurrents by moving the spot 6 μm laterally aside from the center of the cell (Figure 3G) and a residual 10% of photocurrent when the spot was 10 μm apart (corresponding to the lateral size of the illumination spot). Axially, photocurrents exhibited a decay of 50% at around 14 μm from the focal plane, with residual photocurrents below 10% for axial shifts superior to 30 μm (Figure 3H). Overall, the spatial selectivity of photoactivation was estimated to be 12 and 28 μm, corresponding to the lateral and axial FWHM of the photocurrents’ spatial distribution, respectively. These results show that photostimulation of RBCs could be achieved at cellular resolution.
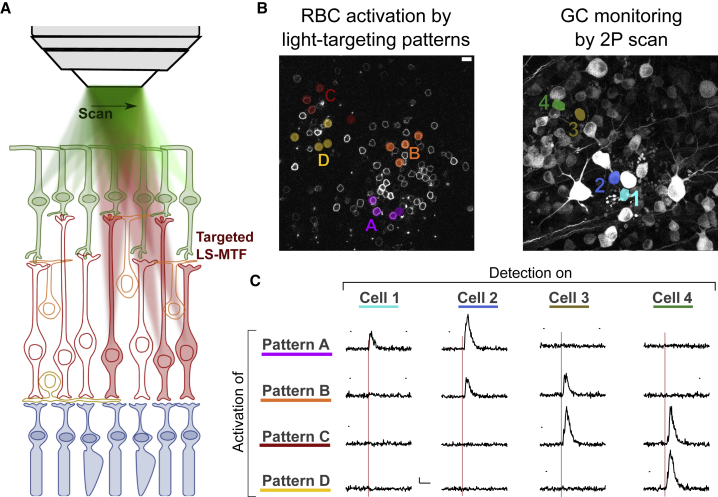
All-optical inter-layer functional connectivity
Here, we proved the capability of the microscope to decouple the imaging from the photostimulation plane to stimulate RBCs while recording the photo-evoked activity on the GC layer. Mice were injected with two AAVs driving expression of CoChR in RBC and of GCaMP6s in RGCs. The same construct as described previously was used to target RBCs. To selectively target RGCs we injected AAV2 in the vitreous of the mouse eye, to express GCaMP6s under the control of SNCG, a promoter specific to RGC (Chaffiol et al., 2017).
We imaged the GC layer by performing 2P scanning Ca2+ imaging at 920 nm, while using the MTF-LS unit to generate one or multiple excitation spots at the RBC layer, which is on average 70 μm deeper (Figure 4A).
Figure 4.
Patterned optogenetic stimulation of RBCs evokes selective activation of GCs
(A) Schematics of simultaneous photoactivation of groups of CoChR-expressing RBCs via light-targeting MTF-LS and functional imaging of GCaMP6-expressing GCs via 2P raster scan.
(B) CoChR/GFP-expressing RBCs (left) and GCaMP6-expressing GCs (right) within the same xy field of view located on retina layers 60 μm axially apart. Different colors correspond to different patterns of photoactivation across RBCs (left) and different monitored GCs (right). Scale bars, 10 μm.
(C) Calcium response induced on different GCs cells upon photoctivation of RBCs with different illumination patterns. Each row corresponds to a different pattern of stimulation in the RBC layer as shown in (B). Each line corresponds to the response of a different GC as shown in (B). The red line indicates the timing of the holographic stimulation (500 ms, 0.06 < I < 0.1 mW/μm2). Vertical scale bar, 50%. Horizontal scale bar, 5 s. Measurements were conducted after LAP4+ACET application.
In control retinas, where BCs only expressed GFP (Figure S3A), we observed responses to holographic illumination (23/123 cells, n = 2 retinas) that were due to out of focus photoreceptor stimulation (Euler et al., 2009; Palczewska et al., 2014) (Figure S3B). To avoid photoreceptor stimulation and ensure calcium responses only evoked by holographic stimulation of RBCs, we therefore blocked the transmission from photoreceptors to BCs by putting LAP4 and ACET, which block the transmission to ON and OFF BCs, respectively (see STAR Methods) (Borghuis et al., 2014). Adding this pharmacological cocktail to the bath in control experiments abolished the responses induced by holographic illumination in all cases. Responses to flashes of visible light (observed in 44/84 cells before drugs administration, n = 2 retinas) were also abolished (Figure S3B). All subsequent experiments were performed under this pharmacological block allowing recording of calcium responses originating exclusively from holographic stimulation of CoChR-expressing RBCs (see STAR Methods).
Stimulation with one holographic spot rarely evoked any detectable calcium response in GCs (0.15% ± 0.07% of 5,540 RBC-GC pairs tested, n = 3 experiments), while the signal of a single rod can evoke response in GCs at scotopic light levels. This could be because single spot stimulations evoke small voltage responses in GCs that do not reach spike threshold (Rousso et al., 2016).
To increase the evoked response to a detectable level, we took advantage of our holographic system to simultaneously stimulate n cells (2 ≤ n ≤ 10) and found that this configuration evoked detectable responses preferentially on the GCs closest to the stimulation pattern (Figures 4B and 4C).
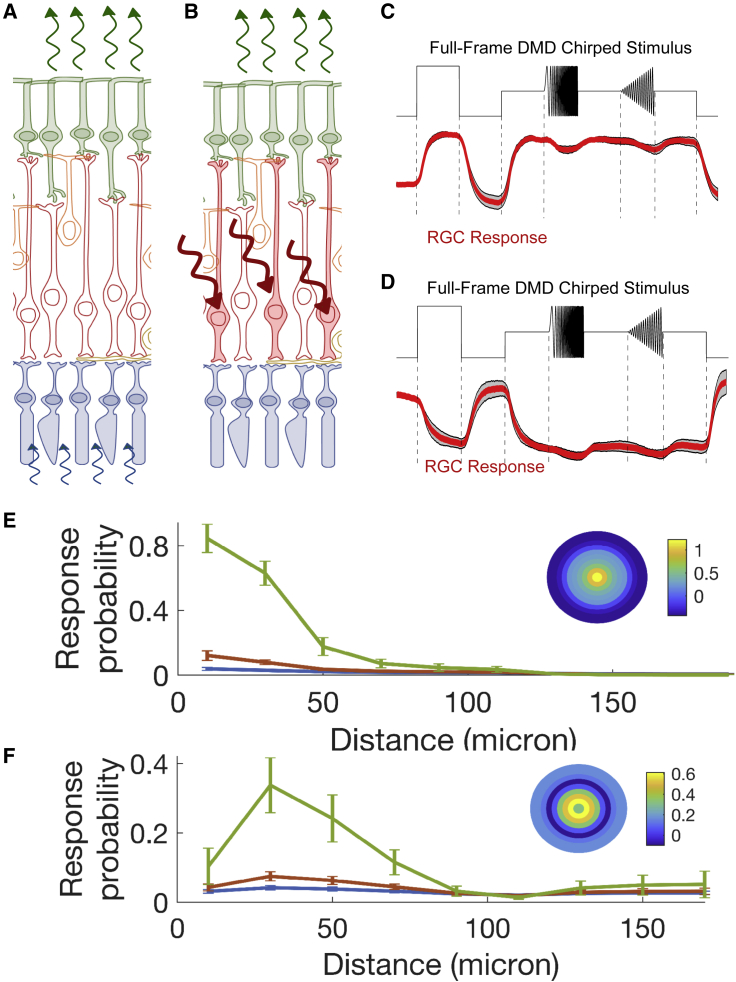
To define the size of the cellular receptive field for different GC types, we first determined the type of the imaged GCs from their responses to visual stimulation (Figure 5A). We performed calcium imaging while stimulating photoreceptors with a DMD full-field stimulation (Baden et al., 2016) (Figures 5C and 5D). We then blocked the photoreceptor-BC transmission, as previously described, and stimulated RBCs with multiple patterns (Figure 5B) while imaging GCs. We assumed that all the GCs of the same type have a cellular receptive field of similar size, a reasonable hypothesis since GCs of the same type are functionally similar (Baden et al., 2016). We thus pooled data from different cells of the same type to estimate the cellular receptive field of each type.
