Abstract
Photosynthesis and the biosynthesis of many important metabolites occur in chloroplasts. In these semi-autonomous organelles, the chloroplast genome encodes approximately 100 proteins. The remaining chloroplast proteins, close to 3,000, are encoded by nuclear genes whose products are translated in the cytosol and imported into chloroplasts. However, there is still no consensus on the composition of the protein import machinery including its motor proteins and on how newly imported chloroplast proteins are refolded. In this study, we have examined the function of orf2971, the largest chloroplast gene of Chlamydomonas reinhardtii. The depletion of Orf2971 causes the accumulation of protein precursors, partial proteolysis and aggregation of proteins, increased expression of chaperones and proteases, and autophagy. Orf2971 interacts with the TIC (translocon at the inner chloroplast envelope) complex, catalyzes ATP (adenosine triphosphate) hydrolysis, and associates with chaperones and chaperonins. We propose that Orf2971 is intimately connected to the protein import machinery and plays an important role in chloroplast protein quality control.
Repression of Orf2971 induces accumulation of chloroplast precursor proteins and impaired chloroplast quality indicating that Orf2971 is required for protein import and chloroplast quality control.
IN A NUTSHELL.
Background: The chloroplast is an important bioreactor as well as a photosynthetic site. Approximately 3,000 plastid proteins encoded in the nucleus are translocated into the chloroplast envelope via the TOC (translocon at the outer chloroplast envelope) and TIC machineries. Most nucleus-encoded preproteins that enter the plastid are unfolded as they traverse the TOC–TIC import complexes. To prevent these unfolded or misfolded proteins from causing chloroplast damage, a quality control mechanism comprising molecular chaperones and proteases ensures that all polypeptides entering chloroplasts are either correctly folded or degraded. However, there is still no consensus on the TIC complex’s components, motor proteins, or mechanism for refolding proteins entering the chloroplast.
Question: What is the precise function of each of the proteins in the TIC complex? What is the composition of the chloroplast protein import machinery motor? How are the newly imported chloroplast proteins refolded and assembled into functional complexes?
Findings: We found that Orf2971, encoded by the largest gene in the Chlamydomonas reinhardtii chloroplast genome and proposed to be an ortholog of Ycf2, is directly associated with the protein import machinery and plays a crucial role in ensuring the quality of proteins targeted to the chloroplast. Orf2971 deficiency induces protein precursor accumulation, partial proteolysis and protein aggregation, increased expression of chaperones and proteases, and autophagy. We hypothesize that Orf2971 is intimately linked to the protein import machinery and plays a critical role in chloroplast protein quality control.
Next steps: The next challenge is to identify the sorting components associated with this complex on the stromal side. Furthermore, additional experimental evidence is required to investigate the relationship between different import machineries, including the analysis of the accumulation of precursor proteins in the various import mutants.
Introduction
Chloroplast function is dependent on a large set of proteins encoded by the nuclear and chloroplast genomes. While the chloroplast genome encodes about 100 proteins, the remaining 3,000 chloroplast-localized proteins are encoded by the nucleus, translated in the cytosol on 80S ribosomes and imported into the organelle (Soll and Schleiff, 2004; Li and Chiu, 2010; Nakai, 2018). Chloroplast nucleus-encoded precursor proteins are specifically recognized, bound, and translocated across the outer envelope membrane of the chloroplast and into the intermembrane space by the TOC translocon machinery (Andres et al., 2010). The protein is then transported across the chloroplast inner envelope membrane by the TIC translocon machinery, entering the stroma where it can be directed to a specific subcompartment within the organelle.
There is still some uncertainty about the specific components of the TIC complex (Li et al., 2020; Nakai, 2020). It was previously assumed that TIC40 directly interacts with TIC110 and stromal chaperones to assist transmembrane transport (Kovacs-Bogdan et al., 2010; Flores-Perez and Jarvis, 2013). Recently, a 1-MD TIC complex consisting of TIC20, TIC56, TIC100, and TIC214 was identified in Arabidopsis thaliana (Kikuchi et al., 2013). Moreover, a Ycf2 (hypothetical chloroplast open reading frames 2) – FTSHi (filamentation temperature sensitive protein H like) ATPase complex was found to be associated with the TIC machinery and proposed to act as a motor that pulls preproteins across the inner envelope membrane (Kikuchi et al., 2018; Schreier et al., 2018). Furthermore, the 1-MDa TIC complex was also identified in Chlamydomonas, indicating conservation of this translocation system during evolution (Ramundo et al., 2020).
Most nucleus-encoded preproteins that enter the plastid are unfolded as they traverse the TOC–TIC import machinery and emerge in the chloroplast stroma (Ruprecht et al., 2010). A quality control system consisting of chaperones and proteases ensures that all polypeptides entering the chloroplast are properly folded or, if not, that they are degraded to prevent accumulation of unfolded or misfolded proteins that can impair normal plastid functions (Kuroda and Maliga, 2003; Ramundo et al., 2014; Perlaza et al., 2019). Additionally, chloroplast stromal proteins that do not meet quality control standards can be packaged in RuBisCO-containing bodies (RCBs) and moved to the central vacuole for proteolysis (Ishida et al., 2008). Quality control also occurs at the chloroplast outer membrane where the degradation of chloroplast envelope proteins by the ubiquitin-proteasome system (UPS) regulates chloroplast development (Ling et al., 2012, 2019; Woodson et al., 2015). Finally, in addition to the initial involvement of chaperones and proteases in ensuring the maintenance of proteins and functional chloroplast processes under a range of environmental conditions, more severe conditions may elicit the “self-consuming” process of autophagy, which facilitates extensive clearing of damaged proteins and other constituents of cells/organelles, setting the stage for the synthesis of new proteins and the assembly of functioning complexes (Ishida et al., 2014).
In this work we show that Orf2971, which is proposed to be the ortholog of Ycf2 and is encoded by the largest gene of the chloroplast genome of Chlamydomonas reinhardtii (Chlamydomonas throughout), is directly connected to the protein import machinery and plays an important role in maintaining the quality of proteins targeted to the chloroplast. In particular, Orf2971 interacts with CrTic214 (also called Orf1995 or Ycf1), a protein involved in chloroplast protein translocation (Kikuchi et al., 2013; Ramundo et al., 2020), and with chaperones such as HSP70 and CPN60, which are involved in protein folding or assembly (Dickson et al., 2000; Schroda and Muhlhaus, 2009; Veyel et al., 2014; Mao et al., 2015; Aigner et al., 2017). The depletion of Orf2971 causes abnormal chloroplast development, increased accumulation of aggregated proteins, induction of chaperones and proteases, and triggers the chloroplast unfolded protein response, which can be followed by autophagy and ultimately cell death.
Results
Chlamydomonas strains conditionally repressed in orf2971 expression
Orf2971 of Chlamydomonas was proposed to be the ortholog of Ycf2 in Arabidopsis (Kikuchi et al., 2018). However, sequence alignments have revealed that the primary structures of these two proteins have diverged greatly, except for the AAA+ ATPase domain (Supplemental Figure S1). The plastid gene ycf2 in Nicotiana tabacum appears to be essential for cell survival based on the persistence of the heteroplasmic state of the gene when attempts were made to generate a knockout of all copies of this gene in N. tabacum chloroplasts (Drescher et al., 2000).
A vitamin-repressible riboswitch system was used to investigate the role of orf2971 in Chlamydomonas (Supplemental Figure S2A; Croft et al., 2005, 2007; Ramundo et al., 2013). The promoter/5′–UTR (untranslated region) of orf2971 was replaced with that of psbD to make expression of orf2971 dependent on NAC2 (Supplemental Figure S2B), while the NAC2 gene was placed under the control of a vitamin-repressible promoter. However, the construct containing the psbD promoter/5′-UTR driving expression of full-length orf2971 in the chloroplast was unable to be fully repressed when the nuclear NAC2 gene was repressed by the addition of vitamin to the medium (Supplemental Figure S2, C–E). To test whether Chlamydomonas survives when the cells are disrupted for orf2971, the aadA cassette was inserted into the middle of orf2971 to generate a null mutant. Failure to obtain homoplasmic transformants for the gene disruption indicated that this gene, like in N. tabacum, is essential for the survival of Chlamydomonas cells (Supplemental Figure S2F).
Based on our inability to fully suppress orf2971 expression through repression of NAC2 expression, we considered that orf2971 might be shorter than predicted or contain multiple promoters, allowing for some expression even in the NAC2-repressed strain. Indeed, transformants with repressible truncated orf2971 were obtained when it was fused to the psbD 5'-UTR at the third in phase initiation codon; the homoplasmic state was verified by PCR (polymerase chain reaction) (Supplemental Figure S2, D and E). The open reading frames in orf2971 that started further downstream of the initial psbD promoter/5′-UTR orf2971 fusion transcript were also analyzed by fusing them to the psbD promoter/5′-UTR with homoplasmic strains (S1–S6), generating six truncated ORFs that initiate downstream of the original construct (initiation codon furthest downstream is localized 1,116 bp from the original 5′-end; Supplemental Figure S2E) that could support viability of the cells and be repressed upon addition of vitamins to the medium (suppression of NAC2). These results demonstrate that the 5′ 1,116-bp region within orf2971 is not essential for viability of Chlamydomonas and probably not essential for the function of orf2971.
