Abstract
Archaeoglobus fulgidus, a hyperthermophilic, archaeal sulfate reducer, is one of the few organisms that can utilize d-lactate as a sole source for both carbon and electrons. The A. fulgidus open reading frame, AF0394, which is predicted to encode a d-(−)-lactate dehydrogenase (Dld), was cloned, and its product was expressed in Escherichia coli as a fusion with the maltose binding protein (MBP). The 90-kDa MBP-Dld fusion protein was more efficiently expressed in E. coli when coexpressed with the E. coli dnaY gene, encoding the arginyl tRNA for the codons AGA and AGG. When cleaved from the fusion protein by treatment with factor Xa, the recombinant Dld (rDld) has an apparent molecular mass of 50 kDa, similar to that of the native A. fulgidus Dld enzyme. Both the purified MBP-Dld fusion protein and its rDld cleavage fragment have lactate dehydrogenase activities specific for d-lactate, are stable at 80°C, and retain activity after exposure to oxygen. The flavin cofactor FAD, which binds rDld apoprotein with a 1:1 stoichiometry, is essential for activity.
Dissimilatory sulfate reducers inhabit aquatic and terrestrial sediments and play an essential role in the biogeochemical sulfur cycle. These strict anaerobes reduce sulfate to H2S (35), which either is released into the environment or is assimilated by other organisms as a substrate for growth. Members of the genus Archaeoglobus are the only known sulfate reducers that are hyperthermophilic, and they belong to the domain Archaea. Archaeoglobus spp. have been isolated from ocean and terrestrial oil deposits, where they likely account for the formation of H2-contaminated “sour oil,” particularly at high temperatures. The key enzymes involved in sulfate reduction, including ATP sulfurylase (7), adenylylsulfate reductase (22, 33), and sulfite reductase (8), are conserved evolutionarily in Archaeoglobus and resemble the enzymes characterized from bacterial sulfate reducers (17, 34).
In Archaeoglobus fulgidus, the type species, sulfate reduction occurs at 83°C, the optimal temperature for its growth. A. fulgidus thrives in anaerobic environments and can use either d- or l-lactate as a sole carbon and electron source. When grown on lactate, dissimilatory sulfate reducers obtain energy by using a lactate dehydrogenase to complete the conversion to pyruvate with the transfer of electrons to the anaerobic respiratory chain. The electrons removed from lactate are transferred through intermediate carriers, such as cytochromes and quinones in the membrane. The cytochrome-dependent lactate dehydrogenase enzymes are used during anaerobic respiration to generate a proton motive force for growth (23).
Lactate dehydrogenases (Ldhs) convert d- or l-lactate to pyruvate and are found in almost every organism (11, 13). Traditionally, they have been classified into families by their ability to transfer electrons to various electron acceptors in vitro, including NAD+ (EC 1.1.1.27 and 1.1.1.28), cytochrome (EC 1.1.2.3 and 1.1.2.4), and cytochrome c553 (EC 1.1.2.5). Ldhs are also classified into subfamilies depending on their substrate specificities (d-lactate versus l-lactate). The best-characterized lactate dehydrogenase is the NAD-dependent yeast enzyme, which catalyzes the reduction of pyruvate to lactate to regenerate NAD and thereby restore the NAD+-NADH balance during fermentation.
In contrast, the d-lactate dehydrogenases (cytochrome) (EC 1.1.2.4.) are poorly understood. Only a few d-lactate dehydrogenases (cytochrome) have been characterized from yeast and bacteria, and these have been shown to be NAD-independent, membrane-associated proteins (5, 12, 26, 28, 29) that use FAD and Zn2+ for electron transfer reactions (5, 11, 12, 29). The 63-kDa Kluyveromyces lactis DLD protein and its close relative, the 64-kDa S. cerevisiae D-LCR (d-lactate ferricytochrome c oxidoreductase) protein, are both mitochondrial enzymes (23, 24). These enzymes are thought to transfer electrons to cytochromes in vivo because they do so in vitro (5, 12, 27, 28).
To understand the pathway of energy production that initiates with d-lactate in A. fulgidus, an open reading frame (ORF) (AF0394) (18) whose predicted product resembles the S. cerevisiae and K. lactis d-lactate dehydrogenases was cloned. Expression of AF0394 as part of a fusion protein with maltose-binding protein (MBP) in E. coli results in a stable product, which has activity in an assay coupling the oxidation of d-lactate with the reduction of the artificial acceptor phenazine methosulfate (PMS).
MATERIALS AND METHODS
Reagents and vectors.
PCR and plasmid DNAs were purified with reagents from Qiagen. Restriction enzymes and DNA-modifying enzymes, from New England Biolabs (Beverly, Mass.), were used under recommended conditions. The antibiotics and chemicals were from Aldrich, Fisher, and Sigma. E. coli JM107 (38) was used as the host for the cloning and manipulation of DNA and for the purification of recombinant proteins. Strains of JM107 carrying plasmids were grown in Luria-Bertani (LB) medium supplemented with ampicillin (100 μg/ml) and/or kanamycin sulfate (40 μg/ml). The pMALc expression vector and factor Xa protease were obtained from New England Biolabs. Vector pUBS520 (Kanr) was a gift from Peter Buchner (Boehringer Mannheim, Mannheim, Germany).
Archaeoglobus growth and DNA preparations.
A. fulgidus VC-16 (DSM4304) was obtained from Karl Stetter (Lehrstuhl für Mikrobiologie, Universität Regensburg) (14). The cells were grown at 83°C in anaerobic sulfate-thiosulfate-lactate (STL) medium gassed with N2. STL medium was modified from medium 3 of Balch et al. (2) as follows: 20 mM PIPES (piperazine-N,N′-bis(2-ethanesulfonic acid) buffer was substituted for NaHCO3, 11.2 mM sodium lactate was substituted for sodium acetate, 0.5 g of yeast extract/liter was substituted for Trypticase and vitamins, and 0.11 mg of NiSO4 · 6H2O/liter was added. The medium was reduced with 1 mM Na2S and 1 mM Na2S2O3 and then inoculated with 5% (vol/vol) of logarithmic-phase A. fulgidus cells. Growth was monitored as the change in absorbance at 600 nm with a Milton-Roy Spectronic 21D spectrophotometer. Large-scale cultures of A. fulgidus were grown in a 45-liter glass carboy, heated with a drum heating belt (Barnstead Thermolyne, Dubuque, Iowa), stirred continuously, and sparged with 0.07 liters of N2/min. Glacial acetic acid was added periodically to maintain a pH of <8.0. The cells were harvested initially with an S3Y100 spiral ultrafiltration cartridge (Amicon, Beverly, Mass.) and then by centrifugation at 16,000 × g for 15 min at 4°C and then lysed by passage through a French pressure cell at 20,000 lb/in2 and centrifuged at 14,000 × g for 30 min. NaCl was added to 150 mM to the supernatant of the low-speed centrifugation, which was then centrifuged at 230,000 × g for 1 h at 4°C in a Sorvall S100AT5 rotor; the supernatant was used for subsequent assays.
