Summary
In living systems, a complex network of protein-protein interactions (PPIs) underlies most biochemical events. The human protein-protein interactome has been surveyed using yeast two-hybrid (Y2H)- and mass spectrometry (MS)-based approaches such as affinity purification coupled to MS (AP-MS). Despite decades of systematic investigations and collaborative multi-disciplinary efforts, there is no “gold standard” for documenting PPIs. A surprisingly large fraction of the human interactome remains uncharted, which we refer to as the “dark interactome.” In this review, we highlight the complexity of the human interactome and discuss the current status of the human reference interactome maps. We discuss why a large proportion of the human interactome has remained refractory to traditional approaches. We propose an experimental model that can enable the identification of the dark interactome in a cell-type-specific manner. We also propose a framework to implement when embarking on studies designed to rigorously identify and characterize protein interactions.
Keywords: protein-protein interaction, interactome, mass spectrometry, AP-MS, Y2H, BioID
A large fraction of the human protein-protein interaction network, or “dark interactome,” remains uncharted. In this review, Sharifi Tabar et al. discuss the dynamic nature of protein-protein interactions, features of current methodologies, and the lack of appropriate experimental models. A strategy to illuminate the dark interactome is proposed.
Introduction
A detailed understanding of biochemical processes is required to dissect the pathogenesis of human diseases. Protein-protein interactions (PPIs) underlying these biochemical processes can be considered as the molecular language of life because biological information is passed on via a myriad of protein interactions throughout the cellular milieu. Thus, the more we understand this molecular language, the more we will understand the molecular basis of diseases. Comparisons of protein expression profiles between healthy and diseased individuals can pave the way for molecular therapies (Fathi et al., 2018; Wang et al., 2021; Xu et al., 2020). International initiatives such as the chromosome-centric human proteome project have generated a “parts list” of proteins across human tissues and cell lines (Adhikari et al., 2020; Betancourt et al., 2021; Jangravi et al., 2013; Wilhelm et al., 2014). However, documenting a list of tissue-specific or differentially expressed proteins does not adequately account for the nuances of biochemical processes involved in health and disease. This is mainly because highly complex diseases such as cancer do not follow the one-gene/one-function rule, and system-level information is required (Beadle and Tatum, 1941; Sharifi Tabar et al., 2022a; Wagner and Zhang, 2011).
Affinity purification coupled to mass spectrometry (AP-MS) and yeast two-hybrid (Y2H) approaches have been widely used to map PPIs and have generated a wealth of information (Hein et al., 2015; Huttlin et al., 2021; Low et al., 2020; Qin et al., 2021; Schmidberger et al., 2016; Sharifi Tabar et al., 2019; Torrado et al., 2017). Such datasets have been used to generate large-scale reference protein interactome maps (Huttlin et al., 2021; Luck et al., 2020). Despite these efforts, a substantial fraction of the human proteome remains uncharacterized. Those missing interactions, which we refer to as the “dark interactome,” frequently include PPIs that are not identifiable using traditional techniques. Technical limitations as well as a lack of appropriate experimental cell models have greatly contributed to the existence of the dark interactome. These limitations arise from the suitability of molecular tools, biochemical reagents, and instrumentation currently being used to study PPIs. For instance, many reports use the same biochemical reagents and buffer recipes in AP-MS-based PPI studies, which, notably, are not appropriate for all proteins within the human proteome.
Moreover, gene expression can be cell-type specific and vary between different cell types in the same tissue (e.g., dopaminergic neurons versus oligodendrocytes in brain) or between different tissues (Vakilian et al., 2015; Wang et al., 2020). Therefore, immortalized cell lines, such as HEK293 and HeLa cells, which have been widely used in interactome studies, are not always appropriate models to identify tissue-specific and cell-type-specific interactions. These standard cell models inherently limit the capture of prey proteins that are absent, differentially located, or weakly expressed in these cells. Thus, new experimental cell models and alternative MS-based approaches are required to explore the dark interactome.
Proximity labeling coupled to MS (PL-MS) approaches such as proximity-dependent biotin identification (BioID) have successfully been used to map the interaction networks of membrane proteins and intrinsically disordered proteins. Such PPIs have been refractory to biochemical isolation and identification using standard methods (Huttlin et al., 2021). Unlike AP-MS, where protein lysate is used as input material, PL-MS is implemented in situ and thus is capable of identifying low-affinity and transient interactions. This is exemplified in studying signaling pathways in response to stimuli or host-pathogen interactions. PL-MS has also successfully been used to study tissue-specific protein interactions (Uezu et al., 2016). In this review, we discuss the challenges of currently used approaches and propose a PL-MS-focused strategy in combination with a new experimental cell model to facilitate identification of the dark interactome.
The complexity of the interactome
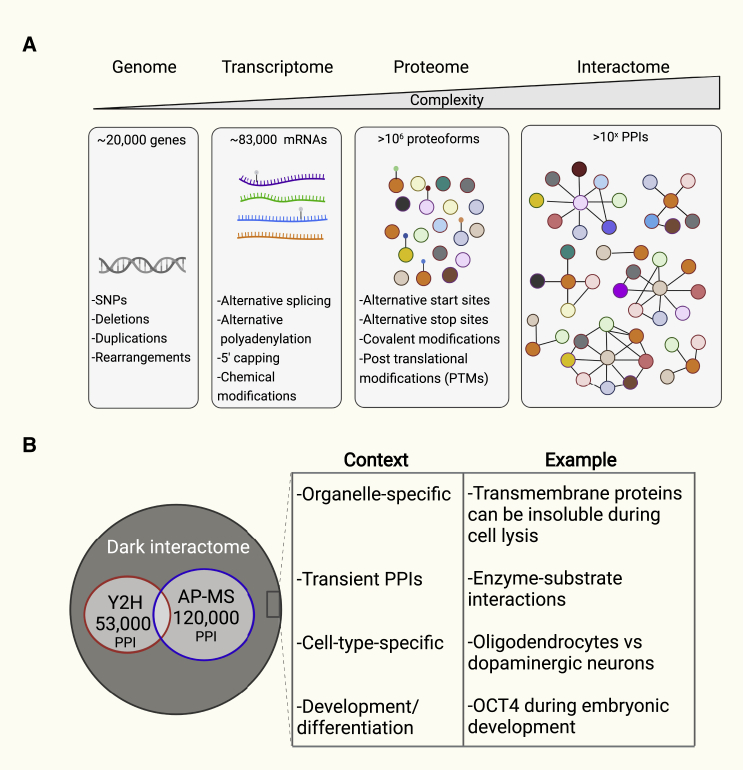
In biological systems, complexity increases from the genome, through the transcriptome, to the proteome. The system becomes exponentially complex at the interactome level (Figure 1A). The UniProt database contains more than 130,000 validated coding non-synonymous single-nucleotide polymorphisms, which can provide a significant additional source of variation at the transcriptome and proteome levels (UniProt, 2021). However, in cancer, this complexity becomes even more elaborate due to alterations at the DNA, RNA, and protein levels. The cancer genome often contains many mutations that arise from errors during protein translation and defects in the DNA repair machinery. The genomic landscape of over 3,000 tumor samples has revealed nearly 300,000 mutations in protein-coding regions (Vogelstein et al., 2013).
Figure 1.
Complexity increases from the genome to the interactome in human cells
(A) Interactome complexity is generated at the genome, transcriptome, and proteome levels, with alternative splicing and post-translational modifications (PTMs) among other features that contribute to protein-protein interaction (PPI) diversity.
(B) Y2H and AP-MS have been used to define PPI networks of human proteins. Tens of thousands of interactions have been reported, but a large portion of the human interactome has remained uncharacterized, known as the dark interactome, which is depicted in dark gray. The table provides the biological and cellular context and examples of proteins within the dark interactome.
