1.
Nucleophosmin 1 (NPM1) mutated acute myeloid leukemia is recognized as distinct entity in the 2016 revision of the WHO classification [1]. In patients with normal karyotype, presence of NPM1 mutations results in improved risk from intermediate to favorable despite the presence of morphologic dysplasia [2, 3]. Monocytic morphology, blasts with cup like nuclear invaginations, and dim CD34 expression are all associated with NPM1‐mutated acute myeloid leukemia (AML); the findings are relatively nonspecific and definite diagnosis relies on demonstration of NPM1 mutations that result in aberrant cytoplasmic localization of the NPM1 protein. The vast majority of NPM1 mutations described affect exon 12 (NM_001355006 transcript, or exon 11 of the alternatively annotated transcript NM_002520). These mutations result in generation of an extra nuclear export signal in the C‐terminus of the NPM1 protein resulting in aberrant cytoplasmic localization [4]. Therefore, the vast majority of amplicon‐based assays currently used in clinical laboratories are designed to target the mutational hotspots in exon 12. However, emerging evidence suggests that other mutations besides the canonical exon 12 mutations may contribute to aberrant NPM1 cytoplasmic localization [5].
At our institution, somatic mutation analysis is routinely performed on all AML patients using a next‐generation sequencing with a large‐scale hybrid capture assay covers all exons of the NPM1 gene [6]. In the context of emerging data regarding nonexon 12 NPM1 mutations, we reviewed our clinical database of AML patients with NPM1 variants to determine if additional cases with pathogenic NPM1 mutations could be identified.
In the database of AML patients sequenced at our institution, canonical frameshift mutations were detected in NPM1 exon 12 (NM_001355006), or exon 11 of the alternatively annotated transcript (NM_002520) in 92 patients. These included 89 patients with W288Cfs, and three patients with W290Cfs. An additional patient had a frameshift mutation F268Lfs*8, which would be expected to abrogate the nucleolar localization signal (NoLS) at NPM1 C‐terminus and to behave similarly to the other canonical mutations.
However, based on the review of data we identified two additional AML patients with NPM1 exon 5 (NM_002520) mutations (Table 1). Patient 1 was a 73‐year‐old female with de novo AML and a 21‐bp in‐frame insertion at c.407_c.408, resulting in a 7‐amino acid insertion between Leu136 and Ser137 (Figure 1A). Patient 1 also had BCOR, NRAS, PTPN11, RAD21 and WT1 mutations. They were treated with azacitidine and venetoclax with complete hematopoietic recovery after cycle 1 of therapy. They continue on azacitdine and venetoclax with normal blood counts. Patient 2 was a 45‐year‐old male with de novo AML and a 21‐bp duplication of c.387_407, resulting in a duplication of the sequence between Glu130 and Leu136 (Figure 1B). Patient 2 also had DNMT3A and IDH1 mutations. They were initially treated on the SWOG S1203 protocol [7] with idarubicin, cytarabine, and vorinostat with primary refractory disease. Second line of therapy was on protocol [8] with azacitidine, cytarabine, and mitoxantrone with residual disease followed by a hematopoietic stem and progenitor cell transplant before relapsing. They received two cycles of hypomethylating agent‐based therapies before dying of refractory disease. In both patients, the extra amino acids in the mutated NPM1 generated an extra nuclear export signal (NES) motif ([9], Figure 1A,B). Both patients had normal karyotypes and no FLT3 mutations. Similar to the cases described by Martelli and colleagues, bone marrow findings were notable for presence of dysplasia most notable in the megakaryocyte lineage (Figure 1C,D) and variable to absent CD34 expression. In order to determine the functional impact of the newly created nuclear export signal, we performed immunohistochemistry (IHC) using the "pan" NPM1 antibody (DAKO clone 376). IHC confirmed the aberrant NPM1 cytoplasmic localization (NPM1c+) (Figure 1E,F).
TABLE 1.
