Abstract
The epithelial-to-mesenchymal transition (EMT) has been recognized as a driving force for tumor progression in breast cancer. Recently, our group identified the RNA Binding Motif Single Stranded Interacting Protein 3 (RBMS3) to be significantly associated with an EMT transcriptional program in breast cancer. Additional expression profiling demonstrated that RBMS3 was consistently upregulated by multiple EMT transcription factors and correlated with mesenchymal gene expression in breast cancer cell lines. Functionally, RBMS3 was sufficient to induce EMT in two immortalized mammary epithelial cell lines. In triple-negative breast cancer (TNBC) models, RBMS3 was necessary for maintaining the mesenchymal phenotype and invasion and migration in vitro. Loss of RBMS3 significantly impaired both tumor progression and spontaneous metastasis in vivo. Using a genome-wide approach to interrogate mRNA stability, we found that ectopic expression of RBMS3 upregulates many genes that are resistant to degradation following transcriptional blockade by actinomycin D (ACTD). Specifically, RBMS3 was shown to interact with the mRNA of EMT transcription factor PRRX1 and promote PRRX1 mRNA stability. PRRX1 is required for RBMS3-mediated EMT and is partially sufficient to rescue the effect of RBMS3 knockdown in TNBC cell lines. Together, this study identifies RBMS3 as a novel and common effector of EMT, which could be a promising therapeutic target for TNBC treatment.
INTRODUCTION
The epithelial-to-mesenchymal transition (EMT) is critical for development of many tissues and organs in the developing embryo [1, 2]. Accumulated experimental evidence has suggested that EMT can be pathologically activated in epithelial cancer cells, rendering cells with malignant and stem cell traits [3–6]. Consequently, EMT has been proposed as a driving force of metastatic progression [7, 8]. Unraveling the molecular mechanism shared among multiple EMT programs could facilitate the clinical application of EMT-targeting strategies [9–11].
The EMT is transcriptionally regulated by several well-characterized EMT transcription factors (EMT-TFs) [12]. While individual EMT-TFs are sufficient to induce EMT, only some EMT-TFs are consistently upregulated during EMT [13]. Despite a shared capacity to induce EMT, EMT-TFs exhibit substantial functional differences in normal physiology and cancer progression. For example, Twist1 is overexpressed in mammary atypical ductal hyperplasia in both mouse models and human in situ ductal carcinomas [14], whereas Snail expression is associated with metastatic progression [15]. Snail and Slug, but not TWIST1 and ZEB1, were shown to mediate resistance to mechanistic target of rapamycin kinase inhibitors [16]. Snail was also the only EMT-TF that was significantly increased in paclitaxel-resistant triple-negative breast cancer (TNBC) cell lines [17]. Previous data have suggested that different EMT-TF-driven programs converge on the activation or repression of a set of “common” EMT-associated genes [13]. ZEB1 is one example of a common EMT-inducing factor that has been demonstrated to be causally necessary for the induction of EMT [18]. Due to the partial redundancy between EMT-TFs, common downstream effectors of the EMT would provide the best therapeutic targets for inhibiting EMT-driven metastasis.
We hypothesized that genes reproducibly associated with EMT across cell lines and human tumor samples would more likely be regulators of EMT. A multi-step pipeline was incorporated to integrate expression data from normal cell models of EMT, breast cancer cell lines, and human breast cancer patients. This analysis identified the RNA Binding Motif Single Stranded Interacting Protein 3 (RBMS3) as a commonly upregulated gene that is reproducibly correlated with EMT in multiple cancers. Notably, previous publications have demonstrated RBMS3 is functionally linked to EMT-associated processes like craniofacial development and hepatic fibrosis [19, 20].
We determined that RBMS3 plays a critical role in EMT and cell motility in human mammary epithelial cells and TNBC cell models. These effects translated to markedly impaired local tumor growth and spontaneous lung metastasis in vivo in the MDA-MB231 cell line with RBMS3 knockdown. Using a genome-wide approach, we identified a group of RNA transcripts that were stabilized due to RBMS3 expression. Specifically, we showed that RBMS3 interacts with and stabilizes PRRX1 mRNA. Finally, we demonstrated that PRRX1 is required for RBMS3-mediated EMT and can partially rescue the effects of RBMS3 knockdown.
RESULTS
RBMS3 expression is correlated with EMT in multiple models
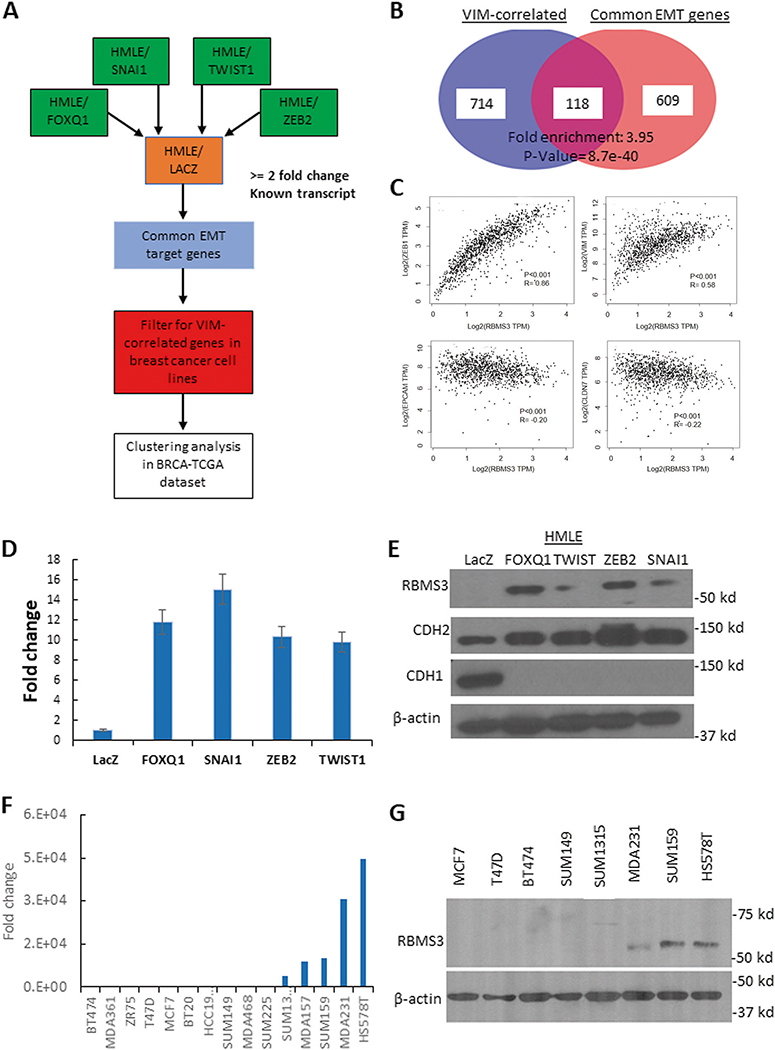
To identify genes that were reproducibly associated with EMT across multiple models, we performed a three-step analytic process (Fig. 1A). First, we performed a microarray analysis of differential gene expression in four human mammary epithelial cell line (HMLE)-based models of EMT. In each model, one of four EMT-TFs (FOXQ1, TWIST1, ZEB2, or SNAI1) were ectopically overexpressed in the HMLE cell line to induce EMT [4]. This result identified 727 genes significantly upregulated in all four HMLE/EMT models relative to the epithelial control cell line (HMLE/LacZ). Separately, using gene expression data from 51 breast cancer cell lines, we calculated the Spearman correlation coefficient between the well-characterized mesenchymal marker vimentin (VIM) with every gene in the database [21, 22]. We selected all genes which had a biologically relevant correlation (rho ≥ 0.5) with VIM, yielding a total of 832 genes. These two datasets were integrated, identifying a core set of 118 genes that were both upregulated by multiple EMT-TFs and significantly correlated with VIM in breast cancer cell lines (Fig. 1B and Supplementary Table 1). Finally, we evaluated the correlations between each gene in the Cancer Genome Atlas breast cancer expression dataset (n = 1088). After calculating the correlation between genes, the correlation matrix was subjected to unsupervised hierarchical clustering by the pairwise correlation coefficients. Most of the genes were significantly positively correlated, forming several smaller clusters (Supplementary Fig. 1A).