Compared with the genome, the transcriptome is more diverse and complex, containing coding (i.e., mRNA) and non-coding RNA species (e.g., ribosomal RNA, tRNA, long non-coding RNA, and microRNA). The majority (93%) of human protein-coding genes are alternatively spliced, and many exhibit alternate transcription start sites, which have been estimated to produce more than 83,000 functional isoforms (Aebersold et al., 2018; Wang et al., 2008). In addition to alternative splicing, RNA modifications such as 3′ alternative polyadenylation, 5′ capping, and chemical modifications (e.g., m6A) can also lead to more complexity and diverse functionality, all affecting mRNA processing and stability (Figure 1A).
Biological complexity is further increased at the protein level by post-translational modifications (PTMs), of which ∼400 different types have been identified and recorded within UniProt (Bludau and Aebersold, 2020; UniProt, 2021). These modifications can individually or in a combinatorial fashion modulate many biological processes through influencing protein stability, localization, and interactions. Collectively, these variations and modifications are estimated to generate more than one million different proteoforms, which can consequently lead to potentially millions of PPIs in both normal and disease states (Bludau and Aebersold, 2020) (Figure 1A).
The interactome comprises both permanent and transient interactions that occur at nanomolar and micromolar affinities, respectively. The human interactome is predominantly transient, with stable complexes occurring less frequently (Hein et al., 2015). Multi-subunit protein complexes such as transcriptional and translational machinery are typically permanent interactions with conserved stoichiometry across various cell types and species. There are over 4,600 stable protein complexes characterized in the human proteome (Bludau and Aebersold, 2020; UniProt, 2021). The majority of proteins are involved in transient interactions for adaptive responses to biochemical or environmental stimuli. Transient interactions are mainly mediated by transmembrane and cytoplasmic proteins and are key features of signaling pathways and regulatory networks (Varnaite and MacNeill, 2016).
The dark interactome
The human interactome has been rigorously subjected to standard biochemical characterization (Huttlin et al., 2021; Luck et al., 2020). In 2020, the Human Reference Interactome, also known as HuRI, reported the largest physical binary interaction map for human proteins using a Y2H approach (Luck et al., 2020). In this project, 17,500 bait and prey proteins each were co-expressed and tested for interaction in a pairwise manner, a total of approximately three billion individual tests. The resultant dataset contained ∼53,000 high-confidence interactions among ∼8,000 proteins (Figure 1B) (Luck et al., 2020); however, it represented less than 11% of all human protein interactions. The vast majority of the interactome remained undetected for several reasons: (1) yeast are not optimal to examine all mammalian proteins and generally lack human biomolecular co-factors; (2) secretory pathways, membrane and highly disordered proteins fail to express and fold properly by Y2H; (3) yeast exhibits less fidelity in reproducing PTMs important for protein folding and interaction of mammalian proteins; (4) proteins within multi-subunit complexes often require the presence of that complex to interact; (5) the presence of fusion tags can influence protein folding and interaction; and (6) some proteins only interact in signaling pathways that are absent in yeast.
In parallel, a reference human interactome was generated for the BioPlex project using AP-MS. Here, human genes were hemagglutinin (HA)- and FLAG-tagged on the C terminus and expressed in HEK293 cells, and associating protein complexes were affinity purified from crude cell lysates and analyzed by MS. Nearly 120,000 direct and indirect interactions were reported (Huttlin et al., 2021) (Figure 1B). Accordingly, there are several cellular and biological contexts in which PPIs fail to be detected, and the examples provided highlight this (Figure 1B). In addition, AP-MS suffers from several limitations that contribute to those missing interactions, as well as the detection of false positive interactions. These include (1) mild cell lysis conditions used to preserve protein complexes in their semi-native conditions result in many nuclear, membrane, and cytosolic proteins being poorly solubilized and remaining insoluble (Beck et al., 2014; Varnaite and MacNeill, 2016); (2) protein interactions with a weak binding affinity may not be identified; (3) some protein interactions are disrupted upon cell lysis because such interactions occur only in a specific signaling pathway or within a unique microenvironment within its correct location (e.g., Golgi); and (4) some interactions are also lost during stringent washing conditions.
Together, it is evident that there is scant interaction information for a considerable portion of the human proteome. Methodological limitations and the lack of appropriate experimental models are the main obstacles. In the following sections, we discuss MS-based approaches for mapping PPIs and discuss a model experimental design that should greatly facilitate the illumination of the dark interactome.
AP-MS is the method of choice to capture high-affinity protein interactions
Two derivatives of AP-MS exist, which are based on similar principles and are used interchangeably in the literature: immunoprecipitation followed by MS (IP-MS) and pull-down followed by MS (PD-MS). Advantages and disadvantages of either approach exist (Table 1), which should be taken into consideration during experimental design. In IP-MS, an IP-grade antibody is immobilized onto a solid phase (i.e., a bead) and mixed with cell lysates to capture the target protein and its associated protein complexes (Figure 2A). Whereas in PD-MS, the gene of interest is fused to an epitope tag (e.g., FLAG, HA, cMyc) and ectopically expressed in target cell types. Overexpression is achieved either by transfection (mammalian expression vector) or transduction (retroviral/lentiviral vector) depending on the plasmid carrying the gene of interest (Figure 2A). Alternatively, to achieve a more physiological level of expression, the epitope tag can be inserted into the endogenous locus of the target gene using CRISPR-Cas9-mediated gene editing. Some of the advantages and disadvantages of IP-MS and PD-MS are explained in more detail below.
-
(1)
Depth. The depth of interactome data obtained for PD-MS is greater than for IP-MS for a number of reasons. Firstly, high-affinity monoclonal antibodies pre-conjugated to beads are commercially available to capture epitope tags. Strong binding of these antibodies to epitope tags enhances the purification of protein complexes and, hence, identification of PPIs. Secondly, the overexpression of the bait protein provides a molecular interface to capture binding partners in abundance. In contrast, IP-MS studies usually result in an incomplete PPI map for two main reasons: the lack of an appropriate high-affinity IP-grade antibody for most proteins, and antibody interaction with the target protein can mask potential PPI motifs or lead to protein conformation changes and loss of PPIs (Al Qaraghuli et al., 2020; Wilson and Stanfield, 1994).
-
(2)
Time. In contrast to IP-MS, in ectopic PD-MS, there is minimal requirement to optimize the antibody-antigen binding, allowing parallel sample preparation for many proteins with a common protocol (Gingras et al., 2007). However, endogenous PD-MS experiments require more time as they utilize CRISPR-Cas9-mediated gene editing.
-
(3)
Cost. Ectopic PD-MS is more cost effective compared with endogenous PD-MS and IP-MS because it is less labor intensive and can be done in a shorter time.
-
(4)
Flexibility. If the PPI study requires examining a range of cell types or cellular or biological contexts (see Figure 1B), then IP-MS is a more flexible approach, as it utilizes lysates from native cells. PD-MS, however, usually requires transfection or transduction of target cells, which may not be optimal in all contexts.
-
(5)
Domain-specific PPIs. A key advantage of ectopic PD-MS is that domain-specific interactome studies can be readily performed for the majority of proteins within the human proteome. In addition, the impact of deletion mutants or disease-associated missense mutations on PPIs can be directly addressed.
Table 1.
