Abstract
The adhE gene of Escherichia coli, located at min 27 on the chromosome, encodes the bifunctional NAD-linked oxidoreductase responsible for the conversion of acetyl-coenzyme A to ethanol during fermentative growth. The expression of adhE is dependent on both transcriptional and posttranscriptional controls and is about 10-fold higher during anaerobic than during aerobic growth. Two putative transcriptional start sites have been reported: one at position −292 and the other at −188 from the translational start codon ATG. In this study we show, by using several different transcriptional and translational fusions to the lacZ gene, that both putative transcriptional start sites can be functional and each site can be redox regulated. Although both start sites are NarL repressible in the presence of nitrate, Fnr activates only the −188 start site and Fis is required for the transcription of only the −292 start site. In addition, it was discovered that RpoS activates adhE transcription at both start sites. Under all experimental conditions tested, however, only the upstream start site is active. Available evidence indicates that under those conditions, the upstream promoter region acts as a silencer of the downstream transcriptional start site. Translation of the mRNA starting at −292, but not the one starting at −188, requires RNase III. The results support the previously postulated ribosomal binding site (RBS) occlusion model, according to which RNase III cleavage is required to release the RBS from a stem-loop structure in the long transcript.
The reduction of acetyl-coenzyme A (CoA) to ethanol is essential for disposing of excess reducing equivalents by Escherichia coli when respiratory pathways fail to maintain redox balance. For instance, genetic blockage of the ethanol pathway prevents fermentative growth on glucose or mannitol as a sole carbon and energy source. The two-step reduction of acetyl-CoA to ethanol is catalyzed by a bifunctional enzyme encoded by the adhE gene (Fig. 1) located at 27.9 min (7, 10, 13, 16, 21, 24). In addition, this protein also functions as a deactivase of pyruvate-formate lyase (13, 14). Expression of adhE is about 10-fold higher during anaerobic than during aerobic growth, and this regulation is independent of ArcA and Fnr. The accumulation of reducing equivalents, possibly the NADH concentration or the NADH/NAD ratio, was suggested to be a signal for transcriptional regulation of adhE (5, 15). Three transcriptional proteins involved in the control of adhE expression have been identified, but none of them is directly responsible for redox regulation. The first protein, NarL (8), represses adhE expression, but this occurs only in the presence of nitrate (5, 15). The second protein, Cra (for catabolite repressor activator), represses adhE expression (19), but this transcriptional regulator is functional only when complexed with fructose-1-phosphate or fructose-1,6-bisphosphate as an effector (for a review, see reference 22). The third protein, Fis (for factor for inversion stimulation) (9, 28), is required for adhE transcription, but Fis is not known to require any effector for function and the expression of the fis gene itself is independent of the respiratory condition of growth (18).
FIG. 1.
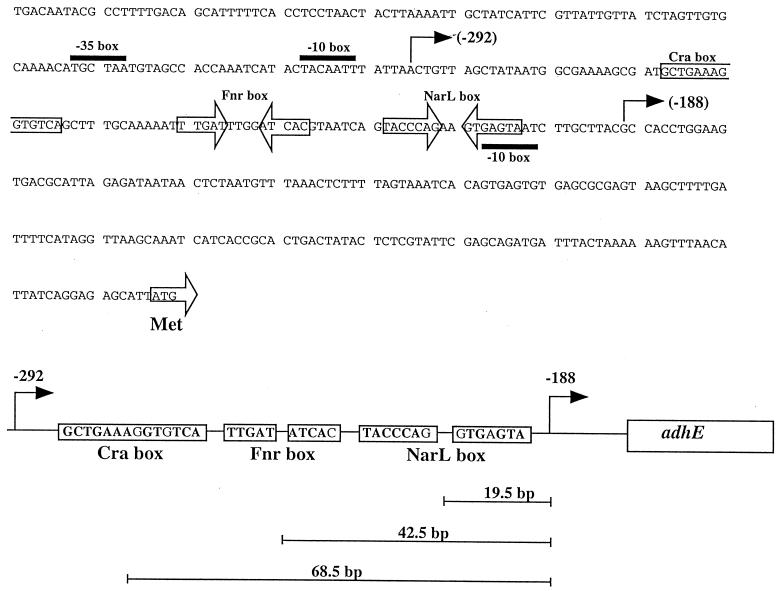
Base sequence and locations of regulatory sites in the adhE promoter region. The distance between putative regulatory sequences and the −188 transcriptional start site is indicated at the bottom.
In addition to transcriptional controls, translation of the adhE mRNA depends on the activity of RNase III. It was suggested that intramolecular base pairing occludes the RBS (ribosomal binding site) and that cleavage of the obstructing secondary structure liberates the translation. The results of primer extension experiments indicated two putative adhE transcriptional start sites: one at position −292 and the other at position −188 from the translational start codon ATG (1). However, the mRNA starting at position −188 was not seriously considered for three reasons: (i) the primer extension might be prematurely terminated by a secondary structure of the full-length adhE mRNA, (ii) the short transcript starting at position −188 might be a breakdown product, and (iii) transcription from this site was not likely to require RNase III cleavage as predicted by the RBS occlusion model. The focus of the present study is to address the functionality of the downstream start site.
MATERIALS AND METHODS
Strains, culture conditions, and reagents.
The relevant characteristics and sources of the bacterial strains and plasmids used in this study are given in Table 1. Luria-Bertani (LB) medium (20) containing 0.1 M MOPS (morpholinepropanesulfonic acid) and 0.2% glucose was adjusted to pH 7.4 (LB-glucose medium). Minimal medium was prepared as previously described (19). Culture optical density at 600 nm (OD600) was determined in a DU640 Beckman spectrophotometer. Aerobic cultures were grown at 37°C with shaking (200 rpm) in 250-ml Erlenmeyer flasks containing 10 ml of medium. Anaerobic cultures were grown at 37°C in 10-ml test tubes filled to the brim. When used, nitrate was added at 40 mM. The BBL (Cockeysville, Md.) Gas-Pack system was used for anaerobic growth on solid media. Antibiotics, purchased from Sigma, were added when appropriate at the following concentrations: ampicillin, 200 μg/ml; tetracycline, 15 μg/ml; and kanamycin, 100 μg/ml. Oligonucleotides were custom synthesized by Oligos Etc., Inc. All enzymes were purchased from New England Biolabs.