Figure 5.
Cellular receptive field for different ganglion cell types
(A and B) Schematic of the visual (A) and holographic (B) stimulation protocols, performed one after the other on the same retina.
(C and D) Representative calcium response (red line) of two different ganglion cell types illuminated with a chirped stimulus (black line). Scale bar, 5 s.
(E and F) Prediction of the response to multiple-spot stimulation by the model for the two different types of ganglion cell shown in (C and E) and (D and F). y axis: probability of evoking a response. x axis: distance between the different spots of the stimulation pattern and the recorded ganglion cell for different numbers of stimulation spots (different colors correspond to different number of spots: blue, one spot; red, two spots; green, five spots). Inset: inferred model filter. Estimated radii (see STAR Methods) were 51 ± 9 μm (E) and 70 ± 13 μm (F). Errors are computed as SD of the response probability following a bootstrap on the training data before model inference.
For the analysis, we realigned all the cells of the same type and placed the GC in the center (Figure S4). We then inferred a logistic model to relate the response with the relative location of the stimulation pattern. This statistical model takes each pattern, measures the relative distance of each spot to the GC’s position, and assigns a weight to each spot depending only on this relative distance. The sum of these weights is then converted into a probability to detect a calcium response. The weights are learned from the data, and together form a filter that tells which region of the visual space is the most sensitive to stimulation. Note that this is equivalent to fitting a linear-nonlinear model (Chichilnisky, 2001) to the data. Here, we assumed that this cellular receptive field is isotropic and centered on the GC.
To fit this model and learn its parameters, we pooled together all the cells of a given type, fitted our model to these data and measured its performance in predicting the responses of a test dataset not used for the learning. The model has a significant prediction performance, close to the upper bound defined theoretically (see STAR Methods, Figures S6 and S5 for other types), demonstrating that the filter inferred is significant.
Our model allows the estimation of the probability of having a calcium response for any pattern of RBC stimulation. We predicted the probability of response systematically for a given number of spots all placed at the same distance (Figures 5E and 5F, different colors). We found five types where we had enough cells to have a reliable estimate of the filter. Further cell types could have been identified by prolonging the imaging section to analyze a larger number of FOVs (Baden et al., 2016) at the cost of the time left for the photostimulation part. For all the identified cell types (Figures 5E, 5F, and S5), there was always a clear fall off of the probability of the response with the increase of the distance of the stimulation pattern. We estimated the size of the cellular receptive field by fitting a Gaussian distribution on the inferred filter. All the size values were between 48 and 76 μm (five tested types, see STAR Methods). The sizes were comparable with the classical, photoreceptor-based receptive field (Baden et al., 2016). This shows that these cells integrated RBC output over a limited region of space in all cases where this could be reliably estimated. The divergence present across the RBC pathway is thus limited and remains compatible with spatially accurate computations.
Discussion
MTF-LS and functional imaging
We have established an all-optical method, for non-invasive inter-layers interrogation of retinal circuits. The system combines 2P multi-target temporally focused holographic illumination with a 2P scanning microscope, to independently activate one or multiple RBCs with single-cell resolution while monitoring the GCs response via 2P calcium scan imaging with single-cell resolution and cell-type specificity. The system also allows stimulation of the photoreceptor layer to classify the different cell types in the GC layer and electrophysiological whole-cell recording of light-induced current on the BCs.
We have demonstrated the capability for single- and multi-target stimulation of the BC layer and functional imaging of photo-evoked responses in the GC layer with single-cell resolution and cell-type specificity.
Multitarget optogenetic at cellular resolution has become possible thanks to the combination of recent advances in opsin engineering, multiplexed light-shaping approaches, and high-power fiber laser development (Chen et al., 2018). Despite its tremendous potential, it remains a very new technology and, so far, only a few studies have been reported using this approach. Combined with 2P Ca2+ imaging and behavioral assays, 2D patterned optogenetics has been used to show that the activation of a few cells can bias behavior by triggering the activity of precisely defined ensembles in the mouse cortex (Carrillo-Reid et al., 2019; Chettih and Harvey, 2019; Dalgleish et al., 2020; Marshel et al., 2019) and olfactory bulb (Gill et al., 2020) or that selective stimulation of a small number of hippocampal place cells is sufficient to bias mice behavior during a spatial memory task (Robinson et al., 2020). 3D light-targeted photostimulation of neuronal ensembles in V1 layers 2/3 and 5 has been recently applied to study how natural dynamics and associated behavior can be elicited by optogenetic recruitment of a critical number of individually defined percept-specific neurons (Marshel et al., 2019). All these “all-optical” experiments have used behavioral assay to probe the effects produced by the manipulation of a sub-group of cells identified on the base of their functional response using Ca2+ imaging. In the present all-optical approach, we use, for the first time, light excitation to both manipulate the circuits input and probe the produced output. Also, for the first time, we used an all-optical approach for the investigation of a retinal circuit. With this approach we were able to probe functional connectivity mapping among RBCs and five different GC types.
Existing approaches to dissect functional connectivity in retinal circuits have used electrophysiology (Asari and Meister, 2012, 2014) or electrophysiology combined with wide-field optogenetics (Park et al., 2015); however, they lack either specific targeting of cells or (spatially) precise stimulation. Previous works have performed stimulation of a single BC with sharp electrodes while recording the evoked activity of GCs with multi-electrode arrays (Asari and Meister, 2012; Baccus and Meister, 2002). However, the yield of these experiments is low as they required electrode stimulation of a single cell at a time and lack cell type specificity of the recorded cells. Several studies have also recorded a single GC with patch clamp while performing an optogenetic stimulation on specific cell types expressing an optogenetic protein (Park et al., 2015). However, the stimulation was full field, and all the cells expressing the optogenetic protein were simultaneously activated.
Connectivity mapping with cellular resolution has been recently demonstrated in vitro (Hage et al., 2022; Printz et al., 2021) and in vivo (McRaven et al., 2020) using sequential photostimulation of opsin-expressing presynaptic cells and patch recording of the evoked postsynaptic response. These studies have enabled the monitoring of hundreds of presynaptic cells, but have been limited in probing the connections to one postsynaptic cell at a time and thus, for this, required the use of whole patch recording. Alternatively, in vivo intra-layer all-optical functional connectivity measurements were achieved to identify inhibitory connections between L1 interneurons in mouse barrel cortex. In this case, multi-target optogenetic stimulation of presynaptic cells (expressing the blue opsin SomCheRiff) was combined with simultaneous voltage recording and photostimulation of the postsynaptic cells (co-expressing the red indicator SomArchon and SomCheRiff) (Fan et al., 2020). However, the lack of optical sectioning and shallow light penetration of single-photon excitation have limited this approach to the study of one to three postsynaptic cells at shallow depths and to the probing of intra-layer connections only.
The main advantages of our approach, are the use of 2P GCaMP imaging, which enables recording with cellular resolution from all (∼50) GCaMP expressing cells in the imaging FOV, and the superior axial resolution of 2P excitation, which enables decoupling the imaging from the photostimulation plane; thus extending this kind of study to map the inter-layers of functional connectivity.
