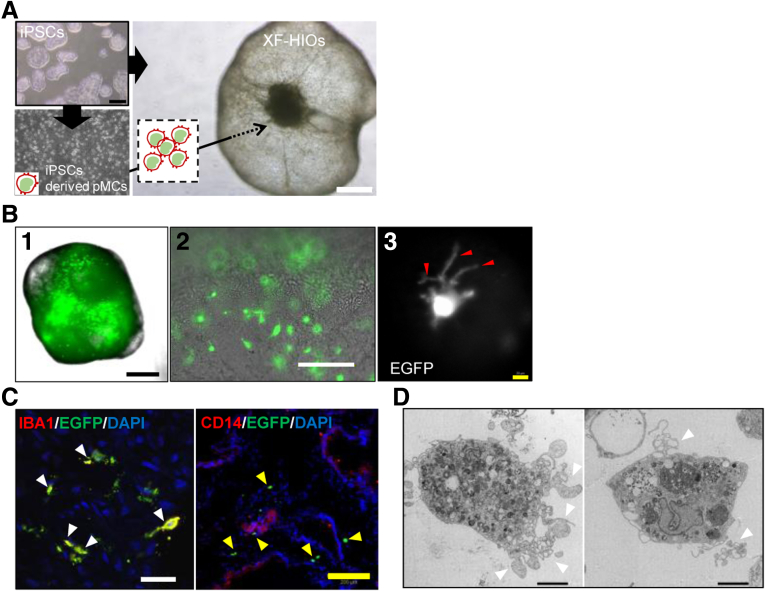
Intestinal macrophages are largely responsible for the innate immune response and also for intestinal homeostasis.1 We have developed novel xenogeneic-free human intestinal organoids (XF-HIOs) that are uniquely structured with an apical-out mucosal epithelium and complex mesenchymal tissue, including smooth muscle and intestinal nerve cells.2,3 To further develop XF-HIOs containing tissue macrophages, we first prepared human-induced pluripotent stem cell (hiPSC)-derived monocyte-like cells (pMCs). These were directly injected into the cystic cavity of an XF-HIO, followed by differentiation into macrophage-like cells (pGMACs) within XF-HIOs in the presence of macrophage colony-stimulating factor (M-CSF) (Figure 1A). We prepared macrophages/monocytes derived from an enhanced green fluorescent protein (EGFP)–hiPSC line, which constitutively expressed EGFP (Supplementary Figure 1A). The pGMACs were observed to evenly disperse inside the XF-HIOs, and image analysis showed pGMACs with short-elongated projections (Figure 1B). Immunofluorescence staining also revealed that ionized calcium-binding adapter molecule 1 was detectable in the monocyte (MC)-XF-HIOs; however, CD14 was not (Figure 1C). This staining pattern is similar to that observed in human intestinal macrophages.4,5 We also identified that ionized calcium-binding adapter molecule 1 and C-X3-C motif chemokine receptor 1 were co-localized in MC-XF-HIOs (Supplementary Figure 1B and C).
Figure 1.
Establishment of hiPSC-derived gut organoid residing macrophages. (A) Human iPSC-derived monocyte-like cells (pMCs) were transplanted into human intestinal organoids (XF-HIOs) and then treated with macrophage colony-stimulating factor to differentiate into monocyte-XF-HIOs (MC-XF-HIOs). Scale bars: 500 μm. (B) Macrophages from enhanced green fluorescent protein labeled human-induced pluripotent stem cells (EGFP–hiPSCs) were dispersed in the organoids (1, 2), and image analysis revealed short-elongated projections (red arrowheads) of gut macrophages (pGMACs) in MC-XF-HIOs. (3). Scale bars: black, 500 μm; white, 100 μm; yellow, 20 μm. (C) Immunostaining for macrophage-specific marker IBA1 merged with EGFP-hiPSCs (white arrowheads) but not CD14 (yellow arrowheads). Scale bars: white, 30 μm; yellow, 200 μm. (D) Representative views of pGMACs by transmission electron microscopy showed a characteristic large nucleus, phagocytic vacuoles, and short pseudopodia (white arrowheads). Scale bar: 5 μm.