Summary of advantages and disadvantages of different AP-MS-based approaches
| IP-MS | PD-MS |
|||
|---|---|---|---|---|
| Endogenous | Ectopic |
|||
| Transduction | Transfection | |||
| Depth | low-medium | high | high | high |
| Time | weeks to months | months | days to weeks | days to weeks |
| Cost | high | high | low | very low |
| Flexibility | high | low | medium | low |
| Domain-specific PPIs | not feasible | not feasible | feasible | |
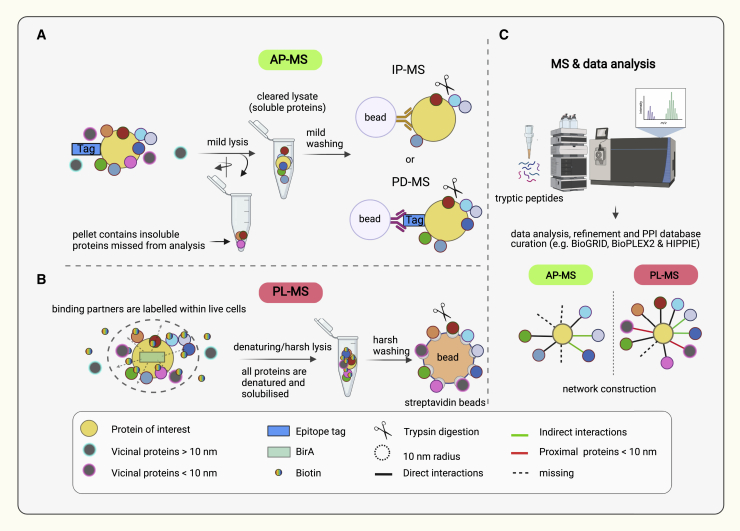
Figure 2.
Schematic of AP-MS and PL-MS approaches
(A) AP-MS using an IP-grade antibody (IP-MS) or epitope tag (PD-MS). Epitope-tagged proteins of interest (yellow circle) are endogenously or ectopically expressed in target cells. After mild lysis, solubilized proteins are separated from insoluble proteins by centrifugation. Soluble proteins are mixed with beads conjugated to specific antibodies against the epitope tag or POI to capture the associated proteins. After several stringent washes, direct and indirect interactions (colored circles) are co-purified with the bait from the complex cell lysate.
(B) In PL-MS, BirA-fused proteins of interest (yellow circle and light green rectangle) are endogenously or ectopically expressed in target cells. Labeling is initiated with the addition of biotin to cultured cells. BirA enzyme mediates the labeling of direct and indirect interactions as well as vicinal proteins within 10 nm distance (represented by dotted circle). After labeling, cells are lysed using a harsh and denaturing lysis buffer to enhance solubilization of proteins. Biotinylated proteins are immobilized on streptavidin beads and then are washed before proceeding to on-bead tryptic digestion.
(C) Peptides generated in (A) and (B) are desalted using C18 columns and then subjected to MS analysis. As depicted in the interaction network, PL-MS results in increased detection of interactions and fewer missed interactions than AP-MS.
PL-MS has the potential to uncover the dark interactome
The main regulators of signaling pathways undergo transient interactions with upstream and downstream effectors in response to stimuli or stress conditions. PL-MS has greatly advanced the identification of these interactions that were inaccessible using AP-MS or Y2H approaches (Bosch et al., 2021; Go et al., 2021; Qin et al., 2021; Samavarchi-Tehrani et al., 2020b). Briefly, PL-MS has been designed to screen for transient and stable protein interactions as well as neighboring proteins (within a 10 nm radius) in a natural cellular environment. In this approach, the protein of interest (POI) is fused to an engineered BirA enzyme, a biotin ligase derived from E. coli. These enzymes utilize ATP to release biotinoyl-AMP intermediates from biotin molecules, which can attach to side-chain amines on lysine residues. In the engineered BirA, arginine residue 118 has been replaced with glycine, enabling efficient biotin labeling of transient interactions in situ and often in a spatiotemporal manner (Figure 2B). Typically, proteins more vicinal to the enzyme active site exhibit higher labeling densities (Rhee et al., 2013).
The application, advantages, and disadvantages of various PL-MS approaches has been reviewed elsewhere (Bosch et al., 2021; Samavarchi-Tehrani et al., 2020b; Trinkle-Mulcahy, 2019). Importantly, PL-MS has been successfully used to identify interactions in diverse cellular and biological contexts, including enzyme-substrate interactions (Gingras et al., 2007) and host-pathogen interactions (Laurent et al., 2020; Samavarchi-Tehrani et al., 2020a). BioID, the most widely used PL method, has been used to map the interactome of proteins with different subcellular localization in a variety of model systems including primary and immortalized cancer cells, yeast, flies, mice, zebrafish, worms, and plants (Qin et al., 2021). One of the main reasons that BioID has been popular in identifying “dark” or refractory interactions is the extraordinarily high binding affinity of biotin to streptavidin (Kd of ∼10−14 mol/L) (Green, 1975). Such a strong complex can withstand the presence of organic solvents, extreme pH, temperature, and detergents and denaturing reagents such as urea, SDS, and Triton X-100 in the lysis and wash buffers (Branon et al., 2018; Holmberg et al., 2005; Roux et al., 2012, 2018).
To enhance the efficiency and speed of labeling while minimizing toxicity, new classes of BirA enzymes have been engineered, including TurboID and miniTurbo (Branon et al., 2018). An innovative PL approach, so-called “off-the-shelf” proximity biotinylation, has been introduced recently. Here, TurboID fused to protein A is targeted to the bait protein using specific antibodies with the method successfully benchmarked on nuclear proteins in both fixed and non-fixed cells (Santos-Barriopedro et al., 2021). Future developments in this method might enable its application to clinical samples and primary cells that are otherwise hard to manipulate for PL-MS studies. Taken together, PL-MS offers tremendous potential for the high-throughput identification of previously inaccessible PPIs within the dark interactome.
A cell-type-specific proteomics strategy to illuminate the dark interactome
Cell-type-specific transcriptome and proteome studies have consistently demonstrated that the cellular protein content varies between different cell types (Alvarez-Castelao et al., 2019; Jiang et al., 2020; Wang et al., 2019; Wilson and Nairn, 2018). Body-wide quantitative proteomics of 12,000 proteins has recently revealed that nearly half are tissue enriched or tissue specific (Jiang et al., 2020). As an example, homeobox protein OTX2 is highly expressed in the brain tissue mainly in the neural progenitor cell, while dopamine transporter 1 (DAT1) and glial fibrillary acidic protein (GFAP) are dominantly expressed in dopaminergic and astrocyte neurons, respectively (Maury et al., 2015). Therefore, protein interaction networks will be markedly different in various cell types within the same tissue and between different tissues within humans. Proteome-wide AP-MS studies (e.g., BioPlex project) have only employed transformed cell lines for mapping the human interactome (Huttlin et al., 2021). Thus, current models are inadequate for generating comprehensive maps, and there is a need for new experimental pipelines to reveal dark interactions.