Fig. 1. RBMS3 is commonly upregulated during EMT and associated with EMT-like gene expression in breast cancer cells.
A A schematic illustration of the multi-step pipeline for identification of common EMT downstream effectors. B Integrative analysis of VIM- associated genes and common EMT genes. C Scatterplot illustrating the correlations between RBMS3 and VIM, ZEB1, EPCAM and CLDN7 in the TCGA breast cancer cohort. D RBMS3 mRNA expression was measured in HMLE/LacZ, /FOXQ1, TWIST1, ZEB2 and SNAI1 cell lines with qPCR. E Western blot analysis for RBMS3, CDH2, and CDH1 expression in control and HMLE/EMT cell models. F RBMS3 mRNA expression was measured by qPCR in a panel of breast cancer cell lines. G Western blot analysis for RBMS3 expression in a subset of breast cancer cell lines.
Within these smaller clusters, we noted a small grouping of genes around the core EMT-TF ZEB1 (Supplementary Fig. 1B). It has been reproducibly shown that ZEB1 is upregulated across diverse models of EMT [23]. We noted that one of the genes in this cluster, RBMS3, had been previously identified by our group as one of 24 genes that were both highly correlated with ZEB1 and transcriptionally regulated by ZEB1 in breast cancer [24]. Additional correlation analysis using the TCGA breast cancer dataset demonstrated that RBMS3 was significantly positively correlated with both ZEB1 and VIM, while showing significant inverse correlations with the epithelial markers CLDN7 and EPCAM (Fig. 1C). A separate analysis confirmed that high RBMS3 expression significantly co-occurred with high expression of multiple EMT-TFs in this dataset (Supplementary Fig. 1C).
To validate RBMS3 upregulation during EMT, we measured the expression of RBMS3 in both the HMLE/EMT models and a subset of human breast cancer cell lines. qPCR and western blotting confirmed that RBMS3 was upregulated in all four models of EMT (Fig. 1D, E). We also confirmed that RBMS3 was specifically highly expressed in the mesenchymal TNBC cell lines MDA-MB231, SUM-159, and HS.578T relative to epithelial ER+ and other TNBC cell lines (Fig. 1F, G). The RBMS3 expression pattern was consistent with the expression of multiple EMT TFs across breast cancer cell lines (Supplementary Fig. 1D). Finally, we evaluated the correlation between RBMS3, ZEB1, and VIM expression in eight other epithelial tumor types with data obtained from The Cancer Genome Atlas. As predicted, RBMS3 was consistently positively correlated with both ZEB1 and VIM in all tumor types examined, confirming that RBMS3 is reproducibly associated with mesenchymal gene expression in human cancers (Supplementary Fig. 1E, F).
RBMS3 induces an EMT and promotes cell migration and invasion
The consistent association between RBMS3 expression and mesenchymal differentiation suggested that RBMS3 may regulate the EMT program. To test this hypothesis, RBMS3 was ectopically expressed in the HMLE cell line. RBMS3 expression markedly increased the expression of the mesenchymal markers FN1, VIM, and CDH2 while suppressing the expression of CDH1 (Fig. 2A, B and Supplementary Fig. 2A). A colony growth assay demonstrated that RBMS3 expression disrupted normal epithelial colony formation, consistent with the cells having undergone EMT (Fig. 2C). By q-PCR, HMLE/RBMS3 cells exhibited a marked decrease in CDH1 mRNA expression concurrent with a substantial increase in the mRNA expression of FN1, VIM, and CDH2 (Supplementary Fig. 2B).
Fig. 2. RBMS3 induces EMT and maintains mesenchymal differentiation.
A Western blot analysis for RBMS3, mesenchymal markers VIM and FN1, and epithelial marker CDH1 in the HMLE/LacZ and HMLE/RBMS3 cell models. B Immunofluorescence analysis for CDH1, and VIM in HMLE/LacZ and /RBMS3 cell lines. Scale bar, 100 μm. C A colony formation assay compares the cell morphology of HMLE/LacZ and HMLE/RBMS3 cells. Scale bar, 300 μm D Cell proliferation in the HMLE/LacZ and /RBMS3 cell lines was measured using the Sulforhodamine B assay. Three-dimensional cell migration (E, G) and invasion (F, G) were significantly increased in the HMLE/RBMS3 cell line compared to HMLE/LacZ. Scale bar: 100 μm.
Functionally, ectopic RBMS3 expression did not have a significant effect on cell proliferation (Fig. 2D). However, ectopic RBMS3 expression significantly increased both three-dimensional migration and invasion in the HMLE cell line (Fig. 2E–G). Enforced expression of RBMS3 also led to a fivefold increase in the proportion of CD44hi/CD24low cells by FACS, a correlate for EMT-induced stemness (Supplementary Fig. 2C). Consistent with this observation, RBMS3 expression also significantly increased the mammosphere formation capacity of HMLE cells (Supplementary Fig. 2D).
To confirm our observations in the HMLE cell model, we ectopically expressed RBMS3 in another immortalized mammary epithelial cell line, MCF10A. We observed similar results, including apparent EMT at both the molecular and morphological levels (Supplementary Fig. 3A, B). In addition, ectopic RBMS3 expression did not change the cell proliferation in the first three days (Supplementary Fig. 3C), but significantly increased cell migration and invasion in MCF10 A cells (Supplementary Fig. 3D, E).