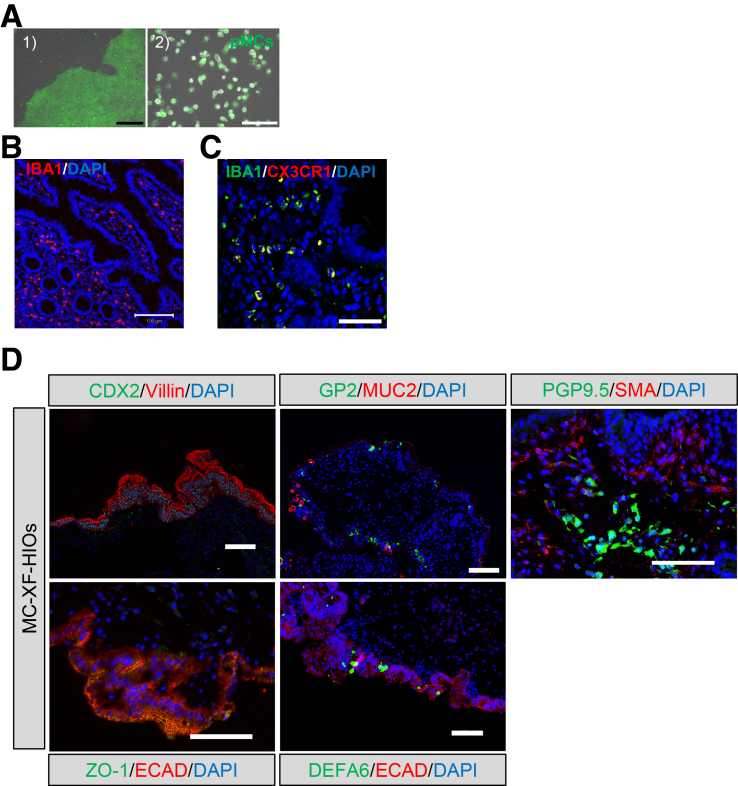
Supplementary Figure 1.
Characterization of hiPSC-derived macrophage-integrated gut organoids. (A) Enhanced green fluorescent protein (EGFP)–human-induced pluripotent stem cells (hiPSCs), which constitutively expressed EGFP under a cytomegalovirus promoter, were cultured under feeder-free conditions in StemFlex medium (1) and differentiated into monocyte-like cells (pMCs) (2). Scale bars: black, 300 μm; white, 100 μm (2) (B) Immunofluorescence staining for ionized calcium-binding adapter molecule 1 (IBA1) in human intestine. Scale bar, 100 μm. (C) Immunofluorescence staining for IBA1 and CX3CR1 in MC-XF-HIOs derived from a non-EGFP hiPSC line (Edom-iPSCs). Scale bar: 50 μm. (D) Immunostaining for caudal type homeobox 2 (CDX2), villin, zonula occludens-1 (ZO-1), E-cadherin (ECAD), glycoprotein 2 (GP2), mucin 2 (MUC2), defensin alpha 6 (DEFA6), protein gene product 9.5 (PGP9.5), and smooth muscle actin (SMA). PGP9.5-positive enteric neuronal cells were surrounded by SMA-positive mesenteric tissue in MC-XF-HIOs. Cell nuclei were counterstained with 4′, 6-diamidino-2-phenylindole, dihydrochloride (DAPI). Scale bars, 100 μm. Anti-CDX2 (1:1000, ab76541; Abcam), anti-villin (1:50, sc-7672; Santa Cruz Biotechnology, Dallas, TX), anti-GP2 (1:1000, HPA016668; Sigma-Aldrich), anti-MUC2 (1:50, sc-7314; Santa Cruz Biotechnology), anti-PGP9.5 (1:10, ab8189; Abcam), anti-SMA (1:400, A2547; Sigma-Aldrich), anti-ECAD (1:50, 610181; BD Pharmingen, San Diego, CA), anti–ZO-1 (1:100, 40-2200; Invitrogen), and anti-DEFA6 (1:500, HPA019462; Sigma-Aldrich) were used as primary antibodies.