To prepare A. fulgidus genomic DNA, 50 ml of cells at 5 × 108/ml were harvested by centrifugation at 10,000 × g for 10 min and resuspended in 0.5 ml of 25% sucrose and 10 mM Tris (pH 7.5). The cells were incubated with 0.25 ml of lysis solution (5% sodium dodecyl sulfate [SDS], 0.125 M EDTA, 0.5 M Tris, pH 9.4) for 1 h at 55°C. Pronase E was added to a final concentration of 2 mg/ml, and incubation was continued at 37°C overnight. Potassium acetate (pH 5.0) was added to 0.5 M, and the sample was incubated for 10 min at 37°C and then for 1 h at 4°C and centrifuged at 10,000 × g for 15 min at 4°C. Two volumes of ethanol were added to the supernatant, and DNA was spooled with a glass rod, washed four times (70% ethanol, 10 mM Tris, 10 mM MgCl2, 1 mM EDTA, pH 8.0), allowed to dry, and resuspended in 200 μl of TE (10 mM Tris and 0.1 mM EDTA, pH 8.0).
Cloning and expression of A. fulgidus genes.
The ORFs AF0394 and AF0868 were amplified by PCR with A. fulgidus chromosomal DNA as a template. The oligonucleotide primers 5′ GCTCTAGAATGAGCTGGATTGATGAG and 5′ ACCTGCAGTCATAGTTTGCGAACAACCTTG included XbaI and PstI sites (underlined) designed to generate an in-frame fusion between AF0394 and malE in the vector pMALc. The oligonucleotide primers 5′ GGAATTCACCATGGTGATAGCCATCGAAAAAGTT and 5′ AACTGCAGTCACAGCATCACCCCCCTGTTGAG included EcoRI and PstI sites (underlined) designed to generate an in-frame fusion between AF0868 and malE. The conditions for PCR were 30 cycles consisting of 30 s at 95°C, 30 s at 55°C, and 3 min at 72°C in a reaction mixture containing 2 U of Deep Vent polymerase, 200 μM (each) deoxynucleotide triphosphate, 250 nM (each) primer, PCR buffer, and 0.3 μg of A. fulgidus chromosomal DNA.
A single PCR product of about 1.4 kb was obtained for both reactions, purified, digested with XbaI (or EcoRI) and PstI, and ligated with plasmid pMALc cut with the same enzymes. DNA was extracted with phenol-chloroform, ethanol precipitated, and resuspended in TE. DNA fragments were purified on a 0.7% agarose gel, excised, purified through a glass wool spin column, precipitated with ethanol, and resuspended in TE. The plasmids were recovered in E. coli after electroporation and selection for recombinants on LB agar with ampicillin (30). After restriction analysis to identify vectors carrying AF0394 and AF0868, plasmids pDR4 and pDR3, respectively, were used for subsequent studies.
Induction of the MBP-Dld fusion protein in E. coli.
To determine if an MBP-Dld fusion protein of the expected size is produced in E. coli, strain JM107 carrying either pDR3 or pDR4 was induced with IPTG (isopropyl-β-d-thiogalactopyranoside) and cell extracts were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) for the appearance of a 90-kDa protein. Overnight cultures were grown at 37°C in LB medium with antibiotics and 0.2% glucose, diluted 1:100 in LB-glucose plus antibiotic, and grown at 33°C for 2 h until the optical density at 600 nm reached 0.25. The culture was divided, half was supplemented with IPTG to a final concentration of 0.2 mM, and the cultures were incubated for 2 h at 33°C. Samples were harvested by centrifugation at 4,000 × g for 15 min, and the pellets were resuspended in buffer A (10 mM Na2HPO4, 30 mM NaCl, 0.25% Tween 20, 10 mM 2-mercaptoethanol, 10 mM EDTA, 10 mM EGTA [pH adjusted to 8.0 with NaOH]) and stored at −80°C.
Although initial experiments showed that plasmids pDR3 and pDR4 make a fusion protein of the expected size, the yield of protein was low. We tested whether a higher yield of the fusion protein could be obtained from an E. coli host that also carried a copy of pUBS520, which expresses the E. coli dnaY gene from the tac promoter. pUBS520 carries a Kanr (kanamycin resistance) determinant and can be maintained in the same host as pDR3 (pDR4), which carries an Ampr determinant (4), if both plasmids are selected simultaneously.
Purification of the MBP-Dld fusion protein.
The purification of the 90-kDa MBP-Dld fusion protein from E. coli was carried out at 4°C. Frozen cells resuspended in buffer A were thawed, freeze fractured with liquid N2, and lysed by passage through a French pressure cell at 19,000 lb/in2 three times. Samples were centrifuged at 230,000 × g for 30 min, and the supernatants were diluted 1:32 with buffer B (10 mM sodium phosphate, 0.5 M NaCl, 1 mM EGTA, pH 7.2) and loaded onto an amylose column equilibrated in buffer B at a rate of about 0.3 ml/min. After the column was washed with 3 volumes of buffer B, the fusion protein was eluted with 25 mM maltose in buffer B. The eluate was concentrated by ultrafiltration with a Microcon YM10 membrane (Amicon), and the maltose was removed by dialysis against 4 changes of buffer C (10 mM Tris, 100 mM NaCl, 1 mM EGTA, pH 8.2), each at a volume 100 times the sample volume. The fusion protein was concentrated again by ultrafiltration, purified by gel filtration on a Superose 12 HR 10/30 fast protein liquid chromatography column, equilibrated with buffer D (100 mM NaCl, 20 mM Tris, pH 8.0), and concentrated by ultrafiltration. To liberate the Dld fragment, 100 μg of MBP-Dld was cleaved by proteolysis with 1 μg of factor Xa for 5 days at 4°C in buffer D with 2 mM CaCl2. The C-terminal recombinant Dld (rDld) fragment was then separated from the N-terminal MBP fragment by passage through a second amylose affinity column. rDld, detected in the column flowthrough, was concentrated by ultrafiltration.
The protein concentration was determined by the Bradford method (3), with bovine serum albumin and gamma globulin as standards. Proteins were boiled in Laemmli buffer and separated by 10% Tricine SDS-PAGE (21, 31). The gels were stained with Coomassie GelCode Blue staining reagent (Pierce).
Enzyme assays.
Samples were assayed for d-lactate dehydrogenase activity by a gel assay. E. coli cells were centrifuged, suspended in buffer D, frozen, thawed, and incubated with lysozyme (200 μg/ml) at 25°C for 10 min and then at 4°C for 30 min. After 12 sonication pulses (5 s; 5 W) at 4°C, the samples were centrifuged at 14,000 × g for 10 min to remove debris. The supernatants were separated through Tris-glycine nondenaturing polyacrylamide gels anaerobically at 25°C (21). The gels were incubated at 83°C in 10 mM Tris (pH 8.3 at 25°C), 10 mM MgSO4, 2 mM d-lactate, 65 μM PMS, and 240 μM 3[4,5-dimethylthiazol-2,yl]-2,5,diphenyl tetrazolium (MTT) in an anaerobic chamber. After 30 min the reaction was stopped by addition of HCl to a concentration of 0.1 M. Enzyme activity was detected by the appearance of dark bands, which form as MTT is reduced to formazan, an insoluble blue compound (10).