Characteristics of two patients with NPM1 exon 5 mutation
| Characteristics | Patient 1 | Patient 2 |
|---|---|---|
| Gender | F | M |
| Age | 73 | 45 |
| Hb (g/dl) | 7.4 | 8.4 |
| PLT (k/μl) | 45 | 106 |
| WBC (k/μl) | 2.9 | 1.3 |
| Diagnosis | Acute myeloid leukemia | Acute myeloid leukemia |
| IHC | NPM1(c+) | NPM1(c+) |
| Karyotype | Normal | Normal |
| Dysplastic features | Multilineage, most notable in megakaryocytes | Multilineage, most notable in megakaryocytes |
| Blasts flow/CD34 expression | 22% | 24% |
| FLT3 | WT | WT |
| DNMT3A (VAF%) | WT |
DNMT3A p.R598* (7%) DNMT3A p.P385Rfs*22 (9%) |
| IDH1/2 | WT | IDH1 p.R132C (12%) |
| BCOR | p.V797Cfs*19 (26%) | WT |
| NRAS | p.G12A (15%) | WT |
| PTPN11 | p.D61H (13%) | WT |
| RAD21 | p.Q433* (17%) | WT |
| WT1 | p.A387Vfs*4 (28%) | WT |
| First‐line therapy | Azacitidine, venetoclax (complete hematologic response) | Idarubicin 25 mg (days 4–6), cytarabine (3200 mg days 4–7), vorinostat (500 mg BID days 1‐3) (primary refractory disease) |
| Second‐line therapy | NA | Azacitidine, high‐dose cytarabine, mitoxantrone (residual disease) |
| Hematopoietic stem and progenitor cell conditioning | NA | Fludarabine, busulfan, alemtuzumab |
| Relapse #1 therapy (cycle 1) | NA | Azacitidine (progression) |
| Relapse #1 therapy (cycle 2) | NA | Decitabine (10 day), donor lymphocyte infusion |
FIGURE 1.
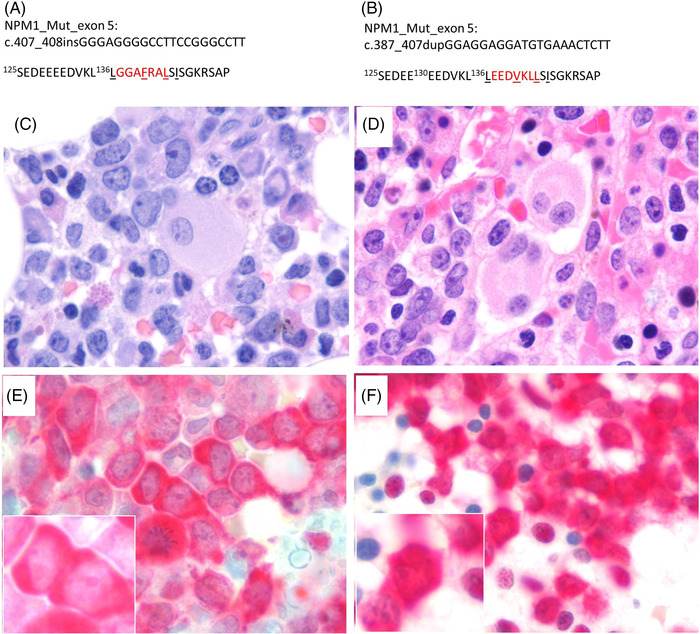
NPM1 mutations in exon 5. (A, B) Predicted protein sequence of the exon 5 mutants in patients 1 and 2. Nucleotide insertions (patient 1) and duplication (patient 2) are shown according to the NPM1 complementary DNA sequence. The newly acquired amino acids are highlighted in red and the predicted nuclear export signal motifs are underlined. (C, D) Hematoxylin and eosin‐stained bone marrow biopsies show extensive bone marrow involvement by leukemic blasts with folded nuclei and finely dispersed chromatin reminiscent of monocytic differentiation (total magnification 1000×). (E, F) Immunostaining performed with NPM1 antibody (Dako clone 376, pan NPM1) confirmed aberrant cytoplasmic as well as retained nucleolar staining (inset images) (total magnification 1000×)
Overall, we confirm that NPM1 exon 5 in‐frame insertions or duplications are an uncommon but recurrent finding in AML. Because AML with mutated NPM1 has been recognized as a distinct entity based on a single gene mutation by the WHO classification of AML [1], identification of NPM1 mutations is critical for accurate AML diagnosis and patient management [10]. Both of our patients with mutations in NPM1 exon 5 were re‐stratified from "Intermediate" to "Favorable" risk based on the 2017 European LeukemiaNet guidelines [2]. This revision in risk has direct implications for decisions regarding hematopoietic stem and progenitor cell transplant, and similar patients could be spared unnecessary upfront transplants in the future. Our observations further demonstrate the importance of either screening for nonexon 12 NPM1 mutations with IHC, or whole NPM1 gene sequencing to identify rare NPM1 mutations outside of exon 12 for better classification of AML patients. Furthermore, similar to exon 12 mutations, exon 5 NPM1 mutations can be followed either by RT‐PCR based assays [11] or highly sensitive error corrected next‐generation sequencing [12] to monitor measurable residual disease to obtain definitive information on the impact of NPM1 exon 5 mutations in AML prognosis.