TABLE 1.
Strains, plasmids, and phages used in this study
| Strain, plasmid, or phage | Relevant genotype | Source or reference |
|---|---|---|
| Strains | ||
| ECL3999 | MC4100 but adhE::kan | 18 |
| ECL4010 | Same as MC4100 but λADHpr291 | This study |
| ECL4011 | Same as MC4100 but λADHop291 | This study |
| ECL4012 | Same as MC4100 but λADHpr656 | This study |
| ECL4013 | Same as MC4100 but λADHop656 | This study |
| ECL4014 | Same as MC4100 but λADHop2656 | This study |
| ECL4053 | Same as MC4100 but λADHop3656 | This study |
| ECL4015 | Same as ECL4011 but rnc::Tn10 | P1(RS6521) × ECL4011 |
| ECL4016 | Same as ECL4012 but rnc::Tn10 | P1(RS6521) × ECL4012 |
| ECL4017 | Same as ECL4013 but rnc::Tn10 | P1(RS6521) × ECL4013 |
| ECL4018 | Same as ECL4010 but rnc::Tn10 | P1(RS6521) × ECL4010 |
| ECL4020 | Same as ECL4011 but fnr-271::Tn10 | P1(VJS1741) × ECL4011 |
| ECL4022 | Same as ECL4013 but fnr-271::Tn10 | P1(VJS1741) × ECL4013 |
| ECL4024 | Same as ECL4011 but cra::Tn10 | P1(LJ2805) × ECL4011 |
| ECL4027 | Same as ECL4011 but fis::kan | P1(RJ708) × ECL4011 |
| ECL4028 | Same as ECL4013 but fis::kan | P1(RJ708) × ECL4013 |
| ECL4031 | Same as ECL4011 but appY::aphA | P1(TC3572) × ECL4011 |
| ECL4032 | Same as ECL4013 but appY::aphA | P1(TC3572) × ECL4013 |
| ECL4033 | Same as ECL4011 but narL::Tn10 | P1(RK5267) × ECL4011 |
| ECL4034 | Same as ECL4013 but narL::Tn10 | P1(RK5267) × ECL4013 |
| ECL4055 | Same as ECL4011 but rpoS::kan | P1(ECL1226) × ECL4011 |
| ECL4056 | Same as ECL4011 but rpoS::kan fnr-271::Tn10 | P1(ECL1226) × ECL4020 |
| ECL4054 | Same as MC4100 but λADHop656TATA | This study |
| LJ2805 | cra::Tn10 | 6 |
| MC4100 | F′ ΔlacU169 rpsL150 | 5 |
| RJ708 | fis::kan | Reid Johnson |
| RK5267 | narL::Tn10 | Valley Stewart |
| RS6521 | rnc::Tn10 | R. W. Simons |
| TC3572 | appY::aphA | 3 |
| VJS1741 | fnr-271::Tn10 | Valley Stewart |
| ECL1226 | rpoS::kan | Laboratory collection |
| Plasmids | ||
| pADH8 | ApradhE+ | 5 |
| pADHop2656 | pRS415/ADHop2656a | This study |
| pADHop291 | pRS415/ADHop291a | This study |
| pADHop656 | pRS415/ADHop656a | This study |
| pADHpr291 | pRS414/ADHpr291a | This study |
| pADHpr656 | pRS414/ADHpr656a | This study |
| pRS414 | AprlacZYA+ (protein fusion) | 25 |
| pRS415 | AprlacZYA+ (operon fusion) | 25 |
| pJAMDH1 | pBR322/adhE+ up to −656 | This study |
| pJMADH2 | pBR322/adhE+ up to −291 | This study |
| pJMADH3 | pBR322/adhE+ up to −656 but Pribnow TACAAT box (from positions −304 to −298) changed to CGGGCC | This study |
| Phages | ||
| λADHop291 | (adhE-lacZ)b | This study |
| λADHop656 | (adhE-lacZ)b | This study |
| λADHpr291 | (adhE-lacZ)b | This study |
| λADHpr656 | (adhE-lacZ)b | This study |
| λADHop2656 | (adhE-lacZ)b | This study |
| λADHop3656 | (adhE-lacZ)b | This study |
Genetic methods and DNA manipulations.
Genetic crosses were performed by P1vir-mediated transduction (20). Standard methods were used for restriction endonuclease digestion and ligation of DNA (20, 23). Plasmid DNA was isolated by using the QIAPrep system from Qiagen, and the DNA fragments were isolated from agarose gels with the QIAquick kit (Qiagen). Transformation of bacteria with plasmid DNA was done by electroporation (23) with an E. coli Pulser (Bio-Rad). PCRs were carried out in a Minicycler (MJ Research), using Pfu DNA polymerase from Stratagene (La Jolla, Calif.).
Construction of adhE operon and protein fusions.