Spectrophotometric enzyme assays were done at 60°C in anaerobic quartz cuvettes (Starna) on a dual-beam Perkin-Elmer Lambda 12 UV/VIS spectrophotometer equipped with a PTP-6 temperature block. Anoxic assay mixtures (1 ml) contained 25 mM Tris (pH 8.5 at 25°C), 50 mM NaCl, 10 mM d-lactate, 65 μM PMS, 120 μM MTT, and 0.5 μg of protein. Enzyme activity was monitored as the increase in absorbance at 578 nm due to the formation of formazan. Reduction of dimethylnaphthoquinone (DMN) was monitored at 270 nm as described elsewhere (25).
Cofactor analysis.
Spectra of rDld were taken from 200 to 900 nm with a Perkin-Elmer Lambda 12 UV/VIS spectrophotometer supported by a Dell Optiplex XMT590 work station and UV-Winlab software. Fluorometric analysis of rDld was done with a Photon Technology Ratiomaster fluorescent spectrophotometer with a PTI Felix software package. Excitation was at 451 nm, and emission was monitored from 455 to 600 nm.
To identify the cofactor, cofactor released from purified protein was analyzed by ascending paper chromatography as described previously (16). rDld (1 μg) was heated to 100°C for 3 min and then centrifuged at 10,000 × g for 10 min to remove denatured protein. The supernatant from the denatured rDld sample, flavin adenine dinucleotide (FAD), flavin mononucleotide (FMN), and riboflavin controls were spotted on Whatman no. 1 paper, air dried, and chromatographed, in the dark, separately, with 5% aqueous Na2HPO4 and n-butanol–acetic acid–H2O (4:1:5). Sample positions were identified with a 365-nm-wavelength UV light.
The FAD/enzyme ratio was determined from the absorbance maxima of Dld at 370 and 450 nm. Because the MBP-Dld recombinant protein was expressed under conditions where the amount of cofactor might be limiting, we tested whether purified MBP-Dld could bind more FAD. MBP-Dld fusion protein (100 μg) was incubated with FAD at 4°C overnight, dialyzed against 3 changes of buffer D (pH 8.5), each at a volume 2,000 times the sample volume, to remove excess FAD, and then examined spectrophotometrically and assayed for activity.
To determine if flavin is essential for activity, apoprotein was prepared and assayed in the presence and absence of different flavin molecules. rDld (10 μg) was denatured by incubation at 100°C for 3 min in sample buffer and then separated by electrophoresis on SDS–10% polyacrylamide. rDld was renatured in the gel after two 1-h incubations in 20 mM Tris (pH 8.5) at 25°C to remove SDS.
Analysis of ions.
To assay for metals associated with the MBP-Dld fusion, 1.8 mg of protein was diluted in deionized water, filtered with a 0.45-μm-pore-size nylon filter, and acid hydrolyzed. Metal and ion concentrations were determined by Leeman inductively coupled plasma (ICP) atomic emission spectroscopy.
To determine if the MBP-Dld fusion protein had been expressed under conditions with low metal and ion availability, we tested whether the catalytic activity of MBP-Dld was increased by the addition of ions that had been detected at stoichiometric or substoichiometric amounts by ICP. Fusion protein (25 μg) was dialyzed for 4 h against elements [0.1 mM (each) CaCl2, MgCl2, ZnCl2, Cd(C2H3O2)2, BeSO4, Fe(NH4)2(SO4)2, and CuCl3] in buffer E (1 mM NaCl, 20 mM Tris, pH 8.4), dialyzed overnight to remove the excess, and assayed for activity. Parallel experiments performed with rDld yielded similar results.
RESULTS
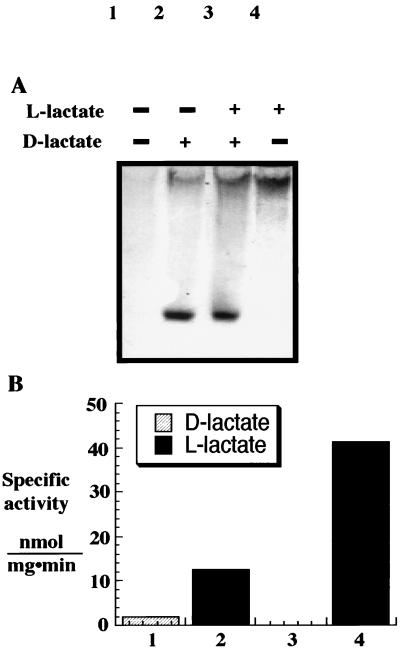
A. fulgidus can use either d- or l-lactate as its sole electron donor and carbon source. This hyperthermophilic sulfate reducer likely makes either two different lactate dehydrogenases, each specific for d-lactate or l-lactate, or one lactate dehydrogenase and a d- or l-lactate racemase. To identify the lactate dehydrogenases of A. fulgidus, cell extracts were prepared, separated by nondenaturing gel electrophoresis, and assayed for activity with the artificial electron acceptors PMS and MTT. When gels were incubated with d-lactate, a single intense band was seen near the bottom of the gel (Fig. 1A, lanes 2 and 3). In contrast, when gels were incubated with l-lactate, a different band was seen near the top of the gel (Fig. 1A, lanes 3 and 4). Lactate dehydrogenase activities in A. fulgidus cell extracts were also assayed spectrophotometrically with d- or l-lactate as an electron donor and PMS plus MTT or DMN as acceptors (Fig. 1B). As with the gel assay, with PMS plus MTT as electron acceptors, two forms of lactate dehydrogenase were detected. In contrast, with DMN as the electron acceptor, only one (l-lactate) dehydrogenase activity was detected. These results show that A. fulgidus makes at least one lactate dehydrogenase specific for d-lactate and another enzyme specific for l-lactate.
FIG. 1.
A. fulgidus produces d- and l-lactate-specific dehydrogenases. (A) Analysis of enzyme activity in extracts of A. fulgidus cells grown on d- and l-lactate (500 mg per lane) after separation on continuous nondenaturing 6% PAGE as described in Materials and Methods. Lanes: 1, no lactate; 2, with d-lactate; 3, with d- and l-lactate; 4, with l-lactate. (B) Activity of lactate dehydrogenases was assayed spectrophotometrically under anaerobic conditions with a 200-mg A. fulgidus sample and either d- or l-lactate with PMS and MTT (lanes 1 and 2) or DMN (lanes 3 and 4).
