Abstract
Like other complex multicellular organisms, plants are composed of different cell types with specialized shapes and functions. For example, most laminar leaves consist of multiple photosynthetic cell types. These cell types include the palisade mesophyll, which typically forms one or more cell layers on the adaxial side of the leaf. Despite their importance for photosynthesis, we know little about how palisade cells differ at the molecular level from other photosynthetic cell types. To this end, we have used a combination of cell-specific profiling using fluorescence-activated cell sorting and single-cell RNA-sequencing methods to generate a transcriptional blueprint of the palisade mesophyll in Arabidopsis thaliana leaves. We find that despite their unique morphology, palisade cells are otherwise transcriptionally similar to other photosynthetic cell types. Nevertheless, we show that some genes in the phenylpropanoid biosynthesis pathway have both palisade-enriched expression and are light-regulated. Phenylpropanoid gene activity in the palisade was required for production of the ultraviolet (UV)-B protectant sinapoylmalate, which may protect the palisade and/or other leaf cells against damaging UV light. These findings improve our understanding of how different photosynthetic cell types in the leaf can function uniquely to optimize leaf performance, despite their transcriptional similarities.
Leaf cell sorting and scRNA-seq approaches are used to transcriptionally profile the palisade mesophyll layer and provide resources for understanding mesophyll biology.
In a Nutshell.
Background: Photosynthesis is arguably the most important biochemical process in plants, and provides humans and other animals with food and oxygen. This process generally occurs in specialized cell types in the leaf. One type of photosynthetic cell, called palisade, forms one or more cell layers at the top of the leaf in many plant species. Despite the importance of these cells, we know surprisingly little at the molecular level as to how palisade differs from other photosynthetic cell types.
Question: We wanted to compare the gene expression profile of palisade cells to other photosynthetic cell types to discover their unique roles in leaf biology. We did this by using a combined approach of bulk palisade cell purification and single cell analysis in the model plant Arabidopsis thaliana.
Findings: Despite their unique morphology and placement in the leaf, gene expression was similar between the palisade and other photosynthetic cell types. Nevertheless, our complementary approaches revealed a role for the palisade in production of an ultraviolet (UV) light-absorbing molecule called sinapoylmalate, which acts as a “sunscreen” to reduce the damaging effects of UV light. This may serve to protect the palisade and/or additional cell types in the leaf. These results provide a better understanding of how different leaf cells function to optimize overall leaf performance.
Next steps: In the course of our studies we generated novel genetic tools to probe the function of the palisade and other photosynthetic cells. Future cell-specific manipulations using these tools will further explore how these cells contribute to leaf function across different environments. This will provide insights into why and how leaves adopt different cellular architectures.
Introduction
For nearly all vascular plants, leaves are the primary sites of light acquisition and photosynthesis. These processes occur primarily in chloroplast-rich mesophyll cells, which make up the bulk of the leaf interior. In laminar leaves, these cells are generally of two types: palisade mesophyll cells, which form one or more layers of columnar cells on the adaxial leaf side, and spongy mesophyll cells, which are arranged below the palisade with a range of irregular to highly ordered morphologies (Haberlandt, 1914; Esau, 1977; Borsuk et al., 2022). The shape and arrangement of these cells are thought to facilitate light capture and CO2 movement (Smith et al., 1997; Terashima et al., 2011). Specifically, columnar palisade cells likely conduct excess light deep into the leaf (Vogelmann, 1993) where it is scattered by the spongy mesophyll, acting as a light trap. Additional anatomical properties of leaves, such as the shape of epidermal cells (Vogelmann et al., 1996; Brodersen and Vogelmann, 2007) and placement of stomata mostly on the abaxial surface, further tailor leaf performance to the environment (Smith et al., 1997; Borsuk and Brodersen, 2019; Borsuk et al., 2022).
Of the two mesophyll types, incident sunlight will generally contact the upper palisade cells first. Not surprisingly then, the shape of these cells is particularly affected by light (Hansen, 1917). In high light, additional palisade layers may be present, and the cells adopt a taller, more cylindrical morphology with tighter packing. Experiments using the model plant Arabidopsis thaliana (Arabidopsis) suggest these changes are dependent on the blue light sensor phototropin 2 (Kozuka et al., 2011). Other factors regulating leaf adaxial–abaxial asymmetry have similarly been identified from mutant screens in Arabidopsis, and lesions in these genes result in alterations to palisade and/or spongy mesophyll identity (Manuela and Xu, 2020). However, far less is known about the unique molecular signature of mature palisade cells compared with other photosynthetic cell types, and to what extent these cells even differ from each other.
To this end, we sought to transcriptionally profile palisade mesophyll. Reporter lines driving fluorescent proteins in specific cell types have previously been used to sort and profile cells of the root and other organs of Arabidopsis using fluorescence-activated cell (FAC) sorting coupled with microarray or RNA-sequencing (RNA-seq) analysis (Bargmann and Birnbaum, 2010). More recently, single-cell RNA-seq (scRNA-seq) technologies have also been applied to plants (Ryu et al., 2019; Shulse et al., 2019; Liu et al., 2020; Kim et al., 2021; Lopez-Anido et al., 2021), which, in principle, negate the need for specialized reporters and can assess gene expression in all cells of an organ simultaneously. Here, we use both approaches to analyze gene expression in the palisade. Although these cells have unique morphology and appearance, gene expression was strikingly similar to that of other photosynthetic cell types, likely a consequence of their shared primary function. Despite this, we describe novel marker genes for the palisade and characterize a previously unappreciated role for the phenylpropanoid pathway in palisade tissue.
Results
FAC sorting of a palisade mesophyll cell population
To better characterize the palisade layer, we sought to find DNA regulatory sequences in Arabidopsis that drive expression of a fluorescent marker protein specifically in these cells. Arabidopsis leaves show a typical laminar structure, with a single palisade layer directly below the adaxial epidermis and spongy mesophyll below the palisade (Figure 1A). Under our growth conditions, first true leaf expansion was mostly complete by 17 days postgermination (Figure 1B), and, unless otherwise indicated, we performed all subsequent experiments at this developmental stage to assess transcription in the mature leaf.
Figure 1.
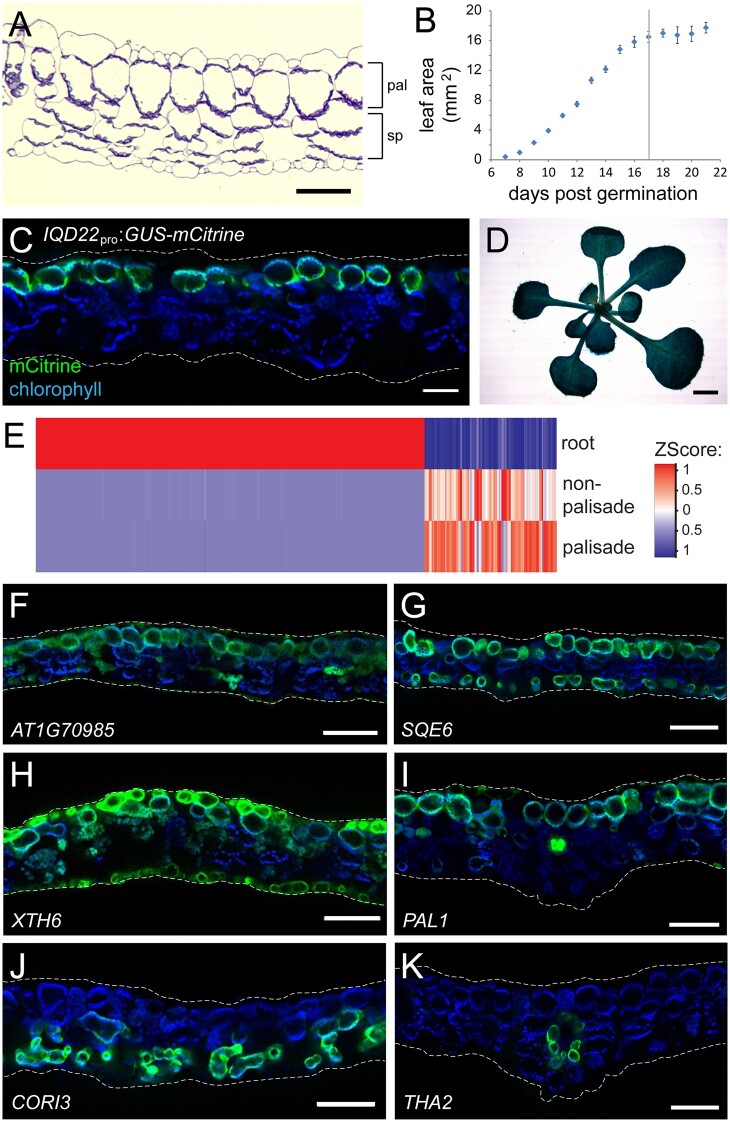
FAC sorting of IQD22pro:GUS-mCit leaf protoplasts can enrich for palisade mesophyll cells. A, Toluidine blue-stained cross-section of a mature Arabidopsis leaf (first true leaf pair), showing a single layer of palisade mesophyll (pal) and spongy mesophyll (sp) below. B, Growth of the first true leaves. In our conditions, the leaves reached their maximum area at 17 days postgermination (vertical line). Mean ± sem. C, Fluorescence image of a leaf cross-section from a 17-day-old IQD22pro:GUS-mCit reporter plant. Chlorophyll autofluorescence is shown in blue and highlights chloroplasts of the mesophyll cells. GUS-mCit protein (green) is expressed in palisade cells. D, Image of the shoot of a GUS-stained IQD22pro:GUS-mCit plant at day 17. E, Heatmap comparing expression of the top 500 most differentially-regulated genes (columns) identified from pairwise comparisons between total root protoplasts and FAC sorted palisade and nonpalisade photosynthetic cell populations from IQD22pro:GUS-mCit leaves. F–K, Fluorescence images of example transcriptional reporters of genes identified as being upregulated (F–I) or downregulated (J and K) in palisade mesophyll compared with nonpalisade photosynthetic cells by FAC sorting. Leaf cross sections of 17-day-old T1 plants are shown (n ≥ 5). Chlorophyll autofluorescence, blue, and gene promoter:GUS-mCit reporter, green. Note reporter activity in the palisade cells in (F–K), as well as additional cell types. (I) and (K) are at the location of the midvein; other panels are lateral. Note that when imaging mCit at high exposures in (F) and (H), some autofluorescence is also observed from the cuticle and chlorophyll. Scale bars: (A) and (C), 50 μm; (D), 2 mm; (F–K), 100 μm. In (A), (C), and (F–K), adaxial is up. In (C) and (F–K), dashed white lines show approximate position of cuticle.
Work by others previously described β-glucuronidase (GUS) enzyme activity in the palisade layer of the cotyledons in the UCR6 GUS enhancer trap line (Geisler et al., 2002). As such, we sought to better characterize these plants. Using inverse PCR (iPCR), we mapped the GUS-coding transposable element insertion site to a region between two genes on chromosome 4. Approximately 4 kb of sequences flanking the insertion site were sufficient to drive expression of the fluorescent reporter protein mCitrine (mCit) in leaf palisade cells, as well as cells in other regions of the plant (Supplemental Figure S1, A and B). We further narrowed down the regulatory sequence to a ∼3.5 kb promoter element upstream of the IQ-DOMAIN 22 (IQD22) gene, and showed that in the leaf this drives accumulation of a GUS-mCit fusion protein in the palisade (IQD22pro:GUS-mCit, Figure 1, C and D and Supplemental Figure S1, C–F).