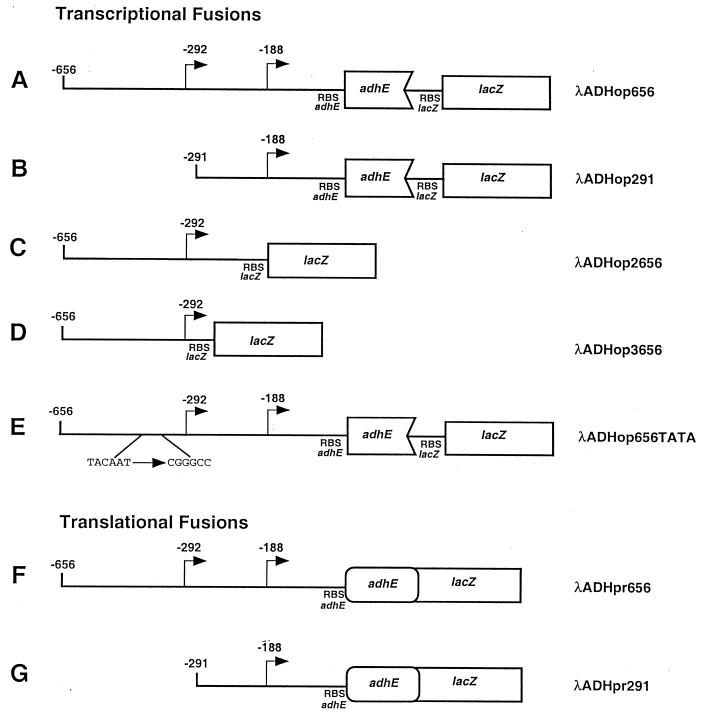
Different protein and operon fusions of adhE to lacZ, Φ(adhE-lacZ), were constructed on a plasmid and then transferred to λ phage by recombination in vivo (25). To construct λADHop656 (operon fusion) and λADHpr656 (protein fusion), a 1.1-kb DNA fragment was excised from pADH8 by BglII and BstYI. The product was ligated into the BamHI site of the plasmid pRS415 (for an operon fusion) and pRS414 (for a protein fusion). The recombinant plasmids were used to transform strain MC4100 (Δlac). Each of these fusions comprises 656 bp upstream of the translational start site of adhE. To construct λADHop291 (operon fusion) and λADHpr291 (protein fusion), primers 291-5 (5′-CAATGAATTCACTGTTAGCTATAATGGCG-3′) and 291-3 (5′-GCCGGATCCAGATCTTTCGGAGC-3′) were used to amplify a 0.8-kb DNA fragment comprising 291 bp upstream of the adhE translational start site. The fragment was gel purified, digested with EcoRI and BamHI, and cloned into pRS415 (operon fusion) and pRS414 (protein fusion). To construct λADHop2656 (operon fusion), primers 2656-5 (5′-AGCGAGATCCACAAGATAATGGCC-3′) and 2656-3 (5′-AGCTGGATCCGTAAGCAAGATTACTCACTTCTGGG-3′) were used to amplify a 0.3-kb DNA fragment comprising a segment from position −656 to position −190 from the adhE translational start site. To construct λADHop3656 (operon fusion), primers 3656-5 (5′-AGCGGATCCACAAGATAATGGCC-3′) and 3656-3 (5′-CGCTGGATCCCATTATAGCTAACAGTTAATAAATTGTAGTATG-3′) were used to amplify a 0.4-kb DNA fragment comprising a segment from position −656 to position −267 from the adhE translational start site. For λADH656TATA, primers 3656-5 and 291-3 were used, but the plasmid pJMADH3 (see below) was used as a template. The fragments were gel purified, digested with BamHI and EcoRI, and cloned into pRS415. The sequence of the inserts was confirmed by automated sequencing (Core Facility at Harvard Medical School). The appropriate fusions in the plasmids were recombined onto λRS45 to yield their correspondent λADH (Fig. 2). Each fusion was inserted into the chromosome of MC4100, and several transductants were isolated for assays of the activity level of β-galactosidase and Ter tests (25). A single-copy lysogen bearing each fusion was isolated for further study.
FIG. 2.
Schematic representations of Φ(adhE-lacZ) operon and Φ(adhE-lacZ) protein fusions in λ cassettes (identified on right). The numbers indicate base pair positions from the translational start site. Transcriptional start sites are denoted by bent arrows.
β-Galactosidase assays.
Exponential- (OD600, 0.4) or stationary-phase (OD600, 2.5) LB cultures (10 ml) of the desired strains were treated with chloramphenicol (final concentration, 40 μg/ml) and incubated for an additional 5 min at 37°C. The cultures were then pelleted, suspended in 2.5 ml of Z buffer (20), and kept on ice. Specific β-galactosidase activity in cells permeabilized with chloroform and sodium dodecyl sulfate was assayed at 28°C and is expressed in Miller units (ΔOD420 per minute per OD600 unit) (20). Each culture was assayed at least in triplicate; typically, these values gave a coefficient of variation (mean divided by the standard deviation) of <5%. At least three independent experiments were carried out under each set of growth conditions.
Ethanol oxidoreductase assays.
Cells were disrupted by sonication, and the extracts were assayed at 25°C, essentially as previously described (17). The assay mixture (1 ml) consisted of 1.6 M ethanol, 0.3 M potassium carbonate buffer (pH 10), and 0.66 mM NAD.
Primer extension.
Total RNA was isolated with the RNAeasy kit (Qiagen) from cells of strain ECL4010 growing anaerobically. Ten nanograms of 5′-end-labeled (23) oligonucleotide JM-1 (5′-CGCCAGGGTTTTCCCAGTCACGACG-3′) corresponding to positions +46 to +28 relative to the ATG of the lacZ gene was mixed with 1 μg of total mRNA in 10 μl of the primer extension buffer (50 mM Tris-HCl [pH 8.3], 50 mM KCL, 10 mM MgCl2, 10 mM dithiothreitol, 1 mM [each] deoxynucleoside triphosphate, 0.5 mM spermidine), heated at 75°C for 3 min, and cooled to room temperature for 10 min. Ten microliters of a prewarmed reverse transcriptase extension mixture (1 U of avian myeloblastosis virus reverse transcriptase [New England Biolabs] in primer extension buffer containing 5.6 mM sodium pyrophosphate) was added to the annealed primer-RNA complex, incubated at 42°C for 30 min, and ethanol precipitated. Nucleic acids were resuspended in formamide loading dye and separated on a 6% denaturing polyacrylamide gel. The size of the primer-extended product was calculated by running a known sequence ladder (M13mp18 DNA).
Construction of adhE plasmids and site-directed mutagenesis.
Plasmids pBR322 bearing adhE alleles were constructed by inserting a PCR fragment containing the entire adhE coding region and the desired length of the regulatory region. The insert of plasmid pJMADH1 was made by using primer 3656-5 (5′-AGCGGGATCCACAAGATAATGGCC-3′) and primer JMADH13 (5′- AGCTGGATTCATTGCCCAGAAGGGGCCGTTTATGTTGCCAGACAG CGC-3′). The insert for plasmid pJMADH2 was made by using primer 291-5Bam (5′-CAATGGATCCACTGTTAGCTATAATGGCG-3′) and primer JMADH13. The inserts were gel purified with the QIAquick kit and cloned into pBR322 predigested with BamHI. Site-directed mutagenesis of the Pribnow TACAAT box was carried out with the QuickChange kit (Stratagene) with plasmid pJMADH1 as a template. The primers TATA5 (5′-GTAGCCACCAAATCATACCGGGCCTTATTAACTGTTAGC-3′) and TATA3′ (5′-GCTAACAGTTAATAAGGCCCGGTATGATTTGGTGGCTAC-3′) were used for site-directed mutagenesis. The new plasmid was designated pJMADH3. Confirmation of the sequences of all the inserts was performed by automated DNA sequencing (Core Facility at Harvard Medical School).