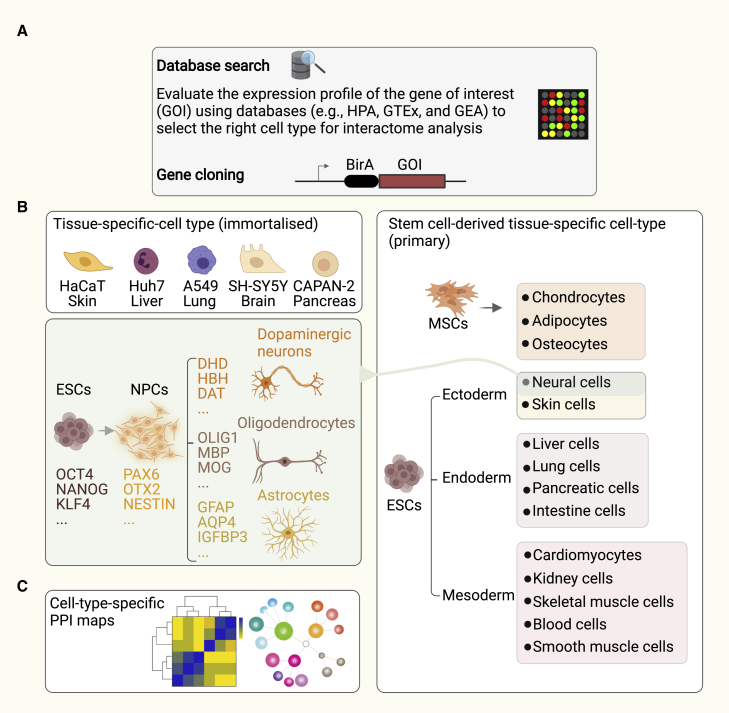
Here, we propose a cell-type-specific approach to shine a light on the dark interactome (Figure 3). In this model, proteins are first categorized based on their expression pattern in relevant tissues and cell types. Freely accessible repositories such as Human Protein Atlas (HPA), The Geno-type-Tissue Expression (GTEx) portal, Gene Expression Atlas (GEA), and Proteomics DataBank (ProteomicsDB) are very useful resources to investigate the expression pattern of the gene of interest (Figure 3A). After classification, the gene of interest is cloned into an appropriate mammalian expression vector for PL-MS analysis (Figure 3A). As a complementary approach, BirA can be endogenously fused to the gene of interest using CRISPR-Cas9-mediated gene editing, which will greatly reduce identification of false positive hits due to bait protein overexpression. In the next step, either primary or immortalized tissue-specific cells are transfected or transduced. Examples of cell types can include stem cells, their derivatives, and cancer-specific cell lines (Figure 3B). Finally, cell-type-specific PPIs are identified and reported for various cell types. This strategy will uncover genuine and functional interacting partners of tissue-enriched and tissue-specific proteins.
Figure 3.
Proposed strategy for uncovering cell-type-specific PPIs
(A) Prior to an interactome study, the expression pattern of the gene of interest (GOI) is investigated using data repositories such as the Human Protein Atlas (HPA), Geno-type-Tissue Expression (GTEx), or Gene Expression Atlas (GEA). An appropriate mammalian expression viral vector is used to express the GOI in frame with a BirA enzyme (e.g., TurboID) for PL-MS.
(B) Transfection or transduction is carried out using an immortalized tissue-specific cell type, tissue-specific or primary stem cell line such as embryonic stem cells (ESCs) and mesenchymal stromal cells (MSCs), or their derivative cell types.
(C) Once MS-based interactome studies are conducted, cell-type-specific maps can be generated.
Cell-type-focused PPI studies are of crucial importance for understanding host-pathogen interactions. Recently, several studies have used either AP-MS or PL-MS approaches to investigate the interaction of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) proteins with host cellular proteins for drug discovery projects (Gordon et al., 2020a, 2020b; Laurent et al., 2020; Samavarchi-Tehrani et al., 2020a). Comparison of the data generated using the same proteomics approach, but on different cell types, reveal that despite significant overlap, many unique PPIs are reported in each study. The discrepancy between datasets could potentially be due to the differences in cell types being used or statistical analyses applied. In these types of studies, A549 alveolar basal epithelial cells would be an appropriate model, as SARS-CoV-2 primarily infects airway epithelial cells. Therefore, choosing an appropriate model cell for host-pathogen protein interactions will provide more meaningful data and shed more light on the dark interactome.
General considerations for interactome studies
A successful interactomics workflow requires the integration of proper experimental design, sample preparation, instrumentation, and bioinformatics analysis. Thus, it is important to understand the objectives and hypotheses of the project and choose the best sample preparation and quantitation approaches accordingly. Important parameters required for successful PPI studies are summarized in Table 2 but expanded upon further below.
-
(1)
Localization of the POI. Depending on the localization of the target protein, the composition of the cell lysis buffer can greatly affect the solubilization efficiency. For example, in AP-MS experiments, including reagents such as digitonin and n-dodecyl-b-D-maltoside (DDM) in the lysis buffer can efficiently solubilize and enable the pull-down of proteins localized in the membrane of the cell and organelles, while for nuclear proteins, it may be optional. In addition, subcellular fractionation can enhance the chance of detecting lowly abundant interactors and reducing any background contaminants. The HPA, and SubCellBarcode repositories are very useful to check the localization of proteins. For PL-MS studies, variations in the pH, redox environment, and nucleophile concentrations can affect the activity of BirA enzymes. For example, TurboID outperforms BioID and miniTurbo in the mitochondrial matrix, nucleus, and endoplasmic reticulum lumen (Branon et al., 2018). Therefore, it is important to choose an enzyme that is active in the target subcellular compartment.
-
(2)
Molecular weight of the POI. Generally, large proteins (>200 kDa) are difficult to pull-down efficiently compared with smaller proteins, and this may compromise the depth and quality of the data. Smaller domains can be effectively used to complement data obtained with the full-length protein. Large proteins are also intractable to in vitro recombinant expression and purification unless domains or sub-regions are used.
-
(3)
Tissue specificity and choice of cell line. Proteins with an enhanced or restricted expression in certain tissues may consequently exhibit a tissue-specific interactome. Therefore, not every laboratory cell line fulfills the purpose. Performing a protein-protein interactome study in a physiologically relevant cell line will reveal more genuine interactions. Of note, some proteins co-localize upon the presence of specific stimuli or stress conditions; therefore, specific experimental design and parameters are required to capture interactions while the protein is behaving in its native context.
-
(4)
Epitope tags. Epitope tagging can result in partial misfolding of the tagged protein, consequently altering its interaction profile either by disrupting or introducing binding artifacts (Wissmueller et al., 2011). For example, partial misfolding and false positive interactions have been reported with GST-tagged Kruppel-like factor 3 (KLF3) (Wissmueller et al., 2011). In addition, N-terminally FLAG-tagged histone deacetylase 1 (HDAC1) exhibits a sharp reduction in enzymatic activity compared with wild-type or C-terminally-tagged HDAC1 protein, indicating the importance of the position of the tag. Furthermore, a recent study has confirmed the interaction of the nucleosome remodeling and deacetylase (NuRD) complex subunit, cyclin-dependent kinase 2-associated protein 1 (CDK2AP1), with the nuclear receptor co-repressor (NCOR) complex, when CDK2AP1 was FLAG tagged (Sharifi Tabar et al., 2022b), However, previous studies failed to show this when green fluorescent protein (GFP) was used as a tag, possibly due to steric hindrance (Spruijt et al., 2010). Taken together, the size and position of the epitope tag may affect the PPI networks, and insertion of the tag at either the C terminus or the N terminus might need to be tested.
-
(5)
Cell culture medium. During PL-MS experiments, it is crucial to check the formulation of cell culture medium as to whether it is supplemented with biotin. The presence of biotin can significantly skew results, especially when exogenous biotin needs to be added at a specific time point to explore temporal interactions such as the cell cycle or host-pathogen interactions. An alternative medium or biotin depletion should be used.
-
(6)
Appropriate controls. Choosing an appropriate control is extremely important in PPIs studies to distinguish POI-mediated enriched proteins from non-specifically enriched proteins. Incubating the cell lysate with isotype antibodies or unbound beads has widely been used as a control in interactome studies. Ideally, the best control would be a gene knockout (KO) cell line model, where the target protein is absent, and non-specific binding of the antibody can be clearly distinguished. However, generating KO controls can be time consuming and technically demanding, which is further complicated by gene essentiality. In case of PL-MS, suitable controls include a construct carrying the target protein only, an “empty vector” containing the BirA enzyme alone, or not adding biotin into the media for detecting any promiscuous labeling of proximal proteins.