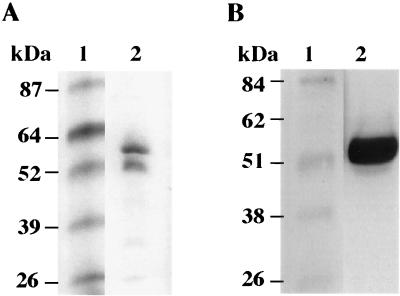
The A. fulgidus protein band with d-lactate dehydrogenase activity was excised from the nondenaturing gel and electrophoresed on a second SDS-PAGE gel to determine the apparent molecular mass of the protein. As shown in Fig. 2A, two proteins, about 50 and 57 kDa, were detected. When this gel was incubated in buffer without SDS at room temperature to renature the proteins and then assayed in the presence of FAD, only the 50-kDa protein had Dld activity (Fig. 2B). Hence, the A. fulgidus Dld should correspond to a gene of about 1,400 bp.
FIG. 2.
The active form of d-lactate dehydrogenase from A. fulgidus has an apparent molecular mass of about 50 kDa and requires FAD. (A) Coomassie blue-stained denaturing gel showing the Dld from A. fulgidus in lane 2 and prestained protein markers in lane 1. Partially purified A. fulgidus Dld was separated on a nondenaturing gel, assayed for Dld activity, excised from the gel, and electrophoresed through a 10% denaturing gel. (B) Partially purified A. fulgidus Dld was heat denatured in Laemmli buffer and electrophoresed by SDS-PAGE, incubated in Tris (pH 8.5), and then assayed with Tris (pH 8.3), MgSO4, PMS, MTT, FAD, and d-lactate at 80°C. Lanes: 1, Benchmark protein standard (Gibco-BRL); 2, Dld sample.
The annotated sequence of the A. fulgidus (18) genome has three ORFs, AF0394, AF0808, and AF0868, predicted to encode proteins related to d-lactate dehydrogenases. A BLAST (Blosum62) alignment (1) of the predicted products of these ORFs with the K. lactis KIDLD gene product shows that AF0394 has 30% identity and 48% similarity with KIDLD, whereas AF0808 has 29% identity and 46% similarity, and AF0868 has 25% identity and 42% similarity. All three ORFs contain domains with an essential histidine (GEHGD) (13, 15) conserved in enzymes that bind lactate. AF0394 and AF0868 contain an NAD-FAD binding motif (GXGX2GX21D/E) (37). Because d-lactate dehydrogenases typically have FAD as a cofactor (6, 9, 12, 27, 29) and AF0808 lacks a strong binding site for FAD, AF0394 and AF0868 were chosen as candidates for the 50-kDa Dld in A. fulgidus.
Expression of A. fulgidus genes in E. coli.
To determine if either AF0394 or AF0868 encodes a protein with Dld activity, each gene was expressed in E. coli. Chromosomal DNA, prepared from A. fulgidus cells, was used as a template to amplify the AF0394 and AF0868 genes by PCR. PCR products of the expected size were obtained and cloned into the expression vector pMALc to generate plasmids pDR4 and pDR3, which fuse AF0394 and AF0868, respectively, with a truncated malE gene, which encodes MBP without a signal peptide. AF0394 and AF0868 are predicted to encode products of 443 and 446 amino acids, respectively, and each should produce a fusion protein of about 90 kDa.
Cultures of E. coli JM107 carrying plasmid pDR3 or pDR4 were induced with IPTG, and proteins present in the induced cells were analyzed by SDS-PAGE. In both cases, a fusion protein band with an apparent mass of 90 kDa was detected. The fusion protein, which is made only by strains with pDR3 (data not shown) and pDR4 (Fig. 3, compare lanes 1, 3, and 5), reacts with anti-MBP antibody by immunoblot analysis (data not shown).
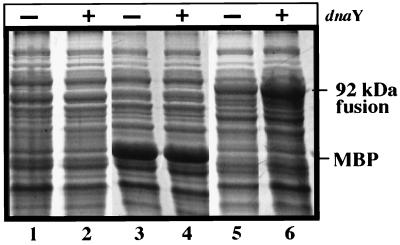
FIG. 3.
Addition of dnaY enhances the expression of the fusion protein. The expression of proteins in E. coli strains carrying pMAL or pDR4 was analyzed in the presence (+) and absence (−) of pUBS520 (dnaY). The cells were harvested 2 h after the addition of IPTG, and equivalent amounts of sample were analyzed by SDS-PAGE. Lanes: 1 and 2, E. coli cells; 3 and 4, E. coli cells with pMAL; 5 and 6, E. coli cells with pDR4. The samples in lanes 2, 4, and 6 contained the dnaY gene, which encodes an arginyl tRNA. The positions of the 92-kDa malE-AF0394 fusion product and the malE product (MBP) are indicated.
The yield of fusion protein in induced samples was disappointingly low. Attempts to increase the yield of fusion protein by decreasing the growth temperature, or changing the conditions of growth and induction, failed to improve the yield of protein. Although the G+C content of DNA in A. fulgidus (49%) is similar to that of the E. coli chromosome (52%), the codon usage in A. fulgidus differs from that in E. coli. For example, the AF0394 gene contains several AGR (R represents A or G) codons for arginine, and E. coli produces limiting amounts of a single isoacceptor arginyl-tRNA species (DnaY) that recognizes AGR codons. Hence, expression of foreign genes rich in AGR codons often is translationally impaired. To overcome this problem, plasmids pDR3 and pDR4 were introduced into an E. coli strain carrying the plasmid pUBS520, which expresses the E. coli dnaY gene at high levels after induction with IPTG (4). In the presence of plasmid pUBS520 (dnaY), the yield of the fusion protein after induction in E. coli increased (Fig. 3 shows results with pDR4; similar results were obtained with pDR3).
AF0394 produces a d-lactate dehydrogenase.
Proteins present in extracts of E. coli cells from induced and uninduced samples were separated on nondenaturing polyacrylamide gels to determine if either fusion protein has Dld activity. When gels were incubated at 83°C with d-lactate, PMS, and MTT, a single, intense band of activity was detected in lanes loaded with extract from E. coli cells expressing the fusion protein from pDR4. When an equivalent amount of the malE-AF0868 fusion protein from pDR3 was assayed in parallel, it was inactive with d-lactate, PMS, and MTT. These results show that AF0394 encodes a protein with lactate dehydrogenase activity.
As described above, the A. fulgidus Dld uses d-lactate, but not l-lactate, as its substrate. To determine the substrate specificity of the MBP-Dld fusion protein, samples expressing the malE-AF0394 protein were separated in nondenaturing gels and incubated with d-lactate, d- and l-lactate, l-lactate, and pyruvate. Whereas the 90-kDa fusion protein reduces MTT to formazan in the presence of d-lactate, l-lactate is neither a substrate for the enzyme nor an inhibitor of its d-lactate dehydrogenase activity.
To confirm that the product of AF0394 is the Dld enzyme identified in A. fulgidus extracts, a protein band corresponding to the protein band with d-lactate-specific dehydrogenase activity was excised from polyvinylidene difluoride membranes and analyzed for N-terminal sequence. Repeated attempts to obtain the N-terminal sequence of Dld were unsuccessful even though good sequence data were obtained from the same sample for a protein that copurifies with Dld. This, and the fact that N-terminal sequence could be obtained for a minor contaminant (glutamyl-tRNA amidotransferase; AF2329) of one of the Dld samples, suggests that the N terminus of Dld is blocked.