Phenotype of orf2971 depletion in the Y28 strain
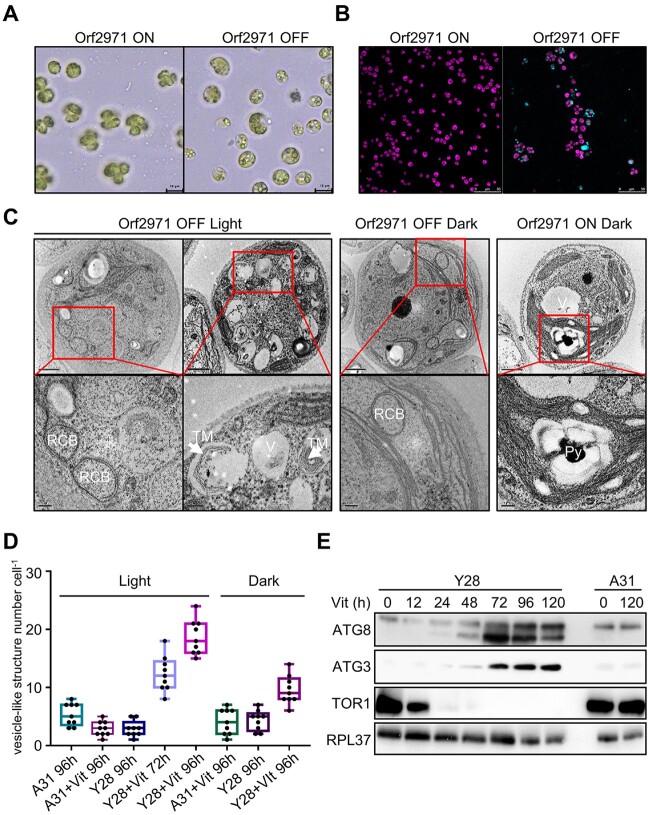
Two homoplasmic transformants (Y26 and Y28) carrying the psbD-orf2971 transgene had a pale green phenotype following the transfer of the cells from vitamin-depleted to vitamin-supplemented growth medium, suggesting that this gene is essential for cell survival (Figure 1A). After exposure to vitamins for 48 h, cell growth ceased, chlorophyll content per cell declined, and photosystem II activity (Fv/Fm) was completely lost (Figure 1, B–D). After 72, 96, and 120 h of vitamin treatment the cells became progressively paler and started to die (Figure 1, E and F). These results indicate that the lack of Orf2971 lowers the threshold level of light perceived as excess light due to dramatically reduced photosynthetic activities. To rule out the effect of light stress and photooxidative damage on the phenotype, we counted cell death not only in the light but also in the dark. Although there was no statistically significant difference in the number of dead cells in the dark versus in the light, the lethal phenotype could not be rescued by growing the Y28 strain in the dark, indicating that the primary cause of this phenotype is not photooxidative damage or impairment of photosynthesis (Figure 1, A–F).
Figure 1.
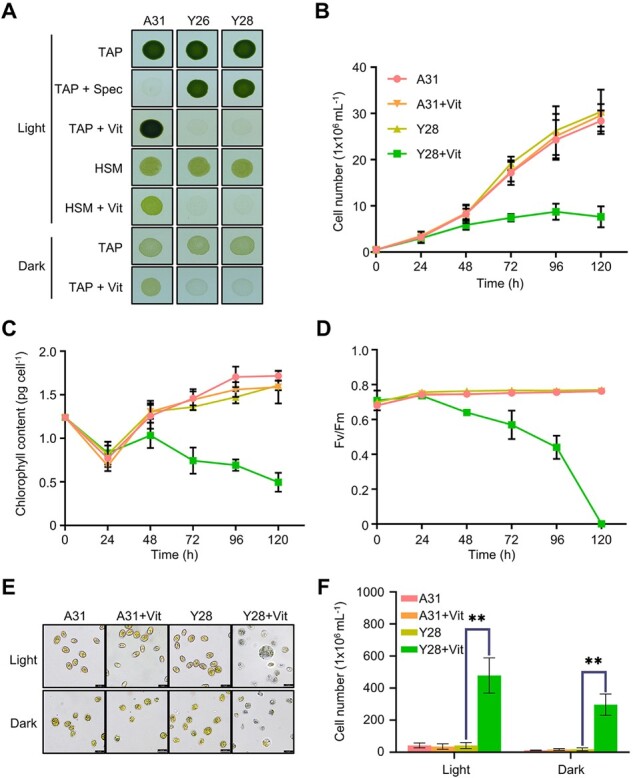
Phenotype of strains with vitamin-repressible orf2971. A, Light- and dark-grown cell cultures of A31 (WT), Y26, and Y28 in the absence or presence of vitamins B12 (20 mg L−1) and thiamine (20 μM) with an irradiance of 60 μmol photons m−2 s−1. Cells were spotted on TAP and HSM plates as indicated for 3 and 7 days, respectively. Vit, vitamin; Spec, spectinomycin. B–D, Growth curves (B), chlorophyll content per cell (C), and Fv/FM (D) measurements in the absence or presence of vitamins. Cell concentration, chlorophyll, and Fv/Fm of A31 and Y28 were measured at different times after the addition of vitamins. Cells were diluted to 0.5 × 106 cells mL−1 when they reached a concentration between 2 and 4 × 106 cells mL−1. The experiment was repeated three times with similar results. E, Light microscopy of cells of A31 and Y28 in the presence (+) or absence (–) of vitamins. Cells were stained with Trypan blue after 4 days in the light (60 μmol photons m−2 s−1) or 8 days in the dark. F, Number of dead cells identified by Trypan blue staining in the presence or absence of vitamin. Averages from three biological replicates and their standard errors are shown. Data are represented as mean ± sd. Statistical analysis was performed by unpaired Student’s t test. **P < 0.005.
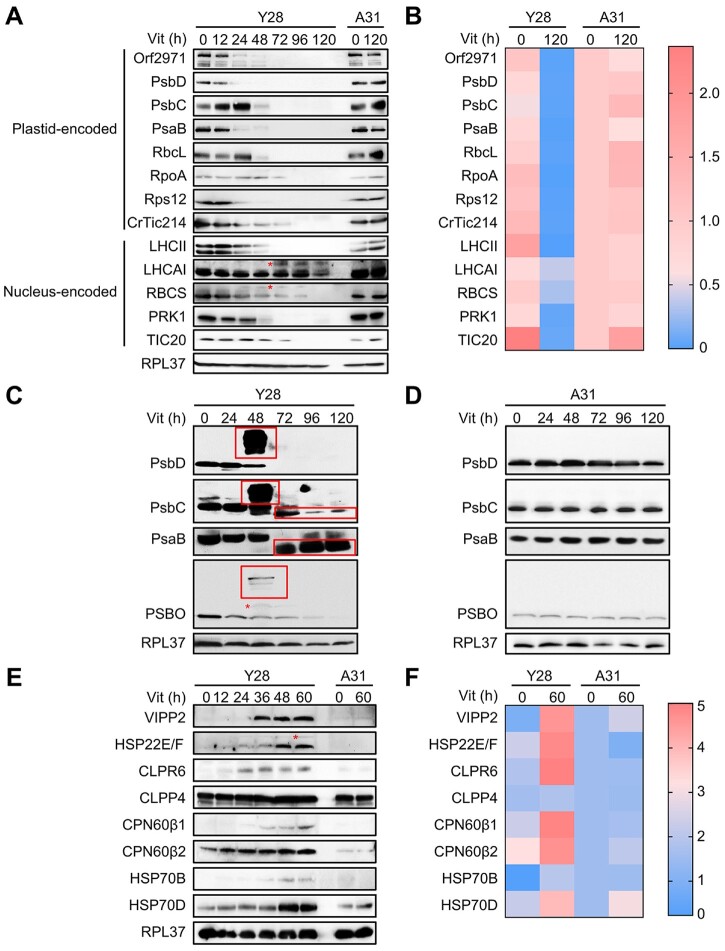
Immunoblot analysis showed that the level of Orf2971 diminished gradually after 24 h of vitamin treatment of the Y28 strain. The levels of most plastid-encoded proteins associated with photosynthetic reaction centers (PsbD, PsbC, and PsaB), RuBisCo (RbcL), transcription (RpoA), and translation (Rps12) decreased upon repression of Orf2971. The levels of CrTic214 (plastid-encoded) and TIC20 (nucleus-encoded), two subunits of the TIC complex also declined. It is therefore not surprising that there was a decrease in the levels of the nucleus-encoded chloroplast proteins associated with photosynthetic functions, LHCII, LHCA1, RBCS, and PRK1 (Figure 2, A and B). The accumulation of putative precursor forms of LHCA1 and RBCS (Figure 2, marked with an asterisk) is consistent with impaired chloroplast protein translocation upon Orf2971 depletion.
Figure 2.
Protein abundance during Orf2971 depletion. A, Immunoblot analysis of Orf2971 and chloroplast proteins during Orf2971 depletion. The asterisks indicate putative preproteins. B, Relative abundance of proteins is shown according to the results of (A). The color map shows a color-coded visual representation of relative protein abundance. The color map scale is shown on the right. C, D, Immunoblot analysis of chloroplast proteins separated by SDS–PAGE without (C) or with (D) urea; protein aggregation and partial proteolysis are detected. The asterisks indicate putative preproteins and the boxes depict aggregated and partially degraded proteins. E, Time course of protein accumulation in Y28 upon repression of Orf2971 with vitamins for 60 h. A31 serves as control for vitamin treatment and all of the proteins tested were chloroplast proteins except for HSP70D and RPL37 (ribosomal protein L37) which served as cytosolic control. F, Relative levels of proteins are shown according to the results of (E). The color map shows a color-coded visual representation of relative protein levels. The color map scale is shown on the right.
Protein aggregation and partial proteolysis triggers protein quality control upon Orf2971 depletion
Immunoblot analysis of PsbC, PsaB, and PSBO revealed a smear in the large molecular weight region after prolonged vitamin treatment when extraction with 6-M urea was omitted. These smears most likely represent protein aggregates (Figure 2C). The reduced size of PsbC and PsaB, after extended times in medium supplemented with vitamins indicates partial degradation of these proteins (Figure 2, C and D). In contrast, VIPP2 (vesicle-inducing protein in plastids 2) and HSP22E/F (heat shock protein 22E/F) were dramatically induced following the time-dependent decrease of Orf2971. The accumulation of chloroplast chaperonins and chaperones (CPN60β1, CPN60β2, HSP70B), chloroplast Clp protease subunits (CLPR6, CLPP4) and the cytosolic HSP70D increased upon Orf2971 depletion (Figure 2, E and F). Moreover, putative precursors of PSBO and HSP22E/F could be detected.
Cellular characterization of the Y28 repressible strain
To examine the cell and chloroplast morphology upon suppressed expression of orf2971, samples of Y28 were analyzed by light and transmission electron microscopy (TEM). Upon the depletion of Orf2971, cells were swollen with enlarged vacuolar structures in the cytosol (Figure 3A). Under these conditions, the Y28 strain could be stained with dansylcadaverine (monodansylcadaverine, MDC), an indicator of autophagy (Figure 3B). Interestingly, unusual circularized membranes were observed. These structures seemed to bud from the chloroplast envelope and circularize as a double membrane, resembling the RCBs associated with chloroplast autophagy (Figure 3C). The number of vesicle-like structures dramatically increased (Figure 3D). At this stage, disorganized thylakoid stacks filled the cell and appeared to be engulfed by vesicle-like structures resembling autophagosomes (Figure 3C, left). In the dark, enlargement and vacuolization of the cells still occurred, although more slowly following the depletion of Orf2971 (Figure 3C, middle; Supplemental Figure S3). In contrast, cell morphology was regular when Orf2971 was not repressed (Figure 3C, right), indicating that this process resulted from the depletion of Orf2971 and that that it might be accelerated by photodamage.