Using FAC sorting, we generated protoplasts from first true leaves of IQD22pro:GUS-mCit plants, and sorted them based on mCit fluorescence in green wavelengths. In addition, we took advantage of the natural autofluorescent properties of chlorophyll in red wavelengths to further separate chloroplast-containing cells from other, nonphotosynthetic cell types. Using this approach, we purified two cell populations: (1) a chlorophyll-containing population that expressed GUS-mCit, representing the palisade cells and (2) a reference population containing chlorophyll but not displaying GUS-mCit fluorescence (Supplemental Figure S2, A–D). This reference population likely includes spongy mesophyll, bundle sheath cells, and other chlorophyll-containing cells of the vasculature (Haberlandt, 1914; Crookston and Ozbun, 1975; Ding et al., 2015). Here, we assume that this reference population is photosynthetic. RNA-seq suggests that our two populations are transcriptionally very similar: when compared with total root protoplasts, the greatest differences in gene expression are between root cells and photosynthetic cell populations, and not between the photosynthetic cells themselves (Figure 1E and Supplemental Figure S2G). This similarity between photosynthetic cell types is supported by our scRNA-seq analysis (see below). Using these methods, we identified several hundred differentially regulated genes in the palisade cells relative to other photosynthetic cell types, including a known abaxial patterning factor (Supplemental Figure S2, H–J).
To validate our approach, we performed a similar analysis of sorted protoplasts generated from the GAL4-VP16 enhancer trap line JR11-2, which expresses a modified GFP reporter protein highest in the lower spongy mesophyll (Supplemental Figure S2, E and F; Gardner et al., 2009). FAC-sorted protoplasts from JR11-2 plants divided into a spongy mesophyll-like population (exhibiting both chlorophyll and GFP fluorescence) and a nonspongy, chlorophyll-containing population (exhibiting chlorophyll autofluorescence but lacking GFP, which includes the palisade cells), showed significant overlap with our results from IQD22pro:GUS-mCit plants, thereby validating our methods (Supplemental Figure S2, G–I). However, we were able to detect a larger number of differentially regulated genes using IQD22pro:GUS-mCit generated protoplasts, perhaps due to greater purification of the palisade cells, and thus we confined subsequent analyses to this line.
Finally, to minimize the effect on transcription of the protoplasting process itself, we investigated the effect of adding transcription inhibitors to our protoplasting medium. We found that inhibitors minimized the induction of genes associated with wounding and defense (Supplemental Figure S2, K and L), and thus were added to all subsequent experiments.
Our final analysis found 238 nuclear-encoded genes to be upregulated and 591 downregulated in the palisade population when compared with other photosynthetic cell types (false discovery rate (FDR) < 0.05 and fold-difference > 2; Supplemental Data Sets S1 and S2). To confirm that these genes are palisade enriched, we fused ∼1.5–2 kb promoter elements from a selection of these to a GUS-mCit reporter sequence. Transgenic plants carrying these reporters showed diverse expression patterns, some of which had higher expression in the palisade than other photosynthetic cells (8 of 17 reporters tested; Supplemental Figure S3). Examples of these include the promoters for: AT1G70985, which drove weak expression mostly in the palisade (Figure 1F); SQUALENE MONOXYGENASE 6 (SQE6), which drove high expression in the palisade and single layer of lowermost spongy mesophyll (Figure 1G); XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 6 (XTH6), which in addition to the palisade had epidermal expression highest on the adaxial side (Figure 1H); and genes related to phenylpropanoid biosynthesis, including PHENYLALANINE AMMONIA-LYASE 1 (PAL1) (Fraser and Chapple, 2011), which drove highest expression in the vasculature, strong expression in palisade, weak in epidermis, including stomata, and almost none in spongy mesophyll (Figure 1I). By comparison, promoter elements of genes downregulated in the palisade population drove higher expression in other chloroplast-containing cells of the leaf. Examples include the promoters for: CORONATINE INDUCED 3 (CORI3), which drove expression in the lower spongy mesophyll and bundle sheath (Figure 1J); THREONINE ALDOLASE 2 (THA2) in bundle sheath (Figure 1K); and other gene promoter elements that drove expression in spongy mesophyll, bundle sheath and/or chloroplast containing cells of the vasculature (Supplemental Figure S4).
Some of our reporters for genes predicted to be palisade-enriched failed to express in these cells (Supplemental Figure S3). This may reflect that we have inadequate promoter/regulatory sequences for these particular genes. For example, we were unable to detect GUS-mCit activity from a plant carrying an LHCB4.3pro:GUS-mCit reporter; however, a previous study suggested that this gene is expressed on the adaxial leaf side (Sawchuk et al., 2008). Similarly, while we were able to identify expression of the phenylpropanoid-related gene FERULIC ACID 5-HYDROXYLASE 1 (FAH1) in FAC sorted palisade cells (see below), regulatory elements required for FAH1 gene function in the leaf lie not within the promoter sequence but rather in a ∼12.5 kb region downstream of the stop codon (Ruegger et al., 1999). Alternatively, our FAC sorted palisade population may contain contaminating cells. Nevertheless, our results suggest that our methods have successfully purified the palisade cells, at least partially.
scRNA-seq of the mature Arabidopsis leaf
Recently, scRNA-seq methods have been developed as an alternative approach to query specific cell populations. Previously reported scRNA-seq analyses of Arabidopsis cotyledons, immature developing leaves, and leaves of mature 6-week-old plants did not distinctly separate the palisade and spongy mesophyll (Liu et al., 2020; Kim et al., 2021; Lopez-Anido et al., 2021), possibly as a result of the high transcriptional similarity we have observed. As such, to complement our FAC sorting experiments, we performed scRNA-seq using the 10× Genomics platform on protoplasts generated from the first true leaves of 17-day-old plants carrying the IQD22pro:GUS-mCit reporter, so as to improve our ability to identify the palisade population.
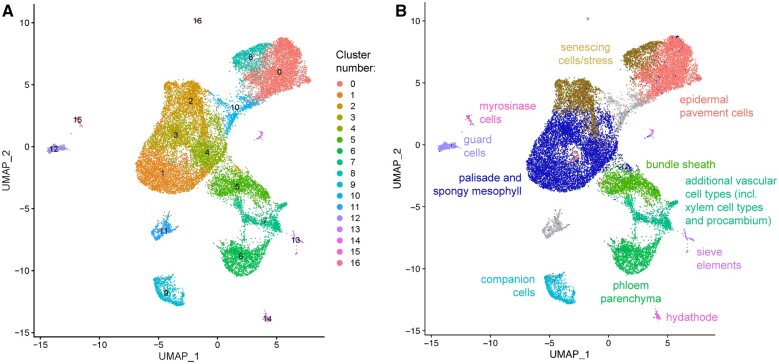
Following doublet removal (DePasquale et al., 2019) and filtering, our scRNA-seq dataset included 23,729 cells, with 3,639 median unique transcripts per cell and 1,637 median genes detected per cell. While these median counts are generally lower or comparable to other similar studies from plants, the number of cells in our dataset are several fold higher (Supplemental Table S1). Using Uniform Manifold Approximation and Projection (UMAP) for dimensionality reduction, we identified 17 cell clusters corresponding to unique cell types and/or transcriptional states (Figure 2A). More stringent filtering of our data set to remove cells with > 10,000 unique molecular identifiers (UMIs) per cell generated similar results, suggesting that the inclusion of some cells with high RNA content is not affecting our overall findings (Supplemental Figure S5). To assign biological identity to these (Figure 2B) we used two approaches. First, we used a biased approach, by mapping known cell type marker genes to the UMAP plot (Figure 3A). Second, we used an unbiased method. For this, we fused ∼2 kb of promoter elements for one or more genes identified through our analyses as being differentially upregulated in a given cluster to the GUS-mCit coding sequence, and imaged leaves of T1 transformed plants in cross-section (Supplemental Data Sets S3–S19, Figure 3, B and C, and Supplemental Figure S6).
Figure 2.
A scRNA-seq atlas of mature Arabidopsis leaf protoplasts. A, Unbiased clustering of RNA-sequenced leaf protoplasts from IQD22pro:GUS-mCit plants. 17 unique cell clusters were assigned based on transcriptional differences (clusters 0–16). B, Cell identities of UMAP-based clusters determined by marker gene analysis and validation.
Figure 3.
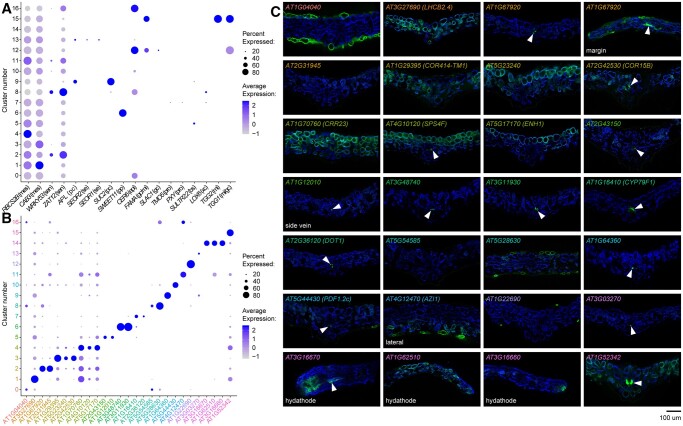
Gene marker analysis of scRNA-seq cell clusters. A, Dot plot showing expression of known tissue-specific marker genes in the scRNA-seq leaf cell clusters (see Figure 2A). Dot color intensity represents average expression of the gene in a given cluster, and dot size the percentage of cells in the cluster expressing that particular marker. Markers are expressed in mesophyll (mes), senescing cells (sen), phloem companion (pc), sieve elements (se), phloem parenchyma (pp), epidermis (epi), GCs (gc), myrosinase idioblasts (mi), procambium (pro), bundle sheath and veins (bs), and xylem contact cells (xc). B, Dot plot showing expression of de novo-identified cluster marker genes. Gene name color is the same as the cell cluster in Figure 2A for which the marker gene was identified. C, Fluorescence images of transcriptional reporters for genes shown in (B). Cross sections of first true leaves of T1 transgenic plants are shown (representative from n ≥ 4 T1 plants). Chlorophyll autofluorescence, blue, and gene promoter:GUS-mCit reporter activity, green. Adaxial is up. All images are at the midvein except where otherwise indicated (margin, side vein, lateral or hydathode). Arrowheads mark vascular and/or bundle sheath expression in some lines.
Using these combined approaches, we associated several clusters with cell types of veins. Known marker genes SWEET11 (Chen et al., 2011), ALTERED PHLOEM DEVELOPMENT (APL) and SUCROSE TRANSPORTER 2 (SUC2) (Truernit and Sauer, 1995; Bonke et al., 2003), SIEVE ELEMENT OCCLUSION-RELATED 1 and 2 (SEOR1 and SEOR2) (Rüping et al., 2010; Anstead et al., 2012), and THIOGLUCOSIDE GLUCOHYDROLASE 2 (TGG2) (Barth and Jander, 2006), expressed in phloem parenchyma, phloem companion cells, sieve elements, and myrosinase idioblasts, respectively, mapped to cluster numbers 6, 9, 13, and 15 (Figure 3A). Procambium markers TARGET OF MONOPTEROS 5 (TMO5) and PHLOEM INTERCALATED WITH XYLEM (PXY) (Fisher and Turner, 2007; De Rybel et al., 2013), and the xylem contact cell-expressed gene LIPOXYGENASE 6 (LOX6) (Chauvin et al., 2012; Nguyen et al., 2018), mapped to distinct regions of cluster 7, suggesting this cluster represents a mixed population of vascular cell types (Figure 3A). Consistent with these findings, promoter elements from mostly uncharacterized genes associated with clusters 6, 7, 9, 13, and 15 (AT3G48740, AT3G11930, AT1G16410/CYTOCHROME P450 79F1 (CYP79F1; see also Tantikanjana, 2001), AT2G36120/DEFECTIVELY ORGANIZED TRIBUTARIES 1 (DOT1), AT1G64360, AT3G03270, and AT1G52342; Figure 3B) all drove GUS-mCit reporter activity in cells associated with minor and/or major veins, and, in the case of CYP79F1, weakly in the bundle sheath (Figure 3C and Supplemental Figure S6). Gene ontology (GO) term enrichment analysis (Supplemental Data Sets S20–S36) suggests that these cell clusters express genes involved with glucosinolate production, water transport and phloem transport, among other processes. Cluster 5, which shared similarities in gene expression with cell types of the vein, expressed the bundle sheath and vein marker SULPHATE TRANSPORTER 2;2 (SULTR2;2) (Takahashi et al., 2000; Kirschner et al., 2018), as well as genes related to glucosinolate production, starch biosynthesis and photosynthesis (Figure 3A and Supplemental Data Set S25). Similarly, reporters for novel cluster 5 marker genes AT2G43150 and AT1G12010 drove expression in bundle sheath and/or cells of the vein (Figure 3, B and C).