Furthermore, the presence of pGMACs under the epithelium of each organoid as indicated by zonula occludens-1 staining was also observed in a three-dimensional image (Supplementary Videos 1A and B). Transmission electron microscopy of a section of MC-XF-HIO showed a pGMAC displayed phagocytic vacuoles, a large nucleus, and several short pseudopodia (Figure 1D). The MC-XF-HIOs have an intestinal tissue structure composed of apical-out epithelial and mesenchymal cells with neuronal cells (Supplementary Figure 1D) and also showed peristaltic-like movements (Supplementary Video 1C), as previously demonstrated in XF-HIOs.2 By sectionalizing their supernatant and fluid contents, this enabled us to mimic human intestinal physiological conditions in vitro. To assess the abilities of MC-XF-HIOs to produce and secrete soluble cytokines and chemokines, we investigated the fluid content (FC) of the organoids using a bead-based Multiplex cytokine assay (Figure 2A). Consistent with the intestinal epithelial barrier (Supplementary Figure 1D), several differences were apparent in the amounts of soluble cytokines in the FC of MC-XF-HIOs (Supplementary Figure 2A). Quantitative reverse transcription polymerase chain reaction (PCR) analysis based on single-cell sorting of pGMACs in MC-XF-HIOs revealed the distinct expression of macrophage polarization markers such as TNF, NOS2, HLA-DB1, IL-6, KLF4, and VEGFA (Supplementary Figure 2B). MC-XF-HIOs expressed pleiotropic types of cytokines. In addition, lipopolysaccharide (LPS) was used as a potential inflammatory stimulus.6 However, the expression of inflammatory cytokines, except for interleukin 4, did not exhibit a statistically significant change after exposure to LPS (Figure 2B). We showed that LPS induced a strong response in pMCs (Supplementary Figure 2C). Two possible reasons exist for the very low or no responses to LPS observed in MC-XF-HIOs. Macrophages in MC-XF-HIOs are CD14 negative cells (Figure 1C). Resident intestinal macrophages characterized as lacking CD14 did not show enhanced cytokine production by LPS.7 We observed that toll-like receptor 4 protein was weakly expressed on the apical surface of MC-XF-HIOs (Supplementary Figure 2D). This observation is consistent with a recent report by Price et al,8 who observed a weaker expression of toll-like receptor 4 in the small intestine in comparison with that in the stomach or colon and very low responses to LPS in human intestinal organoids compared with colon organoids.
Figure 2.
Macrophage-related characterization of MC-XF-HIOs. (A) An illustration that indicates the characteristic structure of a MC-XF-HIO compartmentalizing fluid content (FC). (B) Secretions released in the FC fluid of a single XF-HIO or MC-XF-HIO were assayed for selected interleukin (IL) cytokines, and these were quantified. LPS stimulation of organoids for 24 hours; FC samples were then collected. Data represent the mean ± standard error of 3–6 independent gut organoids generated in at least 3 individual experiments in the presence or absence of LPS. Statistical significance was identified using Student t test (∗P < .05, ∗∗P < .01, NS, not significant. (C) EGFP-expressing pGMACs in MC-XF-HIOs demonstrated red fluorescence (white arrowheads) inside the cells after exposure to pHrodo red Escherichia coli bioparticles. Scale bars: white, 300 μm; gray, 100 μm. (D) A diagram of hiPSC-derived MC-XF-HIOs.
Supplementary Figure 2.
Cytokine and chemokine profiles of MC-XF-HIOs. (A) Total of 29 cytokines and chemokines in the fluid content (FC) and supernatant fluid (SF) of the organoids were assayed using a bead-based Multiplex cytokine assay. Secretions released in the SF medium and FC fluid of a single xenogeneic-free human intestinal organoid (XF-HIO) or macrophage–xenogeneic-free human intestinal organoid (MC-XF-HIO) were assayed for selected cytokines and chemokines, and these were quantified. Lipopolysaccharide (LPS) stimulation of organoids for 24 hours; SF and FC samples were then collected. Data represent the mean ± standard error of the mean of 3–6 independent gut organoids generated in at least 4 individual experiments in the presence or absence of LPS. (B) Representative fluorescence-activated cell sorting (FACS) images of MC-XF-HIOs disassembled into single cells. Expression of key macrophage polarization markers determined by quantitative reverse transcription PCR: M1 macrophage–associated genes (TNF, NOS2, HLA-DB1); M2 macrophage–associated genes (IL6, KLF4, VEGFA). Relative expression was calculated using the ΔΔCT method, with GAPDH as an endogenous control and normalization to human blood monocytes. Samples as human-induced pluripotent stem cell (hiPSC)–derived monocyte-like cells (pMCs) and differentiated macrophages (pMACs) were generated in independent experiments. Originally injected monocytes (pMCs) in XF-HIOs or co-cultured human pluripotent stem cell–derived gut macrophages (pGMACs) were isolated from 5 MC-XF-HIOs in 3 individual experiments using FACS. The data represent the mean ± standard error, and statistical significance was identified using Student t test (∗P < .05) (n = 3). (C) Monocytes derived from hiPSCs were stimulated with the indicated concentrations of LPS for 24 hours and analyzed for interleukin 6 by quantitative reverse transcription PCR. Each assay was performed with 3 biologically independent replicates. Values of interleukin 6 were normalized against the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The data represent the mean ± standard error, and statistical significance was identified using Student t test (∗P < .05, ∗∗P < .01; n = 3) versus 0 ng/mL LPS as a control. (D) Immunofluorescence staining for toll-like receptor 4 (TLR4) in an MC-XF-HIO. Scale bar: 50 μm.