RBMS3 maintains mesenchymal status and migration and invasion in TNBC
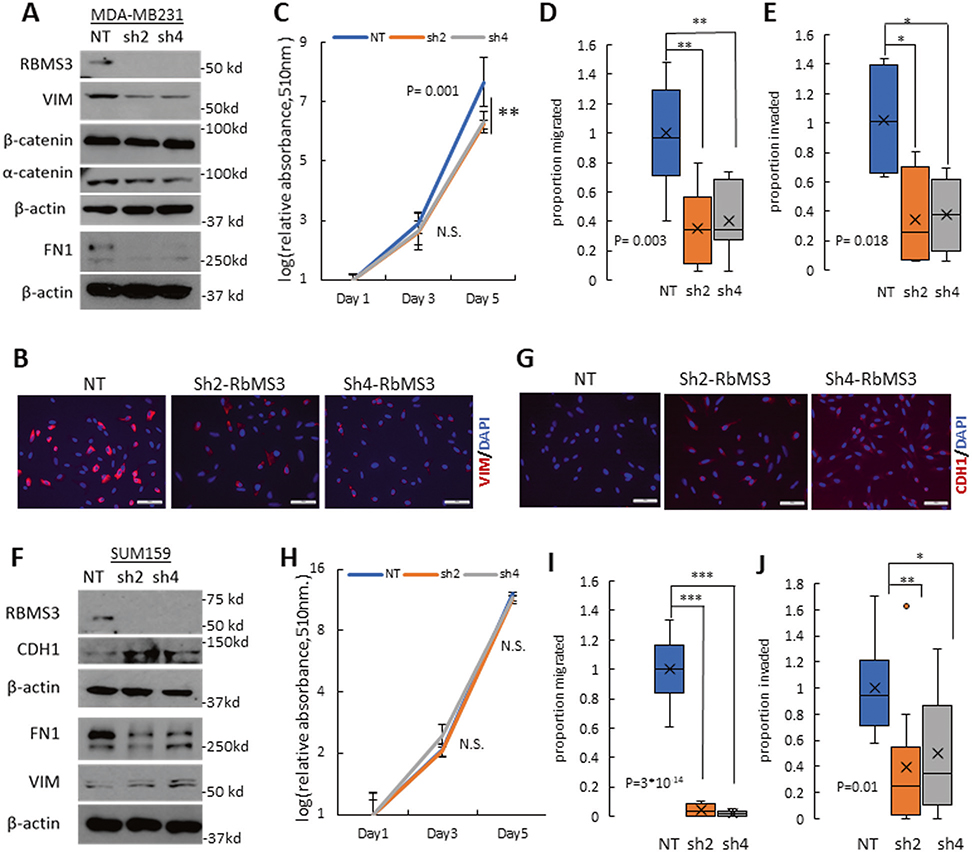
Next, shRNA-mediated RBMS3 knockdown models were generated in the MDA-MB231 and SUM159 breast cancer cell lines, which exhibited high endogenous expression of RBMS3 as shown in Fig. 1. In the MDA-MB231 cell line, knockdown of RBMS3 significantly reduced the expression of FN1 and VIM, modestly decreased the expression of α-catenin, and did not affect the expression of β-catenin (Fig. 3A and Supplementary Fig. 4A). Consistent with previous reports, we were unable to detect CDH1 protein in this cell line [25]. Immunofluorescence analysis confirmed a decrease in the protein expression of VIM upon RBMS3 knockdown (Fig. 3B). In addition, RBMS3 knockdown led to a modest reduction in cell proliferation in vitro (Fig. 3C). RBMS3 knockdown also significantly decreased both three-dimensional cell migration and invasion (Fig. 3D, E and Supplementary Fig. 4B).
Fig. 3. RBMS3 is required for mesenchymal gene expression and cell migration/invasion in TNBC.
A Western blot analysis for RBMS3, FN1, VIM, α-catenin, and β-catenin in MDA-MB231/NT, sh2-RBMS3, and sh4-RBMS3 cell lines. B Immunofluorescence analysis showed decreased expression of Vimentin in MDA MB231 cells with RNMS3 knockdown. Scale bar: 100 μm. C Proliferation assay comparing MDA-MB231/NT, sh2-RBMS3, and sh4-RBMS3 cell lines over 5 days. Migration (D) and invasion (E) assay for MDA231/NT, sh2-RBMS3, and sh4-RBMS3 cell lines. F Western blot analysis for RBMS3, FN1, VIM, and CDH1 in SUM159 /NT, sh2-RBMS3, and sh4-RBMS3 cell lines. G Immunofluorescence analysis showed upregulation of E-cadherin in SUM159 cells with RNMS3 knockdown. Scale bar: 100 μm. H Proliferation assay comparing SUM159/NT, sh2-RBMS3 and sh4-RBMS3 cell lines over 5 days. Migration (I) and invasion (J) assays for SUM-159/NT, sh2-RBMS3, and sh4-RBMS3 cell lines.
In the SUM159 cell line, knockdown of RBMS3 decreased the expression of FN1 and increased the expression of CDH1 (Fig. 3F, G and Supplementary Fig. 4C). RBMS3 knockdown had no significant effect on cell proliferation in vitro (Fig. 3H). However, loss of RBMS3 significantly impaired both cell migration and invasion, consistent with the other cell models (Fig. 3I, J and Supplementary Fig. 4D).
Finally, the effect of RBMS3 knockdown on stemness was evaluated in each cell line. After knockdown of RBMS3, the proportion of CD44high/CD24low cells was not affected in the MDA-MB231 cell line but was slightly decreased in the SUM159 cell line (Supplementary Fig. 4E, F). No significant difference was observed in mammosphere formation capacity in either model, although RBMS3 knockdown cells exhibited a trend towards lower numbers of mammosphere formation (Supplementary Fig. 4G, H). These results indicate that RBMS3 is necessary for maintaining mesenchymal differentiation and cell migration and invasion, but not stemness, in mesenchymal TNBC cell lines.
RBMS3 regulates tumor progression and spontaneous metastasis in vivo
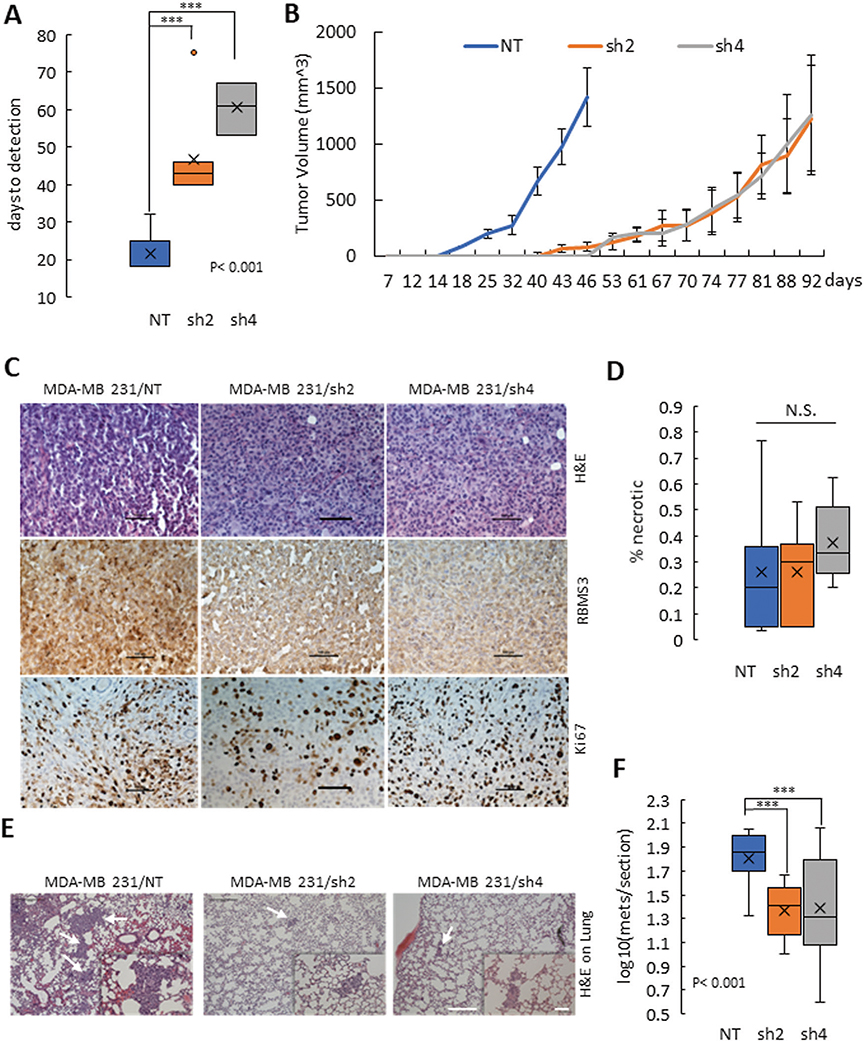
As RBMS3 regulated cell migration and invasion in vitro, we hypothesized that RBMS3 might also regulate tumor progression in vivo. 2.5 × 105 cells of MDA-MB231/NT, /sh2, or /sh4 RBMS3 cells were injected into the mammary fat pad of female Nod-Scid-Gamma (NSG) mice. We observed rapid tumor onset in the control model, with a median time to tumor measurability of 18 days. Both knockdown models exhibited significantly delayed tumor onset (43–61 days) (Fig. 4A). Tumor growth was also impaired by RBMS3 knockdown, as the control mice reached the humane endpoint (average tumor weight ~1.5 g) 46 days after injection, while both knockdown models reached the endpoint at 92 days post-injection (Fig. 4B).