Figure 3.
Autophagy-like phenotype after the depletion of Orf2971. A, Light microscopy of Y28 cells in the presence (Orf2971 OFF) or absence of vitamins (Orf2971 ON). B, Y28 cells treated or untreated with vitamins were stained with autophagy indicator MDC (Dansylcadaverine) to visualize autophagosomes. Cells were imaged by confocal microscopy. The merged channels of chlorophyll fluorescence and MDC stain fluorescence are shown. Cells were grown in the light for 96 h. C, Analysis by transmission electron microscopy of epoxy-embedded thin sections of Y28 (Orf2971 OFF) and A31 (Orf2971 ON) cells following treatment by vitamins for 72 and 96 h. The images in the second row represent the enlarged area in the rectangle of the first row. V, vacuoles; RCB, double membrane Rubisco-containing bodies; TM indicates thylakoid membranes engulfed in the autophagy-like vesicles and Py indicates the pyrenoid. D, Number of vacuoles and vesicle-like structures in A31 and Y28 in the presence or absence of vitamins. Data are represented as mean ± sd. E, Autophagy-related proteins were examined at different time points after addition of vitamins by immunoblotting using antibodies against the indicated proteins.
The progressive vacuolization and budding membrane structures are hallmarks of autophagy. Immunoblot analysis of autophagy-related proteins ATG8 and ATG3 (Mizushima et al., 2011) revealed a strong increase of these proteins following the depletion of Orf2971. The decreased level of TOR1 (target of rapamycin) as ATG8 begins to increase suggests a link between ATG8 and TOR1 during the onset of autophagy (Figure 3E) and is fully compatible with the fact that TOR1 is associated with cell growth and division (generally stimulates anabolic processes) and is inhibitory to algal autophagy (Diaz-Troya et al., 2008; Perez-Perez et al., 2010).
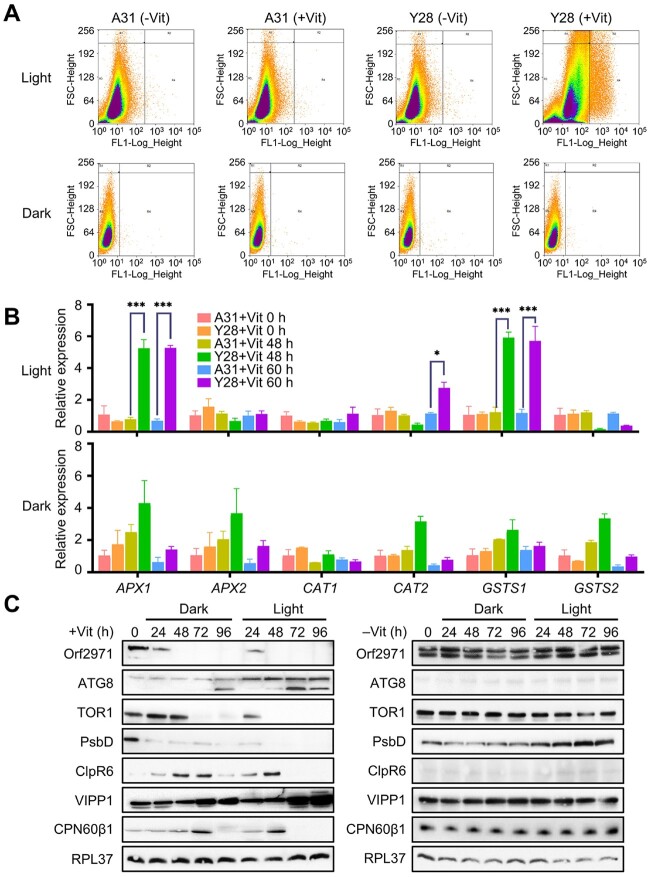
To determine whether the phenotype associated with the protein encoded by orf2971 was caused by photo-oxidative damage, ROS levels in the wild-type (WT) and Y28 were evaluated by flow cytometry following the addition of vitamins to the growth medium. While ROS accumulation upon Orf2971 depletion showed a marked increase in the light, there was little difference of ROS accumulation in the dark (Figure 4A). The expression of the ROS marker genes APX1, CAT2, and GSTS1 increased in the light but not in the dark (Figure 4B). Comparison of the levels of several proteins under light and dark conditions revealed similar changes, but the time of their appearance was delayed by about 24 h in the dark (Figure 4C). Most likely, two factors caused this delay: the absence of photooxidative stress and the slower rate of Orf2971 depletion in the dark. These findings confirm that the phenotype of Y28 is caused by the depletion of Orf2971 and that ROS accelerate this process in the light. In addition, the results also revealed variations in protein accumulation (Figure 4C); for example, unlike the continuous increase of VIPP1 (vesicle-inducing protein in plastids 1) until 96 h, accumulation of some proteins such as CPN60β1 and CLPR6 increased until 60 h in the light or 72 h in the dark, but gradually decreased after that time point, which could be due to the blocking of protein import and shorter half-life of these proteins.
Figure 4.
Accumulation of ROS upon repression of Orf2971. A, Scatter plot of ROS-induced DCF-DA fluorescence from samples with 400,000 cells of A31 and Y28 in the light and 100,000 cells in the dark in the absence or presence of vitamins showing coincidence of elevated ROS. FSC-Height (Y-axis) indicates cell size and cell surface area, FL1-Log-Height (X-axis) indicates ROS fluorescence intensity. B, Levels of expression of ROS-related marker genes in the light or dark upon Orf2971 repression determined by qPCR. Data are represented as mean ± sd. C, Analysis of proteins from Y28 grown in the light or dark upon Orf2971 repression. Proteins were extracted at different time points and immunoblotted with the antisera as indicated on the left.
Orf2971 is part of a large complex associated with TIC
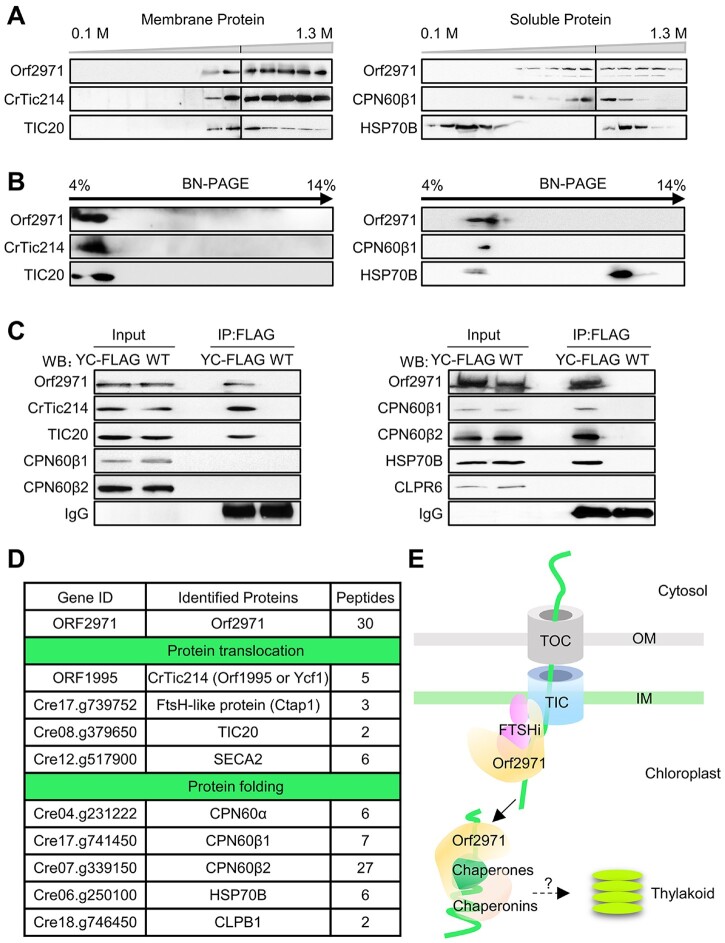
To determine the localization of Orf2971, a total cell extract was separated into soluble and insoluble fractions and examined by immunoblotting of FLAG-tagged Orf2971 (Supplemental Figure S4A). A band with an apparent molecular mass above 200 kDa most likely representing full length Orf2971 was detected in the total extract, and mostly in the pellet fraction; small amounts of Orf2971 protein could also be detected in the soluble fraction (Supplemental Figure S4B). However, Orf2971 is not associated with the thylakoid membrane (Supplemental Figure S4C). To test the strength of the association of Orf2971 with the membranes, they were subjected to different treatments known to detach nonintrinsic membrane proteins. Orf2971 was completely released from the membranes after treatment with 2-M NaCl, 2-M NaBr, 0.1-M Na2CO3, or 2-M KI, while CrTic214 which contains six transmembrane domains remained associated with the membranes (Supplemental Figure S4D). In addition, prediction of transmembrane segments revealed that Orf2971 has two very weak peaks (https://services.healthtech.dtu.dk/service.php?TMHMM-2.0). The expected number of amino acids in transmembrane helices in these two regions, however, does not meet the minimal transmembrane requirement (more than 18 amino acids) as indicated by TMHMM2.0 (Supplemental Figure S4E). The results showed that the first region can be deleted indicating it is not essential for the function of Orf2971 (Supplemental Figure S2). These findings indicate that Orf2971 is not an intrinsic membrane protein and can be readily removed from the membranes by salt treatment.
To examine whether Orf2971 is associated with other proteins in a larger complex, membrane and soluble extracts of the WT and Y28 were separated by sucrose density gradient centrifugation. Analysis of the gradient fractions of the membrane extract by immunoblotting revealed that Orf2971 co-fractionates with CrTic214 and TIC20 (Figure 5A). Analysis of the gradient fractions of the stromal extract showed that Orf2971, chaperonins CPN60, and chaperones HSP70B partially overlap (Figure 5A). A similar co-fractionation was observed from the 2D BN/sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE; Figure 5B).
Figure 5.
Orf2971 is associated with the TIC complex and protein folding components in the chloroplast. A, Membrane fractions (left) and soluble fractions (right) of the gradients were collected from WT and analyzed by SDS–PAGE and immunoblotting. B, Fractions of the gradients of the membrane (left) and soluble phase (right) were collected from WT and analyzed by 2D BN/SDS–PAGE. C, Immunoprecipitation of FLAG-tagged Orf2971 and associated proteins. Insoluble (left) and soluble (right) extracts of FLAG-tagged Orf2971 and WT strains were co-immunoprecipitated with anti-Flag antibody. The immunoprecipitated proteins were separated by SDS–PAGE and detected by immunoblotting with anti-Orf2971 antibody and the indicated antibodies. YC-FLAG, FLAG-tagged Orf2971 strain. D, Classification of proteins interacting with Orf2971 based on co-immunoprecipitation and mass-spectrometry reveals two categories involved in protein translocation across the chloroplast envelope and in the folding of chloroplast proteins. E, Schematic illustration of Orf2971 interactions with chloroplast proteins.