Next, we assessed the phagocytosis of pGMACs in response to foreign antigens on the epithelium of MC-XF-HIOs using pH-dependent dye labeled Escherichia coli bioparticles.9 The bioparticles only fluoresced when localized in the acidic environment of the phagolysosome. A magnified image showed red signals detectable within pGMACs and suggested pGMACs existing in the organoid captured bioparticles in acidified phagolysosomes (Figure 2C).
Here we present the development of hiPSC-derived intestinal organoids inhabited by tissue macrophages that model intestinal immune responses in vitro. One of the important features of the MC-XF-HIO system is that both organoids and macrophages are derived from an identical hiPSC line. We further applied this technique to a novel Crohn’s disease model as a potential platform for studying human intestinal inflammatory disorders (Supplementary Figure 3). The MC-XF-HIO culture system we describe here provides a species-specific in vitro model for temporally and spatially investigating interactions between the gastrointestinal tract and intestinal macrophages (Figure 2D). This represents a powerful addition to the repertoire of methods available to research gut homeostasis and the immune system.
Supplementary Figure 3.
XF-HIOs and MC-XF-HIOs derived from Crohn’s disease–specific iPSC lines. Crohn’s disease–specific induced pluripotent stem cell (iPS) lines (CD-iPSCs) were derived from patients with Crohn’s disease (CD). HPS1508 and HPS2816 cell lines were derived from 2 separate patients with an ileal form of CD. HPS2054 was derived from a patient with an ileocolic form of CD. These 3 cell lines were confirmed to differentiate into xenogeneic-free human intestinal organoids (XF-HIOs) and PSC-derived monocyte-like cells (pMCs). Macrophage–xenogeneic-free human intestinal organoids (MC-XF-HIOs) were generated from the CD-iPSC lines and each pMC. Hematoxylin-eosin staining of CD-iPSC derived MC-XF-HIOs (CD-MC-XF-HIOs). The CD-MC-XF-HIOs are structured outward and oriented toward the epithelial layers. Scale bar corresponds to 200 μm. The XF-HIOs are structured outward and oriented toward the epithelial layers. White and black scale bars correspond to 500 μm and 200 μm, respectively.
Acknowledgments
The authors thank the members of the Life Science Laboratory, Dai Nippon Printing Co (DNP), and members of our laboratory for their helpful discussions. They thank Minoru Ichinose and Shoko Miyajima for preparing histologic samples.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported by a grant from the Ministry of Health, Labour, and Welfare Sciences to HA and AU. HA was supported by research grants from the Japan Agency for Medical Research and Development (AMED; JP20bk0104089h0002, and JP20bm0804017h0001), JSPS KAKENHI (19K22988), and the Danone Institute of Japan Foundation. ST was supported by a JSPS KAKENHI Grant (19K22988). This research was also supported by a grant from AMED under grant numbers JP18dm0207002 and JP21bm0704046 (SS and TS). The funders had no role in the study design, data collection and analysis, decision to publish, or the preparation of the manuscript.
Supplementary Materials and Methods
Ethics Approval
Human material (hiPSCs and intestinal tissues) was obtained with informed consent from patients or their families and the approval of relevant institutions. The use of intestinal tissues was approved by the institutional review board (of the National Center for Child Health and Development (NCCHD) (IRB permission #146, #927) and according to the Declaration of Helsinki.
Cell Lines
The hiPSC line, Edom-iPS, was generated in our laboratory1,2 and cultured in StemFlex medium (Thermo Fisher Scientific, Waltham, MA). An EGFP-Edom-iPSC line, which constitutively expressed EGFP under a cytomegalovirus promoter with a hyperactive PiggyBac vector,3 was used for MC/macrophage differentiation.
Crohn’s disease–specific hiPSC lines were obtained from RIKEN BRC Cell Bank (Ibaraki, Japan) with ethics approval. Three independent cell lines, HPS1508, HPS2816, and HPS2054, were generated from peripheral blood mononuclear cells of Crohn’s disease patients.