As originally proposed in Accanto et al. (2018), we have fabricated a transmissive static phase mask to generate the static holographic disk on the TF grating. This enables holographic patterns not affected by ghost or unmodulated (i.e., zero-order) light components and more compact and flexible on-axis operation compared with reflective SLMs. Compared with other similar approaches of temporally focused spatial multiplexing, such as 3D-SHOT (Pegard et al., 2017), the proposed system enables full illumination of the SLM and better axial confinement for the same spot size (<10 μm for 10–15-μm-diameter spots) (Mardinly et al., 2018; Pegard et al., 2017). Alternatively, methods based on spiral-scanning 2P photoactivation would have required the insertion of a galvanometric unit in the photostimulation pathway. Notably, the use of TF makes the system further beneficial for applications in highly scattering and dense tissues (Bègue et al., 2013; Papagiakoumou et al., 2013).
In this study, distribution of cells in the GC layer is relatively flat, thus enabling to restrict 2P calcium imaging to a single plane. For the investigation of more complex circuits the system can be integrated with multi-plane imaging strategies based on, e.g., beam multiplexing, PSF engineering, or fast beam scanning (Ji et al., 2016).
Retinal circuit investigation
The rod signal is transmitted to two or more RBCs, which in turn input to at least 7 AII amacrine cells, each reaching at least 14 cone bipolar cells (Tsukamoto and Omi, 2013, 2017), which activate GCs. This pathway is therefore strongly divergent. This divergence is essential to amplify weak signals and optimize detection at weak light levels. Ganglion cells can be divided into more than 30 cell types, and it is expected that most of them can be activated through the RBC circuit, except at very dim light levels. We have applied our approach to investigate how GCs integrate the output of RBCs and confirmed that this pathway maintained a high-resolution picture of the visual input. Consistent with previous results from anatomy (Tsukamoto and Omi, 2013, 2017), we found that RBC stimulation evoked responses in all types studied, and that the size of the cellular receptive field was like the size of the classical receptive field.
We have shown that our method enables to probe and study retinal information processing in mice retina. These investigations could be extended to post-mortem human retina for vision restoration. It was recently shown (Sahel et al., 2021) that expressing ChrimsonR in GCs allowed partial restoration of visual function in a subject affected by retinitis pigmentosa. Although this result is extremely promising, the treated subject was able to perceive basic, highly contrasted stimuli and not natural images at high resolution. A possible strategy to further restore retinal computations is either to target and photostimulate cells upstream, e.g., photoreceptors (Busskamp et al., 2010; Khabou et al., 2018) or BCs (Cronin et al., 2014; Doroudchi et al., 2011; Gaub et al., 2014; Hulliger et al., 2020; Lagali et al., 2008; Macé et al., 2015; van Wyk et al., 2015), or to stimulate the GCs with activity patterns resembling those received in heathy conditions. In this context, our method will enable to derive input-output conversion across multiple retina layers to guide the design of optimal stimulation patterns. For these, more-complex preparations, the superior axial resolution achieved with TF will be of key importance to reach single-cell activation.
The superior axial resolution achieved with multi-target temporally focused excitation will be particularly relevant also when used for in vivo (Chen et al., 2019) multi-layered circuit investigation, in highly scattering tissue, and/or dense 3D populations of cells, as is the case when investigating, for example, information transmission across the multiple layers of the visual cortex (Wertz et al., 2015; Yang et al., 2016).
Our approach can be also extended to other brain regions, opening the way for a precise interrogation in vitro and in vivo (Chen et al., 2019) of multi-layered circuits; for example, to investigate how information is transmitted across the multiple layers of the visual cortex (Wertz et al., 2015; Yang et al., 2016). Understanding this information transfer is a promising avenue to dissect complex neural circuits and understand the neural basis of computations.
Limitations of the study
The low sensitivity of GCaMP imaging requires stimulation of groups of 2 to 10 cells and therefore has limited our study to probe the functional connectivity between RBCs and specific GC types. Extending the same approach to measure the functional connectivity between individual cells will be possible by increasing the number of stimulation patterns and using compress sensing connectivity analysis (Hu and Chklovskii, 2009). Alternatively, subthreshold or single-spike functional response of GCs to single RBC stimulation could be probed by replacing GCaMP with voltage imaging. However, simultaneous recording from all GCs in the imaging FOV would require using whole-field 1P illumination with consequent lack of cellular resolution and artefactual BCs activation. The use of 2P voltage imaging using 2P activatable indicators is a promising alternative, although so far this has been limited to the recording of three to four cells at a time (Villette et al., 2019).
A limitation of our work is that the state of the retinal network during our recordings might not reflect the state of the retinal network when it is dark-adapted to a low light level. In particular, our drug cocktail could hyperpolarize RBCs and reduce their output. The coupling between AII amacrine cells will also vary with the light level. As a result, our estimate of the cellular receptive field size might change with the adaptation state of the retina.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| ACET | Tocris | Ref 2728/10 |
| L-AP4 | Tocris | Ref 0103/10 |
| AMES medium | Sigma-Aldrich | Ref A1420 |
| Experimental models: Organisms/strains | ||
| Mouse: C57Bl6J mice | Janvier Laboratories | |
| Recombinant DNA | ||
| In4s-In3-200En-mGluR500P-CoChR | Lu et al., 2016 | See Data S1 for full sequence |
| SNCG-GCaMP6s | Chaffiol et al., 2017 | See Data S1 for full sequence |
| Software and algorithms | ||
| pClamp10 | Molecular Devices | |
| ScanImage 3.8 | Pologruto et al., 2003 | http://wscanimage.org |
| MATLAB R2017a | Mathworks inc, USA | http://www.mathworks.org |
| OriginPro 2016 | OriginLab | https://www.originlab.com |
| μManager | Edelstein et al., 2014 | https://micro-manager.org |
| WaveFront Designer IV | Photonics Department; Institut de la Vision | N/A |
| IgorPro | Wavemetrics | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact: Valentina Emiliani, valentina.emiliani@inserm.fr.
Materials availability
This study did not generate new materials.
Experimental model and subject details
All experiments were done in accordance with the National Institutes of Health Guide for Care and Use of Laboratory Animals. The protocol was approved by the Local Animal Ethics Committee of Paris 5 (CEEA 34) and conducted in accordance with Directive 2010/63/EU of the European Parliament. All mice used in this study were C57Bl6J mice (wild type) from Janvier Laboratories (Le Genest Saint Isle, France). Animals were housed in 2-5 per cage, with a light-dark cycle of 12 + 12 h, a constant temperature of 22°C, and food and water ad libitum. Bedding was changed once a week. Nesting and enrichment materials were provided. Animals were checked on regular intervals by veterinarians and handled by qualified animal care takers within the authorized facility. Animals aged between 4 and 15 weeks were used for the experiments. Mice were housed in enriched environment with no food restriction and maintained in accordance with institutional guidelines under approved authority protocols, checked on regular intervals by veterinarians and handled by qualified animal care takers within an authorized animal facility.
Method details
AAV Production and injections
Recombinant AAVs were produced by the plasmid cotransfection method (Choi et al., 2007) and the resulting lysates were purified via iodixanol gradient ultracentrifugation as previously described. Briefly, 40% iodixanol fraction was concentrated and buffer exchanged using Amicon Ultra-15 Centrifugal Filter Units (Millipore, Molsheim, France). Vector stocks were then tittered for DNase-resistant vector genomes by real-time PCR relative to a standard (Choi et al., 2007).
For injection, animals were anesthetized with Isofluorane (Isoflurin 250 mL, Vetpharma Animal Health) inhalation and pupils were dilated. A 33-gauge needle was inserted into the eye to deliver the vector into the vitreous. 2 μL of vector solution was injected per eye, containing 1 μL of the vector delivering GCaMP6s (containing ∼ 1010 vg) and 1 μL of the vector delivering either CoChR (containing ∼ 1010 vg) or GFP (containing ∼ 1010 vg).