To identify guard cells (GCs), we looked for the overlap in expression between the cuticle-formation gene ECERIFERUM 6 (CER6) (Hooker et al., 2002), the GC-specific gene SLOW ANION CHANNEL-ASSOCIATED 1 (SLAC1) (Vahisalu et al., 2008), and the GC and myrosinase idioblast markers FAMA and THIOGLUCOSIDE GLUCOHYDROLASE 1 (TGG1) (Barth and Jander, 2006; Shirakawa et al., 2014; Figure 3A). This identified cluster 12 as GC-specific. As validation, a promoter element for a de novo identified cluster 12 marker, AT1G22690, drove reporter expression specifically in stomata (Figure 3, B and C).
To identify clusters 11, 14, and 16, we tested reporter activity of various marker genes identified through our bioinformatic approaches. For cluster 14, promoter elements from markers AT3G16670, AT1G62510, and AT3G16660 all drove expression either specifically in the hydathode, or the hydathode and other cell types (Figure 3, B and C). This cluster was enriched for genes related to immune responses among other biological processes, including auxin biosynthesis, known to occur in hydathodes (Biedroń and Banasiak, 2018; Supplemental Data Set S34). In contrast, the marker gene AT4G12470/AZELAIC ACID INDUCED 1 (AZI1), involved in systemic immunity (Cecchini et al., 2015), showed enriched expression in clusters 11 and 16 (Figure 3B). Both these clusters express genes related to oxidative stress and stress hormones jasmonic acid and salicylic acid (Supplemental Data Sets S31 and S36). Interestingly, the promoter element of AZI1 drove reporter activity in a subset of abaxial epidermal cells and mesophyll, and, as such, we were unable to assign a specific cell type or state to these two clusters (Figure 3C). Future study of these populations may resolve their identity, and what specific roles they play in leaf function and defense.
The two largest groups of cells, representing clusters 0 and 8, and clusters 1–4 (Figure 2A), expressed epidermal pavement and mesophyll/photosynthesis-related genes, respectively. In clusters 0 and 8, this included the cuticle formation gene CER6, as well as the newly identified epidermal markers AT1G04040 and AT5G28630 (Figure 3, A–C; a reporter for an additional cluster 8 marker, AT5G54585, had little expression in the first true leaves). In contrast, photosynthesis-related marker genes CHLOROPHYLL A/B BINDING PROTEIN 3 (CAB3) and RUBISCO SMALL SUBUNIT 2B (RBCS2B), which are expressed in various regions of the mature leaf (Susek et al., 1993; Sawchuk et al., 2008), had broad expression domains with strongest expression in clusters 1 and 4, respectively (Figure 3A). Interestingly, clusters 2 and 8 shared expression of stress- and senescence-related genes such as WRKY53 and ZAT12 (Davletova et al., 2005; Bresson et al., 2018; Zentgraf and Doll, 2019; Figure 3A and Supplemental Data Sets S22 and S28). This may indicate that senescence promotes a transcriptional convergence of epidermal pavement and mesophyll cells irrespective of cell type. A reporter for a cluster 2 marker gene, AT1G67920, drove expression in vascular and epidermal cells (Figure 3, B and C), and we hypothesize that this cluster includes mixed cell types undergoing senescence; however, another cluster 2 promoter, AT2G31945, failed to drive expression in the leaf blade, perhaps due to low activity or incomplete regulatory sequences. Similarly, abaxial-positioning in the leaf may influence the transcriptome of some epidermal and mesophyll cells into cluster 10, which expresses the spongy mesophyll marker CORI3, described above, and a newly identified marker gene for regions of the abaxial epidermis, AT5G44430/PLANT DEFENSIN 1.2c (PDF1.2c) (Figures 3, B and C and 4A).
Figure 4.
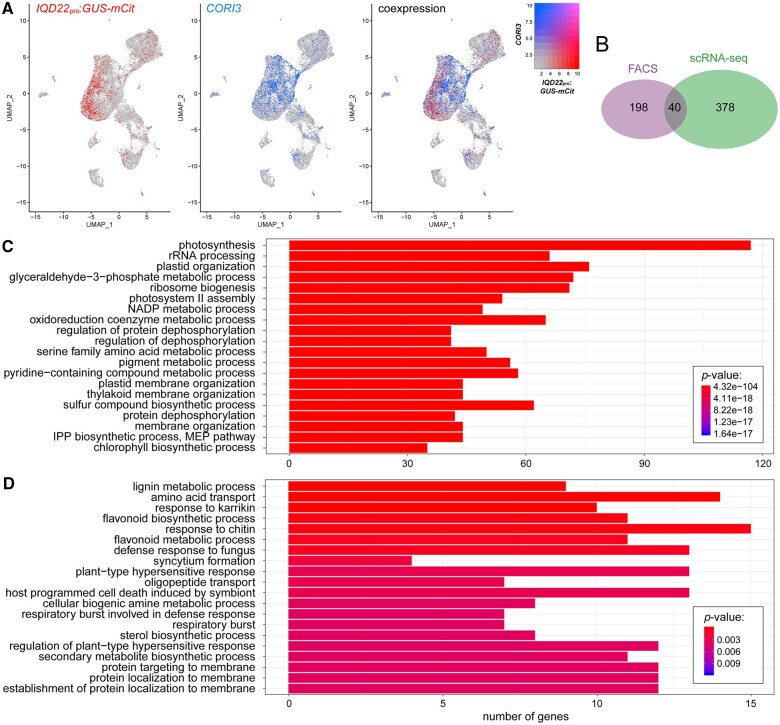
Identification of palisade-expressed genes by scRNA-seq. A, Location of GUS-mCit and CORI3 expressing cells in our scRNA-seq UMAP plot of protoplasts from the mature leaf of IQD22pro:GUS-mCit transgenic plants. Note little overlap of cells expressing these two transcripts. B, Overlap of nuclear-encoded genes identified to be upregulated in the palisade compared with reference photosynthetic cell populations as determined by FAC sorting, and GUS-mCit expressing cells (reads > 1) compared with CORI3 expressing cells (reads > 2) by scRNA-seq methods. P < 0.005, hypergeometric test. C and D, GO term enrichment analysis of genes upregulated in the palisade as identified using scRNA-seq (C) or FAC sorting (D). The top 20 terms by adjusted P-value are shown.
In contrast, other markers we tested for clusters 1, 3, and 4 were unable to resolve if one or more of these clusters corresponded to a specific mesophyll cell type or not. For example, promoter elements for cluster 4 markers AT4G10120/SUCROSE PHOSPHATE SYNTHASE 4F (SPS4F) and AT5G17170/ENHANCER OF SOS3-1 (ENH1) drove expression predominantly in the palisade, as did cluster 3 marker AT5G23240 (Figure 3, B and C), while IQD22pro: GUS-mCit was detected predominantly in cluster 1 (Figure 4A). Other cluster 1, 3, and 4 markers AT3G27690/LHCB2.4, AT1G29395/COLD REGULATED 414 THYLAKOID MEMBRANE 1 (COR414-TM1) and AT1G70760/CHLORORESPIRATORY REDUCTION 23 (CRR23), respectively, drove expression more broadly in the palisade and spongy mesophyll, while the promoter element from another cluster 3 marker, AT2G42530/COLD REGULATED 15B (COR15B), drove expression mostly in bundle sheath. As such, we broadly associate these clusters with mesophyll cell types and possibly some bundle sheath cells. Our and others’ (Liu et al., 2020; Kim et al., 2021; Lopez-Anido et al., 2021) inability to cleanly resolve the mesophyll populations by scRNA-seq, in addition to their close proximity on our UMAP plot, further supports the conclusion that mesophyll cell types are transcriptionally similar to one another, despite their morphological differences.
Interestingly, subclustering of related major clusters may reveal additional cell-types and cell-states. For example, subclustering of epidermal pavement cell clusters 0 and 8 revealed that subpopulations of epidermal cells may be present therein (Supplemental Figure S7). Reporter analysis of marker genes AT2G39310 and AT3G09260/LONG ER BODY (LEB) suggests that one of these subclusters includes abaxial epidermal cells near the margins and proximal midvein.
Phenylpropanoid-related genes are expressed in the palisade
The inability to discretely resolve spongy and palisade mesophyll populations in our scRNA-seq study may be due to (1) high levels of similarity between the two cell types (see above); (2) gene expression within the mesophyll being influenced by additional positional information on the proximal–distal and/or medio–lateral axes, or proximity to other leaf structures such as veins; or (3) complexity in cell states; for example, mesophyll cells undergoing various levels of senescence. Alternatively, it may (4) reflect limitations with our reporter-based verification approach. To circumvent these issues, we instead took advantage of the fact that our sequenced leaf protoplasts carried the IQD22pro:GUS-mCit transgene, which should uniquely label the palisade cells. As a reference population, we compared these cells to those with high expression of the abaxial spongy mesophyll marker we identified above, CORI3 (Figure 4A). Comparing these populations, our scRNA-seq results identified 418 nuclear-encoded genes as being differentially upregulated in IQD22pro:GUS-mCit expressing cells (adjusted P-value < 0.05) (Supplemental Data Set S37). Of these, 40 were also identified as upregulated in the palisade in our FAC-sorting experiments, including IQD22, with a statistically significant overlap (hypergeometric test, P < 0.005; Figure 4B). In addition to IQD22, for eight of these genes (AT4G19200, AT1G11210, ANTHOCYANINLESS 2 (ANL2), HYDROXYCINNAMOYL-COA SHIKIMATE/QUINATE HYDROXYCINNAMOYL TRANSFERASE (HCT), AT1G22500, HAT3, AT1G70985, and LHCB4.3) we had already generated transcriptional reporters (Supplemental Figure S3). The lack of further overlap between the two approaches might reflect differences in the reference cell populations chosen, among other possibilities (see “Discussion”). These results demonstrate that the two approaches detect many of the same transcripts, although the presence of uniquely identified transcripts with each method suggests the two techniques are complementary and might uncover unique information.
GO term enrichment analysis of genes with high expression in the palisade relative to CORI3 transcript-expressing cells identified using scRNA-seq suggested that many of the differentially regulated genes were related to photosynthesis and chloroplast function (Figure 4C). This included many chlorophyll-a/b binding proteins (Supplemental Data Set S37), which have previously been detected at high levels in the palisade of soybean (Glycine max) cotyledons (Chang and Walling, 1992). These results suggest that the palisade may have unique photochemistry. Interestingly, a similar analysis of palisade upregulated genes identified from FAC sorting also suggested high activity of genes associated with lignin and flavonoid metabolism (Figure 4D). Lignin and flavonoid production both originate from precursor molecules synthesized in the phenylpropanoid biosynthetic pathway (Zhang et al., 2021). While these GO terms were not enriched in our list of palisade upregulated genes identified from scRNA-seq, several of these same genes were also found to be upregulated; for example, CINNAMYL-ALCOHOL DEHYDROGENASE 7 (CAD7), O-METHYLTRANSFERASE 1 (OMT1), and HCT (Supplemental Data Sets S1 and S37; Fraser and Chapple, 2011). As such, both data sets support high activity of the phenylpropanoid pathway within the palisade layer compared with other photosynthetic cell types.
The inability of scRNA-seq methods to detect as many phenylpropanoid-related transcripts in the palisade may be due to limitations with this method in detecting low abundance transcripts (Saliba et al., 2014). For example, while both scRNA-seq and FAC sorting methods were able to detect our palisade marker IQD22 and the phenylpropanoid-related transcript HCT, only FAC sorting was able to identify enriched palisade expression of another phenylpropanoid gene, PAL1, even though we observed PAL1 reporter expression within this tissue (Figure 1I and Supplemental Figure S8). In the future, deeper coverage may or may not resolve this issue. Because of these limitations, we chose to use FAC sorting methods for subsequent transcriptional profiling experiments.