Purification of the A. fulgidus Dld from E. coli.
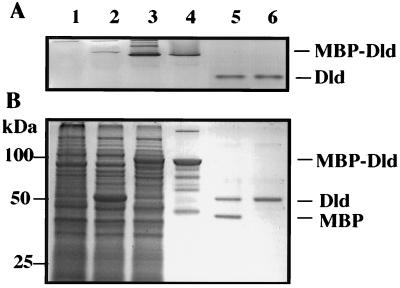
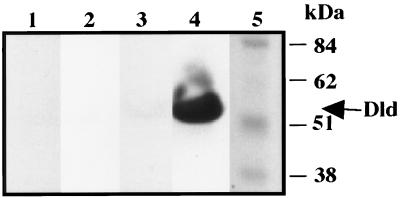
The 90-kDa MBP-Dld fusion protein from E. coli was enriched by amylose affinity chromatography and purified to homogeneity by gel filtration. Purified MBP-Dld was incubated with factor Xa protease to yield MBP (about 40-kDa) and Dld (about 50-kDa) products (Fig. 4, lane 5). The MBP portion of the MBP-Dld fusion protein was removed by passing the cleaved material over an amylose column. The eluate contained a single protein of 50 kDa with Dld activity, designated rDld to indicate that it is a recombinant Dld cleavage product (Fig. 4, lane 6).
FIG. 4.
Purification of rDld. (A) rDld activity was assayed with Tris (pH 8.3), MgSO4, PMS, MTT, and d-lactate after separation of 20-μl aliquots of E. coli samples through 10% nondenaturing gels. Lanes: 1, extract from IPTG-induced E. coli JM107 host; 2, extract from IPTG-induced E. coli JM107 with pMAL; 3, extract from IPTG-induced E. coli JM107 with pDR4 and pUBS520; 4, eluate from the initial amylose affinity column used to enrich for MBP-Dld; 5, sample containing MBP and rDld after gel filtration purification of the amylose eluate and treatment with protease factor Xa; 6, purified rDld was obtained after cleaved material was chromatographed over a second amylose column to remove MBP. (B) Aliquots (200 μl) of the same E. coli samples shown in panel A were separated by SDS-PAGE and stained with Coomassie blue. The molecular masses of the Benchmark protein standards are indicated.
rDld is a Zn+2 flavoprotein.
Like other flavin-containing proteins, purified rDld has an intense yellow color. Spectrophotometric analysis of rDld reveals absorption maxima at 370 and 450 nm with a shoulder at 480 nm, which indicate the presence of a flavin. When excited at 451 nm, the protein fluoresces with an emission maximum of 518 nm, also characteristic of proteins with bound flavin cofactors.
Each of the two yeast Dld enzymes that are related to the A. fulgidus enzyme contains a bound FAD moiety. This, and the presence of the adenine and ribityl binding sites predicted for the A. fulgidus Dld protein, suggest that the flavin bound by A. fulgidus Dld is FAD. To confirm that rDld contains FAD, and not FMN or riboflavin, the cofactor was removed from rDld by boiling and compared with FAD, FMN, and riboflavin standards by thin-layer chromatography. As shown in Table 1, the Rf value for the rDld cofactor is similar to that of FAD, confirming that the active form of rDld contains an FAD cofactor. These results also show that, although the cofactor is tightly associated with the enzyme, it is not covalently attached.
TABLE 1.
Rf values of flavins by ascending paper chromatographya
| Solvent |
Rf value
|
|||
|---|---|---|---|---|
| Flavin standard
|
Dld cofactorb | |||
| FMN | Riboflavin | FAD | ||
| Ac | 0.50 | 0.28 | 0.35 | 0.34 |
| Bd | 0.23 | 0.39 | 0.09 | 0.10 |
Samples were separated on Whatman no. 1 paper as described in the text.
rDld cofactor was released by boiling. Apoprotein was removed by centrifugation.
Solvent system A; 5% disodium hydrogen phosphate in H2O.
Solvent system B; n-butanol–acetic acid–H2O (4:1:5).
The rDld apoprotein was assayed in the presence and absence of flavin molecules to determine if FAD is required for rDld function and if F420, a deazaflavin that A. fulgidus produces, or other flavins, such as FMN, could substitute for FAD. Purified rDld was denatured by boiling and separated on denaturing gels. The enzyme was renatured and assayed in situ with buffer or buffer plus the deazaflavin F420 or flavin. Only rDld renatured in the presence of FAD was active, whereas renatured apoprotein or apoprotein renatured in the presence of FMN, riboflavin, or F420 was inactive (Fig. 5).
FIG. 5.
Reconstitution of active rDld after extraction of cofactor. rDld was electrophoresed under denaturing conditions, allowed to renature with or without flavin cofactor, and then assayed as described in Materials and Methods. Lanes: 1, renaturation with FMN-riboflavin; 2, renaturation with F420; 3, no cofactor added; 4, renaturation with FAD; 5, Benchmark protein standards.
The stoichiometry of FAD (ɛ450 = 11,300 M−1 cm−1) was estimated to be 0.2 FAD to 1 rDld by measuring the absorption maxima for purified protein. Because the amount of FAD per molecule of enzyme subunit is expected to be a positive integer, these results suggested that a portion of the bound flavin was in the reduced state or that a substantial fraction of the sample was rDld apoprotein without bound FAD. When the rDld was treated with persulfate to oxidize any FADH2 to FAD and reexamined, the increased absorbance at 450 nm yielded a ratio of 0.5 FAD to 1 rDld. When aliquots of rDld (ratio 0.2:1) were incubated with excess FAD and then dialyzed to remove unbound FAD, the ratio of FAD to rDld increased to 1:1.
When the rDld loaded with FAD was compared with the untreated rDld sample (0.2 FAD to 1 rDld) in the standard d-lactate dehydrogenase assay, the enzyme activity increased only slightly, suggesting that the activity of rDld might be limited for a second component. When rDld was incubated with excess FAD and dialyzed against a buffer containing the ions zinc, calcium, magnesium, cadmium, beryllium, iron, and copper, the activity of the enzyme increased threefold. Zinc is tightly bound to DLD from yeast and is thought to be essential for catalytic activity. To determine if rDld contained stoichiometric amounts of a metal cofactor, rDld was analyzed by ICP. rDld contains 1 mol of Zn2+ per mol of protein, suggesting that Zn2+ may also be essential for activity. The divalent cations Mg2+ and Ca2+ were also detected at significant levels and may be important for the catalytic activity or stability of the protein.
Kinetic analysis.
The Km of rDld for d-lactate was determined with PMS and MTT as electron acceptors. Double-reciprocal plots of the initial rates versus the concentration of d-lactate were linear at a fixed concentration of these acceptors and yielded a Km of 150 μM at 60°C. The Vmax was estimated to be 1.4 μM min−1 in the direction of d-lactate reduction with PMS plus MTT.