Fig. 4. RBMS3 is required for tumor progression and spontaneous metastasis in vivo.
A Comparison of days to tumor measurability between MDA-MB231/NT, /sh2-RBMS3, and /sh4- RBMS3. B The tumor growth curve over time for the MDA-MB231/NT, /sh2-RBMS3 and /sh4- RBMS3 groups. C Primary tumor sections stained for H&E (up panels), RBMS3 (middle panels), and Ki67 expression (low panels). Scale bar: 100 μm. D Summary of necrosis lesion in tumors derived from MDA-MB231/NT, /sh2-RBMS3 and /sh4-RBMS3 cells. E Representative images of metastatic lesions in the three groups. White arrows identify metastatic lesions. F The number of metastatic lesions per lung section in the MDA-MB231/NT, /sh2-RBMS3, and sh4-RBMS3 groups. For both C and E, scale bar, 500 μm for 40X magnification and 100 μm for 200× magnification.
Intratumoral morphology appeared similar by H&E staining (Fig. 4C, top panels. IHC staining for RBMS3 confirmed lower RBMS3 expression in both knockdown tumors than the nontarget control (Fig. 4C, middle panels). Consistent with the in vitro results, the tumor proliferation was not significantly changed upon RBMS3 knockdown, as determined by Ki-67 staining (Fig. 4C, bottom panels). In addition, tumor necrosis was unaffected by RBMS3 knockdown (Fig. 4D). We next examined the angiogenesis status using α-Smooth Muscle Actin (α-SMA) antibody. The tumors originated from two knockdown cell models showed less intensive α-SMA staining in the middle of the tumors than those tumors from cells with NT clones (Supplementary Fig. 5). Consequently, we hypothesize that the effect of RBMS3 on tumor latency and growth likely resulted from impaired migration, invasion, and angiogenesis rather than a direct effect on cell proliferation.
To determine the effect of RBMS3 knockdown on spontaneous metastasis, the lungs from each mouse were removed, sectioned, H&E stained, and examined via microscopy. We observed significantly fewer metastatic lesions in mice engrafted with RBMS3 knockdown cell lines relative to the control, despite the longer time frame available for metastases to develop in the knockdown models (Fig. 4E, F).
To investigate if RBMS3 expression was associated with metastatic progression in breast cancer patients, a tissue microarray containing samples from primary breast tumors of 75 patients was stained for RBMS3 and scored. We observed that patients with high RBMS3 expression were significantly more likely to have developed both nodal and distant organ metastasis (Supplementary Fig. 6). RBMS3 was not associated with tumor stage, grade or patient age (data not shown).
RBMS3 specifically regulates the expression of PRRX1
As RBMS3 is known to be an RNA binding protein, we investigated whether RBMS3 promoted EMT by stabilizing the RNA expression of EMT-related genes. We treated HMLE/Lac Z and HMLE/RBMS3 cells for 0, 90, 180, and 360 min with actinomycin D (ACTD), which prevents transcription and allows the measurement of mRNA decay over time. RNA was collected, purified, and subjected to mRNA sequencing. Differential expression (DE) analysis was performed to identify genes that were significantly upregulated in the RBMS3 cell model, with 1669 DE genes identified across all four-time points. This gene set was filtered to identify a subset of 179 genes with a mean of normalized counts in control less than 10% of the maximum of expression in cells with RbMS3. Next, we performed unsupervised clustering on the 179 gene set, and identified twelve clusters with sizes ranged from 9 to 30 genes (Fig. 5A and Supplementary Fig. 7). Among the identified 12 clusters, cluster 4 included 21 genes that were monotonically upregulated over 4 fixed time points, indicating a high level of mRNA stability (Fig. 4B). In these 21 genes, at least ten genes were previously reported to be related to EMT including PRRX1, FOXC2, TBX18, DDR2, and MLPH (Supplementary Table 2). PRRX1 was one of the EMT transcription factors shown to be most significantly stabilized by RBMS3 (Fig. 5C). A literature search identified a previous study that implicated RBMS3 as a direct regulator of PRRX1 expression during hepatic fibrosis by binding to PRRX1 mRNA, suggesting that PRRX1 may be a critical functional target of RBMS3 regulation [19].
Fig. 5. RBMS3 regulates the expression and stability of PRRX1 mRNA.
A A schematic illustration of the strategy for identification of genes stabilized by RBM3. B Standardized log2 fold-change, between RBMS3 and control, of genes (n = 21) in cluster 4. The red lines indicate individual gene standardized log2 fold-change trajectory, the blue line is the mean of the genes within the cluster, and the light-blue band represents ± 2 standard deviations from the cluster mean. C Normalized expression counts for PRRX1 in control and RbMS3 treated cells separately across four time points. D qPCR measurements of EMT-TF expression in the HMLE/LacZ and /RBMS3 cell lines demonstrate increased expression of SNAI1 and PRRX1. E Western blotting for PRRX1 in the HMLE/LacZ and /RBMS3 cell lines confirms increased PRRX1 expression. F Western blot analysis for PRRX1 expression in the SUM159 and MDA-MB 231 cell lines shows a decreased expression of PRRX1 upon shRBMS3 knockdown. Correlation analysis of RBMS3 and PRRX1 expression in the CCLE breast cancer dataset (G) and human breast tumor expression dataset (TCGA) (H). I RNA immunoprecipitation against V5-tagged RBMS3 in the HMLE/RBMS3 cell line significantly enriches PRRX1 mRNA. J Actinomycin D treatment over 12 h identifies an increase in PRRX1 mRNA stability in the HMLE/RBMS3 cell line relative to HMLE/LacZ. K Actinomycin D treatment over 12 h demonstrates a decrease in PRRX1 mRNA stability in the MDA-MB231 cell line upon knockdown of RBMS3.