To further validate the interaction between Orf2971 and other plastid proteins, membrane and soluble extracts of the strain transformed with Orf2971-FLAG were used for immunoprecipitation with beads that specifically bind FLAG. The immunoprecipitates of the membrane fraction were examined by immunoblotting. This analysis indicated that Orf2971, CrTic214, and TIC20 co-immunoprecipitate. In a similar way, analysis of the soluble fraction showed that CPN60, HSP70B, and Orf2971 interact with each other (Figure 5C).
The aggregated antenna and the abnormal degradation of CrTic214 after the depletion of Orf2971 also suggest an important role for this protein in chloroplast protein quality control (Supplemental Figure S5, A and B). Upon the depletion of Orf2971, a portion of CrTic214 was detached from the high MW (molecular weight) complexes as revealed by sucrose density gradient centrifugation, indicating that Orf2971 is essential for maintaining the stability of the translocation machinery (Supplemental Figure S5, B and C). It should be noted that in the soluble extracts some chlorophylls and antennas dissociated from the membrane during the fractionation process (Supplemental Figure S5C). It was reported that LHCII trimers bind to PSII–LHCII supercomplexes are more easily dissociated from the PSII core, particularly in Chlamydomonas (Watanabe et al., 2019). In addition, no bands of PSII–LHCII or PSI–LHCII complexes were found in the soluble fraction, and immunoblot analysis of PsbD (membrane protein) and RbcL (soluble protein) also revealed that the soluble and membrane fractions had little contamination with each other (Supplemental Figure S4, B and C).
To further analyze the components of these complexes, proteins of the immunoprecipitates were digested with trypsin, and the resulting peptides analyzed using liquid chromatography tandem mass spectrometry (LC–MS/MS). Based on LC–MS/MS analysis of the proteins that co-IP with Orf2971, the largest number of peptides, apart from Orf2971 itself, matched CrTic214 (Figure 5D; Supplemental Data Set S1). Peptides of TIC20 were also detected. Some peptides of the immunoprecipitates matched the FTSHi proteases, and the uncharacterized FTSH-like protein, encoded by Cre17.g739752, previously named Chloroplast translocon associated protein 1 (Ctap1) (Ramundo et al., 2020). They belong to a family of membrane-bound proteases containing an AAA+ ATPase domain and appear to be involved in “pulling” translocated proteins into the chloroplast stroma (Kikuchi et al., 2018). Additionally, the transmembrane segments of FTSHi (FHL in Chlamydomonas) proteins that interacted with CrTic214 (Ramundo et al., 2020) and Orf2971 were also analyzed (Supplemental Figure S4E). FHL1, like Orf2971, has no obvious transmembrane segments, whereas FHL3 and Ctap1 have one and two obvious transmembrane segments, respectively. This finding suggests that not all subunits of the complex are intrinsic membrane proteins.
Additional co-immunoprecipitated proteins of the stromal extract were the chaperones HSP70B and chaperonins CPN60. According to co-IP and IP-Mass results, the binding of Orf2971 to chaperones and chaperonins appears to be relatively stable (Figure 5, C and D). The depletion of Orf2971 caused aggregation of some nucleus-encoded proteins (Figure 2C), indicating that these proteins could not be targeted to the appropriate chaperones or chaperonins involved in folding. Therefore, the interactions between stromal Orf2971 and chaperones or chaperonins are essential and critical for the correct folding of the imported proteins rather than representing an assembly intermediate. In addition, peptides of SECA2 were present. This protein plays a role in the sorting of chloroplast inner membrane proteins (Skalitzky et al., 2011).
Taken together, we suggest that Orf2971 is either directly or indirectly involved in chloroplast protein translocation and its close interaction with HPS70B and CPN60 further indicates a possible role in targeting the unfolded proteins to chaperones and chaperonins (Figure 5E), consistent with the observed accumulation of unfolded proteins such as PSBO upon Orf2971 depletion (Figure 2).
Altered localization of RBCS2 and accumulation of chloroplast precursor proteins during Orf2971 depletion
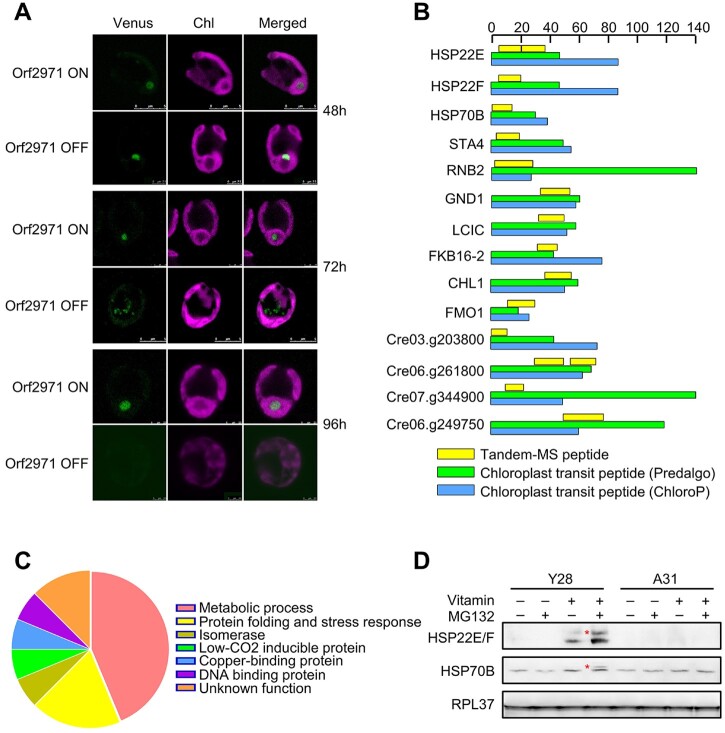
Immunoblot analysis revealed small amounts of precursors of RBCS2, LHCA1, PSBO, and HSP22E/F (Figures 2 and 3). One prediction of the model proposing that Orf2971 plays an important role in chloroplast protein import is that chloroplast precursor proteins would accumulate in the cytosol upon Orf2971 depletion. To test this prediction, we introduced Venus-tagged RBCS2 in Y28. After treatment with vitamins to repress the expression of orf2971 for 48 h, RBCS2 was imported but no longer targeted to the pyrenoid and was found instead in aggregates in the chloroplast. After the repression of Orf2971 for 72 h, RBCS2-Venus accumulated in the cytoplasm. After 96 h, the chloroplast was severely damaged and no RBCS2 signal was detected, which could be a consequence of generalized protein degradation (Figure 6A).
Figure 6.
Detection of localization of nucleus-encoded RBCS2 and chloroplast precursor proteins upon Orf2971 depletion. A, Confocal microscopy of Y28-RBCS2-Venus in the presence (Orf2971 OFF) or absence (Orf2971 ON) of vitamins for different times. B, Schematic representation of the putative chloroplast protein transit peptides (cTP). The scale at the top indicates the number of amino acids. C, Chart summarizing the classifications based on the proteins shown in (A). D, Immunoblot analysis of the HSP22E/F and HPS70B precursor with or without MG132 treatment during Orf2971 depletion. The asterisks indicate putative accumulated preproteins.
However, the accumulation of RBCS2 in the cytosol could also be caused by autophagy which is known to give rise to RCBs. In order to directly prove that protein transport is impaired, MS/MS was used to examine whether chloroplast preproteins accumulate upon Orf2971 depletion, more specifically to identify transit peptides of chloroplast-localized proteins. Because the UPS could rapidly degrade the accumulated chloroplast precursor proteins in the cytosol (Lee et al., 2009, 2016), MG132 was added to inhibit the activity of UPS. In order to make the data more reliable, proteins identified with less than five peptides were discarded. Upon Orf2971 depletion, a total of 16 proteins were detected with sequences derived from their predicted chloroplast protein transit peptides (cTPs) (Supplemental Table S1 and Supplemental Data Set S2). Amongst these proteins seven are involved in metabolic pathways, three are involved in protein folding (HSP22E, HSP22F, and HSP70B). In addition, one isomerase, one low-CO2-inducible protein C, one copper-binding protein, one DNA-binding protein and two proteins with unknown function were identified (Figure 6, B and C). Furthermore, immunoblot analysis supported these proteomic data as the precursors of HSP22E/F and HSP70B accumulated when MG132 was added upon Orf2971 depletion (Figure 6D). Taken together, the presence of peptides containing chloroplast transit peptide sequences, the accumulation of uncleaved preproteins, together with accumulation of protein aggregates in the chloroplast confirm that the depletion of Orf2971 causes both impaired chloroplast protein import and quality.