Generation of XF-HIOs From hiPSCs
We previously4,5 generated highly functional intestinal organoids using defined xenogeneic-free differentiation medium: 85% knockout Dulbecco modified Eagle medium, 15% knockout serum replacement XF (XF-KSR; Gibco, Waltham, MA), 2 mmol/L GlutaMAX-I, penicillin–streptomycin, 50 μg/mL L-ascorbic acid 2-phosphate (Sigma-Aldrich, St Louis, MO), 10 ng/mL heregulin-1β (R&D Systems, Minneapolis, MN), 200 ng/mL recombinant human insulin growth factor-1 (Sigma-Aldrich), and 20 ng/mL human basic fibroblast growth factor (Gibco). Undifferentiated hiPSCs were dissociated and plated on a cell-patterning glass substrate CytoGraph (Dai Nippon Printing, Tokyo, Japan). XF differentiation medium was replaced every 3–4 days. Floating orbicular gut organoids were collected and cultured in a culture dish (Corning, Corning, NY) in XF differentiation medium.
Generation of Human Monocyte-Like Cells From hiPSCs and Differentiation Into Macrophages
The hiPSCs were differentiated into macrophage progenitors following a previously published protocol.6 Monocyte-like cells emerging into the supernatant after approximately 4 weeks were repeatedly harvested once per week by straining (Corning).
XF-HIOs and Macrophages Co-Cultures
To co-culture XF-HIOs with MCs derived from the same iPSCs, we established an injection method for reproducing biological sites and scaffolds. To accurately reproduce reciprocal biological sites of local macrophages and the mesenchymal tissue of XF-HIOs as a cell scaffold, we established manipulative transplantation with a microsyringe. Human iPSC-derived MCs were collected, centrifuged, and resuspended in XF culture medium supplemented with 100 ng/mL M-CSF to give a final cell concentration of 5.0 × 106/mL. The XF-HIOs that grew to approximately 10 mm in diameter were collected, and the prepared MCs were injected into each XF-HIO using a syringe (Nipro, Osaka, Japan) and atraumatic 34-gauge microneedles (Unisis, Tokyo, Japan) under a microscope. XF-HIOs injected with MCs were cultured in 6-well plates (Corning) in XF medium with 100 ng/mL M-CSF for 14 days.
Quantitative Reverse Transcription Polymerase Chain Reaction Analysis
RNA was isolated from organoids using a RNeasy Mini Kit (Qiagen, Hilden, Germany), and cDNA was generated using SuperScript IV VILO Master Mix (Thermo Fisher Scientific). Quantitative reverse transcription PCR were carried out in triplicate using SYBR Green PCR Master mix. All reactions were run for 40 cycles at 95°C for 15 seconds and 60°C for 30 seconds, followed by melting curve analysis. For comparing characteristics of co-cultured iPSC-derived gut macrophages (pGMACs), iPSC-derived pMCs, and human MCs, quantitative reverse transcription PCR was performed using a GeneQuery Human Macrophage Polarization Markers qPCR Array Kit (ScienCell Research Laboratories, Carlsbad, CA). QuantStudio 12K Flex software was adopted to quantify the relative levels of mRNA of target genes after normalization against the housekeeping gene, GAPDH. Healthy human primary small intestine (ileum) cDNA (BioChain Institute, Newark, CA) and human peripheral blood mononuclear cells cDNA (3H Biomedical, Uppsala, Sweden) were used as positive controls.
The hiPSC-derived MCs were stimulated with indicated concentrations of LPS from E coli O111 (Sigma-Aldrich) for 24 hours and analyzed for interleukin 6 by quantitative reverse transcription PCR with SYBR Green chemistry according to the manufacturer’s protocol.
Immunocytochemical Staining
Organoids fixed with 4% paraformaldehyde in phosphate-buffered saline (Gibco) were incubated overnight at 4°C with primary antibodies: anti-IBA1 (Abcam, Cambridge, UK), anti-GFP (Abcam), anti-CD14 (Abcam), anti-ZO-1 (Invitrogen, Waltham, MA), anti-CX3CR1 (Abcam), anti-TLR4 (Novus Biologicals, Littleton, CO), and anti-vimentin (Abcam). Alexa 488- or Alexa 546-conjugated anti-mouse, anti-rabbit, or anti-goat secondary antibodies (BD Biosciences, Franklin Lakes, NJ) were used. Cell fluorescence was analyzed using a Nikon A1 confocal microscope (Nikon, Tokyo, Japan) or a BZ-X700 microscope (Keyence, Osaka, Japan). Three-dimensional fluorescent images were obtained using a confocal FV-1200 microscope (Olympus, Tokyo, Japan), and three-dimensional movies were made using IMARIS software (Bitplane, Zurich, Switzerland).