Phenylpropanoid genes are induced by light in the palisade
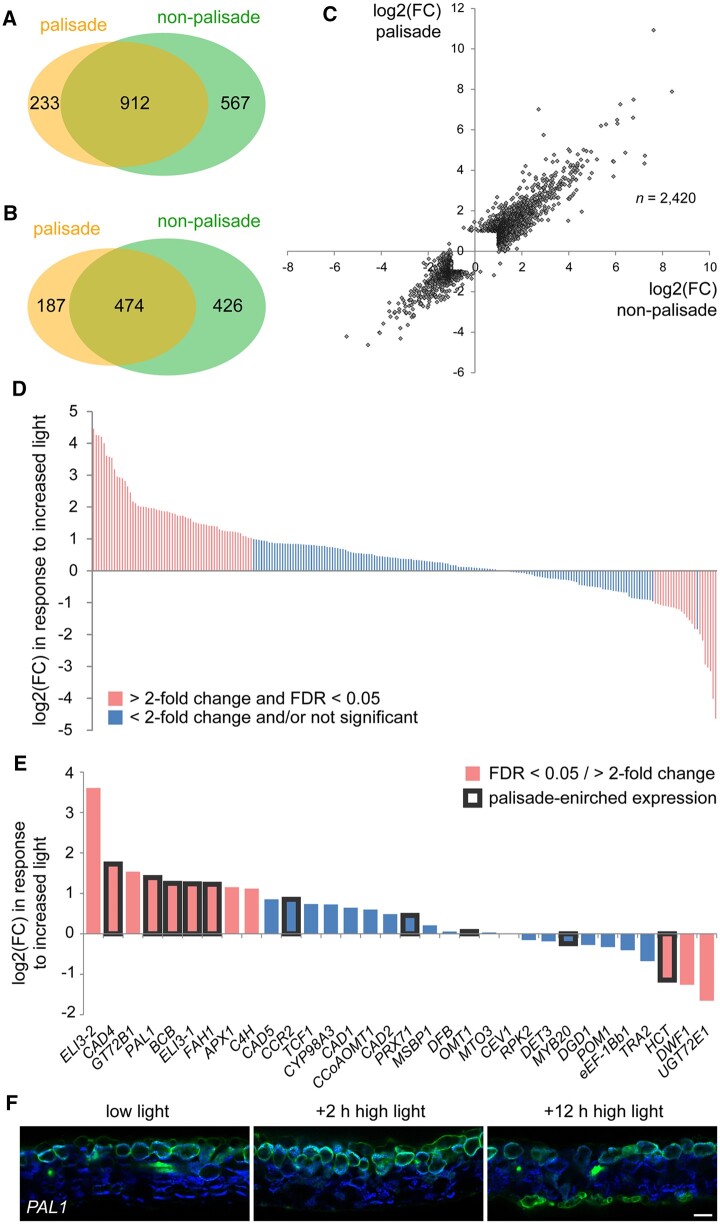
High light is known to regulate both gene expression and palisade cell morphology (Hansen, 1917; Huang et al., 2019). Therefore, to test the effect of light specifically on the palisade layer, we performed RNA-seq of FAC sorted protoplasts from IQD22pro:GUS-mCit plants treated for 2 h with increased light fluence rate. This treatment did not alter the fluorescent reporter used for sorting (Supplemental Figure S9). We observed that high light had a profound effect on both palisade mesophyll and the reference cells, with over a thousand genes being upregulated or downregulated in both populations (Supplemental Data Sets S38–S41). However, like observations we made above, we saw few differences between them, further reinforcing that different mesophyll types have very similar transcriptional programs and also respond similarly to the light environment (Figure 5, A–C). Interestingly, we also observed that many palisade-enriched genes identified via FAC sorting were regulated by high light, including phenylpropanoid pathway genes associated with GO terms related to lignin and flavonoid metabolism (Figure 5, D and E and Supplemental Figure S9E). This is consistent with the observation that ultraviolet (UV) light induces phenylpropanoid pathway activity (Dixon and Paiva, 1995; Demkura and Ballaré, 2012). The genes induced by high light included PAL1, which was predicted to be upregulated ∼2.6-fold in the palisade cells following the 2 h high light treatment. While we were unable to verify this increase in expression with transgenic PAL1pro:GUS-mCit reporter plants due to the qualitative nature of our imaging assay and variation among T1 plants, we did observe a shift in expression from the palisade to the abaxial epidermis following a longer 12-h high light treatment (Figure 5F). This was in addition to the vascular and adaxial epidermal expression we reported above. This greater expression in the abaxial epidermis under prolonged high light may correlate with flavonoid pigment production (Lois, 1994; Hughes and Smith, 2007; Huang et al., 2010), and further supports the conclusion that phenylpropanoid genes are dynamically regulated by light. Because of this observation, as well as the observation that a number of phenylpropanoid genes had palisade-enriched expression under our low light conditions, we further investigated the role of this pathway in the palisade.
Figure 5.
Effect of high light on gene expression in the palisade mesophyll. A and B, A comparison of the genes induced (A) and repressed (B) by high light in FAC sorted palisade cells compared with the photosynthetic reference cell population (FDR < 0.05 and fold change (FC) > 2). IQD22pro:GUS-mCit plants were grown for 17 days at 50 μmol m−2 s−1. High light treated plants were then moved to 300 μmol m−2 s−1 for 2 h prior to protoplasting. The two cell populations have a high degree of overlap in the genes that are regulated (P < 0.005, hypergeometric test). C, A comparison of the FC in gene expression in response to high light for all genes identified as differentially regulated by light in either the palisade or reference cell population. Note that most genes are regulated to a similar degree in the two populations. D, FC in expression when responding to the high light stimulus for genes identified as upregulated in the palisade mesophyll by FAC sorting (bars on x-axis). Genes significantly regulated by light (FDR < 0.05) with FC > 2 are indicated. E, The high light response of genes associated with GO term “lignin biosynthetic process” (GO:0009809) detected in our RNA-seq experiment in the FAC-sorted palisade mesophyll cells. Genes also identified as upregulated in the palisade are boxed in black. F, Representative fluorescence images of first true leaf cross-sections of T1 plants carrying a PAL1pro:GUS-mCit reporter (green). Plants were grown at 50 μmol m−2 s−1 (low light) for 17 days then shifted to 300 μmol m−2 s−1 (high light) for 2 or 12 h prior to imaging (n = 6 T1 plants each condition). Images were taken at the same exposure. Note a shift in reporter expression to the abaxial epidermis after 12-h high light. Chlorophyll autofluorescence, blue. Scale bar: 50 μm. Adaxial is up.
Phenylpropanoid gene expression in the palisade is required for sinapoylmalate production
To better understand the role of phenylpropanoid biosynthesis in palisade tissue, we took advantage of mutant plants carrying a lesion in the FAH1 gene, which encodes ferulate-5-hydroxylase (Chapple et al., 1992; Meyer et al., 1996). Mutations in FAH1 result in a reduction in two end products of the phenylpropanoid pathway: (1) sinapic acid-derived syringyl lignin (S lignin), and (2) sinapate esters, including sinapoylmalate in leaves (Chapple et al., 1992). Importantly, sinapoylmalate functions as a UV-B light absorbant and protectant, and fah1 mutants are sensitive to increased UV (Landry et al., 1995). When visualized under UV light, the leaves of fah1 mutants fluoresce red due to increased absorbance of UV light by chlorophyll pigments (Chapple et al., 1992).
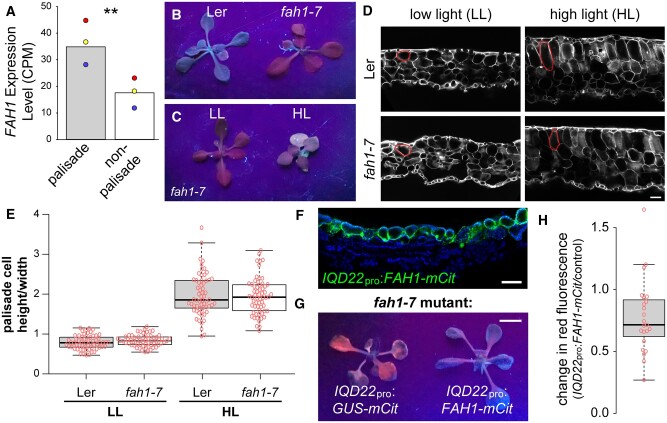
In our FAC sorting studies, we observed that FAH1 had increased expression in palisade cells relative to other photosynthetic cell types, similar to other phenylpropanoid pathway genes (Figure 6A). Leaves of fah1 mutants in our conditions also exhibited the previously reported red fluorescent phenotype under UV light (Figure 6B). We further observed a reduction in red fluorescence when plants were grown under high light, likely due to greater sinapoylmalate production as a result of higher expression of other genes in the pathway under this condition (Figure 6C). This is consistent with our RNA-seq results suggesting higher activity of the phenylpropanoid pathway in high light (Figure 5E and Supplemental Figure S9E).
Figure 6.
FAH1 expression in the palisade is required for sinapoylmalate accumulation. A, FAH1 gene expression by counts per million (CPM) in FAC sorted palisade mesophyll and photosynthetic reference cell populations. Colored dots indicate values from the three paired replicates. B, Red fluorescent phenotype when viewed under UV light of fah1-7 mutants compared with Landsberg erecta (Ler) control plants grown for 17 days at 50 μmol m−2 s−1. C, Red fluorescent phenotype under UV light of fah1-7 mutants grown for 17 days at 50 μmol m−2 s−1 (low light grown, LL), or 7 days 50 μmol m−2 s−1 followed by 10 days at 300 μmol m−2 s−1 (high light grown, HL). D, Representative images of calcofluor white-stained first true leaf cross-sections of LL- and HL-grown fah1-7 mutant and control plants. Example palisade cells are outlined in red. E, Palisade mesophyll cell height to width ratio from the first leaf pair of plants grown as in (C) and (D). F, Representative fluorescence image of a leaf cross-section from a 17-day-old T1 plant carrying a IQD22pro:FAH1-mCit transgene (n = 6). FAH1-mCit, green; chlorophyll autofluorescence, blue. G and H, Representative image (G) and quantification (H) of the relative change in red fluorescence of T1 generation 17-day-old LL-grown fah1-7 mutant plants carrying transgenes driving either FAH1-mCit or a control cDNA (GUS-mCit) under the IQD22 promoter. P = 0.006, paired t test. Scale bar (G): 5 mm. In (D) and (F), scale bars: 50 μm; adaxial is up. In (E) and (H), boxes indicate 25th and 75th percentiles, horizontal lines the median. Tukey whiskers are shown.
To test the role of phenylpropanoid biosynthesis in the palisade, we first examined if changes in palisade shape in response to high light are altered in fah1 mutants. However, we found no defect in the cell height to width ratio under both low and high light conditions (Figure 6, D and E). This suggests that neither sinapoylmalate nor the secondary cell wall component S lignin are required for palisade cell shape changes.
Alternatively, phenylpropanoid biosynthesis in the palisade may be important for sinapoylmalate accumulation. Interestingly, the abaxial surface of Arabidopsis wild-type leaves exhibits greater red fluorescence under UV light, suggesting that sinapoylmalate accumulates to greater levels on the adaxial side (Ruegger et al., 1999). While adaxial accumulation of sinapoylmalate in Arabidopsis has generally been assumed to be within the epidermis (Ruegger et al., 1999), in other species ∼50% of UV-B absorbing compounds in the leaf have been found in mesophyll (Day et al., 1996).