Recombinant Orf2971 protein exhibits ATPase activity
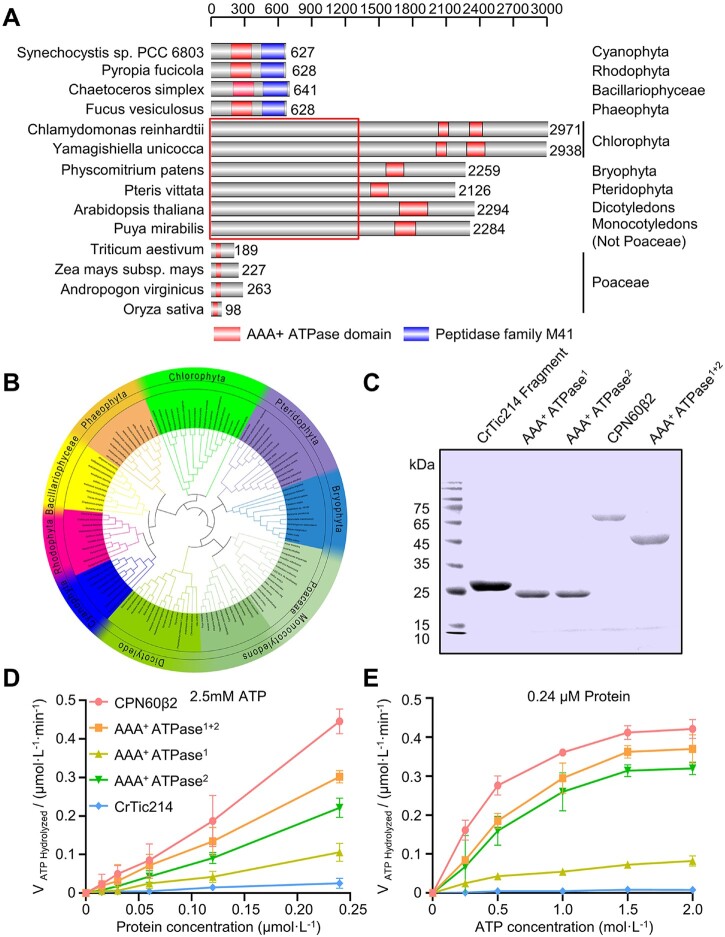
It has been proposed that orf2971 or ycf2 originated from the ftsh gene of cyanobacteria. The ATP-dependent zinc metalloprotease domain (Peptidase family M41) in the C-terminal part was lost in the green lineage and a fairly long N-terminal sequence with unknown function emerged in most of the green lineage except for the Poaceae. Interestingly, we found that some green algae contain two AAA+ ATPase domains. In Poaceae both large N-terminal domains of Ycf2 (Figure 7A) and Ycf1 were lost. An evolutionary relationship (phylogenetic tree) was plotted based on the gene model of ftsh (Cyanobacteria Synechocystis sp PCC6803), orf2971 (C.reinhardtii), and ycf2 (A.thaliana; Figure 7B; Supplemental Data Set S3). Cyanobacterial Ftsh possesses an ATP-dependent zinc metalloprotease domain in its C-terminal part and an AAA+ ATPase domain in the central section. This ftsh gene is conserved in cyanobacteria and in the chloroplast genome of Rhodophyta, Bacillariophyceae, and Phaeophyta which still retain these two domains.
Figure 7.
Protein domain analysis of Orf2971. A, Schematic representation of the conserved AAA+ ATPase domains of representative species. The regions that represent the conserved AAA+ ATPase and M41 peptidase domain are shown, respectively. The box indicates the nonconserved N-terminal parts. B, Phylogenetic analysis of chloroplast encoded FtsH, Orf2971, and Ycf2 of cyanobacterial origin. The sequence data are available in NCBI (https://www.ncbi.nlm.nih.gov/). The inner circle depicts the names of the organisms, the outer circle refers to the category, and the middle circle shows the subfamily of Poaceae. The nine classes of proteins are marked by different colors. C, Heterologous expression of the recombinant AAA+ ATPase domains (ATPase1+2), the first AAA+ ATPase domain (AAA+ ATPase1) and the second AAA+ ATPase domain (AAA+ ATPase2). Proteins extracted from Escherichia coli cells were separated by SDS–PAGE and stained with Coomassie Blue. CPN60β2 and a fragment of CrTic214 were used as positive and negative control, respectively. D, Rate of ATP hydrolysis of the ATPase domain versus the concentration of the ATPase domain analyzed by the malachite green phosphate assay. E, Relationship between ATP hydrolysis and the concentration of ATP analyzed by the malachite green phosphate assay. Data are represented as mean ± sd.
Orf2971 possesses two AAA+ ATPase domains, amino acids 2,012–2,107 (AAA+ ATPase1) and amino acids 2,279–2,392 (AAA+ ATPase2), in its C-terminal region as predicted by NCBI (national center for biotechnology information). Failure to create knockout mutants missing either of these two domains suggests that they have an essential function (Supplemental Figure S2F). To further characterize the functions of these two AAA+ ATPase domains, we conducted experiments to assess the ATP hydrolysis activity of various recombinant proteins through the malachite green phosphate assay. We first expressed the entire C-terminal region of Orf2971 (AAA+ ATPase1+2) containing both the first AAA+ ATPase1 and the second AAA+ ATPase2 domain (Figure 7C). The rate of hydrolysis increased almost linearly with increasing concentration of the three AAA+ ATPase constructs at 2.5-mM ATP (Figure 7D). At 0.24-μM protein concentration the hydrolysis rate increased with ATP concentration until it reached saturation near 1.5-mM ATP (Figure 7E). The ATP hydrolysis activity of the first AAA+ ATPase1 domain was significantly lower than for AAA+ ATPase2 suggesting that the second AAA+ ATPase domain is the major functional unit (Figure 7, D and E).
Proteomic analysis of Y28 upon vitamin-mediated depletion of Orf2971
To determine the changes in protein levels caused by the depletion of Orf2971, quantitative proteomics was performed on Y28 with or without vitamin treatment. The results revealed that upon Orf2971 depletion many proteins involved in metabolic pathways are upregulated, including those related to proteolysis (DEGO, DEG7), the ubiquitin-dependent 26S proteasome catabolic process (POA3, POA6, PBA1, PBG1, CSN6), as well as chaperones involved in protein folding (HSP70B, HSP70D, HSP22E/F, CLPB3) and disulfide oxidoreductase (Supplemental Figures S6 and S7 and Supplemental Data Set S4). Furthermore, the levels of proteins involved in thylakoid biogenesis (VIPP1, VIPP2), vesicle organization and endocytosis (VPS45, VSR1), and autophagy (APG3, APG7) were increased. The accumulation level of VIPP2, HSP22E/F, DEGO, and HSP70D are not shown because these proteins could not be detected when Orf2971 was expressed (Supplemental Data Set S4). It is noteworthy that the abundance of the signal peptide peptidase SPP1A and SPP1B in the chloroplast stroma increased, pointing to a possible feedback regulation in response to the accumulation of chloroplast preproteins. In contrast, the majority of proteins involved in photosynthesis, pigment binding, carbon metabolism, as well as chloroplast envelope proteins were downregulated upon Orf2971 depletion (Supplemental Figures S6 and S7 and Supplemental Data Set S4).
Discussion
There is currently still no unanimous consensus on the composition of the chloroplast protein translocation machinery, especially on the TIC complex and its associated motor. It was previously assumed that TIC40, TIC110, and the stromal chaperones assist transmembrane protein transport (Kovacs-Bogdan et al., 2010; Flores-Perez and Jarvis, 2013). However, a complex consisting of TIC20, TIC56, TIC100, and Tic214 was recently identified both in Arabidopsis and Chlamydomonas (Kikuchi et al., 2013; Ramundo et al., 2020). In addition, a proposed nonphotosynthetic-type TIC system including TIC20-IV (the TIC20 paralog) but not TIC56, TIC100, and TIC214 was also hypothesized (Nakai, 2015).
In this study, we investigated the role of chloroplast Orf2971 in cellular metabolism and growth in Chlamydomonas. The riboswitch system allowed us to study the state of the cells during the progressive depletion of Orf2971 so as to infer its biological function. Our results reveal that Orf2971 is part of a large supercomplex that includes TIC20 and CrTic214 (Ramundo et al., 2020). It is of particular interest that the chloroplast genes crtic214 and orf2971 in Chlamydomonas are coordinately expressed with the nuclear genes of the components of the TOC and TIC translocons during the cell cycle (Ramundo et al., 2020). Accumulation of chloroplast precursors in the cytosol after the depletion of Orf2971 suggests that Orf2971 participates in chloroplast protein translocation or at least that it interacts closely with the protein import machinery. Although the amino acid sequences have diverged greatly during evolution, both Chlamydomonas Orf2971 and Arabidopsis Ycf2 contain one or two conserved AAA+ ATPase domains for ATP hydrolysis to provide energy and proposed to be involved in pulling proteins into the chloroplast stroma (Figure 7A). Orf2971 is associated with some chaperones such as CPN60 and HSP70B in the stroma, indicating that these chaperones could be recruited at a later stage for folding or assembly in Chlamydomonas.
The depletion of Orf2971 affects protein import into chloroplasts. However, it is difficult to detect the precursor proteins, as they are rapidly degraded by the 26S proteasome in the cytoplasm (Lee et al., 2009, 2016). RuBisCO and antenna proteins have a longer half-life than other nucleus-encoded proteins in chloroplasts (Webber et al., 1994; Dinc et al., 2014), indicating that they are resilient to degradation by chloroplast proteases. Immunoblot analysis of RBCS, LHCAI and HSP22E/F revealed precursor forms in the absence of MG132 to inhibit 26S proteasome activity, while HSP70B showed precursor forms only when 26S proteasome activity was inhibited (Figures 2, A and 6, D). These results indicate that some protein precursors are more resistant to breakdown by the 26S proteasome whereas others are not. Taken together, we presume that the amount of protein precursors accumulated may be closely related to their half-life.
It is worth noting that repression Orf2971 in Chlamydomonas leads to an accumulation of stress-induced proteins like VIPP2. However, immunoblot and tandem mass spectrometry analysis failed to detect the precursor form of VIPP2 (Figure 2E; Supplemental Table S1), suggesting that VIPP2 accumulated in the chloroplast, not in the cytosol. When the Orf2971-containing complex is repressed, an alternative complex may be responsible for the import of VIPP2. Alternatively, the remaining low amount of the Orf2971 complex may still allow for some import of the over-expressed VIPP2. Furthermore, our proteomic data showed a slight increase of TIC110 (1.41-fold for 48 h and 1.46-fold for 60 h), a subunit of the classic import machinery upon Orf2971 repression (Supplemental Data Set S4). It is too early to conclude that VIPP2 and other stress response proteins could be imported via an alternative import system. Additional experimental evidence is required, including the analysis of the accumulation of the precursor proteins in various import mutants using more advanced methods such as genetic, biochemical, and proteomic techniques.
Protein aggregation and partial proteolysis occur upon the depletion of Orf2971. This may be related to the fact that the newly imported proteins are no longer targeted to the components involved in the folding or degradation of these proteins. Nucleus-encoded chloroplast proteins are translated in the cytosol and must be unfolded to pass through the TOC and TIC translocons. Failure to be rapidly processed and folded once they emerge into the chloroplast stroma may lead to misfolding and protein aggregation as observed in this study. While the cpUPR (chloroplast unfolded protein response) appears to be a relatively early response to the depletion of Orf2971, the unfolded proteins could elicit chloroplast degradation and programmed cell death (Li et al., 2021). Orf2971 depletion is accompanied by a progressive vacuolization and swelling. Circular double-layer membrane structures appear that resemble autophagosomes (van Doorn and Papini, 2013). Moreover, proteins like VPS45 and VSR1 involved in membrane biogenesis and vesicular trafficking are upregulated (Bryant and James, 2003; Zouhar et al., 2010). The concomitant strong upregulation of chloroplast VIPP1, VIPP2, and HSP22E/F raises the possibility that these proteins also participate in membrane remodeling during chloroplast autophagy (Nordhues et al., 2012). The dramatic increased abundance of proteins involved in the ubiquitin-dependent protein catabolic process and 26S proteasome suggests that ubiquitination participates in chloroplast protein quality control and degradation upon Orf2971 depletion in Chlamydomonas. At present it is unclear how the autophagy and ubiquitin pathways work in concert in chloroplast degradation in response to defects in the chloroplast protein import system.