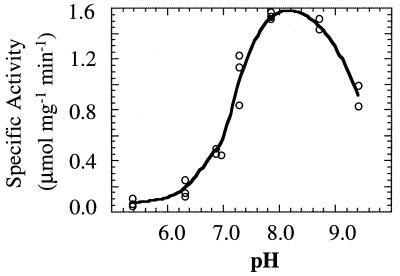
rDld reduces dichlorophenolindophenol (DCIP) and MTT, and reduction is stimulated by the addition of PMS. In contrast, methylene blue, DMN, NAD, and horse heart cytochrome c did not serve as electron acceptors for the purified rDld. When the concentration of d-lactate was fixed at 10 mM (at a d-lactate concentration of 4 mM, the reaction proceeds at maximum velocity), the specific activity of the homogeneous rDld enzyme at 60°C was 1.4 μmol of MTT reduced per mg per min (ɛ578 = 13 mM−1 cm−1), and it had a pH optimum at 8.0 in a 25 mM Tris buffer at 60°C (Fig. 6). At higher concentrations of Tris (>30 mM), the activity was inhibited slightly. The reaction rate was about 60% of the optimum in 25 mM potassium phosphate and 35% of the optimum in 25 mM sodium acetate at pH 8.2 in assays performed at 60°C. The detergents Triton X-100, CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} and n-dodecyl β-d-maltoside stimulated activity about twofold. The addition of Tris-buffered oxalate (1 mM) to a reaction mixture inhibited activity.
FIG. 6.
Effect of pH on rDld activity. rDld was assayed spectrophotometrically at 60°C as described in Materials and Methods. The final pH ranged from 5 to 9.5 (measured at 60°C) in 25 mM Tris.
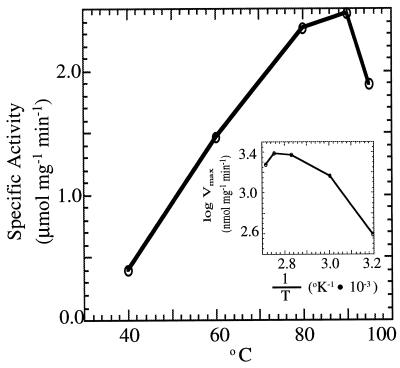
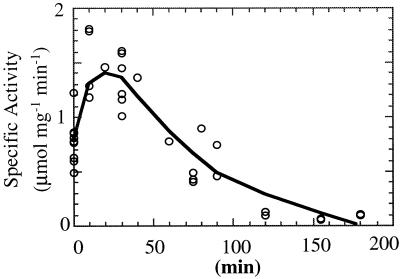
An Arrhenius plot shows that the rDld has maximum catalytic activity at 90°C with the activation energy increasing 5.1 kJ/mol in the range of 80 to 90°C (Fig. 7) (32). The rDld is active from 25 to 95°C, and at 83°C, the half-life of the enzyme was about 105 min (Fig. 8). At 25°C, the activity was about 8% of the activity at 90°C.
FIG. 7.
Effect of temperature on rDld activity. The activity of rDld (0.5 μg) was monitored at 578 nm by MTT reduction at the temperatures indicated. The assay contained saturating amounts of d-lactate (20 mM). (Inset) Data plotted according to the method of Arrhenius.
FIG. 8.
Thermal stability of rDld. rDld (1 mg/ml) was incubated at 83°C in 10 mM Tris (pH 8.0). At the times indicated, aliquots were removed and assayed at 60°C as described in Materials and Methods. Data from three independent experiments are presented. The half-life was estimated at 105 min.
DISCUSSION
The enzymes involved in activation and reduction of sulfate to H2S have been purified, revealing that the enzymology of sulfate reduction in the hyperthermophilic archaeon A. fulgidus is very similar to the enzymology of sulfate reduction in mesophilic eubacteria. During dissimilatory reduction of sulfate, electrons are obtained from substrates such as lactate. Hence, lactate dehydrogenases, which oxidize lactate to pyruvate, are essential for the growth of dissimilatory sulfate reducers on lactate as a sole electron source.
In contrast with what is known about the enzymology of sulfate reduction, the initial steps of electron transfer from substrates, such as lactate, to the sulfate acceptor are poorly understood. The first step in this process, the conversion of lactate to pyruvate, is mediated by membrane-associated lactate dehydrogenases, but the proton and electron acceptors for these reactions are not known. Although both d- and l-lactate dehydrogenases have been described in eubacterial sulfate reducers, they have not been purified, nor is sequence information available for the genes that encode these enzymes. Both soluble and membrane-associated Ldhs that can transfer electrons to either rubredoxin or cytochrome c in other organisms have been described. The second step in archaeal electron transfer involves the conversion of pyruvate to acetate and is mediated by pyruvate-ferredoxin oxidoreductase, which has been purified from eubacterial sulfate reducers and A. fulgidus (20).
The complete sequence for A. fulgidus is available, making it possible for the first time to look for genes encoding lactate dehydrogenases in a sulfate reducer. Based on similarity with the d-lactate dehydrogenase (KIDLD) from K. lactis, several ORFs, AF0394, AF0808, and AF0868, from the A. fulgidus genome were identified as candidates for dld. Because AF0808 lacks motifs predicted to be critical for the activities of other Dld enzymes, such as FAD and lactate binding, AF0394 and AF0868 were chosen for further study.
When extracts with fusion proteins were assayed with d-lactate with PMS as an electron acceptor, only the 90-kDa MBP-AF0394 fusion protein was able to oxidize d-lactate. This activity was specific for d-lactate, showing that AF0394 encodes a d-lactate dehydrogenase (Dld). rDld is active over a wide range of temperatures, and activity is optimal at 90°C, slightly above the optimal temperature for growth (83°C) for A. fulgidus (35).
Expression of the MBP-AF0394 fusion protein initially resulted in low levels of fusion protein in E. coli, despite the fact that transcription of its gene is controlled by the strong, IPTG-inducible tac promoter. A. fulgidus genes, like yeast genes, are rich in codons AGA and AGG for arginyl tRNA, which are rare in E. coli. Because these rare codons might prevent efficient translation, a plasmid carrying the dnaY (tRNA-Arg) gene was introduced into strains expressing the malE-AF0394 fusion proteins. Coexpression of dnaY resulted in an increase in the yield of fusion protein and a 1.5- to 4-fold increase in enzyme activity. Thus, this technique may be more generally useful for the increased production of gene products from archaea in a bacterial host.
FAD has been shown to be the cofactor for Dld from yeast and Megasphaera elsdenii (6, 12, 29). The predicted product of the dld gene has conserved motifs for FAD binding which consist of an N-terminal glycine-rich region and a ribityl binding site. When a C-terminal Dld fragment was purified from the fusion protein, the resulting 50-kDa protein, rDld, had an intense yellow color and spectral features characteristic of FAD. The observations that the flavin cofactor, FAD, copurifies with MBP-Dld and that this protein has activity like that of the A. fulgidus Dld when assayed at temperatures of >80°C shows that the rDld portion of the fusion protein folds properly when expressed in E. coli at 33°C.