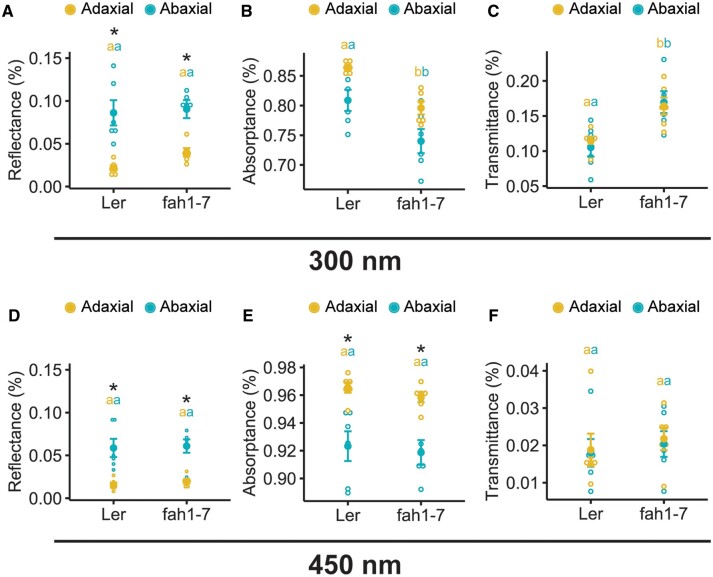
Here, we find that restoring FAH1 activity specifically to palisade cells of fah1 mutants by driving expression of the cDNA under the IQD22 promoter (Figure 6F) rescued the defective red fluorescent phenotype under UV light (Figure 6, G and H). In addition, we found that leaf optical properties are consistent with adaxial sinapoylmalate production. Specifically, compared with wild-type control plants, the leaves of fah1 mutants have decreased UV light absorptance and increased transmittance (Figure 7, A–C). These defects were specific for UV light; for example, fah1 mutants displayed minimal differences to wild-type plants in blue wavelengths (Figure 7, D–F). Importantly, less UV light reflectance was also observed on the adaxial surface compared with the abaxial side of both wild-type and mutant plants (Figure 7A), suggesting higher sinapoylmalate levels on the adaxial side. However, a similar observation was seen for blue light (Figure 7D), and, as such, we cannot rule out that other properties of the leaf contribute to this observation. Despite this, our collective results suggest that FAH1 activity in the palisade is required for adaxial production of sinapoylmalate.
Figure 7.
Leaf optical properties are consistent with higher sinapoylmalate production on the adaxial side. Reflectance (A, D), absorptance (B, E), and transmittance (C, F) of first true leaves of 17-day-old Arabidopsis plants under adaxial (yellow) and abaxial (blue) illumination with UV-B light at 300 nm (A–C) or blue light at 450 nm (D–F). n = 6 leaves per genotype, indicated by open circles; mean ± sem indicated by filled circles and error bars. Genotypes with different lowercase letters have statistically significant differences (P ≤ 0.05, nested ANOVA and Tukey’s HSD test). Asterisks indicate significant (P < 0.05) differences in leaf optical properties when illuminated on the adaxial compared with the abaxial leaf surface.
Discussion
Despite the undisputed importance of palisade mesophyll for optimal leaf performance in many plants, we know surprisingly little about the unique molecular signature of these cells. To this end, we leveraged the power of cell-specific transcriptional profiling using both FAC sorting and scRNA-seq, and identified a previously unappreciated role for the phenylpropanoid pathway in the palisade for production of the UV protectant sinapoylmalate. Our results do not rule out additional production of sinapoylmalate in the adaxial epidermis, where phenylpropanoid gene reporters are also weakly expressed (Figure 1I), and we cannot exclude FAH1 expression in additional cell types to the palisade in younger leaf tissue in our complementation experiment driven by the IQD22 promoter (Figure 6, G and H and Supplemental Figure S1F). Interestingly, our observation that at least one gene in the phenylpropanoid pathway, PAL1, shifts its expression domain toward the abaxial epidermis under prolonged high intensity light (Figure 5F) suggests that different cell types may have more prominent roles to play in sinapoylmalate and flavonoid pigment production depending on the light environment in which the plants are growing (Hughes and Smith, 2007). Some phenylpropanoid pathway gene expression may be driven by the gradient of light down the leaf. However, stronger PAL1 and HCT reporter activity in the palisade relative to the adaxial epidermis—where incident light is highest—suggests some interaction with cell identity or state (Supplemental Figure S3, F and G).
scRNA-seq is a powerful tool for the characterization of all cell types simultaneously in a given organ. A challenge, however, is to translate clusters of transcriptionally unique cells identified through bioinformatic analyses into verified biological cell identity or transcriptional state. Here, we used biased and unbiased approaches to assign cell types to our scRNA-seq atlas of Arabidopsis leaf protoplasts. Using these methods, we identified upwards of ten different cell types in the mature leaf. In actuality, this number is likely higher, as we did not completely resolve all vascular cell types (see, e.g. Kim et al., 2021) and did not detect or resolve trichome cells. Regardless, this number of cell types is not that dissimilar from some animal organs with their own dedicated functions; for example, the human liver has only around five major cell types, while the kidney contains 20–30 (Quaggin and Phimister, 2016; Trefts et al., 2017). As scRNA-seq technology expands to cover additional vascular plant species, it will be interesting to see just how many leaf cell types identified through transcriptional profiling will be conserved or novel to particular plant clades. For example, in addition to different modes of photosynthesis (Yamori et al., 2013), some plants have evolved novel leaf functions and cell types of which we understand little at the transcriptional level. This includes glandular trichomes for the secretion of biochemicals to protect from herbivores (Huchelmann et al., 2017), hydrenchyma tissue for water storage in succulents (Griffiths and Males, 2017), and mechanosensory leaf structures that sense insect prey in plant carnivores (Lloyd, 1942).
We found similar although not completely overlapping sets of genes with palisade-enriched expression using FAC sorting or scRNA-seq. This may reflect differences in our filtering and analysis methods between the two approaches, differences in the reference photosynthetic cell populations used, the presence of contaminating cells in our FAC-sorted populations, or limitations with scRNA-seq methods, particularly in the identification of low abundance transcripts (Saliba et al., 2014). Of note, we were unable to conclusively identify the palisade cells in our scRNA-seq UMAP plot without taking advantage of the presence of the IQD22pro:GUS-mCit reporter transcript in the background, which provided a molecular handle on the cells. Similarly, other scRNA-seq studies of the Arabidopsis leaf have to date been unable to differentiate the palisade cell population from other mesophyll cells (Liu et al., 2020; Kim et al., 2021; Lopez-Anido et al., 2021). Finally, we also identified a greater number of phenylpropanoid-related genes as being more highly expressed in the palisade relative to other photosynthetic cell types using RNA-seq of bulk FAC sorted cells, perhaps due to the low expression of some of these genes (Supplemental Figure S8). These findings show the limitations of scRNA-seq methods, particularly for cell types that are poorly characterized, share transcriptional similarity to other cell types and/or for which few marker genes exist in the literature. Indeed, caution must be taken when interpreting bioinformatic analyses using scRNA-seq that may include mixed cell populations. In such cases, traditional FAC sorting methods may be more informative, provided a robust and specific fluorescent marker for the cell type of interest is available. Nevertheless, in our hands, both FAC sorting and scRNA-seq methods identified some common genes encoding components of the phenylpropanoid pathway and demonstrate the utility of different approaches to query specific cell types.
One surprising finding was the lack of readily identifiable palisade markers, and instead a diversity of gene expression patterns in the leaf mesophyll. For example, promoter elements from SQE6, PAL1 and XTH6 all drove expression in the palisade; however, each had additional expression in nonoverlapping sets of other leaf cell types (Figure 1, F–I). This diversity suggests that few genes with strict palisade-specific expression may exist. This observation, coupled with the high degree of similarity we have likely observed between mesophyll types, may have affected our and others’ ability to easily resolve a palisade-specific cluster using scRNA-seq.
Finally, our data describe resources and novel gene promoter elements for those interested in probing palisade cell biology and other leaf cell types. Future work will be necessary to elucidate how photochemistry of the palisade differs, what molecular factors are necessary for regulating and altering palisade cell shape, and ultimately how palisade shape and chemistry interact with other cells to optimize photosynthesis and leaf performance across diverse environments. Our finding here that the palisade layer likely plays a role in protecting the leaf from photooxidative UV damage by increased sinapoylmalate production highlights the utility of unbiased transcriptional profiling experiments like ours to uncover novel aspects of mesophyll biology.
Materials and methods
Plant materials and growth conditions
Arabidopsis strains used were Columbia-0 (Col-0), Ler, UCR6 (Geisler et al., 2002), JR11-2 (Gardner et al., 2009), and fah1-7 (Meyer et al., 1996). For FAC sorting, JR11-2 was introgressed into the Col-0 background eight times. Seeds were sterilized, stratified, germinated, and grown in constant white light (50 μmol m−2 s−1 unless otherwise indicated; Cool White fluorescent bulbs) on 0.7% agar with 0.5× Linsmaier and Skoog salts and vitamins, on reflective aluminum foil, at 22°C.
Identification of the UCR6 enhancer trap insertion site
iPCR was used to map the insertion site of the DsE enhancer trap transposable element in the UCR6 line using previously described methods (Sundaresan et al., 1995; Geisler et al., 2002). The DsE element includes the GUS reporter gene fused to a 35S minimal promoter of CaMV, as well as the NPTII gene conferring kanamycin resistance. Briefly, gDNA extracted from UCR6 plants was digested with restriction enzyme (RE) BfaI. The digested product was purified and circularized using T4 DNA ligase, which then served as a template for PCR with primers Ds5O and Ds5I (see Sundaresan et al., 1995; all primer sequences are also shown in Supplemental Data Set S42). This reaction generated a single product of ∼1 kb. Sanger sequencing of the PCR product was used to identify the insertion site of the GUS transposable element at position −706 relative to the ATG start site of the IQD22 gene. This was verified on the other side of the DsE element by PCR amplification from UCR6 gDNA template with primers that annealed in the GUS and IQD22 coding sequences (primers CP0190 and CP0194, respectively). On this side, the GUS-coding element was inserted at position −715 relative to the IQD22 start site. To verify that this genomic region was capable of driving palisade-specific expression, we made reporter construct pCP13.11 to replicate the genomic context of the UCR6 GUS insertion (see Supplemental Figure S1A). This construct was made using a multistep process:
Regions spanning the DsE insertion site were cloned into pBluescript KS(+) (pBS) to create plasmids pCP12.44 and pCP12.45. First, ∼4 kb was PCR amplified using primers CP0241 and CP0244 from Col-0 gDNA template, and ligated into pBS at ApaI/SacI RE sites. These primers inserted into the IQD22 promoter sequence an additional 3 bp, generating an ApaI site to facilitate the cloning process. Sanger sequencing of the insert revealed a number of differences between the cloned amplicon and the published TAIR10 sequence; however, direct Sanger sequencing of PCR products from our Col-0 gDNA template showed these same base differences in our wild-type Col-0 strain. Next, we inserted into this plasmid a ∼2 kb PCR product amplified from Col-0 gDNA using primers CP0239 and CP0240 at KpnI/ApaI sites. Finally, we inserted one of two PCR fragments amplified from Col-0 gDNA template: (1) a ∼1.5 kb fragment generated using primers CP0245 and CP0246 at a single SacI site to create plasmid pCP12.44 or (2) a ∼1 kb fragment generated using primers CP0237 and CP0238 at the single KpnI site to create plasmid pCP12.45. The orientation of these inserts was confirmed by RE digest and Sanger sequencing.
From this, we generated Gateway vectors (Invitrogen) including enhancer regions 5′ and 3′ of the UCR6 DsE GUS insertion site, and which also included either the 35S(−45) minimal promoter or Nos terminator (NosT) sequences, respectively. The 35S(−45) minimal promoter was amplified using primers CP0273 and CP0274 and inserted into pBS at NgoMIV/KpnI RE sites. The 5′ flanking region of the UCR6 DsE insertion site was then PCR amplified from pCP12.44 plasmid template using primers CP0296 and CP0298 and inserted at BamHI/SacII RE sites. From this, a 5′ UCR6 enhancer-35S(−45) fragment was then amplified using primers CP0299 and CP0300, and recombined with Gateway vector pDONR P4-P1R to create plasmid pCP12.52. Similarly, for the enhancer region 3′ of the GUS insertion site, we first amplified the NosT sequence using primers CP0216 and CP0217 from Gateway plasmid pK7m34GW template, and inserted this into pBS at BamHI/SpeI RE sites. The 3′ region of the UCR6 DsE insertion site was PCR amplified from pCP12.45 using primers CP0290 and CP0291 and inserted at BamHI/XmaI sites. From this, a NosT-3′ UCR6 enhancer fragment was then amplified using primers CP0301 and CP0302 and recombined with Gateway vector pDONR P2R-P3 to create plasmid pCP12.51. The sequences of both pCP12.52 and pCP12.51 were verified by Sanger sequencing.