In summary, we conditionally repressed the essential plastid-encoded protein Orf2971 using a vitamin-repressible riboswitch and explored its function through genetics, comparative cell biology, proteomics, biochemistry and molecular biology in Chlamydomonas. The results show that Orf2971 is involved, directly or indirectly, in a variety of molecular events that underlie chloroplast protein translocation. The depletion of Orf2971 induces a plastid to nuclear retrograde signaling pathway for maintaining chloroplast homeostasis. Thus, this work provides new avenues for an in-depth study of the mechanisms of chloroplast protein translocation and for identifying new molecular components involved in chloroplast development (Supplemental Figure S8).
Materials and methods
Strains, culture conditions, and stress treatments
The C.reinhardtii WT strain A31 (mt+) was described previously (Ramundo et al., 2013). The algal cells were cultured in TAP medium at 25°C under continuous cool-white fluorescent light (60 µmol photons m−2 s−1) or in the dark. In the Orf2971 depletion experiments, cells were grown to a density of 1 × 107 cells mL−1, and then diluted to 0.5 × 106 cells mL−1 in vitamin-containing medium (20-mM thiamine–HCl and 20-μg L−1 vitamin B12).
Construction of the Orf2971 repressible, FLAG-tagged, and Y28-RBCS-Venus strains
The upstream region of the chloroplast orf2971 gene was amplified from genomic DNA with the primer pairs of Up-F/R, and cloned in the pUC-atpX-aadA vector containing the aadA chloroplast selectable marker (Goldschmidt-Clermont, 1991) digested with EcoRV and ClaI by the Gibson cloning kit (Vazyme, C112-01). The downstream region of the orf2971 CDS was amplified with the primer pairs of S3 (Y28)-F/R; the psbD 5′-UTR was amplified from genomic DNA with the primer pairs of the psbD-promoter/5′-UTR-F/R. In order to fuse the psbD 5′-UTR with the orf2971 CDS, the PCR products were gel purified, mixed together and cloned into the pUC-atpX-aadA vector digested with NotI and BamHI by Gibson cloning. To generate the Orf2971-FLAG tagged strain, the upstream region was amplified with the primer pairs of YC-FLAG-Up-F/R and the downstream region containing the FLAG sequence was amplified with the primer pairs of YC-FLAG-Down-F/R, and cloned into the pUC-atpX-aadA vector. A31 was transformed with the vector as host strain. RBCS2 was amplified with the primer pairs of RBCS-F/R and fused with pMO449 vector (Onishi and Pringle, 2016) digested with HpaI. The Y28-RBCS2-VENUS strain was generated by transformation with the pMO449-RBCS2-VENUS plasmid. All primers are listed in Supplemental Table S2.
Nuclear and chloroplast transformation
Nuclear transformation of the Y28 strain with pMO449-RBCS2-VENUS was performed by electroporation as previously described (Shimogawara et al., 1998). The resulting transformants were recovered in 1-mL fresh TAP plus 40-mM sucrose for 10 min, followed by plating on TAP medium containing 10-µg mL−1 paromomycin at 25°C in constant light (40 µmol photons m−2 s−1). After 1 week, single colonies were picked.
Chloroplast transformation was performed with a Bio-Rad gene particle gun (PDS-1000 He−1). 3–5 × 107 cells were collected and plated on solid TAP medium with 100-µg mL−1 spectinomycin. The gold particles (Seashell Technology S550d) coated with 300–500 ng of plasmid DNA were bombarded into A31. Seven to 10 rounds of subcloning with the same concentration of spectinomycin (750-µg mL−1 spectinomycin) were performed to obtain homoplasmic strains.
Fluorescence measurements and cell counting
Transformants growing on liquid TAP with or without 10-mM Thiamine–HCl and 20-µg L−1 B12 were examined with a Maxi-Imaging PAM chlorophyll fluorometer (Walz, Effeltrich, Germany). Prior to each measurement, cells were adapted for 5 min in the dark. Cell numbers were analyzed by the automated Cell Counter (LIFE Technologies Countess 3, Invitrogen, Carlsbad).
Laser scanning confocal microscopy and transmission electron microscopy
Laser scanning confocal microscopy was performed with Leica TCS SP5. Cells were grown in the light (60 μmol photons m−2 s−1). For Venus, fluorescence excitation and emission were at 514 and 529 nm, respectively. For chlorophyll, fluorescence excitation and emission were at 561 and 685 nm, respectively. The representative images are randomly chosen from 10 cells that showed the same localization. For TEM, 10-mL algae in exponential growth phase were harvested and resuspended in 10-mL HEPES (pH 7.0). About 2.5% glutaraldehyde and 1% OsO4 were added to fix the cells for 60 min on ice. Cells were resuspended in BSA (bovine serum albumin) buffer and further fixed for 30 min. Furthermore, the sample was sliced in 1% aqueous glutaraldehyde. The pellet was washed with water and dehydrated with a gradient concentration of ethanol, then embedded with LR white resin. The sample was cut in slices with an ultra Diatome knife, then stained with methylene blue. Samples were observed by transmission electron microscope.
Protein extraction and immunoblot analysis
Cell breakage and isolation of total protein and protein content measurements were performed as described (Ossenbuhl et al., 2004). The Chlamydomonas antibodies against PsbD (1:10,000), PsbC (1:5,000), PsaB (1:5,000) were from Agrisera (AS06146, AS111787, and AS10695) and diluted according to the supplier’s instructions. The antibodies against RbcL (1:5,000), Orf1995 (1:1,000), LHCII (1:5,000), HSP70D (1:10,000), HSP70B (1:5,000), ATG3 (1:5,000), VIPP1 (1:5,000), and RPL37 (1:10,000) were used as previously reported (Ramundo et al., 2013, 2020). Antibodies against CPN60α (1:5,000), CPN60β1 (1:5,000), CPN60β2 (1:5,000), CLPT (1:1,000), and CLPR6 (1:1,000) were from Professor Cuimin Liu. The antibody against ATG8 (1:1,000) was from Professor Faqiang Li. The antibody against TOR1 (1:5,000) was from Professor Wenfei Wang. The antibody against HSP22E/F (1:1,000) and VIPP2 (1:5,000) was from Silvia Ramundo. Protein separation by SDS–PAGE and immunoblot analysis were performed as described previously (Ramundo et al., 2013).
Isolation of soluble proteins, membranes, and thylakoid membranes
Soluble proteins, membranes and thylakoid membranes were separated according to the method previously described (Wittkopp et al., 2018). Cells were broken by a high-pressure homogenizer. The soluble fraction was treated with saturated ammonium sulfate at 4°C for 30 min and the protein were collected by centrifugation at 2,000g for 20 min at 4°C. Total membranes were pelleted by centrifugation at 100,000g for 40–60 min at 4°C. Total membranes were resuspended in HEPES buffer (5-mm HEPES-KOH, pH 7.5, 10-mm EDTA, 1.8-M sucrose), and overlaid on the surface of 0.5 M–1.3 M–1.8 M sucrose density gradient. Thylakoid membranes were isolated by centrifugation at 256,800g, for 15 min at 4°C and collected at the interface of the 1.3-M and 1.8-M sucrose gradient layers.
Treatment of membranes with chaotropic agents
Analysis of association of Orf2971 and CrTic214 with the membranes was performed according to the method previously described (Boudreau et al., 1997). Total membranes were separated as described above and diluted to a concentration of 0.4-μg Chl μl−1. Each of 250-μL membrane samples was treated with 2-M NaCl, 2-M NaBr, 0.1-M Na2CO3, or 2-M KI for 10 min at room, then diluted five-fold with double-distilled H2O. Soluble and membrane phases were separated by centrifugation 20,000g for 30 min at 4°C. The membrane proteins were analyzed by immunoblotting.
Vitality, autophagy, and ROS test
Cell vitality was tested with Trypan Blue (Sigma-Aldrich, T8154) staining with a final concentration of 0.02%. For MDC staining, cells were stained for 2 h with 0.5-mM MDC dissolved in water (Sigma-Aldrich, 30432). Cells were then washed with HS medium three times to remove excess fluorescent dyes. Cells were observed using a confocal microscope with excitation at 405 nm and fluorescence detection at 451–539 nm. ROS was measured with a fluorogenic dye 2′—7′ dichlorofluorescin diacetate (DCFH-DA). Briefly, 1–3 × 106 cells were harvested and DCFH-DA was added with a final concentration of 10 μmolL−1. After incubation at room temperature for 20 min, DCFH-DA diffused into the cell. It was then deacetylated by cellular esterase to a nonfluorescent compound, which was later oxidized by ROS into 2′,7′–dichlorofluorescein (DCF). DCF is highly fluorescent and was detected by fluorescence spectroscopy with excitation/emission at 488 nm/525 nm.
RNA extraction and real-time PCR analysis
Isolation of total RNA from the algae was performed by using the RNA easy fast plant kit (Tiangen, DP432). cDNA synthesis was completed with a reverse transcription kit (Takara, RR047A). Real-time PCR was performed using TB Green Premix Ex Taq (Takara, RR420A) and the primers are listed in Supplemental Table S1. The real-time PCR results were analyzed by the ΔΔCt comparative quantification method and normalized by the level of CBLP expression. N = 3 biological replicates, error bars = sd, *P < 0.05, ***P < 0.001.
Production of antiserum against Orf2971 protein
Anti-Orf2971 antibody was generated against a synthetic peptide containing amino acids located at the N terminus of Orf2971 (CYFKTNNIQPTKSKSTS, CSDFLSSPKIRKTWFRN, CMVLSNTEMPSKSFGTP). Rabbits were injected with these three peptides for the production of anti-Orf2971 polyclonal antibody (Beijing Protein Innovation, Beijing). To detect Orf2971 expression, total protein was extracted from WT and Y28 strains and analyzed by immunoblotting.
Immunoprecipitation and MS
Immunoprecipitations and MS were performed as previously described (Mackinder et al., 2017). Cells were broken with a high-pressure homogenizer. The soluble and insoluble fractions of WT and Y28 were separated by ultracentrifugation at 100,000g for 30 min at 4°C. The insoluble extract was solubilized with 1% [w/v] digitonin. Insoluble material was removed by centrifugation at 20,000g for 10 min at 4°C. The supernatant was incubated for 1 h at 4°C with magnetic beads conjugated to FLAG antibody (Sigma-Aldrich, F1804). After incubation, the beads were washed five times with immunoprecipitation medium before eluting the bound proteins with elution buffer (50-mM Tris–HCl, pH 7.5, 2% [w/v] SDS). The eluted bound proteins were separated by SDS–PAGE. The gel slices were digested and the peptides were analyzed by MS.