Electron Microscope Analysis
Electron microscopy imaging was performed as previously described.7 Organoid samples were fixed in 2.5% glutaraldehyde and 0.1 mol/L phosphate buffer (Muto Pure Chemicals, Tokyo, Japan) and dehydrated in serial fixation steps. Ultrathin sections on copper grids were examined with a transmission electron microscope (JEM-1400plus; JEOL, Tokyo, Japan) at 100 keV for ultrahigh-resolution imaging. A multibeam scanning electron microscope (Multi-SEM 505; Carl Zeiss, Oberkochen, Germany) was used for whole-section electron microscopy imaging.
Assessment of Phagocytic Activity
To measure the antigen uptake of pGMACs by MC-XF-HIOs, pHrodo Red E coli BioParticles Conjugate (Thermo Fisher Scientific), which fluoresces red in acidic phagosomes, was used. A single MC-XF-HIO was washed 3 times with phosphate-buffered saline and incubated with 500 μg/mL bioparticles. Fluorescence was measured with a BZ-X700 fluorescence microscope (Keyence) in a Keyence imaging platform after 1.5-hour incubation at 37.0°C. Three different experiments were performed.
Multiple Analyte Profile for Cytokine and Chemokine Level Determination
For multiple cytokine and chemokine analysis, XF-HIO samples cultured for 2 weeks from a pMC injection were selected. After 72 hours in culture, the supernatant (SF) and FC of XF-HIOs or MC-XF-HIOs were collected. Both XF-HIOs and MC-XF-HIOs were stimulated with 100 ng/mL LPS from E coli O111 (Sigma-Aldrich) for 24 hours before collection. Cytokine levels were determined in duplicate using a Milliplex MAP Human Cytokine/Chemokine Panel (Merck, Kenilworth, NJ) according to the manufacturer’s instructions. Fluorescence signals were measured by Luminex 200 (Luminex Corp, Austin, TX), and data were analyzed using MilliplexAnalyst (VigeneTech, Carlisle, MA).
Video Recordings
MC-XF-HIOs were observed using a BZ-X700-All-in-One fluorescence microscope. Original videos were recorded at 29 frames per second. The playback speed of the video was 20 times actual speed.
Statistical Analysis
Data are reported as mean ± standard error of the mean from at least 3 independent experiments. Statistical analyses were performed using either an unpaired or two-tailed t test. P values ≤.05 were considered statistically significant.
Supplementary Material
References
- 1.Santaolalla R., et al. Curr Opin Gastroenterol. 2011;27:125–131. doi: 10.1097/MOG.0b013e3283438dea. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uchida H., et al. JCI Insight. 2017;2 doi: 10.1172/jci.insight.86492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sasaki K., et al. StemJournal. 2021;3:1–10. [Google Scholar]
- 4.Green K.Y., et al. Nat Commun. 2020;11:2759. doi: 10.1038/s41467-020-16491-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smythies L.E., et al. J Clin Invest. 2005;115:66–75. doi: 10.1172/JCI19229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Candelli M., et al. Int J Mol Sci. 2021;22:6242. [Google Scholar]
- 7.Smith P.D., et al. J Immunol. 2001;167:2651–2656. doi: 10.4049/jimmunol.167.5.2651. [DOI] [PubMed] [Google Scholar]
- 8.Price A.E., et al. Immunity. 2018;49:560–575. doi: 10.1016/j.immuni.2018.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kapellos T.S., et al. Biochem Pharmacol. 2016;116:107–119. doi: 10.1016/j.bcp.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplementary References
- 1.Sugawara T., et al. Regen Ther. 2020;15:1–9. doi: 10.1016/j.reth.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sono W., et al. Regen Ther. 2020;15:161–168. doi: 10.1016/j.reth.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamada M., et al. STAR Protoc. 2021;2 doi: 10.1016/j.xpro.2021.100811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uchida H., et al. JCI Insight. 2017;2 doi: 10.1172/jci.insight.86492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sasaki K., et al. StemJournal. 2021;3:1–10. [Google Scholar]
- 6.van Wilgenburg B., et al. PLOS One. 2013;8 doi: 10.1371/journal.pone.0071098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shibata S., et al. Front Neural Circuits. 2019;13:29. doi: 10.3389/fncir.2019.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.