RESULTS
Expression and redox regulation of Φ(adhE-lacZ)291 operon and protein fusions bearing only the −188 transcriptional start site.
To test whether the downstream transcriptional start site was functional, we constructed two pairs of adhE-lacZ fusions differing in the lengths of their promoter regions: (i) a Φ(adhE-lacZ)656 operon fusion and a Φ(adhE′-′lacZ)656 protein fusion extending to position −656 from the translational start site (Fig. 2A and F) and (ii) a Φ(adhE-lacZ)291 operon fusion and a Φ(adhE′-′lacZ)291 protein fusion extending to position −291 from the translational start site (Fig. 2B and G). The pair of long fusions contained both the −292 and −188 transcriptional start sites, whereas the pair of short fusions contained only the −188 transcriptional start site. Each of the four fusions was then inserted into the att site of strain MC4100 (adhE+ Δlac) to give strains ECL4013 [Φ(adhE-lacZ)656], ECL4012 [Φ(adhE′-′lacZ)656], ECL4011 [Φ(adhE-lacZ)291], and ECL4010 [Φ(adhE′-′lacZ)291]. The adhE+ Φ(adhE-lacZ) merodiploid reporter strains were grown aerobically or anaerobically in LB-glucose, and their β-galactosidase activity levels were determined.
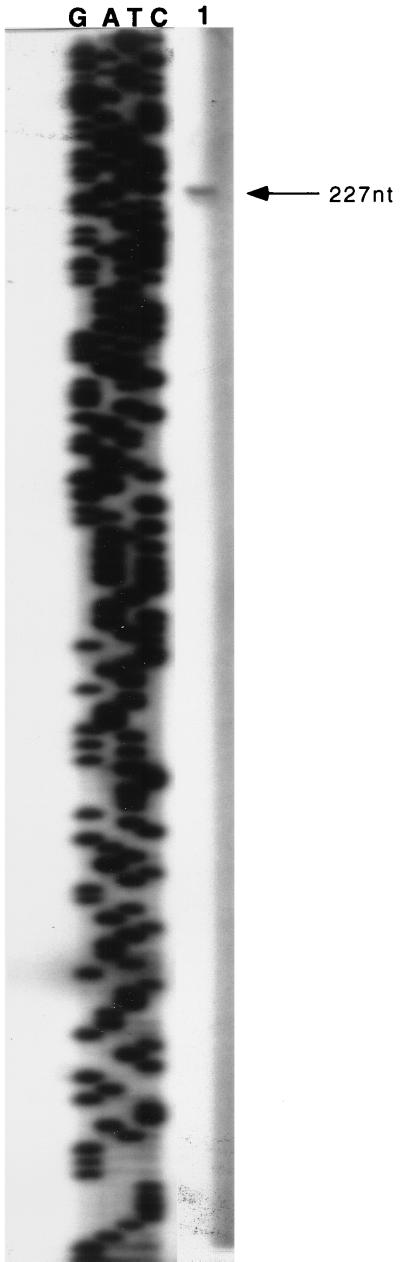
Strain ECL4011 [Φ(adhE-lacZ)291] showed a ninefold-higher β-galactosidase activity level during anaerobic growth than during aerobic growth. These activity levels represent 30% of the respective values found in strain ECL4013 [Φ(adhE-lacZ)656] (Table 2). A similar pattern of activity levels was observed when the strain bearing the short protein fusion was compared with that bearing the long protein fusion (data not shown). Primer extension analysis in the strain ECL4010 [Φ(adhE′-′lacZ)291] showed that, as expected, the transcriptional start site was at position −188 (Fig. 3). The results taken together suggest that the downstream transcriptional start site (−188) can be functional by itself and that the promoter region in this fragment contains the necessary structure for response to redox regulation.
TABLE 2.
β-Galactosidase activities of exponentially growing cells (OD600, 0.4) bearing different Φ(adhE-lacZ) operon fusions in LB-glucose medium
| Strain | Genotype | β-Galactosidase activity (Miller units)a
|
||
|---|---|---|---|---|
| +O2 | −O2 | −O2/+O2 ratio | ||
| ECL4013 | Φ(adhE-lacZ)656 | 1,000 ± 100 | 9,200 ± 530 | 9.3 |
| ECL4028 | Φ(adhE-lacZ)656 fis::kan | 910 ± 60 | 890 ± 100 | 0.9 |
| ECL4022 | Φ(adhE-lacZ)656 fnr::Tn10 | 958 ± 46 | 9,880 ± 240 | 10.3 |
| ECL4011 | Φ(adhE-lacZ)291 | 300 ± 50 | 3,600 ± 250 | 9.0 |
| ECL4027 | Φ(adhE-lacZ)291 fis::kan | 380 ± 40 | 3,000 ± 350 | 8.0 |
| ECL4020 | Φ(adhE-lacZ)291 fnr::Tn10 | 330 ± 45 | 345 ± 32 | 1.1 |
+O2, aerobic; −O2, anaerobic.
FIG. 3.
Primer extension analysis of the Φ(adhE′-′lacZ)291 protein fusion. Primer JM-1, complementary to positions +46 to +28 of the lacZ gene, was annealed with total RNA of strain ECL4010 and extended with avian myeloblastosis virus reverse transcriptase. A known DNA ladder (lanes G, A, T, and C) was used to calculate the length of the transcript. The only transcript observed (at 227 nucleotides [nt]) corresponds to the −188 transcriptional start site of the adhE gene (lane 1).
Effect of Fnr on the anaerobic expression of Φ(adhE-lacZ)291 operon and protein fusions.
A putative Fnr site has been previously noted (11, 13), but in retrospect its location at position −769 seems too far from the putative adhE transcriptional start sites. Moreover, in two previous independent studies, no effect of an fnr null mutation on Φ(adhE-lacZ) and adhE+ expressions was observed in the merodiploids (5, 15). However, we noticed that, centered at −42.5 bp from the −188 transcriptional start site, there is a 9/10 Fnr consensus sequence (TTGAT-N4-ATCAA) (Fig. 1). The location of this box is typical of that found in target promoters positively regulated by Fnr (27).