For all experiments we used GCaMP6s (Chen et al., 2013) under the SNCG promoter (Chaffiol et al., 2017) to specifically target all types of retina ganglion cells and we used AAV2 as viral vector. To express CoChR (Klapoetke et al., 2014; Shemesh et al., 2017), we used a recently published promoter (In4s-In3-200En-mGluR500P) (Lu et al., 2016), which has been proved to allow specific expression of optogenetic proteins in RBCs. To deliver it across the retinal layers we used 7m8 a genetic variant of AAV2 (Dalkara et al., 2013). Finally we used GFP only under the grm6 promoter (Macé et al., 2015) delivered with AAV2-7m8 to target BCs in control experiments. The injections were performed in 4–5 weeks old mice.
Tissue preparation
For all experiments, we used female mice 4-8 weeks after the injection. Animals were dark adapted for at least 1 h, then anesthetized with isofluorane (Isoflurin 250 mL, Vetpharma Animal Health) and killed by cervical dislocation. The eyes were enucleated and placed in AMES medium (Sigma-Aldrich, St Louis, MO; A1420), bubbled with 95% O2 and 5 % CO2 at room temperature. The eyes were dissected under dim red light (>645 nm) and the isolated retinas were flat mounted with GCs up and transferred to the recording chamber in the microscope. The retina was continuously perfused with bubbled Ames’ medium at a rate of 5–7 mL/min during experiments and temperature was maintained around 34 degrees.
Experiment description and pharmacology
At the beginning of the experiments the flat mounted retina was placed under the microscope and left to rest for ∼30 min in the dark. The first step of the experiment was to perform the visual stimulations (see below). Then, to block the photoreceptors (Borghuis et al., 2014), we added to bubbled AMES medium the KAR selective agonist ACET (1uM, catalog no 2728, Tocris bioscience) and the metabotropic glutamate receptor agonist LAP4 (20 μM, catalog no 0103, Tocris Bioscience). The retina was left to rest in the dark for ∼30-45 min. Before starting to stimulate the RBCs expressing CoChR, we tested that the photoreceptor transmission to BCs was effectively blocked by doing visual stimulations on a central FOV of 100 × 100 μm2. The highest intensity of light used to stimulate photoreceptors was 1,53 x 10−3 mW/mm2. As shown in (Shemesh et al., 2017) (Figure S2D) this power is negligible compared to the one necessary to induce any activation of the opsin, which has small responses for ∼2 mW/mm2. If no ganglion cell was responding to the visual stimulation, we proceeded with the holographic stimulation. Holographic stimulation was set to a duration of 500 ms to be compatible with natural stimulation except where differently reported.
Patch-clamp tissue preparation
4 to 5 weeks old mice were injected with 1 or 1.5 μL volume of AAV2-7m8 carrying CoChR (∼1010 vg) under a promoter specific for RBCs(In4s-In3-200En-mGluR500P) (Lu et al., 2016). 4 to 10 weeks after the injection, the animals were anesthetized with isoflurane and killed by cervical dislocation. Eyeballs were enucleated and dissected under white light. To have a better access to the BCs with the patch pipette, we removed the photoreceptor layer using a vibratome (Leica VT1200S slicer). This procedure was previously described in details (Clérin et al., 2014). Briefly, the dissected retina was transferred in the vibratome tank filled with bubbled Ames. The retina was placed photoreceptors down on a gelatin block in the center of the tank and the solution was removed to permit the sealing of the flat-mounted retina. Once the retina was sealed, the tank was filled with bubbling Ames again and the vibratome’s blade was lowered until the GCs level. A slice of ∼80–90 μm was cut and transferred to the recording chamber under the microscope with GCs down. BCs were thus on the upper side without the photoreceptors on top of them, which made them more accessible to patch recordings (Figures 2B and 2C).
Patch-clamp recordings
BCs layer was imaged with a 2P imaging system to select the region with cells expressing the opsin. BCs were visualized with an IR illumination, a water-immersion 40× objective (40× W APO NIR; Nikon), and an IR CCD (see “Optical system”) while approaching the cell with the patch pipette. Light was turned off soon after the whole-cell configuration was established. Patch-clamp electrodes were pulled from borosilicate glass capillaries (1.5 mm outer diameter, 0.86 mm internal diameter; Harvard apparatus) with a horizontal micropipette puller (P1000, Sutter Instruments). Pipettes were filled with the following solution (mM): 130 K-gluconate, 7 KCl, 4 MgATP, 0.3 mM Na-GTP, 10 Na-phosphocreatine, and 10 mM HEPES (pH adjusted to 7.28 with KOH; osmolarity 280 mOsm). Pipette resistance in the bath was 4.5–6 MΩ. An Ag/AgCl pellet was used as reference electrode in the recording chamber. Patched cells were loaded with Alexa 594 (Invitrogen) added to the pipette solution to reconstruct the morphology at the end of each experiment.
Data were acquired with a MultiClamp 700B amplifier (Molecular Devices), a National Instrument board, and the Neuromatic software (www.neuromatic.thinkrandom.com) running on IgorPro interface (Wavemetrics). Voltage and current clamp recordings were low-pass filtered at 6-10 kHz and sampled at 20-50 kHz. Cells were clamped at −40 mV. The cell resting membrane potential (Vm) was measured soon after achieving the whole-cell configuration (Vm = 42 ± 10 mV, from 24 cells). Series resistance (Rs) was determined and compensated from 70 to 80% with the MultiClamp software during acquisition (Rs = 18 ± 7 MΩ, from 24 cells). Cell membrane capacitance (Cm) was 3.8 ± 0.8 pF (from 24 cells). Voltage values shown are not corrected for the liquid junction potential (estimated value: 15 mV).
To determine the axial and lateral resolution of the system we stimulated the cell with a holographic spot and moved it in steps of 2.5 μm laterally or 5 μm axially to estimate the photostimulation selectivity. We determined the peak photocurrent for increasing 2P light intensities and we normalized photocurrents to the maximum value for each recorded cell. Photocurrent saturation curve in Figure 2E was given by empirically fit data with with k equals to 0.06.
Optical system
The optical system was built around a commercial upright microscope (SliceScope; Scientifica) and combined a multi-light-path imaging architecture, a 3D multiplexing temporally focused holographic-based photoactivation apparatus and a spatiotemporally-controlled visual stimulation system.
The imaging system has been already described in Ref. (Ronzitti et al., 2017a). Briefly, it includes three different imaging pathways: a 2P raster scanning, a 1P wide-field epifluorescence, and a wide-field infrared (IR) illumination imaging. 2P imaging was provided by a femtosecond pulsed beam (Coherent Chameleon Vision II, pulse width 140 fs, tuning range 680–1080 nm), relayed on a pair of XY galvanometric mirrors (3 mm aperture, 6215H series; Cambridge Technology), imaged at the back aperture of the microscope objective (40× W APO NIR; Nikon) through an afocal telescope. Galvanometric mirrors were driven by two servo drivers (MicroMax series 671; Cambridge Technology) controlled via a digital/analog converter board (PCI-6110; National Instrument) through ScanImage software (Pologruto et al., 2003). Emitted fluorescence was collected by two photomultiplier tubes (PMT) GaAsP (H10770-40 SEL; Hamamatsu #H10770-40 SEL) coupled to the objective back aperture via a fiber-coupled detection scheme (Ronzitti et al., 2017a) . 2P imaging laser power was tuned by combining an electrically controlled liquid crystal variable phase retarder (Meadowlark Optics #LRC-200-IR1) and a polarizer cube (Meadowlark Optics #BB-050-IR1). For image acquisition, we used ScanImage synchronized with the visual stimulation or CGH-excitation with a custom-made software running in MATLAB. For visual stimulation acquisitions, we divided a 200umx200um FOV in 4 100 × 100 μm2 smaller FOV and we took 64x64 pixel image sequences at 7.8 frames per sec (Imaging power (P) < 7 mW after the objective for all recordings). For image acquisitions during optogenetic stimulation, we recorded a 200 × 200 μm2 FOV in a 128x128 pixel image sequences at 5.92 frames per sec (P ranging from 9 mW to 15 mW). For high resolution morphology scans, we took 512x512 pixel images.