FUNDING INFORMATION
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ETHICS STATEMENT
The study is deemed as exempt from IRB approval.
AUTHOR CONTRIBUTION
Peng Wang and Sandeep Gurbuxani designed research, performed research, analyzed data, and wrote the paper; Michael W. Drazer, Girish Venkataraman, and Daniel A. Arber performed research and wrote the paper. Michael W. Drazer has served as a consultant to Cardinal Health.
Wang P, Segal J, Drazer MW, Venkataraman G, Arber DA, Gurbuxani S. NPM1 exon 5 mutations in acute myeloid leukemia: Implications in diagnosis and minimal residual monitoring. eJHaem. 2022;3:962–965. 10.1002/jha2.445
Contributor Information
Peng Wang, Email: pengwang@bsd.uchicago.edu.
Sandeep Gurbuxani, Email: sandeep.gurbuxani@uchospitals.edu.
REFERENCES
- 1. Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016;127(20):2391–405. [DOI] [PubMed] [Google Scholar]
- 2. Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017;129(4):424–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Falini B, Macijewski K, Weiss T, Bacher U, Schnittger S, Kern W, et al. Multilineage dysplasia has no impact on biologic, clinicopathologic, and prognostic features of AML with mutated nucleophosmin (NPM1). Blood 2010;115(18):3776–86. [DOI] [PubMed] [Google Scholar]
- 4. Falini B, Brunetti L, Sportoletti P, Martelli MP. NPM1‐mutated acute myeloid leukemia: from bench to bedside. Blood 2020;136(15):1707–21. [DOI] [PubMed] [Google Scholar]
- 5. Martelli MP, Rossi R, Venanzi A, Meggendorfer M, Perriello VM, Martino G, et al. Novel NPM1 exon 5 mutations and gene fusions leading to aberrant cytoplasmic nucleophosmin in AML. Blood 2021;138(25):2696–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kadri S, Long BC, Mujacic I, Zhen CJ, Wurst MN, Sharma S, et al. Clinical validation of a next‐generation sequencing genomic oncology panel via cross‐platform benchmarking against established amplicon sequencing assays. J Mol Diagn. 2017;19(1):43–56. [DOI] [PubMed] [Google Scholar]
- 7. Garcia‐Manero G, Othus M, Pagel JM, Radich JP, Fang M, Rizzieri DA, et al. SWOG S1203: a randomized phase III study of standard cytarabine plus daunorubicin (7+3) therapy versus idarubicin with high dose cytarabine (IA) with or without vorinostat (IA+V) in younger patients with previously untreated acute myeloid leukemia (AML). Blood 2016;128(22):901. [Google Scholar]
- 8. Cahill KE, Karimi YH, Karrison TG, Jain N, Green M, Weiner H, et al. A phase 1 study of azacitidine with high‐dose cytarabine and mitoxantrone in high‐risk acute myeloid leukemia. Blood Adv. 2020;4(4):599–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xu D, Farmer A, Collett G, Grishin NV, Chook YM. Sequence and structural analyses of nuclear export signals in the NESdb database. Mol Biol Cell. 2012;23(18):3677–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pozdnyakova O. NPM1: not present? Mark! Blood 2021;138: 2602–3. [DOI] [PubMed]
- 11. Ivey A, Hills RK, Simpson MA, Jovanovic JV, Gilkes A, Grech A, et al. Assessment of minimal residual disease in standard‐risk AML. N Engl J Med. 2016;374(5):422–33. [DOI] [PubMed] [Google Scholar]
- 12. Ritterhouse LL, Parilla M, Zhen CJ, Wurst MN, Puranik R, Henderson CM, et al. Clinical validation and implementation of a measurable residual disease assay for NPM1 in acute myeloid leukemia by error‐corrected next‐generation sequencing. Mol Diagn Ther. 2019;23(6):791–802. [DOI] [PubMed] [Google Scholar]