To test whether Fnr is involved in the expression of the Φ(adhE-lacZ)291 fusion, we transduced an fnr::Tn10 allele into strain ECL4011 [Φ(adhE-lacZ)291] to yield strain ECL4020. When the parent and the transductant were grown aerobically on LB-glucose medium, both strains showed low β-galactosidase activity levels. By contrast, when grown anaerobically, strain ECL4011 (fnr+), but not strain ECL4020 (fnr::Tn10), became induced for β-galactosidase (Table 2). However, consistent with the results of previous studies (5, 15), Fnr did not activate the anaerobic expression of the long fusion in strain ECL4013 [Φ(adhE-lacZ)656 fnr+] and its isogenic derivative ECL4022 [Φ(adhE-lacZ)656 fnr::Tn10] (Table 2). Similar results were obtained when the protein fusions were used (data not shown). We also determined the AdhE activity levels in cell extracts of these two strains grown under aerobic or anaerobic conditions and found the results concordant with the β-galactosidase activity levels (data not shown). Thus, it seems that Fnr can regulate adhE expression from the downstream transcriptional start site only in the physical absence of the upstream region.
Effect of Cra on the anaerobic expression of Φ(adhE-lacZ)291 operon and protein fusions.
From positions −265 to −251 lies the previously identified Cra box (Fig. 1) (19). To determine whether the Φ(adhE-lacZ)291 fusion can be regulated, the cra::kan allele was transduced from strain LJ2805 into strain ECL4011, yielding strain ECL4024 [Φ(adhE-lacZ)291 cra::kan]. Strains ECL4011 and ECL4024 were then grown aerobically or anaerobically in LB-glucose medium, and their β-galactosidase activity levels were compared. No significant effect of Cra was found. A set of experiments with the protein fusions also showed no Cra effect (data not shown).
Effect of NarL on the anaerobic expression of Φ(adhE-lacZ)291 operon and protein fusions.
The NarL box comprises two heptamer inverted repeats (TACYYMT, where Y is C or T and M is A or C) separated by 2 bases (8). Sequence scanning revealed a putative site, TACCCAG-N2-GTGAGTA, located between positions −199 and −213 from the translational start site, differing from the consensus in only 1 base in each heptamer (Fig. 1). In agreement with previous reports (5, 15), the expression of Φ(adhE-lacZ)656 was strongly repressed both aerobically and anaerobically by nitrate in strain ECL4013 (narL+) but not in its isogenic derivative ECL4034 (narL::Tn10) (data not shown). Similarly, the expression of Φ(adhE-lacZ)291 was nitrate repressible both aerobically and anaerobically in strain ECL4011 but not in strain ECL4033 (narL::Tn10) (Table 3). The results were supported by the responses of the two corresponding protein fusions (data not shown).
TABLE 3.
Aerobic and anaerobic β-galactosidase activity of Φ(adhE-lacZ)291 in the presence of 40 mM nitrate and a narL mutationa
| Strain | Relevant genotype | β-Galactosidase activity (Miller units)
|
|||
|---|---|---|---|---|---|
| +O2 | −O2 | +NO3
|
|||
| +O2 | −O2 | ||||
| ECL4011 | Φ(adhE-lacZ)291 | 312 ± 15 | 2,815 ± 49 | 129 ± 10 | 151 ± 29 |
| ECL4033 | Φ(adhE-lacZ)291 narL::Tn10 | 326 ± 23 | 3,153 ± 18 | 188 ± 25 | 2,993 ± 23 |
The cells were grown in LB-glucose medium and assayed in the exponential phase of growth (OD600, 0.4).
Effect of Fis on the anaerobic expression of Φ(adhE-lacZ)291 operon and protein fusions.
To test for a role of Fis, strain ECL4011 [Φ(adhE-lacZ)291] and the isogenic strain ECL4027 [Φ(adhE-lacZ)291 fis::kan] were grown aerobically or anaerobically in LB-glucose medium and assayed for β-galactosidase activity levels. As shown in Table 2, the lack of Fis did not affect the ninefold anaerobic increase in expression of Φ(adhE-lacZ)291. As expected from our previous finding (18), a null mutation in fis greatly diminished the anaerobic expression of the Φ(adhE-lacZ)656 with the full-length promoter region. Concordant results were obtained from the corresponding protein fusions (data not shown). Thus, Fis is required only for transcriptional initiation from the upstream start site at −292.
Effect of RpoS on the expression of Φ(adhE-lacZ)291 operon fusion.
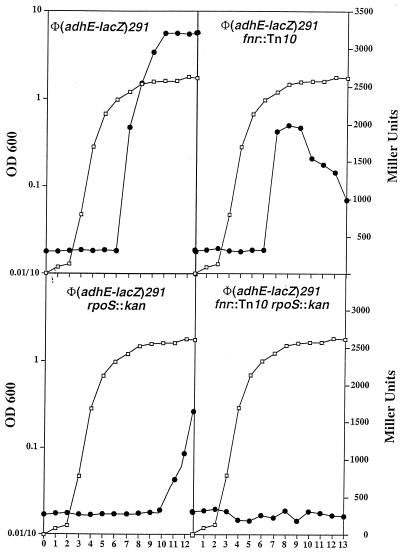
During the course of this study, we were struck by an observation that the level of β-galactosidase activity of an aerobic culture of strain ECL4011 [Φ(adhE-lacZ)291] was severalfold higher in stationary- than in exponential-phase cells. Although this phenomenon could be explained by oxygen limitation in the culture, a growth phase regulation could not be excluded. We therefore monitored the β-galactosidase activity levels in strain ECL4011 [Φ(adhE-lacZ)291] and its fnr::Tn10 and/or rpoS::kan derivatives during the growth cycle. We found that, in the wild-type background, the increase in activity level occurred abruptly at the onset of stationary phase (Fig. 4, top left panel). In the fnr::Tn10 rpoS+ background, there was a sixfold increase in activity at the onset of stationary phase (OD600, 2.0), but the levels reproducibly dropped 50% afterwards, possibly resulting from specific protein degradation in the absence of Fnr-promoted synthesis (Fig. 4, top right panel). By contrast, in the fnr+ rpoS::kan background, there was no increase in the activity level until the culture was 5 h into the stationary phase, eventually reaching five times that found in the exponential-phase cells (Fig. 4, bottom left panel). It appears that the relatively early increase in the enzyme activity level in the fnr::Tn10 rpoS+ cells reflected RpoS transcriptional activation following the onset of stationary phase, whereas the relatively late increase in the enzyme activity level in the fnr+ rpoS::kan cells reflected Fnr transcriptional activation in gradual response to anoxia. This interpretation is consistent with the finding of a low level of β-galactosidase activity throughout the growth cycle in the fnr::Tn10 rpoS::kan double mutant (Fig. 4, bottom right panel).