1P widefield imaging was provided by a LED source (Thorlabs #M470L2). 1P emitted fluorescence was collected through a tube lens (f = 200 mm), on a charge-coupled device (CCD) camera (Hamamatsu Orca-05G) after passing through a dichroic mirror (Semrock #FF510-Di02) and a visible bandwidth filter (Semrock FF01-609/181). 1P- and 2P- emitted fluorescence was separated through a movable dichroic mirror (70 × 50 mm custom size; Semrock #FF705-Di01) and an upstream dichroic mirror (Chroma #ZT670rdc-xxrxt).
IR illumination was provided by a custom-made external IR stalk lamp fitted near the microscope. IR light reflected by the sample was collected with an IR CCD (DAGE- MIT IR-1000).
2P optogenetic photoactivation was performed by generating 10 μm diameter circular spots pinpointing opsin-tagged cells in the sample via a 3D multiplexed spatially controlled phase modulation of the illumination beam wavefront thoroughly detailed in (Accanto et al., 2018). Specifically, a femtosecond pulsed beam delivered by a diode pumped, fiber amplifier system (Satsuma HP, Amplitude Systemes; pulse width 270 fs, tunable repetition rate 500–2000 kHz, gated from single shot up to 2000 kHz with an external modulator, maximum pulse energy 20 μJ, maximum average power 10 W, wavelength λ = 1030 nm) operated at 500 kHz, was widened through an expanding telescope (x5) and transmitted through a custom-designed 5 × 5 mm2 8-grey-levels static phase-mask calculated via Gerchberg and Saxton algorithm and fabricated by etching of fused silica (Double Helix Optics, LLC). The phase mask profile was expanded 2x and was Fourier transformed by a 500 mm lens to project the holographic spot on a blazed reflective diffraction grating (600 L/mm) for temporal focusing. The beam was then collimated through a 1000 mm lens to impinge the sensitive area of a reconfigurable liquid crystal SLM (X13,138-07, Hamamatsu Photonics) placed in the Fourier plane of the diffraction grating. A beam stop was placed to physically block the SLM’s not modulated zero order. The SLM plane was imaged through a telescope on the back focal plane of the objective lens and addressed with a phase modulation calculated with a custom-designed software (Wavefront-Designer IV) to produce a set of diffraction-limited spots able to multiplex the circular spot in 3D at the sample plane and light-target opsin-expressing cells in the bipolar cells layer. For all experiments in Figures 4 and 5, we used photostimulation intensities ranging from 0.06 mW/μm2 to 0.1 mW/μm2.
Visual stimulation was performed by spatiotemporally-controlled full-field visual stimuli generated through a DMD-based amplitude modulation. A 420-nm LED beam (Thorlabs #M420L2) was filtered by a bandwidth excitation filter (Semrock FF01-420/10), conveniently attenuated with density filters and collimated to illuminate the sensitive area of a DMD (Vialux GmbH). The DMD plane was conjugated to the sample plane by a telescope through the rear port of the microscope. Visual stimuli were generated by a Matlab custom-designed software and synchronized with the 2P raster scan retrace. The LED intensity was calibrated to range (as photoisomerization rate, 103 P∗/sec cone) from 0,3-2 (photoisomerization rate) and 1-5 to 39-43 and 120-130 for S and M opsins respectively. For all experiments, the retina was kept at constant intensity level for 30 s from the laser scanning start to the beginning of the visual stimuli. We used full field ‘chirp’ stimulus (Baden et al., 2016) consisting of a bright step of 10 s and two sinusoidal intensity modulations, one with increasing frequency and one with increasing contrast.
Quantification and statistical analysis
Data analysis
Data analysis was performed using MATLAB. Region of interest (ROIs), corresponding to somata in the RGC layer, were identified semi-automatically using a custom software based on a high resolution image of the ganglion cell layer and on a projection of all the images acquired for each stimulation. Electrophysiological recordings were analysed with IgorPro (Wavemetrics) and OriginPro (OriginLab).
Logistic regression
To fit our model, we first binned each stimulation pattern by dividing the space around each ganglion cell: for the analysis of Figure 5 we used annuli of 20 μm thickness.
Each stimulation pattern it was thus transformed into a vector Si, which our model used to predict the probability of response pi
where F is the model filter, g the sigmoid function, and a constant. This is the model used for logistic regression, and this model is also analog to a LN model (Chichilnisky, 2001) with a sigmoid as non-linear function and no temporal integration. The null model we used for comparison has no filter F instead.
We learned the parameters F and with a leave one out strategy, where all but one stimulation patterns are used as a training set and the remaining one is used for testing, and we iterate over all stimulation patterns.
We then maximized log likelihood with a L1 penalty for sparseness of the parameters, and a L2 smoothness constraint between neighbouring values of the filter. The weights of these two cost functions was chosen such that the log-likelihood of the testing set was maximal.
The performance of the model was evaluated using the R2introduced by Tjur for logistic regressions (Tjur, 2009):
Statistical error on R2 have been computed as error of the mean.
These R2 values can be below 1 for two reasons. It could be because the model is missing something, and/or because the prediction performance is intrinsically limited by the noise in the data and the finite size of the dataset.
To test the second hypothesis, we estimated a “maximal” value for the performance. We generated surrogate data from the model itself and estimated the performance of the model at predicting them, a test similar to methods previous employed in the literature to find upper bound on model performance (McIntosh et al., 2016). We found that for all cell types included in the results, this maximal performance was not significantly different from the performance measured on data (Figure S6).
To estimate the size of the filters in Figure 5, we fitted a Gaussian function with an offset over the filters previously inferred,and estimated the cellular receptive field size as the fitted standard deviation.
Classification of ganglion cell types
To divide the ganglion cell population in different types, we followed a process similar to the one described in Baden et al. (2016).
For each cell, we used the normalized fluorescence response to the chirp stimulus as a feature vector. We pooled the feature vectors of all the cells to build a feature matrix of dimensionality n_cells ∗n_time_steps.
We used principal component analysis to reduce the dimensionality of the features, by keeping only the first 10 principal components (enough to explain 90% of the variance of the responses).
As a result, we obtained for each cell a new reduced feature vector of only 10 elements.
We then applied an unsupervised learning algorithm based on gaussian mixture models to group these cells in clusters. This algorithm works as follows. We consider each cell as a data point in the feature space. We choose a number of clusters N. N gaussians are fitted on the data points using Expectation-Maximization. Each cell is assigned to the gaussian that fits best its feature vector. We obtain in this way N different cell clusters. This routine is performed with different values of N. The best value N is found by maximizing the Bayesian Information Criterion.
We repeated the PCA and clustering steps recursively: Principal Component Reduction was performed again on each cluster to obtain new reduced feature vectors. Each cluster was then further subdivided in subgroups by applying the same clustering routine.We built in this way a tree of clusters.
We applied a stopping condition based on the cluster noise, defined as < Std[ R ]c >t where R is the T by C cluster response matrix (time samples by cells in the cluster) and ⟨⟩x and Std[ ]x denote the mean and standard deviation across the indicated dimension, respectively.
When this cluster noise was below a given threshold (t_noise = 0.2), we considered that cluster as a leaf of our tree, and it was no longer subdivided.
Once the whole tree had been generated, we applied a pruning rule to polish our clustering.
We pruned all the leaf clusters that did not meet our acceptance criterion (cluster size smaller than 4 elements and/or cluster noise above 0.25). We repeated this procedure recursively until all the leaves of our tree met these acceptance requirements. The leaf clusters of the pruned tree were taken as the result of our classification algorithm.