Proteomics and data analysis
For the time-course proteomic analysis of Orf2971 depletion, samples treated for different times (48 h and 60 h) with or without vitamins were harvested. To detect preproteins through LC–MS/MS analysis, all cultures were incubated for 3 h in the presence of 30-µM MG132 (Sigma Aldrich, M7449) before collection of cells. The pellets were resuspended in cell lysis buffer (100-mM Tris–HCl, pH 6.8, 4% SDS, and 20-mM EDTA), protease inhibitor cocktail and PMSF were added to prevent protein degradation. The mixture was transferred to a high-speed centrifuge tube and centrifuged at 15,000g at 4°C for 45 min to remove cell debris. The supernatant was collected and four volumes of pre-cooled 10% trichloroacetic acid (TCA)/acetone solution was added, then precipitated overnight at −20°C. The protein precipitate was washed with pre-cooled acetone three times, then dried in a vacuum rotary concentrator and stored at −80°C. The lysates (100-μg protein for each sample) were digested with sequencing grade trypsin (Promega, Madison, WI; 1:50 (w: w)) at 37°C overnight, reduced with 100-mM DTT at 37°C for 1 h and the resulting tryptic peptides were desalted by StageTips and completely dried and analyzed by an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Scientific, Rockford, IL Waltham, MA, USA) coupled online to an Easy-nLC 1200 in the data-dependent mode. The database search was performed for all raw MS files using the software MaxQuant (v.1.6). The C.reinhardtii sequence database downloaded from Uniprot or Phytozome 13.0 was used to search the data. The parameters used for the database search were set up as follows: Type: standard; protease used for protein digestion: rypsin; Label-free quantification: LFQ; minimum score for unmodified peptides. Default values were used for all other parameters. Statistical testing was based on a two-sided t test.
Sucrose density gradient fractionation of protein complexes and 2D BN/SDS–PAGE analysis
Purified membranes (0.8-mg mL−1 chlorophyll) were solubilized with 0.75% (w/v) n‐dodecyl‐β‐d‐maltoside (β‐DM) on ice in the dark for 40 min. Solubilized membranes or soluble proteins were loaded onto a continuous sucrose density gradient (0.1–1.3 M sucrose, 5-mM Tricine‐NaOH, pH 8, 0.05% β‐DM) and the various protein complexes resolved by ultracentrifugation for 16 h at 288,000g, 4°C. 2D BN/SDS–PAGE analysis was performed as previously described (Kikuchi et al., 2011).
Phylogenetic analysis
The amino acid sequences were aligned, and the phylogenetic trees of these proteins were constructed using the statistical method maximum likelihood with the software MEGA-X. The maximum likelihood tree was searched with default values. The units of branch length are nucleotide substitutions per site. The reliability of the maximum likelihood tree nodes was assessed by the bootstrap method with 100 replicates. The resulting phylogenetic trees were edited and depicted with iTOL as midpoint rooted (https://itol.embl.de/). The resulting pairwise distances of phylogenetic trees and the associated multiple sequence alignment are shown in Supplemental data set S3.
ATPase activity assay
Briefly, the C-terminus of Orf2971 containing one or two AAA+ ATPase domains (AAA+ ATPase1 and AAA+ ATPase2) was amplified by PCR using specific primers (Supplemental Table S2). The amplified fragment was cloned directly into the Pet28a vector with a 6 × HIS tag. Proteins were expressed in BL21 (DE3). All proteins were expressed at 17°C to increase the fraction of soluble protein. All purification steps were carried out at 4°C using the Ni-NTA RESIN. The assay procedure was as previously described (Chang et al., 2008). The protein was diluted with the assay buffer (100-mM Tris–HCl, 20-mM KCl, 6-mM MgCl2, pH 7.4.). The malachite green (0.0812%, w/v), polyvinyl alcohol (2.32%, w/v), ammonium molybdate (5.72%, w/v, in 6 M HCl), and ddH2O were mixed in the ratio 2:1:1:2. The diluted protein and ATP were mixed with the assay buffer and incubated at 37°C for 3 h. The Malachite green reagent of 80 μL was added into each well. Immediately following this step, 10 μL 34% sodium citrate was used to halt the nonenzymatic hydrolysis of ATP. The samples were mixed thoroughly and incubated at 37°C for 15 min before measuring the OD620.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Sequence comparison of Orf2971 and Ycf2 of C.reinhardtii and A.thaliana.
Supplemental Figure S2. Characterization of the C. reinhardtii homoplasmic strains and Y28.
Supplemental Figure S3. Analysis by transmission electron microscopy of epoxy-embedded thin sections of Y28 (Orf2971 OFF) and A31(Orf2971 ON) cells following treatment with vitamins for 72, 96 and 120 h in the dark and light.
Supplemental Figure S4. Subcellular localization of Orf2971.
Supplemental Figure S5. Sucrose density gradient ultracentrifugation and SDS–PAGE.
Supplemental Figure S6. Quantitative analysis of the proteome of Y28 in the presence (Orf2971 OFF) or absence (Orf2971 ON) of vitamins after 48 h.
Supplemental Figure S7. Quantitative analysis of the proteome of Y28 in the presence (Orf2971 OFF) or absence (Orf2971 ON) of vitamins after 60 h.
Supplemental Figure S8. Proposed role of Orf2971.
Supplemental Table S1. Tandem mass spectrometric analysis of chloroplast transit peptides (cTP) of preproteins.
Supplemental Table S2. List of oligonucleotide primers
Supplemental Data Set S1. LC–MS/MS identification of proteins coimmunoprecipitated with Orf2971-FLAG.
Supplemental Data Set S2. Proteomic analysis of peptides of chloroplast precursor proteins upon Orf2971 depletion.
Supplemental Data Set S3. Sequence alignments and phylogenetic analysis of chloroplast encoded FtsH, Orf2971, and Ycf2 of cyanobacterial origin.
Supplemental Data Set S4. Proteomic analysis of Y28 upon vitamin-mediated depletion of Orf2971.
Funding
This study was funded by the National Key R&D Program of China (No. 2019YFA0904600); National Key R&D Program of China (No. 2019YFA0906300); National Natural Science Foundation of China (No. 31870217).
Conflict of interest statement. The authors declare no conflict of interest.
Supplementary Material
Contributor Information
Jiale Xing, Photosynthesis Research Center, Key Laboratory of Photobiology, Institute of Botany, Chinese Academy of Sciences, Beijing, China; College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, Beijing, China.
Junting Pan, Photosynthesis Research Center, Key Laboratory of Photobiology, Institute of Botany, Chinese Academy of Sciences, Beijing, China; College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, Beijing, China.
Heng Yi, Photosynthesis Research Center, Key Laboratory of Photobiology, Institute of Botany, Chinese Academy of Sciences, Beijing, China; College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, Beijing, China.
Kang Lv, Photosynthesis Research Center, Key Laboratory of Photobiology, Institute of Botany, Chinese Academy of Sciences, Beijing, China; College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, Beijing, China.
Qiuliang Gan, Photosynthesis Research Center, Key Laboratory of Photobiology, Institute of Botany, Chinese Academy of Sciences, Beijing, China; College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, Beijing, China.
Meimei Wang, Photosynthesis Research Center, Key Laboratory of Photobiology, Institute of Botany, Chinese Academy of Sciences, Beijing, China; College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, Beijing, China.
Haitao Ge, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China.
Xiahe Huang, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China.
Fang Huang, Photosynthesis Research Center, Key Laboratory of Photobiology, Institute of Botany, Chinese Academy of Sciences, Beijing, China.
Yingchun Wang, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China.
Jean-David Rochaix, Departments of Molecular Biology and Plant Biology, University of Geneva, Geneva, Switzerland.
Wenqiang Yang, Photosynthesis Research Center, Key Laboratory of Photobiology, Institute of Botany, Chinese Academy of Sciences, Beijing, China; College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, Beijing, China; Innovative Academy of Seed Design, Chinese Academy of Sciences, Beijing, China; Sino-Danish College, University of Chinese Academy of Sciences, Beijing, China.
J.X., F.H., J.-D.R., and W.Y. designed the research; J.X., J.P., H.Y., X.H., Q.G., and K.L. performed the research; M.W., H.G., Y.W., Y.W., J.-D.R., and W.Y. analyzed the data; and J.X., J.P., J.-D.R., and W.Y. wrote the article.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Wenqiang Yang (wqyang@ibcas.ac.cn).