To confirm the relationship between RBMS3 and PRRX1 expression, the expression of known EMT-TFs in the HMLE/RBMS3 cell line was evaluated by qPCR. PRRX1 was identified as the most significantly upregulated EMT-TF by RBMS3 (Fig. 5D). Upregulation of PRRX1 was confirmed by western blot analysis (Fig. 5E). Next, Western blot analysis demonstrated that RBMS3 knockdown in both SUM-159 and MDA-MB231 cell lines significantly downregulated PRRX1 expression (Fig. 5F). An analysis of the breast cancer cell line expression dataset (CCLE) and human breast tumor expression dataset (TCGA) confirmed that RBMS3 and PRRX1 expression were significantly correlated in both datasets (Fig. 5G and H). A reproducibly strong correlation between RBMS3 and PRRX1 expression was further confirmed in multiple epithelial cancer models (Supplementary Fig. 8). These data supported the hypothesis that PRRX1 may be a target of regulation by RBMS3 in breast and other human cancer types.
Next, we confirmed direct RBMS3 binding to PRRX1 mRNA with RNA immunoprecipitation. Using a ChIP-grade antibody against the V5 tag, we pulled down RBMS3 in the HMLE/RBMS3 cell line and probed for PRRX1 expression. We found significant enrichment for the PRRX1 mRNA, suggesting that RBMS3 interacts with PRRX1 mRNA (Fig. 5I). Finally, we evaluated the effect of RBMS3 expression on PRRX1 stability with an ACTD treatment. We observed that ectopic RBMS3 expression increased the stability of PRRX1 mRNA (Fig. 5J). Conversely, knockdown of RBMS3 reduced PRRX1 stability in the MDA-MB231 cell line (Fig. 5K). These data indicate that RBMS3 positively regulates PRRX1 expression by post-transcriptional stabilization of PRRX1 mRNA.
PRRX1 is required for RBMS3-mediated EMT
To confirm the functional significance of the RBMS3/PRRX1 axis, PRRX1 was knocked down in the HMLE/RBMS3 cell line with shRNA. Knockdown of PRRX1 restored the expression of CDH1 and decreased the expression of VIM and CDH2, suggesting that PRRX1 is required for RBMS3-mediated EMT (Fig. 6A). The western blot analysis was repeated for a second time and quantitation showed a similar pattern (Supplementary Fig. 9A, B). The reestablishment of E-cadherin cell junctions was confirmed by immunofluorescence analysis (Fig. 6B). Moreover, knockdown of PRRX1 caused a marked reversion toward epithelial colony formation (Fig. 6C). We observed no significant change in cell proliferation over 3 days (Fig. 6D), but a significant decrease in both cell migration and invasion upon knockdown of PRRX1 (Fig. 6E–H). To exclude a non-specific effect of PRRX1 knockdown, we also checked the effect of PRRX1 knockdown in HMLE/LacZ cells and observed no changes in EMT marker expression, cell proliferation, or cell migration and invasion (Supplementary Fig. 10A–D).
Fig. 6. PRRX1 is a critical effector of RBMS3-mediated EMT and cell motility in human mammary epithelial cells.
A Western blot analysis for the mesenchymal markers VIM and CDH2, the epithelial marker CDH1, RBMS3, and PRRX1 in the HMLE/RBMS3-NT and -shPRRX1 cell lines. B Immunofluorescence analysis for CDH1 demonstrates restoration of CDH1 expression and membrane localization. Scale bar, 100 μm. C A colony assay shows a reversion to an epithelial colony morphology in HMLE/RBMS3 cells with PRRX1 knockdown. Scale bar, 300 μm. D Cell proliferation in the HMLE/RBMS3-NT and HMLE/RBMS3-shPRRX1 cell lines was measured by Sulforhodamine B assay. Knockdown of PRRX1 reduces the migratory (E, F) and invasive (G, H) capability of HMLE/RBMS3 cells with or without PRRX1 knockdown.
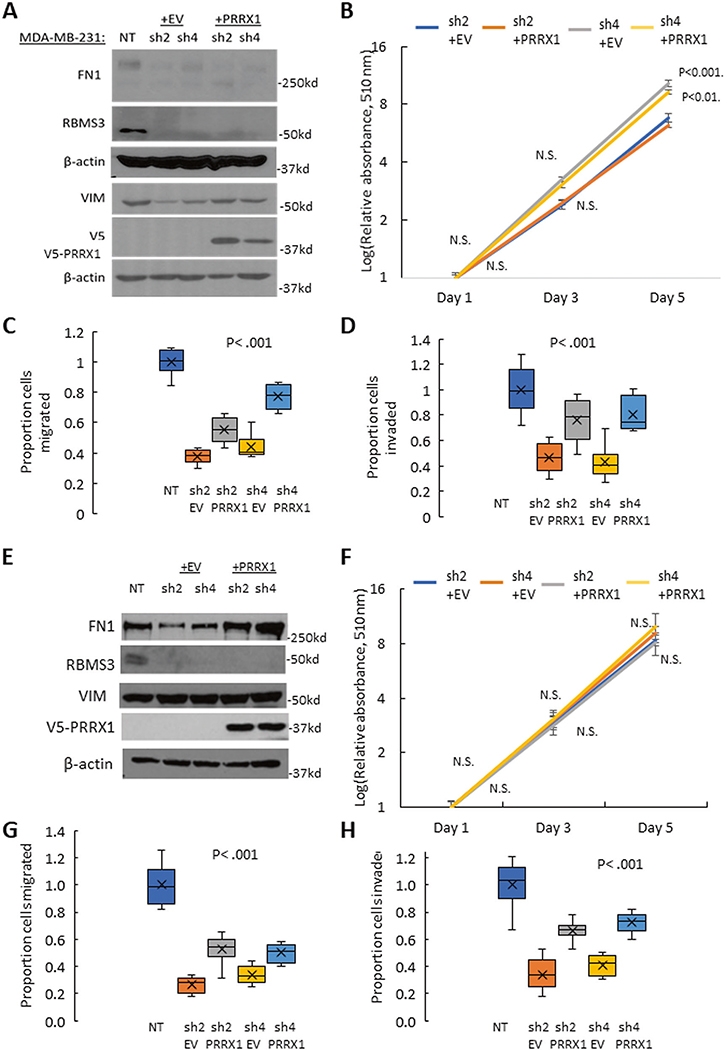
Next, PRRX1 was ectopically re-expressed in MDA-MB231/shRBMS3 and SUM159/shRBMS3 cell models to test whether PRRX1 could rescue the EMT phenotype. Re-expression of PRRX1 increased the expression of VIM, but did not affect FN1 expression in MDA-MB231 cells (Fig. 7A). The western blot analysis was repeated for a second time, and quantitation showed a similar pattern (Supplementary Fig. 9C, D). We also observed no significant change in cell proliferation in the first 3 days (Fig. 7B), but a partially restored migration and invasion capacity in the RBMS3-knockdown cell lines with PRRX1 rescuing (Fig. 7C, D and Supplementary Fig. 11A, B). In parallel, we expressed PRRX1 in SUM159/shRBMS3 cell models to confirm the results obtained in MDA-MB231 cells. We discovered that re-expression of PRRX1 markedly increased FN1 expression (Fig. 7E). Similarly, PRRX1 expression did not alter cell proliferation (Fig. 7F), but partially reversed cell migration and invasion significantly (Fig. 7 G, and H and Supplementary Fig. 11C, D).