Static phase mask Fabrication
In order to produce an 8-level dielectric phase mask, three binary amplitude masks were used. The binary masks were designed by using the resultant phase of the Gerchberg-Saxton optimization algorithm and discretizing the phase into 8 equal levels. Figures S1E and S1F shows the resulting optimized phase (Figure S1E) and it’s discretized 8-level version (Figure S1F).
The binary amplitude masks were then fabricated using a direct write laser system – Heidelberg DWL 66FS which has a lateral resolution of ∼1 μm. Each binary mask is then used to perform contact lithography on the fused silica substrate where a positive photo resist is spun coated on the surface of substrate, followed by transferring the binary pattern onto the photoresist using UV light exposure. The substrate is then developed to remove the unexposed areas followed by anisotropic etching using a reactive ion etcher which was pre-calibrated to measure etch depth as a function of time. The residual photoresist was then washed off. This process was then repeated for each binary amplitude mask. The resulting phase mask was then imaged using a Veeco white light vertical scanning interferometer. A 2x magnification objective was used to maximize the field of view. Figures S1G and S1H shows the comparison between the phase profile of the discretized theoretical mask (Figure S1G) and the height profile of the fabricated mask (Figure S1H) in the same region of interest.
Acknowledgments
We thank Vincent de Sars for the implementation of Wavefront Designer; the IHU FOReSIGHT (grant P-ALLOP3-IHU-000); the Fondation Bettencourt Schueller (Prix Coups d’élan pour la recherche française); the National Institutes of Health (NIH) (grant 1UF1NS107574 - 01); Retina France; AVIESAN-UNADEV; ANR (ANR-18-CE37-0011 (DECORE), ANR-20-CE37-0018-04 (ShootingStar), and ANR-21-CE37-0024 NatNetNoise); Sorbonne Université (emergence program CrInfoNet); AVIESAN-UNADEV (AIDE); and Axa research funding for financial support. G.L.B.S. was supported by a fellowship from Fondation pour la Recherche Médicale.
Author contributions
Conceptualization, G.L.B.S., E.R., S.P., O.M., and V.E.; methodology, G.L.B.S., E.R., E.P., D.D., U.F., O.M., and V.E.; formal analysis, G.L.B.S., E.R., F.T., U.F., and OM; investigation, G.L.B.S., E.R., and V.Z.; resources, G.L.B.S., E.R., E.P., U.F., H.K., D.D., and A.A.; writing – original draft, G.L.B.S., E.R., O.M., and E.V.; writing – review & editing, G.L.B.S., E.R., E.P., V.Z., U.F., O.M., and E.V.; supervision, O.M. and E.V.; funding acquisition, S.P., O.M., and E.V.
Declaration of interests
Anurag Agrawal is an employee of Double Helix Optics, Inc.
Published: August 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2022.100268.
Contributor Information
Olivier Marre, Email: olivier.marre@gmail.com.
Valentina Emiliani, Email: valentina.emiliani@inserm.fr.
Supplemental information
Data and code availability
Data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Accanto N., Molinier C., Tanese D., Ronzitti E., Newman Z.L., Wyart C., Isacoff E., Papagiakoumou E., Emiliani V. Multiplexed temporally focused light shaping for high-resolution multi-cell targeting. Optica. 2018;5:1478. [Google Scholar]
- Asari H., Meister M. Divergence of visual channels in the inner retina. Nat. Neurosci. 2012;15:1581–1589. doi: 10.1038/nn.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asari H., Meister M. The projective field of retinal bipolar cells and its modulation by visual context. Neuron. 2014;81:641–652. doi: 10.1016/j.neuron.2013.11.029. [DOI] [PubMed] [Google Scholar]
- Baccus S.A., Meister M. Fast and slow contrast adaptation in retinal circuitry. Neuron. 2002;36:909–919. doi: 10.1016/s0896-6273(02)01050-4. [DOI] [PubMed] [Google Scholar]
- Baden T., Berens P., Franke K., Román Rosón M., Bethge M., Euler T. The functional diversity of retinal ganglion cells in the mouse. Nature. 2016;529:345–350. doi: 10.1038/nature16468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bègue A., Papagiakoumou E., Leshem B., Conti R., Enke L., Oron D., Emiliani V. Two-photon excitation in scattering media by spatiotemporally shaped beams and their application in optogenetic stimulation. Biomed. Opt. Express. 2013 doi: 10.1364/BOE.4.002869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi A., Cui J., Ma Y.P., Olshevskaya E., Pu M., Dizhoor A.M., Pan Z.H. Ectopic expression of a Microbial-type Rhodopsin restores visual responses in mice with photoreceptor degeneration. Neuron. 2006;50:23–33. doi: 10.1016/j.neuron.2006.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghuis B.G., Looger L.L., Tomita S., Demb J.B. Kainate receptors mediate signaling in both Transient and Sustained OFF bipolar cell pathways in mouse retina. J. Neurosci. 2014;34:6128–6139. doi: 10.1523/JNEUROSCI.4941-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden E.S. Optogenetics and the future of neuroscience. Nat. Neurosci. 2015;18:1200–1201. doi: 10.1038/nn.4094. [DOI] [PubMed] [Google Scholar]
- Boyden E.S., Zhang F., Bamberg E., Nagel G., Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- Busskamp V., Roska B. Optogenetic approaches to restoring visual function in retinitis pigmentosa. Curr. Opin. Neurobiol. 2011;21:942–946. doi: 10.1016/j.conb.2011.06.001. [DOI] [PubMed] [Google Scholar]
- Busskamp V., Duebel J., Balya D., Fradot M., Viney T.J., Siegert S., Groner A.C., Cabuy E., Forster V., Seeliger M., et al. Genetic reactivation of cone photoreceptors restores visual responses in retinitis pigmentosa. Science. 2010;329:413–417. doi: 10.1126/science.1190897. [DOI] [PubMed] [Google Scholar]
- Carrillo-Reid L., Han S., Yang W., Akrouh A., Yuste R. Controlling visually guided behavior by holographic recalling of cortical ensembles. Cell. 2019;178:447–457.e5. doi: 10.1016/j.cell.2019.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffiol A., Caplette R., Jaillard C., Brazhnikova E., Desrosiers M., Dubus E., Duhamel L., Macé E., Marre O., Benoit P., et al. A new promoter allows optogenetic vision restoration with enhanced sensitivity in macaque retina. Mol. Ther. 2017;25:2546–2560. doi: 10.1016/j.ymthe.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaigneau E., Ronzitti E., Gajowa M.A., Soler-Llavina G.J., Tanese D., Brureau A.Y.B., Papagiakoumou E., Zeng H., Emiliani V. Two-photon holographic stimulation of ReaChR. Front. Cell. Neurosci. 2016;10:234. doi: 10.3389/fncel.2016.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Refractive indices of the rat retinal layers. Ophthalmic Res. 1993;25:65–68. doi: 10.1111/j.1755-3768.1993.tb08722.x. [DOI] [PubMed] [Google Scholar]
- Chen I.-W., Papagiakoumou E., Emiliani V. Towards circuit optogenetics. Curr. Opin. Neurobiol. 2018;50:179–189. doi: 10.1016/j.conb.2018.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I.-W., Ronzitti E., Lee B.R., Daigle T.L., Dalkara D., Zeng H., Emiliani V., Papagiakoumou E. In vivo sub-millisecond two-photon optogenetics with temporally focused patterned light. J. Neurosci. 2019;39:1785–1818. doi: 10.1523/JNEUROSCI.1785-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T.-W., Wardill T.J., Sun Y., Pulver S.R., Renninger S.L., Baohan A., Schreiter E.R., Kerr R.A., Orger M.B., Jayaraman V., et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chettih S.N., Harvey C.D. Single-neuron perturbations reveal feature-specific competition in V1. Nature. 2019;567:334–340. doi: 10.1038/s41586-019-0997-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chichilnisky E.J. A simple white noise analysis of neuronal light responses. Network. 2001;12:199–213. [PubMed] [Google Scholar]