FIG. 4.
Effect of mutations in rpoS and/or fnr on the β-galactosidase activity of the Φ(adhE-lacZ)291 operon fusion throughout the growth cycle. The open squares represent the OD600, and the solid circles represent β-galactosidase activity in Miller units.
Since the RpoS effect was unexpected, we tested whether a similar influence was exerted by the regulator on the full-length promoter. We found that strain ECL4013 [Φ(adhE-lacZ)656] also exhibited a 10-fold increase in the β-galactosidase activity level during stationary phase. This induction was lowered by 30 to 40% when an rpoS::kan allele was introduced (data not shown).
β-Galactosidase synthesis specified by the Φ(adhE-lacZ)291 protein fusion is exempt from RNase III requirement.
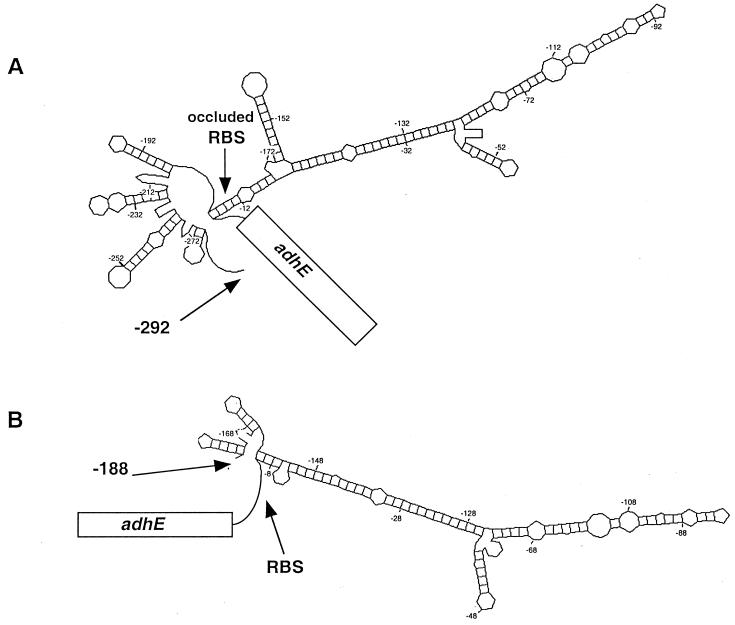
The availability of the fusion without the −292 transcriptional start offered an opportunity to test genetically the proposed model of RBS occlusion by the secondary structure of the 5′ region (1). Since the abbreviated mRNA is no longer expected to form the elaborate 5′ stem-loop complex (Fig. 5), translation of the open reading frame may no longer require the intervention of RNase III. As shown in Table 4, the β-galactosidase activity level specified by the Φ(adhE-lacZ)291 protein fusion is independent of RNase III (compare strain ECL4010 with strain ECL4018). In further support of the RBS occlusion model, we found that when the lacZ open reading frame possessing its own RBS was placed sufficiently downstream of the adhE RBS, as in the case of the Φ(adhE-lacZ)656 operon fusion (Fig. 2A), the synthesis of β-galactosidase also became RNase III independent (compare strain ECL4013 with strain ECL4017). As expected, the β-galactosidase activity level specified by the Φ(adhE-lacZ)656 protein fusion (in which the translation of lacZ depended on the adhE RBS [Fig. 2F]) remained RNase III dependent (compare strain ECL4012 with strain ECL4016).
FIG. 5.
Probable structures of the 5′ untranslated regions of two different adhE transcripts. The MFold program (29) was used to model the thermodynamically most favorable secondary structure of the 5′ segment of adhE transcript starting at position −292 (A) or −188 (B).
TABLE 4.
RNase III dependence of β-galactosidase activity specified by different Φ(adhE-lacZ) fusion constructsa
| Strain | Relevant genotype | β-Galactosidase activity (Miller units) |
|---|---|---|
| ECL4012 | Φ(adhE-lacZ)656 protein fusion | 9,000 ± 360 |
| ECL4016 | Φ(adhE-lacZ)656 protein fusion; rnc::Tn10 | 620 ± 60 |
| ECL4013 | Φ(adhE-lacZ)656 operon fusion | 9,900 ± 280 |
| ECL4017 | Φ(adhE-lacZ)656 operon fusion; rnc::Tn10 | 9,800 ± 150 |
| ECL4010 | Φ(adhE-lacZ)291 protein fusion | 3,400 ± 110 |
| ECL4018 | Φ(adhE-lacZ)291 protein fusion; rnc::Tn10 | 3,500 ± 150 |
The cells were grown anaerobically on LB-glucose medium and assayed in the exponential phase of growth (OD600, 0.4).
Qualitative evidence for the silencing of the downstream start site by the upstream start region.
One phenotype of the adhE::kan mutant strain (ECL3999) is its inability to grow anaerobically on glucose minimal medium (15, 16). To test the abilities of various adhE constructs to complement an adhE null mutation, we used three different pBR322-based plasmids: (i) pJMADH1, containing the adhE structural gene plus its regulatory region up to position −656 from the translational start site; (ii) pJMADH2, similar to pJMADH1 but lacking the region upstream of position −291; and (iii) pJMADH3, similar to pJMADH1 but with the putative Pribnow TACAAT box of the −292 site changed to CGGGCC. As shown in Table 5, the first and second, but not the third, plasmid restored the ability of strain ECL3999 (adhE::kan) to grow anaerobically on minimal glucose medium. It should be mentioned that pJMADH1 transformants appeared as visible colonies after 18 h of incubation but pJMADH2 transformants appeared only after 48 h. Transformants of pJMADH3, however, gave rise to two colonies (probably suppressor mutants) after 72 h of incubation. These results lend support to the notion that the upstream regulatory region silences transcription initiated at position −188, at least under the experimental conditions employed.
TABLE 5.