References
- Aigner H, Wilson RH, Bracher A, Calisse L, Bhat JY, Hartl FU, Hayer-Hartl M (2017) Plant RuBisCO assembly in E. coli with five chloroplast chaperones including BSD2. Science 358: 1272–1278 [DOI] [PubMed] [Google Scholar]
- Andres C, Agne B, Kessler F (2010). The TOC complex: preprotein gateway to the chloroplast. Biochim Biophys Acta 1803: 715–723 [DOI] [PubMed] [Google Scholar]
- Boudreau E, Takahashi Y, Lemieux C, Turmel M, Rochaix JD (1997) The chloroplast ycf3 and ycf4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the photosystem I complex. EMBO J 16: 6095–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant NJ, James DE (2003) The Sec1p/Munc18 (SM) protein, Vps45p, cycles on and off membranes during vesicle transport. J Cell Biol 161: 691–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Bertelsen EB, Wisen S, Larsen EM, Zuiderweg ERP, Gestwicki JE (2008) High-throughput screen for small molecules that modulate the ATPase activity of the molecular chaperone DnaK. Analyt Biochem 372: 167–176 [DOI] [PubMed] [Google Scholar]
- Croft MT, Moulin M, Webb ME, Smith AG (2007) Thiamine biosynthesis in algae is regulated by riboswitches. Proc Natl Acad Sci USA 104: 20770–20775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft MT, Lawrence AD, Raux-Deery E, Warren MJ, Smith AG (2005) Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438: 90–93 [DOI] [PubMed] [Google Scholar]
- Diaz-Troya S, Perez-Perez ME, Florencio FJ, Crespo JL (2008) The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy 4: 851–865 [DOI] [PubMed] [Google Scholar]
- Dickson R, Weiss C, Howard RJ, Alldrick SP, Ellis RJ, Lorimer G, Azem A, Viitanen PV (2000) Reconstitution of higher plant chloroplast chaperonin 60 tetradecamers active in protein folding. J Biol Chem 275: 11829–11835 [DOI] [PubMed] [Google Scholar]
- Dinc E, Ramundo S, Croce R, Rochaix JD (2014) Repressible chloroplast gene expression in Chlamydomonas: a new tool for the study of the photosynthetic apparatus. Biochim Biophys Acta 1837: 1548–1552 [DOI] [PubMed] [Google Scholar]
- Drescher A, Ruf S, Calsa T Jr, Carrer H, Bock R (2000) The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J 22: 97–104 [DOI] [PubMed] [Google Scholar]
- Flores-Perez U, Jarvis P (2013) Molecular chaperone involvement in chloroplast protein import. Biochim Biophys Acta 1833: 332–340 [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M (1991) Transgenic expression of aminoglycoside adenine transferase in the chloroplast: a selectable marker of site-directed transformation of Chlamydomonas. Nucleic Acids Res 19: 4083–4089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida H, Izumi M, Wada S, Makino A (2014) Roles of autophagy in chloroplast recycling. Biochim Biophys Acta 1837: 512–521 [DOI] [PubMed] [Google Scholar]
- Ishida H, Yoshimoto K, Izumi M, Reisen D, Yano Y, Makino A, Ohsumi Y, Hanson MR, Mae T (2008) Mobilization of rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiology 148: 142–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S, Bedard J, Nakai M (2011) One- and two-dimensional blue native-PAGE and immunodetection of low-abundance chloroplast membrane protein complexes. Methods Mol Biol 775: 3–17 [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Bedard J, Hirano M, Hirabayashi Y, Oishi M, Imai M, Takase M, Ide T, Nakai M (2013) Uncovering the protein translocon at the chloroplast inner envelope membrane. Science 339: 571–574 [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Asakura Y, Imai M, Nakahira Y, Kotani Y, Hashiguchi Y, Nakai Y, Takafuji K, Bedard J, Hirabayashi-Ishioka Y, et al. (2018) A Ycf2-FtsHi heteromeric AAA-ATPase complex is required for chloroplast protein import. Plant Cell 30: 2677–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs-Bogdan E, Soll J, Bolter B (2010) Protein import into chloroplasts: the Tic complex and its regulation. Biochim Biophys Acta 1803: 740–747 [DOI] [PubMed] [Google Scholar]
- Kuroda H, Maliga P (2003) The plastid clpP1 protease gene is essential for plant development. Nature 425: 86–89 [DOI] [PubMed] [Google Scholar]
- Lee DW, Kim SJ, Oh YJ, Choi B, Lee J, Hwang I (2016) Arabidopsis BAG1 functions as a cofactor in Hsc70-mediated proteasomal degradation of unimported plastid proteins. Mol Plant 9: 1428–1431 [DOI] [PubMed] [Google Scholar]
- Lee S, Lee DW, Lee Y, Mayer U, Stierhof YD, Lee S, Jurgens G, Hwang I (2009) Heat shock protein cognate 70-4 and an E3 ubiquitin ligase, CHIP, mediate plastid-destined precursor degradation through the ubiquitin-26S proteasome system in Arabidopsis. Plant Cell 21: 3984–4001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HM, Chiu CC (2010) Protein transport into chloroplasts. Annu Rev Plant Biol 61: 157–180 [DOI] [PubMed] [Google Scholar]
- Li HM, Schnell D, Theg SM (2020) Protein import motors in chloroplasts: on the role of chaperones. Plant Cell 32: 536–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Sun W, Tyurin VA, DeLucia M, Ahn J, Kagan VE, van der Wel PCA (2021) Activation of cytochrome C peroxidase function through coordinated foldon loop dynamics upon interaction with anionic lipids. J Mol Biol 433: 167057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Q, Huang W, Baldwin A, Jarvis P (2012) Chloroplast biogenesis is regulated by direct action of the ubiquitin-proteasome system. Science 338: 655–659 [DOI] [PubMed] [Google Scholar]
- Ling Q, Broad W, Trosch R, Topel M, Demiral Sert T, Lymperopoulos P, Baldwin A, Jarvis RP (2019) Ubiquitin-dependent chloroplast-associated protein degradation in plants. Science 363: eaav4467 [DOI] [PubMed] [Google Scholar]
- Mackinder LCM, Chen C, Leib RD, Patena W, Blum SR, Rodman M, Ramundo S, Adams CM, Jonikas MC (2017) A spatial interactome reveals the protein organization of the algal CO2-concentrating mechanism. Cell 171: 133–147.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J, Chi W, Ouyang M, He B, Chen F, Zhang L (2015) PAB is an assembly chaperone that functions downstream of chaperonin 60 in the assembly of chloroplast ATP synthase coupling factor 1. Proc Natl Acad Sci USA 112: 4152–4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y (2011) The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 27: 107–132 [DOI] [PubMed] [Google Scholar]
- Nakai M (2015) YCF1: a green TIC: response to the de Vries et al. Commentary. Plant Cell 27: 1834–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai M (2018) New perspectives on chloroplast protein import. Plant Cell Physiol 59: 1111–1119 [DOI] [PubMed] [Google Scholar]
- Nakai M (2020) Reply: the revised model for chloroplast protein import. Plant Cell 32: 543–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordhues A, Schottler MA, Unger AK, Geimer S, Schonfelder S, Schmollinger S, Rutgers M, Finazzi G, Soppa B, Sommer F, et al. (2012) Evidence for a role of VIPP1 in the structural organization of the photosynthetic apparatus in Chlamydomonas. Plant Cell 24: 637–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M, Pringle JR (2016) Robust transgene expression from bicistronic mRNA in the green alga Chlamydomonas reinhardtii. G3 (Bethesda) 6: 4115–4125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossenbuhl F, Gohre V, Meurer J, Krieger-Liszkay A, Rochaix JD, Eichacker LA (2004) Efficient assembly of photosystem II in Chlamydomonas reinhardtii requires Alb3.1p, a homolog of Arabidopsis ALBINO3. Plant Cell 16: 1790–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Perez ME, Florencio FJ, Crespo JL (2010) Inhibition of target of rapamycin signaling and stress activate autophagy in Chlamydomonas reinhardtii. Plant Physiol 152: 1874–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlaza K, Toutkoushian H, Boone M, Lam M, Iwai M, Jonikas MC, Walter P, Ramundo S (2019) The Mars1 kinase confers photoprotection through signaling in the chloroplast unfolded protein response. eLife 8: e51430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramundo S, Rahire M, Schaad O, Rochaix JD (2013) Repression of essential chloroplast genes reveals new signaling pathways and regulatory feedback loops in Chlamydomonas. Plant Cell 25: 167–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramundo S, Casero D, Muhlhaus T, Hemme D, Sommer F, Crevecoeur M, Rahire M, Schroda M, Rusch J, Goodenough U, et al. (2014). Conditional depletion of the Chlamydomonas chloroplast ClpP protease activates nuclear genes involved in autophagy and plastid protein quality control. Plant Cell 26: 2201–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramundo S, Asakura Y, Salome PA, Strenkert D, Boone M, Mackinder LCM, Takafuji K, Dinc E, Rahire M, Crevecoeur M, et al. (2020) Coexpressed subunits of dual genetic origin define a conserved supercomplex mediating essential protein import into chloroplasts. Proc Natl Acad Sci USA 117: 32739–32749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruprecht M, Bionda T, Sato T, Sommer MS, Endo T, Schleiff E (2010) On the impact of precursor unfolding during protein import into chloroplasts. Mol Plant 3: 499–508 [DOI] [PubMed] [Google Scholar]
- Schreier TB, Clery A, Schlafli M, Galbier F, Stadler M, Demarsy E, Albertini D, Maier BA, Kessler F, Hortensteiner S, et al. (2018). Plastidial NAD-dependent malate dehydrogenase: a moonlighting protein involved in early chloroplast development through its interaction with an FtsH12-FtsHi protease complex. Plant Cell 30: 1745–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda M, Muhlhaus T (2009) A ‘foldosome’ in the chloroplast? Plant Signal Behav 4: 301–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimogawara K, Fujiwara S, Grossman A, Usuda H (1998) High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics 148: 1821–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalitzky CA, Martin JR, Harwood JH, Beirne JJ, Adamczyk BJ, Heck GR, Cline K, Fernandez DE (2011) Plastids contain a second sec translocase system with essential functions. Plant Physiol 155: 354–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll J, Schleiff E (2004) Protein import into chloroplasts. Nat Rev Mol Cell Biol 5: 198–208 [DOI] [PubMed] [Google Scholar]
- van Doorn WG, Papini A (2013) Ultrastructure of autophagy in plant cells: a review. Autophagy 9: 1922–1936 [DOI] [PubMed] [Google Scholar]
- Veyel D, Sommer F, Muranaka LS, Rutgers M, Lemaire SD, Schroda M (2014) In vitro characterization of bacterial and chloroplast Hsp70 systems reveals an evolutionary optimization of the co-chaperones for their Hsp70 partner. Biochem J 460: 13–24 [DOI] [PubMed] [Google Scholar]
- Watanabe A, Kim E, Burton-Smith RN, Tokutsu R, Minagawa J (2019) Amphipol-assisted purification method for the highly active and stable photosystem II supercomplex of Chlamydomonas reinhardtii. FEBS Lett 593: 1072–1079 [DOI] [PubMed] [Google Scholar]
- Webber AN, Nie GY, Long SP (1994) Acclimation of photosynthetic proteins to rising atmospheric CO2. Photosynth Res 39: 413–425 [DOI] [PubMed] [Google Scholar]
- Wittkopp TM, Saroussi S, Yang W, Johnson X, Kim RG, Heinnickel ML, Russell JJ, Phuthong W, Dent RM, Broeckling CD, et al. (2018) GreenCut protein CPLD49 of Chlamydomonas reinhardtii associates with thylakoid membranes and is required for cytochrome b6 f complex accumulation. Plant J 94: 1023–1037 [DOI] [PubMed] [Google Scholar]
- Woodson JD, Joens MS, Sinson AB, Gilkerson J, Salom PA, Weigel D, Fitzpatrick JA, Chory J (2015) Ubiquitin facilitates a quality-control pathway that removes damaged chloroplasts. Science 350: 450–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouhar J, Munoz A, Rojo E (2010) Functional specialization within the vacuolar sorting receptor family: VSR1, VSR3 and VSR4 sort vacuolar storage cargo in seeds and vegetative tissues. Plant J 64: 577–588 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.