Finally, to create reporter construct pCP13.11, we recombined pCP12.51, pCP12.52, and pDONR221 carrying mCit with Gateway destination vector pK7m34GW.
Generation of IQD22pro:GUS-mCit reporter plants
Approximately 3.5 kb of IQD22 promoter sequence was PCR amplified from plasmid pCP12.44 template using primers CP0293 and CP0218, and recombined with Gateway vector pDONR P4-P1R to create plasmid pCP12.46. Clones were verified using Sanger sequencing. To create IQD22pro:GUS-mCit reporter construct pCP13.42 (Supplemental Figure S1A), pCP13.41 was recombined with pDONR221 carrying GUS (Procko et al., 2016), pDONR P2R-P3 carrying mCit (Jaillais et al., 2011) and the destination vector pK7m34GW. Col-0 plants were dipped with Agrobacterium strain GV3101 carrying pCP13.42, and T2 and T3 seeds tested for single insertion, homozygous segregation patterns. A representative line with strong expression (line #3) was chosen for all subsequent analyses.
Generation of marker gene transcriptional reporters
Approximately 1.5–2 kb promoter fragments of marker genes identified from FAC sorting and scRNA-seq were PCR amplified using high-fidelity Phusion enzyme (New England BioLabs, Ipswich, Massachusetts, USA) and recombined with Gateway plasmid pDONR P4-P1R. See Supplemental Data Set S42 for primer sequences. Amplification of the correct promoter elements was verified by RE digests and Sanger sequencing with M13F(-21) and M13R primers. Due to the number of sequences being cloned, the promoter fragments were not completely sequenced, and PCR-induced errors in some constructs or variation from the TAIR10 sequence due to differences in our Col-0 strain (e.g. see above) may exist. The plasmids carrying the promoter elements were then recombined with pDONR 221 carrying GUS, pDONR P2R-P3 carrying mCit, and the destination vector pK7m34GW. Col-0 plants were dipped with Agrobacteria carrying the various reporter constructs. Kanamycin-resistant T1 plants growing on medium containing 50 μg/mL kanamycin were imaged.
Cloning of additional constructs
To create the UAS:GUS reporter shown in Supplemental Figure S2F, a pDONR P4-P1R plasmid carrying 5x UAS sequences was recombined with pDONR221 carrying GUS (Procko et al., 2016), a pDONR-P2RP3 plasmid carrying a coding sequence for a 6xHis-3xFlag tag, and destination vector pB7m34GW. To generate the IQD22pro:FAH1-mCit construct shown in Figure 6, the FAH1 coding sequence was amplified from Col-0 cDNA template using primers CP1550 and CP1551, and recombined with pDONR221. This was then recombined with pCP12.46, pDONR P2R-P3 carrying mCit, and destination vector pK7m34GW to generate plasmid pCP20.124.
Leaf protoplasting, FAC sorting, and RNA extraction
Protoplasts from the first pair of true leaves were generated using previously described methods (Bargmann and Birnbaum, 2010). Leaves were cut into pieces and placed into protoplasting solution (1.25% w/v Cellulase [Yakult, Tokyo, Japan], 0.3% w/v Macerozyme [Yakult, Tokyo, Japan], 0.4 M D-mannitol, 20 mM MES, 20 mM KCl, 10 mM CaCl2, pH 5.7) with gentle agitation for 2 h in a light chamber at 22°C. Each replicate consisted of protoplasts generated from ∼60 leaves. For most experiments, transcription inhibitors actinomycin D and cordycepin were also added to the protoplasting solution at 33 and 100 mg/L, respectively (Leonhardt et al., 2004). The material was then run through a 40 μm cell strainer and the protoplasts pelleted at 500 g for 10 min at 4°C. The protoplast pellet was resuspended in 1 mL ice-cold protoplasting solution without Cellulase and Macerozyme. Protoplasts were immediately sorted. For scRNA-seq, the same method was used, except that RNase inhibitors were added to the protoplasting solution (RNaseOut and SUPERase-In, each at 1:1,000 [Invitrogen]). The protoplast pellet after straining was then washed twice with 0.4 M d-mannitol, 20 mM MES, 20 mM KCl, 33 mg/L actinomycin D, 100 mg/L cordycepin, 1:1,000 RNaseOut, 1:1,000 SUPERase-In, 0.1% BSA, pH 5.7. The first wash also contained 10 mM CaCl2. Following the final wash step, the protoplasts were resuspended and collected on a 10 µm strainer (CellTrics 10 µm, sterile [Sysmex]) to allow small debris to flow through. Protoplasts were subsequently washed from the strainer using protoplast buffer and used as input for Single Cell 3′ Gene Expression v3 (10× Genomics) according to the manufacturer’s protocol with the exception that greater than 16,000 protoplasts were used as input. The scRNA-seq library was sequenced on a NovaSeq 6000 (Illumina).
For FAC sorting, mCit+ and GFP+ protoplasts were sorted using the following gating strategy: cells were gated first using forward and side scatter pulse area parameters (FSC-A and SSC-A), aggregates were then excluded using pulse width (FSC-W and SSC-W) before finally gating populations of interest based on red autofluorescence and mCit/GFP fluorescence compared with nontransgenic Col-0 leaf and root protoplast controls. A BD Influx sorter was used, with 1× PBS for sheath fluid, a 100-μm nozzle, and sheath pressure 16.5 PSI. The 1.5-drop Purity sort mode was used. A total of 100,000 protoplasts were sorted into 1.5 mL tubes containing 500 μL Trizol-LS (Thermo Fisher Scientific, Waltham, Massachusetts, USA). Multiple tubes were collected for each sample, with the total number of protoplasts per sample ranging from 200 to 1,100 K. RNA was then extracted using previously described methods (Mertens et al., 2015). Briefly, H2O was added to each tube to increase the volume to 750 μL. To this, 250 μL Trizol-LS was added and the tubes gently shaken for 5 min before adding 200 μL chloroform. Tubes were shaken for 5 s, stood for 5 min, and then spun at 12 K rpm using an Eppendorf 5417R refrigerated centrifuge with standard 30-tube capacity fixed-angle rotor for 15 min at 4°C. The aqueous phase was moved to a fresh tube, and 1 μL GlycoBlue (Invitrogen, Waltham, Massachusetts, USA) and 1 mL isopropanol added, mixed, and allowed to stand for 10 min. The RNA was then pelleted at 12 K rpm for 10 min at 4°C. Multiple tubes of the same sample type were pooled at this stage. The RNA pellet was washed with ethanol before air drying and resuspending in 22 μL H2O. To this, TURBO DNase was added and inactivated per the manufacturer’s instructions (Invitrogen). Stranded mRNA-seq libraries were prepared using an Illumina TruSeq stranded mRNA library preparation kit according to the manufacturer’s instructions. Libraries were then quantified, pooled, and sequenced at single-end 50-bp reads using the Illumina HiSeq 2500 platform at the Salk Next-Generation Sequencing Core. Raw sequencing data were demultiplexed and converted into FASTQ files using CASAVA (version 1.8.2). Libraries were sequenced at an average depth of 23.7 million reads. All FAC-sorted samples were collected in triplicate.
Imaging
GUS staining was performed using standard protocols and imaged on a Leica MZ FLIII stereo microscope. Fluorescence images were taken with a Zeiss LSM 710 confocal microscope (mCit: 514 nm excitation, 520–540 nm emission; chlorophyll: 633 nm excitation, 650–720 nm emission; Calcofluor white: 405 nm excitation, 410–520 nm emission). For leaf cross-sections for fluorescence imaging, fresh leaf tissue was embedded in 2% low-melting-temperature agarose for ∼1 h in the dark, and cuts were made along the meso-lateral axis at approximately the middle of the leaf blade. For Calcofluor white staining, cross-sections were placed into 0.1% w/v staining solution, washed with water and imaged. To determine palisade shape, cell height and width were measured using ImageJ from leaf cross section images stained with Calcofluor white or imaged for chlorophyll autofluorescence. In Figure 6E, 12 leaves of each genotype in each condition were sectioned, and 5 palisade cells from each section scored (n = 60 cells total for each treatment).
Leaf red fluorescence under UV light
Seedlings were irradiated with a UVL-56 Blak-ray lamp (Ultra-violet Products, Inc., Cambridge, UK). Images were processed in Photoshop (Adobe). For quantification in Figure 6H, kanamycin-resistant T1 plants growing on 50 μg/mL kanamycin were scored. Because the red fluorescence quickly faded under UV light, an experimental plant was always photographed alongside a control and the red intensity within a 1 mm2 region of a first true leaf from each plant recorded using Photoshop and the ratio determined. Plants where the first true leaves were too small to fit a 1 mm2 square were not analyzed.
Leaf optical property measurements
First true leaves were illuminated adaxially and abaxially by a 50W xenon arc lamp (Photon Technology International, Birmingham, New Jersey, USA) emitting wavelengths 200–900 nm. Transmittance (T) was measured using an integrating sphere (4 cm diameter, 3 mm diameter ports) as described by (Gorton et al., 2010) for direct light. Reflectance (R) was measured using a QR600-7-UV-125F reflection/backscatter probe (Ocean Insight) directed at the leaf surface. Spectral information was captured using a USB4000 spectrometer and OceanView software (Ocean Insight). Absorptance (A) was calculated using the relationship 1 = T + R + A. Data were collected at 300 and 450 nm to compare points in the UV and visible spectrum. A nested analysis of variances (ANOVA) was used to test the effect of genotype (main factor) and adaxial or abaxial illumination (nested factor) on leaf T, R, and A. A Tukey’s honest significant difference test (Tukey HSD) was used to examine statistical significance (P ≤ 0.05) for pairwise differences between factors. Test results are provided in Supplemental Data Sets S43 and S44.
Analysis of RNA-seq data from FAC sorted protoplasts
RNA-seq analysis was performed using CyVerse resources (Goff et al., 2011). Raw reads were aligned to the Arabidopsis TAIR10 genome using TopHat version 2.0.9 (strand-specific) (Kim et al., 2013). HTseq version 0.6.1 (mode intersection-strict) (Anders et al., 2014) was used for determining read counts over each gene, and edgeR for differential gene expression (Robinson et al., 2009). In each pairwise comparison, lowly expressed genes with counts < 2 in four or more of the six samples were discarded from the analysis, as were nonnuclear encoded genes. Palisade and nonpalisade reference cells sorted from the same total leaf protoplast pool were paired for statistical analyses where possible. Genes with fold change (FC) > 2 and FDR < 0.05 were classed as differentially regulated.
scRNA-seq analysis
FASTQ files were generated from BCL files and sequencing reads were aligned to the Araport 11 transcriptome to generate the cell by gene matrix using Cell Ranger version 3.1.0 (10× Genomics, Pleasanton, California, USA). Prior to doublet identification with DoubletDecon (DePasquale et al., 2019), the data were initially filtered based on the number of genes detected and organellar RNA levels to remove both low-quality cells and cells with abnormally high numbers of detected genes, a potential indicator of doublet cells (specifically, cells were removed where detected genes < 650, detected genes > 7,500, mitochondrial reads > 5%, and/or chloroplast reads > 20%). This initial filtering step removed 2,287 cells. The remaining 26,270 cells were then used as input for doublet identification using DoubletDecon, which identified 2,696 putative doublets. In total, 4,983 cells were filtered prior to downstream analysis (17.3% of total cells). Cells containing the highest UMI counts were generally included in the DoubletDecon filtered population. Single cells were then normalized for sequencing depth and UMI coverage of individual cells using sctransform (Hafemeister and Satija, 2019), reads aligned to the chloroplast and mitochondrial genomes were regressed, and single cells were embedded into 2D space by UMAP. Cluster markers were identified using the FindAllMarkers function, comparing expression profiles of cells of individual clusters to all other cells, with cluster markers restricted to transcripts expressed in >25% cells of a cluster at a level of log2 FC > 0.25 and adjusted P-value < 0.05. Subclustering analysis was performed identically after subsetting the data for cells of annotated epidermal clusters.