-
(7)
Quality of the beads. Agarose or magnetic beads conjugated to streptavidin, protein A/G, or epitope tag antibodies (e.g., FLAG, HA) are frequently used for affinity capture of protein complexes from a complex cellular milieu. However, it has been noted that they can introduce substantial variation in the quality of interaction data (St-Germain et al., 2020). One reason could be batch-to-batch variation or inappropriate storage conditions of the affinity resins. Therefore, it is crucial to use validated high-quality reagents and perform quality checks over time to monitor performance.
-
(8)
Quantitation method. Quantitative PPI studies can be label based or label free, which is comprehensively reviewed elsewhere (Anand et al., 2017; Neilson et al., 2011). In the label-based quantification approaches, MS-detectable specific chemical tags are added to the proteins or peptides to enhance quantification accuracy and signal-to-noise ratio. These mass tags can be introduced into proteins via metabolic labeling of cells such as stable isotope labeling of amino acids (SILAC) in cell culture or into peptides by chemical means such as tandem mass tag (Bantscheff et al., 2007; Ong and Mann, 2006). Label-free approaches are cost effective and faster as no labeling is performed during sample preparation. Hence, label-free approaches are widely used for quantification purposes. The quantification of label-free samples is generally measured by comparing either peptide (precursor ion) intensity or number of spectral counts between different groups (Neilson et al., 2011).
Table 2.
General considerations for protein-protein interactome studies
| Consideration | Problem | Solution |
|---|---|---|
| Localization |
|
|
|
|
|
| Molecular weight |
|
|
| Epitope tags or fusions |
|
|
| Cell culture medium |
|
|
| Controls |
|
|
| Quality of the beads |
|
|
| Quantitation method |
|
|
Challenges in validation of direct interactions
The determination of direct PPIs is essential for a mechanistic understanding of molecular events and for rational drug design. MS-based approaches identify many novel PPIs but cannot distinguish direct interactions from indirect interactions. In addition, false positive interactions are inevitably included. Thus, discriminating between direct PPIs and false positive hits is challenging and needs careful experimental design. Numerous computational algorithms have been developed that have improved the quality and trustworthiness of PPI networks (Tyanova et al., 2016). However, it is nearly impossible to determine direct interactions using scoring algorithms. Therefore, verification of direct interactions using a robust experimental method is required, for which two such approaches exist. First, cross-reference matching, where the potential novel PPIs identified using MS-based approaches can sometimes be validated through cross-referencing with several large PPI repositories that frequently update their database (examples include The Biological General Repository for Interaction Datasets [BioGrid] [Oughtred et al., 2021], the International Molecular Exchange Consortium [IMEx] (Orchard et al., 2012), and the Human Integrated Protein-Protein Interaction reference [HIPPIE] (Alanis-Lobato et al., 2017). Second, validating a list of highly enriched and functionally relevant candidates using biochemical and biophysical approaches, as it has been well established that not every interaction documented in literature-curated PPI repositories is reliable (Cusick et al., 2009; Mackay et al., 2007; Myers et al., 2006). This is mainly because curated interaction information in the databases is generated using machine-learning algorithms that search for specific terms within the text of any publication. This problem perpetuates when false positive and wrongly reported direct interactions become embedded in the literature due to subsequent citations and integration into databases. For example, the direct interaction of GATA zinc finger domain containing 2A or B (GATAD2A/B) with retinoblastoma binding protein 4 or 7 (RBBP4/7), HDAC1 or methyl-CpG-binding domain protein (MBD3) was reported previously, which was later disproven by several other studies (Low et al., 2020; Sharifi Tabar et al., 2019, 2022b; Torrado et al., 2017). In an attempt to validate ∼20 physical interactions previously in the literature, Mackay et al. could only verify 50% of these interactions using biophysical methods such as nuclear magnetic resonance (NMR) (Mackay et al., 2007). This indicates that extra care must be taken when either reporting or referring to direct PPIs. Confirmatory studies in which PPIs are further supported by robust biochemical and biophysical assays are recommended.
A fundamental question is how to choose the best experimental method to characterize direct PPIs? Verification of direct interactions is not straightforward due to the different biochemical and biophysical properties of proteins. However, verification can be undertaken in a stepwise manner to obtain high-confidence direct interactions. First, a list of highly enriched prey proteins should be selected from the list of the identified proteins. Second, proteins that are functionally relevant and are localized in the same compartment as the bait protein are selected. Third, proteins ranking lowly in repositories of common contaminants in AP-MS experiments (e.g., CRAPome [Mellacheruvu et al., 2013]) are prioritized.
Next, the candidate prey proteins and the bait protein need to be tagged and co-expressed in a model cell line (e.g., HEK293 cells) for a pairwise comparison using co-immunoprecipitation. Initial results can be refined by using smaller fragments or domains. Mutation or deletion of the minimal interaction domains or motif can be used to further corroborate the data. Notably, however, defining direct interactions between overexpressed subunits of a multi-subunit complex in mammalian cells can be compromised by the presence of endogenous complexes (Torrado et al., 2017). Finally, a pairwise comparison using in vitro-transcribed and -translated protein can reliably demonstrate whether the interaction is direct. This approach has successfully been used to characterize direct inter-subunit connections within large protein complexes (Low et al., 2020; Schmidberger et al., 2016; Sharifi Tabar et al., 2019, 2022b; Torrado et al., 2017).
After confirming direct PPIs, high-resolution structural and biochemical information is required to guide any drug discovery or functional evaluation of the interactions. Powerful biophysical methods including surface plasmon resonance (SPR), NMR, isothermal titration calorimetry (ITC), X-ray crystallography, and cryoelectron microscopy (cryo-EM) are the most frequently used techniques and have been reviewed elsewhere (Walport et al., 2021).
Deep learning and artificial intelligence may reveal the dark interactome at scale
The use of artificial intelligence (AI) and deep learning (DL) has revolutionized the field of in silico protein structure prediction. Deep learning (DL)-based algorithms such as AlphaFold2 and RoseTTAFold have claimed to predict protein structures as accurately as X-ray crystallography (Baek et al., 2021; Jumper et al., 2021). Using AlphaFold2, 58% of total human protein residues have been confidently annotated structurally as opposed to X-ray crystallography, which could only resolve up to 17% of residues (Jumper et al., 2021). Furthermore, having a deep understanding of sequence-to-structure relationships and based on the assumption that interacting proteins co-evolve, these algorithms have also been implemented to predict PPIs. For example, Humphreys et al. has screened more than 8 million PPIs in Saccharomyces cerevisiae and accurately predicted ∼1,500 PPIs (Humphreys et al., 2021). One such study in human has predicted ∼3,000 confident PPIs out of a total of more than 65,000 PPIs screened (Burke et al., 2021). In both studies, hundreds of PPIs were reported for the first time. To assist with PPI prediction and determination, AlphaFold Protein Structure Database (AlphaFold DB) features proteomes from 21 model organisms, containing more than 360,000 predicted structures, of which 23,391 are predicted for the human proteome (Varadi et al., 2022).
The ability to screen proteome libraries to find novel interactions between two or more candidates is very promising. Furthermore, these methods can also be used as a quick tool to confirm the effect of mutations on existing PPIs. There are certain limitations though. First, the accuracy of the PPI prediction relies on the presence of orthologs spanning other species. Therefore, proteins that are evolving rapidly and have few orthologs in phylogenetically restricted species may not be detected by these methods. Second, prediction of PPIs in higher eukaryotes may not uncover as many as in lower eukaryotes where there is an increased number of genomes sequenced from closely related species and, hence, a wider availability of orthologs. Third, proteins that form multi-subunit or even higher order complexes may not be represented accurately by binary PPIs (Humphreys et al., 2021). Last, but most importantly, the computational infrastructure required to run AI- and DL-based algorithms for prediction of PPIs is extensive. Even for the relatively simple eukaryotic S. cerevisiae proteome, it would demand 0.1 to 1 million graphics processing unit (GPU) hours (Humphreys et al., 2021), which may restrict the analysis of more complex higher eukaryotic proteomes such as human.