Fig. 7. PRRX1 plays a vital role in RBMS3-mediated EMT and motility in TNBC cells.
A Western blotting demonstrates that re-expression of PRRX1 in MDA-MB231/shRBMS3 cells restores expression of VIM. B Cell proliferation in the MDA-MB231/shRBMS3 with or without PRRX1 rescuing (+EV or +PRRX1) cell lines was measured by Sulforhodamine B assay. Re-expressing PRRX1 in MDA-MB231/shRBMS3 cells partially rescues migratory (C) and invasive (D) capability. E Western blotting demonstrates that re-expression of PRRX1 in SUM159/shRBMS3 cells restores expression of Fibronectin. F Cell proliferation in the SUM159/shRBMS3 with or without PRRX1 overexpression (+EV or +PRRX1) cell lines was measured by Sulforhodamine B assay. Re-expressing PRRX1 in SUM159/shRBMS3 cells partially rescues migratory (G) and invasive (H) capability.
DISCUSSION
Based on these results, we propose a working model for RBMS3 (Fig. 8). RBMS3 is upregulated during EMT in normal epithelial cells and then acts to upregulate PRRX1, supporting the cell transition to a mesenchymal phenotype. In mesenchymal TNBC cells, high expression of RBMS3 modulates cell migration, invasion, and tumor progression by stabilizing and maintaining PRRX1 expression.
Fig. 8. The RBMS3 working model.
Schematic illustration of the RBMS3 function in mammary epithelial cells and breast cancer cells.
Multiple groups have shown that PRRX1 is critical for maintaining EMT and both migratory and invasive capabilities in mesenchymal cancer cell models [26–29]. Our results indicate that PRRX1 is stabilized by RBMS3 and plays an essential role in RBMS3-mediated EMT. More importantly, RBMS3 likely regulates many other mediators of EMT and cell migration and invasion according to our RNA-seq analysis based on cells with ACTD treatment. Recently, a genome-wide study showed that RBMS3 regulates the mRNA stability and expression of several Wnt-pathway regulatory genes, including AXIN1, DKK3, and NFAT5, in ovarian cancer [30]. More in-depth studies in relevant breast and breast cancer cell models are underway in our group to elucidate the global direct RBMS3-regulome and identify additional critical EMT-linked targets.
The physiological functions of RBMS3 are unclear. It was previously reported that loss of RBMS3 dramatically impaired craniofacial development in zebrafish [20]. RBMS3 was also shown to be upregulated during hepatic fibrosis and to contribute to mouse pancreatic development by regulating the stability and expression of Ptf1a [19, 31]. As these developmental and physiologic processes depend on EMT, these findings suggest that RBMS3 may have an evolutionarily-conserved function in promoting EMT in both physiologic and pathologic settings [2].
RBMS3 has been implicated as a tumor suppressor in human gastric cancer, lung squamous carcinoma, esophageal and nasopharyngeal carcinoma [32–35]. It has been observed that EMT factors can act as either tumor suppressors or oncogenes depending on the tissue-specific context [36]. For example, FOXQ1 is a well-recognized oncogene in multiple carcinomas [37, 38], but suppresses EMT, invasion, and metastasis in melanoma cells [39]. Similarly, SNAIL2 and ZEB2 transcription factors act as oncogenes in different human carcinomas but can behave as tumor-suppressor proteins by activating a MITF-dependent melanocyte differentiation program in melanoma [40]. Like other EMT regulatory factors, RBMS3 effects on tumor phenotype are probably context dependent.
Interestingly, a previous functional study suggested that RBMS3 is a tumor suppressor in breast cancer [41]. This study found that overexpressing RBMS3 in the MDA-MB231 cell line decreased invasion and migration in vitro and metastasis in a tail-vein injection in vivo model. This study also suggested that RBMS3 destabilized TWIST1 mRNA, leading to its downregulation. In our hands, we observed no significant change in TWIST1 expression upon ectopic RBMS3 expression (Fig. 5A). Based on our results, we suggest that the effects observed by Zhu et al. were likely due to overexpressing a pro-mesenchymal gene in an already mesenchymal cell, which may have impaired cell viability and driven compensatory reduction in the expression of other mesenchymal genes, like TWIST1. We also note that prior studies failed to identify the reproducible association between RBMS3 and EMT, which may have impacted expectations before performing functional studies.
There is a discrepancy in our study between the significant delay in tumor progression in vivo and minimal decrease in cell proliferation in vitro for MDA MB231 cells with RBMS3 knockdown. We believe multiple factors may contribute to this discrepancy. First, the reduction in migratory and invasive capacity following RBMS3 knockdown could impede early tumor establishment and growth. We also observed that RBMS3-knockdown tumors demonstrated lower levels of angiogenesis than the tumors derived from control cells. Finally, our recent work demonstrated that an RBMS3-associated transcriptional network is inversely correlated with antitumor immune activation in human breast cancer samples [24]. Future work from our group will investigate the relationship between RBMS3 activity and the tumor immune response using fully immunocompetent mouse models.
While RBMS3 has been demonstrated to enhance post-transcriptional mRNA stability across several studies, the precise mechanism of this activity remains unclear. RBMS3, unlike its closely related family members RBMS1 and 2, is restricted to the cytoplasm and has only been characterized as a post-transcriptional RNA regulatory factor [42]. RBMS3 has not been shown to interact with known regulators of mRNA stability, nor to localize to RNA-protein granules. Thus, further study is required to elucidate the mechanism by which RBMS3 mediates increased transcript stability. By understanding this mechanism, it may be possible to target RBMS3 to inhibit EMT and metastatic progression in breast and other cancers.
MATERIALS AND METHODS
Cell culture
Breast cancer cell lines MCF7, T47D, BT474, ZR75, MDA361, BT20, HCC1957, MDA-MB468, MDA-MB157, MDA-MB231, and HS.578T were all obtained from ATCC. They were all grown according to ATCC-recommended culture conditions and were monitored for mycoplasma. SUM-1315, SUM225, SUM149, and SUM-159 cells were gifts from Dr. Stephen P. Ethier at MUSC. The HMLE cell line was a gift from Dr. Robert Weinberg at MIT. All cell lines were authenticated upon receipt by comparing them to the original morphological and growth characteristics and cultured as previously described [38, 43, 44]. All the EMT cell models were generated in our lab by ectopic expression of EMT-TFs in HMLE cells using lentiviral infection [4].
qPCR, western blot, and Immunofluorescence analysis
All these analyses were performed as previously described [38, 43, 44]. Primer sequences are included in Supplementary Table 3. Antibodies are listed in Supplementary Table 4.