GO term enrichment of filtered cluster markers and gene lists from FAC sorting experiments were performed with clusterProfiler (Wu et al., 2021). Significant GO terms were filtered by P-value < 0.05, and functionally redundant GO terms collapsed.
Accession numbers
RNA-seq raw and processed data have been deposited into the Gene Expression Omnibus with accession numbers GSE182414 and GSE184511.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Identification of a palisade mesophyll-specific regulatory element.
Supplemental Figure S2. FAC sorting of palisade and nonpalisade reference cells.
Supplemental Figure S3. The complete set of transcriptional reporters (gene promoter:GUS-mCit) we generated for a selection of genes identified to be upregulated in the palisade tissue by FAC sorting approaches.
Supplemental Figure S4. The complete set of transcriptional reporters (gene promoter:GUS-mCit) we generated for a selection of genes identified to be upregulated in the nonpalisade photosynthetic cell population when compared with palisade cells using FAC sorting approaches.
Supplemental Figure S5. More stringent filtering does not overly affect our scRNA-seq UMAP-based clustering.
Supplemental Figure S6. Expression patterns of de novo-identified scRNA-seq cluster-specific marker genes.
Supplemental Figure S7. Subclustering of our leaf scRNA-seq data uncovers additional cell types or cell transcriptional states.
Supplemental Figure S8. scRNA-seq fails to detect some lowly expressed transcripts enriched in the palisade.
Supplemental Figure S9. Effect of high light on IQD22 reporter and palisade-associated gene expression.
Supplemental Table S1. Comparison of recent scRNA-seq data sets from plants.
Supplemental Data Sets S1 and S2. Genes upregulated and downregulated in the palisade relative to other photosynthetic cell types.
Supplemental Data Sets S3–S19. scRNA-seq UMAP cluster 0–19 markers.
Supplemental Data Sets S20–S36. GO enrichment analysis for clusters 0–16.
Supplemental Data Set S37. Genes upregulated in IQD22pro:GUS-mCit transgene positive cells (TransG+) relative to CORI3 positive (CORI3+) cells using scRNA-seq.
Supplemental Data Set S38. Genes upregulated in the palisade by increased light intensity.
Supplemental Data Set S39. Genes upregulated in nonpalisade photosynthetic cells by increased light intensity.
Supplemental Data Set S40. Genes downregulated in the palisade by increased light intensity.
Supplemental Data Set S41. Genes downregulated in nonpalisade photosynthetic cells by increased light intensity.
Supplemental Data Set S42. Primers used in this study.
Supplemental Data Set S43. ANOVA test results for Figure 7.
Supplemental Data Set S44. Tukey HSD test results for Figure 7.
Supplementary Material
Acknowledgments
We thank J. Mertens, C. Fitzpatrick, L Baum, M. Ku, N. Hah, and L. Ouyang for technical assistance. Some analysis was performed using CyVerse, supported by the NSF (DBI-0735191 and DBI-1743442), and the Flow Cytometry and NGS Core Facilities of the Salk Institute, with funding from NIH-NCI CCSG: P30 014195 and Shared Instrumentation Grant S10-OD023689; and NIH-NCI CCSG: P30 014195, the Chapman Foundation and the Helmsley Charitable Trust, respectively.
Funding
This work was supported by the National Institutes of Health (NIH) fellowship 1F32GM101876 (C.P.), National Science Foundation (NSF) fellowship DGE1752134 (A.B.), and NIH grants 5R35GM122604 (J.C.) and GM43644 (M.E.).
Conflict of interest statement. None declared.
Contributor Information
Carl Procko, Plant Biology Laboratory, Salk Institute for Biological Studies, La Jolla, California 92037, USA.
Travis Lee, Plant Biology Laboratory, Salk Institute for Biological Studies, La Jolla, California 92037, USA; Genomic Analysis Laboratory, Salk Institute for Biological Studies, La Jolla, California 92037, USA; Howard Hughes Medical Institute, Salk Institute for Biological Studies, La Jolla, California 92037, USA.
Aleca Borsuk, School of the Environment, Yale University, New Haven, Connecticut 06511, USA.
Bastiaan O R Bargmann, Biological Sciences, University of California, San Diego, California 92093, USA.
Tsegaye Dabi, Plant Biology Laboratory, Salk Institute for Biological Studies, La Jolla, California 92037, USA; Howard Hughes Medical Institute, Salk Institute for Biological Studies, La Jolla, California 92037, USA.
Joseph R Nery, Genomic Analysis Laboratory, Salk Institute for Biological Studies, La Jolla, California 92037, USA.
Mark Estelle, Biological Sciences, University of California, San Diego, California 92093, USA.
Lisa Baird, Department of Biology, University of San Diego, San Diego, California 92110, USA.
Carolyn O’Connor, Flow Cytometry Core Facility, Salk Institute for Biological Studies, La Jolla, California 92037, USA.
Craig Brodersen, School of the Environment, Yale University, New Haven, Connecticut 06511, USA.
Joseph R Ecker, Plant Biology Laboratory, Salk Institute for Biological Studies, La Jolla, California 92037, USA; Genomic Analysis Laboratory, Salk Institute for Biological Studies, La Jolla, California 92037, USA; Howard Hughes Medical Institute, Salk Institute for Biological Studies, La Jolla, California 92037, USA.
Joanne Chory, Plant Biology Laboratory, Salk Institute for Biological Studies, La Jolla, California 92037, USA; Howard Hughes Medical Institute, Salk Institute for Biological Studies, La Jolla, California 92037, USA.
All authors contributed to experimental design. T.L. performed the scRNA-seq analysis. A.B. performed the leaf optical property measurements. Other experiments performed by C.P. with assistance from B.O.R.B. and C.O. on FAC sorting, L.B. for leaf staining, J.R.N. with sequencing, and T.D. with molecular cloning. C.P., T.L., and A.B. wrote the manuscript. All other authors provided comments.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) are: Carl Procko (procko@salk.edu) and Joanne Chory (chory@salk.edu).
References
- Anders S, Pyl PT, Huber W (2014) HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169 doi:10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anstead JA, Froelich DR, Knoblauch M, Thompson GA (2012) Arabidopsis p-protein filament formation requires both AtSEOR1 and AtSEOR2. Plant Cell Physiol 53: 1033–1042 doi:10.1093/pcp/pcs046 [DOI] [PubMed] [Google Scholar]
- Bargmann BOR, Birnbaum KD (2010) Fluorescence activated cell sorting of plant protoplasts. J Vis Exp 36: 1673 doi:10.3791/1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth C, Jander G (2006) Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. Plant J 46: 549–562 doi:10.1111/j.1365-313X.2006.02716.x [DOI] [PubMed] [Google Scholar]
- Biedroń M, Banasiak A (2018) Auxin-mediated regulation of vascular patterning in Arabidopsis thaliana leaves. Plant Cell Rep 37: 1215–1229 doi:10.1007/s00299-018-2319-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonke M, Thitamadee S, Mähönen AP, Hauser MT, Helariutta Y (2003) APL regulates vascular tissue identity in Arabidopsis. Nature 426: 181–186 doi:10.1038/nature02100 [DOI] [PubMed] [Google Scholar]
- Borsuk AM, Brodersen CR (2019) The spatial distribution of chlorophyll in leaves. Plant Physiol 180: 1406–1417 doi:10.1104/pp.19.00094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsuk AM, Roddy AB, Théroux‐Rancourt G, Brodersen CR (2022) Structural organization of the spongy mesophyll. New Phytol 234: 946–960 doi:10.1111/nph.17971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresson J, Bieker S, Riester L, Doll J, Zentgraf U (2018) A guideline for leaf senescence analyses: from quantification to physiological and molecular investigations. J Exp Bot 69: 769–786 doi:10.1093/jxb/erx246 [DOI] [PubMed] [Google Scholar]
- Brodersen CR, Vogelmann TC (2007) Do epidermal lens cells facilitate the absorptance of diffuse light? Am J Bot 94: 1061–1066 doi:10.3732/ajb.94.7.1061 [DOI] [PubMed] [Google Scholar]
- Cecchini NM, Steffes K, Schlappi MR, Gifford AN, Greenberg JT (2015) Arabidopsis AZI1 family proteins mediate signal mobilization for systemic defence priming. Nat Commun 6: 7658 doi:10.1038/ncomms8658 [DOI] [PubMed] [Google Scholar]
- Chang YC, Walling LL (1992) Spatial and temporal expression of Cab mRNAs in cotyledons of the developing soybean seedling. Planta 186: 262–272 doi:10.1007/bf00196256 [DOI] [PubMed] [Google Scholar]
- Chapple CC, Vogt T, Ellis BE, Somerville CR (1992) An Arabidopsis mutant defective in the general phenylpropanoid pathway. Plant Cell 4: 1413–1424 doi:10.1105/tpc.4.11.1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvin A, Caldelari D, Wolfender JL, Farmer EE (2012) Four 13‐lipoxygenases contribute to rapid jasmonate synthesis in wounded Arabidopsis thaliana leaves: a role for lipoxygenase 6 in responses to long‐distance wound signals. New Phytol 197: 566–575 doi:10.1111/nph.12029 [DOI] [PubMed] [Google Scholar]
- Chen LQ, Qu XQ, Hou BH, Sosso D, Osorio S, Fernie AR, Frommer WB (2011) Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335: 207–211 doi:10.1126/science.1213351 [DOI] [PubMed] [Google Scholar]
- Crookston RK, Ozbun JL (1975) The occurrence and ultrastructure of chloroplasts in the phloem parenchyma of leaves of C4 dicotyledons. Planta 123: 247–255 doi:10.1007/bf00390703 [DOI] [PubMed] [Google Scholar]
- Davletova S, Schlauch K, Coutu J, Mittler R (2005) The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol 139: 847–856 doi:10.1104/pp.105.068254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day TA, Howells BW, Ruhland CT (1996) Changes in growth and pigment concentrations with leaf age in pea under modulated UV-B radiation field treatments. Plant Cell Environ 19: 101–108 doi:10.1111/j.1365-3040.1996.tb00231.x [Google Scholar]
- De Rybel B, Möller B, Yoshida S, Grabowicz I, Barbier de Reuille P, Boeren S, Smith RS, Borst JW, Weijers D (2013) A bHLH complex controls embryonic vascular tissue establishment and indeterminate growth in Arabidopsis. Dev Cell 24: 426–437 doi:10.1016/j.devcel.2012.12.013 [DOI] [PubMed] [Google Scholar]
- Demkura PV, Ballaré CL (2012) UVR8 mediates UV-B-induced Arabidopsis defense responses against Botrytis cinerea by controlling sinapate accumulation. Mol Plant 5: 642–652 doi:10.1093/mp/sss025 [DOI] [PubMed] [Google Scholar]
- DePasquale EAK, Schnell DJ, Van Camp PJ, Valiente-Alandí Í, Blaxall BC, Grimes HL, Singh H, Salomonis N (2019) Doublet decon: deconvoluting doublets from single-cell RNA-sequencing data. Cell Rep 29: 1718–1727.e1718 doi:10.1016/j.celrep.2019.09.082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B, Cayla T, Batailler B, Le Hir R, Revers F, Anstead JA, Thompson GA, Grandjean O, Dinant S (2015) Live imaging of companion cells and sieve elements in Arabidopsis leaves. PLoS ONE 10: e0118122 doi:10.1371/journal.pone.0118122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA, Paiva NL (1995) Stress-induced phenylpropanoid metabolism. Plant Cell 7: 1085–1097 doi:10.1105/tpc.7.7.1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau K (1977) Anatomy of Seed Plants, Ed 2. John Wiley & Sons, New York, NY [Google Scholar]
- Fisher K, Turner S (2007) PXY, a receptor-like kinase essential for maintaining polarity during plant vascular-tissue development. Curr Biol 17: 1061–1066 [DOI] [PubMed] [Google Scholar]