Complementation of strain ECL3999 (adhE::kan) with plasmids bearing different constructs of adhE
| Plasmid | Relevant characteristics | No. of colonies testeda
|
|
|---|---|---|---|
| LB Ap (aerobic) | MM glucose (anaerobic) | ||
| pBR322 | No insert | 500 | 0 |
| pJMADH1 | adhE+ regulatory region up to −656 | 500 | 496b |
| pJMADH2 | adhE+ regulatory region up to −291 | 500 | 491c |
| pJMADH3 | adhE+ regulatory region up to −656 but Pribnow TACAAT box (from positions −304 to −298) changed to CGGGCC | 500 | 2d |
LB Ap, LB medium plus ampicillin; MM glucose, glucose minimal medium.
Colonies were scored after 24 h of incubation at 37°C.
Colonies were scored after 48 h of incubation at 37°C.
Colonies were scored after 72 h of incubation at 37°C.
Quantitative evidence for the silencing of the downstream start site by the upstream start region.
Two approaches were designed to test the hypothesis that the upstream promoter region (up to position −656) acts as a silencer of downstream transcription. First, if the downstream promoter is ordinarily not contributing to the net transcription of adhE, then a Φ(adhE-lacZ) fusion containing the −292, but not the −188, transcriptional start site should express lacZ at an undiminished level. Accordingly, we constructed two operon fusions with abbreviated promoters: (i) Φ(adhE-lacZ)2656, comprising an adhE segment from bp −656 to −290 joined to a lacZ sequence that includes its own RBS, and (ii) Φ(adhE-lacZ)3656, comprising an adhE segment from bp −656 to −267 joined to a lacZ sequence that includes its own RBS (Fig. 2C and D, respectively). Strains bearing a single copy of either fusion exhibited a β-galactosidase activity level that was indistinguishable from that of the Φ(adhE-lacZ)656-bearing strain, when grown aerobically or anaerobically (data not shown).
Second, if the mere presence of the upstream region is silencing the downstream transcriptional start site, as indicated by the plasmid complementation studies, then even the presence of a nonfunctional upstream start site may prevent transcription from the −188 site. Accordingly, we constructed strain ECL4054, bearing a single copy of the Φ(adhE-lacZ)656TATA operon fusion in which the upstream putative Pribnow box (TACAAT from bp −304 to −298 [Fig. 2E]) was changed to CGGGCC. When grown aerobically, this strain showed a β-galactosidase activity level 60% lower than that of the strain bearing the Φ(adhE-lacZ)656 operon fusion with the wild-type promoter sequence. When grown anaerobically, strain ECL4054 [Φ(adhE-lacZ)656TATA] did not show an increase in β-galactosidase activity (data not shown), confirming that even the presence of a nonfunctional upstream promoter region prevents transcription from position −188.
DISCUSSION
The emergence of AdhE from a fusion of an alcohol oxidoreductase with an aldehyde oxidoreductase (13) might be an important turning point in the evolution of ethanol fermentation by E. coli. The fused protein not only would facilitate an intramolecular interconversion of acetyl-CoA to ethanol but also would provide additional domain surfaces to acquire the deactivase activity for pyruvate formate-lyase (14). There may well be other acquired biochemical functions yet to be discovered. For instance, we do not yet know the biological significance of spirosome structures arising from the AdhE molecules (13, 14).
The acquisition of multiple functions by the AdhE protein can be expected to have advanced hand in hand with the elaboration of increasingly complex control of gene expression. The embellishment and enlargement of the promoter, associated with the recruitment of different regulatory proteins, should be a part of this process. The direct roles of Cra (19) and Fis (18, 28) have been established, although the functional significance of Fis control remains a puzzle. The physiological role of Fnr is still in question, and the nature and significance of the RpoS effect on adhE expression require exploration.
In the meantime, the regulatory element responsible for redox control continues to elude identification. Only a few genes, including adhE, are expressed at significantly increased levels anaerobically without intervention by Fnr (11). In some cases, the increased expression is dependent on DNA supercoiling. Examples include tppB, torA, pepN, and tonB (12). In other cases, such as hya and cyx (2–4), the increased expression is dependent on the anaerobic transcriptional factor AppY. We found that the anaerobic expression of adhE is independent of AppY but is influenced by DNA topology (data not shown).
At the posttranslational level, the AdhE protein is rapidly and irreversibly inactivated during aerobic metabolism. The Fe2+ bound to the alcohol oxidoreductase domain of the protein is responsible for this inactivation by catalyzing the oxidative destruction of certain amino acid residues via the Fenton reaction (26). Enzyme inactivation upon the shift from anaerobic to aerobic metabolism accelerates the diversion of the inefficient fermentative mode of energy generation by covalent dismutation to the highly efficient mode of energy generation by respiration.
The key enigma, highlighted by this study, is the presence of two transcriptional start sites in the adhE promoter. The actual existence of the transcript starting at position −188 in cell extracts (1) and the ability of Fnr to activate the anaerobic expression of the 5′-truncated Φ(adhE-lacZ)291 fusion suggest that under certain conditions the silencer effect of the upstream promoter region can be lifted to allow significant transcription at the −188 site. How can such conditions be discovered? A possible approach would be to characterize the suppressor mutation(s) that allowed the anaerobic expression of adhE from the −188 start site (in plasmid pJMADH3 [Table 5]). A successful result might even reveal the teleonomic basis for two adhE transcriptional start sites and the significance of the RNase III requirement for the translation of the long transcript.
Alternatively, the −188 site could be a vestige in the adhE promoter that is evolving away from Fnr control. However, such a hypothesis is not only difficult to test but is also unattractive in view of the parsimonious use of DNA in prokaryotes, which should include the prompt deletion of superfluous DNA. Moreover, Fnr sites in adhE are found in the genomes of other bacterial species, including Actinobacillus pleuropneumoniae, Clostridium acetobutylicum, Lactococcus lactis, and Salmonella typhimurium.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants GM40993 and GM39693.
We thank Reid Johnson, Tove Atlung, Valley Stewart, and Mary Berlyn for strains used in this study.