RNA immunoprecipitation and actinomycin D treatments
Cells were lysed in RIP buffer. Anti-V5 beads, or agarose/Protein G beads incubated with non-specific IgG, were washed in PBS. The cell lysate was added to the beads and incubated overnight at 4 degrees Celsius with agitation. Beads were spun down and washed three times. Trizol was added to the beads and RNA was extracted. cDNA was prepared as described and used for qPCR-based measurement of PRRX1 mRNA levels. To measure RNA stability, cells were treated with 10 μg/mL of Actinomycin D dissolved in DMSO for the indicated times. RNA was extracted with Trizol and converted to cDNA. 18S rRNA was used to normalize gene expression for comparison between conditions.
RNA sequencing and data analysis
To measure the global effect of RBMS3 on mRNA degradation, the HMLE/LacZ and HMLE/RBMS3 cells were treated with 10 μg/mL of Actinomycin D for four different time points (0, 90, 180, and 360 min). At each time point, three replicates were collected for each cell model. RNA was extracted with the RNeasy Plus mini kit (Qiagen) and 10 ng of each sample was subjected to RNA sequencing using a 3′ mRNA sequencing approach (quantSeq) [45].
Normalized counts were computed using a median of ratio approach within DESeq2 [46]. Genes with no detectable expression in control, i.e., zero normalized counts at all time-points in all replicates, were excluded from differential expression analysis. Gene DE analysis was performed separately at each of four-time points with threshold of adjusted p value (p. adj) < 0.05. Only genes showing significant DE across all four time-points were used for downstream analysis. We further narrowed down our focus to the genes that showed relatively low expression in control, defined as the mean of normalized counts less than 10% of the maximum expression of all RBMS3 samples. We performed clustering of those genes with similar log2 fold change trajectories over time using an infinite Gaussian process mixture model implemented in DP_GP [47].
Analysis of CCLE and TCGA data
Gene expression data were downloaded from the Cancer Cell Line Encyclopedia and imported as csv files into R (3.5.1). For correlation and clustering analyses, the correlation matrix was calculated and clustering by the first principal component was performed. Heatmaps were generated using the corrplot package. For analysis of TCGA expression data, the GEPIA2 server was used to calculate Spearman correlation coefficients and generate scatterplots [48].
Colony, proliferation, and invasion/migration assays
Approximately 50,000 cells were plated in a six-well dish. After 24 h, colonies were stained with sulforhodamine B and imaged on a microscope. Cell proliferation was measured by sulforhodamine B assay [49]. The Boyden invasion and migration chamber assay were performed as previously described [38, 43, 44].
In vivo tumor growth and metastasis assay
2.5 × 105 MDA-MB231/NT, /sh2-RBMS3, or /sh4-RBMS3 cells were injected into the mammary fat pad of female NSG mice, with eight mice per group. The MDA-MB231 cell line was selected due to its capacity to form spontaneous metastases in this mouse model [50, 51]. Tumor growth was monitored twice a week with caliper measurements. Mice were sacrificed when they reached a tumor burden of ~1.5 g. Primary tumors and lungs were collected from each mouse and sectioned. Three sections from each pair of mouse lungs were H&E stained and examined by light microscopy to quantify the number of metastases per section. Metastatic lesions were defined as a cluster of >4 metastatic cells.
Statistical analysis
For comparisons between two independent groups, a two-sided two-sample T-test was performed. For groups with more than two comparisons, an ANOVA with posthoc Tukey test was performed. If results were not normally distributed, data were log-transformed and then evaluated with the appropriate parametric test. The Spearman correlation coefficient was calculated for all gene association studies. The Fischer exact test or Chi-square test was used to evaluate the results obtained from the tissue microarray.
DATA AVAILABILITY
Microarray data that support the funding of this study have been deposited in the GEO under accession number GSE181322. RNA-seq data that support the finding of this study have been deposited in the GEO under accession number GSE181237.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr. Larry Matherly at the Karmanos Cancer Institute (KCI) and Wayne State University School of Medicine for helpful discussions and valuable comments. We also want to thank Dr. Robert A. Weinberg at MIT for providing the HMLE cell line.
FUNDING
This work is supported by Grant boost from Wayne State University (GW) and KCI strategic plan cancer immunology and immunotherapy award (GW and HG). The Biostatistics Core, AMTEC and the Biobanking and Correlative Sciences Core of KCI are supported by grant number P30-CA022453–38.
Footnotes
COMPETING INTERESTS
The authors declare no competing interests.
ADDITIONAL INFORMATION
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41388-021-02030-x.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
REFERENCES
- 1.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. [DOI] [PubMed] [Google Scholar]
- 3.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meng F, Speyer CL, Zhang B, Zhao Y, Chen W, Gorski DH, et al. PDGFRalpha and beta play critical roles in mediating Foxq1-driven breast cancer stemness and chemoresistance. Cancer Res. 2015;75:584–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–73. [DOI] [PubMed] [Google Scholar]
- 6.Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol. 2017;14:611–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aiello NM, Kang Y. Context-dependent EMT programs in cancer metastasis. J Exp Med. 2019;216:1016–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeung KT, Yang J. Epithelial-mesenchymal transition in tumor metastasis. Mol Oncol. 2017;11:28–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis FM, Stewart TA, Thompson EW, Monteith GR. Targeting EMT in cancer: opportunities for pharmacological intervention. Trends Pharm Sci. 2014;35:479–88. [DOI] [PubMed] [Google Scholar]
- 10.Marcucci F, Stassi G, De Maria R. Epithelial-mesenchymal transition: a new target in anticancer drug discovery. Nat Rev Drug Discov. 2016;15:311–25. [DOI] [PubMed] [Google Scholar]
- 11.Pasquier J, Abu-Kaoud N, Al Thani H, Rafii A. Epithelial to mesenchymal transition in a clinical perspective. J Oncol. 2015;2015:792182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stemmler MP, Eccles RL, Brabletz S, Brabletz T. Non-redundant functions of EMT transcription factors. Nat Cell Biol. 2019;21:102–12. [DOI] [PubMed] [Google Scholar]