Notwithstanding the above limitations, the future looks promising with the steady evolution of computational resources and increasing accuracy of protein structure prediction algorithms. Along with experimentally determined human protein structures, computationally predicted pairwise PPIs will be imminently available in public PPI repositories. How these algorithms and resources will be used to support and advance interactome-based studies are the subject of ongoing exploration. Researchers will be significantly empowered to find novel therapeutic targets for many diseases and continue to bring the dark interactome into the light.
Concluding remarks
One of the main objectives of molecular therapy is to target disease-specific proteins that contribute to the initiation or progression of diseases. Targeting disease-associated proteins is a complex task because most of them also play an important role in normal biological processes. However, it has been well established that protein interaction partners of disease-associated proteins can vary between the normal and disease states, especially in cancer following genomic alterations (Sharifi Tabar et al., 2022a). This offers a unique opportunity to target disease-specific protein interaction interfaces using small molecule drugs. Therefore, construction of comprehensive reference protein interactome maps will pave the way for the identification of disease-specific interactions and will provide solid foundations for future therapeutics.
Methodological advances have enabled the differentiation of embryonic and adult stem cells into specialized cell types or lineages, and now production of a range of tissue-specific cell types is feasible. In parallel, high-quality single-cell RNA sequencing (RNA-seq) technology has provided a massive amount of genomics data and enhanced our understanding of the cell-type-specific expression of many genes. This means that the identification of tissue-enriched or -specific PPIs is not elusive anymore and can be performed for proteins whose interactions are currently poorly characterized. The new generation of PL enzymes has now enabled efficient in situ labeling of transient and dynamic interactions within minutes in nearly all compartments of living cells (Branon et al., 2018; Qin et al., 2021; Roux et al., 2018). This technology will greatly enhance the identification of many PPIs that have been refractory to traditional approaches.
High-resolution MS with improved speed and accuracy has facilitated the proteome-scale identification of thousands of proteins from a complex cellular milieu. Furthermore, a recent breakthrough in predicting PPIs of protein complexes using AlphaFold Multimer suggests that later versions would improve further the prediction of protein interactions. Ultimately, the consolidation of results from AP-MS, PL-MS, and Y2H studies, as well as integration of DL-based methodologies, will all accelerate further exploration of the uncharted interactome. Together, all these factors provide a unique opportunity to systematically survey the human interactome and discover spatiotemporal and cell-type-specific interactions that have not previously been visible in the dark interactome. Cell-type-specific interactome maps will therefore provide a detailed view of complex biological processes and may explain tissue-specific gene expression and phenotype relationships in normal and disease states.
Acknowledgements
Financial support was provided by National Health and Medical Research Council, Australia (Investigator Grant #1177305 and Project Grant #1128748 to J.E.J.R.); Cancer Council NSW project grants (RG20-07 and RG20-12) and an anonymous foundation to J.E.J.R. and C.G.B.; Tour de Cure Research Project support to C.G.B.; and Centenary Institute ECR Booster grant to M.S.T.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Charles G. Bailey, Email: c.bailey@centenary.org.au.
John E.J. Rasko, Email: j.rasko@centenary.org.au.
References
- Adhikari S., Nice E.C., Deutsch E.W., Lane L., Omenn G.S., Pennington S.R., Paik Y.K., Overall C.M., Corrales F.J., Cristea I.M., et al. A high-stringency blueprint of the human proteome. Nat. Commun. 2020;11:5301. doi: 10.1038/s41467-020-19045-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebersold R., Agar J.N., Amster I.J., Baker M.S., Bertozzi C.R., Boja E.S., Costello C.E., Cravatt B.F., Fenselau C., Garcia B.A., et al. How many human proteoforms are there? Nat. Chem. Biol. 2018;14:206–214. doi: 10.1038/nchembio.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Qaraghuli M.M., Kubiak-Ossowska K., Ferro V.A., Mulheran P.A. Antibody-protein binding and conformational changes: identifying allosteric signalling pathways to engineer a better effector response. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-70680-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alanis-Lobato G., Andrade-Navarro M.A., Schaefer M.H. HIPPIE v2.0: enhancing meaningfulness and reliability of protein-protein interaction networks. Nucleic Acids Res. 2017;45:D408–D414. doi: 10.1093/nar/gkw985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Castelao B., Schanzenbächer C.T., Langer J.D., Schuman E.M. Cell-type-specific metabolic labeling, detection and identification of nascent proteomes in vivo. Nat. Protoc. 2019;14:556–575. doi: 10.1038/s41596-018-0106-6. [DOI] [PubMed] [Google Scholar]
- Anand S., Samuel M., Ang C.S., Keerthikumar S., Mathivanan S. Label-based and label-free strategies for protein quantitation. Methods. Mol. Biol. 2017;1549:31–43. doi: 10.1007/978-1-4939-6740-7_4. [DOI] [PubMed] [Google Scholar]
- Baek M., DiMaio F., Anishchenko I., Dauparas J., Ovchinnikov S., Lee G.R., Wang J., Cong Q., Kinch L.N., Schaeffer R.D., et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science. 2021;373:871–876. doi: 10.1126/science.abj8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantscheff M., Schirle M., Sweetman G., Rick J., Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem. 2007;389:1017–1031. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- Beadle G.W., Tatum E.L. Genetic control of biochemical reactions in neurospora. Proc. Natl. Acad. Sci. USA. 1941;27:499–506. doi: 10.1073/pnas.27.11.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck D.B., Narendra V., Drury W.J., 3rd, Casey R., Jansen P.W.T.C., Yuan Z.F., Garcia B.A., Vermeulen M., Bonasio R. In vivo proximity labeling for the detection of protein-protein and protein-RNA interactions. J. Proteome Res. 2014;13:6135–6143. doi: 10.1021/pr500196b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt L.H., Gil J., Kim Y., Doma V., Çakır U., Sanchez A., Murillo J.R., Kuras M., Parada I.P., Sugihara Y., et al. The human melanoma proteome atlas-Defining the molecular pathology. Clin. Transl. Med. 2021;11:e473. doi: 10.1002/ctm2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bludau I., Aebersold R. Proteomic and interactomic insights into the molecular basis of cell functional diversity. Nat. Rev. Mol. Cell. Biol. 2020;21:327–340. doi: 10.1038/s41580-020-0231-2. [DOI] [PubMed] [Google Scholar]