- Chimier B., Utéza O., Sanner N., Sentis M., Itina T., Lassonde P., Légaré F., Vidal F., Kieffer J.C. Damage and ablation thresholds of fused-silica in femtosecond regime. Phys. Rev. B. 2011;84:094104–094110. [Google Scholar]
- Choi V.W., Asokan A., Haberman R.A., Samulski R.J. Production of recombinant adeno-associated viral vectors. Curr. Protoc. Hum. Genet. 2007 doi: 10.1002/0471142301.ns0417s57. [DOI] [PubMed] [Google Scholar]
- Clérin E., Yang Y., Forster V., Fontaine V., Sahel J.-A., Léveillard T. Vibratome sectioning mouse retina to prepare photoreceptor cultures. J. Vis. Exp. 2014 doi: 10.3791/51954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin T., Vandenberghe L.H., Hantz P., Juttner J., Reimann A., Kacsó A.E., Huckfeldt R.M., Busskamp V., Kohler H., Lagali P.S., et al. Efficient transduction and optogenetic stimulation of retinal bipolar cells by a synthetic adeno-associated virus capsid and promoter. EMBO Mol. Med. 2014;6:1175–1190. doi: 10.15252/emmm.201404077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgleish H.W., Russell L.E., Packer A.M., Roth A., Gauld O.M., Greenstreet F., Thompson E.J., Häusser M. How many neurons are sufficient for perception of cortical activity? Elife. 2020;9 doi: 10.7554/eLife.58889. e58889–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalkara D., Byrne L.C., Klimczak R.R., Visel M., Yin L., Merigan W.H., Flannery J.G., Schaffer D.V., Schaffer D.V. In vivo-directed Evolution of a new adeno-associated virus for Therapeutic outer retinal gene delivery from the vitreous. Sci. Transl. Med. 2013;5:189ra76. doi: 10.1126/scitranslmed.3005708. [DOI] [PubMed] [Google Scholar]
- Doroudchi M.M., Greenberg K.P., Liu J., Silka K.A., Boyden E.S., Lockridge J.A., Arman A.C., Janani R., Boye S.E., Boye S.L., et al. Virally delivered channelrhodopsin-2 safely and effectively restores visual function in multiple mouse models of blindness. Mol. Ther. 2011;19:1220–1229. doi: 10.1038/mt.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euler T., Masland R.H. Light-evoked responses of bipolar cells in a mammalian retina. J. Neurophysiol. 2000;83:1817–1829. doi: 10.1152/jn.2000.83.4.1817. [DOI] [PubMed] [Google Scholar]
- Edelstein Arthur D., Tsuchida M.A., Amodaj N., Pinkard H., Vale R.D., Stuurman N. Advanced methods of microscope control using μManager software. J. Biol. Methods. 2014;1:e10. doi: 10.14440/jbm.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euler T., Hausselt S.E., Margolis D.J., Breuninger T., Castell X., Detwiler P.B., Denk W. Eyecup scope-optical recordings of light stimulus-evoked fluorescence signals in the retina. Pflugers Arch. 2009;457:1393–1414. doi: 10.1007/s00424-008-0603-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L.Z., Kheifets S., Böhm U.L., Wu H., Piatkevich K.D., Xie M.E., Parot V., Ha Y., Evans K.E., Boyden E.S., et al. All-optical electrophysiology reveals the role of lateral inhibition in sensory processing in cortical layer 1. Cell. 2020;180:521–535.e18. doi: 10.1016/j.cell.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaub B.M., Berry M.H., Holt A.E., Reiner A., Kienzler M.A., Dolgova N., Nikonov S., Aguirre G.D., Beltran W.A., Flannery J.G., Isacoff E.Y. Restoration of visual function by expression of a light-gated mammalian ion channel in retinal ganglion cells or ON-bipolar cells. Proc. Natl. Acad. Sci. USA. 2014;111:E5574–E5583. doi: 10.1073/pnas.1414162111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauvain G., Akolkar H., Chaffiol A., Arcizet F., Khoei M.A., Desrosiers M., Jaillard C., Caplette R., Marre O., Bertin S., et al. Optogenetic therapy: high spatiotemporal resolution and pattern discrimination compatible with vision restoration in non-human primates. Commun. Biol. 2021;4:125–215. doi: 10.1038/s42003-020-01594-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill J.V., Lerman G.M., Zhao H., Stetler B.J., Rinberg D., Shoham S. Precise holographic manipulation of olfactory circuits reveals coding features determining perceptual detection. Neuron. 2020;108:382–393.e5. doi: 10.1016/j.neuron.2020.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hage T.A., Bosma-Moody A., Baker C.A., Kratz M.B., Campagnola L., Jarsky T., Zeng H., Murphy G.J. Synaptic connectivity to L2/3 of primary visual cortex measured by two-photon optogenetic stimulation. Elife. 2022;11 doi: 10.7554/eLife.71103. e71103–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig H. Taylor and Francis; 1997. Micro-Optics Elements, Systems and Applications. [Google Scholar]
- Hu T., Chklovskii D.B. Advances in Neural Information Processing Systems 22 - Proceedings of the 2009 Conference. 2009. Reconstruction of sparse circuits using multi-neuronal excitation (RESCUME) [Google Scholar]
- Hulliger E.C., Hostettler S.M., Kleinlogel S. Empowering retinal gene therapy with a specific promoter for human rod and cone ON-bipolar cells. Mol. Ther. Methods Clin. Dev. 2020;17:505–519. doi: 10.1016/j.omtm.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji N., Freeman J., Smith S.L. Technologies for imaging neural activity in large volumes. Nat. Neurosci. 2016;19:1154–1164. doi: 10.1038/nn.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khabou H., Garita-Hernandez M., Chaffiol A., Reichman S., Jaillard C., Brazhnikova E., Bertin S., Forster V., Desrosiers M., Winckler C., et al. Noninvasive gene delivery to foveal cones for vision restoration. JCI Insight. 2018;3:96029. doi: 10.1172/jci.insight.96029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapoetke N.C., Murata Y., Kim S.S., Pulver S.R., Birdsey-Benson A., Cho Y.K., Morimoto T.K., Chuong A.S., Carpenter E.J., Tian Z., et al. Independent optical excitation of distinct neural populations. Nat. Methods. 2014;11:338–346. doi: 10.1038/nmeth.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagali P.S., Balya D., Awatramani G.B., Münch T.A., Kim D.S., Busskamp V., Cepko C.L., Roska B. Light-activated channels targeted to ON bipolar cells restore visual function in retinal degeneration. Nat. Neurosci. 2008;11:667–675. doi: 10.1038/nn.2117. [DOI] [PubMed] [Google Scholar]