- 13.Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci USA. 2010;107:15449–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morel AP, Hinkal GW, Thomas C, Fauvet F, Courtois-Cox S, Wierinckx A, et al. EMT inducers catalyze malignant transformation of mammary epithelial cells and drive tumorigenesis towards claudin-low tumors in transgenic mice. PLoS Genet. 2012;8:e1002723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blanco MJ, Moreno-Bueno G, Sarrio D, Locascio A, Cano A, Palacios J, et al. Correlation of Snail expression with histological grade and lymph node status in breast carcinomas. Oncogene. 2002;21:3241–6. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Ye Q, Cao Y, Guo Y, Huang X, Mi W, et al. Snail determines the therapeutic response to mTOR kinase inhibitors by transcriptional repression of 4E-BP1. Nat Commun. 2017;8:2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park SY, Kim MJ, Park SA, Kim JS, Min KN, Kim DK, et al. Combinatorial TGF-beta attenuation with paclitaxel inhibits the epithelial-to-mesenchymal transition and breast cancer stem-like cells. Oncotarget. 2015;6:37526–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Addison JB, Voronkova MA, Fugett JH, Lin CC, Linville NC, Trinh B, et al. Functional hierarchy and cooperation of EMT master transcription factors in breast cancer metastasis. Mol Cancer Res. 2021;19:784–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fritz D, Stefanovic B. RNA-binding protein RBMS3 is expressed in activated hepatic stellate cells and liver fibrosis and increases expression of transcription factor Prx1. J Mol Biol. 2007;371:585–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jayasena CS, Bronner ME. Rbms3 functions in craniofacial development by posttranscriptionally modulating TGF-beta signaling. J Cell Biol. 2012;199:453–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci USA. 2009;106:13820–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghandi M, Huang FW, Jane-Valbuena J, Kryukov GV, Lo CC, McDonald ER 3rd, et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature. 2019;569:503–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang P, Sun Y, Ma L. ZEB1: at the crossroads of epithelial-mesenchymal transition, metastasis and therapy resistance. Cell Cycle. 2015;14:481–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Block CJ, Dyson G, Campeanu IJ, Watza D, Ratnam M, Wu G. A stroma-corrected ZEB1 transcriptional signature is inversely associated with antitumor immune activity in breast cancer. Sci Rep. 2019;9:17807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lombaerts M, van Wezel T, Philippo K, Dierssen JW, Zimmerman RM, Oosting J, et al. E-cadherin transcriptional downregulation by promoter methylation but not mutation is related to epithelial-to-mesenchymal transition in breast cancer cell lines. Br J Cancer. 2006;94:661–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lv ZD, Kong B, Liu XP, Jin LY, Dong Q, Li FN, et al. miR-655 suppresses epithelial-to-mesenchymal transition by targeting Prrx1 in triple-negative breast cancer. J Cell Mol Med. 2016;20:864–73. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Ocana OH, Corcoles R, Fabra A, Moreno-Bueno G, Acloque H, Vega S, et al. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell. 2012;22:709–24. [DOI] [PubMed] [Google Scholar]
- 28.Takahashi Y, Sawada G, Kurashige J, Uchi R, Matsumura T, Ueo H, et al. Paired related homoeobox 1, a new EMT inducer, is involved in metastasis and poor prognosis in colorectal cancer. Br J Cancer. 2013;109:307–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takano S, Reichert M, Bakir B, Das KK, Nishida T, Miyazaki M, et al. Prrx1 isoform switching regulates pancreatic cancer invasion and metastatic colonization. Genes Dev. 2016;30:233–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu G, Cao L, Zhu J, Tan Z, Tang M, Li Z, et al. Loss of RBMS3 confers platinum resistance in epithelial ovarian cancer via activation of miR-126–5p/beta-catenin/CBP signaling. Clin Cancer Res. 2019;25:1022–35. [DOI] [PubMed] [Google Scholar]
- 31.Lu CK, Lai YC, Chen HR, Chiang MK. Rbms3, an RNA-binding protein, mediates the expression of Ptf1a by binding to its 3’UTR during mouse pancreas development. DNA Cell Biol. 2012;31:1245–51. [DOI] [PubMed] [Google Scholar]
- 32.Chen J, Kwong DL, Zhu CL, Chen LL, Dong SS, Zhang LY, et al. RBMS3 at 3p24 inhibits nasopharyngeal carcinoma development via inhibiting cell proliferation, angiogenesis, and inducing apoptosis. PLoS One. 2012;7:e44636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Chen L, Nie CJ, Zeng TT, Liu H, Mao X, et al. Downregulation of RBMS3 is associated with poor prognosis in esophageal squamous cell carcinoma. Cancer Res. 2011;71:6106–15. [DOI] [PubMed] [Google Scholar]
- 34.Wu Y, Meng D, You Y, Sun R, Yan Q, Bao J, et al. Increased expression of RBMS3 predicts a favorable prognosis in human gallbladder carcinoma. Oncol Rep. 2020;44:55–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang T, Wu Y, Fang Z, Yan Q, Zhang S, Sun R, et al. Low expression of RBMS3 and SFRP1 are associated with poor prognosis in patients with gastric cancer. Am J Cancer Res. 2016;6:2679–89. [PMC free article] [PubMed] [Google Scholar]
- 36.Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol. 2014;16:488–94. [DOI] [PubMed] [Google Scholar]
- 37.Li Y, Zhang Y, Yao Z, Li S, Yin Z, Xu M. Forkhead box Q1: a key player in the pathogenesis of tumors (Review). Int J Oncol. 2016;49:51–58. [DOI] [PubMed] [Google Scholar]
- 38.Zhang H, Meng F, Liu G, Zhang B, Zhu J, Wu F, et al. Forkhead transcription factor foxq1 promotes epithelial-mesenchymal transition and breast cancer metastasis. Cancer Res. 2011;71:1292–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bagati A, Bianchi-Smiraglia A, Moparthy S, Kolesnikova K, Fink EE, Lipchick BC, et al. Melanoma suppressor functions of the carcinoma oncogene FOXQ1. Cell Rep. 2017;20:2820–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caramel J, Papadogeorgakis E, Hill L, Browne GJ, Richard G, Wierinckx A, et al. A switch in the expression of embryonic EMT-inducers drives the development of malignant melanoma. Cancer Cell. 2013;24:466–80. [DOI] [PubMed] [Google Scholar]
- 41.Zhu L, Xi PW, Li XX, Sun X, Zhou WB, Xia TS, et al. The RNA binding protein RBMS3 inhibits the metastasis of breast cancer by regulating Twist1 expression. J Exp Clin Cancer Res. 2019;38:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Penkov D, Ni R, Else C, Pinol-Roma S, Ramirez F, Tanaka S. Cloning of a human gene closely related to the genes coding for the c-myc single-strand binding proteins. Gene. 2000;243:27–36. [DOI] [PubMed] [Google Scholar]
- 43.Meng F, Wu L, Dong L, Mitchell AV, James Block C, Liu J, et al. EGFL9 promotes breast cancer metastasis by inducing cMET activation and metabolic reprogramming. Nat Commun. 2019;10:5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang H, Meng F, Wu S, Kreike B, Sethi S, Chen W, et al. Engagement of I-branching {beta}−1, 6-N-acetylglucosaminyltransferase 2 in breast cancer metastasis and TGF-{beta} signaling. Cancer Res. 2011;71:4846–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Corley SM, Troy NM, Bosco A, Wilkins MR. QuantSeq. 3′ sequencing combined with Salmon provides a fast, reliable approach for high throughput RNA expression analysis. Sci Rep. 2019;9:18895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McDowell IC, Manandhar D, Vockley CM, Schmid AK, Reddy TE, Engelhardt BE. Clustering gene expression time series data using an infinite Gaussian process mixture model. PLoS Comput Biol. 2018;14:e1005896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–W560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Orellana EA, Kasinski AL. Sulforhodamine B (SRB) assay in cell culture to investigate cell proliferation. Bio Protoc. 2016;6:e:1984–e:1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iorns E, Drews-Elger K, Ward TM, Dean S, Clarke J, Berry D, et al. A new mouse model for the study of human breast cancer metastasis. PLoS One. 2012;7:e47995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Puchalapalli M, Zeng X, Mu L, Anderson A, Hix Glickman L, Zhang M, et al. NSG mice provide a better spontaneous model of breast cancer metastasis than athymic (nude) mice. PLoS One. 2016;11:e0163521. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microarray data that support the funding of this study have been deposited in the GEO under accession number GSE181322. RNA-seq data that support the finding of this study have been deposited in the GEO under accession number GSE181237.