- Bosch J.A., Chen C.L., Perrimon N. Proximity-dependent labeling methods for proteomic profiling in living cells: an update. Wiley Interdiscip. Rev. Dev. Biol. 2021;10:e392. doi: 10.1002/wdev.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branon T.C., Bosch J.A., Sanchez A.D., Udeshi N.D., Svinkina T., Carr S.A., Feldman J.L., Perrimon N., Ting A.Y. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 2018;36:880–887. doi: 10.1038/nbt.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke D.F., Bryant P., Barrio-Hernandez I., Memon D., Pozzati G., Shenoy A., Zhu W., Dunham A.S., Albanese P., Keller A., et al. Towards a structurally resolved human protein interaction network. bioRxiv. 2021 doi: 10.1101/2021.11.08.467664. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusick M.E., Yu H., Smolyar A., Venkatesan K., Carvunis A.R., Simonis N., Rual J.F., Borick H., Braun P., Dreze M., et al. Literature-curated protein interaction datasets. Nat. Methods. 2009;6:39–46. doi: 10.1038/nmeth.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fathi A., Mirzaei M., Dolatyar B., Sharifitabar M., Bayat M., Shahbazi E., Lee J., Javan M., Zhang S.C., Gupta V., et al. Discovery of novel cell surface markers for purification of embryonic dopamine progenitors for transplantation in Parkinson's disease animal models. Mol. Cell. Proteomics. 2018;17:1670–1684. doi: 10.1074/mcp.RA118.000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A.C., Gstaiger M., Raught B., Aebersold R. Analysis of protein complexes using mass spectrometry. Nat. Rev. Mol. Cell. Biol. 2007;8:645–654. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- Go C.D., Knight J.D.R., Rajasekharan A., Rathod B., Hesketh G.G., Abe K.T., Youn J.Y., Samavarchi-Tehrani P., Zhang H., Zhu L.Y., et al. A proximity-dependent biotinylation map of a human cell. Nature. 2021;595:120–124. doi: 10.1038/s41586-021-03592-2. [DOI] [PubMed] [Google Scholar]
- Gordon D.E., Hiatt J., Bouhaddou M., Rezelj V.V., Ulferts S., Braberg H., Jureka A.S., Obernier K., Guo J.Z., Batra J., et al. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science. 2020;370:eabe9403. doi: 10.1126/science.abe9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green N.M. Avidin. Adv. Protein. Chem. 1975;29:85–133. doi: 10.1016/s0065-3233(08)60411-8. [DOI] [PubMed] [Google Scholar]
- Hein M.Y., Hubner N.C., Poser I., Cox J., Nagaraj N., Toyoda Y., Gak I.A., Weisswange I., Mansfeld J., Buchholz F., et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell. 2015;163:712–723. doi: 10.1016/j.cell.2015.09.053. [DOI] [PubMed] [Google Scholar]
- Holmberg A., Blomstergren A., Nord O., Lukacs M., Lundeberg J., Uhlén M. The biotin-streptavidin interaction can be reversibly broken using water at elevated temperatures. Electrophoresis. 2005;26:501–510. doi: 10.1002/elps.200410070. [DOI] [PubMed] [Google Scholar]
- Humphreys I.R., Pei J., Baek M., Krishnakumar A., Anishchenko I., Ovchinnikov S., Zhang J., Ness T.J., Banjade S., Bagde S.R., et al. Computed structures of core eukaryotic protein complexes. Science. 2021;374 doi: 10.1126/science.abm4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin E.L., Bruckner R.J., Navarrete-Perea J., Cannon J.R., Baltier K., Gebreab F., Gygi M.P., Thornock A., Zarraga G., Tam S., et al. Dual proteome-scale networks reveal cell-specific remodeling of the human interactome. Cell. 2021;184:3022–3040.e28. doi: 10.1016/j.cell.2021.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangravi Z., Alikhani M., Arefnezhad B., Sharifi Tabar M., Taleahmad S., Karamzadeh R., Jadaliha M., Mousavi S.A., Ahmadi Rastegar D., Parsamatin P., et al. A fresh look at the male-specific region of the human Y chromosome. J. Proteome Res. 2013;12:6–22. doi: 10.1021/pr300864k. [DOI] [PubMed] [Google Scholar]
- Jiang L., Wang M., Lin S., Jian R., Li X., Chan J., Dong G., Fang H., Robinson A.E., GTEx Consortium. Snyder M.P. A quantitative proteome map of the human body. Cell. 2020;183:269–283.e19. doi: 10.1016/j.cell.2020.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent E.M.N., Sofianatos Y., Komarova A., Gimeno J.-P., Tehrani P.S., Kim D.-K., Abdouni H., Duhamel M., Cassonnet P., Knapp J.J., et al. Global BioID-based SARS-CoV-2 proteins proximal interactome unveils novel ties between viral polypeptides and host factors involved in multiple COVID19-associated mechanisms. bioRxiv. 2020 Preprint at. [Google Scholar]
- Low J.K.K., Silva A.P.G., Sharifi Tabar M., Torrado M., Webb S.R., Parker B.L., Sana M., Smits C., Schmidberger J.W., Brillault L., et al. The nucleosome remodeling and deacetylase complex has an asymmetric, dynamic, and modular architecture. Cell Rep. 2020;33 doi: 10.1016/j.celrep.2020.108450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck K., Kim D.K., Lambourne L., Spirohn K., Begg B.E., Bian W., Brignall R., Cafarelli T., Campos-Laborie F.J., Charloteaux B., et al. A reference map of the human binary protein interactome. Nature. 2020;580:402–408. doi: 10.1038/s41586-020-2188-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay J.P., Sunde M., Lowry J.A., Crossley M., Matthews J.M. Protein interactions: is seeing believing? Trends. Biochem. Sci. 2007;32:530–531. doi: 10.1016/j.tibs.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Maury Y., Côme J., Piskorowski R.A., Salah-Mohellibi N., Chevaleyre V., Peschanski M., Martinat C., Nedelec S. Combinatorial analysis of developmental cues efficiently converts human pluripotent stem cells into multiple neuronal subtypes. Nat. Biotechnol. 2015;33:89–96. doi: 10.1038/nbt.3049. [DOI] [PubMed] [Google Scholar]
- Mellacheruvu D., Wright Z., Couzens A.L., Lambert J.P., St-Denis N.A., Li T., Miteva Y.V., Hauri S., Sardiu M.E., Low T.Y., et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers C.L., Barrett D.R., Hibbs M.A., Huttenhower C., Troyanskaya O.G. Finding function: evaluation methods for functional genomic data. BMC. Genom. 2006;7:187. doi: 10.1186/1471-2164-7-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilson K.A., Ali N.A., Muralidharan S., Mirzaei M., Mariani M., Assadourian G., Lee A., van Sluyter S.C., Haynes P.A. Less label, more free: approaches in label-free quantitative mass spectrometry. Proteomics. 2011;11:535–553. doi: 10.1002/pmic.201000553. [DOI] [PubMed] [Google Scholar]
- Ong S.E., Mann M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC) Nat. Protoc. 2006;1:2650–2660. doi: 10.1038/nprot.2006.427. [DOI] [PubMed] [Google Scholar]
- Orchard S., Kerrien S., Abbani S., Aranda B., Bhate J., Bidwell S., Bridge A., Briganti L., Brinkman F.S.L., Brinkman F., et al. Protein interaction data curation: the International Molecular Exchange (IMEx) consortium. Nat. Methods. 2012;9:345–350. doi: 10.1038/nmeth.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orre L.M., Vesterlund M., Pan Y., Arslan T., Zhu Y., Fernandez Woodbridge A., Frings O., Fredlund E., Lehtiö J. SubCellBarCode: proteome-wide mapping of protein localization and relocalization. Mol. Cell. 2019;73:166–182.e7. doi: 10.1016/j.molcel.2018.11.035. [DOI] [PubMed] [Google Scholar]
- Oughtred R., Rust J., Chang C., Breitkreutz B.J., Stark C., Willems A., Boucher L., Leung G., Kolas N., Zhang F., et al. The BioGRID database: a comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein. Sci. 2021;30:187–200. doi: 10.1002/pro.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peach M., Marsh N., Miskiewicz E.I., MacPhee D.J. Solubilization of proteins: the importance of lysis buffer choice. Methods. Mol. Biol. 2015;1312:49–60. doi: 10.1007/978-1-4939-2694-7_8. [DOI] [PubMed] [Google Scholar]