- Lu Q., Ganjawala T.H., Ivanova E., Cheng J.G., Troilo D., Pan Z.-H. AAV-mediated transduction and targeting of retinal bipolar cells with improved mGluR6 promoters in rodents and primates. Gene Ther. 2016;23:680–689. doi: 10.1038/gt.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz C., Otis T.S., DeSars V., Charpak S., DiGregorio D.A., Emiliani V. Holographic photolysis of caged neurotransmitters. Nat. Methods. 2008;5:821–827. doi: 10.1038/nmeth.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macé E., Caplette R., Marre O., Sengupta A., Chaffiol A., Barbe P., Desrosiers M., Bamberg E., Sahel J.-A., Picaud S., et al. Targeting channelrhodopsin-2 to ON-bipolar cells with vitreally administered AAV restores ON and OFF visual responses in blind mice. Mol. Ther. 2015;23:7–16. doi: 10.1038/mt.2014.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardinly A.R., Oldenburg I.A., Pégard N.C., Sridharan S., Lyall E.H., Chesnov K., Brohawn S.G., Waller L., Adesnik H. Precise multimodal optical control of neural ensemble activity. Nat. Neurosci. 2018;21:881–893. doi: 10.1038/s41593-018-0139-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshel J.H., Kim Y.S., Machado T.A., Quirin S., Benson B., Kadmon J., Raja C., Chibukhchyan A., Ramakrishnan C., Inoue M., et al. Cortical layer–specific critical dynamics triggering perception. Science. 2019;365:eaaw5202. doi: 10.1126/science.aaw5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh L.T., Maheswaranathan N., Nayebi A., Ganguli S., Baccus S.A., Baccus S.A. Deep learning models of the retinal response to natural scenes. Adv. Neural Inf. Process. Syst. 2016;29:1369–1377. [PMC free article] [PubMed] [Google Scholar]
- McRaven C., Tanese D., Zhang L., Yang C.-T., Ahrens M., Emiliani V., Koyama M. High-throughput cellular-resolution synaptic connectivity mapping in vivo with concurrent two-photon optogenetics and volumetric Ca 2+ imaging. bioRxiv. 2020 doi: 10.1101/2020.02.21.959650. Preprint at. [DOI] [Google Scholar]
- Nagel G., Szellas T., Huhn W., Kateriya S., Adeishvili N., Berthold P., Ollig D., Hegemann P., Bamberg E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. USA. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewska G., Vinberg F., Stremplewski P., Bircher M.P., Salom D., Komar K., Zhang J., Cascella M., Wojtkowski M., Kefalov V.J., Palczewski K. Human infrared vision is triggered by two-photon chromophore isomerization. Proc. Natl. Acad. Sci. USA. 2014;111:E5445–E5454. doi: 10.1073/pnas.1410162111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papagiakoumou E., Bègue A., Leshem B., Schwartz O., Stell B.M., Bradley J., Oron D., Emiliani V. Functional patterned multiphoton excitation deep inside scattering tissue. Nat. Photonics. 2013;7:274–278. [Google Scholar]
- Papagiakoumou E., de Sars V., Oron D., Emiliani V. Patterned two-photon illumination by spatiotemporal shaping of ultrashort pulses. Opt Express. 2008;16:22039–22047. doi: 10.1364/oe.16.022039. [DOI] [PubMed] [Google Scholar]
- Park S.J.H., Borghuis B.G., Rahmani P., Zeng Q., Kim I.-J., Demb J.B. Function and circuitry of VIP+ interneurons in the mouse retina. J. Neurosci. 2015;35:10685–10700. doi: 10.1523/JNEUROSCI.0222-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegard N.M., Oldenburg I., Sridharan S., Walller L., Adesnik H. 3D scanless holographic optogenetics with temporal focusing. Nat. Commun. 2017:1–14. doi: 10.1038/s41467-017-01031-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pologruto T.A., Sabatini B.L., Svoboda K. ScanImage: flexible software for operating laser scanning microscopes. Biomed. Eng. Online. 2003;2:13. doi: 10.1186/1475-925X-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Printz Y., Patil P., Mahn M., Benjamin A., Litvin A., Levy R., Bringmann M., Yizhar O. Determinants of functional synaptic connectivity among amygdala-projecting prefrontal cortical neurons. bioRxiv. 2021 doi: 10.1101/2021.11.07.467611. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson N.T.M., Descamps L.A.L., Russell L.E., Buchholz M.O., Bicknell B.A., Antonov G.K., Lau J.Y.N., Nutbrown R., Schmidt-Hieber C., Häusser M. Targeted activation of hippocampal place cells Drives memory-Guided spatial behavior. Cell. 2020;183:2041–2042.e10. doi: 10.1016/j.cell.2020.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronzitti E., Conti R., Zampini V., Tanese D., Foust A.J., Klapoetke N., Boyden E.S., Papagiakoumou E., Emiliani V. Submillisecond optogenetic control of neuronal firing with two-photon holographic photoactivation of chronos. J. Neurosci. 2017;37:10679–10689. doi: 10.1523/JNEUROSCI.1246-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronzitti E., Ventalon C., Canepari M., Forget B.C., Papagiakoumou E., Emiliani V. Recent advances in patterned photostimulation for optogenetics. J. Opt. 2017;19:113001. [Google Scholar]
- Roska B., Sahel J.-A. Review - restoring vision. Nature. 2018;557:359–367. doi: 10.1038/s41586-018-0076-4. [DOI] [PubMed] [Google Scholar]
- Rousso D.L., Qiao M., Kagan R.D., Yamagata M., Palmiter R.D., Sanes J.R. Two pairs of ON and OFF retinal ganglion cells are defined by intersectional patterns of transcription factor expression. Cell Rep. 2016;15:1930–1944. doi: 10.1016/j.celrep.2016.04.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahel J.A., Boulanger-Scemama E., Pagot C., Arleo A., Galluppi F., Martel J.N., Esposti S.D., Delaux A., de Saint Aubert J.B., de Montleau C., et al. Partial recovery of visual function in a blind patient after optogenetic therapy. Nat. Med. 2021;27:1223–1229. doi: 10.1038/s41591-021-01351-4. [DOI] [PubMed] [Google Scholar]
- Shemesh O.A., Tanese D., Zampini V., Linghu C., Piatkevich K., Ronzitti E., Papagiakoumou E., Boyden E.S., Emiliani V. Temporally precise single-cell-resolution optogenetics. Nat. Neurosci. 2017;20:1796–1806. doi: 10.1038/s41593-017-0018-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjur T. Coefficients of determination in logistic regression models—a new proposal: the coefficient of discrimination. Am. Stat. 2009;63:366–372. [Google Scholar]
- Tsukamoto Y., Omi N. Functional allocation of synaptic contacts in microcircuits from rods via rod bipolar to AII amacrine cells in the mouse retina. J. Comp. Neurol. 2013;521:3541–3555. doi: 10.1002/cne.23370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y., Omi N. Classification of mouse retinal bipolar cells: type-specific connectivity with special reference to rod-driven AII amacrine pathways. Front. Neuroanat. 2017;11:92. doi: 10.3389/fnana.2017.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wyk M., Pielecka-Fortuna J., Löwel S., Kleinlogel S. Restoring the ON switch in blind retinas: opto-mGluR6, a next-generation, cell-tailored optogenetic tool. PLoS Biol. 2015;13:1–30. doi: 10.1371/journal.pbio.1002143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villette V., Chavarha M., Dimov I.K., Bradley J., Pradhan L., Mathieu B., Evans S.W., Chamberland S., Shi D., Yang R., et al. Ultrafast two-photon imaging of a high-gain voltage indicator in awake behaving mice. Cell. 2019;179:1590–1608.e23. doi: 10.1016/j.cell.2019.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz A., Trenholm S., Yonehara K., Hillier D., Raics Z., Leinweber M., Szalay G., Ghanem A., Keller G., Rózsa B., et al. Single-cell-initiated monosynaptic tracing reveals layer-specific cortical network modules. Science. 2015;349:70–74. doi: 10.1126/science.aab1687. [DOI] [PubMed] [Google Scholar]
- Yang W., Miller J.E.K., Carrillo-Reid L., Pnevmatikakis E., Paninski L., Yuste R., Peterka D.S. Simultaneous multi-plane imaging of neural circuits. Neuron. 2016;89:269–284. doi: 10.1016/j.neuron.2015.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Wang L.P., Brauner M., Liewald J.F., Kay K., Watzke N., Wood P.G., Bamberg E., Nagel G., Gottschalk A., Deisseroth K. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.