Although rDld contains substoichiometric levels of bound FAD, it is possible to increase the ratio of FAD to rDld to 1:1 by incubating the protein with FAD. However, the enzyme activity does not increase despite the increased FAD binding. However, if a cocktail of ions, including those that are important for the growth of A. fulgidus in defined medium, are preincubated with the enzyme plus FAD, the activity of the enzyme increases threefold. These results suggest that a metal cofactor is required in addition to FAD. When rDld was analyzed by ICP, Zn2+, Mg2+, and Ca2+ were detected in stoichiometric quantities. Like the yeast Dlds, rDld contains about 1 Zn2+ atom per mol of protein. Zinc is thought to enable the FAD moieties of flavoproteins to interact with substrates that do not react with free flavin. When rDld is heat treated to remove cofactor(s), some activity is restored when FAD is added back to the renatured protein, indicating that some of the protein contains zinc which is not removed by heat treatment and dialysis. This is consistent with the finding that Zn2+ is tightly bound to the S. cerevisiae Dld (12).
Kinetic analyses show that rDld has a Km of 150 μM for d-lactate, which is comparable to that of the K. lactis Dld, which has a Km of 285 μM (12). Ldhs differ in their abilities to donate electrons to artificial acceptors. Many Llds (EC 1.1.2.3; cytochrome type) can use DMN, DCIP, and methylene blue as artificial electron acceptors, whereas Dlds (EC 1.1.2.4; cytochrome type) prefer PMS as an artificial acceptor. Consistent with this, rDld is active with PMS, but does not use DMN or methylene blue as an acceptor.
Electrons transferred to FAD from d-lactate during oxidation must ultimately be passed to another electron acceptor before they reach the terminal acceptor, sulfate. Potential acceptors include cytochromes or quinones. A. fulgidus produces b- and c-type cytochromes (19, 29a), and a 7-menaquinone (36), which might accept electrons from Dld. Although rDld did not transfer electrons to horse heart cytochrome c, it may transfer electrons to a cytochrome from Archaeoglobus. The partially purified Dld (EC 1.1.2.5) from the eubacterial sulfate reducer Desulfovibrio vulgaris is specific for cytochrome c553, which it produces (28), but the enzyme does not reduce cytochrome c from other sources. Similarly, the yeast Dld is able to reduce its own cytochrome and some commercial preparations in vitro but not all cytochromes from other organisms (12).
The recombinant enzyme has a temperature optimum of 90°C. Although the enzyme was routinely assayed at lower temperatures (60°C) to minimize nonspecific reduction of PMS and MTT, the specific activity of the enzyme was enhanced by preincubating the enzyme at 83°C prior to assay at 60°C. This suggests that temperatures closer to the optimal growth range of A. fulgidus may stimulate rDld activity. As expected for an enzyme involved in electron transfer reactions, Dld activity is found in the membrane fraction of A. fulgidus and the activity of rDld from E. coli is stimulated by detergents. However, a significant portion of the enzyme activity is detected in the soluble fraction, and the addition of NaCl to cell extract releases some of the enzyme from the membrane fraction. These results suggest that Dld is associated with the membrane.
Although A. fulgidus is a strict anaerobe, Dld and rDld are relatively stable under aerobic conditions at 4°C. As expected, Dld is heat stable and rDld is also heat stable. The purified enzyme has a half-life of 1.5 to 2 h at 83°C, which is not due to the hydrolysis of FAD at 83°C, because the addition of FAD does not restore enzyme activity after prolonged heat denaturation.
Several attempts were made to verify that the N terminus of the Dld protein prepared from A. fulgidus corresponds with the predicted product of AF0394. Because we were successful at obtaining sequence for other A. fulgidus proteins prepared in parallel, this suggests that the N-terminus of the Dld protein has been modified. Nevertheless, we conclude that the Dld from A. fulgidus is the product of AF0394 because expression of AF0394 in E. coli generates a protein that is indistinguishable from Dld in size, cofactor requirement, and kinetic properties. Only one enzyme with d-lactate specific activity is detected in A. fulgidus cell extracts, which is consistent with the fact that only one gene with a predicted product with d-lactate dehydrogenase homology is found in the A. fulgidus genome.
This study provides new insights into the structure, function, and mechanism of the archaeal Dld (cytochrome) group of dehydrogenases that have primary catabolic roles in sulfate reducers. Future studies aimed at identifying the proteins with which Dld interacts will provide a better understanding of the electron transfer reactions that are essential for growth on d-lactate during the anaerobic respiration of the hyperthermophilic sulfate reducer A. fulgidus.
ACKNOWLEDGMENTS
We thank Philip Youderian for helpful discussions and critical reading of the manuscript, Jack Millstein and Kraig White for technical assistance, and David Graham for providing sequence information prior to publication.
This work was supported by grants OSR-9350539 and MCB9906433 from the National Science Foundation.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balch W E, Fox G E, Magrum L J, Woese C R, Wolfe R S. Methanogens: reevaluation of a unique biological group. Microbiol Rev. 1979;43:260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Brinkmann U, Mattes R E, Buckel P. High-level expression of recombinant genes in Escherichia coli is dependent on the availability of the dnaY gene product. Gene. 1989;85:109–114. doi: 10.1016/0378-1119(89)90470-8. [DOI] [PubMed] [Google Scholar]
- 5.Brockman H J, Jr, Wood W A. d-Lactate dehydrogenase of Peptostreptococcus elsdenii. Methods Enzymol. 1975;41:309–312. doi: 10.1016/s0076-6879(75)41070-9. [DOI] [PubMed] [Google Scholar]
- 6.Brockman H L, Wood W A. d-Lactate dehydrogenase of Peptostreptococcus elsdenii. J Bacteriol. 1975;124:1454–1461. doi: 10.1128/jb.124.3.1454-1461.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dahl C, Koch H, Keuken O, Truper H G. Purification and characterization of ATP sulfurylase from the extremely thermophilic archaebacterial sulfate-reducer, Archaeoglobus fulgidus. FEMS Microbiol Lett. 1990;67:27–32. [Google Scholar]