- Qin W., Cho K.F., Cavanagh P.E., Ting A.Y. Deciphering molecular interactions by proximity labeling. Nat. Methods. 2021;18:133–143. doi: 10.1038/s41592-020-01010-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee H.W., Zou P., Udeshi N.D., Martell J.D., Mootha V.K., Carr S.A., Ting A.Y. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 2013;339:1328–1331. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux K.J., Kim D.I., Burke B., May D.G. BioID: a screen for protein-protein interactions. Curr. Protoc. Protein Sci. 2018;91:19.23.1–19.23.15. doi: 10.1002/cpps.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux K.J., Kim D.I., Raida M., Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell. Biol. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P., Abdouni H., Knight J.D.R., Astori A., Samson R., Lin Z.-Y., Kim D.-K., Knapp J.J., St-Germain J., Go C.D., et al. A SARS-CoV-2 – host proximity interactome. bioRxiv. 2020 doi: 10.1101/2020.09.03.282103. Preprint at. [DOI] [Google Scholar]
- Samavarchi-Tehrani P., Samson R., Gingras A.C. Proximity dependent biotinylation: key enzymes and adaptation to proteomics approaches. Mol. Cell. Proteomics. 2020;19:757–773. doi: 10.1074/mcp.R120.001941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Barriopedro I., van Mierlo G., Vermeulen M. Off-the-shelf proximity biotinylation for interaction proteomics. Nat. Commun. 2021;12:5015. doi: 10.1038/s41467-021-25338-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidberger J.W., Sharifi Tabar M., Torrado M., Silva A.P.G., Landsberg M.J., Brillault L., AlQarni S., Zeng Y.C., Parker B.L., Low J.K.K., Mackay J.P. The MTA1 subunit of the nucleosome remodeling and deacetylase complex can recruit two copies of RBBP4/7. Protein Sci. 2016;25:1472–1482. doi: 10.1002/pro.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifi Tabar M., Francis H., Yeo D., Bailey C.G., Rasko J.E.J. Mapping oncogenic protein interactions for precision medicine. Int. J. Cancer. 2022;151:7–19. doi: 10.1002/ijc.33954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifi Tabar M., Giardina C., Feng Y., Francis H., Moghaddas Sani H., Low J.K.K., Mackay J.P., Bailey C.G., Rasko J.E.J. Unique protein interaction networks define the chromatin remodelling module of the NuRD complex. FEBS J. 2022;289:199–214. doi: 10.1111/febs.16112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifi Tabar M., Mackay J.P., Low J.K.K. The stoichiometry and interactome of the Nucleosome Remodeling and Deacetylase (NuRD) complex are conserved across multiple cell lines. FEBS J. 2019;286:2043–2061. doi: 10.1111/febs.14800. [DOI] [PubMed] [Google Scholar]
- Spruijt C.G., Bartels S.J.J., Brinkman A.B., Tjeertes J.V., Poser I., Stunnenberg H.G., Vermeulen M. CDK2AP1/DOC-1 is a bona fide subunit of the Mi-2/NuRD complex. Mol. Biosyst. 2010;6:1700–1706. doi: 10.1039/c004108d. [DOI] [PubMed] [Google Scholar]
- St-Germain J.R., Samavarchi Tehrani P., Wong C., Larsen B., Gingras A.C., Raught B. Variability in streptavidin-sepharose matrix quality can significantly affect proximity-dependent biotinylation (BioID) data. J. Proteome Res. 2020;19:3554–3561. doi: 10.1021/acs.jproteome.0c00117. [DOI] [PubMed] [Google Scholar]
- Taverna D., Gaspari M. A critical comparison of three MS-based approaches for quantitative proteomics analysis. J. Mass. Spectrom. 2021;56:e4669. doi: 10.1002/jms.4669. [DOI] [PubMed] [Google Scholar]
- Torrado M., Low J.K.K., Silva A.P.G., Schmidberger J.W., Sana M., Sharifi Tabar M., Isilak M.E., Winning C.S., Kwong C., Bedward M.J., et al. Refinement of the subunit interaction network within the nucleosome remodelling and deacetylase (NuRD) complex. FEBS J. 2017;284:4216–4232. doi: 10.1111/febs.14301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L. Vol. 8. 2019. p. F1000Res. (Recent Advances in Proximity-Based Labeling Methods for Interactome Mapping). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- Uezu A., Kanak D.J., Bradshaw T.W.A., Soderblom E.J., Catavero C.M., Burette A.C., Weinberg R.J., Soderling S.H. Identification of an elaborate complex mediating postsynaptic inhibition. Science. 2016;353:1123–1129. doi: 10.1126/science.aag0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakilian H., Mirzaei M., Sharifi Tabar M., Pooyan P., Habibi Rezaee L., Parker L., Haynes P.A., Gourabi H., Baharvand H., Salekdeh G.H. DDX3Y, a male-specific region of Y chromosome gene, may modulate neuronal differentiation. J. Proteome Res. 2015;14:3474–3483. doi: 10.1021/acs.jproteome.5b00512. [DOI] [PubMed] [Google Scholar]
- Varadi M., Anyango S., Deshpande M., Nair S., Natassia C., Yordanova G., Yuan D., Stroe O., Wood G., Laydon A., et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50:D439–D444. doi: 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnaitė R., MacNeill S.A. Meet the neighbors: mapping local protein interactomes by proximity-dependent labeling with BioID. Proteomics. 2016;16:2503–2518. doi: 10.1002/pmic.201600123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B., Papadopoulos N., Velculescu V.E., Zhou S., Diaz L.A., Jr., Kinzler K.W. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner G.P., Zhang J. The pleiotropic structure of the genotype-phenotype map: the evolvability of complex organisms. Nat. Rev. Genet. 2011;12:204–213. doi: 10.1038/nrg2949. [DOI] [PubMed] [Google Scholar]
- Walport L.J., Low J.K.K., Matthews J.M., Mackay J.P. The characterization of protein interactions - what, how and how much? Chem. Soc. Rev. 2021;50:12292–12307. doi: 10.1039/d1cs00548k. [DOI] [PubMed] [Google Scholar]
- Wang D., Eraslan B., Wieland T., Hallström B., Hopf T., Zolg D.P., Zecha J., Asplund A., Li L.H., Meng C., et al. A deep proteome and transcriptome abundance atlas of 29 healthy human tissues. Mol. Syst. Biol. 2019;15 doi: 10.15252/msb.20188503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.B., Karpova A., Gritsenko M.A., Kyle J.E., Cao S., Li Y., Rykunov D., Colaprico A., Rothstein J.H., Hong R., et al. Proteogenomic and metabolomic characterization of human glioblastoma. Cancer Cell. 2021;39:509–528.e20. doi: 10.1016/j.ccell.2021.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Ding S.L., Li Y., Royall J., Feng D., Lesnar P., Graddis N., Naeemi M., Facer B., Ho A., et al. The Allen mouse brain common coordinate framework: a 3D reference atlas. Cell. 2020;181:936–953.e20. doi: 10.1016/j.cell.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm M., Schlegl J., Hahne H., Gholami A.M., Lieberenz M., Savitski M.M., Ziegler E., Butzmann L., Gessulat S., Marx H., et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- Wilson I.A., Stanfield R.L. Antibody-antigen interactions: new structures and new conformational changes. Curr. Opin. Struct. Biol. 1994;4:857–867. doi: 10.1016/0959-440x(94)90267-4. [DOI] [PubMed] [Google Scholar]
- Wilson R.S., Nairn A.C. Cell-type-specific proteomics: a neuroscience perspective. Proteomes. 2018;6:51. doi: 10.3390/proteomes6040051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissmueller S., Font J., Liew C.W., Cram E., Schroeder T., Turner J., Crossley M., Mackay J.P., Matthews J.M. Protein-protein interactions: analysis of a false positive GST pulldown result. Proteins. 2011;79:2365–2371. doi: 10.1002/prot.23068. [DOI] [PubMed] [Google Scholar]
- Xu J.Y., Zhang C., Wang X., Zhai L., Ma Y., Mao Y., Qian K., Sun C., Liu Z., Jiang S., et al. Integrative proteomic characterization of human lung adenocarcinoma. Cell. 2020;182:245–261.e17. doi: 10.1016/j.cell.2020.05.043. [DOI] [PubMed] [Google Scholar]