- Fraser CM, Chapple C (2011) The phenylpropanoid pathway in Arabidopsis. Arabidopsis Book 9: e0152 doi:10.1199/tab.0152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MJ, Baker AJ, Assie JM, Poethig RS, Haseloff JP, Webb AA (2009) GAL4 GFP enhancer trap lines for analysis of stomatal guard cell development and gene expression. J Exp Bot 60: 213–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler M, Jablonska B, Springer PS (2002) Enhancer trap expression patterns provide a novel teaching resource. Plant Physiol 130: 1747–1753 doi:10.1104/pp.011197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Vaughn M, McKay S, Lyons E, Stapleton AE, Gessler D, Matasci N, Wang L, Hanlon M, Stanzione D, et al. (2011) The iPlant collaborative: Cyberinfrastructure for plant biology. Front Plant Sci 2: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorton HL, Brodersen CR, Williams WE, Vogelmann TC (2010) Measurement of the Optical Properties of Leaves Under Diffuse Light. Photochem Photobiol 86: 1076–1083 [DOI] [PubMed] [Google Scholar]
- Griffiths H, Males J (2017) Succulent plants. Curr Biol 27: R890–R896 doi:10.1016/j.cub.2017.03.021 [DOI] [PubMed] [Google Scholar]
- Haberlandt G (1914) Physiological Plant Anatomy. Macmillan, London [Google Scholar]
- Hafemeister C, Satija R (2019) Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol 20: 296 doi:10.1186/s13059-019-1874-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen HC (1917) Leaf-structure as related to environment. Am J Bot 4: 533–560 [Google Scholar]
- Hooker TS, Millar AA, Kunst L (2002) Significance of the expression of the CER6 condensing enzyme for cuticular wax production in Arabidopsis. Plant Physiol 129: 1568–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Gu M, Lai Z, Fan B, Shi K, Zhou YH, Yu JQ, Chen Z (2010) Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol 153: 1526–1538 doi:10.1104/pp.110.157370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Zhao X, Chory J (2019) The Arabidopsis transcriptome responds specifically and dynamically to high light stress. Cell Rep 29: 4186–4199.e4183 doi:10.1016/j.celrep.2019.11.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huchelmann A, Boutry M, Hachez C (2017) Plant glandular trichomes: natural cell factories of high biotechnological interest. Plant Physiol 175: 6–22 doi:10.1104/pp.17.00727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes NM, Smith WK (2007) Attenuation of incident light in Galax urceolata (Diapensiaceae): concerted influence of adaxial and abaxial anthocyanic layers on photoprotection. Am J Bot 94: 784–790 doi:10.3732/ajb.94.5.784 [DOI] [PubMed] [Google Scholar]
- Jaillais Y, Hothorn M, Belkhadir Y, Dabi T, Nimchuk ZL, Meyerowitz EM, Chory J (2011) Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Genes Dev 25: 232–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36 doi:10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Symeonidi E, Pang TY, Denyer T, Weidauer D, Bezrutczyk M, Miras M, Zöllner N, Hartwig T, Frommer WB, et al. (2021) Distinct identities of leaf phloem cells revealed by single cell transcriptomics. Plant Cell 33: 511–530 doi:10.1093/plcell/koaa060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner S, Woodfield H, Prusko K, Koczor M, Gowik U, Hibberd JM, Westhoff P (2018) Expression of SULTR2;2, encoding a low-affinity sulphur transporter, in the Arabidopsis bundle sheath and vein cells is mediated by a positive regulator. J Exp Bot 69: 4897–4906 doi:10.1093/jxb/ery263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozuka T, Kong SG, Doi M, Shimazaki K, Nagatani A (2011) Tissue-autonomous promotion of palisade cell development by phototropin 2 in Arabidopsis. Plant Cell 23: 3684–3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry LG, Chapple CCS, Last RL (1995) Arabidopsis mutants lacking phenolic sunscreens exhibit enhanced ultraviolet-B injury and oxidative damage. Plant Physiol 109: 1159–1166 doi:10.1104/pp.109.4.1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI (2004) Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant[W ]. Plant Cell 16: 596–615 doi:10.1105/tpc.019000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Zhou Y, Guo J, Li J, Tian Z, Zhu Z, Wang J, Wu R, Zhang B, Sun X, et al. (2020) Global dynamic molecular profiling of stomatal lineage cell development by single-cell RNA sequencing. Mol Plant 13: 1178–1193 doi:10.1016/j.molp.2020.06.010 [DOI] [PubMed] [Google Scholar]
- Lloyd FE (1942) The Carnivorous Plants. Chronica Botanica Company, Waltham, MA [Google Scholar]
- Lois R (1994) Accumulation of UV-absorbing flavonoids induced by UV-B radiation in Arabidopsis thaliana L. Planta 194: 498–503 doi:10.1007/bf00714462 [Google Scholar]
- Lopez-Anido CB, Vatén A, Smoot NK, Sharma N, Guo V, Gong Y, Gil MXA, Weimer AK, Bergmann DC (2021) Single-cell resolution of lineage trajectories in the Arabidopsis stomatal lineage and developing leaf. Dev Cell 56: 1043–1055.e1044 doi:10.1016/j.devcel.2021.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuela D, Xu M (2020) Patterning a leaf by establishing polarities. Front Plant Sci 11: 568730 doi:10.3389/fpls.2020.568730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens J, Paquola ACM, Ku M, Hatch E, Böhnke L, Ladjevardi S, McGrath S, Campbell B, Lee H, Gage FH, et al. (2015) Directly reprogrammed human neurons retain aging-associated transcriptomic signatures and reveal age-related nucleocytoplasmic defects. Cell Stem Cell 17: 705–718 doi:10.1016/j.stem.2015.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K, Cusumano JC, Somerville C, Chapple CC (1996) Ferulate-5-hydroxylase from Arabidopsis thaliana defines a new family of cytochrome P450-dependent monooxygenases. Proc Natl Acad Sci USA 93: 6869–6874 doi:10.1073/pnas.93.14.6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen CT, Kurenda A, Stolz S, Chételat A, Farmer EE (2018) Identification of cell populations necessary for leaf-to-leaf electrical signaling in a wounded plant. Proc Natl Acad Sci USA 115: 10178–10183 doi:10.1073/pnas.1807049115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procko C, Burko Y, Jaillais Y, Ljung K, Long JA, Chory J (2016) The epidermis coordinates auxin-induced stem growth in response to shade. Genes Dev 30: 1529–1541 doi:10.1101/gad.283234.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaggin SE, Phimister EG (2016) Kindling the kidney. New England J Med 374: 281–283 doi:10.1056/NEJMcibr1514351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2009) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 doi:10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger M, Meyer K, Cusumano JC, Chapple C (1999) Regulation of ferulate-5-hydroxylase expression in Arabidopsis in the context of sinapate ester biosynthesis. Plant Physiol 119: 101–110 doi:10.1104/pp.119.1.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüping B, Ernst AM, Jekat SB, Nordzieke S, Reineke AR, Müller B, Bornberg-Bauer E, Prüfer D, Noll GA (2010) Molecular and phylogenetic characterization of the sieve element occlusion gene family in Fabaceae and non-Fabaceae plants. BMC Plant Biol 10: 219 doi:10.1186/1471-2229-10-219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu KH, Huang L, Kang HM, Schiefelbein J (2019) Single-cell RNA sequencing resolves molecular relationships among individual plant cells. Plant Physiol 179: 1444–1456 doi:10.1104/pp.18.01482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saliba AE, Westermann AJ, Gorski SA, Vogel J (2014) Single-cell RNA-seq: advances and future challenges. Nucleic Acids Res 42: 8845–8860 doi:10.1093/nar/gku555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawchuk MG, Donner TJ, Head P, Scarpella E (2008) Unique and overlapping expression patterns among members of photosynthesis-associated nuclear gene families in Arabidopsis. Plant Physiol 148: 1908–1924 doi:10.1104/pp.108.126946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakawa M, Ueda H, Nagano AJ, Shimada T, Kohchi T, Hara-Nishimura I (2014) FAMA is an essential component for the differentiation of two distinct cell types, myrosin cells and guard cells, in Arabidopsis. Plant Cell 26: 4039–4052 doi:10.1105/tpc.114.129874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulse CN, Cole BJ, Ciobanu D, Lin J, Yoshinaga Y, Gouran M, Turco GM, Zhu Y, O’Malley RC, Dickel DE, et al. (2019) High-throughput single-cell transcriptome profiling of plant cell types. Cell Rep 27: 2241–2247.e2244 doi:10.1016/j.celrep.2019.04.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith WK, Vogelmann TC, DeLucia EH, Bell DT, Shepherd KA (1997) Leaf form and photosynthesis. BioScience 47: 785–793 [Google Scholar]
- Sundaresan V, Springer P, Volpe T, Haward S, Jones JD, Dean C, Ma H, Martienssen R (1995) Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev 9: 1797–1810 doi:10.1101/gad.9.14.1797 [DOI] [PubMed] [Google Scholar]
- Susek RE, Ausubel FM, Chory J (1993) Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74: 787–799 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Watanabe-Takahashi A, Smith FW, Blake-Kalff M, Hawkesford MJ, Saito K (2000) The roles of three functional sulphate transporters involved in uptake and translocation of sulphate in Arabidopsis thaliana. Plant J 23: 171–182 doi:10.1046/j.1365-313x.2000.00768.x [DOI] [PubMed] [Google Scholar]
- Tantikanjana T (2001) Control of axillary bud initiation and shoot architecture in Arabidopsis through the SUPERSHOOT gene. Genes Dev 15: 1577–1588 doi:10.1101/gad.887301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima I, Hanba YT, Tholen D, Niinemets Ü (2011) Leaf functional anatomy in relation to photosynthesis. Plant Physiol 155: 108–116 doi:10.1104/pp.110.165472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trefts E, Gannon M, Wasserman DH (2017) The liver. Curr Biol 27: R1147–R1151 doi:10.1016/j.cub.2017.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E, Sauer N (1995) The promoter of the Arabidopsis thaliana SUC2 sucrose-H+ symporter gene directs expression of beta-glucuronidase to the phloem: evidence for phloem loading and unloading by SUC2. Planta 196: 564–570 [DOI] [PubMed] [Google Scholar]
- Vahisalu T, Kollist H, Wang YF, Nishimura N, Chan WY, Valerio G, Lamminmäki A, Brosché M, Moldau H, Kangasjärvi J, et al. (2008) SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature 452: 487–491 doi:10.1038/nature06608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelmann TC (1993) Plant tissue optics. Ann Rev Plant Physiol Plant Mol Biol 44: 231–251 doi:10.1146/annurev.pp.44.060193.001311 [Google Scholar]
- Vogelmann TC, Bornman JF, Yates DJ (1996) Focusing of light by leaf epidermal cells. Physiol Plant 98: 43–56 doi:10.1111/j.1399-3054.1996.tb00674.x [Google Scholar]
- Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Yu G, et al. (2021) clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation 2: 100141 doi:10.1016/j.xinn.2021.100141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamori W, Hikosaka K, Way DA (2013) Temperature response of photosynthesis in C3, C4, and CAM plants: temperature acclimation and temperature adaptation. Photosynth Res 119: 101–117 doi:10.1007/s11120-013-9874-6 [DOI] [PubMed] [Google Scholar]
- Zentgraf U, Doll J (2019) Arabidopsis WRKY53, a node of multi-layer regulation in the network of senescence. Plants 8: 578 doi:10.3390/plants8120578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Yang J, Li H, Chiang VL, Fu Y (2021) Cooperative regulation of flavonoid and lignin biosynthesis in plants. Critic Rev Plant Sci 40: 109–126 doi:10.1080/07352689.2021.1898083 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.