REFERENCES
- 1.Aristarkhov A, Mikulskis A, Belasco J G, Lin E C C. Translation of the adhE transcript to produce ethanol dehydrogenase requires RNase III cleavage in Escherichia coli. J Bacteriol. 1996;178:4327–4332. doi: 10.1128/jb.178.14.4327-4332.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atlung T, Brøndsted L. Role of the transcriptional activator AppY in regulation of the cyx appA operon of Escherichia coli by anaerobiosis, phosphate starvation, and growth phase. J Bacteriol. 1994;176:5414–5422. doi: 10.1128/jb.176.17.5414-5422.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brøndsted L, Atlung T. Anaerobic regulation of the hydrogenase 1 (hya) operon of Escherichia coli. J Bacteriol. 1994;176:5423–5428. doi: 10.1128/jb.176.17.5423-5428.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brøndsted L, Atlung T. Effect of growth conditions on expression of the acid phosphatase (cyx-appA) operon and the appY gene, which encodes a transcriptional activator of Escherichia coli. J Bacteriol. 1996;178:1556–1564. doi: 10.1128/jb.178.6.1556-1564.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y M, Lin E C C. Regulation of the adhE gene, which encodes ethanol dehydrogenase in Escherichia coli. J Bacteriol. 1991;173:8009–8013. doi: 10.1128/jb.173.24.8009-8013.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crasnier-Mednansky M, Park M C, Studley W K, Saier M H., Jr Cra-mediated regulation of Escherichia coli adenylate cyclase. Microbiology. 1997;143:785–792. doi: 10.1099/00221287-143-3-785. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham P R, Clark D P. The use of suicide substrates to select mutants of Escherichia coli lacking enzymes of alcohol fermentation. Mol Gen Genet. 1986;205:487–493. doi: 10.1007/BF00338087. [DOI] [PubMed] [Google Scholar]
- 8.Darwin A J, Stewart V. The NAR modulon systems: nitrate and nitrite regulation of anaerobic gene expression. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. R. G. Austin, Tex: Landes Company; 1996. pp. 343–359. [Google Scholar]
- 9.Finkel S E, Johnson R C. The Fis protein: it's not just for DNA inversion anymore. Mol Microbiol. 1992;6:3257–3265. doi: 10.1111/j.1365-2958.1992.tb02193.x. [DOI] [PubMed] [Google Scholar]
- 10.Goodlove P E, Cunningham P R, Parker J, Clark D P. Cloning and sequence analysis of the fermentative alcohol dehydrogenase-encoding gene of Escherichia coli. Gene. 1989;85:209–214. doi: 10.1016/0378-1119(89)90483-6. [DOI] [PubMed] [Google Scholar]
- 11.Guest J G, Green J, Irvine A S, Spiro S. The FNR modulon and the FNR-regulated gene expression. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. R. G. Austin, Tex: Landes Company; 1996. pp. 343–359. [Google Scholar]
- 12.Jamieson D J, Higgins C F. Two genetically distinct pathways for transcriptional regulation of anaerobic gene expression in Salmonella typhimurium. J Bacteriol. 1986;168:389–397. doi: 10.1128/jb.168.1.389-397.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kessler D, Leibrecht I, Knappe J. Pyruvate formate-lyase deactivase and acetyl-CoA reductase activities of Escherichia coli reside on a polymeric protein particle encoded by adhE. FEBS Lett. 1991;281:59–63. doi: 10.1016/0014-5793(91)80358-a. [DOI] [PubMed] [Google Scholar]
- 14.Kessler D, Herth W, Knappe J. Ultrastructure and pyruvate formate-lyase radical quenching property of the multienzyme AdhE protein of Escherichia coli. J Biol Chem. 1992;267:18073–18079. [PubMed] [Google Scholar]
- 15.Leonardo M R, Cunningham P R, Clark D P. Anaerobic regulation of the adhE gene encoding the fermentative alcohol dehydrogenase of Escherichia coli. J Bacteriol. 1993;175:870–878. doi: 10.1128/jb.175.3.870-878.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lorowitz W, Clark D P. Escherichia coli mutants with a temperature-sensitive alcohol dehydrogenase. J Bacteriol. 1982;152:935–938. doi: 10.1128/jb.152.2.935-938.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McPhedran P, Sommer B, Lin E C C. Control of ethanol dehydrogenase levels in Aerobacter aerogenes. J Bacteriol. 1961;81:852–857. doi: 10.1128/jb.81.6.852-857.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Membrillo-Hernández J, Kwon O, De Wulf P, Finkel S E, Lin E C C. Regulation of adhE (encoding ethanol oxidoreductase) by the Fis protein in Escherichia coli. J Bacteriol. 1999;181:7390–7393. doi: 10.1128/jb.181.23.7390-7393.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mikulskis A, Aristarkhov A, Lin E C C. Regulation of expression of the ethanol dehydrogenase gene (adhE) in Escherichia coli by the catabolite repressor activator protein Cra. J Bacteriol. 1997;179:7129–7134. doi: 10.1128/jb.179.22.7129-7134.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 21.Rudolph F D, Purich D L, Fromm H J. Coenzyme A-linked aldehyde dehydrogenase from Escherichia coli. I. Purification, properties and kinetic studies of the enzyme. J Biol Chem. 1968;243:5536–5545. [PubMed] [Google Scholar]
- 22.Saier M H, Jr, Ramseier T M. The catabolite repressor/activator (Cra) protein of enteric bacteria. J Bacteriol. 1996;178:3411–3417. doi: 10.1128/jb.178.12.3411-3417.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Schmitt B. Aldehyde dehydrogenase activity of a complex particle from Escherichia coli. Biochimie. 1975;57:1001–1004. doi: 10.1016/s0300-9084(75)80355-5. [DOI] [PubMed] [Google Scholar]
- 25.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 26.Tamarit J, Cabiscol E, Ros J. Identification of the major oxidatively damaged proteins in Escherichia coli cells exposed to oxidative stress. J Biol Chem. 1998;273:3027–3032. doi: 10.1074/jbc.273.5.3027. [DOI] [PubMed] [Google Scholar]
- 27.Wing H J, Williams S M, Busby S J. Spacing requirements for transcription activation by Escherichia coli Fnr protein. J Bacteriol. 1995;177:6704–6710. doi: 10.1128/jb.177.23.6704-6710.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu J, Johnson R C. Identification of genes negatively regulated by Fis: Fis and RpoS comodulate growth-phase-dependent gene expression in Escherichia coli. J Bacteriol. 1995;177:938–947. doi: 10.1128/jb.177.4.938-947.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zucker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]