- 8.Dahl C, Kredich N M, Deutzmann R, Truper H G. Dissimilatory sulphite reductase from Archaeoglobus fulgidus: physico-chemical properties of the enzyme and cloning, sequencing and analysis of the reductase genes. J Gen Microbiol. 1993;139:1817–1828. doi: 10.1099/00221287-139-8-1817. [DOI] [PubMed] [Google Scholar]
- 9.Futai M. Membrane D-lactate dehydrogenase from Escherichia coli: purification and properties. Biochemistry. 1973;12:2468–2474. doi: 10.1021/bi00737a016. [DOI] [PubMed] [Google Scholar]
- 10.Garvie E I. Lactic dehydrogenases of strains of the genus Leuconostoc. J Gen Microbiol. 1969;58:85–94. doi: 10.1099/00221287-58-1-85. [DOI] [PubMed] [Google Scholar]
- 11.Garvie E I. Bacterial lactate dehydrogenases. Microbiol Rev. 1980;44:106–139. doi: 10.1128/mr.44.1.106-139.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gregolin C, Singer T P. The lactic dehydrogenase of yeast. III. D(−)lactic cytochrome c reductase, a zinc flavoprotein from aerobic yeast. Biochim Biophys Acta. 1963;67:201–218. doi: 10.1016/0006-3002(63)91818-3. [DOI] [PubMed] [Google Scholar]
- 13.Griffin H G, Simon R S, Gasson M J. Cloning and sequence analysis of the gene encoding L-lactate dehydrogenase from Lactococcus lactis: evolutionary relationships between 21 different LDH enzymes. Gene. 1992;122:193–197. doi: 10.1016/0378-1119(92)90049-u. [DOI] [PubMed] [Google Scholar]
- 14.Hartzell P L, Millstein J, LaPaglia C L. Biofilm formation in a hyperthermophilic archaeon. Methods Enzymol. 1999;310:335–349. doi: 10.1016/s0076-6879(99)10027-2. [DOI] [PubMed] [Google Scholar]
- 15.Holbrook J J, Liljas A, Steindel S J, Rossmann M G. Lactate dehydrogenase. In: Boyer P D, editor. The enzymes. New York, N.Y: Academic Press; 1975. pp. 191–292. [Google Scholar]
- 16.Huennekens F M, Felton S P. Preparation and enzymatic assay of FAD and FMN. Methods Enzymol. 1957;3:950–959. [Google Scholar]
- 17.Karkhoff-Schweizer R R, Huber D P W, Voordouw G. Conservation of the genes for dissimilatory sulfite reductase from Desulfovibrio vulgaris and Archaeoglobus fulgidus allows their detection by PCR. Appl Environ Microbiol. 1995;61:290–296. doi: 10.1128/aem.61.1.290-296.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klenk H, Clayton R A, Tomb J-F, White O, Nelson K E, Ketchum K A, Dobson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleishmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Peterson S, Reich C I, McNeil L K, Badger J H, Glodek A, Zhou L, Overbeek R, Gocayne J D, Weidman J F, McDonald L, Utterback T, Cotton M D, Spriggs T, Artiach P, Kaine B P, Sykes S M, Sadow P W, D'Andrea K P, Bowman C, Fujii C, Garland S A, Mason T M, Olsen G J, Fraser C M, Smith H O, Woese C R, Venter J C. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 19.Kunow J, Linder D, Stetter K O, Thauer R K. F420H2: quinone oxidoreductase from Archaeoglobus fulgidus. Characterization of a membrane-bound multisubunit complex containing FAD and iron-sulfur clusters. Eur J Biochem. 1994;223:503–511. doi: 10.1111/j.1432-1033.1994.tb19019.x. [DOI] [PubMed] [Google Scholar]
- 20.Kunow J, Linder D, Thauer R K. Pyruvate:ferredoxin oxidoreductase from the sulfate-reducing Archaeoglobus fulgidus: molecular composition, catalytic properties, and sequence alignments. Arch Microbiol. 1995;163:21–28. doi: 10.1007/BF00262199. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.Lampreia J, Fauque G, Speich N, Dahl C, Moura I, Truper H G, Moura J J G. Spectroscopic studies on APS reductase isolated from the hyperthermophilic sulfate-reducing archaebacterium Archaeoglobus fulgidus. Biochem Biophys Res Commun. 1991;181:342–347. doi: 10.1016/s0006-291x(05)81424-x. [DOI] [PubMed] [Google Scholar]
- 23.Lodi T, Ferrero I. Isolation of the DLD gene of Saccharomyces cerevisiae encoding the mitochondrial enzyme D-lactate ferricytochrome c oxidoreductase. Mol Gen Genet. 1993;238:315–324. doi: 10.1007/BF00291989. [DOI] [PubMed] [Google Scholar]
- 24.Lodi T, O'Connor D, Goffrini P, Ferrero I. Carbon catabolite repression in Kluyveromyces lactis: isolation and characterization of the KIDLD gene encoding the mitochondrial enzyme D-lactate ferricytochrome c oxidoreductase. Mol Gen Genet. 1994;244:622–629. doi: 10.1007/BF00282752. [DOI] [PubMed] [Google Scholar]
- 25.Moller-Zinkhan D, Borner G, Thauer R K. Function of methanofuran, tetrahydromethanopterin, and coenzyme F420 in Archaeoglobus fulgidus. Arch Microbiol. 1989;152:362–368. [Google Scholar]
- 26.Nygaard A P. Lactate dehydrogenase of yeast. III. A comparative study of the kinetic properties and the stability of two isolated forms of the enzyme. Biochim Biophys Acta. 1960;40:85–92. doi: 10.1016/0006-3002(60)91317-2. [DOI] [PubMed] [Google Scholar]
- 27.Nygaard A P. Induction of D(−)- and L(+)-lactic cytochrome c reductase in yeast. J Biol Chem. 1961;236:1585–1588. [PubMed] [Google Scholar]
- 28.Ogata M, Arihara K, Yagi T. d-Lactate dehydrogenase of Desulfovibrio vulgaris. J Biochem. 1981;89:1423–1431. doi: 10.1093/oxfordjournals.jbchem.a133334. [DOI] [PubMed] [Google Scholar]
- 29.Olson S T, Massey V. Purification and properties of the flavoenzyme D-lactate dehydrogenase from Megasphaera elsdenii. Biochemistry. 1979;18:4714–4724. doi: 10.1021/bi00588a036. [DOI] [PubMed] [Google Scholar]
- 29a.Reed, D., K. Kashefi, and P. Hartzell. Unpublished data.
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 32.Segel I H. Biochemical calculations. New York, N.Y: John Wiley & Sons, Inc.; 1976. [Google Scholar]
- 33.Speich N, Dahl C, Heisig P, Klein A, Friedrich L, Stetter K O, Truper H G. Adenylylsulphate reductase from the sulphate-reducing archaeon Archaeoglobus fulgidus: cloning and characterization of the genes and comparison of the enzyme with other iron-sulphur flavoproteins. Microbiology. 1994;140:1273–1284. doi: 10.1099/00221287-140-6-1273. [DOI] [PubMed] [Google Scholar]
- 34.Speich N, Truper H G. Adenylylsulphate reductase in a dissimilatory sulphate-reducing archaebacterium. J Gen Microbiol. 1988;134:1419–1425. [Google Scholar]
- 35.Stetter K O. Archaeoglobus fulgidus gen. nov., sp. nov.: a new taxon of extremely thermophilic Archaebacteria. Syst Appl Microbiol. 1988;10:172–173. [Google Scholar]
- 36.Tindall B J, Stetter K O, Collins M D. A novel, fully saturated menaquinone from the thermophilic sulphate-reducing archaebacterium Archaeoglobus fulgidus. J Gen Microbiol. 1989;135:693–696. [Google Scholar]
- 37.Wierenga R K, Terpstra P, Hol W G J. Prediction of the occurrence of the ADP-binding βaβ-fold in proteins using an amino acid sequence fingerprint. J Mol Biol. 1986;187:101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]
- 38.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]